Abstract
Heat stress results in misfolding and aggregation of cellular proteins. Heat shock proteins (Hsp) enable the cells to maintain proper folding of proteins, both in unstressed as well as stressed conditions. Hsp70 genes encode for a group of highly conserved chaperone proteins across the living systems encompassing bacteria, plants, and animals. In the cellular chaperone network, Hsp70 family proteins interconnect other chaperones and play a dominant role in various cell processes. To assess the functionality of rice Hsp70 genes, rice genome database was analyzed. Rice genome contains 32 Hsp70 genes. Rice Hsp70 superfamily genes are represented by 24 Hsp70 family and 8 Hsp110 family members. Promoter and transcript expression analysis divulges that Hsp70 superfamily genes plays important role in heat stress. Ssc1 (mitochondrial Hsp70 protein in yeast) deleted yeast show compromised growth at 37 °C. Three mitochondrial rice Hsp70 sequences (i.e., mtHsp70-1, mtHsp70-2, and mtHsp70-3) complemented the Ssc1 mutation of yeast to differential extents. The information presented in this study provides detailed understanding of the Hsp70 protein family of rice, the crop species that is the major food for the world population.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-012-0395-6) contains supplementary material, which is available to authorized users.
Keywords: Hsp70 superfamily, Hsp70, Hsp110/SSE1, Rice, Transcript expression, Yeast complementation
Introduction
Temperature is a highly significant environmental factor in the growth and development of plants. All organisms respond to heat stress by synthesizing heat shock proteins (Hsps). Hsp70 family of proteins is present constitutively and upregulated in response to various stressors like heat, cold, anoxia, and metal (Li et al. 1999; Sung et al. 2001). Constitutively present Hsp70 proteins perform housekeeping functions of a cell and are called the “heat shock cognates (Hsc).” Hsp70 family proteins are involved in proper folding of nascent synthesized proteins, prevention of protein aggregation, translocation of proteins across membranes, and targeting proteins towards degradation; thus maintaining protein homeostasis (proteostasis) (Hartl et al. 2011). Under conditions of stress, Hsp70 maintain protein quality by preventing aggregation of stress-damaged proteins.
Hsp70 are ATP-dependent chaperones having a conserved ~44-kD N-terminal ATPase domain (also called nucleotide binding domain; NBD), a ~18-kD substrate binding domain (SBD) and a ~10-kD variable C-terminal “lid.” The flexible C-terminal lid assists in holding the substrates at SBD. ATPase domain performs regulatory role and SBD associates to hydrophobic regions exposed in non-native substrates (Dragovic et al. 2006). The hydrophobic region of the client proteins binds transiently to the SBD and is regulated by NBD. Hsp70 proteins require two co-chaperones in the form of J-domain proteins and nucleotide exchange factors (NEFs) for their functions. Binding of J-domain proteins stimulates ATP hydrolysis that facilitates trapping of exposed hydrophobic region of substrates and closed conformation of SBD. NEFs participate in ATP–ADP exchange by catalyzing the release of ADP from Hsp70, leading to conversion of open conformation Hsp70 and substrate release. Proteins of families like GrpE in Escherichia coli, Mge1 in yeast mitochondria, Hsp binding protein (Hsp-BP), or Fes1 and Hsp110 are reported to act as NEFs (Zhang et al. 2010; Liberek et al. 1991; Miao et al. 1997; Kabani et al. 2002). Hsp110 family proteins have high sequence and structural homology to Hsp70 and are therefore included in Hsp70 superfamily. Hsp110 have an N-terminal NBD similar to Hsp70 and C-terminal SBD. They are larger than Hsp70 in size due to either inserted acidic region in SBD or C-terminal extension (Liu and Hendrickson 2007).
Hsp70 is a multigenic family. Various members constituting this family are present in different cellular compartments. In Saccharomyces cerevisiae, 14 Hsp70 genes representing nine cytosolic, three mitochondrial (Ssc, Ecm, and Ssq), and two ER (Kar2 and Lhs1) isoforms have been identified (Walsh et al. 2004). In E. coli, there are three Hsp70 genes. Arabidopsis has 18 Hsp70 superfamily members, of which 14 belong to Hsp70 family and 4 belong to Hsp110/SSE family (Lin et al. 2001). In spinach, at least 12 Hsp70 genes are identified (Guy and Li 1998). The C terminus of organellar Hsp70 proteins is highly conserved and unique for each organelle. The motif for the cytosolic group is EEVD, for the mitochondrion is PEAEYEEAKK and for the plastid is PEGDVIDADFTDSK (Guy and Li 1998). The C terminus EEVD motif of cytosolic Hsp70 proteins interacts with co-chaperones that contain several degenerate 34 amino acid repeats, called tetratricopeptide repeats.
Hsp70 proteins perform diverse biological functions either with their co-chaperones or in collaboration with other chaperones. Whereas DnaK system efficiently solubilized and refolded small protein aggregates (Diamant et al. 2000), large protein aggregates are solubilized in concert with Hsp100/ClpB (Diamant et al. 2000; Acebron et al. 2009). Expression of cytosolic Hsp70 genes is linked to acquired thermotolerance because of their property to reactivate the protein aggregates formed under various stress conditions (Lee and Schoffl 1996; Yang et al. 2009; Qi et al. 2011; Zhou et al. 2012; Sung and Guy 2003). It is noted that Hsp70 may function as a negative feedback regulator of HSF activity (Shi et al. 1998; Kim and Schoffl 2002). In this process, Hsp70 binds to denatured proteins and releases HSF leading to its activation during heat stress (Shi et al. 1998). Hsp70 proteins in plants are involved in additional specific functions. Chloroplast Hsp70 (cpHsp70) in Arabidopsis are implicated in chloroplast development (Latijnhouwers et al. 2010). Mutants of cpHsp70 develop variegated cotyledons, malformed leaves, growth retardation and impaired root growth in addition to being heat sensitive (Latijnhouwers et al. 2010). Hsp70 shows complex developmental regulation during the vegetative and reproductive phases of growth. In tomato, Hsc70 transcripts were detected in mature anthers (Duck and Folk 1994). Several plant cytosolic Hsp70 genes are expressed during seed development, maturation, and/or germination (DeRocher and Vierling 1995; Sung et al. 2001). While levels of Hsp70 proteins are noted to be abundant in dry seeds, their levels drastically decline within 72 h after the onset of imbibitions (Sung et al. 2001).
Plant Hsp70s are also responsive to varied environmental signals. Several members of the Hsp70 family in spinach are regulated by a light/dark signal independent of circadian rhythm (Li and Guy 2001). Hsp70 members are involved in post-translational translocation of proteins across membranes in case of mitochondria and chloroplasts (Su and Li 2010). In rice, ER Hsp70 (BiP) is found to associate with nascent polypeptides that emerge into the ER lumen during prolamin translocation and facilitate peptide folding and assembly (Muench et al. 1997). Over-expression of BiP relieves “ER stress” (Leborgne-Castel et al. 1999).
We analyzed Hsp70 family of rice to understand the functional network of chaperones in this important food species. Rice mitochondrial Hsp70 has been proposed as suppressor of heat and H2O2-induced programmed cell death (Qi et al. 2011). We identified 32 Hsp70 genes in rice genome. Transcript expression analysis revealed that various Hsp70 are regulated under different stress conditions and the genes are expressed constitutively or regulated developmentally. Yeast mitochondrial Hsp70 mutant transformed with rice mitochondrial Hsp70 proteins suggested an evolutionary conserved mechanism of action of these proteins. This study provides the functional relevance of Hsp70 component of the Hsp70/J proteins bichaperone machine of rice.
Materials and methods
Identification of Hsp70 proteins in rice genome
For Hsp70 retrieval in rice genome annotation project (RGAP; http://rice.plantbiology.msu.edu/), putative function search using “Hsp70” as query returned no results. Replacing Hsp70 with “DnaK” showed 31 entries in rice genome. Using protein sequence of one of the genes (i.e., Os01g62290) as query in blast search resulted in 34 genes with maximum E value of 9.6e−39. The protein sequences of these genes subjected to domain search in SMART (http://smart.embl-heidelberg.de/smart) showed Hsp70 domain in 32 proteins. Same 32 genes were recovered following blast search and subsequent SMART analysis when database was queried with protein sequence of Arabidopsis Hsp70, At1g16030. The exon–intron organization of Hsp70 genes was analyzed in Spidey (www.ncbi.nlm.nih.gov/spidey/) taking genomic sequence from RGAP and cDNA sequences from KOME (http://cdna01.dna.affrc.go.jp/cDNA/) and from RGAP if the full-length (FL) cDNA was not present in KOME. Multiple sequence alignment of amino acid sequences of Hsp70 proteins was performed using the ClustalX 2.0 (Larkin et al. 2007) with default parameters. The NJ tree with 1,000 bootstrap was constructed in ClustalX 2.0 and viewed using Treeview1.6.6. Subcellular localization of proteins was analyzed at WoLFPSORT, Predotar, PSORT, Softberry, and TargetP database. Subcellular compartment was decided based on consensus localization in two algorithms and Hsp70-specific C-terminal sequence.
To find segmental duplication events, genome duplications of rice in MSU database were identified with a maximum permitted distance between collinear gene pairs of 100 kB (http://rice.plantbiology.msu.edu/segmental_dup/index.shtml).
Motif analysis
In order to find conserved motifs in rice Hsp70 gene family members, “Multiple EM for Motif Elicitation” (MEME) version 3.5.4 (Bailey et al. 2006) was used. The parameters used for the analysis were: number of repetitions—any; maximum number of motifs—12 and optimum width of motif—≥2 and ≤300.
Isolation of RNA, RT-PCR analysis, and cloning of FL-cDNA
Total RNA was isolated from stressed and non-stressed (control) 3-week-old rice seedlings (Oryza sativa L; cultivar Pusa basmati 1 (PB1)) using TRI reagent (Sigma, USA) as per the manufacturer’s instructions. Concentration of RNA was quantified spectrophotometrically and quality was analyzed by gel electrophoresis. For cDNA synthesis, 5 μg of total RNA of each sample was reverse transcribed using reverse transcriptase (RevertAid H Minus Reverse transcriptase, MBI, Fermentas). RT-PCR amplification was performed for Hsp70 genes using gene-specific primers listed in Supplementary Table 1.
For yeast mutant complementation, Hsp70 genes were amplified using FL-cDNA clones procured from Rice Genome Resource Centre, Japan (primers used are listed in Supplementary Table 1). PCR was carried out using Phusion™ Hi-Fi DNA polymerase in presence of 1 % DMSO in a 50-μl reaction. The amplified products after digestion with requisite enzymes (Supplementary Table 1) were cloned in p426 vector (Mumberg et al. 1995) under the control of glyceraldehydes-3-phosphate dehydrogenase (GPD) promoter and transformed in mutant yeast cells. Empty vector was also transformed in yeast cells as vector control. The phenotype of the yeast was scored by incubating the plates spotted with tenfold serial dilutions of yeast cells. Prior to spotting the OD600 of yeast cells was normalized to 0.2.
Results
Hsp70 superfamily has Hsp70 and Hsp110 proteins
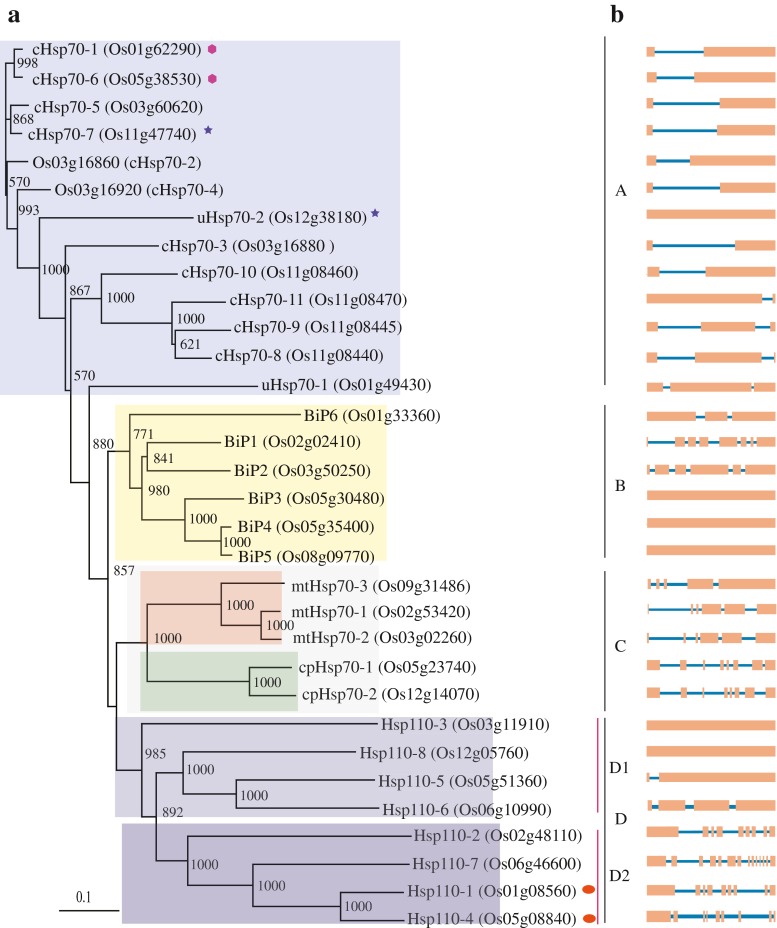
In rice genome, 32 sequences showed Hsp70 domain. Except for chromosomes 4, 7, and 10, Hsp70 genes were scattered over all chromosomes in rice genome (Table 1). Analysis of phylogenetic tree generated from aligned amino acid sequences of rice Hsp70 proteins showed four well-supported lineages (Fig. 1). While Hsp70 genes clustered on clades A, B, and C, clade D contained Hsp110 family genes. Hsp110 family is a subfamily of Hsp70 superfamily, structurally very similar to Hsp70 having N-terminal ATPase domain and C-terminal peptide binding domain and functions as NEFs for Hsp70 family proteins. Analysis of C-terminal Hsp70 sequences and in silico localization ascertained that the clades A and B genes are cytoplasmic and ER, respectively (Table 1 and Supplementary Table 2). Mitochondrial and chloroplastic Hsp70 genes cluster on clade C. Transmembrane domain (TMD) and ER retention signal HDEL were noticed in ER sequences of clade B except in BiP6 gene that appears diverged from other members of clade B. Six of 13 genes in clade A are classical cytoplasmic (Hsp70 having characteristic C-terminal EEVD sequence) while two lineages (uHsp70-1 and uHsp70-2) appeared evolutionary distant (Fig. 1). On clade A, four genes present on chromosome 11 are arranged in tandem (Supplementary Fig. 1) suggesting localized gene duplication event. In addition, three pairs of genes are located on segmental duplicated regions of rice chromosomes (Fig. 1). Two of the segmental duplicates are Hsp70 (cHsp70-1 and cHsp70-6 and cHsp70-7 and uHsp70-2) and one pair (Hsp110-1 and Hsp110-4) is Hsp110 family. Except for one pair (cHsp70-7 and uHsp70-2), the duplicated genes have close phylogenetic relationship (Fig. 1). Motif analysis showed that C terminus of Hsp110 family is more diverged from Hsp70 family while the ATP binding domain has high motif similarity (Fig. 2). Closely related Hsp70 proteins in the phylogenetic tree have similar motif composition (Fig. 2). All the diverged genes in their respective clades differed in the domain structure, mass or intron–exon arrangement (Fig. 1; Table 1). Examination of the intron–exon organization showed that nine genes in clade A contain one intron, three genes have two introns while one gene is intron-less. The position of the intron is conserved in all one intron containing clade A genes except for cHsp70-11. Intron numbers are highly variable in clade B genes. Among the six genes in this clade, three genes are intron-less while other three genes contain two, five, and seven introns. The number and position of introns in two chloroplast Hsp70 genes in clade C is highly conserved. However, out of three mitochondrial Hsp70 genes in clade C, one gene has four introns and two genes have five introns each. Hsp110 family clade D was bifurcated into two subclades: subclade D2 with four genes having multiple introns and molecular weight matched with Hsp110 family genes. In D2, one gene (Hsp110-2) is ER protein having HDEL sequence and a TMD and other three genes are nuclear/cytoplasmic. D1 clade differed from D2 clade in having lesser or no introns and lower molecular weight (47–62 kD) proteins because of the absence of C-terminal extension. Three proteins in this clade have TMD and ER localization whereas one protein (Hsp110-3) is nuclear/cytoplasmic. Orthologs of Hsp70 superfamily genes with features different from the conserved C-terminal sequence were searched in other sequenced genomes and a tree was generated with the retrieved protein sequences (Supplementary Fig. 2). The tree indicated that genes with similar characteristics are present in other genomes as well with the exception of uHsp70-1. Most of the eukaryotic genomes contain multiple members of Hsp70 family existing in same sub-cellular compartment. Consistent with this, Hsp70 family of rice putatively encode for 11 cytosolic, 5 ER luminal, 3 mitochondrial, and 2 chloroplast proteins. The cytoplasmic Hsp70 clade is much complex in rice. Five genes in this group of rice are smaller in size and lack the conserved C-terminal EEVD sequence, a situation similar to yeast. In yeast, four cytosolic Hsp70 genes (Ssa) with EEVD and three genes (two Ssb and one Ssz1) without EEVD sequence have distinct non-overlapping functions. Yeast Ssb genes are associated with ribosomes and function in translation (Nelson et al. 1992). However, it is not possible to predict the functions of rice Hsp70 genes that are different from canonical cytosolic Hsp70 genes on the basis of this analysis only.
Table 1.
Hsp70 domain containing genes of rice
| Gene ID | Protein name | Localization | Amino acids | Molecular weight (kD) | FL-cDNA |
|---|---|---|---|---|---|
| Hsp70 family | |||||
| Os01g62290 | cHsp70-1 | Nuc/cytoplasm | 648 | 71 | AK243277 |
| Os03g16860 | cHsp70-2 | Nuc/cytoplasm | 650 | 71 | AK072830, AK099275 |
| Os03g16880 | cHsp70-3 | Nuc/cytoplasm | 560 | 61 | AK106371 |
| Os03g16920 | cHsp70-4 | Nuc/cytoplasm | 653 | 72 | AK287481 |
| Os03g60620 | cHsp70-5 | Nuc/cytoplasm | 649 | 71 | AK069740, AK102784 |
| Os05g38530 | cHsp70-6 | Nuc/cytoplasm | 646 | 71 | AK243004 |
| Os11g47760 | cHsp70-7 | Nuc/cytoplasm | 649 | 71 | AK065431 |
| Os11g08440 | cHsp70-8 | Nuc/cytoplasm | 577 | 63 | AK063977, AK103083 |
| Os11g08445 | cHsp70-9 | Nuc/cytoplasm | 658 | 71 | N/A |
| Os11g08460 | cHsp70-10 | Nuc/cytoplasm | 562 | 62 | AK106272 |
| Os11g08470 | cHsp70-11 | Nuc/cytoplasm | 467 | 61 | N/A |
| Os12g38180 | uHsp70-2 | unpredicted | 215 | 24 | AK099797 |
| Os02g02410 | BiP1 | ER | 665 | 73 | AK119653 |
| Os03g50250 | BiP2 | ER | 669 | 73 | N/A |
| Os05g30480 | BiP3 | ER | 669 | 72 | N/A |
| Os05g35400 | BiP4 | ER | 687 | 74 | AK106696 |
| Os08g09770 | BiP5 | ER | 676 | 74 | N/A |
| Os01g33360 | BiP6 | ER | 608 | 66 | N/A |
| Os02g53420 | mtHsp70-1 | Mitochondria | 670 | 73 | AK065228 |
| Os03g02260 | mtHsp70-2 | Mitochondria | 676 | 73 | AK103835 |
| Os09g31486 | mtHsp70-3 | Mitochondria | 684 | 73 | AK069787 |
| Os05g23740 | cpHsp70-1 | Chloroplast | 689 | 74 | AK060410 |
| Os12g14070 | cpHsp70-2 | Chloroplast | 698 | 74 | AK121949 |
| Os01g49430 | uHsp70-1 | unpredicted | 539 | 58 | N/A |
| Hsp110/SSE family | |||||
| Os01g08560 | Hsp110-1 | Cytoplasm | 845 | 93 | AK100676 |
| Os02g48110 | Hsp110-2 | ER | 902 | 99 | AK100997 |
| Os03g11910 | Hsp110-3 | Cytoplasm | 578 | 62 | AK102685 |
| Os05g08840 | Hsp110-4 | Cytoplasm | 853 | 94 | AK122040 |
| Os05g51360 | Hsp110-5 | ER | 437 | 47 | AK120015 |
| Os06g10990 | Hsp110-6 | Cytoplasm | 470 | 50 | AK071518, AK104048 |
| Os06g46600 | Hsp110-7 | Cytoplasm | 753 | 83 | N/A |
| Os12g05760 | Hsp110-8 | ER | 461 | 48 | N/A |
Fig. 1.
Phylogenetic analysis of Hsp70 superfamily of rice. a Amino acid sequences of rice Hsp70 superfamily genes aligned in clustalX (2.0) were used for generating NJ tree with bootstrap. Phylogenetic tree was visualized in Treeview 1.6.6. The numbers for the interior nodes indicate the bootstrap values for 1,000 replications. The scale at the bottom is the number of amino acid substitutions per site. Segmental duplicated pairs are marked with similar color symbols. b Exon–intron arrangement of Hsp70 superfamily genes. The exons and introns are represented by box and lines, respectively. The order of genes in (b) match with the order shown in (a)
Fig. 2.
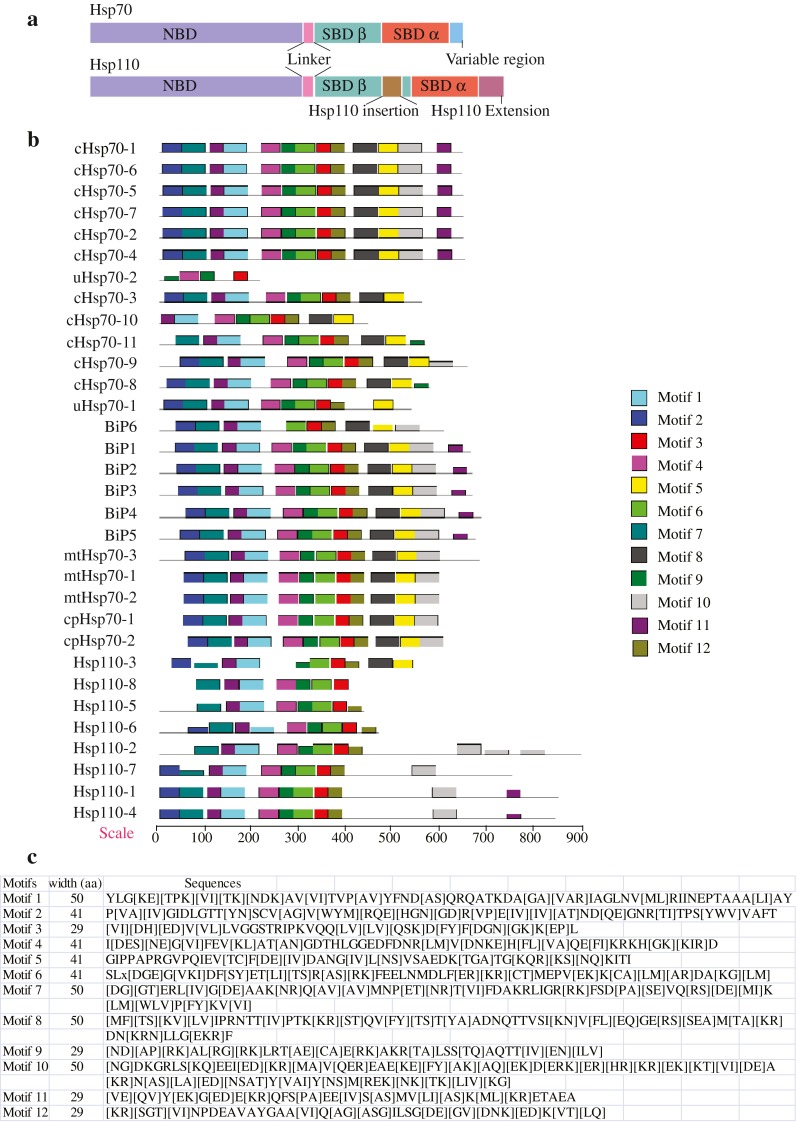
Analysis of conserved motifs present in Hsp70 family proteins. a Domain architecture of Hsp70 and Hsp110 proteins. b Analysis of conserved motifs present in Hsp70 family proteins. Motifs were identified by MEME software using complete amino acid sequences of Hsp70 proteins. The protein sequences are arranged in the order as shown in tree in Fig. 1. Color code of the motifs is depicted on right side. c Multilevel consensus sequences of MEME-derived motifs. Single letters in the motif match that letter; groups of letters in square brackets match any of the letters in the group
Analysis of regulatory cis-elements in the promoters of Hsp70 superfamily genes
2 kB promoter upstream of ATG of Hsp70 genes was analysed to find the cis-regulatory elements that may possibly govern their expression. Six Hsp70 genes (cHsp70-11, BiP6, uHsp70-1, BiP2, BiP3, and BiP5) were not found in the database at Osiris (Morris et al. 2008). Promoters of five genes (cHsp70-1, cHsp70-2, cHsp70-4, cHsp70-7, and mtHsp70-1) were enriched in heat shock elements (HSEs) of perfect type (nTTCnnGAAnnTTCn, P value of <10−4). Gap-type HSEs were detected in two genes (cHsp70-6 and cHsp70-10) and step-type HSEs were detected in cHsp70-2 (Table 2). Promoters of eight Hsp70 genes were manually found to have a variant form of perfect HSEs in which a mismatch in the basic module is permitted (Table 2). Other cis-elements with higher frequency of their occurrence in Hsp70 promoters were motifI, SiteII, DRE, and ABRE. SiteII motifs have been implicated in the expression of genes in meristematic tissue and/or proliferating cells (Welchen and Gonzalez 2006), dehydration-responsive element/C-repeat (DRE/CRT) in drought, high light and cold stress responsive genes (Dubouzet et al. 2003), and ABRE and motifI in abiotic stress/ABA response genes (Mundy et al. 1990).
Table 2.
Enriched regulatory cis-elements in the promoters of Hsp70 superfamily genes
| Regulatory cis-elements | Number of promoters | Predicted sites in promoters | p value | Genes |
|---|---|---|---|---|
| HSE-perfect type | 5 | 5 | 10−4 | cHsp70-1, cHsp70-2, cHsp70-4, cHsp70-7, and mtHsp70-1 |
| Nonenriched TFBS | ||||
| ABREmotif | 12 | 18 | 0.0136 | cHsp70-1, cHsp70-8, cHsp70-10, BiP-1, BiP-4, mtHsp70-1, cpHsp70-1, cpHsp70-2, Hsp110-1, Hsp110-2, Hsp110-5, and Hsp110-8 |
| DRECRTCOREAT | 14 | 19 | 0.034 | cHsp70-1, cHsp70-2, cHsp70-3, cHsp70-4, cHsp70-5, cHsp70-6, cHsp70-7, cHsp70-8, cHsp70-10, BiP-4, mtHsp70-3, cpHsp70-2, uHsp70-2, and Hsp110-5 |
| HSE gaptype | 2 | 2 | 0.0205 | cHsp70-6 and cHsp70-10 |
| HSE steptype | 1 | 1 | 0.485 | cHsp70-2 |
| SITEIIATCYTC | 21 | 64 | 0.0069 | cHsp70-1 cHsp70-2 cHsp70-3, cHsp70-4, cHsp70-5, cHsp70-6, cHsp70-7, cHsp70-8, cHsp70-10, BiP-1, BiP-4, mtHsp70-1, mtHsp70-2, mtHsp70-3, cpHsp70-2, uHsp70-1, Hsp110-1, Hsp110-3, Hsp110-4, Hsp110-5, and Hsp110-8 |
| MotifI | 3 | 3 | 0.0175 | cHsp70-5, Hsp110-6, and Hsp110-7 |
| Imperfect HSE | cHsp70-3, cHsp70-5, cHsp70-6, cHsp70-10, BiP-2, BiP-3, BiP-5, mtHsp70-3, and cpHsp70-1 | |||
p values represent statistically over-represented TF binding sites in selected set of promoters
HSE-perfect type nTTCnnGAAnnTTCn, HSE-gaptype nTTCnnGAAnnnnnnnTTCn, HSE steptype nTTCnnnnnnnTTCnnnnnnnTTCn, ABRE-motif TGACGT, DRE/CRT motif RCCGAC, SiteII element TGGGCY, MotifI RTACGTGGR
Hsp70 genes show constitutive as well as stress induced expression
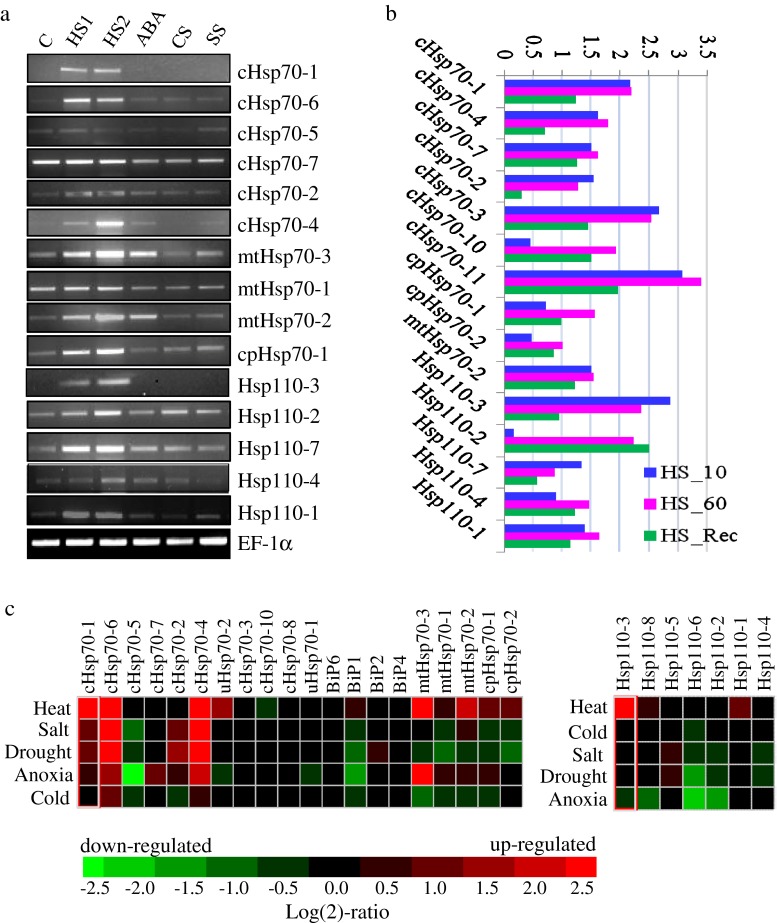
Transcript expression profiles of Hsp70 genes were assessed by semi-quantitative RT-PCR. In addition, heat stress microarray data of rice (Sarkar et al. 2009) and public domain microarray data (genevestigator: www.genevestigator.com/ and rice oligonucleotide array database: www.ricearray.org/) were analyzed for stress induced expression and reproductive stage expression. From RT-PCR, it appears that ten Hsp70 and four Hsp110 genes (Hsp110-2, Hsp110-7, Hsp110-1, and Os05g8840) are expressed constitutively (Fig. 3). These genes can therefore be considered as Hsc70. The transcript expression of 12 genes was upregulated upon HS and among these, expression of two Hsp70 and one Hsp110 genes (Hsp110-3) was strictly induced by HS. Expression of two mitochondrial Hsp70 genes (mtHsp70-2 and mtHsp70-3) was upregulated by ABA. Upregulation of both these genes was higher in heat stress in comparison to the mtHsp70-1 gene. In response to anoxia stress, up-regulated expression of four Hsp70 genes was detected. cHsp70-4 and cHsp70-6 genes were responsive to most of the stresses analyzed. Upregulation of two genes (cHsp70-6 and cHsp70-4) was noticed in CS in microarray profiles. Expression of ten Hsp70 genes was not affected under stress and development (Fig. 3; Supplementary Fig. 3). Among the Hsp110 members, Hsp110-5, Hsp110-6, and Hsp110-8 genes showed negligible expression in the microarray. However, proof of their transcript expression is supported by FL-cDNA for Hsp110-6 (in flower and shoot library) and Hsp110-5 genes (unknown library).
Fig. 3.
Expression analysis of Hsp70 genes of rice. a Semi-quantitative RT-PCR of selective Hsp70 genes under stress conditions. Rice seedlings were given stress under different conditions as mentioned: HS1 heat shock at 42 °C for 10 min, HS2 heat shock at 42 °C for 1 h, ABA 100 μM abscisic acid for 3 h, CS cold stress at 6 °C for 6 h, SS salt stress in 150 mM NaCl for 6 h, C control maintained at 26 °C. b Microarray based expression profiles during heat stress and recovery conditions as described (Sarkar et al. 2009). HS_10 heat stress at 42 °C for 10 min, HS_60 heat stress at 42 °C for 1 h, HS_Rec 30 min recovery after heat stress at 42 °C for 1 h. c Heat map showing in silico expression data retrieved from Genevestigator (www.genevestigator.com/)
During developmental stages, cHsp70-7 gene was highly expressed throughout the life cycle of rice plant with varying expression levels (Supplementary Fig. 3). cHsp70-4 gene was exclusively expressed during seed development stages with maximum expression in mature seed (Supplementary Fig. 3). The expression level of this gene was reduced sharply in germinating seeds. In case of chloroplast localized Hsp70 genes, both the genes were expressed in seed development stages. However, expression level of cpHsp70-2 gene was higher than cpHsp70-1. Expression of mitochondrial Hsp70 genes was also differential during development: transcript expression signals for mtHsp70-2 and mtHsp70-1 were higher in seed development stages as compared with mtHsp70-3. On the other hand, expression of mtHsp70-1 gene was higher than mtHsp70-2 and mtHsp70-3 during panicle development stages. Hsp110-1, Hsp110-2, Hsp110-4, and Hsp110-7 genes showed varying intensity of expression in all stages of development. In contrast, expression of Hsp110-3 gene was restricted to seed development stages.
Rice Hsp70: J-protein bichaperone machine complement corresponding yeast mutants
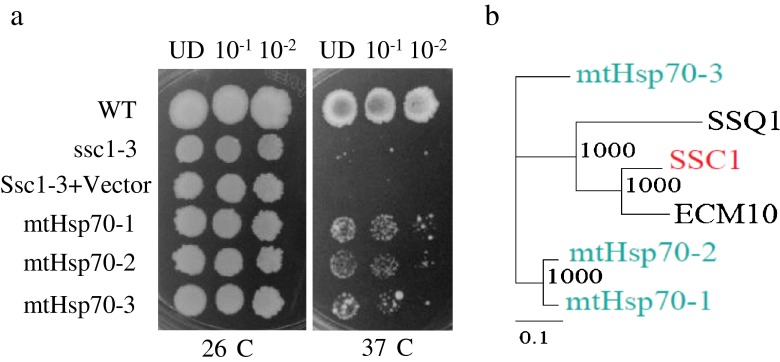
To assess functionality of Hsp70 genes of rice, yeast mutant ssc1–3 defective in mtHsp70 gene was employed. Mutation in mtHsp70 gene renders yeast cells heat sensitive and cells exhibit growth defect at 37 °C (Gambill et al. 1993). Three putative mitochondrial Hsp70 genes of rice namely mtHsp70-1, mtHsp70-2, and mtHsp70-3, were expressed in ssc1–3 yeast cells under the control of GPD promoter and growth phenotype was scored at 37 °C. ssc1-3 cells expressing these mitochondrial genes showed partial protection against HS as against ssc1–3 mutant cells (Fig. 4). Expression of mtHsp70-3 was least effective in complementing the of growth defect of ssc1–3 mutant cells (Fig. 4). From this analysis, it emerges that selective rice mitochondrial Hsp70 genes can function in yeast mitochondrial chaperone system. The Ssc1 protein of yeast is the core component of mitochondrial import motor that facilitates translocation of polypeptides across mitochondrial membrane in ATP-dependent reaction in partnership with 2 J-domain co-chaperones. It is proposed that rice Hsp70 can work in conjunction with yeast Hsp40 to protect yeast cells under heat stress.
Fig. 4.
Expression of rice mitochondrial Hsp70 proteins partially complement yeast Hsp70 mutant. a The mitochondrial Hsp70 genes of rice were cloned under the GPD promoter in p426 vector and transformed in yeast mutant cells (ssc1-3/PK83: MATα ssc1-3 ade2 lys2 ura3 trp1) having mutation in mitochondrial Hsp70 gene (Ssc1). The cells were grown to log phase, growth was normalized to 0.2 OD, and tenfold serial dilutions of wild type, yeast mutant cells transformed with vector and transformed with rice Hsp70 were spotted on selection medium and plates were incubated at 26 and 37 °C. UD is normalized OD600 0.2. Tree depicts rice and yeast mitochondrial Hsp70 genes. b Subset of the phylogenetic tree showing relationship amongst rice and yeast proteins analyzed in this assay
Discussion
The objective of this study was identification and functional characterization of Hsp70 genes in rice genome. This was considered important because Hsp70 proteins are associated with numerous cellular roles under unstressed as well as stressed conditions. Hsp70 proteins occupy central position in the cellular chaperone network interacting with chaperones of other families. We identified that 32 Hsp70 domain containing genes in rice genome constitute Hsp70 superfamily. This includes 24 Hsp70 and 8 Hsp110 genes. As against this, only 14 Hsp70 and 4 Hsp110 genes are reported in Arabidopsis (Lin et al. 2001). While there are 5 cytosolic Hsp70 genes in Arabidopsis, the number of cytosolic genes is 11 in rice. Rice has six canonical (having EEVD at C terminus) and five nonclassical cytosolic Hsp70 genes. The nonclassical Hsp70 genes are not reported in Arabidopsis, indicating that this expansion in Hsp70 family may have occurred after the divergence of monocot–dicot lineage. It is known that segmental and tandem duplication has played a role in the evolution and expansion of gene families in plants (Cannon et al. 2004). The nonclassical rice genes are tandem duplicates. These genes are found in sorghum genome as well, suggesting a monocot-specific clade. However, a definite interpretation in this regard can be only drawn after larger number of genomes are sequenced and analyzed. This is the first comprehensive study of rice Hsp70 superfamily, though isolated studies on mitochondrial Hsp70 and BiPs of Hsp70 family of rice are published (Qi et al. 2011; Oono et al. 2010). Multiple Hsp70 members were identified in various cellular compartments in rice in our analysis. Overall, 11 putative nucleo/cytosolic, 3 mitochondrial, 6 ER, and 2 chloroplastic Hsp70 proteins were identified. Localization of two proteins remained ambiguous due to multiple prediction sites in different localization tools used herein. Eleven Hsp70 proteins predicted to be localized to cytosol or nucleus by different tools were considered as nucleo/cytosolic considering the reports that cytosolic Hsp proteins relocate to nucleus in response to heat stress (Kose et al. 2012). Under heat stress, nuclear localized Hsp70 prevented fragmentation of DNA and provided thermotolarance (Cho and Choi 2009). Qi et al. (2011) reported two mtHsp70 proteins and Oono et al. (2010) reported ten BiPs proteins in rice. In this study, we found that these ten BiPs consist of six Hsp70 and four Hsp110 proteins.
Hsp110 family genes are considered essential for the functioning of Hsp70 genes. This study indicates that rice Hsp110 family contains eight proteins. Four proteins reportedly form Hsp110 family in Arabidopsis (Lin et al. 2001) Yeast Hsp110 family consists of three proteins. Both these species contain one ER localized Hsp110 protein and remainder are cytosolic Hsp110 proteins. In Arabidopsis, Hsp110 members Hsp70-14 and Hsp70-15 represent highly homologous proteins localized predominantly to cytosol. AtHsp70-15 has been implicated in stomatal closure response in unstressed as well as heat stress conditions (Jungkunz et al. 2011). No study has been published on Hsp110 family of rice so far.
Transcript of Hsp70 superfamily genes are expressed constitutively and during development and the expression is enhanced significantly under various stress conditions, suggesting that Hsp70 genes have critical role(s) during growth and stress. The stress induced expression profile of Hsp70 superfamily genes showed diverse response. This diversity in their expression mosaic may be attributed to the regulatory cis-elements present in the promoter region of these genes. Maximum number of Hsp70 genes were upregulated by heat stress. In accordance, HSE elements were enriched in the promoters of the Hsp70 family genes. Consequently, Hsp70 genes have been implicated in development of thermotolerance. Over-expression of mtHsp70 of rice protected rice suspension culture cells from heat stress induced programmed cell death (Qi et al. 2011). Cis-elements like ABRE, DRE, and SiteII coincided with the expression of Hsp70 genes in other stresses and with tissue specific expression.
This study indicates that multiple Hsp70 proteins are present in each of the cellular compartments. It appears that the Hsp70 proteins of the same cellular compartment may not be functionally redundant as the protein isoforms present in specific cellular compartments are noted to be differentially expressed. For instance, of the six classical cytosolic Hsp70 genes, only cHsp70-7 transcript was expressed at all the developmental and anatomical stages. The transcript of this gene was constitutively high and not affected much by heat stress. On the other hand, it also is the case that functional redundancy has been observed in most gene families in spite of expressional diversification (Kafri et al. 2006). In Arabidopsis chloroplast localized two Hsp70 proteins show differential transcript expression. However, only one of these (i.e., cpHsp70-1) is shown to be essential (Su and Li 2010). Hsp70 proteins of mitochondria, chloroplast and ER function as motors driving the translocation of proteins across membranes into the organelles (Su and Li 2010). Rice ER contains six Hsp70 proteins (BiP1-BiP6), and these are functionally diverse under normal growth conditions. Among the six BiP isoforms, BiP1 appears to be highly expressed constitutively in all tissues and is heat induced as well. In contrast, other BiPs are not expressed constitutively (Wakasa et al. 2012). BiPs are upregulated in response to accumulation of misfolded proteins in ER, in unfolded protein response (UPR) pathway. BiP4 and BiP5 were specifically up-regulated upon DTT treatment, a UPR inducer. It is further noted that knockdown of BiP1 leads to induction of BiP2, BiP3, BiP4, and BiP5 (Wakasa et al. 2012), suggesting distinct functional diversity. We observed that BiP1 was highly expressed in seed development stages in agreement with earlier report (Wakasa et al. 2011), indicating its role in protein quality control in seed maturation. In concord, both over-expression and knockdown of BiP1 resulted in ER stress and compromised seed quality (Yasuda et al. 2009; Wakasa et al. 2011). Overall, it will be worth analyzing as to how many genes are dispensable in Hsp70 family in rice. The mutants of all the Hsp70 genes are not available in genetic mutant resources of rice. Arabidopsis genetic resources are more extensively available but Arabidopsis mutants always cannot be used in such analysis especially for genes that have monocot-specific lineages.
The functionality of a subset of Hsp70 genes of rice was analyzed in this study by expressing three Hsp70 mitochondrial proteins in ssc1–3 mutant cells of yeast. Of the three mitochondrial Hsp70 genes in yeast, Ssc1 is the most abundant and essential protein of mitochondrial matrix involved in import and subsequent folding of the precursor proteins (Gambill et al. 1993). ssc1-3 mutants have mutation in the ATPase domain that renders the cells temperature sensitive and hinders the growth at nonpermissive temperature (Gambill et al. 1993). Partial functional complementation of growth defect of ssc1–3 yeast mutant cells by rice mitochondrial Hsp70 genes suggests that rice Hsp70 proteins can possibly interact with yeast mitochondrial protein import machinery though not as effectively as the native yeast proteins. Functional distinction could be ascribed to the interaction of Hsp70 with its co-chaperones. Tutar et al. (2006) also showed that growth of a yeast mutant in cytosolic Hsp70s was faster when supported by its own Ssa1p than Arabidopsis or primate Hsc70. Partial complementation signifies that though Hsp70 are evolutionarily highly conserved, species specificity still plays a role. Taken together, the results demonstrate that rice Hsp70 machinery can interact with yeast Hsp70 chaperone machinery.
Electronic supplementary material
Tandem arrangement of Hsp70 genes on chromosome11. (PPTX 337 kb)
Phylogenetic relation of rice Hsp70 genes with yeast and plant Hsp70 genes. Amino acid sequences of rice and other plant Hsp70 genes aligned in ClustalX (2.0) were used for generating bootstrap (1,000 iterations) NJ tree. The tree was visualized in Treeview 1.6.6. Pink-and blue-colored fonts represent rice and yeast Hsp70 genes, respectively and black font denotes other plant species. The abbreviations used are: Os Oryza sativa, At Arabidopsis thaliana, Cr Chlamydomonas reinhardtii, Sb Sorghum bicolor, Vv Vitis vinifera, Rc Ricinus communis, Bd Brachypodium distachyon, Pt Populus trichocarpa. The accession numbers are: CrHsp70G-XP_001690543.1, Bd1g69700-XP_003558484.1, VvHsp70-CAN68225.1, RcHsp70-XP_002516783.1, Pt081540-XP_002311619.1, Pt10s09880-XP_002315776.1, Sb42680-XP_002465635.1, Bd4g39820-XP_003576944.1, Bd3562113-XP_003562113.1, CrHsp70B-XP_001696432.1, BdHsc70-XP_003561778.1, Sb39510-XP_002468096.1, Sb39500-XP_002468095.1, Sb39390-XP_002465468.1, CrBip1-XP_001701685.1, CrBip2-XP_001701884.1 (JPEG 625 kb)
Microarray based expression meta-analysis of Hsp70 genes during development stages of rice plant (a) and in various tissues (b). Microarray-based analysis was performed in silico at rice oligonucleotide array database (www.ricearray.org/). (PPTX 2377 kb)
List of primers used in this study. (XLSX 9 kb)
Predictions of cellular localization of Hsp70 genes. (XLSX 13 kb)
Acknowledgments
We thank Thomas Langer, University of Cologne, Germany, for providing us yeast mutants of Hsp70. We thank the financial support from the Centre for Plant Molecular Biology and Indo-Finland project from the Department of Biotechnology, Government of India.
References
- Acebron SP, Martin I, del Castillo U, Moro F, Muga A. DnaK-mediated association of ClpB to protein aggregates. A bichaperone network at the aggregate surface. FEBS Lett. 2009;583:2991–2996. doi: 10.1016/j.febslet.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10. doi: 10.1186/1471-2229-4-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho EK, Choi YJ. A nuclear-localized HSP70 confers thermoprotective activity and drought-stress tolerance on plants. Biotechnol Lett. 2009;31:597–606. doi: 10.1007/s10529-008-9880-5. [DOI] [PubMed] [Google Scholar]
- DeRocher A, Vierling E. Cytoplasmic HSP70 homologues of pea: differential expression in vegetative and embryonic organs. Plant Mol Biol. 1995;27:441–456. doi: 10.1007/BF00019312. [DOI] [PubMed] [Google Scholar]
- Diamant S, Ben-Zvi AP, Bukau B, Goloubinoff P. Size-dependent disaggregation of stable protein aggregates by the DnaK chaperone machinery. J Biol Chem. 2000;275:21107–21113. doi: 10.1074/jbc.M001293200. [DOI] [PubMed] [Google Scholar]
- Dragovic Z, Broadley SA, Shomura Y, Bracher A, Hartl FU. Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 2006;25:2519–2528. doi: 10.1038/sj.emboj.7601138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313X.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Duck NB, Folk WR. Hsp70 heat shock protein cognate is expressed and stored in developing tomato pollen. Plant Mol Biol. 1994;26:1031–1039. doi: 10.1007/BF00040686. [DOI] [PubMed] [Google Scholar]
- Gambill BD, Voos W, Kang PJ, Miao B, Langer T, Craig EA, Pfanner N. A dual role for mitochondrial heat shock protein 70 in membrane translocation of preproteins. J Cell Biol. 1993;123:109–117. doi: 10.1083/jcb.123.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy CL, Li QB. The organization and evolution of the spinach stress 70 molecular chaperone gene family. Plant Cell. 1998;10:539–556. doi: 10.1105/tpc.10.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Jungkunz I, Link K, Vogel F, Voll LM, Sonnewald S, Sonnewald U. AtHsp70-15-deficient Arabidopsis plants are characterized by reduced growth, a constitutive cytosolic protein response and enhanced resistance to TuMV. Plant J. 2011;66:983–995. doi: 10.1111/j.1365-313X.2011.04558.x. [DOI] [PubMed] [Google Scholar]
- Kabani M, McLellan C, Raynes DA, Guerriero V, Brodsky JL. HspBP1, a homologue of the yeast Fes1 and Sls1 proteins, is an Hsc70 nucleotide exchange factor. FEBS Lett. 2002;531:339–342. doi: 10.1016/S0014-5793(02)03570-6. [DOI] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Schoffl F. Interaction between Arabidopsis heat shock transcription factor 1 and 70 kDa heat shock proteins. J Exp Bot. 2002;53:371–375. doi: 10.1093/jexbot/53.367.371. [DOI] [PubMed] [Google Scholar]
- Kose S, Furuta M, Imamoto N. Hikeshi, a nuclear import carrier for Hsp70s, protects cells from heat shock-induced nuclear damage. Cell. 2012;149:578–589. doi: 10.1016/j.cell.2012.02.058. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M, Xu XM, Moller SG. Arabidopsis stromal 70-kDa heat shock proteins are essential for chloroplast development. Planta. 2010;232:567–578. doi: 10.1007/s00425-010-1192-z. [DOI] [PubMed] [Google Scholar]
- Leborgne-Castel N, Jelitto-Van Dooren EP, Crofts AJ, Denecke J. Overexpression of BiP in tobacco alleviates endoplasmic reticulum stress. Plant Cell. 1999;11:459–470. doi: 10.1105/tpc.11.3.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Schoffl F. An Hsp70 antisense gene affects the expression of HSP70/HSC70, the regulation of HSF, and the acquisition of thermotolerance in transgenic Arabidopsis thaliana. Mol Gen Genet. 1996;252:11–19. doi: 10.1007/s004389670002. [DOI] [PubMed] [Google Scholar]
- Li QB, Guy CL. Evidence for non-circadian light/dark-regulated expression of Hsp70s in spinach leaves. Plant Physiol. 2001;125:1633–1642. doi: 10.1104/pp.125.4.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QB, Haskell DW, Guy CL. Coordinate and non-coordinate expression of the stress 70 family and other molecular chaperones at high and low temperature in spinach and tomato. Plant Mol Biol. 1999;39:21–34. doi: 10.1023/A:1006100532501. [DOI] [PubMed] [Google Scholar]
- Liberek K, Marszalek J, Ang D, Georgopoulos C, Zylicz M. Escherichia coli DnaJ and GrpE heat shock proteins jointly stimulate ATPase activity of DnaK. Proc Natl Acad Sci U S A. 1991;88:2874–2878. doi: 10.1073/pnas.88.7.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M. Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones. 2001;6:201–208. doi: 10.1379/1466-1268(2001)006<0201:GAOTHS>2.0.CO;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao B, Davis JE, Craig EA. Mge1 functions as a nucleotide release factor for Ssc1, a mitochondrial Hsp70 of Saccharomyces cerevisiae. J Mol Biol. 1997;265:541–552. doi: 10.1006/jmbi.1996.0762. [DOI] [PubMed] [Google Scholar]
- Morris RT, O'Connor TR, Wyrick JJ. Osiris: an integrated promoter database for Oryza sativa L. Bioinformatics. 2008;24:2915–2917. doi: 10.1093/bioinformatics/btn537. [DOI] [PubMed] [Google Scholar]
- Muench DG, Wu Y, Zhang Y, Li X, Boston RS, Okita TW. Molecular cloning, expression and subcellular localization of a BiP homolog from rice endosperm tissue. Plant Cell Physiol. 1997;38:404–412. doi: 10.1093/oxfordjournals.pcp.a029183. [DOI] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene. 1995;156:119–122. doi: 10.1016/0378-1119(95)00037-7. [DOI] [PubMed] [Google Scholar]
- Mundy J, Yamaguchi-Shinozaki K, Chua NH. Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc Natl Acad Sci U S A. 1990;87:1406–1410. doi: 10.1073/pnas.87.4.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA. The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell. 1992;71:97–105. doi: 10.1016/0092-8674(92)90269-I. [DOI] [PubMed] [Google Scholar]
- Oono Y, Wakasa Y, Hirose S, Yang L, Sakuta C, Takaiwa F. Analysis of ER stress in developing rice endosperm accumulating beta-amyloid peptide. Plant Biotechnol J. 2010;8:691–718. doi: 10.1111/j.1467-7652.2010.00502.x. [DOI] [PubMed] [Google Scholar]
- Qi Y, Wang H, Zou Y, Liu C, Liu Y, Wang Y, Zhang W. Over-expression of mitochondrial heat shock protein 70 suppresses programmed cell death in rice. FEBS Lett. 2011;585:231–239. doi: 10.1016/j.febslet.2010.11.051. [DOI] [PubMed] [Google Scholar]
- Sarkar NK, Kim YK, Grover A. Rice sHsp genes: genomic organization and expression profiling under stress and development. BMC Genomics. 2009;10:393. doi: 10.1186/1471-2164-10-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Mosser DD, Morimoto RI. Molecular chaperones as HSF1-specific transcriptional repressors. Genes Dev. 1998;12:654–666. doi: 10.1101/gad.12.5.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su PH, Li HM. Stromal Hsp70 is important for protein translocation into pea and Arabidopsis chloroplasts. Plant Cell. 2010;22:1516–1531. doi: 10.1105/tpc.109.071415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Guy CL. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol. 2003;132:979–987. doi: 10.1104/pp.102.019398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL. Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol. 2001;126:789–800. doi: 10.1104/pp.126.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tutar Y, Song Y, Masison DC (2006) Primate chaperones Hsc70 (constitutive) and Hsp70 (induced) differ functionally in supporting growth and prion propagation in Saccharomyces cerevisiae. Genetics 172:851–861 [DOI] [PMC free article] [PubMed]
- Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. Plant J. 2011;65:675–689. doi: 10.1111/j.1365-313X.2010.04453.x. [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Hayashi S, Takaiwa F (2012) Expression of OsBiP4 and OsBiP5 is highly correlated with the endoplasmic reticulum stress response in rice. Planta 236:1519–1527 [DOI] [PubMed]
- Walsh P, Bursac D, Law YC, Cyr D, Lithgow T. The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep. 2004;5:567–571. doi: 10.1038/sj.embor.7400172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welchen E, Gonzalez DH. Overrepresentation of elements recognized by TCP-domain transcription factors in the upstream regions of nuclear genes encoding components of the mitochondrial oxidative phosphorylation Machinery. Plant Physiol. 2006;141:540–545. doi: 10.1104/pp.105.075366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang KZ, Xia C, Liu XL, Dou XY, Wang W, Chen LQ, Zhang XQ, Xie LF, He L, Ma X, Ye D. A mutation in Thermosensitive Male Sterile 1, encoding a heat shock protein with DnaJ and PDI domains, leads to thermosensitive gametophytic male sterility in Arabidopsis. Plant J. 2009;57:870–882. doi: 10.1111/j.1365-313X.2008.03732.x. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Hirose S, Kawakatsu T, Wakasa Y, Takaiwa F. Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant Cell Physiol. 2009;50:1532–1543. doi: 10.1093/pcp/pcp098. [DOI] [PubMed] [Google Scholar]
- Zhang JX, Wang C, Yang CY, Wang JY, Chen L, Bao XM, Zhao YX, Zhang H, Liu J. The role of Arabidopsis AtFes1A in cytosolic Hsp70 stability and abiotic stress tolerance. Plant J. 2010;62:539–548. doi: 10.1111/j.1365-313X.2010.04173.x. [DOI] [PubMed] [Google Scholar]
- Zhou W, Zhou T, Li MX, Zhao CL, Jia N, Wang XX, Sun YZ, Li GL, Xu M, Zhou RG, Li B. The Arabidopsis J-protein AtDjB1 facilitates thermotolerance by protecting cells against heat-induced oxidative damage. New Phytol. 2012;194:364–378. doi: 10.1111/j.1469-8137.2012.04070.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tandem arrangement of Hsp70 genes on chromosome11. (PPTX 337 kb)
Phylogenetic relation of rice Hsp70 genes with yeast and plant Hsp70 genes. Amino acid sequences of rice and other plant Hsp70 genes aligned in ClustalX (2.0) were used for generating bootstrap (1,000 iterations) NJ tree. The tree was visualized in Treeview 1.6.6. Pink-and blue-colored fonts represent rice and yeast Hsp70 genes, respectively and black font denotes other plant species. The abbreviations used are: Os Oryza sativa, At Arabidopsis thaliana, Cr Chlamydomonas reinhardtii, Sb Sorghum bicolor, Vv Vitis vinifera, Rc Ricinus communis, Bd Brachypodium distachyon, Pt Populus trichocarpa. The accession numbers are: CrHsp70G-XP_001690543.1, Bd1g69700-XP_003558484.1, VvHsp70-CAN68225.1, RcHsp70-XP_002516783.1, Pt081540-XP_002311619.1, Pt10s09880-XP_002315776.1, Sb42680-XP_002465635.1, Bd4g39820-XP_003576944.1, Bd3562113-XP_003562113.1, CrHsp70B-XP_001696432.1, BdHsc70-XP_003561778.1, Sb39510-XP_002468096.1, Sb39500-XP_002468095.1, Sb39390-XP_002465468.1, CrBip1-XP_001701685.1, CrBip2-XP_001701884.1 (JPEG 625 kb)
Microarray based expression meta-analysis of Hsp70 genes during development stages of rice plant (a) and in various tissues (b). Microarray-based analysis was performed in silico at rice oligonucleotide array database (www.ricearray.org/). (PPTX 2377 kb)
List of primers used in this study. (XLSX 9 kb)
Predictions of cellular localization of Hsp70 genes. (XLSX 13 kb)