Abstract
Cells promote polarized growth by activation of Rho-family protein Cdc42 at the cell membrane. We combined experiments and modeling to study bipolar growth initiation in fission yeast. Concentrations of a fluorescent marker for active Cdc42, Cdc42 protein, Cdc42-activator Scd1, and scaffold protein Scd2, exhibited anti-correlated fluctuations and oscillations with a five-minute average period at polarized cell tips. These dynamics indicate competition for active Cdc42, or its regulators, and the presence of positive and delayed negative feedbacks. Cdc42 oscillations and spatial distribution were sensitive to the amounts of Cdc42-activator Gef1 and to the activity of Cdc42-dependent kinase Pak1, a negative regulator. Feedbacks regulating Cdc42 oscillations and spatial self-organization appear to provide a flexible mechanism for fission yeast cells to explore polarization states and control their morphology.
The conserved guanosine triphosphatase (GTPase) Cdc42 establishes cell polarity by regulating the cytoskeletal asymmetry required for normal cell function, differentiation, and motility (1, 2). In budding yeast, Cdc42 breaks the symmetry of spherical cells by clustering in one area of the membrane, the site of bud growth, through a “winner-take-all” positive-feedback mechanism (3-6). However, such a mechanism cannot explain how multiple growing zones form simultaneously in other cells. Fission yeast cells initially grow in a monopolar fashion, from the tip that existed before division (the old end), and activate bipolar growth that includes the new end as well, once a minimal cell length has been achieved (“New End Take Off”, NETO) (7). Fission yeast is thus an ideal system to study how Cdc42 is distributed at multiple sites.
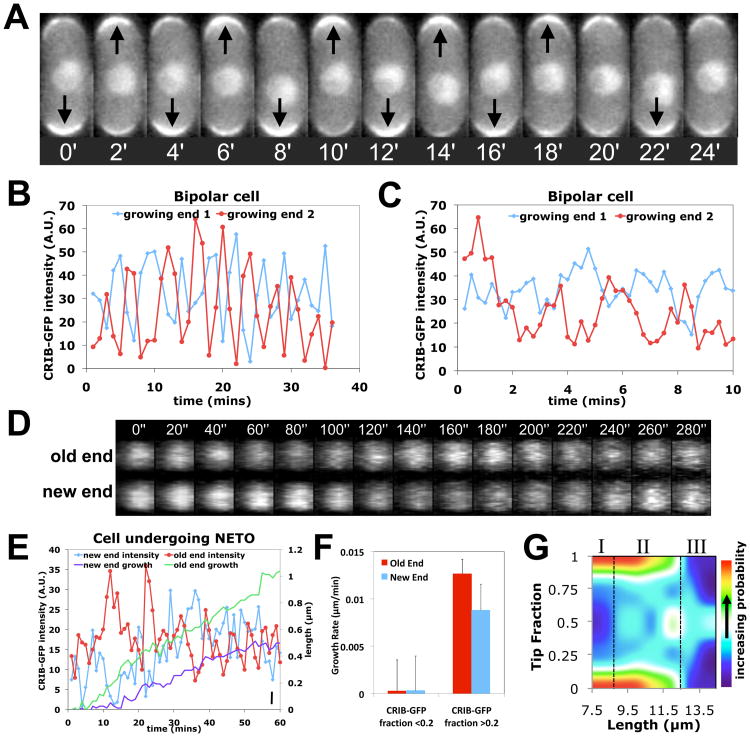
To characterize Cdc42 during the transition to bipolar growth, we used a fluorescent fusion protein (Cdc42/Rac Interactive Binding peptide-GFP, CRIB-GFP) that binds specifically to activated, GTP-bound Cdc42 (8). In larger bipolar cells, CRIB-GFP intensities at cell ends showed out-of-phase oscillations with an average period of five minutes (Fig.1, A and B; movies S1 and S2, tables S2 and S3). Oscillations were detectable in more than 50% of cells (table S2), when imaging every 15 seconds instead of 1 min (Fig.1C), and in 3D (Fig.1D). The rest of the cells displayed anti-correlated fluctuations without obvious periodicity. For shorter cells, non-growing ends still had detectable CRIB-GFP fluorescence, albeit at lower intensities than the older, growing ends (fig. S1). The tip intensities still underwent anti-correlated oscillations and fluctuations, but around asymmetric averages, unlike longer cells (table S2).
Fig 1. Oscillations and fluctuations of CRIB-GFP fluorescence at fission yeast cell tips.
A. CRIB-GFP fluorescence at cell tips in a bipolar cell (2 minute intervals). B. Old (red) and new end (blue) CRIB-GFP intensity in a bipolar cell (1 minute intervals). A.U.=Arbitrary Units. C. As in B, for 15 sec intervals. D. 3-D reconstruction of confocal Z-stack images of a bipolar cell showing CRIB-GFP at the old and new end (20 second intervals). E. Old (red) and new end (blue) CRIB-GFP intensity and cell growth at the old (green) and new (purple) ends in a cell undergoing NETO. The cell was 8.3 μm long at time 0. Bar, bottom right: 1 pixel = 0.1μm. F. Instantaneous growth rate, binned by CRIB-GFP tip fraction (ratio of intensity at one tip over sum of tip intensities) and tip type (old or new). Error bars indicate SEM. G. Heat-map of CRIB-GFP tip fraction versus cell length in wild-type cells (smoothed data, n=653). Note three regions: asymmetric, short cells (I); intermediate-length region with large intensity variations (II); symmetric, longer cells (III).
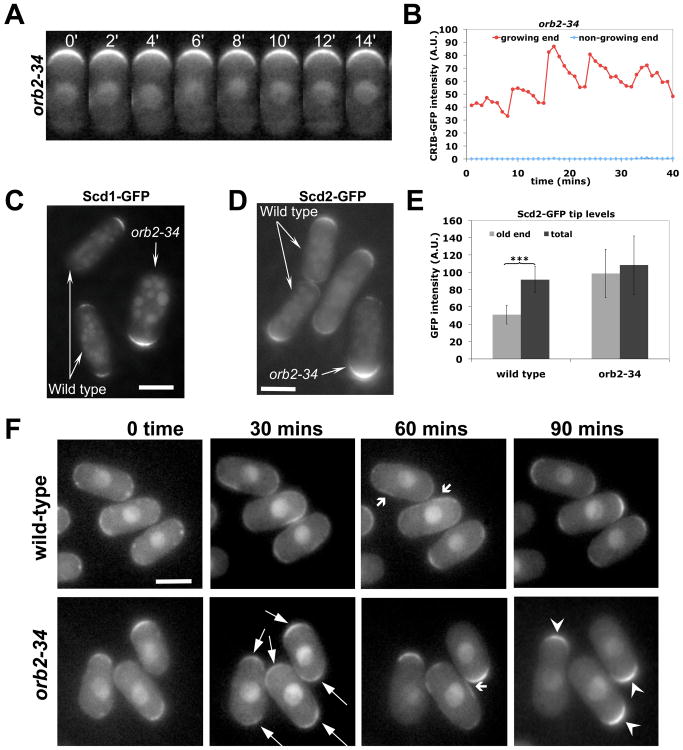
Fig 3. Negative regulation of Cdc42 by the kinase Pak1.
A. Fluorescence of CRIB-GFP in orb2-34 (pak1/shk1 mutant allele) cells (25°C) at growing cell tip (2 min intervals). B. CRIB-GFP fluorescence at growing (red) and non-growing (blue) tips in an orb2-34 cell (1 min intervals). C. Scd1-GFP, in wild-type and orb2-34 cells visualized in the same field. D. Scd2-GFP, in wild type and orb2-34 cells visualized in the same field. E. Scd2-GFP tip intensity in wild-type (old end, total) and orb2-34 mutant (growing end, total) visualized in the same field (***: p<0.0001, Student's t-test). Error bars indicate SD. F. Recovery following mild LatA treatment (10μM, 10 mins) in wild-type and orb2-34 cells. Both show symmetric (thin arrows) and transiently ectopic (wide arrows) CRIB-GFP after treatment. orb2-34 mutants progressively revert to monopolar distribution (arrow heads).
We visualized fluorescently labeled scaffold protein Scd2, which is proposed to mediate Cdc42 activation by binding to the Cdc42 GEF (guanine nucleotide exchange factor) Scd1 and Cdc42 (9, 10). Scd2-GFP intensity at the cell tips oscillated and fluctuated much like CRIB-GFP intensity (fig. S2; table S2), as did Scd1-3×GFP and Cdc42-GFP (fig. S3). Thus CRIB-GFP oscillations and fluctuations appear to reflect the activated Cdc42 protein complex.
To understand how GTP-Cdc42 levels might influence the “new end take off” (NETO) transition, we measured instantaneous cell growth rates along with CRIB-GFP intensity in cells undergoing NETO, which occurs in cells longer than 9 μm (7). Intensities at both new and old ends fluctuated strongly over time (Fig.1E). The instantaneous growth rate was correlated with abundance of CRIB-GFP at both old and new ends: cell tips with CRIB-GFP tip fraction below 0.2 grew slower than tips with fraction above 0.2 (Fig.1F). Varied degrees of asymmetry were also observed at intermediate lengths in a population of asynchronous cells (Fig.1G, region II). These findings indicate that NETO is a noisy transition driven by GTP-Cdc42 redistribution.
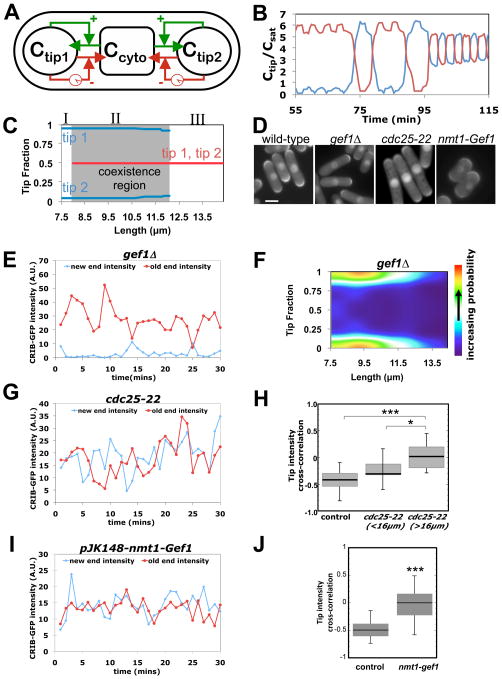
To determine the essential requirements for the transition from oscillating monopolar (asymmetric) to oscillating bipolar (symmetric) states during cell elongation, we developed a coarse-grained mathematical model (Fig. 2A) (11-14). Instead of describing specific molecular interactions, we took into consideration several experimental observations to predict system behavior. We assumed tips compete for Cdc42 or its effectors and regulators, on the basis of observed GTP-Cdc42 anti-correlations. We also assumed that positive and delayed negative feedbacks combine to generate oscillations, as they do in the bacterial Min system (15). We added noise to represent random concentration fluctuations and capture the observed variability (fig. S4A). The model reproduced the observed time courses: dominant-tip oscillations in short cells (with anti-correlated lagging tip) and out-of-phase oscillations at both tips in long cells (Fig 2B). Allowing different rate constants at the two tips caused them to oscillate around slightly different averages, as observed in many cells (fig. S4B). The model also predicted varied Cdc42 asymmetry in cells with similar length (Fig. 2C, “co-existence region”; fig. S5), as in Fig. 1G.
Fig 2. Test of mathematical model describing Cdc42 oscillations.
A. Model schematic showing GTP-Cdc42 distribution between tips and the cytoplasm, autocatalytic amplification (green), and delayed dissociation (red). B. Simulation showing GTP-Cdc42 fraction at each tip as cell progresses from monopolar to bipolar growth. C. Model predicts three regions similar to Fig. 1G. Asymmetric states (I); “coexistence” of symmetric and asymmetric states (II); symmetric states (III). D. CRIB-GFP in wild-type, gef1Δ, cdc25-22 at permissive temperature, 25°C, and Gef1 over-expressing, nmt1-gef1 (+thiamine, in YE medium) cells. Bar: 5μm E. CRIB-GFP tip intensities, in a gef1Δ cell (1 min intervals). F. CRIB-GFP tip fractions versus cell length, in gef1Δ cells (as Fig. 1G, n=381). G. CRIB-GFP tip intensities, in a cdc25-22 cell at 25°C. H. Anti-correlation of CRIB-GFP tip intensities decreases with respect to wild-type cells and with increasing cell length in cdc25-22 mutants (*: p = 0.03, ***: p = 0.00039, Student's t-test). I. CRIB-GFP tip intensities of cell moderately over-expressing Gef1 (+thiamine, in YE medium). J. CRIB-GFP tip anti-correlation decreases in cells moderately over-expressing Gef1 (***: p=5.7 ×10−6, Student's t-test). Whiskers in H and J indicate the full range of data.
According to the mathematical model, changes in abundance or activity of Cdc42, or of its regulators, can shift the system to more asymmetric or symmetric states. It indicated that cells with lower rate of Cdc42 activation (or decreased total amounts of GTP-Cdc42 or Cdc42 GEFs), favor asymmetric states because the lagging tip is influenced more severely by the accumulation of GTP-Cdc42 at the dominant tip and by the resulting depletion of the cytoplasmic pool (fig. S5A and S6A). To test this, we analyzed CRIB-GFP in gef1Δ cells, which lack one of the two Cdc42 GEFs and thus exhibit decreased amounts of global active GTP-Cdc42 (16), but otherwise grow at a normal rate (table S5). Most (75%; n=12) gef1Δ cells had lower amounts of CRIB-GFP at the new tips in time-lapse recordings (Fig.2D and 2E; see movie S3). CRIB-GFP tip-fractions in gef1Δ cells (n=381) were asymmetric (Fig. 2F), consistent with the model.
In our model, increasing cell size increases the total amount of active Cdc42 or of Cdc42 GEFs. In the presence of noise, this is predicted to decouple oscillations because one tip can no longer deplete the pool available to the other. Thus, we studied cdc25-22 cells that delay entry into mitosis, due to a mutation in a cell-cycle control gene, and become longer than wild-type cells at permissive temperatures (17). Consistent with this prediction, we found less anti-correlation between CRIB-GFP tip signals in longer cdc25-22 cells (Fig. 2D, G and H; table S3).
To test if Cdc42 GEF availability influences the anti-correlation of Cdc42 oscillations, we overexpressed Gef1. This eliminated the anti-correlation of CRIB-GFP signal at the cell tips (Fig. 2, I and J, movie S4). Increased amounts of Gef1 also led to increased symmetry of CRIB-GFP and Cdc42-target formin For3 (18) (Fig. 2, D and I, fig. S6, B, C and D). This agrees with the model, which predicts that increasing Cdc42 activation rate (or total amounts of active Cdc42 or Cdc42 GEFs) (fig. S6A) leads to more symmetrical GTP-Cdc42 distribution.
Autocatalytic activation within the Cdc42 complex (9, 10, 19, 20) and actin-mediated transport (21) are likely contributors to positive feedback, as in budding yeast (3-6). Much less is known about negative feedback (22), a required mechanism for oscillations. To identify possible mechanisms, we analyzed CRIB-GFP in morphological mutants (Fig. S7A and S7B), including orb2-34 and tea1Δ that are monopolar (23, 24), rdi1Δ (encoding the only Rho-GDI) and rga4Δ (encoding the only known Cdc42 GAP) (8, 25). orb2-34 mutants oscillated asymmetrically, but with a longer period and a decreased amplitude of CRIB-GFP oscillations (table S2; Fig. 3, A and B). Conversely, rdi1Δ and rga4Δ mutants displayed normal, mostly symmetrical oscillations; tea1Δ mutants fluctuated asymmetrically (Fig. S7C).
Amounts of CRIB-GFP, Cdc42 GEF Scd1-GFP, and scaffold Scd2-GFP at the one growing tip of orb2-34 cells were increased compared to that of control cells (Fig. 3, C, D and E; fig. S8, A and B). No localization change was seen for Gef1-3xYFP or Rga4-GFP (fig. S8C; (8)). The intensity of Scd2-GFP signal (Fig. 3E) or Scd1-GFP at the growing tip in orb2-34 cells roughly equaled the total fluorescence at both tips (new end plus old end) in control cells (Fig. 3E). Amounts of total protein were not changed (fig. S7D). Thus, orb2 regulates intracellular distribution of Scd1 and Scd2. In the model, this behavior is expected when the maximal active Cdc42 allowable at each tip increases (fig. S8E). In mutant cells unable to suppress maximal tip accumulation, the growing tip could function as a “sink”, entrapping Scd1 and Scd2.
To confirm that orb2-34 mutants remain monopolar because of their inability to redistribute Scd1, Scd2 or other regulators, we destabilized the actin-dependent (21) localization of Scd1 by exposing cells for 10 min to Latrunculin A (LatA). In orb2-34 and wild-type cells, CRIB-GFP, Scd1-GFP and Scd2-GFP became symmetric in the first hour after LatA removal (Fig. 3F; fig. S9) consistent with reports of brief actin depolymerization promoting bipolar growth in monopolar cells cdc10ts and ssp1Δ (13, 26). 90 min after LatA removal however, orb2-34 cells re-accumulated these markers at one tip, which could be different than the tip originally growing, while wild-type cells remained largely symmetric (Fig. 3F; fig. S9). This agrees with the model's prediction of lack of a symmetric attractor for cells with reduced negative feedback and convergence to a symmetric attractor for wild-type cells after a perturbation (fig. S4A).
orb2-34, a mutant allele of pak1 (also known as shk1) (23) contains a point mutation (G517E; see (12)) in the Pak1 kinase domain that decreases its activity (fig. S7, E and F). Pak1, a Cdc42-dependent kinase (27, 28), localizes to tips in an Sdc1- and Scd2-dependent manner (21). Negative regulation of Cdc42 could thus be linked to its own activation, as expected from a negative feedback loop. It might occur through Scd2, a substrate of Pak1 (20), consistent with findings in S. cerevisiae where the Pak1 homologue Cla4 negatively regulates the interaction of the scaffold protein Bem1, an Scd2 homologue with Cdc42 GEF Cdc24 (29).
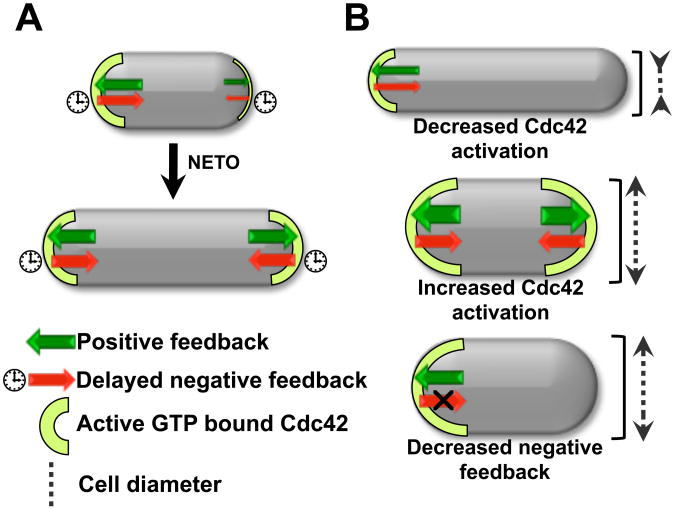
Increased accumulation of Cdc42 GEFs at the membrane, by Gef1 overexpression or loss of negative inhibition (orb2-34 mutants), dampens Cdc42 fluctuations (Fig. S10A). These mutants are wider, possibly because increasing tip-bound Cdc42 results in growth over a wider area (16) (fig. S10, B and C). We suggest that wild-type cells regulate diameter by maintaining Cdc42 activity at the tips within a normal range, and activate bipolar growth by Cdc42 redistribution (Fig. 4A). Oscillations and fluctuations may regulate cell morphology and help the switch to bipolar growth. Prior to NETO, accumulation of Cdc42 at the old end provides a kinetic barrier to bipolar symmetry by depleting the resources available to the new end. Oscillations and fluctuations may relieve this depletion, giving the new end a chance to take off by allowing the system to reach an otherwise-inaccessible state of bipolarity (Fig. S11 and S12). Mutations affecting Cdc42 regulation may alter the system's dynamics, promoting a different pattern of Cdc42 distribution, changing cell diameter and symmetry (Fig. 4B).
Fig 4. Model of self-organization of Cdc42 at the cell tips and control of cell morphogenesis.
A. In wild-type cells, Cdc42 recruitment/activation balances Cdc42 removal/deactivation, limiting GTP-Cdc42 tip level and thus setting cell diameter at a normal range. Increased GEF availability promotes bipolar growth activation at the new cell tip as cell size increases. B. Changes in the system's dynamics alter Cdc42 distribution. Decreased Cdc42 activation (gef1Δ mutants) increases Cdc42 asymmetry and decreased cell diameter. GEF overexpression increases Cdc42 activation at both tips leading to increased diameter. Decreased negative feedback (orb2-34 mutants) leads to the accumulation of most Cdc42 activity at one single tip, resulting in monopolar growth. Increased active Cdc42 levels at the growing end results in increased cell diameter.
Fission yeast Cdc42 oscillations and fluctuations might represent exploratory behavior, a general strategy among self-organizing biological systems (30). Despite the associated energy cost, biological systems may benefit because they acquire the ability to quickly reach states that would otherwise be difficult to access. Fluctuations of Cdc42 activity allows fission yeast to rapidly respond to changing intracellular conditions, such as cell volume and length. In an environment with changing external cues, such as nutrient or pheromone gradients, Rho GTPase fluctuations may allow eukaryotic cells to adapt and redirect the direction of growth.
Supplementary Material
Acknowledgments
We thank E. Karsenti, C. Luetje, S. Lemmon, G. D'Urso, and T.K. Harris for critically reading the manuscript and M. Ioannidou and K. Zhang for help with data analysis. We thank B. Skibbens and Yi Hu for facilitating experiments, and K. Shiozaki, P. Perez and S. Martin for various strains. This work was supported by NSF grant 0745129, NIH grant 1R01GM095867, and a University of Miami SEEDS “You Choose” Leadership award to F.V., and by NIH grant R21GM083928 and a Lehigh University Class of 68 Faculty Fellowship to D.V. T.D. was supported by a Sigma Xi grant-in-aid and as a GAANN fellow through US ED.
Footnotes
Supplementary Materials: Materials and Methods
SOM text
References and Notes
- 1.Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nat Rev Mol Cell Biol. 2008 Sep;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- 2.Perez P, Rincon SA. Rho GTPases: regulation of cell polarity and growth in yeasts. Biochem J. 2010 Mar 15;426:243–253. doi: 10.1042/BJ20091823. [DOI] [PubMed] [Google Scholar]
- 3.Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nat Cell Biol. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- 4.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. J Cell Biol. 2004 Sep 13;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozubowski L, et al. Symmetry-breaking polarization driven by a Cdc42p GEF-PAK complex. Curr Biol. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R. Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Dev Cell. 2009;17:823–835. doi: 10.1016/j.devcel.2009.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mitchison JM, Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- 8.Tatebe H, Nakano K, Maximo R, Shiozaki K. Pom1 DYRK regulates localization of the Rga4 GAP to ensure bipolar activation of Cdc42 in fission yeast. Curr Biol. 2008;18:322–330. doi: 10.1016/j.cub.2008.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endo M, Shirouzu M, Yokoyama S. The Cdc42 binding and scaffolding activities of the fission yeast adaptor protein Scd2. J Biol Chem. 2003 Jan 10;278:843–852. doi: 10.1074/jbc.M209714200. [DOI] [PubMed] [Google Scholar]
- 10.Wheatley E, Rittinger K. Interactions between Cdc42 and the scaffold protein Scd2: requirement of SH3 domains for GTPase binding. Biochem J. 2005 May 15;388:177–184. doi: 10.1042/BJ20041838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Model (see 12) has two populations of Cdc42 associated with each tip, Ctip1 and Ctip2, and one in the cytoplasm, Ccyto. We assumed competition for Cdc42 but similar results are obtained for Cdc42 GEFs or GTP-Cdc42. The total amount, Ctot ≡ Ctip1 + Ctip2 + Ccyto, increases in proportion to cell volume V. Association to the tips obeys dCtipj/dt=λ+Ccyto/V − k−Ctipj, j=1, 2. Autocatalytic activation, , with n ≥ 2, generates asymmetry by allowing one tip to deplete the cytoplasmic pool and preventing the other tip from gathering Cdc42. Saturation at level Csat recovers bipolarity for long cells. Oscillations result by assuming Cdc42 triggers its own removal (delayed negative feedback): . Here, ε determines the delayed dissociation strength, τ is delay time, and h gives the nonlinearity of the effect.
- 12.Material and methods are available as Supporting material in Science online.
- 13.Csikász-Nagy A, Gyorffy B, Alt W, Tyson JJ, Novák B. Spatial controls for growth zone formation during the fission yeast cell cycle. Yeast. 2008 Jan;25:59–69. doi: 10.1002/yea.1571. [DOI] [PubMed] [Google Scholar]
- 14.Howell AS, et al. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009 Nov 13;139:731–743. doi: 10.1016/j.cell.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.H Meinhardt, de Boer PA. Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site. Proc Natl Acad Sci U S A. 2001 Dec 4;98:14202–14207. doi: 10.1073/pnas.251216598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Coll PM, Trillo Y, Ametzazurra A, Perez P. Gef1p, a new guanine nucleotide exchange factor for Cdc42p, regulates polarity in Schizosaccharomyces pombe. Mol Biol Cell. 2003;14:313–323. doi: 10.1091/mbc.E02-07-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fantes P. Epistatic gene interactions in the control of division in fission yeast. Nature. 1979;279:428–430. doi: 10.1038/279428a0. [DOI] [PubMed] [Google Scholar]
- 18.Martin SG, Rincon SA, Basu R, Perez P, Chang F. Regulation of the formin for3p by cdc42p and bud6p. Mol Biol Cell. 2007 Oct;18:4155–4167. doi: 10.1091/mbc.E07-02-0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang EC, et al. Cooperative interaction of S. pombe proteins required for mating and morphogenesis. Cell. 1994 Oct 7;79:131–141. doi: 10.1016/0092-8674(94)90406-5. [DOI] [PubMed] [Google Scholar]
- 20.Chang E, et al. Direct binding and In vivo regulation of the fission yeast p21-activated kinase shk1 by the SH3 domain protein scd2. Mol Cell Biol. 1999 Dec;19:8066–8074. doi: 10.1128/mcb.19.12.8066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kelly FD, Nurse P. Spatial control of Cdc42 activation determines cell width in fission yeast. Mol Biol Cell. 2011 Aug 17; doi: 10.1091/mbc.E11-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ozbudak EM, Becskei A, van Oudenaarden A. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev Cell. 2005 Oct;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 23.Verde F, Wiley DJ, Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotonic dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc Natl Acad Sci U S A. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mata J, Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997 Jun 13;89:939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- 25.Das M, et al. Regulation of cell diameter, For3p localization, and cell symmetry by fission yeast Rho-GAP Rga4p. Mol Biol Cell. 2007 Jun;18:2090–2101. doi: 10.1091/mbc.E06-09-0883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rupes I, Jia Z, Young PG. Ssp1 promotes actin depolymerization and is involved in stress response and new end take-off control in fission yeast. Mol Biol Cell. 1999;10:1495–1510. doi: 10.1091/mbc.10.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marcus S, et al. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci U S A. 1995 Jun 20;92:6180–6184. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ottilie S, et al. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995 Dec 1;14:5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gulli MP, et al. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol Cell. 2000 Nov;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 30.Karsenti E. Self-organization in cell biology: a brief history. Nat Rev Mol Cell Biol. 2008 Mar;9:255–262. doi: 10.1038/nrm2357. [DOI] [PubMed] [Google Scholar]
- 31.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- 32.Liew AWC, Law NF, Cao XQ, Yan H. Statistical power of Fisher test for the detection of short periodic gene expression profiles. Pattern Recognition. 2009;42:549–556. [Google Scholar]
- 33.Qyang Y, et al. The p21-activated kinase, Shk1, is required for proper regulation of microtubule dynamics in the fission yeast, Schizosaccharomyces pombe. Mol Microbiol. 2002 Apr;44:325–334. doi: 10.1046/j.1365-2958.2002.02882.x. [DOI] [PubMed] [Google Scholar]
- 34.Yang P, Pimental R, Lai H, Marcus S. Direct activation of the fission yeast PAK Shk1 by the novel SH3 domain protein, Skb5. J Biol Chem. 1999 Dec 17;274:36052–36057. doi: 10.1074/jbc.274.51.36052. [DOI] [PubMed] [Google Scholar]
- 35.Buchwald P. A general bilinear model to describe growth or decline time-profiles. Math Biosci. 2007;205:108–136. doi: 10.1016/j.mbs.2006.08.013. [DOI] [PubMed] [Google Scholar]
- 36.Buchwald P, Sveiczer A. The time-profile of cell growth in fission yeast: model selection criteria favoring bilinear models over exponential ones. Theor Biol Med Model. 2006;3 doi: 10.1186/1742-4682-3-16. art 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang F, Martin SG. Shaping fission yeast with microtubules. Cold Spring Harb Perspect Biol. 2009 Jul;1:a001347. doi: 10.1101/cshperspect.a001347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS Letters. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. [DOI] [PubMed] [Google Scholar]
- 39.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007 Apr 20;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oosawa F, Kasai M. A theory of linear and helical aggregations of macromolecules. J Mol Biol. 1962 Jan;4:10–21. doi: 10.1016/s0022-2836(62)80112-0. [DOI] [PubMed] [Google Scholar]
- 41.Flyvbjerg H, Jobs E, Leibler S. Kinetics of self-assembling microtubules: an “inverse problem” in biochemistry. Proc Natl Acad Sci U S A. 1996 Jun 11;93:5975–5979. doi: 10.1073/pnas.93.12.5975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novak B, Tyson JJ. Design principles of biochemical oscillators. Nat Rev Mol Cell Biol. 2008 Dec;9:981–991. doi: 10.1038/nrm2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrell JE, Jr, Machleder EM. The biochemical basis of an all-or-none cell fate switch in Xenopus oocytes. Science. 1998 May 8;280:895–898. doi: 10.1126/science.280.5365.895. [DOI] [PubMed] [Google Scholar]
- 44.Wang H, Vavylonis D. Model of For3p-mediated actin cable assembly in fission yeast. PLoS One. 2008;3:e4078. doi: 10.1371/journal.pone.0004078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wu JQ, Pollard TD. Counting cytokinesis proteins globally and locally in fission yeast. Science. 2005 Oct 14;310:310–314. doi: 10.1126/science.1113230. [DOI] [PubMed] [Google Scholar]
- 46.Drake T, Vavylonis D. Cytoskeletal dynamics in fission yeast: a review of models for polarization and division. HFSP Journal. 2010;4:122–130. doi: 10.2976/1.3385659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Coll PM, Rincon SA, Izquierdo RA, Perez P. Hob3p, the fission yeast ortholog of human BIN3, localizes Cdc42p to the division site and regulates cytokinesis. EMBO J. 2007 Apr 4;26:1865–1877. doi: 10.1038/sj.emboj.7601641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snell V, Nurse P. Genetic analysis of cell morphogenesis in fission yeast--a role for casein kinase II in the establishment of polarized growth. EMBO J. 1994 May 1;13:2066–2074. doi: 10.1002/j.1460-2075.1994.tb06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Verde F, Mata J, Nurse P. Fission yeast cell morphogenesis: identification of new genes and analysis of their role during the cell cycle. J Cell Biol. 1995;131:1529–1538. doi: 10.1083/jcb.131.6.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rincon S, Coll PM, Perez P. Spatial regulation of Cdc42 during cytokinesis. Cell Cycle. 2007;6:1687–1691. doi: 10.4161/cc.6.14.4481. [DOI] [PubMed] [Google Scholar]
- 51.Coll PM, Rincon SA, Izquierdo RA, Perez P. Hob3p, the fission yeast ortholog of human BIN3, localizes Cdc42p to the division site and regulates cytokinesis. Embo J. 2007;26:1865–1877. doi: 10.1038/sj.emboj.7601641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kim H, et al. The kelch repeat protein, Tea1, is a potential substrate target of the p21-activated kinase, Shk1, in the fission yeast, Schizosaccharomyces pombe. J Biol Chem. 2003 Aug 8;278:30074–30082. doi: 10.1074/jbc.M302609200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.