Summary
NusA is a core regulator of transcript elongation that is well conserved in bacteria and archaea. NusA interacts with elongating complexes and the nascent RNA transcript in ways that stimulate pausing and termination but can be switched to anti-pausing and anti-termination by other accessory proteins. The regulatory complexity of NusA likely depends on the presence of multiple discrete NusA domains, but it remains unclear which NusA domains possess which regulatory activity and how they interact with elongating RNA polymerase. We used a series of truncated NusA proteins to measure the effect of the NusA domains on transcriptional pausing and termination. We find that the N-terminal domain of NusA is necessary and sufficient for enhancement of transcriptional pausing and the other NusA domains contribute to NusA binding to elongating complexes. Stimulation of intrinsic termination requires higher concentrations of NusA, and involves both the N-terminal domain and other NusA domains. Using a tethered chemical protease in addition to protein-RNA crosslinking, we show that the NusA N-terminal domain contacts the RNA exit channel of RNA polymerase. Finally, we report evidence that the NusA N-terminal domain recognizes duplex RNA in the RNA exit channel.
Keywords: NusA, RNA polymerase (RNAP), transcription regulation, pausing, termination
Introduction
NusA is a conserved, multifunctional regulator of transcript elongation by RNA polymerase (RNAP) in bacteria and archaea. The nusA gene was discovered through mutations in E. coli that prevent λN-dependent bacteriophage λ growth.1 E. coli NusA alone enhances pausing,2–6 enhances intrinsic termination,2; 5 modulates π-dependent termination,7–13 and interacts with error-prone DNA polymerase IV.14; 15 However, NusA also helps suppress pausing and termination in a process called antitermination, when bound to elongating complexes (ECs) together with NusB, NusE, NusG, and one or more other regulators (e.g., λN).7; 16–20 In E. coli, tight binding of NusA to RNAP in antitermination complexes appears restricted to the ribosomal RNA operons. However, E. coli NusA can be crosslinked to RNAP in vivo in all active transcription units once σ is released;21 thus, NusA must maintain a near universal equilibrium association with ECs.
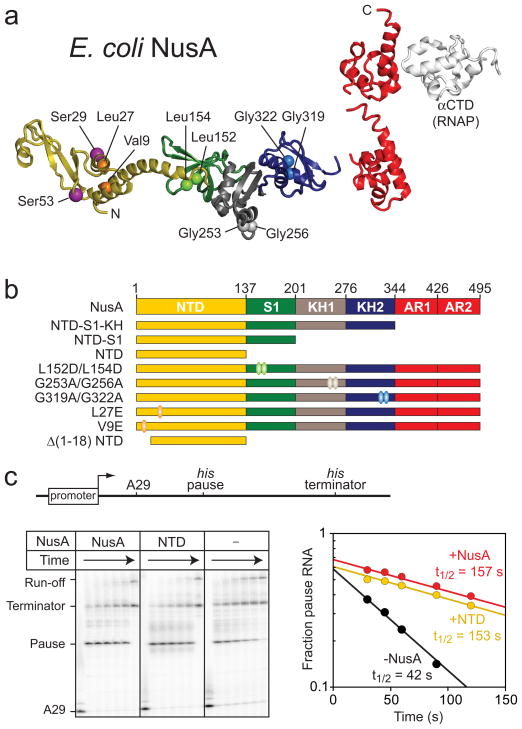
NusA is a multidomain protein (Fig. 1a).22–24 E. coli NusA is composed of an N-terminal domain (NTD), three RNA-binding domains (S1, KH1, KH2 domains), and two C-terminal acidic repeat domains (AR1 and AR2). The NTD interacts with RNAP, likely with the β or β′ subunit, whereas the AR domains contact the C-terminal domain of the RNAP α subunit (αCTD).25–28 The AR domains are present in most γ-proteobacteria, but are absent in archaea and most bacterial lineages and are dispensable for growth of E. coli.29
Fig. 1.
Structure of NusA and the pause enhancing activity. (a) Structural Model of E. coli NusA. The NTD, S1, KH1, and KH2 domains were modeled using the program MODELLER70 and the crystal structure of T. maratima NusA (PDB ID 1l2f).24 AR1, AR2, and the αCTD are from NMR structures of the E. coli NusA and RNA polymerase α subunit domains (PDB IDs 1wcl and 2jzb, respectively; see Methods). (b) NusA constructs used in this study. C-terminal truncation: NTD-S1-KH, NTD-S1, and NTD; domain disruption: L152/L154 (S1 mutant), G253/G256 (KH1 mutant), G319/G322 (KH2 mutant), V9E (NTD mutant), L27E (NTD mutant), and Δ(1-18) NTD. The ovals indicate the residues of substitution. (c) Pause stimulation by NusA. Top, Schematic of the template DNA containing T7 promoter followed by his pause and terminator; Bottom, ECs (~30 nM) halted at position A29 were elongated through the his pause site at position U71 in the absence or presence of the full-length NusA or the NTD (4 μM). Samples were removed at 0, 30, 45, 60, 90, 120 s and quenched. RNA products were separated on an 8% denaturing polyacrylamide gel. The fraction of pause band in each lane was plotted as a function of reaction time to derive the rate of pause escape (kobs).
Elucidation of the roles of NusA domains in its different regulatory activities and their sites of contact to the EC is crucial to understanding the mechanism of NusA action. In antitermination complexes, stable incorporation of NusA requires a network of contacts that includes the NusA CTD, KH, and S1 domains, as well as contacts to both nascent RNA and to RNAP. 26; 30; 31 This interaction network appears to sequester the pause- and termination-enhancing functions of NusA. At least in the case of λQ antitermination complexes, these interactions help NusA create an extended RNA exit channel that shields the nascent transcript against formation of secondary structures that can promote termination or pausing.32
However, the roles of NusA domains in the pause- and termination enhancing activities of NusA and their contacts to non-antitermination-modified ECs are less clear. The magnitude of NusA effects at different pause and termination sites varies greatly, and the concentrations of NusA required to affect termination are typically higher than those required to affect pausing.33–35 Among pause sites, the greatest NusA stimulation occurs when a nascent RNA structure (termed a pause hairpin) stabilizes the paused state.36 Among hairpin-stabilized pauses, NusA may have lesser effects when the pause hairpin is more stable (e.g., contains an RNA tetraloop).37; 38 Much is known about the hairpin-stabilized his pause that synchronizes transcription and translation in the his operon leader region. At the his pause, RNAP forms an off-line paused transcription complex (PTC) in which an active-site rearrangement inhibits nucleotide addition.39 The PTC is stabilized by multiple interactions that include that of the pause hairpin with the RNAP exit channel. NusA likely increases pause dwell time by interacting with the pause hairpin loop and a potion of the RNAP RNA channel called the β flap-tip helix.38; 40.
We show here that all of the pause-enhancing effects of NusA (and much of the intrinsic termination-enhancing effects) reside in the NusA NTD. Two sites on RNAP for interaction of NusA NTD have been proposed. Borukhov et al.41 suggest that NusA NTD interacts with the β′ clamp helices that bind σ70 region 2; this is consistent with the competitive binding of NusA and σ70 to RNAP42 and with NusA-tethered FeBABE cleavage of the region of RNAP.27 Yang et al.43 propose that B. subtilis NusA NTD interacts instead with the β-flap tip that binds σ70 region 4. In a similar model, Vassylyev44 proposes that NusA NTD interacts with the β flap-tip helix by displacement of the N-terminal α-helix of NusA. Interaction of NusA NTD with the β flap is consistent with the requirement for an intact β flap-tip helix for NusA enhancement of pausing and with crosslinking to NusA of a nascent RNA pause hairpin known to contact the βflap tip.38 Based on the mapping of RNAP polypeptide cleavages caused by FeBABE tethered to specific sites on NusA NTD and crosslinking of RNA to NusA, we provide unambiguous evidence that the E. coli NusA NTD contacts the RNA-exit channel as proposed for B. subtilis NusA and RNAP.43 Our experiments with NusA fragments and mutants confirm that this interaction alone enhances transcriptional pausing, whereas other NusA domains help NusA bind ECs.
Results
NusA NTD possesses all pause-enhancing and most termination-enhancing activity of NusA
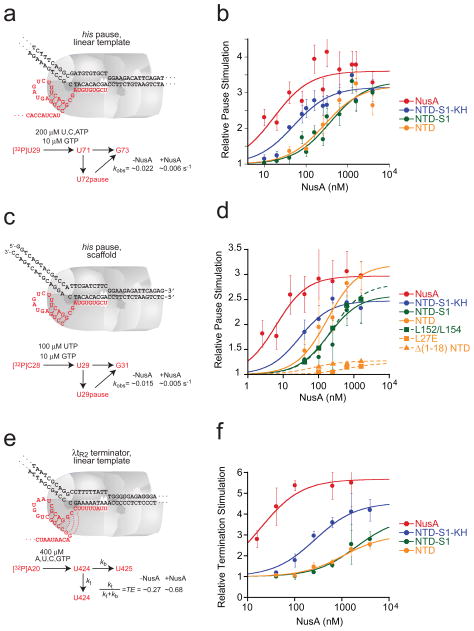
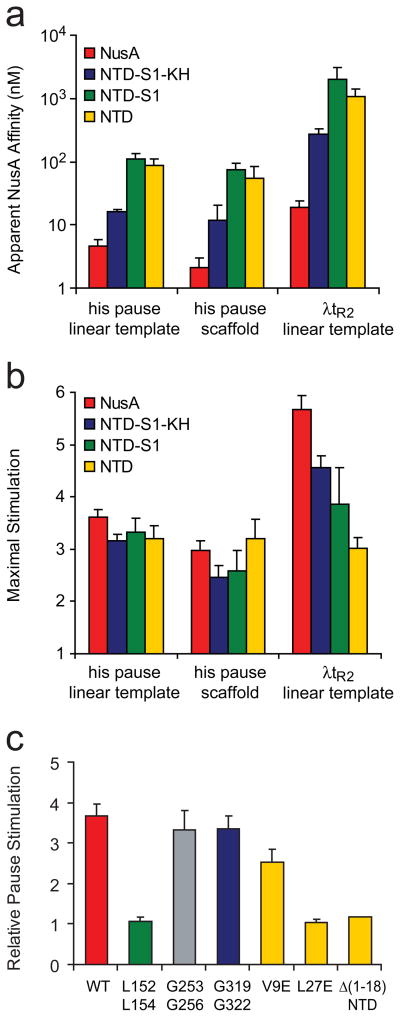
To determine which domains of E. coli NusA are required for enhancement of pausing and termination, we prepared a series of C-terminally truncated NusA proteins (Fig. 1b; Methods): NTD-S1-KH (NusA1-348), NTD-S1 (NusA1-200), and NTD (NusA1-137). First, we measured enhancement of hairpin-stabilized pausing by each NusA fragment using a standard his pause template and in vitro transcription assay (Fig. 1c; Methods).36 NusA increased the dwell time of RNA polymerase at the his pause ~3-fold, with an apparent affinity of ~10 nM (Fig 2a; Table 1). Although removal of NusA AR and KH domains increased the concentration of NusA fragment required for full stimulation, at sufficiently high concentration all fragments including NusA NTD gave ~3-fold enhancement of pause dwell time (Fig. 2b; Table 1). The S1 domain did not appear to contribute to NusA-PTC binding, since pause enhancement by NTD-S1 and NTD exhibited similar concentration dependence.
Fig. 2.
Effects of the C-terminal truncations of NusA on transcriptional pausing and termination. (a) Structure of the his PTC formed on the pIA171 his pause template (pause at +71)73 and kinetic scheme. (b) Relative pause stimulation (k0/kobs) at the promoter-initiated PTC with the C-terminally truncated NusA fragments. Pause assays were performed at 10 μM GTP (GMP is the nucleotide added during pause escape) and 200 μM ATP, UTP, and GTP. The data were fitted to an equation for noncompetitive inhibition of transcript elongation (Methods). (c) Structure of the EC at the reconstituted pause site and the kinetic scheme. (d) Relative pause stimulation (k0/kobs) of the reconstituted PTC with the C-terminally truncated NusAs. (e) Structure of the EC at the λtR2 terminator and the kinetic scheme. (f) Relative termination stimulation (kt,obs/kt0) by the C-terminally truncated NusA fragments.
Table 1.
Apparent NusA affinity and maximal stimulation.
| his pause, linear template | his pause, scaffold | λtR2 terminator, linear template | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| k0/kNusA | KI (nM) | k0/kNusA | KI (nM) | kNusA/k0 | KI (nM) | |
| NusA | 3.61 ± 0.15 | 4.6 ± 1.2 | 2.97 ± 0.19 | 2.1 ± 0.9 | 5.68 ± 0.25 | 19.0 ± 5.2 |
| NTD-S1-KH | 3.16 ± 0.13 | 16.0 ± 1.8 | 2.47 ± 0.23 | 11.8 ± 8.7 | 4.56 ± 0.23 | 269 ± 64 |
| NTD-S1 | 3.33 ± 0.27 | 112 ± 23 | 2.58 ± 0.40 | 74.6 ±19.4 | 3.85 ± 0.71 | 2017 ± 1066 |
| NTD | 3.20 ± 0.25 | 86.3 ± 22.2 | 3.20 ± 0.38 | 55.4 ± 27.8 | 3.01 ± 0.22 | 1081 ± 302 |
| L152D/L154D | - | - | 2.81 ± 0.43 | 87.0 ± 40.6 | - | - |
| L27E NusA | - | - | 1.23 ± 0.08 | - | - | - |
| Δ;(3-18) NTD | - | - | 1.27 ± 0.02 | - | - | - |
The pause escape rates without NusA (k0) on linear template and scaffold are 0.022 ± 0.005 and 0.015 ± 0.002 s−1 respectively. The termination efficiency without NusA (TE0) was 0.28 ± 0.02. We were unable to accurately measure the apparent KI of L27E NusA and σ(3-18) NTD of low enzyme activity.
-, not determined.
The ability of the NusA AR domains to increase NusA-PTC avidity is consistent with the known interaction of these domains with the αCTD (Fig. 1a).25–28 The KH domains form RNA-binding folds,45 co-crystallize with ssRNA,46, and are likely positioned near or interacting with nascent RNA upstream of the pause hairpin, regardless of where NTD contacts RNAP. To ask if the contribution of the KH domains to apparent NusA affinity for the PTC involves their interaction with nascent RNA upstream of the RNA hairpin, we tested the ability of the NusA fragments to delay pause escape of reconstituted his PTCs that lack RNA upstream of the pause hairpin (Fig. 2c).47 Even when upstream RNA was absent, the KH domains decreased the concentration of NusA fragment required for half-maximal pause enhancement by a factor of ~10 (Fig 2d; Table 1). Thus, the effect of the KH domains on pausing must not involve binding to nascent RNA upstream of the pause hairpin.
We next tested the NusA fragments for enhancement of the intrinsic termination using a linear DNA template on which the extensively studied λtR2 terminator is located at ~+450 (Fig 2e; Methods).48 Similar to the effects on hairpin-stabilized pausing, all NusA fragments were capable of stimulating termination when added to sufficient concentration (Fig. 2f). Mah et al. previously reported that NusA fragments similar to NusA NTD and NTD-S1 (NusA1-137 and NusA1-240, respectively) were completely inactive for enhancement of the intrinsic termination (λtR′),26 which is seemingly inconsistent with our results. However, this disparity probably results from the low concentration of NusA (50 nM) used by Mah et al., which is significantly less than the apparent binding constant of NusA NTD (~1 μM) and NTD-S1 (~2 μM). Both the AR and the KH domains decreased the concentration required for half-maximal effect, whereas S1 had little effect. Thus, for hairpin-stabilized pausing and intrinsic termination, a sufficient concentration of NusA NTD can give all or much of the effect of NusA.
To compare the effects of NusA and the NusA fragments in the different pause (Fig. 2b and 2c) and termination assays (Fig. 2c), we calculated the KI and the maximal effects by treating the regulators as noncompetitive inhibitors of nucleotide addition (Figs. 3a and b; Table 1; Methods).40 Consistent with earlier reports,{Gill, 1991 #133;Whalen, 1988 #134;Sigmund, 1988 #135} the half-maximal effects on intrinsic termination required 10–50 times higher concentrations than half-maximal effects on the his pause. However, the pattern and approximate magnitude of these effects resembled that observed for the his pause. For both pause assays, high concentrations of NTD gave the full effect. Even high concentrations of NTD, however, only partially recapitulated the maximal effect of full-length NusA on termination (~3-fold enhancement vs. ~6-fold for full-length NusA). This partial loss of termination-enhancement was attributable to partial contributions of the AR, KH, and S1 domains (Fig. 3b). Given the relatively high concentrations of NTD-S1 and NTD required for maximal effect on termination, we cannot rule out the possibility that aggregation of the NusA fragments prevents them from achieving full activity at high concentration.
Fig. 3.
Apparent affinity and maximal stimulation by NusA for transcriptional pausing and termination. (a) Apparent NusA affinity for promoter-initiated and reconstituted pausing and termination. (see Methods). (b) Maximal stimulation by NusA for promoter-initiated and reconstituted pausing (k0/kNusA) and termination (kt,NusA/kt0). The apparent NusA affinity and the maximal stimulation were calculated from data shown in Figs. 2b, d, and f (Methods). (c) The effects of substitutions in each domain of NusA on transcriptional pausing. The pause dwell time was measured by the pause assay with linear template at 100 nM NusA and normalized by the pause dwell time without NusA.
We conclude that (1) all the interactions of NusA required to enhance hairpin-stabilized pausing and at least a significant portion on the interactions that enhance intrinsic termination are made by the NTD of NusA; (2) the AR, KH, and S1 domains contribute principally to EC binding rather than to pause- or termination-enhancement per se, and (3) at least for pausing, this contribution to NusA-EC binding does not involve interaction with nascent RNA upstream of the pause hairpin.
Domain-disrupting substitutions confirm the key role of NusA NTD in pause enhancement
The use of N-terminal protein fragments precluded testing the effects of individual NusA domains when all NusA domains are present. To circumvent this limitation, we prepared variant NusA proteins in which amino-acid substitutions would disrupt the function of KH1, KH2, S1, or NTD. For KH1 and KH2, we targeted the perfectly conserved GxxG motif that is the hallmark of KH domains49 with substitutions designed to disrupt the KH fold (G253A, G256A in KH1; G319A, G322A in KH2; Fig. 1a and b). G253D and G319D substitutions in KH1 and KH2 were shown previously to interfere with RNA binding by NusA; the KH1 substitutions also compromised λN antitermination and cellular function.31 For S1, we targeted conserved residues in the hydrophobic core with substitutions designed to disrupt proper folding of the domain (L152D, L154D). For NTD, we similarly targeted residues in the hydrophobic core, but with individual substitutions (V9E and L27E) in full-length NusA and with an N-terminal deletion (Δ1-18) in isolated NusA NTD.
Initially, we tested pause-enhancement by the mutant NusA proteins at a subsaturating NusA concentration (100 nM) that would be sensitive to significant changes in either NusA-PTC affinity or the intrinsic pause-enhancing activity of NusA (Fig. 3c). In this assay, the KH1 and KH2 mutants increased pause dwell times similarly to wild-type NusA, supporting the view that interactions of the KH domains with the nascent RNA or with RNAP are not important for the pause enhancement by NusA (Fig. 3c; Table 1). In contrast, the S1 and NTD mutants exhibited significant defects in NusA action (Fig. 3c; Table 1). To test whether this defect was caused by weakened binding of NusA to the PTC or reduced intrinsic activity of NusA, we measured pause lifetimes at increasing NusA concentrations using reconstituted PTCs lacking upstream RNA (Fig. 2d; Table 1). The S1 mutant (L152D, L154D), compared to the wild-type NusA, showed ~10-fold higher apparent KI (32 versus 3.5 nM) but a similar maximal pause lifetime (130 versus 137 s; Table 1). In contrast, we could detect no significant activity of NusA(L27E) or NusA NTD(Δ1-18) even at high concentrations. We conclude that, in the context of full-length NusA, (1) the RNA-binding surfaces of KH1 and KH2 make little or no contribution to NusA-enhancement of pausing; (2) disruption of the S1 affects NusA binding affinity, suggesting it contacts the PTC in intact NusA; and (3) all pause-enhancing activity of NusA resides in the NTD.
The concave surface of NusA NTD interacts with the RNA exit channel
Given our prior demonstration that the β flap-tip helix (Ecβ900-909) at the mouth of the RNA-exit channel is essential for pause-enhancement by NusA,38 the recent evidence that B. subtilis NusA NTD interacts with the β-flap tip,43 and our finding here that the NTD of NusA is sufficient for pause enhancement, it seemed likely that NusA NTD interacts with the RNA exit channel. To reconcile these data with the proposal that E. coli NusA NTD interacts with the β′clamp helices41 and to determine which face of NusA NTD interacts with RNAP, we examined cleavage of RNAP by free radicals generated from Fe-EDTA tethered to opposite sides of the NusA NTD.
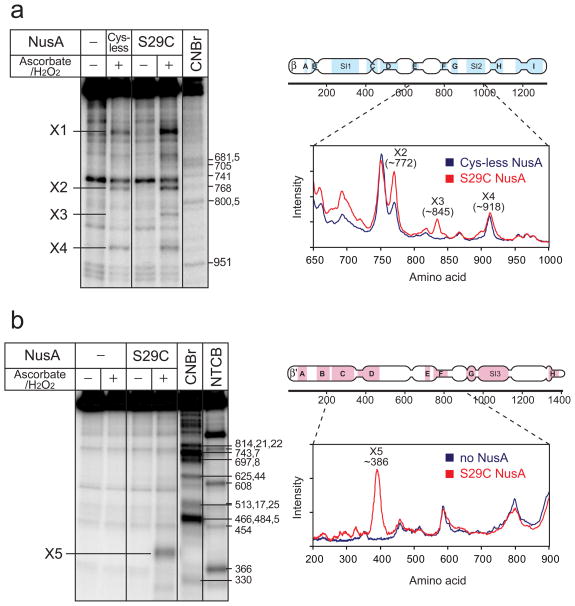
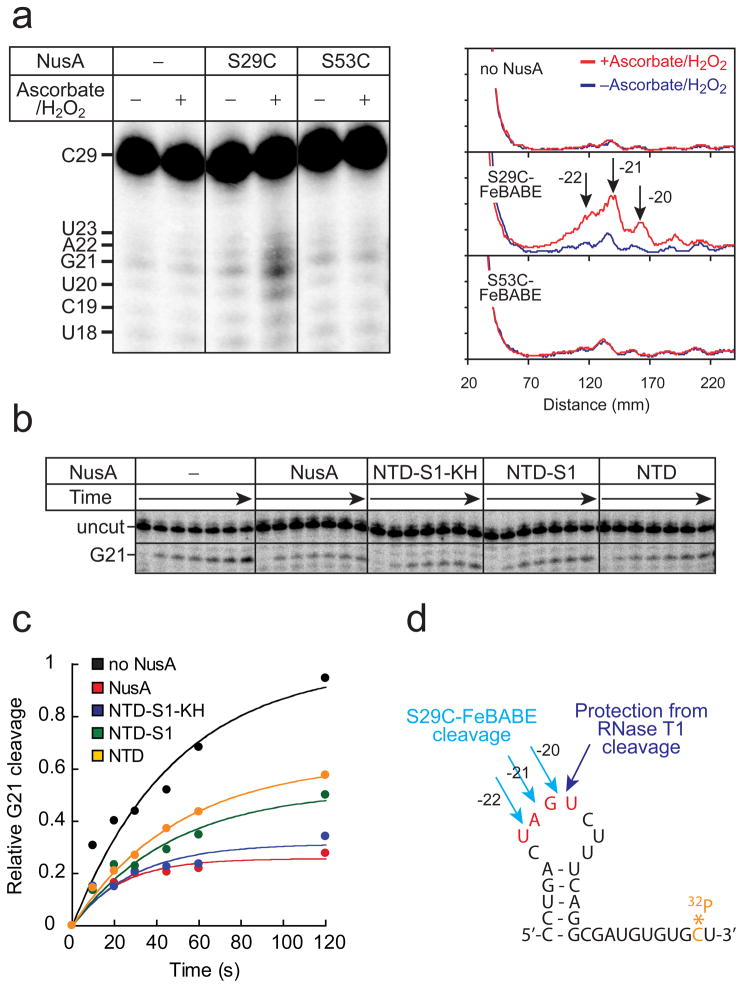
We used the tethered ·OH-generating reagent Fe(III)-(S)-1-(p-bromoacetamido-benzyl)-EDTA (FeBABE), which can be conveniently attached to Cys residues and cleaves nucleic acids or proteins in its vicinity when ·OH-generation is activated.50 We first engineered Cys-less NusA by conversion of C251, C454, and C489 to Ser. We then introduced single Cys substitutions at two non-conserved, surface exposed Ser residues on opposite faces of the NTD (S29C and S53C; Fig. 1a). All three mutant NusAs (Cys-less, S29C, and S53C) were treated with FeBABE. After removal of excess FeBABE by dialysis, the resulting FeBABE-conjugated NusAs were as active as wild-type NusA in pause-enhancement (Supplemental Fig. 1). To analyze cleavage sites on RNAP, we used recombinant E. coli RNAP containing β or β′ subunits bearing N- or C-terminal 32P-labeled HMK tags. We tested the effects of each mutant NusA protein on each 32P-end-labeled RNAP separately, using identical reaction conditions to generate ·OH (Methods).
S29C FeBABE-NusA caused unambiguous, NusA-dependent cleavage of both β- and β′ 32P-labeled RNAPs, whereas no NusA-dependent cleavage was observed for either Cys-less or S53C NusA (Fig. 4 and Supplemental Fig. 2). This result strongly argues that the concave surface of NusA NTD bearing S29 (facing up in Fig. 1a), but not the opposite convex surface of NTD bearing S53 (facing down in Fig. 1a), is oriented toward RNAP upon NusA binding.
Fig. 4.
Cleavage of the β and β′ subunits of the core RNAP by FeBABE-tethered NusA. (a) Cleavage of the β subunit of the core RNAP by S29C-FeBABE NusA and mapping of the cleavage site. RNAP containing the C-terminal 32P-β subunit (100 nM) was incubated with or without Cys-less NusA or S29C NusA (100 nM), which were previously subjected to FeBABE conjugation. The cleavage reaction was activated by addition of ascorbate and hydrogen peroxide. The protein ladder was generated by cleaving 32P-labeled RNAP using the Met-specific cleavage reagent, CNBr. Densitometry traces of lane 2 and 4 are shown on the right. From the experimental error in multiple measurements coupled with an assumption of 1% uncertainty in the migration of β fragments relative to the β markers, we calculated the X1-X4 cleavage sites to be at β subunit 459 ± 56, 772 ± 14, 845 ± 20, and 918 ± 14, respectively. (b) Cleavage of the β′ subunit of the core RNAP by S29C-FeBABE NusA and mapping of the cleavage site. RNAP containing the N-terminal 32P-β′ subunit, as described for panel (a) but with inclusion of the Cys-specific cleavage reagent NTCB. The X5 cleavage was calculated as described for panel (a) to be at β′ subunit 386 ± 12.
To determine the location of NTD interaction with RNAP, we analyzed the cleavage sites on the β and β′ subunit by SDS-PAGE of the fragments generated by S29C-tethered FeBABE-NusA to those caused by treatment with CNBr or NTCB, which cleave at Met and Cys respectively. Four sites on β exhibited S29C-dependent cleavage (X1-X4; 459 ± 56, 772 ± 14, 845 ± 20, and 918 ± 14, respectively; Fig. 4a). A single cleavage site, X5, corresponding to 386 ± 12, was observed on β′ (Fig. 4b; Methods). X3 on β and X5 on β′ were dependent on the presence of S29C-FeBABE NusA and on activation of ·OH production. X1, X2, and X4 were likely caused by nonspecific free-radical cleavage because they were evident with Cys-less NusA (Fig. 4a) and were not detected with βN-terminal 32P-labeled RNAP (Supplemental Fig. 2). The X3 and X5 cleavages are adjacent to the RNA exit channel in the β flap and β′ dock domains (Figs. 5a and 5b). Indeed, these segments of the flap and dock interact with one another in the T. thermophilus EC crystal structure, with direct contacts of β′R675-βL729/730, -β′R679-βA733, and -β′R681-βD636 (equivalent to Ec β′R675-βL729/730, -β′R679-βA733, and -β′R681-βD636).51
Fig. 5.
Model of S29C-FeBABE cleavage sites on the RNAP and the his PTC. The model of the EC is based on the structure of the T. thermophilus EC (see Methods).51 (a) top view of the EC. The α and ω subunits of RNAP are shown in grey. The β′ subunit is shown in pink spacefill. The β subunit is shown as a light blue secondary structure cartoon. DNA is black; RNA is red. (b) A view into the RNA exit channel of RNAP showing the locations of the cleavages from NusA(C29)-FeBABE mapped to the β flap (dark blue) and the β′dock (purple) domains. RNA and DNA are removed for clarity. NusA is depicted adjacent to the RNA exit channel in the orientation predicted by the mapping data, but without attempt to dock NusA on the RNAP surface. E coli RNAP contains an insertion of 100 amino acids in the flap domain (βi9)74; 75 whose location is indicated. (c) same view as (b), but showing a model of the his pause hairpin (see Methods), the sites of cleavage mapped to the hairpin loop (cyan), and a possible path of RNA exit after hairpin formation (red dashes).
NusA NTD-exit channel interactions persist in the his PTC
These results suggest that the NusA NTD interacts with the RNA exit channel of RNAP in the absence of nucleic acids since the flap domain and the clamp domain are parts of the RNA exit channel. To ask whether this interaction persists or is altered in ECs and PTCs, we compared the FeBABE cleavage patterns of RNAP alone to that of PTCs formed on scaffolds with and without a pause hairpin (Fig. 2c, Methods). In these experiments, we observed the same cleavages of the β flap and β′ dock observed in free RNAP (X3 and X5; Supplemental Fig. 3). Thus, the interaction of the NusA NTD with RNAP does not appear to be affected by the presence of RNA in the RNA exit channel, including the presence of a pause RNA hairpin.
NusA NTD contacts or binds near the his pause hairpin
To test the idea that the concave face of NusA NTD interacts with the RNA exit channel in the vicinity of the pause hairpin, we performed two experiments to map the interaction of NusA NTD with the pause hairpin. First, we examined whether the NusA(S29C)-FeBABE cleaves the nascent RNA. To this end, PTCs containing [3′-32P]pause RNA were prepared (Methods) and the RNA cleavage sites generated by localized ·OH were mapped by denaturing gel electrophoresis (Fig. 6a left panel). Cleavage occurred near position -21, which corresponds to the loop of the pause hairpin when NusA(S29C) was used, but no cleavage occurred when NusA(S53C) was used or when NusA was absent (Fig. 6a right panel).
Fig. 6.
Cleavage of the pause hairpin RNA by FeBABE-tethered NusA and protection from RNase T1 digestion. (a) Cleavage of the pause hairpin RNA by FeBABE-tethered NusA and mapping of the cleavage site. rPTCs were prepared as described in Methods. The 32P-labeled rPTC (50 nM) was incubated with S29C-, or S53C-FeBABE NusA (300 nM) for 30 min at 37 °C before cleavage reaction. Densitometry traces are shown on the right. (b) Protection of the pause hairpin by NusA from RNase T1 Digestion. RNase T1 cleavage on the loop of the hairpin in the presence of NusAs at 2 μM. PTCs were prepared as described in Fig. 6a. (c) Relative protection by NusA from RNase T1 digestion. Relative protections of the C-terminally truncated NusAs were calculated by [uncut]max/([uncut]max+[G21]max) and normalized by NusA. (d) Sequence of the pause hairpin RNA and the cleavage sites by S29C-FeBABE NusA and RNase T1.
Second, we examined the ability of NusA or NusA fragments to protect the pause hairpin from cleavage by RNase T1. The loop of the his pause RNA has been shown to be susceptible to RNase T1 and NusA was shown previously to protect the loop of the trp pause hairpin from RNase T1 cleavage.40; 52 In the absence of NusA, RNase T1 cleaved the loop of the his pause hairpin as expected (G21; Figs. 6b and 6c). NusA and all the NusA fragments inhbited the rate of G21 cleavage by RNase T1 (Figs. 6c and 6d). The extent of protection decreased upon removal of the KH domains, consistent with their contribution to overall affinity of NusA for the his PTC.
Taken together, these results confirm that NusA NTD interacts with the RNA exit channel in immediate proximity to the β flap, β′ dock, and pause RNA hairpin. To visualize these interactions, we constructed an improved model of the pause hairpin in the RNA exit channel using the location of exiting RNA in the T. thermophilus EC crystal structure and an NMR structure of a 5-bp stem 8-nt loop RNA from the human PGY/MDR1 mRNA (PDB IDs 2o5i and 2gvo, respectively; Methods; Fig. 5c).51; 53 We refrained from modeling the direct contact of NusA NTD with the exit channel as the limited number of distance constraints allows too many possible solutions. Nonetheless, the approximate positioning of NusA NTD suggests that the acidic head domain of NusA NTD (head, Fig. 5c)24 could be positioned to make contact with the basic charges on the zinc-binding domain in the β′ clamp (ZBD, Fig. 5c). Additionally, modeling of the pause hairpin suggests the loop region would be protected by the NusA NTD, consistent with the observed ·OH cleavage from FeBABE tethered to NusA(S29C) and the observed protection against RNase T1 digestion. Finally, the model predicts a plausible path by which the RNA upstream from the pause hairpin may exit RNAP under the β flap domain (Fig. 5c).
NusA-nascent RNA crosslinking confirms an RNA exit path across KH2
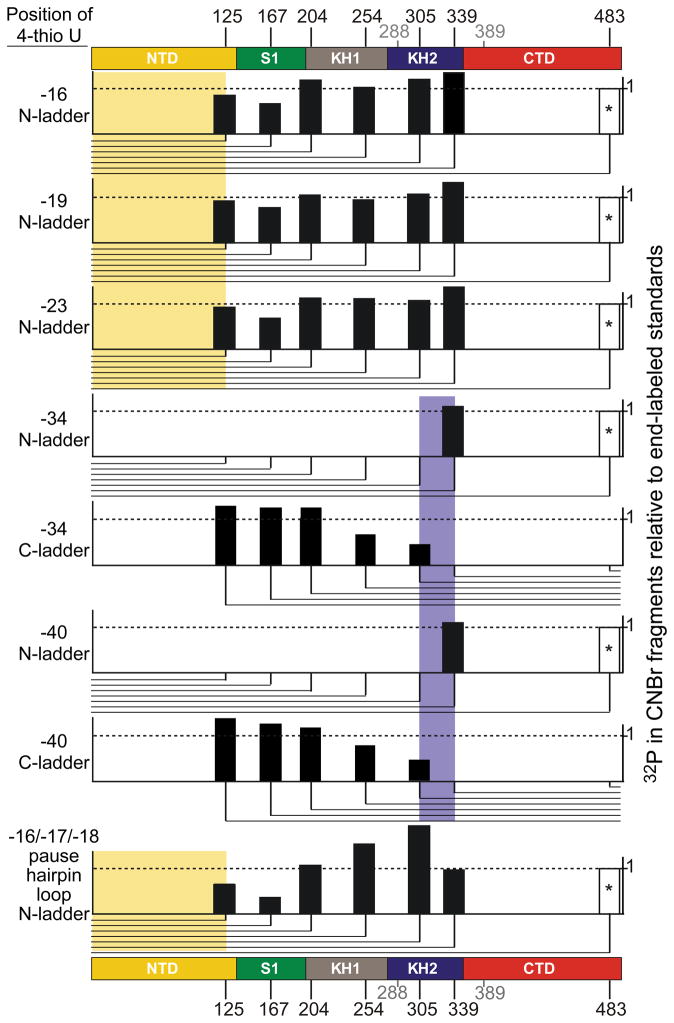
Based on a crystal structure of M. tuberculosis NusA complexed with a portion of antitermination sequence (an 11mer mimic of the rrn boxAC spacer RNA) that is thought to retain interaction with NusA during transcript elongation, Beuth et al.46 proposed a specific set of contacts between exiting RNA and the NusA KH domains. To ask if RNA exiting E. coli ECs and the his PTC are consistent with this proposal, we mapped contacts between exiting RNA and NusA using the photoactive nucleotide analog 4-thio-uridine (4-thioU). 4-thioU and [α-32P]CMP were incorporated into the nascent RNA by halting ECs using stepwise transcription, reacting the halted ECs with 4-thioUTP and [α-32P]CTP, washing to remove these NTPs, and then advancing the ECs to different locations along the RNA with additional rounds of stepwise transcription (Methods; Supplemental Figs. 4a and b). The halted ECs were then crosslinked to excess NusA (2 μM) by UV irradiation. The resulting 32P-labeled NusA-RNA conjugates were briefly treated with RNase A and were analyzed by SDS-PAGE (Methods; Supplemental Fig. 4c). 4-thioU at -19, -23, -34, or -40 in ECs crosslinked to the NTD as well as the full-length NusA. These results suggest that 4-thioUs at these positions are in the vicinity of the NTD. However, they do not indicate that the RNA transcripts at these positions interact with the NTD in the full-length NusA because we can not rule out that 4-thioU might contact the NTD nonspecifically in the absence of normal binding sites (e.g. the RNA binding domains). To address this issue, we mapped the crosslinking sites in the full-length NusA using partial CNBr cleavage (Methods; Supplemental Fig. 4d). The 32P-labeled NusA-RNA conjugates were eluted from the gel and then subjected to partial CNBr cleavage. The resulting fragments were analyzed by SDS-PAGE. When 4-thioU was present at -16, -19, or -23 is ECs, crosslinks occurred to the NusA NTD (Fig. 7; Supplemental Fig. 4d). When 4-thioU was present at -34 or -40, the crosslink shifted to KH2 in a fragment overlapping the KH2-RNA contacts reported by Beuth et al.46 When 4-thioU was instead placed in the loop region of the his pause hairpin (-16-18), crosslinking to the NusA NTD was detected (Fig. 7; Supplemental Fig. 4e). The path of exiting RNA mapped by these crosslinks is entirely consistent with a view that the NusA NTD interacts with the RNA exit channel, the pause hairpin loop, and exiting RNA out to ~nt 23, whereas the KH2 domain interacts with exiting RNA upstream from these positions (see Discussion).
Fig. 7.
RNA crosslinking to NusA. ECs containing 4-thioU and adjacent [α-32P]CMP at various positions of nascent RNA (listed on left) were formed by stepwise elongation, crosslinked to NusA by UV irradiation, and subjected to crosslink mapping using partial CNBr digestion (see Methods). The positions of possible CNBr cleavage are indicated at the top and bottom. We detected no CNBr cleavage at Met288 and Met 389 (gray) even with purified 32P-end-labeled NusA. The amount of radioactivity detected in each partial digestion product normalized to the average of radioactivity in the corresponding bands of partial CNBr digestions of 32P-end-labeled NusA are depicted as bars at the appropriate positions, with fragment orientations (N-terminal partial fragments or C-terminal partial fragments) indicated by the lines below each plot. The location of each crosslink (shaded in color corresponding to the domain in which the crosslink occurred) was assigned to the interval between the last fragment in the ladder containing significant radioactivity and the next fragment lacking radioactivity (or the end of the ladder in the case on crosslinks to the NTD).76 *, the partial digestion product for 483 is inferred but was too close to the parent band to be quantified.
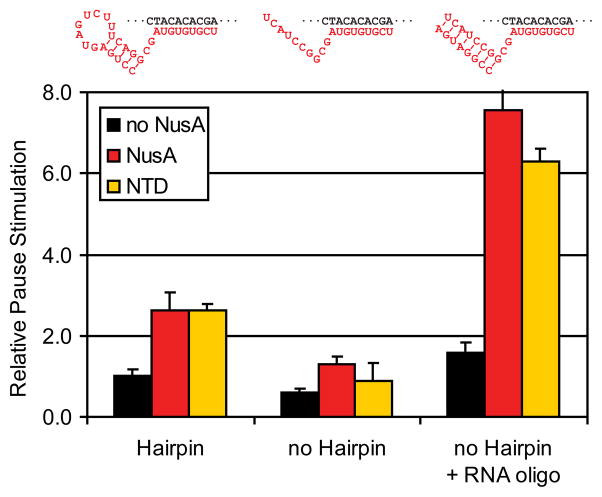
Pause hairpin function requires an RNA:RNA duplex in the RNAP exit channel
Given the apparent interaction of the NusA NTD with the pause hairpin loop, we wondered if specific loop structures are required for maximal NusA effects on transcript elongation. The magnitude of NusA effects on both the lifetime of pauses and the efficiency of termination varies significantly. For instance, replacement of the 8-nt his pause hairpin loop with a highly structured “CUUG” tetraloop greatly diminishes the effect of NusA on pause lifetime.37; 38 To investigate how loop structure affects NusA function, we examined the ability of a “loop-less” RNA duplex to substitute for the his pause hairpin in a pausing assay with reconstituted ECs. An RNA structure was required for NusA enhancement of pausing, consistent with previous findings; however, an 8-bp RNA:RNA duplex created by pairing of an 8mer oligoribonucleotide proved to be an even better substrate for NusA action than a stem-loop structure (Fig. 8). Further, this effect was fully recapitulated by NusA NTD alone. We conclude that NusA has no requirement for a particular loop structure or indeed any single-stranded loop at all. Rather, NusA NTD appears to interact best with a simple A-form duplex.
Fig. 8.
Effect of the pause hairpin on pause enhancement by NusA. Relative effect on pause lifetime was measured using reconstituted his PTCs (Methods).47 PTCs were reconstituted from DNA scaffolds and 27mer or 17mer RNAs (for hairpin and no hairpin PTCs, respectively; Table 2). For no hairpin+ RNA oligo, RNA8, which is complementary RNA17, was added to 1 μM to 200 nM no-hairpin PTC prior to the addition of NTPs to the assay. The measured pause lifetimes were normalized to that of the hairpin PTC without NusA to calculate relative pause stimulation.
Discussion
With six linearly arranged, independently folding domains, E. coli NusA is an unusual transcription regulator. Each NusA domain (NTD, S1, KH1, KH2, AR1, and AR2) may contact different parts of the EC, creating a complex network of both physical and regulatory interactions. This complexity of interactions undoubtedly underlies the remarkable diversity of NusA effects that include stimulation of intrinsic pausing and termination,2; 5; 7; 33; 35; 54–56 enhancement of RNA folding,57 antitermination when partnered with other regulators,1; 19; 20; 58–60 and stimulation of both translesion DNA synthesis14 and stress-induced mutagenesis.15 Here we establish that the NusA NTD alone stimulates pausing and termination through an interaction with the RNA exit channel of RNAP. This finding establishes that NusA can modulate RNAP activity via the interactions of just one of its six separate domains.
NusA NTD activity and interaction with the RNAP exit channel
The biochemical effects of fragments and mutants of NusA establish that the NusA NTD alone possesses all of the pause- and much of the termination-stimulatory activities of NusA. Successive truncations that remove an increasing number of NusA domains starting from the C-terminus increase the concentration of NusA fragment required to stimulate hairpin-stabilized pausing and intrinsic termination, but do not reduce the maximal effect on pausing (Figs. 2b, d and f; Fig. 3a, and 3b). Thus the effect of NusA on pausing is caused solely by interactions of the NTD with the PTC. This conclusion was confirmed by the loss of pause-stimulatory activity at even high concentrations of NusA(L27E) or NusA NTD(Δ1-18) (Fig. 2d).
The contributions of NusA domains to stimulation of intrinsic termination appear to differ from the exclusive effect of the NTD on pausing. Although C-terminal truncations increase the concentration required for maximal effect in the same pattern observed for termination, the absolute concentrations required for maximal effects on termination were consistently greater by a factor of ~10 (Fig. 3a). Further, successive removal of domains from the C-terminus progressively decreased the maximal stimulation of termination while having minimal or no effects on the maximal stimulation of pausing (Fig. 3b). Intrinsic termination occurs in a multistep process, during which the EC passes through a state of partial terminator hairpin formation that closely resembles the geometry of the his PTC (hairpin stem 11 nt from the RNA 3′ nt). This is followed by a rate-limiting extension of the hairpin stem toward the RNA 3′ end that extracts the nascent RNA from the EC RNA:DNA hybrid.61 This bipartite formation of terminator hairpins may explain the pattern of effects of NusA fragments. The effect of the NTD could reflect stabilization of the partial terminator hairpin intermediate via mechanisms similar to that seen on pausing, whereas the S1, KH, and AR domains may assist completion of the hairpin stem and extraction the nascent RNA. Alternatively, the very high concentrations of NusA NTD required to stimulate termination may mask detection of its full termination-stimulatory activity, due either to failure to saturate the effect of NusA NTD or to aggregation of NusA NTD at high concentration.
The pause-enhancing activity of NusA NTD must be caused by interactions of the concave surface of the NusA NTD with the RNA exit channel and exiting RNA. Cleavage of the β flap, the β′ dock, and the loop of the his pause hairpin by ·OH generated from FeBABE attached to NusA C29, but not to NusA C53, dictate this conclusion (Figs. 4 and 6). Further, interaction of NusA NTD with RNAP as well as the effect of NusA on transcriptional pausing require the RNAP β flap-tip helix.38; 43 The charge distribution of NusA NTD also favors orientation of the NTD concave surface toward exiting RNA. The NTD concave faces of both T. maritima24 and E. coli (data not shown) NusA are neutral with patches of positive charge, whereas the convex surfaces bears strong negative charge. Although the NTD can be plausibly docked to the RNAP RNA exit channel (e.g., see Refs. 43; 44), the available data are insufficient to distinguish among possible models for NTD-RNAP interaction. Nonetheless, the constraints imposed by the β flap, β′ dock, pause hairpin-loop ·OH cleavages unambiguously rule out the proposed interaction of NusA NTD with the β′ clamp helices.41 ·OH is incapable of penetrating through a protein62; thus, the proposed location of NusA S29 near β′ clamp helices41 cannot be reconciled with the observed cleavage of the β′ dock because the β′ dock is separated from the β′ clamp helices by the intervening flap domain. The long-established competition of NusA and σ70 from interaction with RNAP42 must occur by competition of the NusA NTD and σ70 region 4 for binding to the β flap-tip helix.
NusA orientation set by NTD-RNAP flap/dock proximity explains several NusA effects
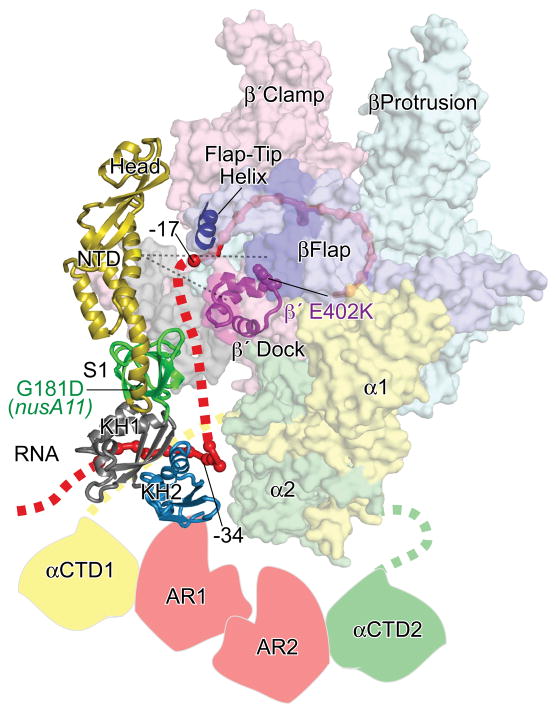
Placement of the NTD against the RNA exit channel coupled with the known interaction of the NusA AR domains with the αCTD25 dictates an orientation of NusA across the surface of RNAP that is consistent with other several other observations (Fig. 9). The KH2 domain of M. tuberculosis NusA interacts with the rrn boxAC spacer RNA across the same KH2 face that we found crosslinked to −34 to −40 of exiting RNA (Fig. 7).46 Connecting the modeled position of this upstream RNA-KH2 segment with the end of the nascent RNA observed in the RNA exit channel of an T. thermophilus EC crystal structure51 generates a plausible path for RNA across the surface of NusA and across the β′ dock domain (Fig. 9). Consistent with this model, Andrecka et al. map the path of RNA exiting yeast RNAPII using FRET triangulation such that RNA −26 to −29 is in contact with the dock domain.63
Fig. 9.
Model of E. coli NusA interactions with an elongating complex. NusA NTD is shown in the same orientation depicted in Fig. 5, and the additional NusA domains are shown in the orientation determined by the model shown in Fig. 1. The locations of αCTD and NusA AR domains are hypothetical and included to illustrate known interactions. The locations of the nusA11(G181D) and cognate suppressor β′E402K are depicted as CPK and labeled.64 The location of the 11mer boxAC spacer RNA from the co-crystal structure of this RNA with the M. tuberculosis NusA S1/KH1/KH2 fragment46 is depicted as a solid red backbone cartoon modeling in the homologous position between the KH1 and KH2 domains. The dotted red line depicts a plausible path of nascent RNA exiting RNAP with the −17 and −34 positions inferred from the crystal structure of the T. thermophilus EC51 and the crosslinking results shown in Fig. 7.
This placement of NusA also is consistent with a genetic interaction of the NusA S1 domain with the β′ dock domain. Ito et al.64 find that the most common β′ suppressor of temperature sensitivity caused by the nusA11(G181D) mutation is β′E402K in the dock domain directly adjacent to the site at which we mapped β′ cleavage by NusAC29-FeBABE (β′378-294). Thus, the resultant suggestion that the S1 domain interacts with the β′dock domain is consistent with the predicted placement of NusA ( Fig. 9) as well as with our finding that S1 contributes to NusA affinity for the his PTC (Fig. 2d).
Finally, this placement of NusA suggests that NusA would shield a segment of RNA from about −17 to −30 from interaction with macromolecules. Indeed, Shankar et al. report that NusA shields nascent RNA from RNaseA digestion at positions −19 to −27 when NusA binding is stabilized by the Q82 antitermination protein.32 NusA alone may interact with exiting RNA too transiently to generate strong protection, but protection of this RNA segment may become strong when NusA interactions with an EC are stabilized.
Taken together these observations and our findings support a model of NusA interaction that places the NusA across a surface of RNAP that spans from the NusA NTD interacting with the RNA exit channel, to the S1 and KH domains interacting with and adjacent to the β′ dock domain, respectively, to the AR domains interacting with the αCTDs (Fig. 9).
Does NusA promote effects of nascent RNA structure directly or indirectly?
A remaining question is how NusA promotes the effects of nascent RNA hairpins, either in stabilizing pauses or in stimulating intrinsic termination. Gusarov et al. argue that NusA acts indirectly by displacing upstream single-stranded RNA from the RNAP surface, thereby promoting hairpin formation.35 A simple alternative is that NusA directly stabilizes pause and terminator hairpins through contacts to duplex RNA. Given the complexity of effects that can result from interactions of NusA, RNA, and RNAP, ranging from promoting nascent hairpin formation to variable extents to inhibiting hairpin formation,32; 33; 54 it is unlikely these models can be distinguished until high resolution crystal structures of EC-NusA complexes that capture these various effects are obtained. Further, it seems plausible that both direct and indirect effects could be involved.
However, for the simpler case of how the NusA NTD increases the effect of nascent RNA pause hairpins, our data are most consistent with a direct effect on RNAP structure. When the stability of the his pause hairpin is increased by replacement of the 8-nt loop with a CUUG tetraloop structure, the effect of NusA on pause lifetime is reduced (from ~2.5x to ~1.3x).37; 38 This could reflect either direct or indirect pause hairpin stabilization by NusA. However, when the his pause hairpin is stabilized by extension of the hairpin stem from 5 bp to 8 bp, either with38 or without (Fig. 8) a hairpin loop, the effect of NusA increased (from ~2.5x to ~6x). If NusA were acting simply to promote formation of the hairpin stem, there is no obvious reason that the effect of NusA would be increased by lengthening the stem. Further, the effect of NusA is about the same whether or not RNA is present upstream of the pause hairpin.47 This is inconsistent with the indirect model of NusA action, since competition with hairpin formation should be less when upstream RNA is available to replace RNAP contacts made by the 5′ arm of the pause hairpin. Taken together, these results suggest NusA increases the effect of the pause hairpin by enhancing the effect of the hairpin on the structure of ECs, by altering the position of the clamp domain as suggested earlier,38; 39; 52 by inhibiting translocation of nascent RNA, or by a combination of these effects.
Finally, our finding that NusA acts directly on RNA duplex stem of the pause hairpin rather than on the pause hairpin loop (Fig. 8) provides a possible explanation for variability in the magnitude of NusA effects on pausing and termination. A hairpin loop is not without structure, and some loops maintain the trajectory of the A-form phosphodiester backbone along at least one arm of the hairpin (e.g., see the 8-nt loop model in Fig. 5c). Thus, some larger loops may resemble duplex RNA and facilitate NusA NTD interaction. Others, such as the CCUG tetraloop may be too compact for this effect or, in the case of some larger loop sequences, may lack well ordered structures. Thus, the magnitude of the NusA effect could depend on the extent to which the combined stem-loop structure conforms to a A-form helix that may be the preferred ligand for NusA NTD. Here again, high-resolution crystal structures of NusA NTD interacting with a PTC are needed to understand both the nature of the NusA NTD interaction and the mechanism by which it stimulates transcriptional pausing.
Methods
Construction and purification of the HMK-tagged RNAP and NusA proteins
To label RNAPs with 32P, the β and β′ subunits were tagged with the recognition sequence (RRASV) for the catalytic domain of heart muscle kinase (HMK). βN-, βC-, β′N-, and β′C-HMK-tagged RNAPs were overexpressed from a T7 RNAP based expression plasmid and purified by chitin-affinity chromatography followed by heparin-affinity chromatography as described previously.65 The fragments of NusA (NTD, NTD-S1, NTD-S1-KH, Δ(1-18)NTD) were amplified by polymerase chain reaction (PCR) and cloned into an overexpression plasmid, pNG5. Cys-less NusA (C251S, C454S, C489S), two single-Cys NusAs (S29C NusA: S29C, C251S, C454S, C489S; S53C NusA: S53C, C251S, C454S, C489S), and the domain disruption mutant NusAs (L152D/L154D, G253A/G256A, G319A/G322A, L27E, and V9E) were constructed by site directed PCR mutagenesis. All NusA proteins contain the N-terminal His6 tag except Δ(1-18)NTD, which has the C-terminal His6 tag and was purified as described previously.47
In vitro transcription assays
For the promoter initiated pause assay, 32P-labeled A29 complexes (30 nM) were prepared from template DNA containing T7A1 promoter in transcription buffer (20 mM Tris-HCl, pH 8.0, 20 mM NaCl, 14 mM MgCl2, 14 mM 2-mercaptoethanol, 0.1 mM EDTA) and then were incubated with NusA for 10 min at 37 °C. Transcription was resumed upon addition of NTPs (200 μM UTP, CTP, and ATP, 10 μM GTP). Aliquots were removed at the indicated times and mixed with 2X loading dye (7 M urea, 90 mM Tris-borate buffer, pH 8.3, 0.02% bromphenol blue, and 0.02% xylene cyanol), and RNA products were separated on denaturing polyacrylamide gel. The reconstituted pause assays were performed as described previously.47 Briefly, ECs were reconstituted with DNA scaffolds (t DNA and nt DNA) and RNA27 and were advanced to position C28 (1 nt before the pause) in the presence of [α-32P]CTP (Table 2). These ECs were incubated with NusA for 10 min at 37°C before resuming transcription upon addition of 100 μM UTP and 10 μM GTP. For termination assays, the linear DNA templates, which contain the T7A1 promoter followed by the λ tR2 terminator, were generated by PCR amplification from the plasmid pCPG λtR2.66 Halted A20 complexes were prepared as described previously.65 Transcription reactions were incubated at 37 °C for 10 min after addition of all four NTPs (400 μM each) to the halted complex.
Table 2.
Strains, plasmids, oligonucleotides
| Stock # | Name | Description | Source or Note |
|---|---|---|---|
| Strains | |||
| BL21 λDE3 | E. coli B F- ompT gal [dcm−] [lon−] hsdS− (rB− mB−) [λDE3(lacUV5::T7 RNAP) imm21 int−–Δnin5 ΔEcoRI(21226-26104) BamHI27972−] | Studier et al., 1990 | |
| BLR | BL21λDE3 recA | Invitrogen | |
| Plasmids | |||
| 775 | pIA171 | T7A1 promoter A29 hisL (his pause) | Ederth et al., 2002 |
| 168 | pCPG λtR2 | T7A1 promoter A20 λtR2 | Reynolds et al. 1992 |
| 3737 | pIA423 | expresses α, β, β′-CBP/intein from T7 promoter | Artsimovitch et al., 2003 |
| 2728 | pRM528 | βN HMK in pIA423 | This work |
| 2677 | pRM477 | βC HMK in pIA423 | This work |
| 2675 | pRM475 | β′N HMK in pIA423 | This work |
| 4201 | pSK101 | β′C HMK in pIA423 | This work |
| 4455 | pVS7 | expresses α, β, β′-His6 from T7 promoter | Seth Darst |
| 4149 | pNG5 | Expresses N-HMK/His6 NusA from T7 promotor | This work |
| 4712 | pKH12 | ΔCTD in pNG5 | This work |
| 4711 | pKH11 | Δ(KH1-KH2-CTD) (NusA1-200) in pNG5 | This work |
| 4116 | pIA378 | Δ(S1-KH1-KH2-CTD) (NusA1-137) in pNG5 | This work |
| 4701 | pKH1 | C251S, C454S, C489S in pNG5 | This work |
| 4702 | pKH2 | S29C in pKH1 | This work |
| 4703 | pKH3 | S53C in pKH1 | This work |
| 4150 | pNG6 | L152D, L154D in pNG5 | This work |
| 4151 | pNG7 | G253A, G256A in pNG5 | This work |
| 4152 | pNG8 | G319A, G322A in pNG5 | This work |
| 4720 | pKH30 | V9E in pNG5 | This work |
| 4721 | pKH32 | L27E in pNG5 | This work |
| 4722 | pKH20 | Expresses C-His6 NusA NTD with Δ(3-18) from T7 promotor | This work |
| Oligonucleotides | |||
| 5069 | NT DNA | GGTCAGTACGTCCATTCGATCTTCGGAAGAG ATTCAGAG | Reconstitution |
| 5420 | T DNA | CTCTGAATCTCTTCCAGCACACATCAGGACG TACTGACC | Reconstitution |
| 4867 | RNA27 | CCUGACUAGUCUUUCAGGCGAUGUGUG | Reconstitution |
| 4865 | RNA29 | CCUGACUAGUCUUUCAGGCGAUGUGUGCU | Reconstitution |
| 4878 | RNA12 | AGGCGAUGUGUG | Reconstitution |
| 4876 | RNA14 | AGGCGAUGUGUGCU | Reconstitution |
| 6593 | RNA17 | UCAUCCGGCGAUGUGUG | Reconstitution |
| 6598 | RNA8 | CCGGAUGA | Reconstitution |
Quantification of apparent NusA affinity and maximal stimulation
The 32P-labeled RNA were electrophoresed through denaturing polyacrylamide gels and quantified to measure the rate of the pause escape (ke) as described previously.47 The observed pause escape rate (kobs) in the presence of NusA is the composite rate of free ECs and NusA-bound ECs. Since NusA has been shown to be a noncompetitive inhibitor for nucleotide addition,{Landick, 1987 #132} the observed rate of the pause escape can be described as kobs = (k0KI + kNusA[NusA])/(KI + [NusA]), where k0, and kNusA are the pause escape rate of free ECs, and NusA-bound ECs respectively. The plot k0/kobs (relative pause stimulation) versus [NusA] were fitted using Kaleidagraph (Synergy Software), yielding the apparent KI and the maximum pause stimulation (k0/kNusA) (Fig. 2; Table 1).
The termination efficiency (TE) is defined as TE=kt/(kt+kb), where kt and kb are the rate of termination and terminator bypass, respectively.67 TE was measured from the relative intensity of termination and run-off RNA products: TE=[termination]/([termination]+[run-off]). Relative stimulation of the termination rate (kt,obs/kt0) by NusA was calculated assuming NusA affects the termination rate (kt) but not the bypass rate (kb): kt,obs/kt0=TEobs(1−TEobs)/(TE0(1−TEobs)), where kt,obs and TEobs are apparent termination rate and efficiency at a specific concentration of NusA respectively, and kt0 and TE0 are termination rate and efficiency without NusA, respectively. Similar to the apparent pause escape rate, relative stimulation of the termination rate can be described as kt,obs/kt0=(KI+(kt,NusA/kt0)[NusA])/(KI+[NusA]), where kt,NusA is the termination rate of NusA-bound TECs. Fitting the plot kt,obs/kt0 versus [NusA] yields the apparent binding constant (KI) and the maximal stimulation (kt,NusA/kt0) by NusA.
Tethered FeBABE cleavage
NusA (20 μM) and FeBABE (400 μM) were incubated in conjugation buffer (10 mM MOPS (pH 8.0), 1 mM EDTA, 100 mM NaCl, 5% glycerol) at 37 °C for 1 hr and dialyzed against storage buffer (10 mM Tris·HCl (pH 8.0), 0.1 mM EDTA, 100 mM NaCl, 10 mM MgCl2, 50% glycerol) to remove unconjugated FeBABE. 32P-labeled RNAP (or EC) and FeBABE-NusA (100 nM each) were incubated at 30 °C for 30 min in cleavage buffer (10 mM MOPS, pH 8.0, 1 mM EDTA, 125 mM NaCl, 10 mM MgCl2, 10% glycerol) prior to cleavage reaction, which was initiated by adding ascorbate and hydrogen peroxide (final 4 mM each). Cleavage was stopped by adding one volume of 2X SDS sample buffer (125 mM Tris·HCl (pH 6.8), 2% SDS, 20% glycerol, 2% 2-mercaptoethanol) after incubation at 30 °C for 5 min. The cleavage products were analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). To map the cleavage site, protein ladders were generated by cleaving 32P-labeled RNAP using the Met-specific cleavage reagent, cyanogen bromide (CNBr) and the Cys-specific cleavage reagent, 2-nitro-5-thiocyanobenzoic-acid (NTCB) as described previously.38 To map the interaction of NusA NTD with the pause hairpin (Figs 6a–d), 32P-labeled rPTCs were prepared by two steps. EC27 (2 nt before pause) was first reconstituted from DNA scaffold, RNA27, and RNAP.47 The EC27 was then elongated to the pause in the presence of [α-32P]CTP (rPTC).
RNase T1 Digestion
rPTCs (15 nM) containing 32P-labeled RNA and NusA or NusA fragments (2 μM) were incubated at 25 °C for 30 min in transcription buffer. E. coli tRNA (final 250 ng/μL) was added prior to the cleavage, which was initiated by addition of RNase T1 (final 50 units/mL). The reactions were terminated by addition of equal volumes of 2X loading dye during the time course.
Crosslinking and mapping the crosslinking site on NusA
ECs containing the photoactive nucleotide analog 4-thio-uridine (4-thioU) at various positions on RNA were prepared by stepwise transcription, where ECs were immobilized through His6-tagged RNAP on Ni2+-NTA beads as described previously.68; 69 4-thioU and adjacent [α-32P]-CMP were incorporated into RNA during elongation from A29 to C31 for EC45(−16), 48(−19), 52(−23), 63(−34), and 69(−40) or during elongation from U52 to C57 for PTC(−16,17,18) (Supplementary Figs. 4a and b). Crosslinking between RNA and NusA and mapping the crosslinking site on the full-length NusA were performed as previously described.39 ECs containing 4-thioU were incubated with NusA or the NTD (2 μM) at 25 °C for 10 min and irradiated on ice for 10 min with a UV-transilluminator LM-26E (365 nm max; UVP, Upland, CA). Samples were then treated with RNaseA (5 U/ml) at 25 °C for 5–10 min, mixed with an equal volume of SDS sample buffer, heated at 65 °C for 10 min, and analyzed by SDS-PAGE. To map the crosslinking site on the full-length NusA, crosslinked NusA was eluted from gel pieces and subjected to CNBr or NTCB cleavage. The cleavage pattern of the crosslinked NusA was compared to that of N-terminal 32P-HMKNusA by SDS-PAGE.
Molecular modeling
The model of E. coli NusA (Fig. 1) was created using the program MODELLER70 (http://www.salilab.org/modeller) by fitting the sequence of E. coli NusA to the crystal structure of T. maratima NusA (PDB ID 1l2f).24 The structure of the his pause hairpin (Fig. 5c) was modeled from the NMR structure of the human PGY/MDR1 mRNA (PDB ID 2gvo)53 using model 1 from the deposited coordinates, superimposing the 3′ O on the 3′ O of nascent RNA −11 of the post-translocated T. thermophilus EC (PDB ID 2o5i)51 (corresponding to nascent RNA −12 of the pretranslocated his PTC),52 and the rotating the hairpin about the 3′ O to minimize clash the β′ clamp and β flap domains while minimizing the distance between the 3′ arm of the hairpin and the path of exiting RNA in the T. thermophilus EC. The position of box RNA relative to KH2 (Fig. 8b) was determined by superimposing the E. coli NusA model the crystal structure of an M. tuberculosis NusA S1-KH1-KH2 fragment complexed with an 11mer oligoribonucleotide whose sequence corresponds to the spacer between the boxA and boxC elements of the boxBAC rrn element in the M. tuberculosis rrn transcript (PDB ID 2asb).46 All structural pictures (Figs. 1, 5, and 9) were created using PyMOL (DeLano Scientific LLC, Palo Alto; http://www.pymol.org).
Supplementary Material
Acknowledgments
We thank members of the Landick lab for many helpful discussions during the course of the work and for comments on the manuscript, Nitin Goel for technical assistance in construction of mutant NusA proteins. This work was supported by grants from the NIGMS to D.G.V. (GM074252) and R.L. (GM038660).
Abbreviations used
- RNAP
RNA polymerase
- EC
elongation complex
- NTD
N-terminal domain
- CTD
C-terminal domain
- PTC
paused transcription complex
- CNBr
cyanogen bromide
- NTCB
2-nitro-5-thiocyanoobenzoic acid
- HMK
heart muscle kinase
- EDTA
ethylenediaminetetraacetic acid
- BABE
(S)-2-[4-(2-bromoacetamido)benzyl]ethylenediaminetetraacetic acid
- FeBABE
iron(III) complex of BABE
References
- 1.Friedman DI. A bacterial mutant affecting lambda development. In: Hershey AD, editor. The bacteriophage lambda. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1971. pp. 733–738. [Google Scholar]
- 2.Farnham PJ, Greenblatt J, Platt T. Effects of NusA protein on transcription termination in the tryptophan operon of Escherichia coli. Cell. 1982;29:945–51. doi: 10.1016/0092-8674(82)90457-3. [DOI] [PubMed] [Google Scholar]
- 3.Fisher R, Yanofsky C. A complementary DNA oligomer releases a transcription pause complex. J Biol Chem. 1983;258:9208–12. [PubMed] [Google Scholar]
- 4.Landick R, Yanofsky C. Stability of an RNA secondary structure affects in vitro transcription pausing in the trp operon leader region. J Biol Chem. 1984;259:11550–5. [PubMed] [Google Scholar]
- 5.Sigmund CD, Morgan EA. Nus A protein affects transcriptional pausing and termination in vitro by binding to different sites on the transcription complex. Biochemistry. 1988;27:5622–7. doi: 10.1021/bi00415a034. [DOI] [PubMed] [Google Scholar]
- 6.Chan CL, Landick R. The Salmonella typhimurium his operon leader region contains an RNA hairpin-dependent transcription pause site. Mechanistic implications of the effect on pausing of altered RNA hairpins. J Biol Chem. 1989;264:20796–804. [PubMed] [Google Scholar]
- 7.Greenblatt J, McLimont M, Hanly S. Termination of transcription by nusA gene protein of Escherichia coli. Nature. 1981;292:215–20. doi: 10.1038/292215a0. [DOI] [PubMed] [Google Scholar]
- 8.Lau LF, Roberts JW, Wu R. RNA polymerase pausing and transcript release at the lambda tR1 terminator in vitro. J Biol Chem. 1983;258:9391–7. [PubMed] [Google Scholar]
- 9.Schmidt MC, Chamberlin MJ. Binding of rho factor to Escherichia coli RNA polymerase mediated by nusA protein. J Biol Chem. 1984;259:15000–2. [PubMed] [Google Scholar]
- 10.Chen CY, Richardson JP. Sequence elements essential for rho-dependent transcription termination at lambda tR1. J Biol Chem. 1987;262:11292–9. [PubMed] [Google Scholar]
- 11.Kainz M, Gourse RL. The C-terminal domain of the alpha subunit of Escherichia coli RNA polymerase is required for efficient rho-dependent transcription termination. J Mol Biol. 1998;284:1379–90. doi: 10.1006/jmbi.1998.2272. [DOI] [PubMed] [Google Scholar]
- 12.Carlomagno MS, Nappo A. NusA modulates intragenic termination by different pathways. Gene. 2003;308:115–28. doi: 10.1016/s0378-1119(03)00472-4. [DOI] [PubMed] [Google Scholar]
- 13.Cardinale CJ, Washburn RS, Tadigotla VR, Brown LM, Gottesman ME, Nudler E. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–8. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen SE, Godoy VG, Walker GC. Transcriptional modulator NusA interacts with translesion DNA polymerases in Escherichia coli. J Bacteriol. 2009;191:665–72. doi: 10.1128/JB.00941-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen SE, Walker GC. The transcription elongation factor NusA is required for stress-induced mutagenesis in Escherichia coli. Curr Biol. 2010;20:80–5. doi: 10.1016/j.cub.2009.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward DF, Gottesman ME. The nus mutations affect transcription termination in Escherichia coli. Nature. 1981;292:212–5. doi: 10.1038/292212a0. [DOI] [PubMed] [Google Scholar]
- 17.Das A, Wolska K. Transcription antitermination in vitro by lambda N gene product: requirement for a phage nut site and the products of host nusA, nusB, and nusE genes. Cell. 1984;38:165–73. doi: 10.1016/0092-8674(84)90537-3. [DOI] [PubMed] [Google Scholar]
- 18.Grayhack EJ, Yang XJ, Lau LF, Roberts JW. Phage lambda gene Q antiterminator recognizes RNA polymerase near the promoter and accelerates it through a pause site. Cell. 1985;42:259–69. doi: 10.1016/s0092-8674(85)80121-5. [DOI] [PubMed] [Google Scholar]
- 19.Horwitz RJ, Li J, Greenblatt J. An elongation control particle containing the N gene transcriptional antitermination protein of bacteriophage lambda. Cell. 1987;51:631–41. doi: 10.1016/0092-8674(87)90132-2. [DOI] [PubMed] [Google Scholar]
- 20.Squires CL, Greenblatt J, Li J, Condon C. Ribosomal RNA antitermination in vitro: requirement for Nus factors and one or more unidentified cellular components. Proc Natl Acad Sci U S A. 1993;90:970–4. doi: 10.1073/pnas.90.3.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mooney RA, Davis SE, Peters JM, Rowland JL, Ansari AZ, Landick R. Regulator trafficking on bacterial transcription units in vivo. Mol Cell. 2009;33:97–108. doi: 10.1016/j.molcel.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Worbs M, Bourenkov GP, Bartunik HD, Huber R, Wahl MC. An extended RNA binding surface through arrayed S1 and KH domains in transcription factor NusA. Mol Cell. 2001;7:1177–89. doi: 10.1016/s1097-2765(01)00262-3. [DOI] [PubMed] [Google Scholar]
- 23.Gopal B, Haire LF, Gamblin SJ, Dodson EJ, Lane AN, Papavinasasundaram KG, Colston MJ, Dodson G. Crystal structure of the transcription elongation/anti-termination factor NusA from Mycobacterium tuberculosis at 1.7 A resolution. J Mol Biol. 2001;314:1087–95. doi: 10.1006/jmbi.2000.5144. [DOI] [PubMed] [Google Scholar]
- 24.Shin DH, Nguyen HH, Jancarik J, Yokota H, Kim R, Kim SH. Crystal structure of NusA from Thermotoga maritima and functional implication of the N-terminal domain. Biochemistry. 2003;42:13429–37. doi: 10.1021/bi035118h. [DOI] [PubMed] [Google Scholar]
- 25.Mah TF, Kuznedelov K, Mushegian A, Severinov K, Greenblatt J. The alpha subunit of E. coli RNA polymerase activates RNA binding by NusA. Genes Dev. 2000;14:2664–75. doi: 10.1101/gad.822900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mah TF, Li J, Davidson AR, Greenblatt J. Functional importance of regions in Escherichia coli elongation factor NusA that interact with RNA polymerase, the bacteriophage lambda N protein and RNA. Mol Microbiol. 1999;34:523–37. doi: 10.1046/j.1365-2958.1999.01618.x. [DOI] [PubMed] [Google Scholar]
- 27.Traviglia SL, Datwyler SA, Yan D, Ishihama A, Meares CF. Targeted protein footprinting: where different transcription factors bind to RNA polymerase. Biochemistry. 1999;38:15774–8. doi: 10.1021/bi9917232. [DOI] [PubMed] [Google Scholar]
- 28.Eisenmann A, Schwarz S, Prasch S, Schweimer K, Rosch P. The E. coli NusA carboxy-terminal domains are structurally similar and show specific RNAP- and lambdaN interaction. Protein Sci. 2005;14:2018–29. doi: 10.1110/ps.051372205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schauer AT, Carver DL, Bigelow B, Baron LS, Friedman DI. lambda N antitermination system: functional analysis of phage interactions with the host NusA protein. J Mol Biol. 1987;194:679–90. doi: 10.1016/0022-2836(87)90245-2. [DOI] [PubMed] [Google Scholar]
- 30.Mogridge J, Mah TF, Greenblatt J. A protein-RNA interaction network facilitates the template-independent cooperative assembly on RNA polymerase of a stable antitermination complex containing the lambda N protein. Genes Dev. 1995;9:2831–45. doi: 10.1101/gad.9.22.2831. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Mah TF, Greenblatt J, Friedman DI. Evidence that the KH RNA-binding domains influence the action of the E. coli NusA protein. J Mol Biol. 2002;318:1175–88. doi: 10.1016/s0022-2836(02)00238-3. [DOI] [PubMed] [Google Scholar]
- 32.Shankar S, Hatoum A, Roberts JW. A transcription antiterminator constructs a NusA-dependent shield to the emerging transcript. Mol Cell. 2007;27:914–27. doi: 10.1016/j.molcel.2007.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Theissen G, Pardon B, Wagner R. A quantitative assessment for transcriptional pausing of DNA-dependent RNA polymerases in vitro. Anal Biochem. 1990;189:254–61. doi: 10.1016/0003-2697(90)90117-r. [DOI] [PubMed] [Google Scholar]
- 34.Linn T, Greenblatt J. The NusA and NusG proteins of Escherichia coli increase the in vitro readthrough frequency of a transcriptional attenuator preceding the gene for the beta subunit of RNA polymerase. J Biol Chem. 1992;267:1449–54. [PubMed] [Google Scholar]
- 35.Gusarov I, Nudler E. Control of intrinsic transcription termination by N and NusA: the basic mechanisms. Cell. 2001;107:437–49. doi: 10.1016/s0092-8674(01)00582-7. [DOI] [PubMed] [Google Scholar]
- 36.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc Natl Acad Sci U S A. 2000;97:7090–5. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chan CL, Landick R. Dissection of the his leader pause site by base substitution reveals a multipartite signal that includes a pause RNA hairpin. J Mol Biol. 1993;233:25–42. doi: 10.1006/jmbi.1993.1482. [DOI] [PubMed] [Google Scholar]
- 38.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–3. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 39.Toulokhonov I, Zhang J, Palangat M, Landick R. A central role of the RNA polymerase trigger loop in active-site rearrangement during transcriptional pausing. Mol Cell. 2007;27:406–19. doi: 10.1016/j.molcel.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 40.Landick R, Yanofsky C. Isolation and structural analysis of the Escherichia coli trp leader paused transcription complex. J Mol Biol. 1987;196:363–77. doi: 10.1016/0022-2836(87)90697-8. [DOI] [PubMed] [Google Scholar]
- 41.Borukhov S, Lee J, Laptenko O. Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol Microbiol. 2005;55:1315–24. doi: 10.1111/j.1365-2958.2004.04481.x. [DOI] [PubMed] [Google Scholar]
- 42.Gill SC, Weitzel SE, von Hippel PH. Escherichia coli sigma 70 and NusA proteins. I. Binding interactions with core RNA polymerase in solution and within the transcription complex. J Mol Biol. 1991;220:307–24. doi: 10.1016/0022-2836(91)90015-x. [DOI] [PubMed] [Google Scholar]
- 43.Yang X, Molimau S, Doherty GP, Johnston EB, Marles-Wright J, Rothnagel R, Hankamer B, Lewis RJ, Lewis PJ. The structure of bacterial RNA polymerase in complex with the essential transcription elongation factor NusA. EMBO Rep. 2009;10:997–1002. doi: 10.1038/embor.2009.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vassylyev DG. Elongation by RNA polymerase: a race through roadblocks. Curr Opin Struct Biol. 2009;19:691–700. doi: 10.1016/j.sbi.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibson TJ, Thompson JD, Heringa J. The KH domain occurs in a diverse set of RNA-binding proteins that include the antiterminator NusA and is probably involved in binding to nucleic acid. FEBS Lett. 1993;324:361–6. doi: 10.1016/0014-5793(93)80152-k. [DOI] [PubMed] [Google Scholar]
- 46.Beuth B, Pennell S, Arnvig KB, Martin SR, Taylor IA. Structure of a Mycobacterium tuberculosis NusA-RNA complex. EMBO J. 2005;24:3576–87. doi: 10.1038/sj.emboj.7600829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kyzer S, Ha KS, Landick R, Palangat M. Direct versus limited-step reconstitution reveals key features of an RNA hairpin-stabilized paused transcription complex. J Biol Chem. 2007;282:19020–8. doi: 10.1074/jbc.M701483200. [DOI] [PubMed] [Google Scholar]
- 48.Reynolds R, Bermudez-Cruz RM, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA polymerase. I. Analysis of 13 rho-independent terminators. J Mol Biol. 1992;224:31–51. doi: 10.1016/0022-2836(92)90574-4. [DOI] [PubMed] [Google Scholar]
- 49.Adinolfi S, Bagni C, Castiglione Morelli MA, Fraternali F, Musco G, Pastore A. Novel RNA-binding motif: the KH module. Biopolymers. 1999;51:153–64. doi: 10.1002/(SICI)1097-0282(1999)51:2<153::AID-BIP5>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 50.Greiner DP, Miyake R, Moran JK, Jones AD, Negishi T, Ishihama A, Meares CF. Synthesis of the protein cutting reagent iron (S)-1-(p-bromoacetamidobenzyl)ethylenediaminetetraacetate and conjugation to cysteine side chains. Bioconjug Chem. 1997;8:44–8. doi: 10.1021/bc9600731. [DOI] [PubMed] [Google Scholar]
- 51.Vassylyev D, Vassylyeva M, Perederina A, Tahirov T, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 52.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–36. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 53.Joli F, Bouchemal N, Laigle A, Hartmann B, Hantz E. Solution structure of a purine rich hexaloop hairpin belonging to PGY/MDR1 mRNA and targeted by antisense oligonucleotides. Nucleic Acids Res. 2006;34:5740–51. doi: 10.1093/nar/gkl617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schmidt MC, Chamberlin MJ. nusA protein of Escherichia coli is an efficient transcription termination factor for certain terminator sites. J Mol Biol. 1987;195:809–18. doi: 10.1016/0022-2836(87)90486-4. [DOI] [PubMed] [Google Scholar]
- 55.Sigmund CD, Morgan EA. Effects of Escherichia coli Nus A protein on transcription termination in vitro are not increased or decreased by DNA sequences sufficient for antitermination in vivo. Biochemistry. 1988;27:5628–35. doi: 10.1021/bi00415a035. [DOI] [PubMed] [Google Scholar]
- 56.Yakhnin AV, Babitzke P. NusA-stimulated RNA polymerase pausing and termination participates in the Bacillus subtilis trp operon attenuation mechanism invitro. Proc Natl Acad Sci U S A. 2002;99:11067–72. doi: 10.1073/pnas.162373299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pan T, Artsimovitch I, Fang XW, Landick R, Sosnick TR. Folding of a large ribozyme during transcription and the effect of the elongation factor NusA. Proc Natl Acad Sci U S A. 1999;96:9545–50. doi: 10.1073/pnas.96.17.9545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Whalen W, Ghosh B, Das A. NusA protein is necessary and sufficient in vitro for phage lambda N gene product to suppress a rho-independent terminator placed downstream of nutL. Proc Natl Acad Sci U S A. 1988;85:2494–8. doi: 10.1073/pnas.85.8.2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Van Gilst MR, von Hippel PH. Assembly of the N-dependent antitermination complex of phage lambda: NusA and RNA bind independently to different unfolded domains of the N protein. J Mol Biol. 1997;274:160–73. doi: 10.1006/jmbi.1997.1389. [DOI] [PubMed] [Google Scholar]
- 60.Greenblatt J, Li J. The nusA gene protein of Escherichia coli. Its identification and a demonstration that it interacts with the gene N transcription anti-termination protein of bacteriophage lambda. J Mol Biol. 1981;147:11–23. doi: 10.1016/0022-2836(81)90076-0. [DOI] [PubMed] [Google Scholar]
- 61.Larson MH, Greenleaf WJ, Landick R, Block SM. Applied force reveals mechanistic and energetic details of transcription termination. Cell. 2008;132:971–82. doi: 10.1016/j.cell.2008.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cheal SM, Ng M, Barrios B, Miao Z, Kalani AK, Meares CF. Mapping protein-protein interactions by localized oxidation: consequences of the reach of hydroxyl radical. Biochemistry. 2009;48:4577–86. doi: 10.1021/bi900273j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrecka J, Lewis R, Bruckner F, Lehmann E, Cramer P, Michaelis J. Single-molecule tracking of mRNA exiting from RNA polymerase II. Proc Natl Acad Sci U S A. 2008;105:135–40. doi: 10.1073/pnas.0703815105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ito K, Nakamura Y. Localization of nusA-suppressing amino acid substitutions in the conserved regions of the beta′ subunit of Escherichia coli RNA polymerase. Mol Gen Genet. 1996;251:699–706. doi: 10.1007/BF02174119. [DOI] [PubMed] [Google Scholar]
- 65.Artsimovitch I, Svetlov V, Murakami K, Landick R. Co-overexpression of E. coli RNA polymerase subunits allows isolation and analysis of mutant enzymes lacking lineage-specific sequence insertions. J Biol Chem. 2003;278:12344–12355. doi: 10.1074/jbc.M211214200. [DOI] [PubMed] [Google Scholar]
- 66.Reynolds R, Chamberlin MJ. Parameters affecting transcription termination by Escherichia coli RNA. II. Construction and analysis of hybrid terminators. J Mol Biol. 1992;224:53–63. doi: 10.1016/0022-2836(92)90575-5. [DOI] [PubMed] [Google Scholar]
- 67.von Hippel P. An integrated model of the transcription complex in elongation, termination, and editting. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 68.Wang D, Landick R. Nuclease cleavage of the upstream half of the nontemplate strand DNA in an E. coli transcripiton elongation complex causes upstream translocation and transcriptional arrest. J Biol Chem. 1997;272:5989–5994. doi: 10.1074/jbc.272.9.5989. [DOI] [PubMed] [Google Scholar]
- 69.Wang D, Meier T, Chan C, Feng G, Lee D, Landick R. Discontinuous movements of DNA and RNA in E. coli RNA polymerase accompany formation of a paused transcription complex. Cell. 1995;81:341–350. doi: 10.1016/0092-8674(95)90387-9. [DOI] [PubMed] [Google Scholar]
- 70.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 71.Studier FW, Rosenberg AH, Dunn JJ, Dubendorff JW. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 72.Ederth J, Artsimovitch I, Isaksson LA, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem. 2002;277:37456–63. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 73.Ederth J, Artsimovitch I, Isaksson L, Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J Biol Chem. 2002;277:37456–37463. doi: 10.1074/jbc.M207038200. [DOI] [PubMed] [Google Scholar]
- 74.Opalka N, Mooney RA, Richter C, Severinov K, Landick R, Darst SA. Direct localization of a beta-subunit domain on the three-dimensional structure of Escherichia coli RNA polymerase. Proc Natl Acad Sci U S A. 2000;97:617–22. doi: 10.1073/pnas.97.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: sequence analysis. J Mol Biol. 2010;395:671–85. doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Grachev MA, Lukhtanov EA, Mustaev AA, Zaychikov E, Abdukayumov M, Rabinov I, Richter V, Skoblov Y, Christyakov P. Studies on the functional topography of Escherichia coli RNA polymerase: A method for localization of the sites of affinity labeling. Eur J Biochem. 1989;180:577–585. doi: 10.1111/j.1432-1033.1989.tb14684.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.