Abstract
Protein tyrosine phosphatase, PTPL1, (also known as PTPN13, FAP-1, PTP-BAS, PTP1E) is a non-receptor type PTP and, at 270 kDa, is the largest phosphatase within this group. In addition to the well-conserved PTP domain, PTPL1 contains at least 7 putative macromolecular interaction domains. This structural complexity indicates that PTPL1 may modulate diverse cellular functions, perhaps exerting both positive and negative effects. In accordance with this idea, while certain studies suggest that PTPL1 can act as a tumor-promoting gene other experimental studies have suggested that PTPL1 may function as a tumor suppressor. The role of PTPL1 in the cancer cell is therefore likely to be both complex and context dependent with possible roles including the modulation of growth, stress-response, and cytoskeletal remodeling pathways. Understanding the nature of molecular complexes containing PTPL1, its interaction partners, substrates, regulation and subcellular localization are key to unraveling the complex personality of this protein phosphatase.
Keywords: PTPL1, FAP1, PTPN13, Cancer, Tumor suppressor, Tumor promoter
1 Introduction
The disruption of normal cell signaling is central to cancer development. Cancer cells depend on stress-response pathways for survival while pathways that control cell motility may be critical for metastasis. Many such signaling pathways rely on the phosphorylation of proteins on tyrosine residues. The reversal of such modifications by the activities of protein tyrosine phosphatases (PTPs) is, therefore, key to the proper regulation of these pathways (reviewed in [1]). Of the known PTPs, only a few, such as PTP1B [2], its close homolog TC-PTP [2], and SHP2 [3] have been studied extensively and found to have roles in cancer and other disease processes (reviewed in [4, 5]). Links to cancer initiation and progression remain unclear for other PTPs, such as PTPL1 (a.k.a. PTPN13, FAP-1, PTP-BAS, PTP1E). There is some evidence suggesting that PTPL1 may act as a tumor promoting gene, while other studies point towards a role for PTPL1 as a tumor suppressor. This dichotomy suggests that PTPL1 may act differently depending upon the disease context. In the present review, we propose that PTPL1 may affect cancer by modulating mechanisms of known oncogenic importance: stress-response, cell-growth and motility pathways.
2 Overview of PTPL1
PTPL1 was identified almost simultaneously by three independent groups, each of which gave it a different name: PTP1E, PTP-BAS and PTPL1, respectively [6-8]. Within a year, a yeast-two-hybrid screen revealed that PTPL1 interacts with the Fas Receptor (CD95/APO-1), a member of the tumor necrosis factor-receptor (TNFR) superfamily, and PTPL1 was given an additional name: Fas-Associated Phosphatase-1 (FAP-1) [9].
2.1 Structure
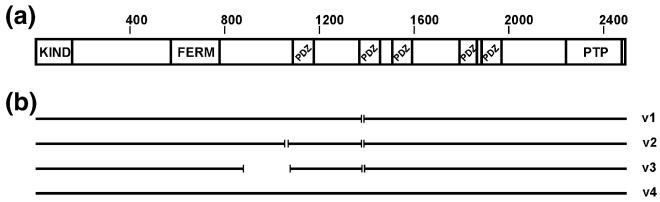
PTPN13 (Genebank Gene ID: 5783) maps to the human chromosomal locus 4q21.3 and encodes a non-transmembrane PTP with a calculated molecular mass of about 270 kDa (for a comprehensive review of nomenclature see ref [10]). We will refer to the protein in this manuscript as PTPL1. Human, murine and bovine PTPL1 have high levels of amino acid sequence homology, while the Xenopus protein is similar, but more distantly related to the mammalian proteins (Fig. 1). The protein structure comprises of an amino-terminal band 4.1/ezrin/radixin/moesin (FERM) domain, which is found in many cytoskeleton-associated proteins [11]. The central portion of PTPL1 contains five PSD-95/Discs-large/ZO-1 (PDZ) domains while the carboxy-terminal catalytic domain contains the well-conserved PTP active site motif (Fig. 2(a)). At the extreme amino-terminus of PTPL1 is a putative kinase non-catalytic C-lobe domain (KIND). The latter domain has been identified in silico by sequence homology and its possible function has not yet been studied experimentally [12].
Fig. 1.
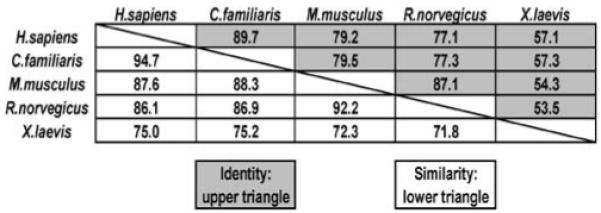
Homology between different PTPL1 homologues. Protein sequences from H. sapiens (NP_006255.1 v4), C. familiaris (XP_860003.1), M. musculus (NP_035334.1), R. norvegicus (XP_213997.4) and X. laevis (AAR97566) were entered into MatGAT (Matrix Global Alignment Tool) to calculate sequence similarity and identity (http://bitincka.com/ledion/matgat/). Values are percent identity (upper triangle) and percent similarity (lower triangle)
Fig. 2.
Schematic representation of PTPL1 structure and identified splice variants. (a) FERM, PDZ and PTP domains of PTPL1 were identified by using PFAM (http://www.sanger.ac.uk/Software/Pfam/) where the sequence from the longest variant of PTPL1, variant 4, was used. The KIND domains is not recognized by PFAM, therefore its location is arbitrary. (b) Different variants of PTPL1 are depicted with the straight lines where each gap in the sequence represents a splice site
The FERM domain of PTPL1 binds to phosphatidylinositol 4,5-biphosphate leading to the enrichment of PTPL1 at a juxtamembrane localization [13]. However, the PTPL1 protein is also detected throughout the cytoplasm [13]. In HeLa cells, PTPL1 localizes to the centrosomes during metaphase and to the midbody during cytokinesis [14]. Even though the PTP domains of protein tyrosine phosphatases share a high degree of sequence similarity [15], the crystal structure of the PTP domain of PTPL1 reveals a secondary phosphotyrosine binding pocket next to the active site, which is similar to that found in the structure of PTP1B [16]. Consistent with a functional role for this pocket in substrate recognition, mono-phosphorylated substrates are dephosphorylated by PTPL1 more slowly when compared to bi- or tri-phosphorylated substrates [16]. The five PDZ domains within PTPL1 are responsible for direct binding to interacting proteins, which could either directly recruit phosphatase substrates or provide a multi-protein scaffold for the indirect binding of substrate proteins.
2.2 Transcriptional and post-translational regulation
Specific mechanisms of PTPL1 transcriptional and post-translational regulation remain cryptic. PTPN13 lies in a head-to-head conformation with MAPK10/JNK3 and they share a 633 bp bi-directional promoter. This contains several putative transcription factor binding sites, including motifs for E2F, Sp1, GATA-1 and AP-1 [17]. An additional promoter sequence may also lie upstream within the MAPK10/JNK3 gene itself [18]. We have also shown that the PTPL1 promoter contains binding sites (GGAA) for the ets-family of transcription factors that are activated by the EWS-FLI1 fusion protein in the Ewing’s Sarcoma Family of Tumors [19].
Post-transcriptional processing of PTPL1 mRNA may lead to the production of four splice variants (Fig. 2(b)). Compared to the fourth variant, splice variants 1, 2 and 3 are missing nucleotides encoding five amino acids (VLFDK) within the second PDZ domain. The presence of these five amino acids eliminates protein binding to the second PDZ (PDZ2) domain of PTPL1 [20]. No study has yet evaluated the expression of these splice variants of PTPL1 in cancer cells nor have expression levels been correlated with either disease progression or metastasis. Additional regulation of PTPL1 may result from molecular interactions with lipids and/or proteins and subsequent changes in its intracellular localization.
Post-translational modification of PTPL1 occurs on serine/threonine and possibly tyrosine residues. Two candidate residues were recently identified following epidermal growth factor stimulation of HeLa cells and phosphoproteomic mass spectroscopy, T1336 and S1339 [21]. Mutation of T1336A led to a significant loss of phosphatase activity, while the S1339A mutation did not result in any significant change in activity. The kinase that phosphorylates these specific sites was not identified [21]. However, a search for protein kinase-A (PKA) substrates in Xenopus oocytes identified PTPL1 as a candidate. In vitro PKA phosphorylation of human PTPL1, immunoprecipitated from HEK293 cells, led to a 50% reduction of phosphatase activity [22]. However, the specific residues phosphorylated by PKA were not identified. These two publications provide preliminary evidence for post-translational regulation by phosphorylation. However, they differ with regard to the effects of phosphorylation, with mutation of T1336 to alanine leading to reduced activity of PTPL1 while phosphorylation by PKA also appeared to inactivate the phosphatase. It is of course possible that the inactivation of PTPL1 by PKA could be mediated via an as yet unmapped site which is distinct from T1336 and that the activity of PTPL1 could be modulated both positively and negatively by multiple signalling pathways.
A recent publication suggested that PTPL1 is phosphorylated on Y2224 within Motif 1 of the PTP domain, which is conserved in 80% of the PTPs [23]. Unfortunately, data was not presented to support this claim and additional investigations are clearly necessary to further understand the role of tyrosine phosphorylation in any post-translational regulatory mechanisms of PTPL1.
2.3 Formation of macromolecular complexes
Based on the presence of a number of well-characterized interaction motifs, PTPL1 has the potential to bind to both proteins and lipids. PTPL1 may therefore function, in part, as a docking protein. These intermolecular interactions may contribute to the regulation of PTPL1 phosphatase activity. The putative scaffolding function of PTPL1 may also lead to the formation of multi-protein complexes that could act to recruit phospho-tyrosine containing substrates to the catalytic domain of PTPL1.
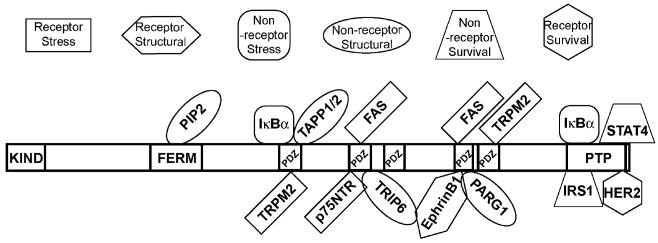
A complete understanding of various PTPL1 domains in mediating both protein-lipid and protein-protein interactions will be critical in resolving the functional role of PTPL1 in cancer (Fig. 3). The FERM domain, which is located near the amino-terminus of PTPL1, binds to phosphatidylinositol biphosphates (PIP2) and triphosphates (PIP3), which facilitate juxtamembrane targeting [13]. These associations are consistent with the behaviour of other FERM-domain-containing proteins that interact with the cell membrane by binding to PIP2 [24]. The FERM domain can localize PTPL1 to the inner-leaflet of the cell membrane where it may interact with these proteins. Additionally, the association of PTPL1 with PIP2 and/or TAPP1/2, supports the hypothesis that PTPL1 is not only localized to the cell membrane but could also alter its localization in response to different extracellular stimuli.
Fig. 3.
Schematic representation of PTPL1 interacting molecules and substrates. Each interacting molecule was assigned to a category respective of its main function in the cell as described in the text. The molecules can be of any group: a receptor molecule in stress-response pathways, a receptor molecule in cytoskeletal/motility pathways, a non-receptor molecule in stress-response pathways, a non-receptor molecule in cell cytoskeletal/motility pathway, a non-receptor molecule in cell survival pathways, or a receptor molecule in cell survival pathways. Each molecule is placed on PTPL1 to where they are described to bind as in the text
The first PTPL1 PDZ domain (PDZ1) binds a family of PIP2-binding adaptor proteins known as tandem-PH-domain-containing proteins 1 and 2 (TAPP1/2) [25]. Endogenous PTPL1 and TAPP1 co-localized predominantly in the cytoplasm, while an H2O2-stimulated increase of intracellular PIP2 levels led to higher levels of co-localization between endogenous TAPP1 and PTPL1 at the cell membrane [25]. In addition to the TAPP1/2 interactions, PDZ1 has been shown to bind the inhibitor of nuclear factor kappa-B alpha (IκBα) [26] as well as the transient receptor potential (TRP) superfamily member TRPM2 [27].
The second PDZ domain of PTPL1 (PDZ2) binds the TNFR superfamily member Fas [28]. The Serine-Leucine-Valine (SLV) tripeptide at the carboxy-terminus of Fas is sufficient for binding to PDZ2 of PTPL1 [28]. Cells pretreated with Fas antibodies, to activate Fas-mediated apoptosis, and then injected with the SLV tripeptide, which inhibits the binding of PTPL1 to Fas, showed significantly increased levels of apoptosis when compared to cells injected with a control peptide [29]. The consequences of Fas tyrosine phosphorylation will be discussed in Section 3. In addition to Fas, PDZ2 also binds the three carboxy-terminal residues of the neurotropin receptor p75NTR, which is also a member of the TNFR superfamily [30]. Finally, PDZ2 binds to thyroid hormone receptor-interacting protein 6 (TRIP6, a.k.a. zyxin related protein-1, ZRP1) [31].
While the third PDZ domain within PTPL1 (PDZ3) has not yet been associated with protein binding, the fourth PDZ domain of PTPL1 (PDZ4) has as many binding partners as the second PDZ domain and these include Fas, which also binds PDZ2 [28]. A novel protein PARG1 (PTPL1-associated RhoGAP1) was identified through a yeast-two-hybrid screen for PTPL1-interacting molecules [32]. And lastly, EphrinB1, a transmembrane ligand for B-class Ephrin receptor tyrosine kinases also binds PDZ4 [33]. PTPL1’s final PDZ domain (PDZ5) has thus far only been shown to bind to TRPM2, which also binds PDZ1 [27]. Although a number of interacting molecules have been described, few have been definitively identified as PTPL1 substrates.
2.4 Substrates
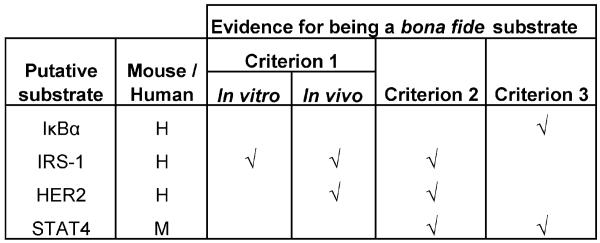
The highly transient nature of PTP-substrate complexes has made the identification of PTP substrates very challenging (Fig. 4). This process has become easier within the last decade by an approach for substrate identification called “substrate-trapping,” first described by Flint et al. [34]. Substrate trapping identification of putative substrates requires rigorous experimental criteria for confirmation, as proposed by Tiganis and Bennett [35], to avoid false positive results. Thus far, IκBα is the only PTPL1 binding partner [26] that is also a putative PTPL1 substrate, identified in vitro using a cys to ser trapping mutant [36]. A recent report suggests the insulin receptor substrate-1 (IRS-1) as a putative PTPL1 substrate [23]. A substrate-trapping mutant of PTPL1 precipitated phospho-IRS-1 in insulin-like growth factor stimulated cells that over-expressed exogenous IRS-1 [23]. In a similar experimental design, a substrate-trapping mutant of PTPL1 co-immunoprecipitated with HER2 in epidermal growth factor stimulated HEK293 cells that over-expressed exogenous HER2, thus making it a putative substrate [21]. STAT4 was identified as a putative substrate of PTP-BL, the mouse homolog of PTPL1 [37]. Additional work will be necessary in order to definitively prove that IκBα, IRS-1, HER2, and STAT4 are bona fide substrates of PTPL1.
Fig. 4.
Putative substrates of PTPL1. According to the guidelines suggested by Tiganis and Bennett [35] for the identification of bona fide PTP substrates, the evidence of a substrate has to be confirmed by three experimental criteria. Criterion 1 states that the presence of an enzyme-substrate complex has to be demonstrated both in vivo and in vitro. Criterion 2 states that the substrate tyrosine levels could be modulated by introducing substrate-trapping mutants of the PTP or altering endogenous PTP protein levels. Criterion 3 states that the phospho-substrate could be dephosphorylated in vitro by the PTP. The experimental evidence provided that fulfills any of these criteria for each putative PTPL1 substrate described in the text has been marked
2.5 Effects of gene targeting experiments for PTP-BL in mice
Two mouse models using homologous recombination for targeted deletion of PTP-BL (the murine PTPL1 homolog) have now been published. The first one is a partial ‘knock-out’ of PTP-BL, resulting in the expression of a truncated protein [38]. These PTP-BLΔP/ΔP mice express a protein containing the FERM and 5 PDZ domains, but which lacks the catalytic PTP domain. These mice develop normally but were found to have a mild deficiency in motor neuron repair [38] and a significant reduction in the growth of retinal glia cultures from lens-lesioned mice when compared to wild-type mice [39]. The second model created was a complete knock-out of PTP-BL. These PTP-BL deleted mice demonstrated increased STAT4 phosphorylation in CD4+-T cells upon IL-12 and T-cell receptor activation [37]. The PTP-BL deleted mice also demonstrated enhanced maturation of T-cells and increased immune modulated killing following the inoculation of bacteria into their lungs [37]. These studies suggest that while PTPL1 is not critical for normal murine development, this phosphatase may participate in very different aspects of cellular physiology. While the overall effects of PTPL1 deletion in ‘knock-out’ mice do not give significant clues to any role in cancer, a number of biochemical and epigenetic studies do indicate that PTPL1 may have a potential function in this disease.
3 Functions of PTPL1 and its role in cancer
Tumors often grow under conditions of stress, such as hypoxia and nutrient deprivation that would normally trigger apoptosis. However, cancer cells adapt to these conditions using a number of strategies to ensure survival. Protein tyrosine phosphorylation transduces signals for cellular stress-response pathways that modulate cell survival, therefore dephosphorylation of substrates by PTPL1 may regulate this endpoint. The dysregulation of these cell survival pathways leads to cancer and ultimately metastasis. An examination of PTPL1 interacting proteins and emerging substrates suggests that PTPL1 has multiple and potentially divergent roles in cancer (Figs. 3 and 4). The induction of PTPL1 by an oncogene and relative increase of PTPL1 levels in tumor tissues supports a role in tumor promotion. In contrast, epigenetic studies are more consistent with a role for PTPL1 as a tumor suppressor.
3.1 PTPL1 as a tumor promoter
While there is no direct evidence that PTPL1 is an oncogene per se, PTPL1 is implicated in oncogenesis via modulation of cell growth and stress-responses. PTPL1 is a direct transcriptional target of the Ewing’s Sarcoma Family of Tumors (ESFT) oncogene, EWS-FLI1. PTPL1 interactions with Fas may lead to direct modulation of stress-related pathways in favor of tumor cell survival. PTPL1 interaction or modulation of p75NTR, transient receptor potential M2 (TRPM2), and NFκB may also support tumor cell survival under stress conditions. In these scenarios, PTPL1 may not necessarily be an oncogene, but could provide a significant advantage for tumor growth.
ESFT contains a pathognomonic chromosomal translocation (t(11;22)) and expresses the EWS-FLI1 fusion protein [40]. PTPL1 is highly expressed in ESFT cells both in comparison to other childhood tumors and in EWS-FLI1 expression models [41-43]. Our findings in ESFT suggest that PTPL1 is a direct transcriptional target of EWS-FLI1 and both promotes cell growth and oncogenesis [19]. Reduction of PTPL1 protein levels, using an antisense strategy, leads to highly significant reductions in both anchorage-dependent and anchorage-independent cell growth [19]. PTPL1-depleted ESFT cells also displayed an increased sensitivity to etoposide induced apoptosis when compared to control cells, although there was no significant difference in basal levels of apoptosis [19]. Our results correlated well with a pilot study using an ESFT tissue microarray, which observed a trend towards increased PTPL1 staining in metastatic ESFT samples compared to localized disease [19]. Ongoing studies by our group are focused on the identification of bona fide PTPL1 substrates and PTPL1-modulated pathways of motility and oncogenesis.
Activation of Fas (CD95/APO-1) by engagement of the tumor necrosis factor (TNF) Superfamily trimeric Fas ligand (FasL/CD95L) is a well recognized homeostatic system for cell growth regulation, particularly in the immune system [44]. Activation of Fas by FasL leads to caspase-8 mediated apoptosis. Many types of cancer cells express both Fas and FasL, yet, these cells do not die by apoptosis. This is because these cells have developed strategies to avoid cell death, thereby maintaining their malignant growth. PTPL1 expression was reported to inhibit Fas-mediated apoptosis in both pancreatic adenocarcinoma [45, 46] and melanoma [47] cell lines thus acting as a survival mechanism in tumor cells expressing both Fas and FasL. Additional tumor models of ovarian cancer [48], colon cancer [49], head/neck cancer [50], hepatocellular carcinoma [51], and hepatoblastoma [52] also showed a correlation between tumor cell survival in the presence of both Fas/FasL and expression of PTPL1. While some functional and correlative evidence exists for the regulation of Fas by PTPL1, at least one potential mechanism is explored below.
Fas, under conditions of stress, could initiate cell death to prevent the survival of a damaged cell. The modulation of Fas-mediated cell death by PTPL1 may derive from its ability to regulate the cellular localization of Fas. The maintenance of Fas in the plasma membrane may be regulated by tyrosine phosphorylation and could thus be reversed by PTPL1 expression. PTPL1 could enhance tumor growth by blocking death signaling. Experiments in melanoma and human embryonic lung fibroblasts demonstrate that expression of wild-type PTPL1 reduced Fas expression at the cell surface, while expression of either a PTP-domain-deleted PTPL1 or a PDZ-domain-deleted PTPL1, increased Fas levels at the cell surface [47]. Following either hyperosmotic stress or Fas ligand binding, the epidermal growth factor receptor (EGFR) tyrosine kinase was shown to phosphorylate cytosolic Fas, which is a prerequisite for Fas targeting to the membrane [53]. Mutation of a C-terminal tyrosine residue of Fas, Y275 (residue 291 on full-length Fas), resulted in decreased PTPL1 binding and increased Fas protein on the surface [47]. This altered tyrosine residue also results in loss of membrane localization when mutated together with a second C-terminal tyrosine residue (Y232 on full-length Fas) [53]. Dephosphorylation of Fas by PTPL1 might impede its membrane targeting, which therefore could result in increased cancer cell survival.
Additional evidence that PTPL1 plays a role in tumor cell survival following cellular stress is provided by studies of its interactions with p75NTR, TRPM2, and IκBα. The mechanisms of p75NTR function in the modulation of cell death signaling are somewhat confused and require resolution [54, 55]. Tyrosine phosphorylation of p75NTR induces its ubiquitination [56] and its potential proteosomal degradation, while dephosphorylation of p75NTR by PTPL1 may stabilize the protein leading to an increase in its intracellular level, thus promoting cell survival. TRPM2, a member of the transient receptor potential (TRP) superfamily that binds to PDZ1 and/or PDZ5 within PTPL1, is phosphorylated in response to either oxidative stress or TNFα treatment [27]. Phosphorylation then results in an influx of extracellular Ca+2 and leads to apoptosis [27]. Cells transfected with PTPL1 display reduced TRPM2 phosphorylation and a corresponding reduction in Ca+2 influx leading to cell survival [27]. IκBα, which has also been identified as a PTPL1 substrate, is a negative regulator of the NFκB stress-response pathway. While the prevalent mechanism of regulation of IκBα is through Ser32/36 phosphorylation, tyrosine phosphorylation of IκBα has also been observed on Y42 [57] and Y305 [58]. PTPL1 may modulate NFκB signaling by dephosphorylating IκBα. Interestingly, Y42 phosphorylation of IκBα is a stimulus-dependent phenomenon that only occurs after hypoxia and to some extent, reoxygenation, and results in activation of NFκB [59]. On the other hand, Y305 phosphorylation results in the inhibition of NFκB signaling [58], therefore dephosphorylation of this residue activates NFκB.
In addition to the functional data described above, many investigators have shown relatively higher levels of PTPL1 expression in multiple carcinomas compared to the normal adjacent tissue as detected by immunohistochemistry [60]. Our data in ESFT patient samples also suggest that PTPL1 could be important in promoting tumor growth. The presence of high levels of PTPL1 in tumor tissues would argue against PTPL1 as a tumor suppressor, however, the highly expressed PTPL1 could be a mutant protein. Mutational analysis is necessary to determine if there is a correlation between immunohistochemical protein levels and mutations. Overall, additional data is required to clarify the conditions when PTPL1 functions as a tumor promoter.
3.2 PTPL1 in metastasis
PTPL1 binds to a number of lipids and proteins that can regulate cell shape and motility and might therefore play an important role in cancer metastasis. These include PIP2, TAPP1/2, EphrinB1 and PARG1. The FERM domain of PTPL1 binds PIP2 as an intermediate step in plasma membrane localization [24]. TAPP1/2 are cytoplasmic adaptor proteins that can shuttle to the cell membrane via PIP2 binding and remodel the actin cytoskeleton [61]. EphrinB1 is a member of a class of transmembrane proteins that modulate many physiological functions including, but not limited to, cell motility [62]. EphrinB1 contains a PDZ binding motif and binds to PTPL1 [33]. Furthermore, EphrinB1 was dephosphorylated in vitro by PTP-BL [63]. Heterologous co-expression of EphrinB1 and a cys to ser PTP-BL substrate trapping mutant in HeLa cells also led to increased EphrinB1 tyrosine phosphorylation [63]. While no specific motility studies were performed, the effect of PTPL1 upon Ephrin B1 might contribute to cell metastases through altered cell motility. Finally, PARG1, the PTPL1 Associated RhoGAP1, binds to PTPL1 through a PDZ domain and regulates cytoskeletal remodeling/motility leading to altered cell morphology [64]. Thus PIP2, TAPP1/2, EphrinB1 and PARG1 together are involved in the maintenance of the cytoskeleton, which is critical for cell motility. The interaction of PTPL1 with proteins important to cell motility may explain why there is an increase in PTPL1 in metastatic Ewing’s Sarcoma [19] as well as in some other solid tumors [60]. Even though these PTPL1 binding proteins have not been described as substrates, it is plausible to argue that PTPL1 can act as a scaffold in the cell, providing the optimal environment for protein interactions.
PTPL1 acting upon the cytoskeleton may reduce metastatic potential. Thyroid receptor interacting protein 6 (TRIP6, a.k.a. Zyxin-related protein 1, ZRP-1) modulates lysophosphatidic acid (LPA)-induced cell migration. In ovarian carcinoma cells, the endogenous expression of PTPL1 reduces phosphorylation of TRIP6, leading to decreased cell motility [65]. PTPL1 levels were directly reduced in ovarian carcinoma using shRNA and these PTPL1 reduced cells demonstrated increased motility [21]. A functional interaction with TRIP6 is supported by the previous detection of binding to PTPL1 in a yeast 2-hybrid assay [31]. The reduction of cell motility may be more indicative of a tumor suppressor rather than a tumor promoter.
3.3 PTPL1 as a tumor suppressor
Tumor suppressor genes encode proteins that either prevent or reverse oncogenic transformation. PTPL1 may either directly or indirectly interact with key proteins or dephosphorylate key substrates in order to reduce tumor growth. While many of the experiments, discussed previously, show that PTPL1 can support cell survival by disrupting Fas signaling, ectopic expression of PTPL1 leads to apoptotic cell death in colon adenocarcinoma [66]. Certain cell lines with high PTPL1 levels were quite sensitive to Fas induced apoptosis, however, overall PTPL1 levels did not correlate with cell death [67]. Additional evidence for PTPL1 as a tumor suppressor comes from epigenetic studies, and insulin-like growth factor (IGF) signal modulation.
PTPL1’s role as a tumor suppressor is supported by epigenetic experiments including the observation that the PTPL1 promoter is hypermethylated and also from mutation analysis, both of which are common mechanisms of inactivation for classical tumor suppressor genes. Hypermethylation of the PTPL1 promoter was found in lymphomas and a small number of carcinomas, including gastric and hepatocellular tumors [68]. Similarly, another study on hepatocellular carcinoma identified down regulation of PTPL1 mRNA in about 50% of cases due to promoter hypermethylation or allelic loss [69]. A large-scale study that looked at the mutations in the tyrosine phosphatome from colorectal cancers identified PTPN13 (the PTPL1 gene) as among the most frequently mutated PTPs [70]. The authors identified 19 mutations, non-sense and missense, in PTPN13, seven of which were in the PTP domain [70]. The physiological function of some of these mutant proteins led to altered phosphatase activity [21]. When the five missense mutations in the catalytic domain of PTPL1 were studied in vitro, four mutations led to amino-acid substitutions but without altered phosphatase activity. The fifth led to the M2307T [16], where M2307 is a highly conserved residue among PTPs [15]. This M2307T produced a protein with a seven-fold higher Km and two-fold lower kcat potentially leading to a decrease in the catalytic activity of PTPL1 [16].
Further evidence for PTPL1 as a tumor suppressor comes from a series of studies in breast cancer cells. PTPL1 was identified as one of the transcripts upregulated in breast cancer cells after tamoxifen treatment [71]. Stable antisense-mediated reduction of PTPL1 protein levels partially protects breast cancer cells from tamoxifen-induced apoptosis [72]. The reduction in PTPL1 levels increased PI3-K activity and AKT phosphorylation after pretreatment with tamoxifen and stimulation with IGF [72]. Recently, IRS-1 was identified as a putative PTPL1 substrate, as discussed previously [23]. A sustained phospho-IRS-1 signal results from coexpression of IRS-1 and a substrate-trapping mutant of PTPL1 or after siRNA-mediated reduction of PTPL1 protein levels [23]. Enhanced cell survival as a result of reducing PTPL1 is consistent with PTPL1 acting as a tumor suppressor.
4 Conclusions
There is data to support PTPL1 acting as both tumor promoter and suppressor in a variety of cancer models. It appears that the role of PTPL1 in cancer depends on the cellular context in which PTPL1 is studied. Some studies, most notably in colon carcinoma, have provided conflicting results regarding the functional role of PTPL1. PTPL1 modulates the cellular stress response and may potentiate cell survival. PTPL1 may also affect cytoskeletal remodeling and motility pathways in ways that either contribute to cancer metastases or reduce cell motility.
In order to clarify the role of PTPL1 under different conditions or tumor types, we need a better understanding of the biochemical role of PTPL1 in the cell. The identification of PTPL1 substrates is critical in understanding the signaling pathways modulated by PTPL1. More detailed studies on PTPL1 knock-out mice may also illuminate potential redundant gene functions explaining the minimal phenotype. Valuable insights into the role of PTPL1 in cancer and metastasis could also be gleaned through analysis of PTPL1 knock-out mice crossed with animals either lacking specific tumor suppressor genes or transgenic/knock-in animals expressing dominant oncogenes resulting in an increased incidence of spontaneous tumors. One caveat with respect to studies in mice, is that murine Fas lacks the tripeptide repeat present in the human protein that accounts for PTPL1 binding [73]. Finally, as is the case for many other protein tyrosine phosphatases, an understanding of physiological function of PTPL1 is greatly hampered by the lack of specific pharmacological inhibitors. The structural homology between the catalytic domain of PTPL1 and PTP1B may provide a basis for the development of these critical tools based on compounds that are relatively specific inhibitors of the latter enzyme [74].
PTPL1 is among the largest of the intracellular protein tyrosine phosphatases and may have multiple functions. Future work should clarify how PTPL1 functions, in the context of different tumors, to identify when PTPL1 will act as a tumor promoter or suppressor protein. It is hoped that these investigations will clarify the role of PTPL1 as a predictor of disease outcome or a potential therapeutic target in human cancer.
Acknowledgements
The authors would like to thank Toretsky Lab members for helpful discussions and Ms. Audrey Kubetin for editorial assistance.
Abbreviations
- ESFT
Ewing’s Sarcoma Family of Tumors
- FERM
band 4.1/ezrin/radixin/moesin
- IκBα
Inhibitor of nuclear factor kappa-B alpha
- KIND
Kinase non-catalytic C-lobe domain
- PARG1
PTPL1-associated RhoGAP1
- PDZ
PSD-95/Discs-large/ZO-1
- PIP
Phosphatidylinositol biphosphates
- PIP3
Phosphatidylinositol triphosphates
- PKA
Protein kinase-A
- PTP
Protein Tyrosine Phosphatase
- TAPP
Tandem-PH-domain-containing proteins
- TNFR
Tumor necrosis factor-receptor
- TRIP6
Thyroid Hormone Receptor-interacting Protein 6
Contributor Information
Ogan D. Abaan, Department of Oncology, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC 20057, USA
Jeffrey A. Toretsky, Department of Oncology, Lombardi Comprehensive Cancer Center, Georgetown University, Washington, DC 20057, USA
References
- 1.Alonso A, Sasin J, Bottini N, Friedberg I, Osterman A, Godzik A, et al. Protein tyrosine phosphatases in the human genome. Cell. 2004;117(6):699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 2.Dube N, Tremblay ML. Involvement of the small protein tyrosine phosphatases TC-PTP and PTP1B in signal transduction and diseases: from diabetes, obesity to cell cycle, and cancer. Biochimica et Biophysica Acta. 2005;1754(1-2):108–117. doi: 10.1016/j.bbapap.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 3.Mohi MG, Neel BG. The role of Shp2 (PTPN11) in cancer. Current Opinion in Genetics & Development. 2007;17(1):23–30. doi: 10.1016/j.gde.2006.12.011. [DOI] [PubMed] [Google Scholar]
- 4.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nature Reviews Cancer. 2006;6(4):307–320. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 5.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nature Reviews. Molecular Cell Biology. 2006;7(11):833–846. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 6.Banville D, Ahmad S, Stocco R, Shen SH. A novel protein-tyrosine phosphatase with homology to both the cytoskeletal proteins of the band 4.1 family and junction-associated guanylate kinases. Journal of Biological Chemistry. 1994;269(35):22320–22327. [PubMed] [Google Scholar]
- 7.Maekawa K, Imagawa N, Nagamatsu M, Harada S. Molecular cloning of a novel protein-tyrosine phosphatase containing a membrane-binding domain and GLGF repeats. FEBS Letters. 1994;337(2):200–206. doi: 10.1016/0014-5793(94)80273-4. [DOI] [PubMed] [Google Scholar]
- 8.Saras J, Claesson-Welsh L, Heldin CH, Gonez LJ. Cloning and characterization of PTPL1, a protein tyrosine phosphatase with similarities to cytoskeletal-associated proteins. Journal of Biological Chemistry. 1994;269(39):24082–24089. [PubMed] [Google Scholar]
- 9.Sato T, Irie S, Kitada S, Reed JC. FAP-1: A protein tyrosine phosphatase that associates with Fas. Science. 1995;268(5209):411–415. doi: 10.1126/science.7536343. [DOI] [PubMed] [Google Scholar]
- 10.Andersen JN, Jansen PG, Echwald SM, Mortensen OH, Fukada T, Del Vecchio R, et al. A genomic perspective on protein tyrosine phosphatases: Gene structure, pseudogenes, and genetic disease linkage. FASEB Journal. 2004;18(1):8–30. doi: 10.1096/fj.02-1212rev. [DOI] [PubMed] [Google Scholar]
- 11.Chishti AH, Kim AC, Marfatia SM, Lutchman M, Hanspal M, Jindal H, et al. The FERM domain: a unique module involved in the linkage of cytoplasmic proteins to the membrane. Trends in Biochemical Science. 1998;23(8):281–282. doi: 10.1016/s0968-0004(98)01237-7. [DOI] [PubMed] [Google Scholar]
- 12.Ciccarelli FD, Bork P, Kerkhoff E. The KIND module: a putative signalling domain evolved from the C lobe of the protein kinase fold. Trends in Biochemical Science. 2003;28(7):349–352. doi: 10.1016/S0968-0004(03)00116-6. [DOI] [PubMed] [Google Scholar]
- 13.Bompard G, Martin M, Roy C, Vignon F, Freiss G. Membrane targeting of protein tyrosine phosphatase PTPL1 through its FERM domain via binding to phosphatidylinositol 4,5-biphosphate. Journal of Cell Science. 2003;116(Pt 12):2519–2530. doi: 10.1242/jcs.00448. [DOI] [PubMed] [Google Scholar]
- 14.Herrmann L, Dittmar T, Erdmann KS. The protein tyrosine phosphatase PTP-BL associates with the midbody and is involved in the regulation of cytokinesis. Molecular Biology of the Cell. 2003;14(1):230–240. doi: 10.1091/mbc.E02-04-0191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, et al. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Molecular and Cellular Biology. 2001;21(21):7117–7136. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Villa F, Deak M, Bloomberg GB, Alessi DR, van Aalten DM. Crystal structure of the PTPL1/FAP-1 human tyrosine phosphatase mutated in colorectal cancer: Evidence for a second phosphotyrosine substrate recognition pocket. Journal of Biological Chemistry. 2005;280(9):8180–8187. doi: 10.1074/jbc.M412211200. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida S, Harada H, Nagai H, Fukino K, Teramoto A, Emi M. Head-to-head juxtaposition of Fas-associated phosphatase-1 (FAP-1) and c-Jun NH2-terminal kinase 3 (JNK3) genes: Genomic structure and seven polymorphisms of the FAP-1 gene. Journal of Human Genetics. 2002;47(11):614–619. doi: 10.1007/s100380200094. [DOI] [PubMed] [Google Scholar]
- 18.Irie S, Li Y, Kanki H, Ohyama T, Deaven LL, Somlo S, et al. Identification of two Fas-associated phosphatase-1 (FAP-1) promoters in human cancer cells. DNA Sequence. 2001;11(6):519–526. doi: 10.3109/10425170109041336. [DOI] [PubMed] [Google Scholar]
- 19.Abaan OD, Levenson A, Khan O, Furth PA, Uren A, Toretsky JA. PTPL1 is a direct transcriptional target of EWS-FLI1 and modulates Ewing’s Sarcoma tumorigenesis. Oncogene. 2005;24(16):2715–2722. doi: 10.1038/sj.onc.1208247. [DOI] [PubMed] [Google Scholar]
- 20.Kachel N, Erdmann KS, Kremer W, Wolff P, Gronwald W, Heumann R, et al. Structure determination and ligand interactions of the PDZ2b domain of PTP-Bas (hPTP1E): splicing-induced modulation of ligand specificity. Journal of Molecular Biology. 2003;334(1):143–155. doi: 10.1016/j.jmb.2003.09.026. [DOI] [PubMed] [Google Scholar]
- 21.Zhu JH, Chen R, Yi W, Cantin GT, Fearns C, Yang Y, et al. Protein tyrosine phosphatase PTPN13 negatively regulates Her2/ErbB2 malignant signaling. Oncogene. 2008 doi: 10.1038/sj.onc.1210922. in press. [DOI] [PubMed] [Google Scholar]
- 22.Nedachi T, Conti M. Potential role of protein tyrosine phosphatase nonreceptor type 13 in the control of oocyte meiotic maturation. Development. 2004;131(20):4987–4998. doi: 10.1242/dev.01368. [DOI] [PubMed] [Google Scholar]
- 23.Dromard M, Bompard G, Glondu-Lassis M, Puech C, Chalbos D, Freiss G. The putative tumor suppressor gene PTPN13/PTPL1 induces apoptosis through insulin receptor substrate-1 dephosphorylation. Cancer Research. 2007;67(14):6806–6813. doi: 10.1158/0008-5472.CAN-07-0513. [DOI] [PubMed] [Google Scholar]
- 24.Balla T. Inositol-lipid binding motifs: signal integrators through protein-lipid and protein-protein interactions. Journal of Cell Science. 2005;118(Pt 10):2093–2104. doi: 10.1242/jcs.02387. [DOI] [PubMed] [Google Scholar]
- 25.Kimber WA, Deak M, Prescott AR, Alessi DR. Interaction of the protein tyrosine phosphatase PTPL1 with the PtdIns(3,4)P2-binding adaptor protein TAPP1. Biochemical Journal. 2003;376(Pt 2):525–535. doi: 10.1042/BJ20031154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maekawa K, Imagawa N, Naito A, Harada S, Yoshie O, Takagi S. Association of protein-tyrosine phosphatase PTP-BAS with the transcription-factor-inhibitory protein Ikappa-Balpha through interaction between the PDZ1 domain and ankyrin repeats. Biochemical Journal. 1999;337(Pt 2):179–184. [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W, Tong Q, Conrad K, Wozney J, Cheung J, Miller BA. Regulation of the TRP channel TRPM2 by the tyrosine phosphatase PTPL1. American Journal of Physiology Cell Physiology. 2007;292:C1746–C1758. doi: 10.1152/ajpcell.00569.2006. [DOI] [PubMed] [Google Scholar]
- 28.Saras J, Engstrom U, Gonez LJ, Heldin CH. Characterization of the interactions between PDZ domains of the protein-tyrosine phosphatase PTPL1 and the carboxyl-terminal tail of Fas. Journal of Biological Chemistry. 1997;272(34):20979–20981. doi: 10.1074/jbc.272.34.20979. [DOI] [PubMed] [Google Scholar]
- 29.Yanagisawa J, Takahashi M, Kanki H, Yano-Yanagisawa H, Tazunoki T, Sawa E, et al. The molecular interaction of Fas and FAP-1. A tripeptide blocker of human Fas interaction with FAP-1 promotes Fas-induced apoptosis. Journal of Biological Chemistry. 1997;272(13):8539–8545. doi: 10.1074/jbc.272.13.8539. [DOI] [PubMed] [Google Scholar]
- 30.Irie S, Hachiya T, Rabizadeh S, Maruyama W, Mukai J, Li Y, et al. Functional interaction of Fas-associated phosphatase-1 (FAP-1) with p75(NTR) and their effect on NF-kappaB activation. FEBS Letters. 1999;460(2):191–198. doi: 10.1016/s0014-5793(99)01324-1. [DOI] [PubMed] [Google Scholar]
- 31.Murthy KK, Clark K, Fortin Y, Shen SH, Banville D. ZRP-1, a zyxin-related protein, interacts with the second PDZ domain of the cytosolic protein tyrosine phosphatase hPTP1E. Journal of Biological Chemistry. 1999;274(29):20679–20687. doi: 10.1074/jbc.274.29.20679. [DOI] [PubMed] [Google Scholar]
- 32.Saras J, Franzen P, Aspenstrom P, Hellman U, Gonez LJ, Heldin CH. A novel GTPase-activating protein for Rho interacts with a PDZ domain of the protein-tyrosine phosphatase PTPL1. Journal of Biological Chemistry. 1997;272(39):24333–24338. doi: 10.1074/jbc.272.39.24333. [DOI] [PubMed] [Google Scholar]
- 33.Lin D, Gish GD, Songyang Z, Pawson T. The carboxyl terminus of B class ephrins constitutes a PDZ domain binding motif. Journal of Biological Chemistry. 1999;274(6):3726–3733. doi: 10.1074/jbc.274.6.3726. [DOI] [PubMed] [Google Scholar]
- 34.Flint AJ, Tiganis T, Barford D, Tonks NK. Development of “substrate-trapping” mutants to identify physiological substrates of protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 1997;94(5):1680–1685. doi: 10.1073/pnas.94.5.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tiganis T, Bennett AM. Protein tyrosine phosphatase function: the substrate perspective. Biochemical Journal. 2007;402(1):1–15. doi: 10.1042/BJ20061548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nakai Y, Irie S, Sato TA. Identification of IkappaBalpha as a substrate of Fas-associated phosphatase-1. European Journal of Biochemistry. 2000;267(24):7170–7175. doi: 10.1046/j.1432-1327.2000.01818.x. [DOI] [PubMed] [Google Scholar]
- 37.Nakahira M, Tanaka T, Robson BE, Mizgerd JP, Grusby MJ. Regulation of signal transducer and activator of transcription signaling by the tyrosine phosphatase PTP-BL. Immunity. 2007;26(2):163–176. doi: 10.1016/j.immuni.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 38.Wansink DG, Peters W, Schaafsma I, Sutmuller RP, Oerlemans F, Adema GJ, et al. Mild impairment of motor nerve repair in mice lacking PTP-BL tyrosine phosphatase activity. Physiological Genomics. 2004;19(1):50–60. doi: 10.1152/physiolgenomics.00079.2004. [DOI] [PubMed] [Google Scholar]
- 39.Lorber B, Hendriks WJ, Van der Zee CE, Berry M, Logan A. Effects of LAR and PTP-BL phosphatase deficiency on adult mouse retinal cells activated by lens injury. European Journal of Neuroscience. 2005;21(9):2375–2383. doi: 10.1111/j.1460-9568.2005.04065.x. [DOI] [PubMed] [Google Scholar]
- 40.Uren A, Toretsky JA. Ewing’s sarcoma oncoprotein EWS-FLI1: The perfect target without a therapeutic agent. Future Oncology. 2005;1(4):521–528. doi: 10.2217/14796694.1.4.521. [DOI] [PubMed] [Google Scholar]
- 41.Baer C, Nees M, Breit S, Selle B, Kulozik AE, Schaefer KL, et al. Profiling and functional annotation of mRNA gene expression in pediatric rhabdomyosarcoma and Ewing’s sarcoma. International Journal of Cancer. 2004;110(5):687–694. doi: 10.1002/ijc.20171. [DOI] [PubMed] [Google Scholar]
- 42.Khan J, Wei JS, Ringner M, Saal LH, Ladanyi M, Westermann F, et al. Classification and diagnostic prediction of cancers using gene expression profiling and artificial neural networks. Nature Medicine. 2001;7(6):673–679. doi: 10.1038/89044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lessnick SL, Dacwag CS, Golub TR. The Ewing’s sarcoma oncoprotein EWS/FLI induces a p53-dependent growth arrest in primary human fibroblasts. Cancer Cell. 2002;1(4):393–401. doi: 10.1016/s1535-6108(02)00056-9. [DOI] [PubMed] [Google Scholar]
- 44.Houston A, O’Connell J. The Fas signalling pathway and its role in the pathogenesis of cancer. Current Opinion in Pharmacology. 2004;4(4):321–326. doi: 10.1016/j.coph.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 45.Ungefroren H, Voss M, Jansen M, Roeder C, Henne-Bruns D, Kremer B, et al. Human pancreatic adenocarcinomas express Fas and Fas ligand yet are resistant to Fas-mediated apoptosis. Cancer Research. 1998;58(8):1741–1749. [PubMed] [Google Scholar]
- 46.Ungefroren H, Kruse ML, Trauzold A, Roeschmann S, Roeder C, Arlt A, et al. FAP-1 in pancreatic cancer cells: functional and mechanistic studies on its inhibitory role in CD95-mediated apoptosis. Journal of Cell Science. 2001;114(Pt 15):2735–2746. doi: 10.1242/jcs.114.15.2735. [DOI] [PubMed] [Google Scholar]
- 47.Ivanov VN, Lopez Bergami P, Maulit G, Sato TA, Sassoon D, Ronai Z. FAP-1 association with Fas (Apo-1) inhibits Fas expression on the cell surface. Molecular and Cellular Biology. 2003;23(10):3623–3635. doi: 10.1128/MCB.23.10.3623-3635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meinhold-Heerlein I, Stenner-Liewen F, Liewen H, Kitada S, Krajewska M, Krajewski S, et al. Expression and potential role of Fas-associated phosphatase-1 in ovarian cancer. American Journal of Pathology. 2001;158(4):1335–1344. doi: 10.1016/S0002-9440(10)64084-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yao H, Song E, Chen J, Hamar P. Expression of FAP-1 by human colon adenocarcinoma: implication for resistance against Fas-mediated apoptosis in cancer. British Journal of Cancer. 2004;91(9):1718–1725. doi: 10.1038/sj.bjc.6602136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wieckowski E, Atarashi Y, Stanson J, Sato TA, Whiteside TL. FAP-1-mediated activation of NF-kappaB induces resistance of head and neck cancer to Fas-induced apoptosis. Journal of Cellular Biochemistry. 2007;100(1):16–28. doi: 10.1002/jcb.20922. [DOI] [PubMed] [Google Scholar]
- 51.Lee SH, Shin MS, Lee HS, Bae JH, Lee HK, Kim HS, et al. Expression of Fas and Fas-related molecules in human hepatocellular carcinoma. Human Pathology. 2001;32(3):250–256. doi: 10.1053/hupa.2001.22769. [DOI] [PubMed] [Google Scholar]
- 52.Lee SH, Shin MS, Lee JY, Park WS, Kim SY, Jang JJ, et al. In vivo expression of soluble Fas and FAP-1: possible mechanisms of Fas resistance in human hepatoblastomas. Journal of Pathology. 1999;188(2):207–212. doi: 10.1002/(SICI)1096-9896(199906)188:2<207::AID-PATH337>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 53.Eberle A, Reinehr R, Becker S, Haussinger D. Fluorescence resonance energy transfer analysis of proapoptotic CD95-EGF receptor interactions in Huh7 cells. Hepatology. 2005;41(2):315–326. doi: 10.1002/hep.20564. [DOI] [PubMed] [Google Scholar]
- 54.Barker PA. p75NTR is positively promiscuous: novel partners and new insights. Neuron. 2004;42(4):529–533. doi: 10.1016/j.neuron.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 55.Mamidipudi V, Wooten MW. Dual role for p75 (NTR) signaling in survival and cell death: Can intracellular mediators provide an explanation? Journal of Neuroscience Research. 2002;68(4):373–384. doi: 10.1002/jnr.10244. [DOI] [PubMed] [Google Scholar]
- 56.Ohrt T, Mancini A, Tamura T, Niedenthal R. c-Cbl binds to tyrosine-phosphorylated neurotrophin receptor p75 and induces its ubiquitination. Cell Signal. 2004;16(11):1291–1298. doi: 10.1016/j.cellsig.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 57.Imbert V, Rupec RA, Livolsi A, Pahl HL, Traenckner EB, Mueller-Dieckmann C, et al. Tyrosine phosphorylation of I kappa B-alpha activates NF-kappa B without proteolytic degradation of I kappa B-alpha. Cell. 1996;86(5):787–798. doi: 10.1016/s0092-8674(00)80153-1. [DOI] [PubMed] [Google Scholar]
- 58.Kawai H, Nie L, Yuan ZM. Inactivation of NF-kappaB-dependent cell survival, a novel mechanism for the proapoptotic function of c-Abl. Molecular and Cellular Biology. 2002;22(17):6079–6088. doi: 10.1128/MCB.22.17.6079-6088.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fan C, Yang J, Engelhardt JF. Temporal pattern of NF kappa B activation influences apoptotic cell fate in a stimuli-dependent fashion. Journal of Cell Science. 2002;115(24):4843–4853. doi: 10.1242/jcs.00151. [DOI] [PubMed] [Google Scholar]
- 60.Foehr ED, Lorente G, Vincent V, Nikolich K, Urfer R. FAS associated phosphatase (FAP-1) blocks apoptosis of astrocytomas through dephosphorylation of FAS. Journal of Neurooncology. 2005;74(3):241–248. doi: 10.1007/s11060-004-7202-x. [DOI] [PubMed] [Google Scholar]
- 61.Hogan A, Yakubchyk Y, Chabot J, Obagi C, Daher E, Maekawa K, et al. The phosphoinositol 3,4-bisphosphate-binding protein TAPP1 interacts with syntrophins and regulates actin cytoskeletal organization. Journal of Biological Chemistry. 2004;279(51):53717–53724. doi: 10.1074/jbc.M410654200. [DOI] [PubMed] [Google Scholar]
- 62.Kullander K, Klein R. Mechanisms and functions of Eph and ephrin signalling. Nature Reviews. Molecular and Cellular Biology. 2002;3(7):475–486. doi: 10.1038/nrm856. [DOI] [PubMed] [Google Scholar]
- 63.Palmer A, Zimmer M, Erdmann KS, Eulenburg V, Porthin A, Heumann R, et al. EphrinB phosphorylation and reverse signaling: regulation by Src kinases and PTP-BL phosphatase. Molecular Cell. 2002;9(4):725–737. doi: 10.1016/s1097-2765(02)00488-4. [DOI] [PubMed] [Google Scholar]
- 64.Myagmar BE, Umikawa M, Asato T, Taira K, Oshiro M, Hino A, et al. PARG1, a protein-tyrosine phosphatase-associated RhoGAP, as a putative Rap2 effector. Biochemical and Biophysical Research Communications. 2005;329(3):1046–1052. doi: 10.1016/j.bbrc.2005.02.069. [DOI] [PubMed] [Google Scholar]
- 65.Lai YJ, Lin WC, Lin FT. PTPL1/FAP-1 negatively regulates TRIP6 function in lysophosphatidic acid-induced cell migration. Journal of Biological Chemistry. 2007;282(33):24381–24387. doi: 10.1074/jbc.M701499200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyazaki T, Atarashi Y, Yasumura S, Minatoya I, Ogawa K, Iwamoto M, et al. Fas-associated phosphatase-1 promotes Fas-mediated apoptosis in human colon cancer cells: novel function of FAP-1. Journal of Gastroenterology and Hepatology. 2006;21(1 Pt 1):84–91. doi: 10.1111/j.1440-1746.2005.04155.x. [DOI] [PubMed] [Google Scholar]
- 67.Tillman DM, Harwood FG, Gibson AA, Houghton JA. Expression of genes that regulate Fas signalling and Fas-mediated apoptosis in colon carcinoma cells. Cell Death Differentiation. 1998;5(5):450–457. doi: 10.1038/sj.cdd.4400369. [DOI] [PubMed] [Google Scholar]
- 68.Ying J, Li H, Cui Y, Wong AH, Langford C, Tao Q. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20(6):1173–1175. doi: 10.1038/sj.leu.2404193. [DOI] [PubMed] [Google Scholar]
- 69.Yeh SH, Wu DC, Tsai CY, Kuo TJ, Yu WC, Chang YS, et al. Genetic characterization of fas-associated phosphatase-1 as a putative tumor suppressor gene on chromosome 4q21.3 in hepatocellular carcinoma. Clinical Cancer Research. 2006;12(4):1097–1108. doi: 10.1158/1078-0432.CCR-05-1383. [DOI] [PubMed] [Google Scholar]
- 70.Wang Z, Shen D, Parsons DW, Bardelli A, Sager J, Szabo S, et al. Mutational analysis of the tyrosine phosphatome in colorectal cancers. Science. 2004;304(5674):1164–1166. doi: 10.1126/science.1096096. [DOI] [PubMed] [Google Scholar]
- 71.Freiss G, Puech C, Vignon F. Extinction of insulin-like growth factor-I mitogenic signaling by antiestrogen-stimulated Fas-associated protein tyrosine phosphatase-1 in human breast cancer cells. Molecular Endocrinology. 1998;12(4):568–579. doi: 10.1210/mend.12.4.0088. [DOI] [PubMed] [Google Scholar]
- 72.Bompard G, Puech C, Prebois C, Vignon F, Freiss G. Protein-tyrosine phosphatase PTPL1/FAP-1 triggers apoptosis in human breast cancer cells. Journal of Biological Chemistry. 2002;277(49):47861–47869. doi: 10.1074/jbc.M208950200. [DOI] [PubMed] [Google Scholar]
- 73.Cuppen E, Nagata S, Wieringa B, Hendriks W. No evidence for involvement of mouse protein-tyrosine phosphatase-BAS-like Fas-associated phosphatase-1 in Fas-mediated apoptosis. Journal of Biological Chemistry. 1997;272(48):30215–30220. doi: 10.1074/jbc.272.48.30215. [DOI] [PubMed] [Google Scholar]
- 74.Stuible M, Zhao L, Aubry I, Schmidt-Arras D, Bohmer FD, Li CJ, et al. Cellular inhibition of protein tyrosine phosphatase 1B by uncharged thioxothiazolidinone derivatives. Chembiochem. 2007;8(2):179–186. doi: 10.1002/cbic.200600287. [DOI] [PubMed] [Google Scholar]