Abstract
Archaeal and eukaryotic B-family DNA polymerases (pols) mainly replicate chromosomal DNA but stall at lesions, which are often bypassed with Y-family pols. In this study, a B-family pol Vent (exo−) from the euryarchaeon Thermococcus litoralis was studied with three types of DNA lesions—N2-alkylG, O6-alkylG, and an abasic (AP) site—in comparison with a model Y-family pol Dpo4 from Sulfolobus solfataricus, to better understand the effects of various DNA modifications on binding, bypass efficiency, and fidelity of pols. Vent (exo−) readily bypassed N2-methyl(Me)G and O6-MeG, but was strongly blocked at O6-benzyl(Bz)G and N2-BzG, whereas Dpo4 efficiently bypassed N2-MeG and N2-BzG and partially bypassed O6-MeG and O6-BzG. Vent (exo−) bypassed an AP site to an extent greater than Dpo4, corresponding with steady-state kinetic data. Vent (exo−) showed ~110-, 180-, and 300-fold decreases in catalytic efficiency (kcat/Km) for nucleotide insertion opposite an AP site, N2-MeG, and O6-MeG but ~1800- and 5000-fold decreases opposite O6-BzG and N2-BzG, respectively, as compared to G, whereas Dpo4 showed little or only ~13-fold decreases opposite N2-MeG and N2-BzG but ~260−370-fold decreases opposite O6-MeG, O6-BzG, and the AP site. Vent (exo−) preferentially misinserted G opposite N2-MeG, T opposite O6-MeG, and A opposite an AP site and N2-BzG, while Dpo4 favored correct C insertion opposite those lesions. Vent (exo−) and Dpo4 both bound modified DNAs with affinities similar to unmodified DNA. Our results indicate that Vent (exo−) is as or more efficient as Dpo4 in synthesis opposite O6-MeG and AP lesions, whereas Dpo4 is much or more efficient opposite (only) N2-alkylGs than Vent (exo−), irrespective of DNA-binding affinity. Our data also suggest that Vent (exo−) accepts nonbulky DNA lesions (e.g., N2- or O6-MeG and an AP site) as manageable substrates despite causing error-prone synthesis, whereas Dpo4 strongly favors minor-groove N2-alkylG lesions over major-groove or noninstructive lesions.
INTRODUCTION
Cellular DNA is unremittingly exposed to various endogenous and exogenous damaging agents, which can produce numerous DNA modifications including alkylation, base loss, deamination, oxidation, stand breakage, cross-links, and bulky adduct formation.1 The resulting DNA lesions, if unrepaired and misprocessed, can induce mutations and cell death depending on the type and extent of damage, potentially contributing to carcinogenesis and aging.2 One of the key enzymes in this process is DNA polymerase (pol), which can incorrectly replicate DNA lesions or stop at the sites of some lesions during DNA replication.3 Therefore, the understanding of mechanistic interactions between pols and various DNA lesions is vital to elucidate the basis for DNA damage-induced mutagenesis or cell death.
Plentiful pols have been found from living organisms throughout all three domains of life and are classified into six families (A, B, C, D, X, and Y).4 Archaeal systems for DNA replication, which are closely related to the eukaryotic ones, have been perceived as a simplified experimental models for eukaryotic DNA replication.5 Similar to eukaryotes, most Bfamily pols from archaea are believed to be utilized primarily for chromosomal DNA replication, although euryarchaea employ an additional replicative D-family polymerase,6 and the crenarchaeon Sulfolobus solfataricus P2 possesses extra two B-family polymerases, Dpo2 and Dpo3, which have been shown to be capable of bypassing oxidative, deaminated, and/or UVinduced DNA lesions.7 Y-family pols have been found in some but not all archaea, especially the genera Sulfolobus and Methanosarcina,8,9 which cope with and bypass replication-blocking DNA lesions. In particular, S. solfataricus P2 Dpo4 has been studied as a model of Y-family translesion pols.10
Guanine (G) is the most frequently damaged base in DNA and is susceptible at the N2, N7, O6, and C8 positions to modifications by various potential carcinogens including formaldehyde,11 acetaldehyde,12 metabolites of polycyclic aromatic hydrocarbons (e.g., benzo[a]pyrene),13 heterocyclic amines (e.g., 2-amino-3-methylimidazo[4,5-f ]quinoline),14 tobacco-specific nitosamines [e.g., 4-(methylnitrosoamino)-1-(3-pyridyl)-1-butanone],15 and other oxidizing and alkylating agents.1,16 N2-Methyl(Me)G and N2-hydroxymethylG are formed as minor and major DNA adducts, respectively, in formaldehyde-treated cells and are also induced by alkylating agents, although the levels of these two N2-adducts are much lower than those of N7-MeG and O6-MeG.11 In addition, guanine bases, in natural or modified forms, are frequently lost from the DNA backbone through spontaneous, chemical, or enzymatic hydrolysis.1,17 Thus, guanine bases can be transformed into various types of DNA lesions including minor- and major-groove adducts and abasic (apurinic/apyrimidinic or AP) sites, which can act as triggers for replication blockage and genetic mutation. While Y-family pols are thought to play the major role in translesion DNA synthesis (TLS) past DNA lesions, recent studies suggest a partial role of B-family pols in a subset of TLS, in that human pol δ [with proliferating cell nuclear antigen (PCNA)] can copy DNA past some DNA lesions such as O6-methyl(Me)G and AP sites.18−21 However, it is not yet certain which types of DNA lesions are the cognate substrates for individual pols. Direct comparative kinetics of TLS upon different types of DNA lesions by archetypal B- and Y-family pols can help in inferring the most-favored or cognate lesion substrates of those pols.
Thermococcus litoralis, a hyperthermophilic euryarchaeon, possesses a high-fidelity B-family pol termed Vent with a proof-reading 3′ to 5′ exonuclease activity,22 which is extremely thermostable; thus, its wild-type and exonuclease-free (exo−) forms have been widely used in polymerase chain reaction (PCR) applications. Vent may also be worthwhile as a distinctive euryarchaeal model of the replicative B-family pols, because these euryarchaeal species of the Thermococcales order appear to encode a B-family pol and a D-family pol, which are suggested to synthesize the leading strand and lagging strand, respectively (but no apparent Y-family pol homologue), differing from crenarchaea.23,24 There are some possibilities for lesion bypass in organisms that lack Y-family pols, such as robust DNA repair mechanisms to remove lesions or existing or novel non-Y-family polymerases to perform lesion bypass. Vent might be a good model in this respect, although Vent (exo−) is known to be blocked at DNA photoproducts.25 Understanding the mode of lesion bypass by Vent (exo−) might also have practical applicability for PCR of damaged DNA. To better understand the effects of modified DNAs on DNA binding and lesion bypass by a euryarchaeal B-family pol Vent (exo−) as compared to the model Y-family pol Dpo4, we performed primer-extension, steady-state kinetic, and pol-DNA binding experiments in parallel using two recombinant pols with three different series of DNA lesions, N2-alkyl (⩵Me or benzyl-(Bz))G, O6-alkyl (⩵Me or Bz)G, and synthetic abasic (tetrahydrofuran) lesions. Here, we show that Vent (exo−) modestly bypasses nonbulky minor- and major-groove MeG adducts and abasic sites, as or more efficiently as Dpo4 opposite the latter two lesions but in a more error-prone manner, whereas Dpo4 extraordinarily favors the minor-groove N2-alkylG lesions over the major-groove and abasic lesions for catalysis, despite little difference in DNA-binding affinities. The implications of in vitro biochemical properties, with respect to the favored DNA lesion substrates of a euryarchaeal B-family pol Vent (exo−), are discussed in comparison with the crenarchaeal Y-family pol Dpo4.
EXPERIMENTAL PROCEDURES
Materials
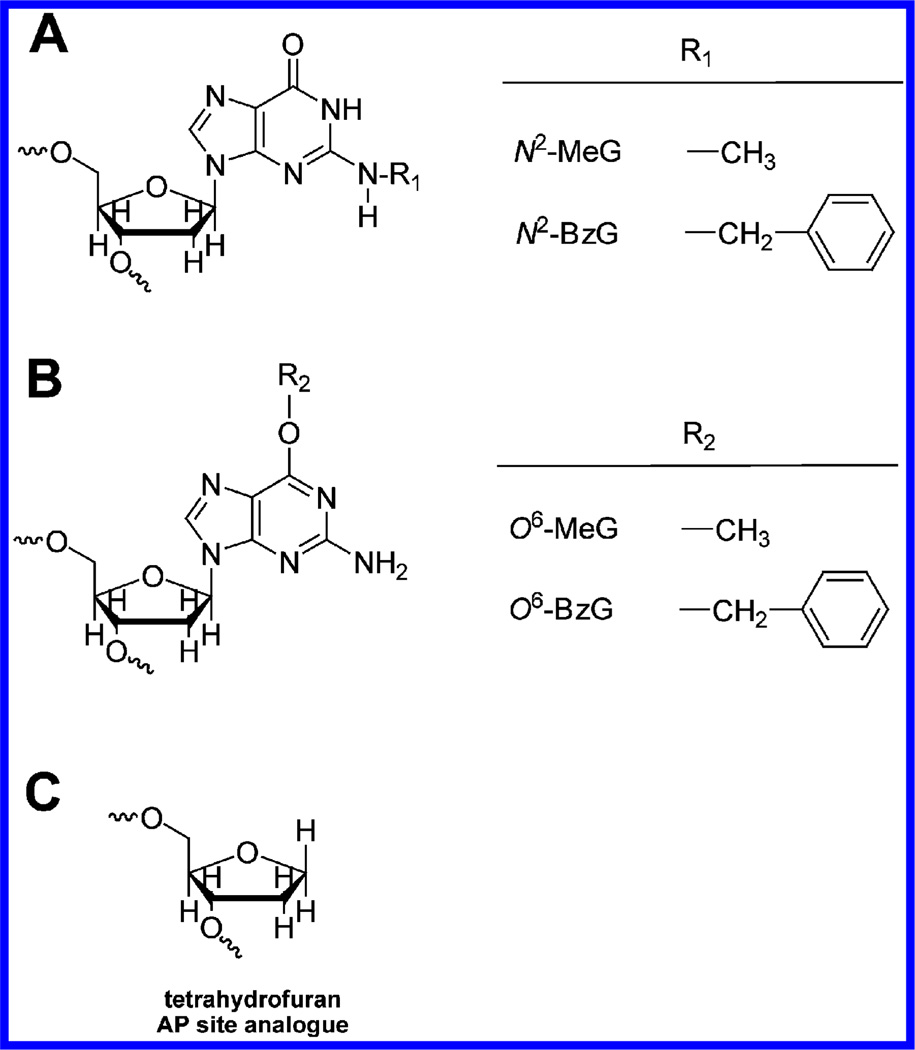
Unlabeled deoxynucleoside triphosphate (dNTPs), T4 polynucleotide kinase, and Vent (exo−) were purchased from New England Biolabs (Ipswich, MA). [γ-32P]ATP (specific activity 3,000 Ci mmol−1) was purchased from PerkinElmer Life Sciences (Boston, MA). Biospin columns were purchased from Bio-Rad (Hercules, CA). A 24-mer (5′-GCC TCG AGC CAG CCG CAG ACG CAG-3′), two 25-mers (5′-GCC TCG AGC CAG CCG CAG ACG CAG Y-3′; Y = C or A), and two 36-mer (3′-CGG AGC TCG GTC GGC GTC TGC GTC XCT CCT GCG GCT-5′; X = G or tetrahydrofuran (abasic site analogue)) oligonucleotides were obtained from Midland Certified Reagents (Midland, TX). Four 36-mers (3′-CGG AGC TCG GTC GGC GTC TGC GTC XCT CCT GCG GCT-5′; X = N2-MeG, N2-BzG, O6-MeG, or O6-BzG) were prepared as described previously.21,26 Recombinant Dpo4 was expressed in Escherichia coli and purified as described previously.27
Reaction Conditions for Polymerase Activity Assays
Standard DNA polymerization reactions were performed in 50 mM Tris-HCl buffer (pH 7.5 at 37 °C) containing 5 mM dithiothreitol, 50 mM NaCl, 100 µg mL−1 bovine serum albumin (w/v), and 10% glycerol (v/v) with 100 nM primer/template at 37 °C, unless indicated otherwise. A reaction temperature of 37 °C was chosen instead of the physiological temperature (~80 °C) of S. solfataricus and T. litoralis, to allow the annealed oligonucleotide primer/template DNA substrate to remain stable, to prevent the reaction mixtures from evaporation during reaction, and to facilitate comparison with the previous kinetic data for many other polymerases assayed at 37 °C. These kinetic approaches at a suboptimal temperature were successfully applied with bacteriophage T7 (exo−) pol and S. solfataricus Dpo4 previously.28,29 Primers (24-mer) were 5′ end-labeled with [γ-32P]ATP using T4 polynucleotide kinase and annealed with templates (36-mer). All reactions were initiated by the addition of dNTP and MgCl2 (5 mM final concentration) to preincubated polymerase/DNA mixtures.
Primer Extension Assays and Gel Electrophoresis
A 32P-labeled primer, annealed to a template, was extended in the presence of all four dNTPs. Each reaction was initiated by adding 4 µL of dNTP-Mg2+ solution (final concentration of 100 µM of each dNTP) to a preincubated polymerase/DNA mixture at 37 °C, yielding a total reaction volume of 8 µL. After 15 min, reactions were quenched with a solution of 20 mM EDTA (pH 8.0) in 95% formamide (v/v). Aliquots were separated by electrophoresis on denaturing gels containing 8.0 M urea and 16% acrylamide (w/v) (from a 19:1 acrylamide:bisacrylamide solution, AccuGel, National Diagnostics, Atlanta, GA) with 80 mM Tris-borate buffer (pH 7.8) containing 1 mM EDTA. Gels were exposed to a phosphorimager screen, and the bands (representing extension of the primer) were visualized with a phosphorimaging system (Bio-Rad, Personal Molecular Imager, Hercules, CA) using the manufacturer's Quantity One Software.
Steady-State Kinetic Reactions
A 32P-labeled primer, annealed to a template, was extended in the presence of varying concentrations of a single dNTP. Polymerase concentrations and reaction times were chosen so that maximal product formation would be ≥20% of the substrate concentration. The primer template was extended with the dNTP in the presence of 0.1−30 nM polymerase for 5 or 10 min. All reactions (8 µL) were done at 10 dNTP concentrations and quenched with 10 volumes of a solution of 20 mM EDTA in 95% formamide (v/ v). Products were resolved using 8 M urea, 16% polyacrylamide (w/v) electrophoresis gels and quantitated by phosphorimaging analysis. Graphs of product formation vs dNTP concentration were fit using nonlinear regression (hyperbolic fits) in GraphPad Prism Version 4.0 (San Diego, CA) for the determination of kcat and Km values.
Electrophoretic Mobility Shift Assays
Increasing concentrations of each polymerase were incubated with 10 nM 32P-24-mer/36-mer primer/template DNA at 22 °C for 15 min in the binding buffer, which contained 50 mM Tris-HCl buffer (pH 7.5 at 22 °C), 50 mM NaCl, 5 mM MgCl2, 5 mM dithiothreitol, 100 µg mL−1 bovine serum albumin (w/v), and 10% glycerol (v/v). The mixtures were directly loaded on nondenaturing 4% polyacrylamide gels (w/v) and electrophoresed at 8 V cm−1 for 40 min at 22 °C in the running buffer [40 mM Tris-acetate (pH 7.5 at 22 °C) containing 5 mM magnesium acetate and 0.1 mM EDTA]. Gels were imaged using a Bio-Rad Personal Molecular Imager instrument, and the fractions of polymerase-bound DNA were quantitated by Quantity One software (Figure S1 in the Supporting Information). The data were fit to a quadratic equation, [E·DNA] = 0.5(KdDNA + E0 + D0) − 0.5[(KdDNA + E0 + D0)2 − 4E0D0]1/2, where E0 = polymerase concentration, D0 = DNA concentration, and [E·DNA] = concentration of polymerase-DNA complex, in GraphPad Prism software.
RESULTS
Extension of Primers Opposite G, N2-Alkylguanine, O6-Alkylguanine, and an AP Site in the Presence of All Four dNTPs by Vent (exo−) and Dpo4
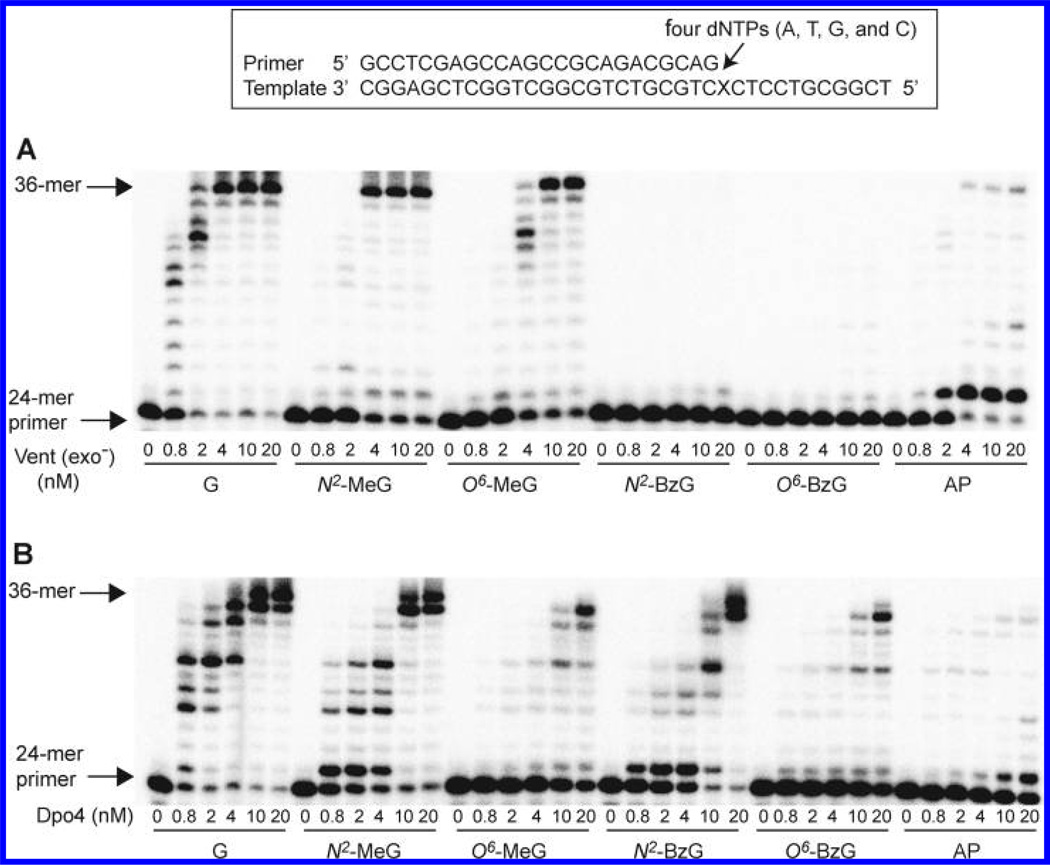
The 24-mer primers were extended at and beyond G, N2-MeG, N2-BzG, O6-MeG, O6-BzG, or an AP site (tetrahydrofuran) (Figure 1) at position 25 of the template in the presence of all four dNTPs with increasing concentrations of Vent (exo−) and Dpo4 (Figure 2), to estimate the qualitative differences in TLS across N2-alkylguanines, O6-alkylguanines, and an AP site. We utilized the exo− form of Vent, which showed about a 3-fold decrease in fidelity as compared to wild-type Vent,22 to analyze the TLS activities and DNA-binding properties of Vent without any possible interferences of exonuclease activity during kinetic assays, and to directly compare kinetic data with other polymerases. The “ladder” bands of polymerized intermediates appeared in polymerization across G at 0.8 nM enzyme concentrations of Vent (exo−) and Dpo4, indicating apparently low processivity (<10 nucleotides) of both polymerases with unmodified DNA substrates in this reaction condition. Vent (exo−) fully extended the primers past N2-MeG and O6-MeG to yield 36-mer products, at 4 and 10 nM enzyme concentrations, respectively, which were similar or 2.5-fold higher enzyme concentrations than with the unmodified DNA substrate, indicating that Vent (exo−) can bypass both N2-MeG and O6-MeG with little or no difficulty as compared to G. However, the primer extension by Vent (exo−) was severely blocked opposite O6-BzG and N2-BzG. In contrast, Dpo4 fully extended primers opposite N2-MeG and N2-BzG with a gradual decrease in extent as compared to unmodified G but was partially blocked at O6-MeG and O6-BzG. Interestingly, Vent (exo−) effectively extended primers opposite an AP site but yielded largely onebase extension products with some full-length extension. In contrast, Dpo4 extended primers opposite an AP site with much difficulty and yielded mainly one-base extension products and traces of 36-mer products at 20 nM enzyme concentration, to a much lesser extent than with N2-BzG.
Figure 1.
DNA lesions used in this work. (A) N2-Alkyldeoxyguanosine adducts. (B) O6-Alkyldeoxyguanosine adducts. (C) Tetrahydrofuran analogue of abasic site.
Figure 2.
Extension of a 32P-labeled 24-mer primer opposite G, N2-alkylguanines, O6-alkylguanines, and an AP site at position 25 by Vent (exo−) and Dpo4. The primer (24-mer) was annealed with each of the six different 36-mer templates containing an unmodified G, N2-MeG, N2-BzG, O6-MeG, O6-BzG, or AP site (tetrahydrofuran) placed at the 25th position from the 3′-end (see Figure 1). Reactions were done for 15 min (extended in the presence of all four dNTPs) with a constant concentration of DNA substrate (100 nM primer/template) and increasing concentrations of polymerases (0, 0.8, 2, 4, 10, and 20 nM) as indicated. The reaction products were analyzed by denaturing gel electrophoresis with subsequent phosphorimaging analysis. (A) Vent (exo−). (B) Dpo4.
Binding of Vent (exo−) and Dpo4 to DNA Substrate Containing a G, N2-Alkylguanine, O6-Alkylguanine, or an AP Site
Electrophoretic mobility shift assays were performed to estimate the relative binding affinities of Vent (exo−) and Dpo4 to six primer-template DNA substrates, each containing a template base G, N2-MeG, N2-BzG, O6-MeG, O6-BzG, or AP site next to the primer-template junction. Electrophoretic mobility shift assays can provide valid comparisons of binding affinities of polymerases to different DNA substrates, although not reflecting the true equilibrium. The fraction of DNA complexed with each polymerase was regarded as an indicator of polymerase-DNA binding to estimate an apparent dissociation constant (Kd DNA) (Table 1). Vent (exo−) bound the adducted DNA substrates with similar or slightly higher (up to 1.6-fold) binding affinities than the unmodified DNA. Similarly, Dpo4 bound the adducted DNA substrates with similar or slightly higher (up to 1.5-fold) binding affinities than unmodified DNA.
Table 1.
Apparent Kd Values for DNA Substrate Binding to Vent (exo−) and Dpo4
|
KdDNA (nM) |
||
|---|---|---|
| primer/template | Vent | Dpo4 |
| 24-mer/36-G-mer | 39 ± 8 | 52 ± 9 |
| 24-mer/36-N2-MeG-mer | 33 ± 6 | 39 ± 5 |
| 24-mer/36-N2-BzG-mer | 32 ± 7 | 34 ± 4 |
| 24-mer/36-O6-MeG-mer | 29 ± 6 | 37 ± 3 |
| 24-mer/36-O6-BzG-mer | 25 ± 5 | 35 ± 5 |
| 24-mer/36-AP-mer | 35 ± 9 | 55 ± 16 |
Steady-State Kinetics of Nucleotide Incorporation into Primers Opposite a G, N2-Alkylguanine, O6-Alkylguanine, or AP Site by Vent (exo−) and Dpo4
To quantitatively analyze the efficiency and fidelity for nucleotide insertion opposite three series of DNA lesions by Vent (exo−) and Dpo4, steady-state kinetic parameters were determined for the incorporation of single nucleotides into a 24-mer primer opposite G, N2-MeG, N2-BzG, O6-MeG, O6-BzG, or an AP site (Tables 2 and 3 and Figure S2 in the Supporting Information) by each polymerase. The specificity constant kcat/Km, which provides a quantitative measure for the catalytic efficiency of nonprocessive polymerases in nucleotide insertion opposite the template base, can be cautiously applied in these studies to compare the catalytic efficiency and fidelity of two polymerases opposite different DNA lesion substrates,30 in that Vent (exo−) and Dpo4 are relatively distributive polymerases with poor processivities (about 7 and 16 nucleotides respectively).31,32 Thus, this approach may not greatly underestimate the steadystate kinetic parameters.33 The kcat/Km for single nucleotide incorporation opposite a template lesion by each polymerase was compared with that for correct nucleotide incorporation opposite the lesion to calculate the misinsertion frequency [f mis = (kcat/Km)dNTP/(kcat/Km)dCTP] and was also compared with that for the correct nucleotide incorporation opposite the unmodified control G to estimate the relative efficiency of nucleotide insertion. Insertion of dCTP opposite an abasic site was tentatively assumed to be more likely to be a more accurate synthesis (e.g., an action of REV1)34 in that depurination is much more prone than depyrimidination in DNA,1 and guanine is most easily oxidized into oxidative lesions that are often removed enzymatically.17
Table 2.
Steady-State Kinetic Parameters for dNTP Incorporation Opposite DNA Lesions by Vent (exo−)
| template base | dNTP | Km (µM) | kcat (s−1) | kcat/Km (mM−1 s−1) | f ins a | relative effciencyb |
|---|---|---|---|---|---|---|
| G | A | 2300 ± 400 | 0.0098 ± 0.0008 | 0.0043 | 0.00039 | 3.9 × 10−4 |
| T | 2700 ± 400 | 0.043 ± 0.004 | 0.016 | 0.0015 | 1.5 × 10−3 | |
| G | 1200 ± 200 | 0.012 ± 0.0006 | 0.01 | 0.00091 | 9.1 × 10−4 | |
| C | 6.2 ± 1.7 | 0.065 ± 0.003 | 11 | 1 | 1 | |
| N2-MeG | A | 3200 ± 400 | 0.0068 ± 0.0004 | 0.0021 | 0.1 | 1.9 × 10−4 |
| T | 1800 ± 300 | 0.0021 ± 0.0001 | 0.0012 | 0.057 | 1.1 × 10−4 | |
| G | 84 ± 32 | 0.0052 ± 0.0005 | 0.062 | 3 | 5.6 × 10−3 | |
| C | 1300 ± 200 | 0.027 ± 0.001 | 0.021 | 1 | 1.9 × 10−3 | |
| N2-BzG | A | 2500 ± 400 | 0.0053 ± 0.0005 | 0.0022 | 52 | 2.0 × 10−4 |
| T | 2300 ± 500 | 0.0004 ± 0.000004 | 0.00017 | 4 | 1.5 × 10−5 | |
| G | 1600 ± 300 | 0.0006 ± 0.00005 | 0.0004 | 10 | 3.6 × 10−5 | |
| C | 2400 ± 600 | 0.0001 ± 0.000002 | 0.000042 | 1 | 3.8 × 10−6 | |
| O6-MeG | A | 2800 ± 900 | 0.0029 ± 0.0005 | 0.0011 | 0.058 | 1.0 × 10−4 |
| T | 380 ± 60 | 0.014 ± 0.0007 | 0.037 | 1.9 | 3.3 × 10−3 | |
| G | 2300 ± 200 | 0.0067 ± 0.0004 | 0.0029 | 0.15 | 2.6 × 10−4 | |
| C | 1200 ± 200 | 0.022 ± 0.002 | 0.019 | 1 | 1.7 × 10−3 | |
| O6-BzG | A | 1500 ± 300 | 0.0017 ± 0.0001 | 0.0011 | 0.18 | 1.0 × 10−4 |
| T | 960 ± 30 | 0.003 ± 0.0003 | 0.0031 | 0.51 | 2.8 × 10−4 | |
| G | 310 ± 150 | 0.0002 ± 0.00003 | 0.00064 | 0.1 | 5.8 × 10−5 | |
| C | 130 ± 60 | 0.0008 ± 0.00008 | 0.0061 | 1 | 5.5 × 10−4 | |
| AP site | A | 430 ± 70 | 0.043 ± 0.002 | 0.099 | 40 | 9.0 × 10−3 |
| T | 2100 ± 900 | 0.017 ± 0.003 | 0.0081 | 3.2 | 7.4 × 10−4 | |
| G | 2300 ± 700 | 0.043 ± 0.007 | 0.019 | 7.6 | 1.7 × 10−3 | |
| C | 830 ± 250 | 0.0021 ± 0.0002 | 0.0025 | 1 | 2.3 × 10−4 |
Misinsertion frequency, calculated by dividing kcat/Km for dNTP incorporation by the kcat/Km for dCTP incorporation opposite each template base.
Relative insertion efficiency, calculated by dividing kcat/Km for dNTP incorporation opposite each template base by the kcat/Km for dCTP incorporation opposite template G.
Table 3.
Steady-State Kinetic Parameters for dNTP Incorporation Opposite DNA Lesions by Dpo4
| template base | dNTP | Km (µM) | kcat (s−1) | kcat/Km (mM−1 s−1) | fins a | relative effciencyb |
|---|---|---|---|---|---|---|
| G | A | 880 ± 80 | 0.0028 ± 0.00009 | 0.0032 | 0.0001 | 1 × 10−4 |
| T | 940 ± 220 | 0.021 ± 0.002 | 0.022 | 0.00071 | 7.1 × 10−4 | |
| G | 430 ± 50 | 0.0045 ± 0.0002 | 0.01 | 0.00032 | 3.2 × 10−4 | |
| C | 12 ± 3 | 0.38 ± 0.02 | 31 | 1 | 1 | |
| N2-MeG | A | 790 ± 30 | 0.0027 ± 0.00004 | 0.0034 | 0.00015 | 1.1 × 10−4 |
| T | 1300 ± 200 | 0.025 ± 0.002 | 0.019 | 0.00083 | 6.1 × 10−4 | |
| G | 250 ± 30 | 0.0049 ± 0.0002 | 0.019 | 0.00083 | 6.1 × 10−4 | |
| C | 4.1 ± 1.8 | 0.092 ± 0.006 | 23 | 1 | 7.4 × 10−1 | |
| N2-BzG | A | 590 ± 90 | 0.0016 ± 0.00008 | 0.0026 | 0.0011 | 8.4 × 10−5 |
| T | 110 ± 80 | 0.00019 ± 0.00005 | 0.0016 | 0.00067 | 5.2 × 10−5 | |
| G | 50 ± 16 | 0.0016 ± 0.0001 | 0.03 | 0.0125 | 9.7 × 10−4 | |
| C | 26 ± 7 | 0.0062 ± 0.004 | 2.4 | 1 | 7.7 × 10−2 | |
| O6-MeG | A | 2200 ± 300 | 0.0036 ± 0.0002 | 0.0016 | 0.013 | 5.2 × 10−5 |
| T | 3800 ± 1200 | 0.063 ± 0.01 | 0.017 | 0.14 | 5.5 × 10−4 | |
| G | 12 ± 5 | 0.00032 ± 0.00002 | 0.027 | 0.23 | 8.7 × 10−4 | |
| C | 45 ± 12 | 0.0055 ± 0.0004 | 0.12 | 1 | 3.9 × 10−3 | |
| O6-BzG | A | 1200 ± 200 | 0.0012 ± 0.00007 | 0.001 | 0.0091 | 3.2 × 10−5 |
| T | 870 ± 40 | 0.0023 ± 0.00004 | 0.0026 | 0.024 | 8.4 × 10−5 | |
| G | 77 ± 23 | 0.00055 ± 0.00004 | 0.0065 | 0.059 | 2.1 × 10−4 | |
| C | 52 ± 20 | 0.0057 ± 0.0005 | 0.11 | 1 | 3.5 × 10−3 | |
| AP site | A | 1100 ± 200 | 0.021 ± 0.003 | 0.02 | 0.24 | 6.5 × 10−4 |
| T | 8900 ± 800 | 0.025 ± 0.002 | 0.0028 | 0.034 | 9.0 × 10−5 | |
| G | 600 ± 140 | 0.0033 ± 0.0002 | 0.0055 | 0.066 | 1.8 × 10−4 | |
| C | 27 ± 8 | 0.0022 ± 0.0002 | 0.083 | 1 | 2.7 × 10−3 |
Misinsertion frequency, calculated by dividing kcat/Km for dNTP incorporation by the kcat/Km for dCTP incorporation opposite each template base.
Relative insertion efficiency, calculated by dividing kcat/Km for dNTP incorporation opposite each template base by the kcat/Km for dCTP incorporation opposite template G.
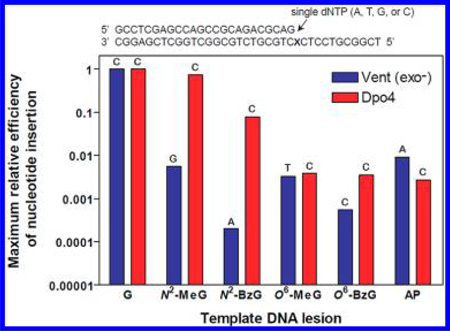
Efficiencies of nucleotide insertion varied, depending on template DNA lesions and pols. Dpo4 showed the highest kcat/ Km for correct dCTP insertion opposite N2-MeG and N2-BzG, similar to or ~13-fold lower than that opposite unmodified G. By contrast, Vent (exo−) showed about 180- and 5000-fold decreases from the highest kcat/Km for nucleotide (dGTP or dATP, respectively) insertion opposite N2-MeG and N2-BzG, respectively, as compared to that opposite G. Vent (exo−) also showed about 500- and 260000-fold decreases in kcat/Km for dCTP insertion opposite N2-MeG and N2-BzG, respectively, as compared to G, which were far greater (2−4 orders of magnitude) reductions than Dpo4. However, unlike the cases with the N2-alkylG adducts, Vent (exo−) and Dpo4 exhibited similar decreases (about 300-fold) for the highest kcat/Km in nucleotide (dTTP or dCTP, respectively) insertion opposite O6-MeG, as compared to the kcat/Km for correct dCTP insertion opposite G. There were only slight (2- or 6-fold, respectively) differences in relative efficiencies between Vent (exo−) and Dpo4 for dCTP insertion opposite O6-MeG and O6-BzG as compared to unmodified G. Vent (exo−) showed a ~1800-fold decrease in the highest kcat/Km for nucleotide (dCTP) insertion opposite O6-BzG as compared to G, which was a 6-fold greater reduction than for Dpo4, indicating a somewhat less tolerability of Vent (exo−) to major-groove adduct bulk than Dpo4. Taken together, these results indicate that Vent (exo−) favors nonbulky MeG lesions over bulky BzG adducts as template lesions for catalysis, whereas Dpo4 greatly prefers minor-groove N2-alkylG adducts to major-groove O6-alkylG adducts.
Interestingly, Vent (exo−) bypassed an AP site with ~4-fold higher relative insertion efficiency than Dpo4. Opposite an AP site, Dpo4 displayed the highest kcat/Km for insertion of dCTP among four dNTPs, which was ~370-fold lower than that opposite G but was almost identical to that opposite O6-alkylG adducts. By contrast, Vent (exo−) displayed the highest kcat/Km for insertion of dATP among four dNTPs opposite an AP site, which was about 110-fold lower than the kcat/Km for dCTP insertion opposite G but several-fold higher than that opposite N2-MeG or O6-MeG.
Selectivity of nucleotide insertion also greatly varied depending on the types of template DNA lesions and pols. Vent (exo−) preferentially incorporated the incorrect nucleotides opposite most of the lesions with varying misinsertion frequency ( fmis): dGTP ( f mis = 3) opposite N2-MeG, dATP ( f mis = 52) opposite N2-BzG, dTTP (f mis = 1.9) opposite O6-MeG, and dATP (f mis = 40) opposite AP site but with a slightly low misinsertion frequency ( f mis = 0.5 for dTTP insertion) opposite O6-BzG. In sharp contrast, Dpo4 preferentially incorporated the correct dCTP opposite all N2- and O6-alkylG adducts and AP site in a relatively error-free manner, although the misinsertion frequency was not very low (f mis = 0.2) in cases of the dGTP insertion opposite O6-MeG and the dATP insertion opposite AP site.
Steady-State Kinetics of Next-Base Extension Following an AP Site Paired with A or C by Vent (exo−) and Dpo4
Because the subsequent next-base extensions were severely retarded after relatively proficient one-base insertion opposite AP site by Vent (exo−) (Figure 2), further kinetic assays were performed to study the next-base extension steps in AP site bypass. Steady-state kinetic analyses were done with either A or C positioned opposite AP site to quantitate the efficiency for next-base extension following the preferentially inserted A or C opposite AP site by Vent (exo−) and Dpo4. The kinetic parameter kcat/Km for Dpo4 for next-base extension from the preferred AP site:C pair was about 3000-fold lower than that from the correct G:C pair (Table 4). By contrast, Vent (exo−) showed a ~830-fold decrease of kcat/Km for nextbase extension from the preferred AP site:A pair as compared to that from the correct G:C pair, indicating that Vent (exo−) has relatively low next-base extension ability past an AP site but is more competent than Dpo4.
Table 4.
Steady-State Kinetic Parameters for Next Base Extension from G:C and AP Site:A (or C) Template:Primer Termini by Vent (exo−) and Dpo4
| extension with dGTP (the next correct nucleotide against template C) |
|||||
|---|---|---|---|---|---|
| polymerase | base-pair at 3′-primer terminus (template:primer) | Km (µM) | kcat (s−1) | kcat/Km (mM−1 s−1) | relative extension effciencya |
| Vent (exo ) | G:C | 8.0 ± 1.5 | 0.042 ± 0.002 | 5.3 | 1 |
| AP:A | 1100 ± 200 | 0.0070 ± 0.0005 | 0.0064 | 1.2 × 10−3 | |
| AP:C | 15 ± 2 | 0.00014 ± 0.000005 | 0.0093 | 1.7 × 10−3 | |
| Dpo4 | G:C | 20 ± 3 | 0.089 ± 0.004 | 4.5 | 1 |
| AP:A | 390 ± 60 | 0.0026 ± 0.0001 | 0.0067 | 1.3 × 10−3 | |
| AP:C | 390 ± 70 | 0.00058 ± 0.00003 | 0.0015 | 3.3 × 10−4 | |
Relative extension efficiency, calculated by dividing kcat/Km for each dGTP incorporation opposite the next template C from the AP site:A (or C) pair by the kcat/Km for dGTP incorporation opposite the next template C from the normal G:C pair.
DISCUSSION
In this work, we examined the effects of adduct type and size of base lesions (i.e., N2- and O6-alkylations of guanines with a methyl or benzyl group and a base loss) in a DNA substrate on the DNA-binding affinity and the efficiency and fidelity of DNA synthesis by a euryarchaeal replicative B-family polymerase Vent (exo−) as compared to a crenarchaeal TLS Y-family polymerase Dpo4, by performing three main sets of experiments: (i) “standing-start” full-length primer-extension assays, (ii) electrophoretic mobility shift assays, and (iii) steady-state kinetic analyses of single nucleotide incorporation. Vent (exo−) was able to bypass the nonbulky base lesions (i.e., base-deficient AP site, major-groove O6-MeG, and minor-groove N2-MeG adducts) with modest efficiency, which is similar or slightly higher opposite the former two lesions but much lower opposite the latter than Dpo4. The adduct bulk, rather than the adduct orientation, seemed to be a more limiting factor for efficient lesion bypass by Vent (exo−), in contrast to Dpo4. Interestingly, Vent (exo−) exhibited a high proclivity for inserting incorrect nucleotides varyingly opposite those DNA lesions, whereas Dpo4 preferred correct dCTP insertion opposite all DNA lesions tested here. However, the type and bulk of DNA modifications did not affect the DNA-binding affinities much with these polymerases.
The replicative B-family pol Vent (exo−) from the hyper-thermophlic euryarchaeon T. litoralis appeared to possess substantial bypass synthesis abilities opposite the small-sized major-groove G adducts and base-lost DNA lesions, which is as or more competent as the TLS Y-family pol Dpo4 from the hyperthermophlic crenarchaeon S. solfataricus, in that the relative efficiencies of Vent (exo−) for nucleotide insertion against O6-MeG and AP site were almost equal to or slightly better than those of Dpo4 (Tables 2 and 3). Vent (exo−) seems to have modest ability to bypass the nonbulky G adducts, in either minor- or major-groove positions, because Vent (exo−) showed similar efficiency for nucleotide insertion opposite N2- MeG and O6-MeG (Table 2) and easily extended primers to full-length past those adducts (Figure 2). A proficient nucleotide insertion ability of Vent (exo−) opposite an AP site is quite noticeable (which was more competent than Dpo4) (Figure 2 and Tables 2 and 3). Moreover, the next-base extension ability of Vent (exo−) past an AP site was slightly more efficient than that of Dpo4 (Table 4). These results are in contrast with the B-family pol Dpo1 from S. solfataricus, which is blocked before an AP site and stalled at O6-MeG more severely than Dpo4.7,35 Similar to our data, polB (from the euryarchaeon Pyrococcus abyssi of the Thermococcales order) could replicate through AP sites to full-length products at a molar excess of enzyme at 55 °C, while polD stopped after nucleotide insertion at AP sites.36 DNA damage presumably occurs at a higher rate in hyperthermophiles growing in extremely high temperature than in mesophiles, as the endogenous level of AP sites has been shown to be much higher in P. abyssi than E. coli.36 The rather feasible TLS activities of Vent on nonbulky DNA lesions such as O6- methylG and AP site might be desirable for the hyperthermophilic T. litoralis to cope with those nonbulky endogenous DNA lesions frequently encountering in chromosomal DNA replication, although the 3′ to 5′ exonuclease function of wild-type Vent might partition the nascent DNA lesion:base pair into the exonuclease domain to remove the base opposite the lesion and thus diminish its TLS ability. Correspondingly, AP site bypass was enhanced by abolishing the proofreading function in P. abyssi polB,36 and thus, the down-regulation of exonuclease activities of archaeal polymerases might facilitate TLS over DNA lesions, although this phenomenon has not been described yet. We also note a very recent report that the genome of T. litoralis encodes a B-family pol Vent and a heterodimeric D-family pol but lacks a dinB- or dpo4-like Y-family pol gene,37 although euryarchaeal methanogens and haloarchaea are known to encode dinB homologue genes.9 Therefore, it can be speculated that some euryarchaea (e.g., Thermococcales) lacking Y-family pols employ the existing B- and/or D-family pols for TLS to bypass the abasic and methylated, nonbulky endogenous DNA lesions in cells, in place of a missing Y-family pol. However, bulky DNA lesions might not be overcome by B- and D-family polymerases and thus might be dealt with by novel polymerase or repair mechanisms that have not been elucidated. It would also be interesting in future studies to compare TLS functions opposite a nonbulky oxidative lesion, for example, 8-oxo-7,8-dihydroG, by Vent from the anaerobe T. litoralis and other pols from an aerobe S. solfataricus, as well as other defense systems against oxidative stress.
Both the orientation and the size of guanine adducts variably altered the nucleotide selectivity of Vent (exo−) for insertion opposite each of those lesions (Table 2), in sharp contrast to the correct dCTP preference of Dpo4 (Table 3). Consequently, replicative bypass of Vent (exo−) opposite both minor- and major-groove guanine lesions was relatively error-prone, whereas those of Dpo4 were relatively error-free. The property of preferential G misinsertion even opposite the small N2-MeG seems to be peculiar to Vent (exo−) (Table 2), because other replicative pols [e.g., bacteriophage T7 (exo−) and human immunodeficiency virus-1 reverse transcriptase (HIV-1 RT)] retain the ability to insert the correct dCTP much more efficiently than the other dNTPs opposite N2-MeG.26 The relatively similar preferences for both T and C insertions of Vent (exo−) opposite O6-alkylGs (Table 2) are similarly observed with many DNA pols such as human pols δ, η, and κ and HIV-1 RT,21,38 which might be explained by the structure of an A-family replicative pol I fragment (BF) from Bacillus stearothermophilus exhibiting a Watson−Crick-like conformation of T (or C) and O6-MeG pair in the active site.39 The high preference of dCTP for insertion opposite O6-MeG and O6-BzG by Dpo4 can be explained by a stable wobble pairing structure between C and O6-alkylGs with this polymerase,40,41 which is different from a human pol ι exhibiting the preferential selectivity of dTTP opposite O6-MeG with a Hoogsteen pairing mode.42 The more efficient bypass opposite O6-MeG by Dpo4 than BF might to be attributed to the more spacious active site of Dpo4. In contrast to BF, Vent (exo−) seems to accommodate O6-MeG in the active site as well as Dpo4, because the bypass efficiency of Vent (exo−) opposite O6-MeG was comparable to that of Dpo4 (Tables 2 and 3). However, the severe reduction of bypass ability opposite O6-BzG observed with Vent (exo−) as compared to Dpo4 (Tables 2 and 3) suggests only limited bypass ability of Vent (exo−) opposite bulky major-groove adducts, which is likely due to the more tight active site of B-family Vent (exo−).
Only Vent (exo−) but not Dpo4 obeyed the A-rule hypothesis43,44 when incorporating a nucleotide (A) opposite the noninstructional AP site (Tables 2 and 3), which was similarly observed with human B-family pols α and δ20 and A-family pols γ and θ.45,46 Although the mechanistic basis for the A-rule remains unclear, the superior base-stacking ability and weak solvation property of adenine have been proposed to have some role in selective insertion of adenine opposite a baseabsent AP site by polymerases.47 The ternary complex structure of a B-family pol from bacteriophage RB69 with the AP site:5-nitro-1-indolyl-nucleotide pair suggests a paramount role of the stacking interactions in the preferential insertion of dATP opposite an AP site.48 Interestingly, recent work reported that the large fragment of an A-family pol from Thermus aquaticus utilizes a tyrosine residue for adenine pairing in the base-vacant space.49 Dpo4 preferentially inserted C (but not A nor G) opposite AP site in our sequence context (Table 3), which does not correspond with either an A-rule or a 5′-rule (“loop-out” of an AP site) mechanism proposed previously for Dpo4 bypass of AP sites in different sequence contexts in the preceded kinetic and structural works.35,50 Thus, a Y-family pol Dpo4 seems to be capable of utilizing variable modes of nucleotide selection in replicative bypass opposite AP sites, which is dependent on local DNA sequence contexts and which appears to contrast with B-family pol Vent obedient to the A-rule.
Y-family pols appear to deal with particular DNA lesions as cognate substrates, catalyzed very efficiently in TLS. Pol η deals with cyclobutane thymidine dimer as efficiently and accurately as unmodified TT dimers.51 Our results on guanine lesion bypass with Dpo4 are in good agreement with previous reports indicating that human Y-family pols (except for pol ι) are able to handle minor-groove N2-G adducts more efficiently and accurately than major-groove O6-G adducts as template lesions, albeit with some dissimilar acceptability to lesion bulk.21,52−55 Pol κ and REV1 efficiently insert C opposite minor-groove N2-G adducts from the small N2-methylG up to the bulky N2-methyl(6-benzo[a]pyrenyl)G,52,54 whereas pol η competently inserts C opposite N2-G adducts up to N2-methyl(2-naphthyl)-G but not N2-methyl(9-anthracenyl)G.55 In contrast to minorgroove DNA lesions, there seems to be a B-family polymerase present to deal with major-groove O6-MeG and abasic lesions as moderately favored substrates in T. litoralis cells as evidenced by our data (Figure 2 and Tables 2 and 3). Similarly, the human B- and Y-family pols δ/PCNA, ι, η, and REV1, in either single or combination, are able to facilitate replicative bypass past AP sites,20 although human pols ι and θ are very efficient in the bypass of O6-MeG and AP sites, respectively.21,46 We also note a recent report suggesting the AP site as a cognate lesion of yeast REV1.56 The capacity of polymerases to accommodate the stable, productive conformation of the specific template lesion and incoming nucleotide in the active site might be critical for efficient catalysis relating to cognate lesions as substrates, as shown with pol η and a cyclobutane thymidine dimer.57,58 In the same context, the versatile properties of Dpo4 in replicative bypass of N2-guanyl adducts can be deduced from the productive conformational changes observed in structural and kinetic studies.59,60 However, the binding of polymerases to specific lesion-containing DNAs appears to contribute little to their cognate substrate selection for efficient TLS, in that different DNA modifications (i.e., O6- and N2-G alkylation and base loss) did not significantly affect dissociation constants for DNA/polymerase complex with Dpo4 or Vent (exo−) (Table 1). These results correspond with the previous reports on O6- and N2-G adducts with bacteriophage T7 (exo−) and HIV-1 RT,26,61 although AP sites (at pause sites) decreased the DNAbinding affinity of Dpo4 several fold in a different sequence context.35
In conclusion, our results suggest that T. litoralis pol Vent (exo−), one of the euryarchaeal B-family pols, can manage nonbulky DNA lesions such as an AP site, O6-MeG, and N2- MeG as template lesions for modest catalysis, albeit in error-prone manners, as efficiently or better than S. solfataricus Dpo4 opposite the former two lesions. This property of Vent might enable the error-prone TLS opposite methylated and base-depleted DNA lesions during T. litoralis genome replication in high-temperature environments, which could cause genetic mutations harmful to genome integrity but advantageous for survival.
Supplementary Material
Acknowledgments
Funding
This work was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science, and Technology (2009-0077313) (to J.-Y.C.) and United States Public Health Service (USPHS) Grant R01 ES010375 (to F.P.G.).
ABBREVIATIONS
- G
guanine
- T
thymine
- A
adenine
- C
cytosine
- Me
methyl
- Bz
benzyl
- AP
apurinic/apyrimidinic
- dNTP
deoxynucleoside triphosphate
- pol
DNA polymerase
- exo−
exonuclease-free
- TLS
translesion synthesis
- PCNA
proliferating cell nuclear antigen
- PCR
polymerase chain reaction
- HIV-1 RT
human immunodeficiency virus-1 reverse transcriptase
Footnotes
ASSOCIATED CONTENT
Supporting Information
Estimation of the apparent Kd DNA for Dpo4 and Vent (exo−) to 24-mer/36-G-mer DNA by electrophoretic mobility shift assays and comparison of the relative efficiencies of single nucleotide incorporation opposite various template DNA lesions by Vent (exo−) and Dpo4. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
REFERENCES
- 1.Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. 2nd ed. Washington, DC: American Society for Microbiology Press; 2006. [Google Scholar]
- 2.Hoeijmakers JH. DNA damage, aging, and cancer. N. Engl. J. Med. 2009;361:1475–1485. doi: 10.1056/NEJMra0804615. [DOI] [PubMed] [Google Scholar]
- 3.Guengerich FP. Interactions of carcinogen-bound DNA with individual DNA polymerases. Chem. Rev. 2006;106:420–452. doi: 10.1021/cr0404693. [DOI] [PubMed] [Google Scholar]
- 4.Rothwell PJ, Waksman G. Structure and mechanism of DNA polymerases. Adv. Protein Chem. 2005;71:401–440. doi: 10.1016/S0065-3233(04)71011-6. [DOI] [PubMed] [Google Scholar]
- 5.Rivera MC, Lake JA. The ring of life provides evidence for a genome fusion origin of eukaryotes. Nature. 2004;431:152–155. doi: 10.1038/nature02848. [DOI] [PubMed] [Google Scholar]
- 6.Böhlke K, Pisani FM, Rossi M, Antranikian G. Archaeal DNA replication: Spotlight on a rapidly moving field. Extremophiles. 2002;6:1–14. doi: 10.1007/s007920100222. [DOI] [PubMed] [Google Scholar]
- 7.Choi J-Y, Eoff RL, Pence MG, Wang J, Martin MV, Kim EJ, Folkmann LM, Guengerich FP. Roles of the four DNA polymerases of the crenarchaeon Sulfolobus solfataricus and accessory proteins in DNA replication. J. Biol. Chem. 2011;286:31180–31193. doi: 10.1074/jbc.M111.258038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boudsocq F, Iwai S, Hanaoka F, Woodgate R. Sulfolobus solfataricus P2 DNA polymerase IV (Dpo4): an archaeal DinB-like DNA polymerase with lesion-bypass properties akin to eukaryotic polη. Nucleic Acids Res. 2001;29:4607–4616. doi: 10.1093/nar/29.22.4607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin LJ, Yoshinaga A, Lin Y, Guzman C, Chen YH, Mei S, Lagunas AM, Koike S, Iwai S, Spies MA, Nair SK, Mackie RI, Ishino Y, Cann IK. Molecular analyses of an unusual translesion DNA polymerase from Methanosarcina acetivorans C2A. J. Mol. Biol. 2010;397:13–30. doi: 10.1016/j.jmb.2010.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- 11.Lu K, Craft S, Nakamura J, Moeller BC, Swenberg JA. Use of LC-MS/MS and stable isotopes to differentiate hydroxymethyl and methyl DNA adducts from formaldehyde and nitrosodimethylamine. Chem. Res. Toxicol. 2012;25:664–675. doi: 10.1021/tx200426b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Terashima I, Matsuda T, Fang TW, Suzuki N, Kobayashi J, Kohda K, Shibutani S. Miscoding potential of the N2-ethyl-2′-deoxyguanosine DNA adduct by the exonuclease-free Klenow fragment of Escherichia coli DNA polymerase I. Biochemistry. 2001;40:4106–4114. doi: 10.1021/bi002719p. [DOI] [PubMed] [Google Scholar]
- 13.Meehan T, Straub K. Double-stranded DNA steroselectively binds benzo[a]pyrene diol epoxides. Nature. 1979;277:410–412. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 14.Turesky RJ, Markovic J. DNA adduct formation of the food carcinogen 2-amino-3-methylimidazo[4,5-f]quinoline at the C-8 and N2 atoms of guanine. Chem. Res. Toxicol. 1994;7:752–761. doi: 10.1021/tx00042a007. [DOI] [PubMed] [Google Scholar]
- 15.Wang L, Spratt TE, Liu XK, Hecht SS, Pegg AE, Peterson LA. Pyridyloxobutyl adduct O6-[4-oxo-4-(3-pyridyl)butyl]guanine is present in 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanon-treated DNA and is a substrate for O6-alkylguanine-DNA alkyltransferase. Chem. Res. Toxicol. 1997;10:562–567. doi: 10.1021/tx9602067. [DOI] [PubMed] [Google Scholar]
- 16.Dipple A. DNA adducts of chemical carcinogens. Carcinogenesis. 1995;16:437–441. doi: 10.1093/carcin/16.3.437. [DOI] [PubMed] [Google Scholar]
- 17.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 18.Meng X, Zhou Y, Zhang S, Lee EY, Frick DN, Lee MY. DNA damage alters DNA polymerase delta to a form that exhibits increased discrimination against modified template bases and mismatched primers. Nucleic Acids Res. 2009;37:647–657. doi: 10.1093/nar/gkn1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schmitt MW, Matsumoto Y, Loeb LA. High fidelity and lesion bypass capability of human DNA polymerase delta. Biochimie. 2009;91:1163–1172. doi: 10.1016/j.biochi.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Choi J-Y, Lim S, Kim EJ, Jo A, Guengerich FP. Translesion synthesis across abasic lesions by human B-family and Y-family DNA polymerases α, δ, η, ι, κ, and REV1. J. Mol. Biol. 2010;404:34–44. doi: 10.1016/j.jmb.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Choi J-Y, Chowdhury G, Zang H, Angel KC, Vu CC, Peterson LA, Guengerich FP. Translesion synthesis across O6-alkylguanine DNA adducts by recombinant human DNA polymerases. J. Biol. Chem. 2006;281:38244–38256. doi: 10.1074/jbc.M608369200. [DOI] [PubMed] [Google Scholar]
- 22.Mattila P, Korpela J, Tenkanen T, Pitkanen K. Fidelity of DNA synthesis by the Thermococcus litoralis DNA polymerase—An extremely heat stable enzyme with proofreading activity. Nucleic Acids Res. 1991;19:4967–4973. doi: 10.1093/nar/19.18.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kelman Z, White MF. Archaeal DNA replication and repair. Curr. Opin. Microbiol. 2005;8:669–676. doi: 10.1016/j.mib.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Henneke G, Flament D, Hübscher U, Querellou J, Raffin JP. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J. Mol. Biol. 2005;350:53–64. doi: 10.1016/j.jmb.2005.04.042. [DOI] [PubMed] [Google Scholar]
- 25.Smith CA, Baeten J, Taylor JS. The ability of a variety of polymerases to synthesize past site-specific cis-syn, trans-syn-II, (6–4), and Dewar photoproducts of thymidylyl-(3′–>5′)-thymidine. J. Biol. Chem. 1998;273:21933–21940. doi: 10.1074/jbc.273.34.21933. [DOI] [PubMed] [Google Scholar]
- 26.Choi J-Y, Guengerich FP. Analysis of the effect of bulk at N2-alkylguanine DNA adducts on catalytic efficiency and fidelity of the processive DNA polymerases bacteriophage T7 exonuclease− and HIV-1 reverse transcriptase. J. Biol. Chem. 2004;279:19217–19229. doi: 10.1074/jbc.M313759200. [DOI] [PubMed] [Google Scholar]
- 27.Zang H, Goodenough AK, Choi J-Y, Irimia A, Loukachevitch LV, Kozekov ID, Angel KC, Rizzo CJ, Egli M, Guengerich FP. DNA adduct bypass polymerization by Sulfolobus solfataricus DNA polymerase Dpo4: Analysis and crystal structures of multiple base pair substitution and frameshift products with the adduct 1,N2-ethenoguanine. J. Biol. Chem. 2005;280:29750–29764. doi: 10.1074/jbc.M504756200. [DOI] [PubMed] [Google Scholar]
- 28.Patel SS, Wong I, Johnson KA. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991;30:511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- 29.Fiala KA, Suo Z. Pre-steady-state kinetic studies of the fidelity of Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2106–2115. doi: 10.1021/bi0357457. [DOI] [PubMed] [Google Scholar]
- 30.Joyce CM. Techniques used to study the DNA polymerase reaction pathway. Biochim. Biophys. Acta. 2010;1804:1032–1040. doi: 10.1016/j.bbapap.2009.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kong H, Kucera RB, Jack WE. Characterization of a DNA polymerase from the hyperthermophile archaea Thermococcus litoralis. Vent DNA polymerase, steady state kinetics, thermal stability, processivity, strand displacement, and exonuclease activities. J. Biol. Chem. 1993;268:1965–1975. [PubMed] [Google Scholar]
- 32.Fiala KA, Suo Z. Mechanism of DNA polymerization catalyzed by Sulfolobus solfataricus P2 DNA polymerase IV. Biochemistry. 2004;43:2116–2125. doi: 10.1021/bi035746z. [DOI] [PubMed] [Google Scholar]
- 33.Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim. Biophys. Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eoff RL, Choi J-Y, Guengerich FP. Mechanistic studies with DNA polymerases reveal complex outcomes following bypass of DNA damage. J. Nucleic Acids. 2010:1–12. doi: 10.4061/2010/830473. Article ID 830473 (DOI: 10.4061/2010/830473) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiala KA, Hypes CD, Suo Z. Mechanism of abasic lesion bypass catalyzed by a Y-family DNA polymerase. J. Biol. Chem. 2007;282:8188–8198. doi: 10.1074/jbc.M610718200. [DOI] [PubMed] [Google Scholar]
- 36.Palud A, Villani G, L'Haridon S, Querellou J, Raffin JP, Henneke G. Intrinsic properties of the two replicative DNA polymerases of Pyrococcus abyssi in replicating abasic sites: Possible role in DNA damage tolerance? Mol. Microbiol. 2008;70:746–761. doi: 10.1111/j.1365-2958.2008.06446.x. [DOI] [PubMed] [Google Scholar]
- 37.Gardner AF, Kumar S, Perler FB. Genome sequence of the model hyperthermophilic archaeon Thermococcus litoralis NS-C. J. Bacteriol. 2012;194:2375–2376. doi: 10.1128/JB.00123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woodside AM, Guengerich FP. Effect of the O6 substituent on misincorporation kinetics catalyzed by DNA polymerases at O6-methylguanine and O6-benzylguanine. Biochemistry. 2002;41:1027–1038. doi: 10.1021/bi011495n. [DOI] [PubMed] [Google Scholar]
- 39.Warren JJ, Forsberg LJ, Beese LS. The structural basis for the mutagenicity of O6-methyl-guanine lesions. Proc. Natl. Acad. Sci. U.S.A. 2006;103:19701–19706. doi: 10.1073/pnas.0609580103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eoff RL, Irimia A, Egli M, Guengerich FP. Sulfolobus solfataricus DNA polymerase Dpo4 is partially inhibited by “wobble” pairing between O6-methylguanine and cytosine, but accurate bypass is preferred. J. Biol. Chem. 2007;282:1456–1467. doi: 10.1074/jbc.M609661200. [DOI] [PubMed] [Google Scholar]
- 41.Eoff RL, Angel KC, Egli M, Guengerich FP. Molecular basis of selectivity of nucleoside triphosphate incorporation opposite O6-benzylguanine by Sulfolobus solfataricus DNA polymerase Dpo4: Steady-state and pre-steady-state kinetics and x-ray crystallography of correct and incorrect pairing. J. Biol. Chem. 2007;282:13573–13584. doi: 10.1074/jbc.M700656200. [DOI] [PubMed] [Google Scholar]
- 42.Pence MG, Choi J-Y, Egli M, Guengerich FP. Structural basis for proficient incorporation of dTTP opposite O6-methylguanine by human DNA polymerase. J. Biol. Chem. 2010;285:40666–40672. doi: 10.1074/jbc.M110.183665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Strauss B, Rabkin S, Sagher D, Moore P. The role of DNA polymerase in base substitution mutagenesis on noninstructional templates. Biochimie. 1982;64:829–838. doi: 10.1016/s0300-9084(82)80138-7. [DOI] [PubMed] [Google Scholar]
- 44.Shibutani S, Takeshita M, Grollman AP. Translesional synthesis on DNA templates containing a single abasic site. A mechanistic study of the “A rule”. J. Biol. Chem. 1997;272:13916–13922. doi: 10.1074/jbc.272.21.13916. [DOI] [PubMed] [Google Scholar]
- 45.Pinz KG, Shibutani S, Bogenhagen DF. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J. Biol. Chem. 1995;270:9202–9206. doi: 10.1074/jbc.270.16.9202. [DOI] [PubMed] [Google Scholar]
- 46.Seki M, Masutani C, Yang LW, Schuffert A, Iwai S, Bahar I, Wood RD. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J. 2004;23:4484–4494. doi: 10.1038/sj.emboj.7600424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 48.Zahn KE, Belrhali H, Wallace SS, Doublié S. Caught bending the A-rule: crystal structures of translesion DNA synthesis with a non-natural nucleotide. Biochemistry. 2007;46:10551–10561. doi: 10.1021/bi7008807. [DOI] [PubMed] [Google Scholar]
- 49.Obeid S, Blatter N, Kranaster R, Schnur A, Diederichs K, Welte W, Marx A. Replication through an abasic DNA lesion: structural basis for adenine selectivity. EMBO. J. 2010;29:1738–1747. doi: 10.1038/emboj.2010.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ling H, Boudsocq F, Woodgate R, Yang W. Snapshots of replication through an abasic lesion; structural basis for base substitutions and frameshifts. Mol. Cell. 2004;13:751–762. doi: 10.1016/s1097-2765(04)00101-7. [DOI] [PubMed] [Google Scholar]
- 51.Washington MT, Prakash L, Prakash S. Mechanism of nucleotide incorporation opposite a thymine-thymine dimer by yeast DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 2003;100:12093–12098. doi: 10.1073/pnas.2134223100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Choi J-Y, Guengerich FP. Kinetic analysis of translesion synthesis opposite bulky N2- and O6-alkylguanine DNA adducts by human DNA polymerase REV1. J. Biol. Chem. 2008;283:23645–23655. doi: 10.1074/jbc.M801686200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi J-Y, Guengerich FP. Kinetic evidence for inefficient and error-prone bypass across bulky N2-guanine DNA adducts by human DNA polymerase ι. J. Biol. Chem. 2006;281:12315–12324. doi: 10.1074/jbc.M600112200. [DOI] [PubMed] [Google Scholar]
- 54.Choi J-Y, Angel KC, Guengerich FP. Translesion synthesis across bulky N2-alkyl guanine DNA adducts by human DNA polymerase κ. J. Biol. Chem. 2006;281:21062–21072. doi: 10.1074/jbc.M602246200. [DOI] [PubMed] [Google Scholar]
- 55.Choi J-Y, Guengerich FP. Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 2005;352:72–90. doi: 10.1016/j.jmb.2005.06.079. [DOI] [PubMed] [Google Scholar]
- 56.Pryor JM, Washington MT. Pre-steady state kinetic studies show that an abasic site is a cognate lesion for the yeast Rev1 protein. DNA Repair (Amst.) 2011;10:1138–1144. doi: 10.1016/j.dnarep.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Biertumpfel C, Zhao Y, Kondo Y, Ramon-Maiques S, Gregory M, Lee JY, Masutani C, Lehmann AR, Hanaoka F, Yang W. Structure and mechanism of human DNA polymerase η. Nature. 2010;465:1044–1048. doi: 10.1038/nature09196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Silverstein TD, Johnson RE, Jain R, Prakash L, Prakash S, Aggarwal AK. Structural basis for the suppression of skin cancers by DNA polymerase η. Nature. 2010;465:1039–1043. doi: 10.1038/nature09104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang H, Eoff RL, Kozekov ID, Rizzo CJ, Egli M, Guengerich FP. Versatility of Y-family Sulfolobus solfataricus DNA polymerase Dpo4 in translesion synthesis past bulky N2-alkylguanine adducts. J. Biol. Chem. 2009;284:3563–3576. doi: 10.1074/jbc.M807778200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Guengerich FP. Effect of N2-guanyl modifications on early steps in catalysis of polymerization by Sulfolobus solfataricus P2 DNA polymerase Dpo4 T239W. J. Mol. Biol. 2010;395:1007–1018. doi: 10.1016/j.jmb.2009.11.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woodside AM, Guengerich FP. Misincorporation and stalling at O6-methylguanine and O6-benzylguanine: Evidence for inactive polymerase complexes. Biochemistry. 2002;41:1039–1050. doi: 10.1021/bi011496f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.