Abstract
Previous efforts to uncover the genetic underpinnings of ongoing ecological speciation of the M and S forms of the African malaria vector Anopheles gambiae revealed two centromere-proximal islands of genetic divergence on X and chromosome 2. Under the assumption of considerable ongoing gene flow between M and S, these persistently divergent genomic islands were widely considered to be “speciation islands”. In the course of microarray-based divergence mapping, we discovered a third centromere-associated island of divergence on chromosome 3, which was validated by targeted re-sequencing. To test for genetic association between the divergence islands on all three chromosomes, SNP-based assays were applied in four natural populations of M and S spanning West, Central and East Africa. Genotyping of 517 female M and S mosquitoes revealed nearly complete linkage disequilibrium between the centromeres of the three independently assorting chromosomes. These results suggest that despite the potential for inter-form gene flow through hybridization, actual (realized) gene flow between M and S may be substantially less than commonly assumed, and may not explain most shared variation. Moreover, the possibility of very low gene flow calls into question whether diverged pericentromeric regions-- characterized by reduced levels of variation and recombination-- are in fact instrumental rather than merely incidental to the speciation process.
Keywords: centromeres, divergent adaptation, ecological speciation, malaria vector, reproductive isolation, speciation islands
Introduction
Identifying the genetic changes causal to the speciation process—their nature, number, size and genomic distribution-- remains a central and largely unsolved puzzle in evolutionary biology (Coyne & Orr 2004; Noor & Feder 2006). One approach to uncovering candidate speciation genes is to study ecotypes not yet completely reproductively isolated, before causal differences are obscured by genetic drift and buildup of intrinsic incompatibilities (Via 2009). During ecological speciation, ecotypes arise from divergent natural selection on resource or habitat use, a process that may lead to reproductive isolation (Schluter 2001; Rundle & Nosil 2005). In the presence of gene flow, ecological speciation is expected to require strong divergent selection and may be facilitated by reduced recombination between traits affecting adaptive divergence and assortative mating (Felsenstein 1981; Via 2001; Coyne & Orr 2004). Under this model, the early stages of speciation with gene flow should be characterized by heterogeneous genomic divergence that has been termed “the genetic mosaic of speciation” (Via & West 2008; see also Nosil et al. 2009). Specifically, genomic regions that directly contribute to local adaptation and reproductive isolation should be highly diverged genetically, given strong ecologically based differential selection and consequent lack of introgression with other genetic backgrounds. The remainder of the genome, at least initially, should be subject to the homogenizing effects of gene flow (Wu & Ting 2004; Storz 2005; Butlin 2008; Via & West 2008; Nosil et al. 2009). As such, genome scanning of partially isolated ecotypes or subspecies provides a means to uncover “genomic islands of divergence” (Turner et al. 2005; Harr 2006) that may contain the genes directly contributing to ecologically based barriers to gene flow.
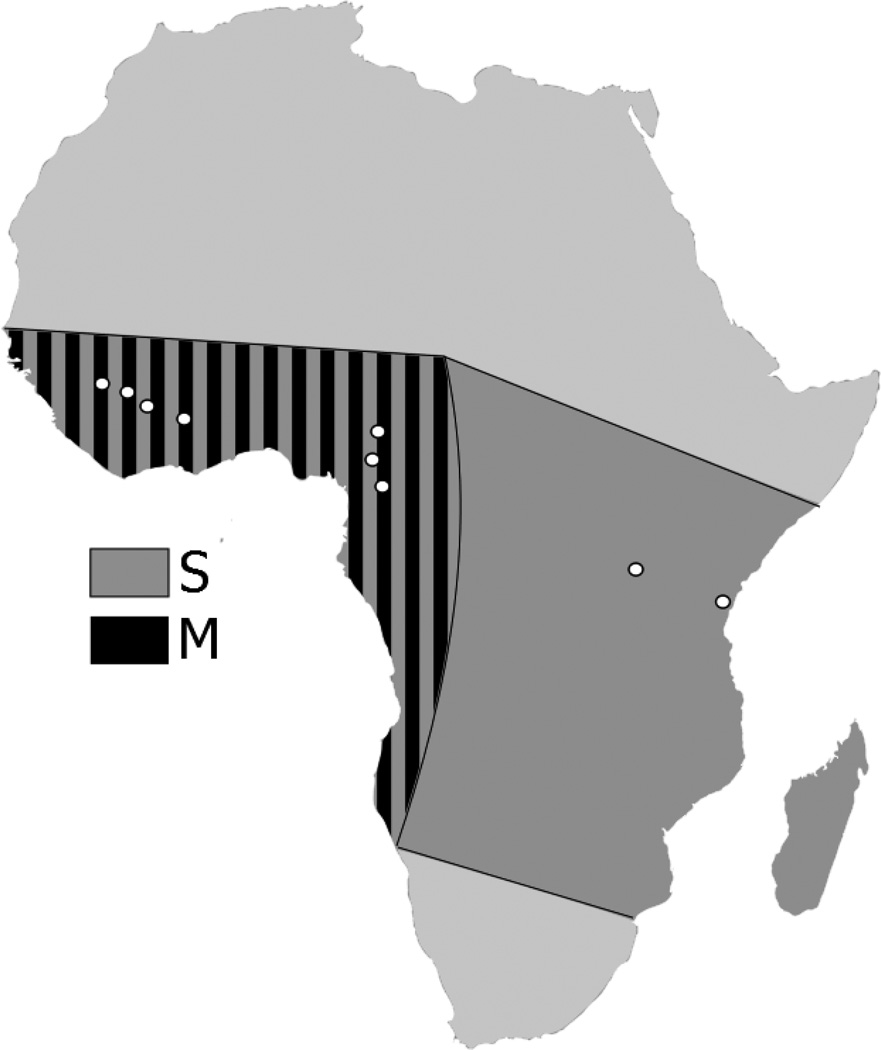
The mosquito Anopheles gambiae is the principal African vector of human malaria. It is the nominal member of the An. gambiae complex, a group of at least seven isomorphic sibling species of relatively recent and rapid origin (Powell et al. 1999). Diversification of most species in this complex is thought to reflect ecologically based divergent selection acting on the ability to exploit characteristic aquatic breeding sites (Coluzzi 1982; Coluzzi et al. 2002). Within An. gambiae sensu stricto (hereafter, An. gambiae), the process of ecological diversification and lineage splitting is ongoing (Lehmann & Diabate 2008; Manoukis et al. 2008; Costantini et al. 2009; Simard et al. 2009). Two nascent species, the M and S molecular forms, are recognized (see reviews of their identification, distribution, genetic differentiation, and ecology by Della Torre et al. 2005; Lehmann & Diabate 2008). The presumed ancestral S form is distributed across sub-Saharan Africa, and breeds only in association with the rainy season in temporary pools and puddles. The derived M form overlaps with the S form in West and Central Africa, but is absent to the east of the Great Rift Valley (Figure 1). The M form, reproductively active throughout the year, breeds in bodies of water that are more stable and more closely associated with human activity and disturbance of natural landscapes. Morphological differences are absent, and other phenotypic differences between the M and S forms are not understood in detail, but recent transplantation studies in the field have suggested that larval predator avoidance behavior and rate of development are key distinguishing factors (Lehmann & Diabate 2008). The S form develops more rapidly and outcompetes the M form in the absence of predators, consistent with a taxon adapted to short-lived aquatic habitats. However, superior predator avoidance behavior favors the M form in more permanent habitats with higher predator densities.
Figure 1.
Coarse-scale distribution of M and S forms of An. gambiae across the African continent, after Della Torre et al. (2005). Approximate location of sampling sites is indicated by white circles.
No intrinsic postmating barriers to gene flow have been reported in F1 hybrids of the M and S forms (Diabate et al. 2007), but as expected during ecological speciation, assortative mating contributes significantly to premating isolation of M and S. Field studies conducted in Mali have shown that strictly sympatric and synchronously breeding populations of M and S cross-mate at a rate of only ~1% (Tripet et al. 2001), and that mating swarms are spatially segregated with no detectable mixing (Lehmann & Diabate 2008; Diabate et al. 2009). Moreover, the incidence of F1 hybrids (as detected by an X-linked SNP) is exceedingly low in the interior of West Africa and undetected in west-central Africa (52 M/S hybrids among ~18,000 An. gambiae in the former region, none among >12,000 in the latter; Della Torre et al. 2005; Costantini et al. 2009; Simard et al. 2009). Nonetheless, the degree of reproductive isolation appears to vary geographically, as mixed mating swarms were found at a low but detectable rate in Burkina Faso (Diabate et al. 2006) and M/S hybrids have been recorded at much higher rates in westernmost West Africa (up to 7% in The Gambia and 19–24% in Guinea Bissau; Caputo et al. 2008; Oliveira et al. 2008).
If the M and S forms are in the process of ecological speciation with gene flow, it is expected that nucleotide divergence across their genomes should be highly heterogeneous, differing significantly only in gene regions directly implicated in ecological and reproductive isolation. In 2005, a groundbreaking scan of the M and S form genomes was performed (Turner et al. 2005). To map divergence, sympatric (and chromosomally homosequential) samples of An. gambiae M or S DNA from Cameroon were individually hybridized to oligonucleotide microarrays representing ~13,000 predicted genes (AgamP3.4 genebuild). Only two small genomic islands of significant divergence, in low recombination centromere-proximal regions on chromosome 2L and the X (coincident with the rDNA locus by which they are defined; Della Torre et al. 2005), were discovered and subsequently confirmed in a distant geographic location (Mali) (Stump et al. 2005; Turner et al. 2005; Turner & Hahn 2007). Termed “speciation islands”, these were predicted to contain the genes responsible for isolation between the M and S forms, and this interpretation has been widely accepted. A third non-centromeric region on chromosome 2R was diverged between M and S in Cameroon but not Mali (Turner & Hahn 2007), and thus is unlikely to contribute to a general mechanism of reproductive isolation between forms.
In the course of oligonucleotide array-based genome scans to map nucleotide divergence between alternative arrangements of An. gambiae chromosomal inversions in Mali, we serendipitously discovered a previously unmapped region of high divergence between the M and S forms abutting the centromere of chromosome 3L. By targeted resequencing of M and S from Mali and elsewhere in Africa, we confirm strong differentiation of this centromere-proximal region and provide evidence for its non-neutral evolution. Furthermore, we find strong genetic association among SNPs in all three pericentromeric islands on three independently segregating chromosomes (X, 2 and 3) across M and S populations from multiple geographic locations. We consider the implications of these findings for the role of gene flow and centromeres in the M and S speciation process.
Materials and Methods
Mosquito collection, identification, and DNA isolation
All mosquito collections consisted of indoor-resting An. gambiae s.l. adults. Figure 1 provides an overview of sampling locations for An. gambiae s.s. More specifically, mosquito genomic DNA hybridized to Affymetrix Anopheles/Plasmodium GeneChip microarrays (White et al. 2009) came from five villages in southern Mali, principally Kela (Coulibaly et al. 2007): Banambani (12°48’N, 08°03’W), Bancoumana (12°20’N, 08°20’W), Fanzana (13°20’N, 06°13’W), Kela (11°88’N, 08°45’W), and Moribabagou (12°69’N, 07°87’W). Mosquito genomic DNA used in targeted sequencing to validate the microarray results included additional population samples from these five localities, augmented by samples from two southern Malian villages: Douna (13°21’N, 5°90’W) and N’Gabakoro (12°68’N, 7°84’W). For SNP genotyping, population samples were derived from all seven Malian localities; two localities sampled in Burkina Faso in 2005: Monemtenga (12°06’N, 01°17’W) and Samandeni (11°27’N, 04°27’W); three localities sampled in Cameroon: Mfou (3°58’N, 11°56’E) in 2005, Tarem (6°41’N, 11°45’E) and Manchoutvi (5°52’N, 11°06’E) in 2007; and two localities sampled in Kenya in 1987: Asembo (0°11’S, 34°23’E) in western Kenya and Jego (4°39’S, 39°11’E) on the east coast. Anopheles quadriannulatus was collected from one locality in southern Zimbabwe in 1986 near Chilongo (Chiredzi: 21°3’S 31°4’E ; Collins et al. 1988).
Anophelines were identified as An. gambiae s.l. using morphological keys (Gillies & De Meillon 1968). DNA was isolated from individual carcasses using the DNeasy Extraction Kit (Qiagen, Valencia, CA). Sibling species and An. gambiae s.s. molecular forms were identified using diagnostic rDNA-PCR assays (Scott et al. 1993; Santolamazza et al. 2004).
Microarray hybridization and analysis
Arrays were hybridized with genomic DNA from single mosquitoes, in sets of five biological replicates per each of three different 2R homokaryotypes (15 arrays, of 5 mosquitoes x 3 homokaryotypes: 2R+, 2Rbc, and 2Rjbcu). All homokaryotypes were M form except 2Rjbcu, which was S form. Labeling and hybridization of genomic DNA followed White et al. (2007). Hybridization and scanning of the arrays was performed at the Indiana University School of Medicine.
Analysis of these data, and re-analysis of the array data from Turner et al. (2005), began by importing Affymetrix CEL files containing the raw probe intensities into Bioconductor (http:/www.bioconductor.org). After background adjustment and quantile normalization, probe level data were exported into Excel. All Anopheles probes from the Anopheles/Plasmodium GeneChip were mapped against the AgamP3 assembly and filtered to remove those that exactly matched multiple genomic locations or had secondary one-off mismatches. For each of the 151,213 retained probes, a two-tailed t-test was performed to compare the background adjusted and normalized probe intensities obtained from M versus S hybridizations. Probes whose signal intensities differed at P<0.01 between forms were considered to have single feature polymorphisms (SFPs) between the two groups (Turner et al. 2005; White et al. 2007). To test for significant clustering of SFPs across chromosome arms, we performed a sliding window analysis with a window size of 300 probes and a step-size of 20 probes. Each window was tested (χ2) for an excess of SFPs compared to the number expected based on the arm-specific frequency of significant probes. Significance was evaluated after Bonferroni correction for multiple tests (conservative because windows are correlated).
PCR, DNA sequencing, and analysis
Primers targeting exons (ideally ≥750 bp in length) were designed based on the AgamP3.4 assembly using Primer3 (Rozen & Skaletsky 2000), and custom synthesized. Introns and intergenic regions were avoided due to the likelihood of heterozygous indels. Primer pair sequences, genome coordinates, and VectorBase IDs for targeted genes are provided in Supplementary Table S1.
PCR reactions were carried out in 25µl reactions containing 200µmol/L each dNTP, 2.5mmol/L MgCl2, 2mmol/L Tris-HCL (pH 8.4), 5 mmol/L KCl, 5 pmol of each primer, 2.5U of Taq polymerase, and ~5 ng of template DNA. Thermocycler conditions were 94°C for 2 min; 35 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 1 min; a final elongation at 72°C for 10 min; and a hold at 4°C. To ensure that only the desired product was amplified, 5µl was visualized on a 1.25% agarose gel stained with ethidium bromide. To remove excess primers and dNTPs, 2U of Exonuclease 1 (USB Corporation, Cleveland, OH), 1U of Shrimp Alkaline Phosphatase (USB), and 1.8µl of ddH20 were added to 8µl of PCR product. This mixture was incubated at 37°C for 15 min, followed by 15 min at 80°C to inactivate the enzymes. The resulting products were directly sequenced on both strands using an Applied Biosystems 3730xl DNA Analyzer and Big Dye Terminator v3.1 chemistry as recommended by the manufacturer. Electropherograms were trimmed and visually inspected for heterozygous SNPs and indels using SeqMan II (DNASTAR, Madison, WI). Sequences have been deposited with GenBank under accession numbers FJ863321-FJ864270.
Measures of diversity and divergence were calculated using DnaSP 4.20.2 (Rozas et al. 2003). These included π, the average pairwise difference between sequences (Tajima 1983); θ, Watterson’s estimate of the population mutation parameter 4Neµ (Watterson 1975); Tajima’s D, a measure of skew in the frequency spectra of polymorphism (Tajima 1989); FST, the extent of population differentiation based on sequence data (Hudson et al. 1992); and Da, the net nucleotide substitutions per site between populations (Nei 1987). Significance of FST and Tajima’s D values was tested by conducting 10,000 coalescent simulations in Arlequin 3.1 (Excoffier et al. 2005).
To test whether selection was responsible for low diversity in the centromere-proximal region of 3L relative to more distal reference loci on this arm, we used polymorphism data and divergence from the sibling species An. quadriannulatus in a multilocus Hudson-Kreitman-Aguadé (HKA) framework (Hudson et al. 1987). As a first step, a standard multilocus HKA test implemented in the HKA program (http://lifesci.rutgers.edu/~heylab/heylabsoftware.htm) was performed to derive estimates of the starting values for the divergence time parameter (T) and θ. These were used to initiate a maximum likelihood extension of the HKA test, implemented with the program MLHKA (Wright & Charlesworth 2004). Maximum likelihood was used to compare the fit of the data to a neutral model of evolution at all 16 reference and 3L divergence island genes (where the selection parameter was fixed to neutral expectation, k = 1, at each gene) over models of selection at all or a subset of the 11 genes in the 3L divergence island (where k was allowed to vary freely at candidate loci). Markov chain lengths of at least 107 were used when multiple genes were hypothesized to be under selection (otherwise, chain lengths were 105), and the program was run five times with different random number seeds to test for convergence. After running MLHKA under a model in which all 11 loci in the island were hypothesized to be under selection, additional nested models were run in which selection was hypothesized only at subsets of the 11 loci that strongly deviated from neutrality in the 11-locus runs in the direction of reduced polymorphism (inferred if maximum likelihood estimates of k were ≤ 0.5).
SNP discovery and genotyping in divergence islands on 3L and 2L
We identified SNP differences between M and S forms in the 3L and 2L divergence islands which were potentially representative of the pericentromeric region and could be exploited by rapid and inexpensive PCR-RFLP assays, suitable for adoption in field-based laboratories. For the 3L island, the M and S form sequences determined from Malian samples in the course of validating sequence divergence led us to target the potential fixed SNP difference found in exon 3 of AGAP010313 at position 3L:296,923 in the AgamP3.4 assembly. Whereas the S form and three other species in the complex shared the sequence AATATC containing this position, the corresponding sequence in the M form—GATATC—is recognized by the restriction enzyme EcoRV. Universal primers were designed to amplify a 335 bp segment surrounding the (G/A)ATATC polymorphism using Primer3 (3LMSFwd: 5’-CACAGTTTGAATGGCGAAGA-3’; 3LMSRev: 5’-CCTAGTCGGTACAGCGGTTCT-3’). Accordingly, digestion with EcoRV is expected to cleave the 335 bp PCR product into two 175 bp and 160 bp fragments only in the M form.
For development of a PCR-RFLP assay targeting the 2L island, a different approach was taken for discovery of potential fixed SNPs based on the An. gambiae M and S genome sequencing project whose source material was non-inbred colonies (Mali-NIH, M form; Pimperena, S form) derived from Mali. Manual assembly of trace files (available for search and download at www.vectorbase.org) was performed on exons within the estimated bounds of this divergence island, roughly spanning coordinates 1~1.7Mb. Putatively fixed SNP differences were screened for possible restriction enzyme recognition sequences. A suitable SNP was discovered in the fourth exon of AGAP004679 at position 2L:209,536 in the AgamP3.4 assembly. Sequences from the M form were fixed for the Hpa1 recognition sequence GTTAAC, while corresponding sequences from the S-genome were fixed for ATTAAC. Using Primer3, flanking universal primers were designed to amplify a 399 bp region containing the SNP (2LMSFwd: 5’-GCATGGCAGAAAGCTGGTAT-3’; 2LMSRev: 5’-GGTCAATGCCTTCCACTGTT-3’). Digestion with HpaI should cleave this 399 bp fragment into two products of 248 bp and 151 bp exclusively in the M form.
Both sets of PCR reactions were performed under the same conditions given above for generation of sequencing templates. Upon completion, 4U of EcoRV (1U of HpaI) (New England Biolabs: NEB, Ipswich, MA), 1.22µl of 10x NEB Buffer 3 (NEB Buffer 4), and 0.8µl of ddH20 were added to 10µl of PCR product and incubated at 37°C overnight. Digestions were visualized on 1.5% agarose gels stained with ethidium bromide.
Although we found no exception to the fixed SNP differences between M and S during implementation of the 2L and 3L island PCR-RFLP assays, rare mutations (<0.1%) elsewhere in the restriction enzyme recognition sequences of the M form prevented digestion, making it appear as if a hybrid genotype had been detected in specimens heterozygous for this mutation. (Specifically, two M specimens from Samandeni, Burkina Faso carried the 2L sequence GTTAAY and two other M specimens, one from Monemtenga, Burkina Faso and the other from Moribabougou, Mali, carried the 3L sequence GRTATC, where Y = C or T and R = A or G.) A false call of “hybrid” was avoided by sequence determination of the uncleaved PCR product from all suspected hybrids, a precaution that is advised if these assays are used to investigate hybridization rates in populations. In addition, DNA extraction protocols should avoid the spermatheca, as sperm from the opposite form might also lead to falsely elevated estimates of hybrid genotypes. Finally, it must be cautioned that the single-SNP genotyping approach as a guide to population (M or S) origin of each divergence island is founded on the assumption of no (or very rare) recombination between the SNP marker and other loci in the island. This assumption appears reasonable based on low recombination rates near centromeres, and our parallel testing of additional SNP markers in the same natural populations (B. White, M. Kern and N. Besansky, unpublished data).
Results
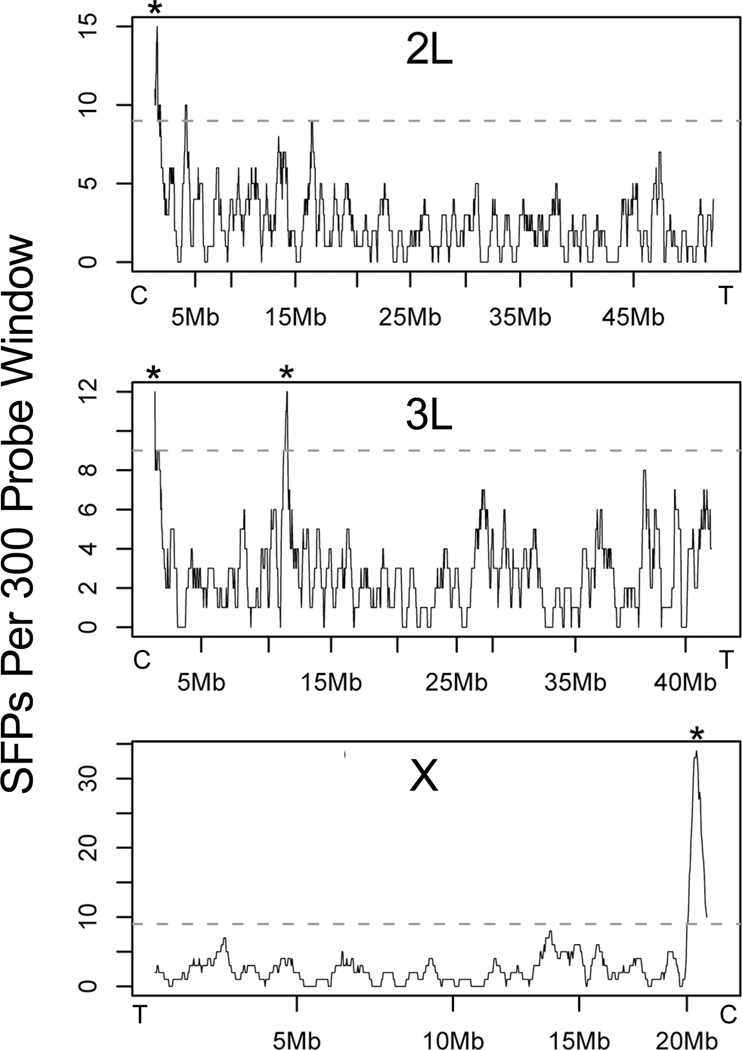
In the course of experiments aimed at mapping sequence divergence between rearrangements of chromosome 2, a third island of genomic divergence between M and S forms of An. gambiae from Mali, West Africa was serendipitously discovered. For these mapping experiments, we hybridized genomic DNA from five mosquitoes of each form to an oligonucleotide microarray platform that interrogates the entire genome with 151,213 unique 25-mer probes from predicted coding regions (for detailed methods and results, see White et al. 2007; White et al. 2009). In this approach, genomic DNA of each genetic class to be compared is fluorescently labeled and hybridized to separate microarrays. If signal intensity at a given probe—a function of the degree of nucleotide identity between target and probe—differs significantly between genetic classes, a single feature polymorphism (SFP) is declared. Any statistically significant clustering of SFPs along chromosome arms reflects the approximate location and extent of divergence between genetic classes. Although our original focus was on alternative gene arrangements, the genomic DNA hybridized to the microarrays also represented M or S form mosquitoes from the same locations in Mali, allowing for divergence mapping between the two genetic backgrounds. Sliding window analysis of divergence between M and S forms along three collinear chromosome arms (X, 2L and 3L) indicated regions in which there was a statistically significant clustering of SFPs (Figure 2). Two of these regions (on X and 2L) coincide with the divergence islands found previously in Cameroon samples of M and S forms (Turner et al. 2005), using the same microarray platform. In addition, two regions of elevated divergence on 3L were revealed. Targeted re-sequencing of the distal 3L island in samples from multiple geographic locations indicated that sequence divergence between M and S forms is geographically variable, suggesting that this gene region is unlikely to contribute to the mechanism of isolation that drove their initial divergence (unpublished data). In the present study, we focus on the centromere-proximal region of genomic differentiation on 3L, hereafter (for brevity) called the 3L divergence island.
Figure 2.
Divergence between M and S form samples from Mali based on the proportion of single feature polymorphisms (SFPs) per 300 probe window across three collinear chromosome arms. Horizontal dashed lines represent significance thresholds for each chromosome arm, after Bonferroni correction. Each of four significantly diverged regions is denoted by an asterisk. Centromeric and telomeric ends of each chromosome arm are indicated as “C” and “T”, respectively.
Sequence analysis of diversity and divergence on 3L
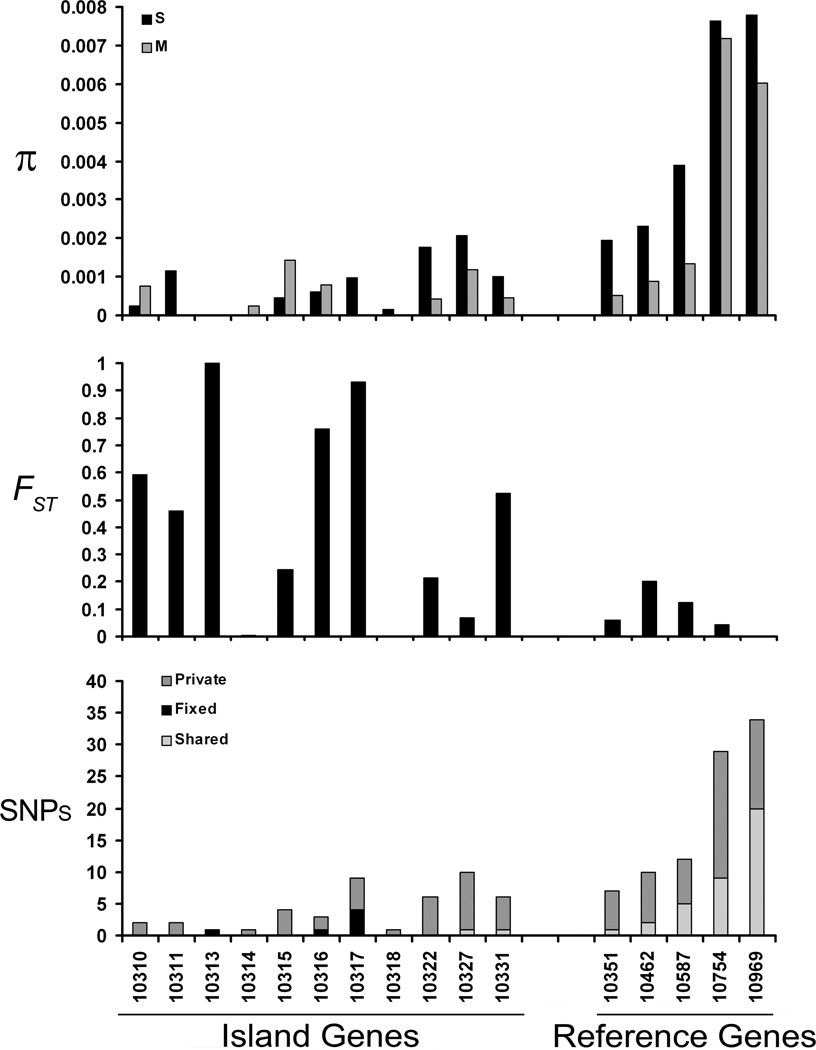
To verify the microarray pattern of higher centromere-proximal divergence on 3L, sequence was determined from 10 to 17 additional M and S form specimens collected in Mali, at 11 loci within the estimated bound of the island and five reference loci distal to this region that were undifferentiated based on the array data. Summary statistics of diversity and divergence are given in Table 1 and plotted in Figure 3.
Table 1.
Summary diversity and divergence statistics for 16 sequenced loci on chromosome 3.
| Locus ID | Len | Pos (Mb) | Form | N | S | π (%) | θ (%) | D | F:S | FST | Da (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 10310 | 630 | 0.08 | M | 24 | 1 | 0.075 | 0.043 | -- | 0:0 | 0.594 | 0.071 |
| S | 26 | 1 | 0.024 | 0.042 | -- | ||||||
| 10311 | 484 | 0.12 | M | 22 | 0 | 0 | 0 | -- | 0:0 | 0.457 | 0.062 |
| S | 36 | 2 | 0.116 | 0.100 | 0.327 | ||||||
| 10313 | 765 | 0.29 | M | 20 | 0 | 0 | 0 | -- | 1:0 | 1 | 0.131 |
| S | 36 | 0 | 0 | 0 | -- | ||||||
| 10314 | 441 | 0.35 | M | 20 | 1 | 0.023 | 0.064 | -- | 0:0 | 0.005 | 0 |
| S | 22 | 0 | 0 | 0 | -- | ||||||
| 10315 | 627 | 0.37 | M | 24 | 2 | 0.143 | 0.085 | 1.475 | 0:0 | 0.245 | 0.03 |
| S | 26 | 2 | 0.046 | 0.084 | −0.960 | ||||||
| 10316 | 636 | 0.39 | M | 24 | 1 | 0.08 | 0.042 | -- | 1:0 | 0.758 | 0.218 |
| S | 28 | 1 | 0.061 | 0.04 | -- | ||||||
| 10317 | 591 | 0.41 | M | 22 | 0 | 0 | 0 | -- | 4:0 | 0.931 | 0.678 |
| S | 24 | 5 | 0.096 | 0.227 | −1.667 | ||||||
| 10318 | 522 | 0.42 | M | 24 | 0 | 0 | 0 | -- | 0:0 | 0 | 0 |
| S | 26 | 1 | 0.015 | 0.05 | -- | ||||||
| 10322 | 702 | 0.67 | M | 20 | 3 | 0.043 | 0.12 | −1.723 | 0:0 | 0.214 | 0.033 |
| S | 26 | 3 | 0.175 | 0.112 | 1.388 | ||||||
| 10327 | 838 | 0.87 | M | 22 | 2 | 0.12 | 0.098 | 0.570 | 0:1 | 0.069 | 0.013 |
| S | 34 | 9 | 0.207 | 0.263 | −0.643 | ||||||
| 10331 | 742 | 1.24 | M | 22 | 2 | 0.046 | 0.074 | −0.871 | 0:1 | 0.522 | 0.086 |
| S | 34 | 5 | 0.099 | 0.165 | −1.056 | ||||||
| 10351 | 726 | 1.95 | M | 22 | 2 | 0.051 | 0.038 | 0.593 | 0:1 | 0.061 | 0.009 |
| S | 26 | 6 | 0.195 | 0.217 | −0.301 | ||||||
| 10462 | 809 | 3.93 | M | 22 | 3 | 0.087 | 0.102 | −0.669 | 0:2 | 0.203 | 0.043 |
| S | 26 | 9 | 0.232 | 0.291 | −0.382 | ||||||
| 10587 | 786 | 6.55 | M | 22 | 5 | 0.133 | 0.175 | −0.702 | 0:5 | 0.126 | 0.038 |
| S | 22 | 12 | 0.390 | 0.454 | −0.237 | ||||||
| 10754 | 756 | 9.84 | M | 12 | 16 | 0.719 | 0.701 | 0.115 | 0:9 | 0.045 | 0.036 |
| S | 22 | 22 | 0.764 | 0.798 | −0.162 | ||||||
| 10969 | 767 | 14.05 | M | 22 | 22 | 0.601 | 0.787 | −0.888 | 0:20 | 0 | 0 |
| S | 22 | 33 | 0.779 | 1.180 | −1.230 |
Locus ID, last 5 digits of VectorBase Gene ID (AGAP0XXXXX); Len, length in base pairs; Pos, physical position (in Mb) on chromosome 3L; N, number of chromosome sequenced; S, number of segregating sites; π, average number of pairwise differences per site (%); θ, expected heterozygosity per site (%) based on the number of segregating sites; D Tajima’s D based on total number of mutations (not calculated if S<2); F:S, ratio of fixed differences to shared polymorphisms; Da: average net nucleotide divergence per site (%); FST, estimate of differentiation. Bolded loci reside in the predicted divergence island.
Figure 3.
Estimates of nucleotide diversity within M and S form samples from Mali and genetic differentiation between them at each of 11 loci in the divergence island and 5 distally located reference loci on 3L. Only the last five digits VectorBase gene IDs are given.
For both forms, the per-site average pairwise nucleotide diversity of 11 genes located within the 3L island (π = 0.00076 and 0.00048 for S and M form, respectively) was more than six-fold lower compared to five genes outside the island (π = 0.00472 and 0.00318 for S and M). When diversity was estimated from the number of segregating sites (θ), the same pattern of more than six-fold decreased variation was observed in the centromere-proximal genes relative to the reference genes for both forms (θ = 0.00098 and 0.00048 for S and M form inside; θ = 0.00588 and 0.00361 for S and M form outside). For both measures of diversity in both M and S forms, the difference was significant (Wilcoxon Rank-Sum Test: for π, one-tailed P = 0.001 and 0.011 for S and M; for θ, one-tailed P = 0.002 and 0.015 for S and M). Of note, levels of polymorphism in the M form were consistently lower than those of the S form both within and outside of the island (Wilcoxon Sign-Rank test, P = 0.0006), as observed on 2R in Cameroon (Turner & Hahn 2007).
Levels of divergence between M and S also contrasted in centromere-proximal versus reference loci. Both the proportion of net nucleotide substitutions per site, Da, and the magnitude of FST values were more than five-fold higher for the set of loci within the predicted 3L divergence island compared to those residing distally. Within the island, mean FST was 0.436, compared to the reference loci for which the corresponding value was 0.087, a significant difference (Wilcoxon Rank Sum, P = 0.031). Similarly, the pattern of fixed differences and shared polymorphisms between forms reflected elevated divergence at the 3L island relative to control loci. In the set of 11 island loci, we observed 6 fixed nucleotide differences and only two shared polymorphisms between M and S compared with no fixed differences and 37 shared polymorphisms across five distal loci (Fisher’s exact test; P = 3.4 × 10−6).
Taken together, these results validate the microarray data. They indicate heterogeneous diversity and divergence on 3L involving a substantial reduction in gene flow between forms near the centromere, particularly within 500 kb of the centromere where all six fixed differences (and no shared polymorphisms) were located.
Selection in the 3L divergence island
Centromere-proximal regions are commonly subject to very low rates of crossing over, including those of An. gambiae (Pombi et al. 2006). Strong correlations between low recombination and low nucleotide diversity have been noted previously, in An. gambiae and a variety of organisms (Begun & Aquadro 1992; Lercher & Hurst 2002; Stump et al. 2005; Wright et al. 2005; Begun et al. 2007). In principle, this correlation can be caused by selection or neutral forces such as a lower mutation rate in low recombination regions. Significantly negative Tajima’s D values at centromere-proximal genes, indicative of a skew toward low frequency mutations, would be consistent with hitchhiking due to positive selection (Tajima 1989). However the paucity of segregating sites near the 3L centromere either prevented calculation of this statistic or severely reduced its power (Table 1).
An alternative approach to test for selection is to evaluate intraspecific diversity relative to interspecific divergence in low recombination (centromere-proximal) versus free recombination (distal) regions, in light of the neutral equilibrium prediction that diversity and divergence levels should be positively correlated (Hudson et al. 1987). This expectation was evaluated separately for the M and S forms using the sibling species An. quadriannulatus A as the outgroup. We employed a maximum likelihood implementation of the HKA test (Wright & Charlesworth 2004) to assess the fit of the data to a neutral model in which the ratios of polymorphism to divergence are the same at all 16 genes, versus the fit to models in which genes located in the divergence island are under selection (Table 2). A model with all eleven centromere-proximal genes under selection did not fit the data significantly better than the neutral model (for M, P=0.26; for S, P=0.31). However, models with a subset of divergence island genes under selection fit the data significantly better than the neutral model. Consistent with recent divergent selection acting on the two forms, only one (AGAP010313) of five genes inferred to be under selection in either form was in common to both.
Table 2.
Maximum-likelihood analysis of polymorphism on 3L in M and S forms relative to divergence from An. quadriannulatus
|
k for island genes |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | ||||||
| 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | ||||||
| 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | ||||||
| 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 2 | 3 | ||||||
| Form | Model | ln L | Contrast | LR (df) | P | 0 | 1 | 3 | 4 | 5 | 6 | 7 | 8 | 2 | 7 | 1 |
| M | A. Neutral | −63.2 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| B. Selection, 11 loci | −56.4 | A vs. B | 13.5 (11) | NS | 1.2 | 2.0 | 0 | 0 | 0.37 | 0.94 | 0 | 0.98 | 0.54 | 0.58 | 1.8 | |
| C. Selection, 4 loci | −56.0 | A vs. C | 14.4 (4) | 0.006 | 1 | 1 | 0 | 0 | 0.57 | 1 | 0 | 1 | 1 | 1 | 1 | |
| D. Selection, 3 loci | −55.8 | A vs. D | 14.6 (3) | 0.002 | 1 | 1 | 0 | 0 | 1 | 1 | 0 | 1 | 1 | 1 | 1 | |
| C vs. D | NS | |||||||||||||||
| S | A. Neutral | −69.4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | |||
| B. Selection, 11 loci | −63.1 | A vs. B | 12.7 (11) | NS | 0.69 | 0 | 0 | 2.3 | 1.5 | 0.17 | 1.5 | 0.28 | 0.66 | 1.0 | 2.7 | |
| C. Selection, 4 loci | −62.4 | A vs. C | 14.0 (4) | 0.007 | 1 | 0 | 0 | 1 | 1 | 0.13 | 1 | 1.2 | 1 | 1 | 1 | |
| D. Selection, 3 loci | −62.0 | A vs. D | 14.7 (3) | 0.002 | 1 | 0 | 0 | 1 | 1 | 0.05 | 1 | 1 | 1 | 1 | 1 | |
| C vs. D | NS | |||||||||||||||
LR, likelihood ratio statistic; P, probability of likelihood ratio test assuming the χ2 distribution (NS, not significant); k, selection parameter for genes identified by the last 5 digits of the VectorBase IDs.
Due in part to its high degree of specialization on humans, An. gambiae is considered one of the most recently derived members of a sibling species complex containing at least six other members (Coluzzi et al. 2002). To infer the ancestral state of the six SNPs fixed between M and S at three loci in the 3L divergence island (AGAP010313, −10316, −10317; Table 1), sequences were determined at these loci from three other species in the complex: An. quadriannulatus, An. arabiensis, and An. merus (Table 3). At each of the six positions, all three outgroup species carried the same presumably ancestral nucleotide. This suggests that differences fixed between M and S may have arisen de novo. Following the pattern recorded previously at the X chromosome divergence island in M and S (Stump et al. 2005), the ancestral state was not invariably associated with one molecular form—not even at the level of a single gene. Instead, the S form carried the ancestral state at four positions while the M form carried the ancestral state in the other two cases. Within a segment of AGAP010317 with four fixed SNPs between M and S, the ancestral state was found in M at one position and in S at three others.
Table 3.
Character state at sites with fixed differences between M and S forms compared to three other species in the An. gambiae complex.
| Site | M form | S form | QUA | ARA | MER |
|---|---|---|---|---|---|
| 296923 | G | A | A | A | N/A |
| 387240 | G | A | G | G | G |
| 413508 | A | G | A | A | A |
| 413550 | G | A | A | A | A |
| 413551 | T | A | A | A | A |
| 413944 | T | C | C | C | C |
Site, physical position in the AgamP3.4 assembly of the PEST reference genome on chromosome 3L; QUA, ARA, and MER are An. quadriannulatus An. arabiensis, and An. merus, respectively; N/A, not sequenced.
Range-wide genotyping of divergence islands on X, 2L and 3L
To test for genetic association between divergence islands on the three independently assorting chromosomes, we developed SNP-based PCR-RFLP genotyping assays for 2L and 3L analogous to existing assays for the X island (Fanello et al. 2002; Santolamazza et al. 2004). Using all three assays, we genotyped each divergence island in 497 An. gambiae collected from Mali, Burkina Faso, and Cameroon where the two forms are sympatric, along with 20 additional specimens from Kenya where only the S form is found. In 512 of 517 An. gambiae adult females analyzed (99%), complete genetic association existed between the three unlinked divergence islands: 275 M-form and 237 S-form mosquitoes carried the expected homozygous alleles on all three chromosomes and could be considered “pure” parental types (Table 4). All five exceptional genotypes (putative hybrids) were sampled in Burkina Faso. Of the five, three were heterozygous for M and S alleles at each of the three islands and likely represent F1 hyrbids between the two forms. The two other exceptions carried genotypes consistent with backcross progeny (from a mating between an F1 hybrid and pure parental form): one specimen was S-like at the X and 2L islands, but heterozygous in the 3L island; another specimen was M-like for the X and 3L islands, but heterozygous for the 2L island. In addition to An. gambiae, we genotyped 46 An. arabiensis from Mali and found only S-like alleles in all three divergence islands. This result is consistent with our previous suggestion that differences fixed between M and S seem to have arisen de novo, not through shared ancestral polymorphism or interspecific gene flow.
Table 4.
Diploid genotypes recorded at three divergence islands (X/2L/3L) in adult female An. gambiae s.s. collected in four African countries
| Parental genotypes |
Assorted genotypes |
||||
|---|---|---|---|---|---|
| Country | MM/MM/MM | SS/SS/SS | MS/MS/MS | SS/SS/MS | MM/MS/MM |
| Mali | 108 | 40 | 0 | 0 | 0 |
| Burkina | 114 | 57 | 3 | 1 | 1 |
| Cameroon | 53 | 120 | 0 | 0 | 0 |
| Kenya | 0 | 20 | 0 | 0 | 0 |
| Total | 275 | 237 | 3 | 1 | 1 |
| 512 (99.03%) parentals | 5 (0.97%) non-parentals | ||||
Discussion
Before this study, only two small islands of genomic divergence near the centromeres of independent chromosomes, the X and chromosome 2, were known to distinguish M and S forms of An. gambiae over a wide geographic area (Turner et al. 2005). Based on variable rates of hybridization observed between M and S in nature, the widely accepted interpretation has been that “these ‘speciation islands’ remain differentiated despite considerable gene flow, and are therefore expected to contain the genes responsible for reproductive isolation” (Turner et al. 2005). Our discovery of a third centromere-proximal region of elevated nucleotide differentiation on chromosome 3 is not surprising, and does not—by itself—alter the prevailing assumptions about hybridization and its consequences for the architecture of genomic divergence. However, the discovery of nearly perfect genetic association of SNPs near the centromeres of all three independently assorting chromosomes does prompt a critical reassessment of inter-form gene flow. In particular, does the potential for gene flow presented by appreciable numbers of M/S hybrids necessarily imply successful genetic introgression (i.e., actual or realized inter-form gene flow), and if not, what can we conclude about the role of centromere-associated sequences as speciation islands?
Speciation in the presence of substantial current gene flow?
Our results raise the fundamental question of how the centromeres of three independently assorting chromosomes remain associated given the potential for inter-form gene flow. Perhaps the simplest answer is the complete absence or extreme rarity of inter-form mating. Although M/S hybrids have not been observed in Cameroon despite intense sampling effort, this explanation does not apply more broadly, given that cross-mating has been observed in Mali (based on insemination by the opposite form) and putative F1 and backcross hybrids have been repeatedly sampled in nature from various parts of West Africa. Alternative explanations allow hybridization between forms, but invoke postmating barriers to reduce or eliminate gene flow. Among possible intrinsic (genetic) postmating barriers to gene flow, incompatibility of F1 or backcross hybrids probably can be ruled out. Controlled crosses of wild-collected specimens revealed no difference between hybrid versus parental classes in egg production, hatch rate, larval development rate, sex ratio or sex organ morphology (Diabate et al. 2007). Moreover, intrinsic incompatibility of F2 hybrids also appears unlikely, given the recovery and viability of all genotypic classes in the expected proportions (unpublished data). Indeed, recovery of all hybrid genotypes casts doubt on other intrinsic mechanisms that could explain the association of unlinked divergence islands in the face of hybridization, such as meiotic drive (Henikoff et al. 2001) and complex centromeric translocations involving all three chromosomes. Remaining among possible postmating barriers to gene flow are extrinsic hybrid inviability due to ecologically-based divergent selection, and/or reduced mating success of hybrids. These general mechanisms are central to ecological speciation and may provide the most plausible hypotheses to explain the observed genetic associations. Unfortunately, ecological studies of M and S forms are rare, and evidence for either ecological maladaptation or sexual isolation of M/S hybrids is lacking. Accordingly, the degree to which selection against hybrid genotypes might limit introgression remains unclear. The development of informative population genetic models of this process is complicated by the likelihood that non-equilibrium conditions (demography, selection, and hybridization between M and S) operate in a spatially and temporally heterogeneous fashion which may or may not reflect historical conditions.
Pericentric islands of divergence: instrumental or incidental to speciation?
Centromeric regions are associated with strongly reduced recombination in a variety of organisms including An. gambiae (Kong et al. 2002; Pombi et al. 2006; Slotman et al. 2006). The connection between reduced recombination, adaptation and speciation has been highlighted recently with respect to chromosomal inversions (Noor et al. 2001; Rieseberg 2001; Ortiz-Barrientos et al. 2002; Navarro & Barton 2003; Butlin 2005; Hoffmann & Rieseberg 2008), tempting an analogy to pericentric regions (Stump et al 2005). Yet, the analogy between centromeres and inversions can only go so far. At least in Dipteran flies (e.g., drosophilids, simulids, and culicids) where chromosomal rearrangements have played an important role in evolutionary diversification (Hoffmann et al. 2004; Coghlan et al. 2005; Hoffmann & Rieseberg 2008), chromosome number is small—only three pairs in mosquitoes. As such, the random chance that a given chromosomal arrangement (of sufficient size) happens to capture a set of locally adapted genes is relatively high. The cytologically visible inversions naturally maintained as polymorphisms in Drosophila and Anopheles species tend to span several Mb, implying that many hundreds of genes within (and to some extent, outside of) the breakpoints are affected by reduced recombination, though only between and not within arrangement classes. In contrast, pericentromeric regions subject to suppressed recombination are generally much smaller in size and less gene-dense. For example, in Drosophila and Anopheles euchromatin, gene density is 11 and 5 per 100 kb, respectively, while the corresponding number in pericentromeric regions is 2 per 100 kb (Sharakhova et al. 2007, and refs therein). Recombination (crossing over) in these regions is persistently suppressed, contributing to low nucleotide diversity. Taken together, these factors-- low levels of standing variation, low recombination, relatively small size and low gene density—suggest that pericentromeric regions are a less likely source of gene-based local adaptations than chromosomal inversions. Moreover, these low recombination regions are subject to shorter coalescence times because they are subject to selection at linked sites, thereby increasing the likelihood that they would appear as outliers in a scan of genomic divergence regardless of their relationship to speciation. This point is notable, because the coalescent simulations performed by Turner et al (2005) ruled out reduced levels of variation and recombination rate near centromeres as sole factors responsible for observed differences between M and S, implicating selection. However, depending upon the level of gene flow within and between forms, any selective sweeps experienced by these pericentromeric regions could have been independent of, or incidental to, the speciation process. In other words, the “speciation islands” could be by-products, rather than drivers, of the isolation process, in which case the component genes would not necessarily include those causal to speciation.
Pericentric heterochromatin, unlike chromosomal inversions, is relatively enriched in repetitive DNA. It remains possible that changes in repeat content or sequence, which can be rapid near centromeres (Henikoff et al. 2001), may play a role in speciation of M and S and may be responsible for the strong genetic association between the pericentromeric islands on X, 2 and 3. It may also be important to consider epigenetic effects of centromeric heterochromatin in the process of speciation. Although incompletely understood, it is known that the spatial arrangement of chromosomes in the nucleus—in particular, the proximity of chromosomal regions to the nuclear periphery-- can influence levels of transcriptional activity as well as the timing of DNA replication, and the rate of recombination and repair (Akhtar & Gasser 2007; Misteli 2007). In this regard, it may be significant that the pericentric heterochromatin of all five chromosome arms in An. gambiae attaches to the nuclear envelope, in distinction to its congener An. funestus (Sharakhov et al. 2001). Although the spatial localization and morphology of chromosomal regions that attach to the nuclear envelope may evolve relatively rapidly and appear to be species-specific in another set of cryptic and closely related anopheline species (the Anopheles maculipennis complex; Stegnii 1987), differences in the nuclear architecture of An. gambiae M and S chromosomes have not been examined to our knowledge.
Reproducibility of divergence mapping by microarray
The same Affymetrix GeneChip platform adopted in this study was applied by Turner et al (2005) for divergence mapping between M and S genomes, using samples from Mali or Cameroon, respectively. Two differences between the studies in non-centromeric regions (an island of divergence on 2R only in Cameroon, and an island on 3L only in Mali) have been validated by population re-sequencing, and can be explained by geographically variable signatures of selection (Turner & Hahn 2007; B. White et al., unpublished). No other discrepancies were found when both data sets were mapped to a common An. gambiae (AgamP3) assembly (data not shown). Both the apparent absence of the 3L pericentromeric divergence island and the apparently smaller size of the X chromosome island as reported by Turner et al (2005) were artifacts resulting from use of the previous An. gambiae genome assembly (MOZ2) available at that time, which was deficient especially in regions of pericentric heterochromatin (Sharakhova et al. 2007). Thus, all three pericentromeric divergence islands (on chromosomes X, 2 and 3) are present and indistinguishable in size between Cameroon and Mali samples, regardless of differences in geographic location, chromosomal inversion composition, and population structure. Based on the improved AgamP3 assembly and AgamP3.4 gene build, the best available GeneChip-derived estimates for the number and size of consistently diverged pericentric islands between M and S is three: the X island at ~4 Mb (49 genes), the 2L island at ~2.5 Mb (35 genes), and the 3L island at ~1.7 Mb (30 genes). Taken together, the three islands comprise ~3% of the ~260 Mb genome (Besansky & Powell 1992) and ~1% of the ~13,000 predicted genes.
Sensitivity of divergence mapping by microarray
Notably absent are indications of significant divergence at the pericentromeric regions on the right arms of chromosomes 2 and 3. (The X chromosome is referred to as acrocentric, as one entirely heterochromatic arm does not polytenize; hence only one pericentromeric region is distinguishable). This observation is almost certainly an artifact, given that the whole centromere acts as a single locus; it could be explained by incompletely assembled pericentromeric regions (Sharakhova et al. 2007). For this same reason, estimates of length and gene numbers in pericentromeric regions should be considered as underestimates, and we cannot dismiss the possibility that other islands of differentiation exist but have gone unmapped for technical reasons. Nevertheless, based on available tools-- the Anopheles/Plasmodium microarray platform and the current genome assembly-- there is no sign of other genomic islands of divergence between the M and S forms.
Based on their studies of speciation genetics in host races of the pea aphid, Via & West (2008) recently introduced the mechanism of “divergence hitchhiking” to explain unexpectedly large (~10 centiMorgan) regions of differentiation in the presence of gene flow. In essence, reduced inter-race mating works together with negative selection to reduce the effective rate of recombination between races in chromosomal regions containing strongly selected isolating genes; within races, normal recombination can continue (Smadja et al. 2008). If such a scenario were operational in the divergence of M and S forms of An. gambiae, whereby loci involved in reproductive isolation are contained in differentiated regions as large as 10 cM (≈5 Mb; Zheng et al. 1996; Stump et al. 2007), the signal(s) should have been unmistakable by microarray, given that this array was sensitive enough to map a differentiated region on 2R in Cameroon spanning only ~37 kb (Turner et al. 2005). On the other hand, smaller islands of genomic divergence between M and S could have been missed both in our study and that of Turner et al. (2005). The Anopheles/Plasmodium array scans the genome only in predicted coding regions, and its ~150,000 unique Anopheles probes are not tiled across the genome. Thus, in high recombination regions of the genome, single “speciation genes” could escape detection for two reasons: because they were not represented on the array, or because the number of differentiated probes per 300-probe sliding window was too small to detect statistically.
Whither the speciation islands?
Due to limitations of microarray design and statistical power, the real possibility exists of undetected speciation genes elsewhere in the genome. If so, they are expected to be in freely recombining regions that are not recognizable as “speciation islands”, and microarray-based divergence mapping would be a poor approach to locate them. Where does this leave the centromere-proximal “speciation islands” sensu Turner et al. (2005)? We have argued that special features inherent to centromere-proximal regions caution against the assumption that any of them must be speciation islands on the basis of significant differentiation, even if selection is implicated. Does this mean that none of the three pericentric regions are potential focal points of M and S ecological speciation? Not necessarily. At 4 Mb, the divergence island near the centromere of the X chromosome is the largest by far (49 genes). Some theoretical models suggest that the X chromosome plays a disproportionate role in behavioral isolation, and there is empirical evidence suggesting that species differences map disproportionately to the X chromosome (Coyne & Orr 2004). In the case of An. gambiae and its sibling species An. arabiensis, the X chromosome plays a large role in hybrid male and female sterility (Curtis 1982; Slotman et al. 2004, 2005). Consistent with a role for the X island in differences between M and S, this one (alone among the three islands) carried a significant excess of genes differentially expressed between forms (Cassone et al. 2008). Accordingly, further dissection of the X island seems warranted, although the process is greatly hindered by the absence of known phenotypic differences.
Our results suggest that realized gene flow between M and S forms of An. gambiae may be far less than previously assumed, and that unraveling the genetic basis of their isolation will be more difficult than was envisioned in 2005 (Butlin & Roper 2005). Nevertheless, the importance of this task goes beyond the goals of speciation genetics. In the case of the medically important M and S forms, it is also hoped that improved understanding of speciation mechanisms occurring at the genetic level will help guide our understanding of gene flow and population structure, without which vector control strategies will be difficult to implement successfully across sub-Saharan Africa.
Supplementary Material
Acknowledgements
The authors are grateful to M.W. Hahn for advice, and to T. Turner for sharing raw microarray data for reanalysis. A. Caccone and anonymous reviewers suggested significant improvements to the manuscript. We thank M. Kern for assistance genotyping specimens and N. Lobo for running sequencing plates. We also thank the scientists and entomological teams for assistance with mosquito collections in Cameroon and Burkina Faso, including C. Kamdem, J.-P. Agbor, N.’F. Sagnon and W. Guelbeogo. M. Coulibaly and S. Traoré collected specimens from Mali for a previous project and kindly made these available for continued analysis. F. Collins generously supplied all An. quadriannulatus and An. merus in addition to the An. gambiae from Kenya. Z. Chunxiao and R. Jerome performed the microarray hybridizations under the direction of H. Edenberg at the Center for Medical Genomics, Indiana University School of Medicine. This work was funded by NIH grant AI63508 to NJB. BJW was supported by an Arthur J. Schmidt Fellowship from the University of Notre Dame. The Center for Medical Genomics is funded in part by the Indiana Genomics Initiative of Indiana University (INGEN); INGEN is funded in part by The Lily Endowment, Inc.
References
- Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Aquadro CF. Levels of naturally occurring DNA polymorphism correlate with recombination rates in D. melanogaster . Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- Begun DJ, Holloway AK, Stevens K, Hillier LW, Poh YP, Hahn MW, Nista PM, Jones CD, Kern AD, Dewey CN, Pachter L, Myers E, Langley CH. Population genomics: Whole-genome analysis of polymorphism and divergence in Drosophila simulans . PLoS Biol. 2007;5:e310. doi: 10.1371/journal.pbio.0050310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besansky NJ, Powell JR. Reassociation kinetics of Anopheles gambiae (Diptera: Culicidae) DNA. J Med Entomol. 1992;29:125–128. doi: 10.1093/jmedent/29.1.125. [DOI] [PubMed] [Google Scholar]
- Butlin R, Roper C. Evolutionary genetics: microarrays and species origins. Nature. 2005;437:199–201. doi: 10.1038/437199a. [DOI] [PubMed] [Google Scholar]
- Butlin RK. Recombination and speciation. Mol Ecol. 2005;14:2621–2635. doi: 10.1111/j.1365-294X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Butlin RK. Population genomics and speciation. Genetica. 2008 doi: 10.1007/s10709-008-9321-3. [DOI] [PubMed] [Google Scholar]
- Caputo B, Nwakanma D, Jawara M, Adiamoh M, Dia I, Konate L, Petrarca V, Conway DJ, della Torre A. Anopheles gambiae complex along the gambia river, with particular reference to the molecular forms of An. gambiae s.s. . Malar J. 2008;7:182. doi: 10.1186/1475-2875-7-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassone BJ, Mouline K, Hahn MW, White BJ, Pombi M, Simard F, Costantini C, Besansky NJ. Differential gene expression in incipient species of Anopheles gambiae . Mol Ecol. 2008;17:2491–2504. doi: 10.1111/j.1365-294X.2008.03774.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coghlan A, Eichler EE, Oliver SG, Paterson AH, Stein L. Chromosome evolution in eukaryotes: a multi-kingdom perspective. Trends Genet. 2005;21:673–682. doi: 10.1016/j.tig.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Collins FH, Mehaffey PC, Rasmussen MO, Brandling-Bennett AD, Odera JS, Finnerty V. Comparison of DNA-probe and isoenzyme methods for differentiating Anopheles gambiae and Anopheles arabiensis (Diptera: Culicidae) J Med Entomol. 1988;25:116–120. doi: 10.1093/jmedent/25.2.116. [DOI] [PubMed] [Google Scholar]
- Coluzzi M. Spatial distribution of chromosomal inversions and speciation in anopheline mosquitoes. In: Barigozzi C, editor. Mechanisms of speciation. New York: Alan R. Liss, Inc; 1982. pp. 143–153. [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415–1418. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Costantini C, Ayala D, Guelbeogo WM, Pombi M, Some CY, Bassole IHN, Ose K, Fotsing J-M, Sagnon NF, Fontenille D, Besansky NJ, Simard F. Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae . BMC Ecology. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly MB, Pombi M, Caputo B, Nwakanma D, Jawara M, Konate L, Dia I, Fofana A, Kern M, Simard F, Conway DJ, Petrarca V, Della Torre A, Traore S, Besansky NJ. PCR-based karyotyping of Anopheles gambiae inversion 2Rj identifies the Bamako chromosomal form. Malar J. 2007;6:133. doi: 10.1186/1475-2875-6-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sinauer Associates, Sunderland, MA. 2004 [Google Scholar]
- Curtis CF. The mechanism of hybrid male sterility from crosses in the Anopheles gambiae and Glossina morsitans complexes. In: Steiner WM, Tabachnick WJ, Rai KS, Narang S, editors. Recent developments in the genetics of insect disease vectors. Champaign, Illinois: Stipes Publishing Company; 1982. pp. 290–312. [Google Scholar]
- Della Torre A, Tu Z, Petrarca V. On the distribution and genetic differentiation of Anopheles gambiae s.s. molecular forms. Insect Biochem Mol Biol. 2005;35:755–769. doi: 10.1016/j.ibmb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Kengne P, Brengues C, Baldet T, Ouari A, Simard F, Lehmann T. Mixed swarms of the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in sympatric area from Burkina Faso. J Med Entomol. 2006;43:480–483. doi: 10.1603/0022-2585(2006)43[480:msotmm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dabire RK, Millogo N, Lehmann T. Evaluating the effect of postmating isolation between molecular forms of Anopheles gambiae (Diptera: Culicidae) J Med Entomol. 2007;44:60–64. doi: 10.1603/0022-2585(2007)44[60:eteopi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Diabate A, Dao A, Yaro AS, Adamou A, Gonzalez R, Manoukis NC, Traore SF, Gwadz RW, Lehmann T. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae . Proc R Soc B. 2009 doi: 10.1098/rspb.2009.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Fanello C, Santolamazza F, della Torre A. Simultaneous identification of species and molecular forms of the Anopheles gambiae complex by PCR-RFLP. Med Vet Entomol. 2002;16:461–464. doi: 10.1046/j.1365-2915.2002.00393.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. Skepticism towards Santa Rosalia, or why are there so few kinds of animals? Evolution. 1981;35:124–138. doi: 10.1111/j.1558-5646.1981.tb04864.x. [DOI] [PubMed] [Google Scholar]
- Gillies MT, De Meillon B. The Anophelinae of Africa south of the Sahara. South African Institute for Medical Research, Johannesburg. 1968 [Google Scholar]
- Harr B. Genomic islands of differentiation between house mouse subspecies. Genome Res. 2006;16:730–737. doi: 10.1101/gr.5045006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Ann Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K. A high-resolution recombination map of the human genome. Nat Genet. 2002;31:241–247. doi: 10.1038/ng917. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Diabate A. The molecular forms of Anopheles gambiae: a phenotypic perspective. Infect Genet Evol. 2008;8:737–746. doi: 10.1016/j.meegid.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–340. doi: 10.1016/s0168-9525(02)02669-0. [DOI] [PubMed] [Google Scholar]
- Manoukis NC, Powell JR, Touré MB, Sacko A, Edillo FE, Coulibaly MB, Traoré SF, Taylor CE, Besansky NJ. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae . Proc Natl Acad Sci U S A. 2008;105:2940–2945. doi: 10.1073/pnas.0709806105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence--accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. Columbia University Press, New York. 1987 [Google Scholar]
- Noor MA, Feder JL. Speciation genetics: evolving approaches. Nat Rev Genet. 2006;7:851–861. doi: 10.1038/nrg1968. [DOI] [PubMed] [Google Scholar]
- Noor MA, Grams KL, Bertucci LA, Reiland J. Chromosomal inversions and the reproductive isolation of species. Proc Natl Acad Sci U S A. 2001;98:12084–12088. doi: 10.1073/pnas.221274498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nosil P, Funk DJ, Ortiz-Barrientos D. Divergent selection and heterogeneous genomic divergence. Mol Ecol. 2009;18:375–402. doi: 10.1111/j.1365-294X.2008.03946.x. [DOI] [PubMed] [Google Scholar]
- Oliveira E, Salgueiro P, Palsson K, Vicente JL, Arez AP, Jaenson TG, Caccone A, Pinto J. High levels of hybridization between molecular forms of Anopheles gambiae from Guinea Bissau. J Med Entomol. 2008;45:1057–1063. doi: 10.1603/0022-2585(2008)45[1057:hlohbm]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ortiz-Barrientos D, Reiland J, Hey J, Noor MA. Recombination and the divergence of hybridizing species. Genetica. 2002;116:167–178. [PubMed] [Google Scholar]
- Pombi M, Stump AD, Della Torre A, Besansky NJ. Variation in recombination rate across the X chromosome of Anopheles gambiae . Am J Trop Med Hyg. 2006;75:901–903. [PubMed] [Google Scholar]
- Powell JR, Petrarca V, della Torre A, Caccone A, Coluzzi M. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia. 1999;41:101–113. [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the www for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics methods and protocols: Methods in molecular biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Rundle HD, Nosil P. Ecological speciation. Ecol Lett. 2005;8:336–352. [Google Scholar]
- Santolamazza F, Della Torre A, Caccone A. Short report: a new polymerase chain reaction-restriction fragment length polymorphism method to identify Anopheles arabiensis from An. gambiae and its two molecular forms from degraded DNA templates or museum samples. Am J Trop Med Hyg. 2004;70:604–606. [PubMed] [Google Scholar]
- Schluter D. Ecology and the origin of species. Trends Ecol Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- Scott JA, Brogdon WG, Collins FH. Identification of single specimens of the Anopheles gambiae complex by the polymerase chain reaction. Am J Trop Med Hyg. 1993;49:520–529. doi: 10.4269/ajtmh.1993.49.520. [DOI] [PubMed] [Google Scholar]
- Sharakhov IV, Sharakhova MV, Mbogo CM, Koekemoer LL, Yan G. Linear and spatial organization of polytene chromosomes of the african malaria mosquito Anopheles funestus . Genetics. 2001;159:211–218. doi: 10.1093/genetics/159.1.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharakhova MV, Hammond MP, Lobo NF, Krzywinski J, Unger MF, Hillenmeyer ME, Bruggner RV, Birney E, Collins FH. Update of the Anopheles gambiae PEST genome assembly. Genome Biol. 2007;8:R5. doi: 10.1186/gb-2007-8-1-r5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem GC, Etouna J, Ose K, Fotsing J-M, Fontenille D, Besansky NJ, Costantini C. Ecological niche partitioning between the M and S molecular forms of Anopheles gambiae in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M, Della Torre A, Powell JR. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. arabiensis . Genetics. 2004;167:275–287. doi: 10.1534/genetics.167.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotman M, Della Torre A, Powell JR. Female sterility in hybrids between Anopheles gambiae and A. arabiensis, and the causes of Haldane's rule. Evolution. 2005;59:1016–1026. [PubMed] [Google Scholar]
- Slotman MA, Reimer LJ, Thiemann T, Dolo G, Fondjo E, Lanzaro GC. Reduced recombination rate and genetic differentiation between the M and S forms of Anopheles gambiae s.s . Genetics. 2006;174:2081–2093. doi: 10.1534/genetics.106.059949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smadja C, Galindo J, Butlin R. Hitching a lift on the road to speciation. Mol Ecol. 2008;17:4177–4180. doi: 10.1111/j.1365-294x.2008.03917.x. [DOI] [PubMed] [Google Scholar]
- Stegnii VN. Systemic reorganization of the architectonics of polytene chromosomes in the onto- and phylogenesis of malarial mosquitoes. II. Species specificity in the pattern of chromosome relations with the nuclear envelope of nutrient ovarian cells. Genetika. 1987;23:1194–1199. [PubMed] [Google Scholar]
- Storz JF. Using genome scans of DNA polymorphism to infer adaptive population divergence. Mol Ecol. 2005;14:671–688. doi: 10.1111/j.1365-294X.2005.02437.x. [DOI] [PubMed] [Google Scholar]
- Stump AD, Fitzpatrick MC, Lobo NF, Traore S, Sagnon N, Costantini C, Collins FH, Besansky NJ. Centromere-proximal differentiation and speciation in Anopheles gambiae . Proc Natl Acad Sci U S A. 2005;102:15930–15935. doi: 10.1073/pnas.0508161102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stump AD, Pombi M, Goeddel L, Ribeiro JMC, Wilder JA, Della Torre A, Besansky NJ. Genetic exchange in 2La inversion heterokaryotypes of Anopheles gambiae . Insect Mol Biol. 2007;16:703–709. doi: 10.1111/j.1365-2583.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- Tajima F. Evolutionary relationship of DNA sequences in finite populations. Genetics. 1983;105:437–460. doi: 10.1093/genetics/105.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripet F, Toure YT, Taylor CE, Norris DE, Dolo G, Lanzaro GC. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae . Mol Ecol. 2001;10:1725–1732. doi: 10.1046/j.0962-1083.2001.01301.x. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW. Locus- and population-specific selection and differentiation between incipient species of Anopheles gambiae . Mol Biol Evol. 2007;24:2132–2138. doi: 10.1093/molbev/msm143. [DOI] [PubMed] [Google Scholar]
- Turner TL, Hahn MW, Nuzhdin SV. Genomic islands of speciation in Anopheles gambiae . PLoS Biol. 2005;3:e285. doi: 10.1371/journal.pbio.0030285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S. Sympatric speciation in animals: The ugly duckling grows up. Trends Ecol Evol. 2001;16:381–390. doi: 10.1016/s0169-5347(01)02188-7. [DOI] [PubMed] [Google Scholar]
- Via S. Natural selection in action during speciation. Proc Natl Acad Sci U S A 106 Suppl. 2009;1:9939–9946. doi: 10.1073/pnas.0901397106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via S, West J. The genetic mosaic suggests a new role for hitchhiking in ecological speciation. Mol Ecol. 2008;17:4334–4345. doi: 10.1111/j.1365-294X.2008.03921.x. [DOI] [PubMed] [Google Scholar]
- Watterson GA. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 1975;7:256–276. doi: 10.1016/0040-5809(75)90020-9. [DOI] [PubMed] [Google Scholar]
- White BJ, Cheng C, Sangare D, Lobo NF, Collins FH, Besansky NJ. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae . Genetics. 2009;183:275–288. doi: 10.1534/genetics.109.105817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Hahn MW, Pombi M, Cassone BJ, Lobo NF, Simard F, Besansky NJ. Localization of candidate regions maintaining a common polymorphic inversion (2La) in Anopheles gambiae . PLoS Genet. 2007;3:e217. doi: 10.1371/journal.pgen.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SI, Bi IV, Schroeder SG, Yamasaki M, Doebley JF, McMullen MD, Gaut BS. The effects of artificial selection on the maize genome. Science. 2005;308:1310–1314. doi: 10.1126/science.1107891. [DOI] [PubMed] [Google Scholar]
- Wright SI, Charlesworth B. The HKA test revisited: A maximum-likelihood-ratio test of the standard neutral model. Genetics. 2004;168:1071–1076. doi: 10.1534/genetics.104.026500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- Zheng L, Benedict MQ, Cornel AJ, Collins FH, Kafatos FC. An integrated genetic map of the African human malaria vector mosquito, Anopheles gambiae . Genetics. 1996;143:941–952. doi: 10.1093/genetics/143.2.941. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.