Abstract
Arabidopsis COLD SHOCK DOMAIN PROTEIN 3 (AtCSP3) shares an RNA chaperone function with E. coli cold shock proteins and regulates freezing tolerance during cold acclimation. Here, we screened for AtCSP3-interacting proteins using a yeast two-hybrid system and 38 candidate interactors were identified. Sixteen of these were further confirmed in planta interaction between AtCSP3 by a bi-molecular fluorescence complementation assay. We found that AtCSP3 interacts with CONSTANS-LIKE protein 15 and nuclear poly(A)-binding proteins in nuclear speckles. Three 60S ribosomal proteins (RPL26A, RPL40A/UBQ2, and RPL36aB) and the Gar1 RNA-binding protein interacted with AtCSP3 in the nucleolus and nucleoplasm, suggesting that AtCSP3 functions in ribosome biogenesis. Interactions with LOS2/enolase and glycine-rich RNA-binding protein 7 that are cold inducible, and an mRNA decapping protein 5 (DCP5) were observed in the cytoplasm. These data suggest that AtCSP3 participates in multiple complexes that reside in nuclear and cytoplasmic compartments and possibly regulates RNA processing and functioning.
Electronic supplementary material
The online version of this article (doi:10.1007/s12192-012-0398-3) contains supplementary material, which is available to authorized users.
Keywords: Cold shock protein, RNA chaperone, Nucleus, Cytoplasm, Arabidopsis thaliana, mRNA processing
Introduction
The cold shock domain (CSD) is a highly conserved nucleic acid-binding domain that is widely distributed in organisms ranging from bacteria to mammals (Graumann and Marahiel 1996). CSD proteins, which were originally identified as cold shock proteins (CSPs) in bacteria, are multifunctional DNA/RNA-binding regulatory proteins that function at the transcriptional and post-transcriptional level (Phadtare 2004). Bacterial CSPs consist solely of a CSD. Nine members of the CSP gene family (cspA to cspI) have been identified in E. coli, four of which (cspA, cspB, cspG, and cspI) are induced by cold shock (Wang et al. 1999). A quadruple mutation in cspA, cspB, cspG and cspE results in a growth defect at low temperature (Xia et al. 2001). Bacterial CSPs are proposed to function as RNA chaperones that destabilize the secondary structures of RNA molecules to facilitate transcription and translation (Jiang et al. 1997).
CSD proteins have been extensively studied in eukaryotic cells. Human Y-box protein, YB-1, was first identified as a transcription factor that binds to the Y-box of MHC class II promoters (Didier et al. 1988). YB-1, which contains a single CSD with N-terminal and C-terminal auxiliary domains, preferentially binds single-stranded pyrimidine-rich sequences (Graumann and Marahiel 1998) and is a major component of cytoplasmic messenger ribonucleoprotein (mRNP). YB-1 represses translation and stabilizes mRNAs in the early embryo and somatic cells (Evdokimova et al. 2001). Another class of CSD proteins in animals is Lin28. This protein was first identified in C. elegans as a regulator of developmental timing. Lin28 is a 25-kDa cytoplasmic protein that contains one CSD and a pair of retroviral-type CCHC zinc fingers (Moss and Tang 2003). Human Lin28 binds precursor forms of let-7 microRNAs (miRNAs) and can inhibit pri-let-7 processing (Viswanathan et al. 2008).
Plant CSD proteins typically contain a single N-terminal CSD and variable copies of C-terminal retroviral-like CCHC zinc fingers that are interspersed by glycine-rich regions (Sasaki and Imai 2011). The first plant CSD protein to be functionally characterized was Triticum aestivum (wheat) cold shock protein 1 (WCSP1) (Karlson et al. 2002). Both WCSP1 mRNA and protein levels steadily increase in crown tissue during cold acclimation. WCSP1 partially complements the cold sensitive phenotype of the E. coli cspA, cspB, cspE, cspG quadruple mutant, suggesting that WCSP1 shares a function with E. coli CSPs for cold adaptation. Arabidopsis contains four CSD proteins (AtCSP1 to AtCSP4). Expression of the four AtCSP genes is differentially regulated in response to cold and developmental cues (Nakaminami et al. 2009; Sasaki et al. 2007; Karlson and Imai 2003). AtCSP3 shows nucleic acid melting activity and complements a cold-sensitive phenotype of the E. coli CSP quadruple mutant (Kim et al. 2009). A loss-of-function mutant of AtCSP3 (atcsp3-2) is more sensitive to freezing than is the wild-type plant under non-acclimated or cold-acclimated conditions. Overexpression of AtCSP3 in transgenic plants confers enhanced freezing tolerance (Kim et al. 2009). Microarray analysis revealed that AtCSP3 regulates the expression of several stress-inducible genes that are not classified as CBF regulon genes. These data suggest that AtCSP3 regulates freezing tolerance in Arabidopsis during cold acclimation independently of the CBF/DREB1 pathway.
To decipher the mechanism underlying AtCSP3-mediated gene regulation, we performed interactome analysis of AtCSP3 using yeast two-hybrid and bi-molecular fluorescence complementation (BiFC) assays. AtCSP3 was shown to interact with a variety of proteins that are involved in RNA processing and metabolism in specific compartments within the nucleus and cytoplasm.
Results
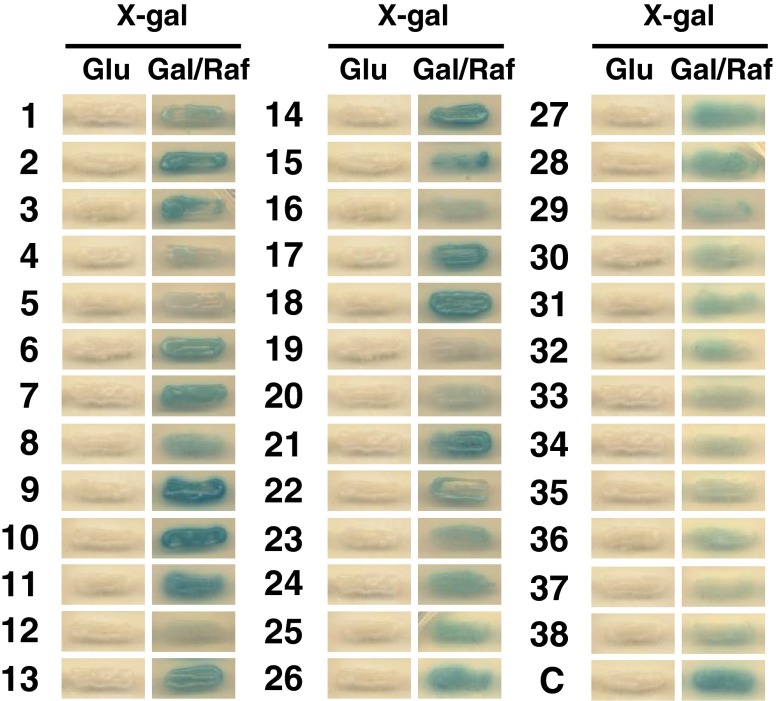
To identify proteins that interact with AtCSP3, we performed an interactome screening using a yeast two-hybrid system. A cDNA library constructed from cold-acclimated Arabidopsis seedlings was used as prey and full-length AtCSP3 was used as bait. Positive clones were selected for Leu auxotrophy and then subjected to a β-galactosidase assay. After screening of 3 × 107 clones, 788 positive clones were selected. Sequence analysis of the positive clones further eliminated overlapping and frame-shifted clones and finally identified 38 independent AtCSP3-interacting proteins (CIPs) (Fig. 1, Table 1). Among these proteins were two members of the DEAD/DEAH-box helicases (RH15 and PRH75) (Aubourg et al. 1999), three 60S ribosomal proteins (RPL36aB, RPL26A, and RPL40A), and three putative nuclear poly(A)-binding proteins (PABN1, PABN 2, and PABN 3) (Table 1). In addition, many other candidate interactors were found to be associated with RNA function. LOS2 (Lee et al. 2002) and AtGRP7 (Kim et al. 2007) have been implicated in the cold response, and SKIP1 has been shown to confer salt and drought tolerance in Arabidopsis (Lim et al. 2010) and Oryza sativa (rice) (Hou et al. 2009) (Table 1), suggesting an association between AtCSP3 complexes and stress tolerance.
Fig. 1.
Interaction between AtCSP3 and candidate proteins (#1–38) revealed by the yeast two-hybrid assay. Positive interactors were selected by histidine auxotrophy (growth on His- plate) and galactose-induced β-galactosidase activity. Arabidopsis thailiana (Col-0) plants were grown at 22 °C under 16-h light/8-h dark conditions. Cold-acclimated (5 days at 4 °C) three-week-old seedlings were harvested for cDNA library preparation. The CloneMiner cDNA Library Construction Kit (Invitrogen) was utilized to construct a cDNA library based on the entry vector pDONR222. The library was subsequently transferred into pJG4-5 (Kaminaka et al. 2006) using LR Clonase (Invitrogen). For bait construction, a PCR-amplified AtCSP3 cDNA was cloned into pENTR/D-TOPO (Invitrogen) and then transferred into the destination vector, pEG202, using LR clonase II (Invitrogen). The Arabidopsis cDNA library (1 × 105 independent clones) in pJG4-5 was transformed into Saccharomyces cerevisiae EGY48 cells (Kaminaka et al. 2006) containing pEG202-AtCSP3 (bait) and pJK103 (reporter). As a positive control (C), interaction between AtLSD1 proteins was utilized (Kaminaka et al. 2006). Identification of candidate proteins is listed in Table 1
Table 1.
List of candidate proteins interacting with the AtCSP3 by yeast two-hybrid
| No | Locus ID | Gene name | Subcellular localization in BiFC |
|---|---|---|---|
| 1 | At1g48920 | rRNA processing protein, nucleic acid binding protein (AtNUC-L1/PARL1) | Nucleolus and nucleoplasm |
| 2 | At4g10970 | Unknown protein | Nucleolus and nucleoplasm |
| 3 | At3g53460 | Chloroplast 29 kDa ribonucleoprotein (CP29) | N.D. |
| 4 | At5g60980 | Nuclear transport factor 2 family protein (NTF2) | N.D. |
| 5 | At1g15690 | Vacuolar-type H + -pumping pyrophosphatase 1/ATPase (AVP1) | N.D. |
| 6 | At3g03920 | Gar1 RNA-binding region family protein | Nucleolus |
| 7 | At1g26110 | Decapping protein 5 (DCP5) | Cytoplasm (P-body) |
| 8 | At5g11200 | DEAD/DEAH box helicase, putative (RH15/ELF4A) | Nucleus and cytoplasm |
| 9 | At5g51120 | Poly A binding protein, nuclear (AtPABN1) | Nuclear speckle |
| 10 | At5g10350 | Poly A binding protein, nuclear (AtPABN2) | Nuclear speckle |
| 11 | At5g65260 | Poly A binding protein, nuclear (AtPABN3) | Nuclear speckle |
| 12 | At3g23390 | 60S ribosomal protein L36a/L44 (RPL36aB) | Nucleolus and nucleoplasm |
| 13 | At4g32420 | Peptidyl-prolyl cis–trans isomerase cyclophilin-type family protein (AtCYP95) | N.D. |
| 14 | At5g13650 | Elongation factor family protein (EFG/EF2) | N.D. |
| 15 | At3g49910 | 60S ribosomal protein L26 (RPL26A) | Nucleolus and nucleoplasm |
| 16 | At5g42050 | Unknown protein/N-rich protein (NPR) | N.D. |
| 17 | At3g16470 | Jasmonate response (JR1) | N.D. |
| 18 | At5g62190 | DEAD/DEAH box helicase binding protein (PRH75) | Nucleolus and nucleoplasm |
| 19 | At1g28050 | Zinc finger (B-box type) family protein (CONSTANS-like protein 15/COL15) | Nuclear speckle |
| 20 | At4g23890 | Unknown protein | N.D. |
| 21 | At3g59810 | U6 snRNA-associated Sm-like protein LSm 6 | N.D. |
| 22 | At3g46780 | Plastid transcriptionally active 16 (PTAC16) | N.D. |
| 23 | At2g37220 | Chloroplast 29 kDa ribonucleoprotein | N.D. |
| 24 | At5g13590 | Unknown protein | N.D. |
| 25 | At2g36170 | Ubiquitin extension protein 2 (UBQ2)/60S ribosomal protein L40 (RPL40A) | Nucleolus and nucleoplasm |
| 26 | At5g05210 | Nuclear matrix protein/protection of teleomeres 1 (POT1) | N.D. |
| 27 | At2g36530 | Copper ion binding/phosphopyruvate hydratase/enolase 2 (ENO2/LOS2) | Nucleus and cytoplasm |
| 28 | At1g35720 | Annexin 1 (ANNAT1) | N.D. |
| 29 | At1g77180 | Chromatin protein family/Ski-interacting protein (SKIP) | Cytoplasm |
| 30 | At2g35860 | Fasciclin-like arabinogalactan protein 16 precursor (FLA16) | N.D. |
| 31 | At3g09690 | Hydrolase, alpha/beta fold family protein | N.D. |
| 32 | At5g23230 | Nicotinamidase 2 (NIC2) | N.D. |
| 33 | At5g37850 | Salt overly sensitive 4 (SOS4) | N.D. |
| 34 | At3g14415 | Peroxisomal (S)-2-hydroxy-acid oxidase/glycolate oxidase (MOA2.13) | N.D. |
| 35 | At5g02810 | Pseudo-response regulator 7 (PRR7/APRR7) | N.D. |
| 36 | At1g53160 | Squamosa promoter binding protein-like 4 (SPL4) | N.D. |
| 37 | At5g23290 | Prefoldin protein 5 (PDF5) | N.D. |
| 38 | At2g21660 | Cold, circadian rhythm and RNA binding 2 (CCR2/ATGRP7) | Cytoplasm |
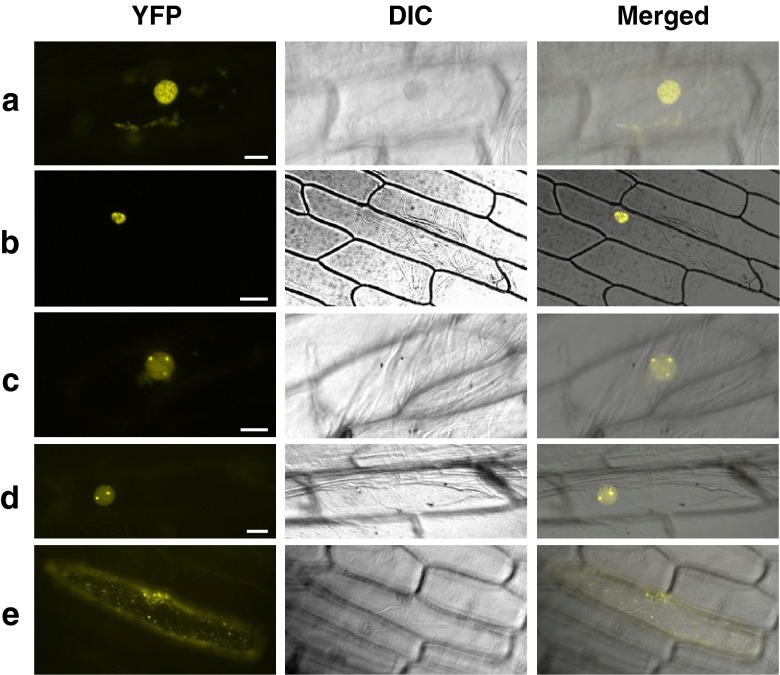
To confirm the data obtained using yeast two-hybrid screening, we utilized the bi-molecular fluorescence complementation (BiFC) assay. For the BiFC assay, AtCSP3 and candidate proteins were fused to the N- and C-terminal fragments of yellow fluorescent protein (YFP), respectively and transiently co-expressed in onion epidermal cells. Interaction was visualized by reconstitution of YFP fluorescence under a fluorescence microscope. Among the 38 genes identified by yeast two-hybrid screening, representative 16 genes were subjected to BiFC analysis. Microscopic observation of onion cells co-transformed with nYFP-AtCSP3 and the cYFP-CIPs revealed reconstituted YFP signals for all 16 combinations, confirming the interaction of these CIPs with AtCSP3 in plant cells. Onion cells co-expressing nYFP-AtCSP3 and cYFP-COL15 (CONSTANS-LIKE gene 15) exhibited YFP signals within a granular sub-compartment of the nucleus, which resembled nuclear speckles (Ali et al. 2003) (Fig. 2a). The interaction of AtCSP3 with AtPABN3 (poly(A)-binding protein 3) localized to a similar nuclear compartment (Fig. 2b).
Fig. 2.
BiFC analysis of AtCSP3-interacting proteins. Representative reconstituted YFP fluorescence (left), differential interference contrast (DIC) (center) and merged (right) images of onion epidermal cells co-bombarded with nYFP-AtCSP3 and cYFP-COL15 (a), cYFP-AtPABN3 (b), cYFP-At4g10970 (c), cYFP-RPL40A/UBQ2 (d), cYFP-DCP5 (e). Reconstitution of functional YFP as detected by YFP fluorescence occurs in nuclear speckles (a and b), the nucleolus and nucleoplasm (c and d), cytoplasm (e). Bars are 50 μm (b) and 20 μm (a and c–e). BiFC assay was carried out as previously described (Shimizu et al. 2005). For vector construction, amplified AtCSP3 cDNA was cloned into pSAT4-nEYFP-N1 to fuse with the N-terminal part of YFP. Similarly, cDNAs for AtCSP3-interacting proteins (CIPs) were cloned into pSAT1-cEYFP-N1 to make a fusion with the C-terminal YFP. Primers used for plasmid construction are listed in Supplementary Table 1. For transient expression, gold particles (1.0 μm) coated with plasmid DNA (2.5 μg) were introduced into onion epidermal cells using a PDS1000/He particle gun (Bio-Rad, USA) according to the manufacturer’s instructions. Onion (Allium cepa) epidermal peels were placed on MS agar medium and used for bombardment with a rupture setting of 1,100 psi. The bombarded samples were incubated for 16 h at 22 °C and were observed by a Leica FW 4000 microscope
Yeast two-hybrid screening identified three ribosomal proteins as CIPs (Table 1). BiFC confirmed the interaction between the 60S ribosomal protein, L40/ubiquitin extension protein 2 (RPL40A/UBQ2) and AtCSP3 in the nucleus (Fig. 2c). A few dot-like structures with intense signal were observed, suggesting that AtCSP3-RPL40A localized within the nucleolus and nucleoplasm (Fig. 2c). The other two 60S ribosomal proteins, L36a/L44 (RPL36aB) and L26 (RPL26A), exhibited a similar pattern of YFP staining, in what appeared to be the nucleolus and nucleoplasm (data not shown). Two known nucleolar proteins, Gar1 and AtNUC-L1, also appeared to interact with AtCSP3 in the nucleolus and nucleoplasm (data not shown). As shown in Fig. 2d, the interaction between AtCSP3 and unknown protein (At4g10970) reconstituted YFP signal in the putative nucleolus and nucleoplasm, suggesting a nucleolus-associated function of At4g10970. In contrast, the AtCSP3-DCP5 interaction was detected in cytoplasmic granules that resembled RNA processing bodies (P-bodies) (Fig. 2e). Nuclear and cytoplasmic localization was also observed for AtCSP3-RH15 interactions (Table 1). Taken together, BiFC analysis identified interactions of AtCSP3 with 16 different proteins and revealed their subcellular localizations.
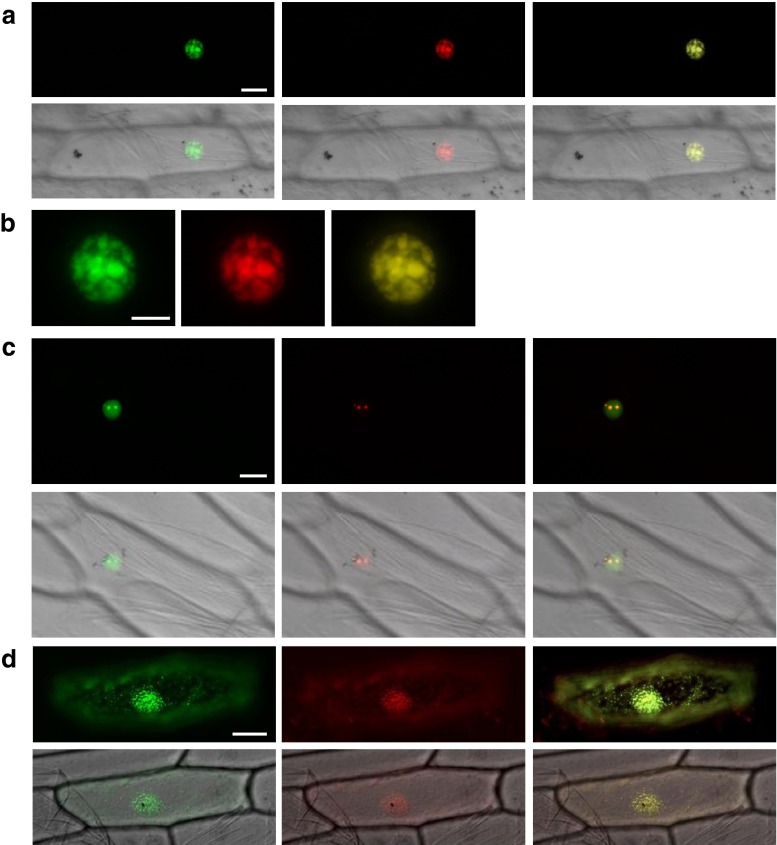
It is important to determine if the site of AtCSP3-CIPs interaction is consistent with the subcellular localization of AtCSP3 and CIPs. AtCSP3 has been shown to localize to the nucleus and cytoplasm (Kim et al. 2009). We therefore determined the subcellular localization of CIPs using the transient expression of GFP-fused proteins. Transient expression in onion cells of PABN3-GFP revealed that PABN3 is localized to nuclear granules, similar to those observed in the BiFC assay for AtCSP3-PABN3 (Fig. 1b). Co-expression of PABN3-GFP and SRp34-RFP (Lorkovic et al. 2004), a marker for nuclear speckle localization, indicated that the observed nuclear granules are speckles (Fig. 3a and b). These data demonstrated that PABN3 is localized to nuclear speckles, where it forms a complex with AtCSP3. Co-expression of COL15-GFP and SRp34-RFP also revealed that nuclear speckle-localization of COL15 (data not shown).
Fig. 3.
tlubcellular localization of AtPABN3, RPL40/UBQ2, and AtDCP5. (a–d) GFP (left), RFP (middle), and merged (right) images of transiently co-transformed onion epidermal cells. DIC images (lower column) reveal the whole cell shapes (a, c, and d). (a and b) Co-localization of AtPABN3-GFP protein with the nuclear speckle marker SRp34-RFP. (c) Co-localization of RPL40/UBQ2-GFP protein with the nucleolar marker AtFbr1-RFP. (d) Co-localization of AtDCP5 protein with the cytoplasmic P-body marker AtXRN4-RFP. Bars are 20 μm (a and c), 10 μm (b), and 50 μm (d). For green fluorescent protein (GFP)-fused constructs, amplified cDNAs were cloned into either the NcoI-SalI or SalI-BsrGI site of the sGFP (S65T) vector (Niwa et al. 1999). For nuclear speckles and P-body markers, SRp34 and XRN4 cDNAs were cloned into pENTR/D-TOPO and then transferred into the destination vector, pH7RWG2.0 (Karimi et al. 2002), using LR clonase II. Primers used for plasmid construction are listed in Supplementary Table 1
Expression of RPL40A-GFP in onion cells detected GFP signal within the nucleolus-like structure and nucleoplasm (Fig. 3c). When RPL40A-GFP was co-expressed with AtFbr1-RFP (Sasaki et al. 2007), a marker gene for nucleolar localization (Fig. 3c), both GFP and RFP signals showed complete overlap (Fig. 3c). The other two 60S ribosomal proteins, L36a/L44 (RPL36aB)-GFP and L26 (RPL26A)-GFP, were also confirmed to localize to the nucleolus and nucleoplasm.
Furthermore, we determined the subcellular localization of DCP5-GFP. XRN4-RFP was used as a marker for P-body localization in plant cells (Kastenmayer and Green 2000). Fig. 3d presents the co-localization of DCP5-GFP and XRN4-RFP, and shows that DCP5 is localized to P-bodies.
Discussion
The interactome analysis indicated that the nuclear speckle is one of the sites at which AtCSP3 functions. A major activity that occurs within the speckles is posttranscriptional splicing (Lamond and Spector 2003). Experimental data suggest that transcription does not occur in speckles (Lamond and Spector 2003). AtCSP3 interacted with COL15 and PABNs within the nuclear speckles (Table 1). Several COL proteins are regulated by the circadian clock (Wenkel et al. 2006; Datta et al. 2006) and associated with responses to light (Datta et al. 2006). Recently, it was revealed that COL1 is involved in the response to cold as well (Mikkelsen and Thomashow 2009). Although the biochemical functions of CO and COL proteins are ambiguous, CO and COL15 have been proposed to interact with the CAAT-binding complex and to be involved in transcriptional regulation (Wenkel et al. 2006). However, it is not known if COLs are involved in RNA processing. Our observation that AtCSP3 interacted with COL15 suggests a role for COL15 in RNA processing within the nuclear speckles and provides novel insight into COL functions.
The three nuclear poly(A)-binding proteins were found to interact with AtCSP3 within the nuclear speckles. In contrast to the cytoplasmic poly(A)-binding proteins, which function in translation initiation and regulation of mRNA decay, the function of nuclear poly(A)-binding proteins is unclear. Human PABPN1 (PABII/PABP2) localizes to the nuclear speckles as a consequence of its binding to poly(A) RNA (Calado and Carmo-Fonseca 2000). PABPN1 interacts with poly(A) polymerase (PAP) to regulate poly(A) tail length (Wahle 1995), a feature that determines both the stability and translation efficiency of mRNA (Coller et al. 1998).
The identification of Gar1, AtNUC-L1, and three ribosomal proteins as CIPs suggested the possible involvement of AtCSP3 in ribosome biogenesis and rRNA metabolism (Pontvianne et al. 2007; Pendle et al. 2005). Gar1, a component of the H/ACA-box small nucleolar ribonucleoprotein (snoRNP) complex, regulates snoRNP assembly and RNA modification in yeast (Henras et al. 2004). Eukaryotic H/ACA-box snoRNPs guide the pseudouridylation of rRNA by base-pairing with target RNA and participate in pre-rRNA processing (Kiss 2001). AtNUC-L1/Parl1 is a nucleolin that is induced by sugars (Kojima et al. 2007) and implicated in auxin-dependent growth and patterning in Arabidopsis (Petricka and Nelson 2007). Nucleolin plays important roles in various steps of ribosome biogenesis (Petricka and Nelson 2007). RPL26 and nucleolin physically interact and bind to a double-stranded RNA structure within the 5′-UTR of p53 (tumor suppressor gene) mRNA to regulate the translation of p53 after exposure to stresses such as UV (Chen et al. 2012).
Two DEAD-box RNA helicases, AtRH7 (PRH75) and AtRH15, exhibited interactions with AtCSP3. The function of these RNA helicases is currently unknown; however, plant DEAD-box RNA helicases have been implicated in several RNA processing events, such as pre-mRNA splicing, rRNA maturation, ribosome assembly, and polyadenylation of mRNA (Aubourg et al. 1999). In addition, a recent report revealed that LOS4, a DEAD-box RNA helicase from Arabidopsis, is involved in mRNA export and regulates plant development and cold stress responses (Gong et al. 2005). Although RNA helicases and RNA chaperones both unwind RNA duplexes, their biochemical mechanisms are different in that the former enzymes require ATP hydrolysis, whereas the latter do not. It is therefore noteworthy that these proteins physically interact. It will be interesting to determine if the complex that forms between the RNA helicase and RNA chaperone activates or stabilizes the RNA unwinding activity.
Arabidopsis DCP5 is a homolog of human RNA-associated protein 55 (RAP55), which is involved in mRNA decay in the P-body (Yang et al. 2006). Xenopus RAP55 (xRAP55) is an RNA-binding component of storage mRNPs in the cytoplasm and acts as a translational repressor (Tanaka et al. 2006). Arabidopsis DCP5 also functions in translational repression and P-body formation and plays an indirect role in mRNA decapping (Xu and Chua 2009). Localization of the AtCSP3-DCP5 complex within the P-body presents the possibility that AtCSP3 is involved in mRNA inactivation or degradation. Interestingly, xRAP55 co-immunoprecipitated with FRGY2, a cold shock domain/Y-box protein in frog (Tanaka et al. 2006).
SKIP1 (Ski-interacting protein) contains a SNW/SKIP domain and functions as a cofactor in transcription and splicing events. Human SKIP1 interacts with PABPN1 in nuclear speckles (Kim et al. 2001). SKIP1 and PABPN1 co-operatively control MyoD-dependent transcription in the skeletal muscle (Kim et al. 2001). SKIP1s from Arabidopsis and rice function as putative transcription factors in an abiotic stress signaling pathway (Hou et al. 2009; Lim et al. 2010). Therefore, it will be interesting to determine if SKIP1, PABN, and AtCSP3 form a complex that regulates abiotic stress tolerance in Arabidopsis.
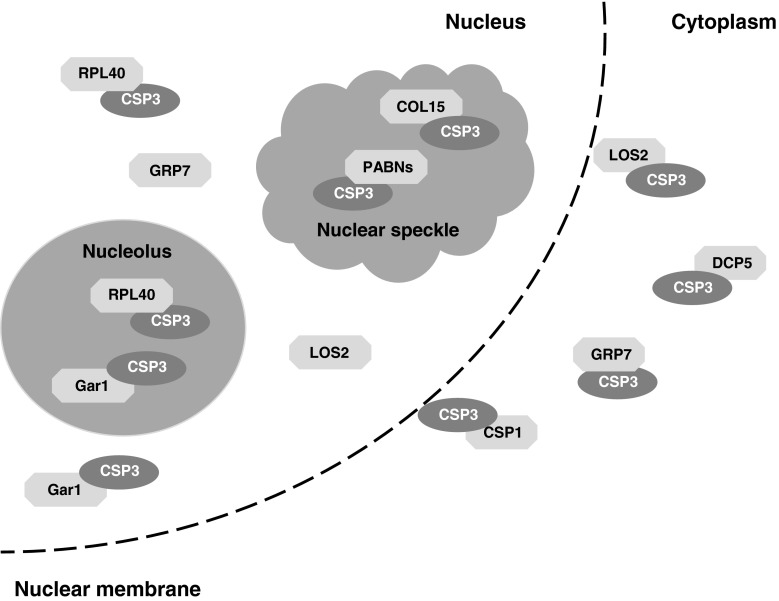
Based on the data presented in this report, we propose a model in which AtCSP3 has versatile functions in RNA processing and metabolism (Fig. 4). Our findings suggest that AtCSP3 forms a complex that functions in mRNA processing and is likely important for its role in gene expression. AtCSP3 may also act by regulating pre-mRNA splicing, polyadenylation, RNA stability, and RNA export by influencing mRNA processing during stress and developmental changes. Taken together, this study revealed that AtCSP3 functions in nuclear and cytosolic RNPs. These data suggest that RNA metabolism is involved in the regulation of the cold response in plants.
Fig. 4.
A model for the AtCSP3 interactome. AtCSP3 is a component of multiple complexes that reside in nuclear and cytoplasmic compartments. The sub-nuclear structures are not presented according to a linear size-scale
Bacterial CSPs, which solely consist of a CSD, are thought to function as a single protein (Phadtare and Severinov 2010). Eukaryotic CSD proteins, such as YB-1, Lin28, and AtCSP3, contain auxiliary domains in addition to a CSD. The eukaryotic CSD proteins may have acquired auxiliary domains to facilitate interaction with other proteins and thereby develop complicated regulatory functions in eukaryotic systems. The CSD may serve as an RNA chaperone module in these proteins. YB-1 has been implicated in transcriptional and translational regulation through its ability to interact with other proteins (Kohno et al. 2003); however, recent research revealed its novel functions in splicing (Allemand et al. 2007) and mRNA degradation (Dhawan et al. 2012). Together with our data, it is now clear that CSD proteins are involved in a wide range of cellular processes associated with RNA metabolism and function.
Electronic supplementary material
(PPTX 143 kb)
Acknowledgments
This research was supported in part by grants from the Japan Society for the Promotion of Science (KAKENHI Scientific Research B 19380063) and NARO project 112g0 (Wheat and Soybean Biotechnology) to R.I.
Abbreviations
- BiFC
Bi-molecular fluorescence complementation
- COL
CONSTANS-LIKE protein
- CBF
C-repeat-binding factor
- CSD
Cold shock domain
- CSPs
Cold shock domain protein
- DCP
Decapping protein
- Gar
Glycine-arginine rich domain
- GFP
Green fluorescent protein
- GRP
Glycine-rich RNA-binding protein
- LOS
Low expression of osmotically responsive gene
- PABN
Nuclear type of poly(A)-binding protein
- RPL
Ribosomal protein
- YFP
Yellow fluorescent protein
Footnotes
An erratum to this article is available at http://dx.doi.org/10.1007/s12192-014-0567-7.
References
- Ali GS, Golovkin M, Reddy AS. Nuclear localization and in vivo dynamics of a plant-specific serine/arginine-rich protein. Plant J Cell Mol Biol. 2003;36(6):883–893. doi: 10.1046/j.1365-313X.2003.01932.x. [DOI] [PubMed] [Google Scholar]
- Allemand E, Hastings ML, Murray MV, Myers MP, Krainer AR. Alternative splicing regulation by interaction of phosphatase PP2Cgamma with nucleic acid-binding protein YB-1. Nat Struct Mol Biol. 2007;14(7):630–638. doi: 10.1038/nsmb1257. [DOI] [PubMed] [Google Scholar]
- Aubourg S, Kreis M, Lecharny A. The DEAD box RNA helicase family in Arabidopsis thaliana. Nucleic Acids Res. 1999;27(2):628–636. doi: 10.1093/nar/27.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calado A, Carmo-Fonseca M. Localization of poly(A)-binding protein 2 (PABP2) in nuclear speckles is independent of import into the nucleus and requires binding to poly(A) RNA. J Cell Sci. 2000;113(Pt 12):2309–2318. doi: 10.1242/jcs.113.12.2309. [DOI] [PubMed] [Google Scholar]
- Chen J, Guo K, Kastan MB. Interactions of nucleolin and ribosomal protein L26 (RPL26) in translational control of human p53 mRNA. J Biol Chem. 2012;287(20):16467–16476. doi: 10.1074/jbc.M112.349274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller JM, Gray NK, Wickens MP. mRNA stabilization by poly(A) binding protein is independent of poly(A) and requires translation. Genes Dev. 1998;12(20):3226–3235. doi: 10.1101/gad.12.20.3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta S, Hettiarachchi GH, Deng XW, Holm M. Arabidopsis CONSTANS-LIKE3 is a positive regulator of red light signaling and root growth. Plant Cell. 2006;18(1):70–84. doi: 10.1105/tpc.105.038182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan L, Liu B, Pytlak A, Kulshrestha S, Blaxall BC, Taubman MB. Y-box binding protein 1 and RNase UK114 mediate monocyte chemoattractant protein 1 mRNA stability in vascular smooth muscle cells. Mol Cell Biol. 2012;32(18):3768–3775. doi: 10.1128/MCB.00846-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85(19):7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evdokimova V, Ruzanov P, Imataka H, Raught B, Svitkin Y, Ovchinnikov LP, Sonenberg N. The major mRNA-associated protein YB-1 is a potent 5' cap-dependent mRNA stabilizer. EMBO J. 2001;20(19):5491–5502. doi: 10.1093/emboj/20.19.5491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Z, Dong CH, Lee H, Zhu J, Xiong L, Gong D, Stevenson B, Zhu JK. A DEAD box RNA helicase is essential for mRNA export and important for development and stress responses in Arabidopsis. Plant Cell. 2005;17(1):256–267. doi: 10.1105/tpc.104.027557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann P, Marahiel MA. Some like it cold: response of microorganisms to cold shock. Arch Microbiol. 1996;166(5):293–300. doi: 10.1007/s002030050386. [DOI] [PubMed] [Google Scholar]
- Graumann PL, Marahiel MA. A superfamily of proteins that contain the cold-shock domain. Trends Biochem Sci. 1998;23(8):286–290. doi: 10.1016/S0968-0004(98)01255-9. [DOI] [PubMed] [Google Scholar]
- Henras AK, Capeyrou R, Henry Y, Caizergues-Ferrer M. Cbf5p, the putative pseudouridine synthase of H/ACA-type snoRNPs, can form a complex with Gar1p and Nop10p in absence of Nhp2p and box H/ACA snoRNAs. RNA. 2004;10(11):1704–1712. doi: 10.1261/rna.7770604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Xie K, Yao J, Qi Z, Xiong L. A homolog of human ski-interacting protein in rice positively regulates cell viability and stress tolerance. Proc Natl Acad Sci U S A. 2009;106(15):6410–6415. doi: 10.1073/pnas.0901940106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W, Hou Y, Inouye M. CspA, the major cold-shock protein of Escherichia coli, is an RNA chaperone. J Biol Chem. 1997;272(1):196–202. doi: 10.1074/jbc.272.1.196. [DOI] [PubMed] [Google Scholar]
- Kaminaka H, Nake C, Epple P, Dittgen J, Schutze K, Chaban C, Holt BF, 3rd, Merkle T, Schafer E, Harter K, Dangl JL. bZIP10-LSD1 antagonism modulates basal defense and cell death in Arabidopsis following infection. EMBO J. 2006;25(18):4400–4411. doi: 10.1038/sj.emboj.7601312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7(5):193–195. doi: 10.1016/S1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Karlson D, Imai R. Conservation of the cold shock domain protein family in plants. Plant Physiol. 2003;131(1):12–15. doi: 10.1104/pp.014472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlson D, Nakaminami K, Toyomasu T, Imai R. A cold-regulated nucleic acid-binding protein of winter wheat shares a domain with bacterial cold shock proteins. J Biol Chem. 2002;277(38):35248–35256. doi: 10.1074/jbc.M205774200. [DOI] [PubMed] [Google Scholar]
- Kastenmayer JP, Green PJ. Novel features of the XRN-family in Arabidopsis: evidence that AtXRN4, one of several orthologs of nuclear Xrn2p/Rat1p, functions in the cytoplasm. Proc Natl Acad Sci U S A. 2000;97(25):13985–13990. doi: 10.1073/pnas.97.25.13985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res. 2007;35(2):506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MH, Sasaki K, Imai R. Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. J Biol Chem. 2009;284(35):23454–23460. doi: 10.1074/jbc.M109.025791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YJ, Noguchi S, Hayashi YK, Tsukahara T, Shimizu T, Arahata K. The product of an oculopharyngeal muscular dystrophy gene, poly(A)-binding protein 2, interacts with SKIP and stimulates muscle-specific gene expression. Hum Mol Genet. 2001;10(11):1129–1139. doi: 10.1093/hmg/10.11.1129. [DOI] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 2001;20(14):3617–3622. doi: 10.1093/emboj/20.14.3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays. 2003;25(7):691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kojima H, Suzuki T, Kato T, Enomoto K, Sato S, Tabata S, Saez-Vasquez J, Echeverria M, Nakagawa T, Ishiguro S, Nakamura K. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49(6):1053–1063. doi: 10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4(8):605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- Lee H, Guo Y, Ohta M, Xiong L, Stevenson B, Zhu JK. LOS2, a genetic locus required for cold-responsive gene transcription encodes a bi-functional enolase. EMBO J. 2002;21(11):2692–2702. doi: 10.1093/emboj/21.11.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim GH, Zhang X, Chung MS, Lee DJ, Woo YM, Cheong HS, Kim CS. A putative novel transcription factor, AtSKIP, is involved in abscisic acid signalling and confers salt and osmotic tolerance in Arabidopsis. New Phytol. 2010;185(1):103–113. doi: 10.1111/j.1469-8137.2009.03032.x. [DOI] [PubMed] [Google Scholar]
- Lorkovic ZJ, Hilscher J, Barta A. Use of fluorescent protein tags to study nuclear organization of the spliceosomal machinery in transiently transformed living plant cells. Mol Biol Cell. 2004;15(7):3233–3243. doi: 10.1091/mbc.E04-01-0055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen MD, Thomashow MF. A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J Cell Mol Biol. 2009;60(2):328–339. doi: 10.1111/j.1365-313X.2009.03957.x. [DOI] [PubMed] [Google Scholar]
- Moss EG, Tang L. Conservation of the heterochronic regulator Lin-28, its developmental expression and microRNA complementary sites. Dev Biol. 2003;258(2):432–442. doi: 10.1016/S0012-1606(03)00126-X. [DOI] [PubMed] [Google Scholar]
- Nakaminami K, Hill K, Perry SE, Sentoku N, Long JA, Karlson DT. Arabidopsis cold shock domain proteins: relationships to floral and silique development. J Exp Bot. 2009;60(3):1047–1062. doi: 10.1093/jxb/ern351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H. Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J Cell Mol Biol. 1999;18(4):455–463. doi: 10.1046/j.1365-313X.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ. Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell. 2005;16(1):260–269. doi: 10.1091/mbc.E04-09-0791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Nelson TM. Arabidopsis nucleolin affects plant development and patterning. Plant Physiol. 2007;144(1):173–186. doi: 10.1104/pp.106.093575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phadtare S. Recent developments in bacterial cold-shock response. Curr Issues Mol Biol. 2004;6(2):125–136. [PubMed] [Google Scholar]
- Phadtare S, Severinov K. RNA remodeling and gene regulation by cold shock proteins. RNA Biol. 2010;7(6):788–795. doi: 10.4161/rna.7.6.13482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontvianne F, Matia I, Douet J, Tourmente S, Medina FJ, Echeverria M, Saez-Vasquez J. Characterization of AtNUC-L1 reveals a central role of nucleolin in nucleolus organization and silencing of AtNUC-L2 gene in Arabidopsis. Mol Biol Cell. 2007;18(2):369–379. doi: 10.1091/mbc.E06-08-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Imai R. Pleiotropic roles of cold shock domain proteins in plants. Front Plant Sci. 2011;2:116. doi: 10.3389/fpls.2011.00116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Kim MH, Imai R. Arabidopsis COLD SHOCK DOMAIN PROTEIN2 is a RNA chaperone that is regulated by cold and developmental signals. Biochem Biophys Res Commun. 2007;364(3):633–638. doi: 10.1016/j.bbrc.2007.10.059. [DOI] [PubMed] [Google Scholar]
- Shimizu H, Sato K, Berberich T, Miyazaki A, Ozaki R, Imai R, Kusano T. LIP19, a basic region leucine zipper protein, is a Fos-like molecular switch in the cold signaling of rice plants. Plant Cell Physiol. 2005;46(10):1623–1634. doi: 10.1093/pcp/pci178. [DOI] [PubMed] [Google Scholar]
- Tanaka KJ, Ogawa K, Takagi M, Imamoto N, Matsumoto K, Tsujimoto M. RAP55, a cytoplasmic mRNP component, represses translation in Xenopus oocytes. J Biol Chem. 2006;281(52):40096–40106. doi: 10.1074/jbc.M609059200. [DOI] [PubMed] [Google Scholar]
- Viswanathan SR, Daley GQ, Gregory RI. Selective blockade of microRNA processing by Lin28. Science. 2008;320(5872):97–100. doi: 10.1126/science.1154040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E. Poly(A) tail length control is caused by termination of processive synthesis. J Biol Chem. 1995;270(6):2800–2808. doi: 10.1074/jbc.270.6.2800. [DOI] [PubMed] [Google Scholar]
- Wang N, Yamanaka K, Inouye M. CspI, the ninth member of the CspA family of Escherichia coli, is induced upon cold shock. J Bacteriol. 1999;181(5):1603–1609. doi: 10.1128/jb.181.5.1603-1609.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkel S, Turck F, Singer K, Gissot L, Le Gourrierec J, Samach A, Coupland G. CONSTANS and the CCAAT box binding complex share a functionally important domain and interact to regulate flowering of Arabidopsis. Plant Cell. 2006;18(11):2971–2984. doi: 10.1105/tpc.106.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia B, Ke H, Inouye M. Acquirement of cold sensitivity by quadruple deletion of the cspA family and its suppression by PNPase S1 domain in Escherichia coli. Mol Microbiol. 2001;40(1):179–188. doi: 10.1046/j.1365-2958.2001.02372.x. [DOI] [PubMed] [Google Scholar]
- Xu J, Chua NH. Arabidopsis decapping 5 is required for mRNA decapping, P-body formation, and translational repression during postembryonic development. Plant Cell. 2009;21(10):3270–3279. doi: 10.1105/tpc.109.070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WH, Yu JH, Gulick T, Bloch KD, Bloch DB. RNA-associated protein 55 (RAP55) localizes to mRNA processing bodies and stress granules. RNA. 2006;12(4):547–554. doi: 10.1261/rna.2302706. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PPTX 143 kb)