Abstract
Transposable elements (TEs) are ubiquitous residents in eukaryotic genomes. They can cause dramatic changes in gene expression and lead to gross rearrangements of chromosome structure, providing the basis for rapid evolution. The virilis species group of Drosophila contains certain species that can be crossed under experimental conditions and their phylogeny is thoroughly investigated. We have shown that Drosophila virilis, the most primitive karyotypically and probably the ancestral species of the group, is in the process of colonization by a very unusual retroelement Penelope which apparently repeatedly invaded the species of the group in the past. However, the molecular mechanisms and evolutionary consequences of such invasions are poorly understood. In this commentary, we discuss the implications of our recent investigation into the response of the RNA silencing system to Penelope invasion of a new host genome which can be achieved in different ways.
Keywords: Drosophila, Penelope retroelement, evolution, invasion, small RNAs
The virilis Species Group of Drosophila and Penelope Retroelement
We recently reported the results of experiments that explored a possibility to introduce a potentially mobile copy of the Penelope retroelement into the genomes of two distant Drosophila species.1 This investigation represents an important step in our long-term studies of D. virilis transposons and their possible role in evolution of closely related species belonging to the “virilis” group,2-4 with special emphasis on Penelope which was previously implicated in hybrid dysgenesis (HD) syndrome in D. virilis.5,6 In Drosophila, Penelope has only been found in the virilis group and in D. willistoni, however, these TEs termed PLEs (Penelope-like elements) in recent years were described in many organisms including fishes, reptiles and rotifers.7-9 PLEs characteristically differ from the other groups of retroelements by the presence of GIY-YIG-endonuclease domain and an ability to retain their introns in the course of proliferation.7 Penelope-like elements (PLEs) represent an ancient enigmatic superfamily of retroelements that apparently shares a common ancestor with telomerase reverse transcriptases.10 It is of note that Penelope endonuclease domain was first described in group I mobile introns from bacteria and organelles.7
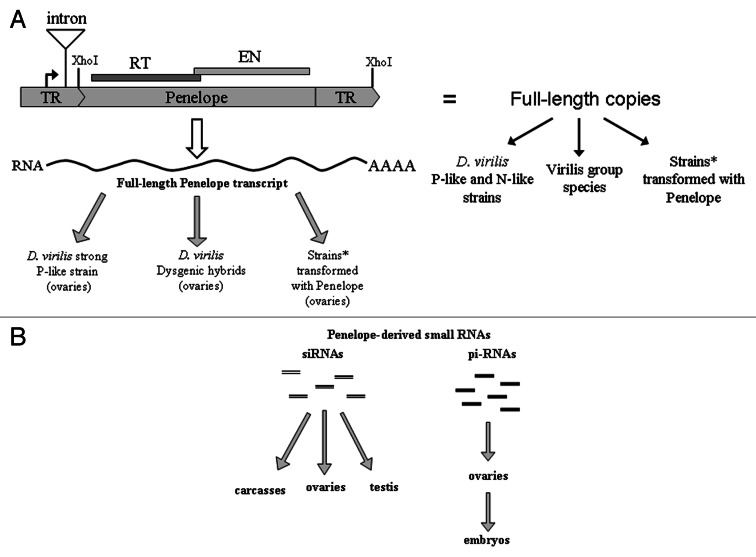
Penelope family in D. virilis and related species is represented by highly variable structure of individual copies.5,11Figure 1A depicts a typical structure of Penelope element “unit” successfully used in developing transgenic strains1 which contains 2.8kb sequence flanked by XhoI sites and apparently all necessary elements for expression and transposition in the genome of host species.
Figure 1. The distribution of full-length Penelope copies and Penelope-derived transcripts in various strains and species of Drosophila. (A) The presence of intact and potentially active Penelope copies and canonical (2.8 kb) Penelope transcripts in different Drosophila strains and species. The position of intron is indicated. Black arrow indicates the transcription start. *In our experiments we obtained strains transformed with Penelope using both D. virilis and D. melanogaster. TR-terminal repeats which can be in tandem or inverse orientation in different Penelope copies. RT-reverse transcriptase; EN-endonuclease. Cleavage sites of XhoI endonuclease are indicated. (B) Penelope-homologous small RNAs detected in the virilis group species, certain D. virilis strains and a few strains transformed with Penelope. Maternally transmitted Penelope-derived piRNAs may target Penelope mRNA through transcript cleavage.
The virilis group comprises 12 species that are traditionally divided into two phylads: the D. virilis phylad and the D. montana phylad.12 In the course of thorough phylogenetic analysis three divergent clades of Penelope were detected in the species of the group.11,13 Importantly, divergence times of the Penelope elements found in certain species were smaller than the age of the species, suggesting that horizontal transfer and multiple invasions by this TE took place in the course of the virilis group species evolution.13
In our model system we have a unique opportunity to investigate the behavior of this retroelement in two distant species, i.e., Drosophila virilis and Drosophila melanogaster separated by 50–60 million years of divergent evolution.12 While various clades of Penelope were found in D. virilis strains,11,13 no trace of Penelope was detected in the sequenced D.melanogaster genome14 which is naïve in terms of Penelope presence.
Penelope elements can invade and amplify in a new host genome in a number of different ways, as described below.
Invasion of D. virils natural populations
Transposons can be transmitted horizontally and spread through interbreeding.15 However, there are multiple mechanisms underlying TE silencing and limiting the invasion process.16-18 Piwi-clade Argonaute proteins were shown to have a prominent role in transposon silencing in vivo in various plants and animals.19,20 Mechanisms of piRNAs biogenesis are best understood in D. melanogaster where they arise from TEs that have landed within certain genomic loci designated as piRNA clusters.20,21 The TEs are controlled in the germ line by short, antisense, TE-derived RNAs (23–29 nt) that are found in complexes with three Piwi-clade Argonaute proteins.21 The piRNA clusters drive the production of primary piRNAs that prime a larger pool of secondary piRNAs through repeated cycles (“ping-pong amplification loop”) of destruction of sense and antisense TE transcripts.20,21
As we mentioned above basing on phylogenetic analysis, various species of the virilis group have been colonized by Penelope at different stages of their divergent evolution.11,13 Previously, we provided substantial evidence that cosmopolitan species D. virilis is at the present time in the process of colonization by Penelope family of retroelements. We were lucky to directly demonstrate recent Penelope invasion of a natural D. virilis population (Middle Asia, Tashkent).2 Thus, the D. virilis strains collected in this area in 1968 were free of Penelope sequences, while all individuals collected from the same population in 1997 carried multiple Penelope copies located exclusively in euchromatic chromosomes arms.2 In situ analysis exploring Penelope probe detected asymmetrical hybridization frequently observed in unpaired regions of polytene chromosomes in the progeny of freshly caught flies in the 1997 population, indicating that an exceptionally high level of heterozygosity was present in the contemporary Tashkent population.2 Interestingly, the ongoing invasion of Penelope occurs in D. virilis, a cosmopolitan species which is itself in the process of global demographic expansion, probably related to human movements.22 Careful analysis of multiple laboratory and geographical strains of D. virilis demonstrated different content of full-length (Fig. 1A) and potentially functional copies of Penelope elements.1-3,5 Most of the studied D. virilis geographical strains including Tashkent strain recently invaded by this TE exhibit neutral cytotypes i.e., do not exhibit high level of gonadal sterility when crossed with tester strains.1,5,6 Therefore, apparently the appearance of Penelope elements may rapidly change the M cytotype characteristic of original Penelope-free strains to neutral status. Furthermore, practically all Penelope-containing strains are characterized by the presence of Penelope-derived piRNAs in their ovaries1 and these piRNAs are faithfully transmitted to the progeny (Fig. 1B). It is of note that, among the analyzed strains, there are several exceptional ones which exhibit a neutral cytotype but seem not to contain Penelope sequences and Penelope-homologous piRNAs. However, some of these strains do contain defective Penelope copies. In summary, large scale analysis of D. virilis strains enables us to conclude that natural invasion of Penelope leads to rapid change of strain cytotype, which usually correlates with the presence of Penelope-derived piRNAs in the ovaries.1
Penelope transpositions due to dysgenic crosses in D. virilis
Dysgenic crosses between certain strains of D. virilis differing by the presence of full-length copies of this element represent another approach enabling rapid transposition and amplification of Penelope sequences in the genome.5,6 Contrary to the previously described syndromes in D. melanogaster (P-M and I-R syndromes), where P and I elements are activated independently in different systems,23,24 the HD syndrome in D. virilis, which includes male and female gonadal sterility, multiple point and chromosomal mutations and other abnormalities observed in the progeny of dysgenic cross, probably results from simultaneous activation of several unrelated TEs.5,6,25 It was shown that besides Penelope, retroelements Ulysses, Helena and Telemac, as well as DNA transposon Paris, are also mobilized and may cause mutations in the same dysgenic cross.25
Recently with the development of deep-sequencing techniques it was clearly demonstrated that P-M hybrid dysgenesis in D. melanogaster also activates both P-elements and other resident transposons and disrupts the piRNA biogenesis machinery.26 Furthermore, interspecific hybrids between D. melanogaster and D. simulans are characterized by widespread derepression of multiple both maternally and paternally inherited TE families.27 It is of note that in the early eighties, soon after the discovery of mobile elements in Drosophila, we were the first to directly demonstrate transposition of a family of dispersed mobile repeats (“pDv elements”) in interspecific hybrids between D. virilis and two other species of the virilis group (i.e., D. lummei and D. littoralis).28
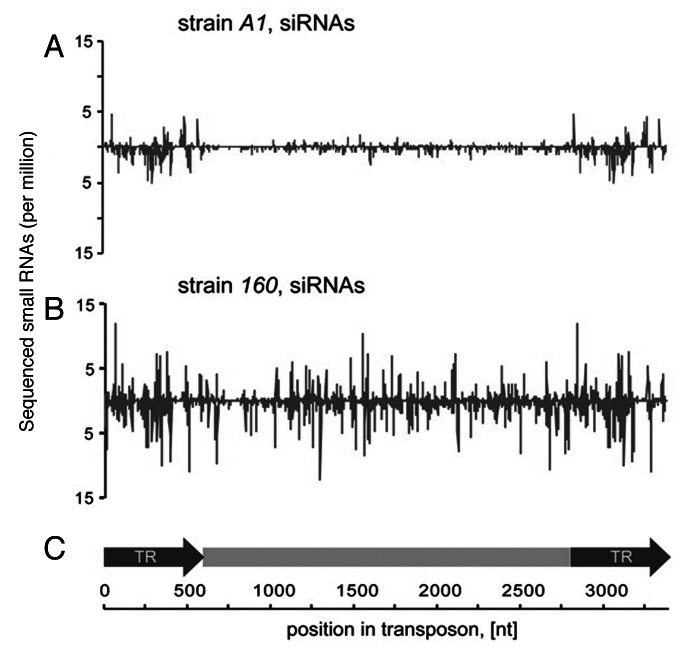
Previous studies suggested that a key driver in D. virilis HD syndrome is the Penelope retroelement, which was proposed not only to become mobile itself, but also to mobilize other TEs mentioned above in the dysgenic hybrids.5,6 It was suggested that Penelope expression somehow interferes with RNAi machinery involved in silencing of other unrelated TEs.29 Although recent reports have implicated RNA silencing in repression of hybrid dysgenesis in D. virilis29and in D. melanogaster,26,30,31 in D. virilis the evidence in favor of direct and critical role of Penelope-homologous small RNAs in HD syndrome is not so straightforward.29,32 In the first report on the role of Penelope-derived small RNAs in the HD the authors did not discriminate between Penelope-derived siRNAs and piRNAs and, hence, encountered some difficulties in localization of the “master locus” producing Penelope-derived small RNAs.29 Furthermore, although in a subsequent investigation we did observe correlation between Penelope transcription in the ovaries of dysgenic hybrids and manifestation of dysgenic traits, we could not always correlate differences in maternally deposited Penelope piRNA in the hybrids between various D. virilis strains with the sterility of the progeny.32 Our investigation also demonstrated that Penelope transposes with low frequency in D. virilis strain 160 used as a P-like in the dysgenic crosses.33 It was shown that small RNAs homologous to Penelope found in this particular strain, belong predominantly to the siRNA category (Fig. 2), and consist of sense and antisense species observed in approximately equal proportion (Fig. 3). The number of Penelope copies in the latter strain has significantly increased during the last decades, probably because Penelope-derived siRNAs are not maternally inherited, while the low level of Penelope-piRNAs, which are transmitted from mother to the embryo, is not sufficient to silence this element completely in strain 160. Therefore, we speculated that intrastrain transposition of Penelope is controlled predominantly at the post-transcriptional level.33
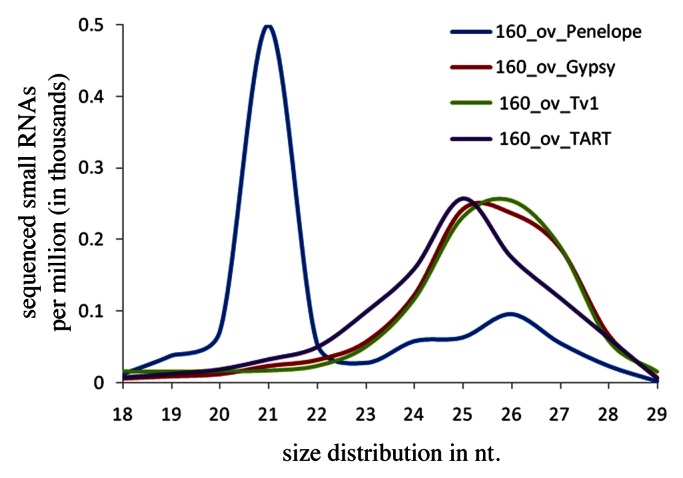
Figure 2. Size profile (in nt) of small RNAs derived from Penelope, Gypsy, Tv1 and TART retroelements32 in the ovaries of D. virilis P-like strain 160.
Figure 3. Distribution of Penelope-derived siRNA (21–23nt) along the transposon body in D. melanogaster strain (A1) transformed by Penelope (A) and in D. virilis strain 160 (B). The structure of the consensus Penelope element, containing two terminal repeats used in transformation experiments is shown at the bottom of the figure. The figure is adapted from reference 32.
High transposition frequency of various TEs induced by dysgenic crosses may play an important role in evolution of the virilis species group. Some time ago, we observed multiple chromosomal rearrangements in the progeny of dysgenic crosses between certain D. virilis strains.3 Remarkably, many rearrangement breakpoints at cytological level coincide with the chromosomal locations of Penelope and Ulysses insertions in the parental strains and with breakpoints of inversions previously established for other species of the group.3
Further analysis revealed the presence of full-length-Penelope copies and Penelope-derived piRNAs in the ovaries of practically all species of the virilis group (Fig. 1A), which, however, correlates with apparent transcriptional inactivation of Penelope in all the species with the exception of D. virilis.1
Direct introduction of Penelope into the genomes of D. virilis and D. melanogaster.
Fortunately, in our model system we could explore a unique opportunity to experimentally imitate evolution and investigate the behavior of this unusual retroelement directly introduced into the strains of two distantly related species, D. virilis and D. melanogaster. The recipient strain of D. melanogaster was Penelope-free, while the D. virilis strain used in the transformation experiments contains only inactive heterochromatic copies of this retroelement (clade II), probably representing the remnants of previous ancient invasions.1,11,13 More than a decade ago we introduced full-length Penelope copies (Fig. 1A) into the D. melanogaster genome exploring P-mediated transformation.34 The introduced copies were actively transcribed and eventually amplified in the D. melanogaster genome.34 Three years ago we investigated the pattern of Penelope-derived small RNAs in the strains transformed by Penelope. The analysis demonstrated the presence of Penelope-derived siRNAs (21–22 nt) in the carcasses of the transgenic strains studied.32 The mechanisms by which TEs become recognized by the siRNA pathway are not fully understood, however, previously in the transgenic strains we described multiple rearranged Penelope copies containing long inverted repeats which could give rise to double-stranded RNAs (dsRNAs) that are processed into siRNAs.34 Remarkably, in D. melanogaster transformed strains the detected siRNAs precisely correspond to these inverted regions within Penelope body.32 On the other hand, in the D. virilis strain 160 Penelope-homologous siRNAs were randomly distributed along the Penelope body and probably result from bidirectional transcription of the TE (Fig. 3A and B). At the next step we performed broad scale analysis of multiple (35 total) iso-female derivatives of several D. melanogaster strains transformed with Penelope in 2001. While in the carcasses of these strains Penelope-derived siRNAs (21nt) were frequently seen, in the ovaries of a few strains we detected by Northern hybridization 25–27 nt small RNAs homologous to the Penelope transcript that were subsequently shown to belong to the piRNA category1 (Fig. 1B).
The appearance of piRNAs in the ovaries of transgenic strains probably resulted from accidental transposition of Penelope copy into one of the piRNA genomic clusters of D. melanogaster. To prove our assumption, we determined the localization of Penelope inserts in the genomes of derivative transgenic iso-female strains characterized by the presence of piRNA in the gonads. Interestingly, in two of these strains we detected Penelope insertion within the major D. melanogaster germ line-specific piRNA 42AB cluster.20 It was also shown that Penelope expression is severely inhibited in the transgenic strains containing Penelope-homologous piRNAs in the ovaries.1
Furthermore, we crossed the strains containing Penelope-derived piRNAs in the ovaries with the strains lacking Penelope and provided evidence that detected piRNAs are maternally deposited and can silence euchromatic transcriptionally active copies of Penelope in trans. Such Trans-Silencing Effect is described for the X-TAS system and P-element insertions in telomeric regions.35
To further elucidate the fate of Penelope in terms of its expression and RNA biogenesis at the early stage of reinvasion in the D. virilis genome, we imitated the phenomenon by introducing full-size Penelope copies into a typical M-like strain of D. virilis devoid of functional copies of this transposon5,11 using the piggy-Bac-based transgenesis system.36 We established several independent transgenic strains containing inserts of a full-length Penelope (Fig. 1A). In situ hybridization experiments demonstrated, besides the sites of the original white-containing constructs, additional sites of Penelope insertion in the chromosomes of the transformed strains. Northern blotting and RT-PCR technique revealed a significant level of Penelope transcription in all obtained transgenic D. virilis strains, both in the ovaries and in the carcasses.1 A detailed cytological analysis of Penelope localization in the transgenic strains demonstrated that, in contrast to the original construct insertion loci that are present in all nuclei of the salivary glands and serve as an internal control, additional hybridization sites are usually seen in only a fraction of salivary gland nuclei and, hence, transgenic larvae represent mosaics in terms of Penelope presence. Therefore, in these experiments we demonstrated that initial transposition of Penelope after its introduction takes place predominantly in somatic tissues.1 Interestingly, a decade ago, when several D. melanogaster strains transformed with the same transposon were developed, we also often observed similar mosaics in salivary gland nuclei immediately after transformation by in situ hybridization and single fly Southern blot hybridization analysis of the transformed strains performed at that time.1 It is of note that multiple somatic transpositions were previously reported for various non-LTR retroelements.37
Conclusions
We explored a unique chance to monitor the consequences of a very unusual retroelement Penelope introduction into the genomes of D. virilis and D. melanogaster. We compared the outcome of natural invasion of Penelope into D. virilis species, the consequences of dysgenic crosses, and the results of direct introduction of this TE into the genomes of the two species by transformation with full-length Penelope. At the initial stages of transposition Penelope tends to produce rearranged copies, which may give rise to siRNAs from the inverted repeats. These Penelope-derived siRNA are not able to efficiently silence Penelope, which leads to active somatic transpositions and Penelope amplification in the genome of a new host species. However, with time Penelope copies can find their way to pi-clusters and become transcriptionally silenced by Penelope-homologous piRNAs. The colonization of host genomes by Penelope may rapidly change the cytotype and induce gross rearrangements of the chromosomes providing the basis for rapid evolution.
Acknowledgments
Work was supported by the Russian Foundation for Basic Research, project Nº 09-04-00643 and 09-04-00660, project from “Genofond dynamics” program, Grant of the Program of Molecular and Cellular Biology RAN to M.B.E. We are grateful to Dr. Nikolai Rozhkov from CSHL for performing the experiments depicted in Figure 2.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Footnotes
Previously published online: www.landesbioscience.com/journals/mge/article/24542
References
- 1.Rozhkov NV, Schostak NG, Zelentsova ES, Yushenova IA, Zatsepina OG, Evgen’ev MB. Evolution and dynamics of small RNA response to a retroelement invasion in Drosophila. Mol Biol Evol. 2013;30:397–408. doi: 10.1093/molbev/mss241. [DOI] [PubMed] [Google Scholar]
- 2.Evgen’ev MM, Zelentsova H, Mnjoian L, Poluectova H, Kidwell MG. Invasion of Drosophila virilis by the Penelope transposable element. Chromosoma. 2000;109:350–7. doi: 10.1007/s004120000086. [DOI] [PubMed] [Google Scholar]
- 3.Evgen’ev MB, Zelentsova H, Poluectova H, Lyozin GT, Veleikodvorskaja V, Pyatkov KI, et al. Mobile elements and chromosomal evolution in the virilis group of Drosophila. Proc Natl Acad Sci U S A. 2000;97:11337–42. doi: 10.1073/pnas.210386297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evgen’ev MB, Arkhipova IR. Penelope-like elements--a new class of retroelements: distribution, function and possible evolutionary significance. Cytogenet Genome Res. 2005;110:510–21. doi: 10.1159/000084984. [DOI] [PubMed] [Google Scholar]
- 5.Evgen’ev MB, Zelentsova H, Shostak N, Kozitsina M, Barskyi V, Lankenau DH, et al. Penelope, a new family of transposable elements and its possible role in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A. 1997;94:196–201. doi: 10.1073/pnas.94.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vieira J, Vieira CP, Hartl DL, Lozovskaya ER. Factors contributing to the hybrid dysgenesis syndrome in Drosophila virilis. Genet Res. 1998;71:109–17. doi: 10.1017/S001667239800322X. [DOI] [PubMed] [Google Scholar]
- 7.Arkhipova IR, Pyatkov KI, Meselson M, Evgen’ev MB. Retroelements containing introns in diverse invertebrate taxa. Nat Genet. 2003;33:123–4. doi: 10.1038/ng1074. [DOI] [PubMed] [Google Scholar]
- 8.Gladyshev EA, Arkhipova IR. Telomere-associated endonuclease-deficient Penelope-like retroelements in diverse eukaryotes. Proc Natl Acad Sci U S A. 2007;104:9352–7. doi: 10.1073/pnas.0702741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalle Nogare DE, Clark MS, Elgar G, Frame IG, Poulter RT. Xena, a full-length basal retroelement from tetraodontid fish. Mol Biol Evol. 2002;19:247–55. doi: 10.1093/oxfordjournals.molbev.a004078. [DOI] [PubMed] [Google Scholar]
- 10.Arkhipova IR. Telomerase, retrotransposons, and evolution. In:Telomerases:Chemistry, Biology and Clinical Applications. Edited by N.F.Lue and C. Autexier. JohnWiley and Sons, Inc.:Hoboken, NJ. 2012; 265-299. [Google Scholar]
- 11.Lyozin GT, Makarova KS, Velikodvorskaja VV, Zelentsova HS, Khechumian RR, Kidwell MG, et al. The structure and evolution of Penelope in the virilis species group of Drosophila: an ancient lineage of retroelements. J Mol Evol. 2001;52:445–56. doi: 10.1007/s002390010174. [DOI] [PubMed] [Google Scholar]
- 12.Spicer G, Bell C. Molecular phylogeny of the Drosophila virilis species group (Diptera: Drosophilidae) inferred from mitochondrial 12S and 16S ribosomal RNA genes. Ann Entomol Soc Am. 2002;95:156–61. doi: 10.1603/0013-8746(2002)095[0156:MPOTDV]2.0.CO;2. [DOI] [Google Scholar]
- 13.Morales-Hojas R, Vieira CP, Vieira J. The evolutionary history of the transposable element Penelope in the Drosophila virilis group of species. J Mol Evol. 2006;63:262–73. doi: 10.1007/s00239-005-0213-1. [DOI] [PubMed] [Google Scholar]
- 14.Kapitonov VV, Jurka J. Molecular paleontology of transposable elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 2003;100:6569–74. doi: 10.1073/pnas.0732024100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kidwell MG. Horizontal transfer. Curr Opin Genet Dev. 1992;2:868–73. doi: 10.1016/S0959-437X(05)80109-1. [DOI] [PubMed] [Google Scholar]
- 16.Le Rouzic A, Capy P. The first steps of transposable elements invasion: parasitic strategy vs. genetic drift. Genetics. 2005;169:1033–43. doi: 10.1534/genetics.104.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hua-Van A, Le Rouzic A, Boutin TS, Filée J, Capy P. The struggle for life of the genome’s selfish architects. Biol Direct. 2011;6:19. doi: 10.1186/1745-6150-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blumenstiel JP. Evolutionary dynamics of transposable elements in a small RNA world. Trends Genet. 2011;27:23–31. doi: 10.1016/j.tig.2010.10.003. [DOI] [PubMed] [Google Scholar]
- 19.Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68. doi: 10.1016/j.cell.2009.01.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, et al. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- 21.Guzzardo PM, Muerdter F, Hannon GJ. The piRNA pathway in flies: highlights and future directions. Curr Opin Genet Dev. 2013;23:1–9. doi: 10.1016/j.gde.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mirol PM, Routtu J, Hoikkala A, Butlin RK. Signals of demographic expansion in Drosophila virilis. BMC Evol Biol. 2008;8:59. doi: 10.1186/1471-2148-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bucheton A. I transposable elements and I-R hybrid dysgenesis in Drosophila. Trends Genet. 1990;6:16–21. doi: 10.1016/0168-9525(90)90044-7. [DOI] [PubMed] [Google Scholar]
- 24.Kidwell MG. Evolution of hybrid dysgenesis determinants in Drosophila melanogaster. Proc Natl Acad Sci U S A. 1983;80:1655–9. doi: 10.1073/pnas.80.6.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrov DA, Schutzman JL, Hartl DL, Lozovskaya ER. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc Natl Acad Sci U S A. 1995;92:8050–4. doi: 10.1073/pnas.92.17.8050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khurana JS, Wang J, Xu J, Koppetsch BS, Thomson TC, Nowosielska A, et al. Adaptation to P element transposon invasion in Drosophila melanogaster. Cell. 2011;147:1551–63. doi: 10.1016/j.cell.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kelleher ES, Edelman NB, Barbash DA. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012;10:e1001428. doi: 10.1371/journal.pbio.1001428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evgen’ev MB, Yenikolopov GN, Peunova NI, Ilyin YV. Transposition of mobile genetic elements in interspecific hybrids of Drosophila. Chromosoma. 1982;85:375–86. doi: 10.1007/BF00330360. [DOI] [PubMed] [Google Scholar]
- 29.Blumenstiel JP, Hartl DL. Evidence for maternally transmitted small interfering RNA in the repression of transposition in Drosophila virilis. Proc Natl Acad Sci U S A. 2005;102:15965–70. doi: 10.1073/pnas.0508192102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science. 2008;322:1387–92. doi: 10.1126/science.1165171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chambeyron S, Popkova A, Payen-Groschêne G, Brun C, Laouini D, Pelisson A, et al. piRNA-mediated nuclear accumulation of retrotransposon transcripts in the Drosophila female germline. Proc Natl Acad Sci U S A. 2008;105:14964–9. doi: 10.1073/pnas.0805943105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rozhkov NV, Aravin AA, Zelentsova ES, Schostak NG, Sachidanandam R, McCombie WR, et al. Small RNA-based silencing strategies for transposons in the process of invading Drosophila species. RNA. 2010;16:1634–45. doi: 10.1261/rna.2217810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rozhkov NV, Zelentsova ES, Shostak NG, Evgen’ev MB. Expression of Drosophila virilis retroelements and role of small RNAs in their intrastrain transposition. PLoS One. 2011;6:e21883. doi: 10.1371/journal.pone.0021883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pyatkov KI, Shostak NG, Zelentsova ES, Lyozin GT, Melekhin MI, Finnegan DJ, et al. Penelope retroelements from Drosophila virilis are active after transformation of Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:16150–5. doi: 10.1073/pnas.252641799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Josse T, Maurel-Zaffran C, de Vanssay A, Teysset L, Todeschini AL, Delmarre V, et al. Telomeric trans-silencing in Drosophila melanogaster: tissue specificity, development and functional interactions between non-homologous telomeres. PLoS One. 2008;3:e3249. doi: 10.1371/journal.pone.0003249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holtzman S, Miller D, Eisman R, Kuwayama H, Niimi T, Kaufman T. Transgenic tools for members of the genus Drosophila with sequenced genomes. Fly (Austin) 2010;4:349–62. doi: 10.4161/fly.4.4.13304. [DOI] [PubMed] [Google Scholar]
- 37.Kazazian HH., Jr. Mobile DNA transposition in somatic cells. BMC Biol. 2011;9:62. doi: 10.1186/1741-7007-9-62. [DOI] [PMC free article] [PubMed] [Google Scholar]