Abstract
Several tissue-specific regulatory genes have been found to play essential roles in both organogenesis and carcinogenesis. In the prostate, the Nkx3.1 homeobox gene plays an important role in normal differentiation of the prostatic epithelium while its loss of function is an initiating event in prostate carcinogenesis in both mouse models and human patients. Thus, the Nkx3.1 homeobox gene provides a paradigm for understanding the relationship between normal differentiation and cancer, as well as studying the roles of homeobox genes in these processes. Here, we review recent findings concerning the roles of Nkx3.1 in development and discuss how its normal function is disrupted in processes of early prostate carcinogenesis.
Keywords: prostate, homeobox, nkx3.1
Introduction
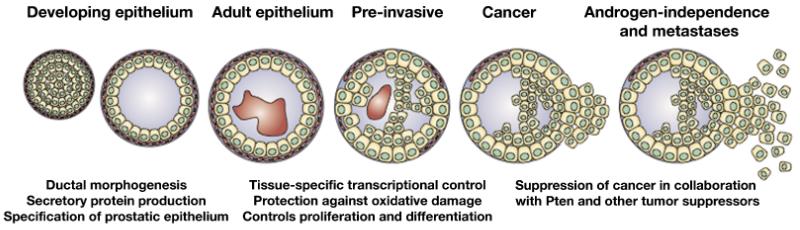
The Nkx3.1 homeobox gene provides an excellent example of a gene with critical functions for both embryogenesis and oncogenesis (Fig. 1). During murine development, Nkx3.1 is the earliest known marker of prostate formation and it continues to be expressed at all stages of prostate differentation and in adulthood. Nkx3.1 plays an essential role in normal prostate development, because its loss of function leads to defects in prostatic protein secretions and in ductal morphogenesis. Moreover, loss-of-function of Nkx3.1 also contributes to prostate carcinogenesis, because Nkx3.1 mutant mice are predisposed to prostate carcinoma and because loss-of-function of Nkx3.1 cooperates with that of other tumor suppressor genes in cancer progression. Furthermore, by a variety of mechanisms NKX3.1 expression is reduced in noninvasive and early stage human prostate cancer, suggesting that its decreased expression is one of the earliest steps in the majority of human prostate cancers. In this review, we describe the molecular properties of the Nkx3.1 homeobox gene, focusing on how this protein contributes to prostate development and prostate cancer.
Fig. 1.
Varying roles of NKX3.1 in prostate development, in normal adult prostate, and in prostate cancer.
Structure and biochemical properties of NKX3.1
The Nkx3.1 homeobox gene
Nkx3.1 is a member of the NK subfamily of homeobox genes, that have been implicated in processes of cell fate specification and organogenesis in many species, including Drosophila, where they were first identified (Kim and Nirenberg, 1989). In Drosphila, NK-3 corresponds to the bagpipe gene, which is required for visceral mesoderm development (Kim and Nirenberg, 1989; Azpiazu and Frasch, 1993) whereas in mammalian species, Nkx3.1 has been implicated primarily in prostate development, while other members of the Nkx family have roles in mesoderm formation and genesis of other organs.
Mouse and human Nkx3.1 share 63% amino acid identity, and are 100% identical within the homeodomain. Mouse Nkx3.1 is localized to chromosome 14, in a region syntenic to human chromosome 8p. The mouse Nkx3.1 gene contains two coding exons; the homeodomain is located within the second exon. The gene has a short 5′ untranslated region (UTR) and an extensive 3′ UTR (Sciavolino et al., 1997). In humans, there are at least five alternatively spliced forms of the NKX3.1 gene, which encode variants of the N-terminal coding region upstream of the homeodomain (Korkmaz et al., 2000). However, the in vivo expression and functional significance of these alternative NKX3.1 transcripts has not yet been elucidated nor have alternative transcripts been identified for the mouse gene.
DNA-binding specificity of Nkx3.1
The Nkx family is characterized by a tyrosine at position 54 of the homeodomain. Divergent amino acid residues around the homeodomain are thought to contribute to differential DNA binding specificity relative to that of the canonical Antennapedia-class homeodomains (Damante and Di Lauro, 1991; Damante et al., 1994; Chen and Schwartz, 1995). Indeed, binding-site selection experiments have identified “TAAGTA” as a consensus preferred site for human NKX3.1 (Steadman et al., 2000); this differs from the consensus sequences identified for most other homeoproteins, which typically contain a “TAAT” core. The NKX3.1 consensus site is also distinct from the consensus sites identified for NK-2 class homeodomains such as Nkx2.1 (CAAGTG) and Nkx2.5 (TNAAGTG) (Damante and Di Lauro, 1991; Damante et al., 1994; Chen and Schwartz, 1995). However, mouse and human NKX3.1 will bind to both the Nkx2.1 and canonical “TAAT” sites in electrophoretic mobility shift experiments, albiet with weaker affinity than the “TAAGTA” site (Sciavolino et al., 1997; Steadman et al., 2000). Notably, however, endogenous binding sites for Nkx3.1 within the promoter/enhancer regions of physiological downstream target genes have not been conclusively identified. In particular, although it has been proposed that the murine androgen receptor gene contains a functional Nkx3.1 binding site (Lei et al., 2006), it remains to be determined whether these in vitro binding sites are bona fide protein target sites in vivo.
Structure features of the NKX3.1 protein
The NKX3.1 homeodomain forms three α-helices, the first two of which form the parallel scaffold that stabilizes the third, perpendicular helix that contacts the major groove of DNA (Gruschus et al., 1997; Ju et al., 2006). Mutational studies have shown that hydrogen bonding with DNA occurs at lysine and isoleucine at homeodomain positions 46 and 47, glutamine and asparagine at homeodomain 50 and 51, and tyrosine at 54 (Harvey, 1996).
The stability of the NKX3.1 protein is influenced by post-translational modification of the homeodomain and flanking regions. Phosphorylation of the N-terminal flanking region at threonines 89 and 93 by protein kinase CK2 prolongs protein half-life, as evidenced by mutation of both threonines to alanines, which decreases NKX3.1 half-life by 50% (Li et al., 2006). On the other hand, phosphorylation at the C-terminal region at serine 185 determines ubiquitination and protein degradation as evidenced by mutation of serine 185 to alanine, which prolongs the protein half-life by fourfold (Markowski et al, submitted for publication). Serines 195 and 196 are targets for phosphorylation induced by inflammatory cytokines such as TNF-α. NKX3.1 expression is reduced in tissues containing inflammatory cells (Bethel et al., 2006) and both TNF-α and IL-1β accelerate ubiquitination and degradation of NKX3.1 in vitro (Markowski et al, submitted for publication). The most likely site for ubiquitination is the homeodomain, but this has not been rigorously demonstrated (Guan et al., 2008).
Posttranslational modification of NKX3.1 also occurs via phosphorylation at serine 48, a residue that is critical for stable phosphorylation of NKX3.1 (Gelmann et al., 2002). Phosphorylation at serine 48 is affected by arginine 52, which is the target of a common genetic polymorphism. The C145T polymorphism, which is present in 11% of the population, causes a nonconservative arginine to cysteine change at amino acid 52. The R52C amino acid replacement decreases phosphorylation at serine 48 and impairs DNA binding in vitro (Gelmann et al., 2002).
In addition to mediating DNA binding, the homeodomain of NKX3.1 can mediate interactions with various proteins. For example, NKX3.1 interacts with serum response factor (SRF) to enhance binding to the cognate DNA serum response element and activate transcription (Carson et al., 2000). NKX3.1, via its homeodomain, can interact directly with the MADS box of SRF in the absence of DNA, suggesting that the homeodomain mediates protein–protein as well as protein–DNA interactions (Ju et al., 2006). Moreover, NKX3.1 has been shown to interact with a variety of other proteins and in each case the interactions required the homeodomain, although they may be enhanced by additional peptide sequences from the N-terminus, as in the cases of topoisomerase I (Bowen et al., 2007) and the transcription factor SP-1 (Simmons and Horowitz, 2006), or the C-terminus as was shown for the prostate-derived ETS factor (PDEF) (Chen and Bieberich, 2005).
Transcriptional repression and activation functions of Nkx3.1
Studies of the Nkx3 class of homeoproteins indicate that they act as transcriptional repressors upon binding to DNA, as exemplified by cell culture assays showing that human NKX3.1 displays repressor activity on a synthetic reporter containing a NKX3.1 consensus sites (Steadman et al., 2000). Similarly, Nkx3.2 acts as a repressor in transient transfection assays in cell culture as well as in retroviral expression studies in chick embryos (Murtaugh et al., 2001). More generally, in vivo analyses of Nkx homeoproteins in ventral neural tube patterning suggest that most, if not all, Nkx homeoproteins function as transcriptional repressors (Muhr et al., 2001). In particular, all Nkx homeoproteins contain a conserved decapeptide motif known as the TN domain (Lints et al., 1993), which shows similarity to the engrailed homology-1 (eh1) domain identified in the Drosophila Engrailed homeoprotein (Smith and Jaynes, 1996). The eh1 domain is essential for repressor activity of Engrailed, and can interact with members of the Groucho (Gro/TLE) family of transcriptional corepressors (Jimenez et al., 1997). Analyses of Drosphila NK-3 have shown that it can interact with Groucho corepressors (Choi et al., 1999).
Drosophila NK-3 has also been reported to interact with the serine-threonine kinase HIPK2 (for homeoprotein-interacting protein kinase 2) (Kim et al., 1998), which enhances its DNA-binding activity and increases its repressor activity (Choi et al., 1999). Co-immunoprecipitation experiments in transfected HeLa cells show that NK-3 interacts with both Groucho and HIP-K2, and that these proteins are likely to exist in a repressive complex together with the histone deacetylase HDAC1 (Choi et al., 1999). Notably, mouse Nkx3.1 has been reported to interact with HDAC1 (Lei et al., 2006), which has been linked to its activity in cancer promotion. Strikingly, the association of NKX3.1 with HDAC1 is dependent on NKX3.1 levels such that distinct histones of different NKX3.1 target genes are differentially affected by reduced levels of NKX3.1 (Mogal et al., 2007).
Protein interactions determine a number of Nkx3.1 transcriptional effects. Nkx3.1 acts as a transcriptional repressor through recruitment of Gro/TLE corepressors. However, in some contexts, the transcriptional repressor activities of Nkx3.1 may be modulated by its ability to repress the transcriptional activator of other proteins, such as PDEF (Prostate-derived Ets Factor), a member of the Ets transcription factor (Oettgen et al., 2000), which was identified as an Nkx3.1-interacting protein (Chen et al., 2002). In cell culture assays, PDEF can activate transcription from the human PSA promoter, while NKX3.1 antagonizes this activity (Oettgen et al., 2000; Chen et al., 2002), which requires both the homeodomain and the far C-terminus of NKX3.1 for binding in yeast two-hybrid assays (Chen and Bieberich, 2005). Similarly, Nkx3.1 has been shown to interact with SP-1 transcriptional regulatory proteins to repress their ability to activate transcription of the PSA promoter, although this interaction is mediated by homeodomain and N-terminus of Nkx3.1 (Simmons and Horowitz, 2006).
However, in certain tissue contexts Nkx3.1 possesses transcriptional activator activity. In particular, Nkx3.1 has been shown to interact with SRF and to activate transcription from the smooth-muscle γ-actin promoter in transfected cells (Carson et al., 2000; Gelmann et al., 2002). This is thought to be due to binding of Nkx3.1 to promoter DNA, which facilitates recruitment of SRF to adjacent sites resulting in increased transcriptional activity (Carson et al., 2000). It has been suggested that Nkx3 homeoproteins may act as transcriptional activators in visceral mesoderm derivatives through an interaction with SRF, which displaces co-repressors such as members of the Groucho/TLE family. Together, these findings highlight the importance of tissue and cellular context for modulating the transcriptional functions of Nkx3.1.
Expression and functions of Nkx3.1 in development
Nkx3.1 expression and mutant phenotype in nonurogenital tissues
During organogenesis, Nkx3.1 is expressed transiently in a wide range of tissues outside of the urogenital system, which is most evident in the paraxial mesoderm, where Nkx3.1 is expressed in the ventral portion of the somites, and then becomes restricted to the sclerotome (Kos et al., 1998; Tanaka et al., 1999). However, despite this striking expression in newly formed paraxial mesoderm, null mutants for Nkx3.1 fail to display any phenotype in sclerotomal derivatives (Bhatia-Gaur et al., 1999; Schneider et al., 2000; Tanaka et al., 2000). One possible reason that the Nkx3.1 null mice do not display phenotypes in this and other expression domains is partial genetic redundancy between Nkx3.1 and Nkx3.2, which have overlapping expression domains in the paraxial mesoderm. (Tribioli et al., 1997; Tribioli and Lufkin, 1999).
Indeed, other non-urogenital sites of Nkx3.1 expression include regions of the tongue, teeth, dorsal aorta, the dorsal region of Rathke’s pouch, and the arcuate and interlobular arteries of the kidney (Sciavolino et al., 1997; Treier et al., 1998; Tanaka et al., 1999; Schneider et al., 2000; Chen et al., 2005). Most of these other non-urogenital tissues that express Nkx3.1 also display no phenotype in the mutant mice (Tanaka et al., 2000; R. Bhatia-Gaur et al., unpublished observations), although a slight phenotype has been described for the lobular arteries of the kidneys (Tanaka et al., 2000). In fact, thus far, the only significant non-urogenital phenotype that has been described for Nkx3.1 null mutants occurs in the minor salivary glands, which display an increased accumulation of mucous secretions, accompanied by a significant decrease in the ductal branching of the palatine glands (Schneider et al., 2000; Tanaka et al., 2000), an effect reminiscent of the Nkx3.1−/− phenotype in urogenital tissues (Bhatia-Gaur et al., 1999).
Roles of Nkx3.1 in prostate development
In both mice and humans, formation of the prostate during embryogenesis occurs through epithelial budding from the urogenital sinus, an endodermal derivative of the hindgut. However, the adult morphology of the prostate greatly differs between mouse and human. At adulthood, the rodent prostate gland consists of four distinct lobes: anterior (also known as the coagulating gland), dorsal and lateral (collectively referred to as the dorsolateral lobe), and ventral, which are arranged circumferentially around the bladder and display characteristic patterns of ductal branching, histological features, and secretory protein production (Sugimura et al., 1986; Hayashi et al., 1991). In contrast, the adult human prostate lacks discernable lobular organization, with the prostatic ducts radiating peripherally to completely surround the urethra at the base of the bladder.
In the adult mouse urogenital system Nkx3.1 is specifically expressed at high levels in all lobes of the prostate as well as the bulbourethral glands, which are ductal tissues of endodermal origin. In contrast, Nkx3.1 is not expressed in ductal urogenital tissues that are not of endodermal origin, such as the seminal vesicle or in nonductal endodermal tissues, such as the bladder and urethra (Bhatia-Gaur et al., 1999). Within the prostate, Nkx3.1 protein is localized in nuclei of luminal epithelial cells, which produce the prostatic secretory proteins (Bhatia-Gaur et al., 1999), as well as in some basal epithelial cells (Chen et al., 2005). In adult human tissues NKX3.1 is expressed in isolated clusters of ureteral transitional epithelium, small bronchial mucous glands, and testis, in addition to the prostatic epithelium (Bieberich et al., 1996).
Expression analysis of Nkx3.1 has provided insights into the earliest stages of prostate formation (Sciavolino et al., 1997; Bhatia-Gaur et al., 1999). In the mouse, the prostatic buds first emerge at the rostral end of the urogenital sinus toward the end of gestation, at approximately 17.5 dpc, under the influence of inductive signals from the surrounding mesenchyme. Within the urogenital system, Nkx3.1 expression is first detected in the lateral aspects of the urogenital sinus epithelium at 15.5 dpc, before prostate formation (Bhatia-Gaur et al., 1999). Notably, Nkx3.1 expression precedes formation of the prostatic buds by 2 days, and appears to correspond to the regions where prostatic buds will emerge, suggesting that regions of the urogenital sinus epithelium may have a differential capacity to form prostate. At present, the significance of the restricted lateral expression of Nkx3.1 is unknown, especially because the corresponding protein is not detectable until 17.5 dpc, in the newly emerging prostatic buds (N. Desai et al., unpublished observations).
Subsequently, the prostatic epithelial buds undergo extensive ductal outgrowth and branching into the surrounding mesenchyme during the first 3 weeks of post-natal development (Sugimura et al., 1986; Timms et al., 1994). These emerging prostatic buds are marked by expression of Nkx3.1 in the prostatic epithelium (Sciavolino et al., 1997; Bhatia-Gaur et al., 1999). Expression of Nkx3.1 is enriched toward the distal ends of the outgrowing ducts, corresponding to regions of active morphogenesis (Bhatia-Gaur et al., 1999). Before canalization of the prostatic ducts, Nkx3.1 is expressed uniformly in the epithelial cells, but later expression is largely restricted to the luminal cells.
Relationship of Nkx3.1 to epithelial-mesenchymal interactions in prostate morphogenesis
In the prostate, the role of epithelial–mesenchymal interactions has been defined through elegant tissue recombination studies performed by Cunha and colleagues (Cunha et al., 1987; Hayward et al., 1997). This approach employs dissection and enzymatic dissociation of epithelium and mesenchyme from embryonic urogenital sinus and/or from other tissues, which are then recombined in vitro and transplanted under the kidney capsules of adult male nude mouse hosts, followed by assessment of prostatic differentiation by histology and by production of appropriate secretory proteins. Prostate-like structures form when urogenital sinus epithelial (UGE) is combined with urogenital sinus mesenchyme (UGM), whereas in absence of either, mature prostatic cell types fail to differentiate. Expression of Nkx3.1 is a highly specific marker of prostate differentiation in tissue recombination assays (Bhatia-Gaur et al., 1999). In particular, tissue recombinants made with the prostate-inducing mesenchyme (UGM) and bladder epithelium express Nkx3.1, although Nkx3.1 is not normally expressed in the bladder.
Relationship of Nkx3.1 to androgen signaling in prostate formation
Androgen signaling is essential for all aspects of prostatic growth and differentiation, because the prostate is not formed in the absence of androgens (reviewed in Cunha et al., 1987; Marker et al., 2003). During murine embryogenesis, androgen receptors are located in the UGM, whereas postnatally they are found in both the mesenchyme and epithelium. Although the initial appearance of Nkx3.1 expression in the prostatic epithelium precedes that of the androgen receptor, the subsequent expression of Nkx3.1 is dependent on androgen signaling, as shown in tissue recombination assays (Bhatia-Gaur et al., 1999). Moreover, expression of Nkx3.1 is significantly down-regulated following castration (surgical removal of the source of androgens) (Bieberich et al., 1996; Sciavolino et al., 1997). Similarly, expression of NKX3.1 in human prostate cancer cells is down-regulated following depletion of androgens from cell culture media (He et al., 1997; Prescott et al., 1998). Although the maintenance of Nkx3.1 expression is dependent on androgen signaling, it is not clear whether this regulation occurs directly through the Nkx3.1 promoter or whether it is the consequences of androgen signaling are indirect.
Requirement for Nkx3.1 in prostate ductal morphogenesis and epithelial differentiation
The predominant effects of Nkx3.1 loss-of-function are manifested in the prostate. Thus, Nkx3.1−/− mice display reduced ductal branching morphogenesis as well as defects in secretory protein production (Bhatia-Gaur et al., 1999). Notably, this phenotype was observed in heterozygotes as well as in homozygotes, indicating haploinsufficiency for this defect. These observations suggest that Nkx3.1 has a role in early post-natal ductal morphogenesis, as well as in production of secretory proteins in the mature prostate, reflecting a general requirement for maintenance of the normal differentiated state of the prostate epithelium as well as control of epithelial cell proliferation. Haploinsufficiency as seen for Nkx3.1 also pertains to other members of the NK family of homeodomain proteins. For example, inheritance of NKX2.5 mutations that introduce premature stop codons and no functional protein is associated with autosomal dominant inheritance of cardiac developmental abnormalities (Kasahara and Benson, 2004).
Roles for Nkx3.1 in prostate cancer
Formation of prostatic intraepithelial neoplasia (PIN) in Nkx3.1 mutant mice
A notable feature of the phenotype of the Nkx3.1 mutant mice is the histological appearance of prostatic epithelial hyperplasia and dysplasia, which becomes increasingly more apparent with advancing age (Bhatia-Gaur et al., 1999; Schneider et al., 2000; Tanaka et al., 2000; Abdulkadir et al., 2002; Kim et al., 2002a). This epithelial phenotype correlates with a significantly increased proliferative index (Bhatia-Gaur et al., 1999; Magee et al., 2003). After 1 year of age a majority of the homozygous mutant mice develop histological features that resemble human PIN, which is thought to be the primary precursor of prostate cancer. Histologic findings in these mice include perturbation of the basal cell layer, attenuation of the stroma, and heterogeneous expression of markers of epithelial differentiation (Kim et al., 2002a). However, although aged Nkx3.1 mutant mice are highly prone to develop PIN, they do not develop invasive carcinoma.
The consequences of loss-of-function of Nkx3.1 for prostate cancer initiation are independent of its role in promoting ductal morphogenesis. Prostate-restricted targeting of a conditional Nkx3.1 allele using a PSA-Cre transgene results in adult-specific deletion of Nkx3.1 and formation of PIN (Abdulkadir et al., 2002). Furthermore, the susceptibility of Nkx3.1 mutant mice to develop PIN but not carcinoma, as well as the precursor-product relationship of PIN to prostate carcinoma, has been further demonstrated using a tissue recombination system. In particular, serial transplantation of prostatic tissue recombinants from Nkx3.1 mutant mice results in neoplastic progression characterized by increasingly dysplastic histopathological alterations (Kim et al., 2002a). Furthermore, the tumor suppressor properties of Nkx3.1 are further underscored by its ability to slow cell proliferation of cultured cells and reduce tumorigenicity in xenograft assays (Kim et al., 2002a; Lei et al., 2006).
One mechanism by which Nkx3.1 may suppress cancer initiation is through protection against oxidative damage (Ouyang et al., 2005). Using gene expression profiling, it was shown that Nkx3.1 mutant mice display deregulated expression of several anti-oxidant and pro-oxidant enzymes, including glutathione peroxidase 2 and 3 (GPx2, GPx3), peroxiredoxin 6 (Prdx6), and sulfhydryl oxidase Q6 (Qscn6). Importantly, formation of PIN is associated with increased oxidative damage of DNA, as evident by increased levels of 8-hydroxy-2′-deoxyguanosine (8-OHdG). These findings suggested that Nkx3.1 provides protection against oxidative damage, while its inactivation leads to increased susceptibility to oxidative damage.
One possible mechanism for these effects is via the interaction of NKX3.1 via its homeodomain with the DNA resolving enzyme topoisomerase I. Thus, NKX3.1 activates topoisomerase I on a stoichiometric basis and markedly enhances enzyme activity by accelerating topoisomerase I binding to DNA. This interaction occurs in vitro with purified proteins and as well as in tissue extracts from Nkx3.1 knock-out mice (Bowen et al., 2007). The interaction of NKX3.1 and topoisomerase I enhances DNA repair, while loss of NKX3.1 expression may predispose human prostate cancer cells further to DNA damage (C. Bowen and E. P. Gelmann, unpublished observations).
Indeed, the importance of NKX3.1 in the early stages of prostate cancer is not restricted to the mutant mice. In human PIN and primary prostate cancer NKX3.1 expression is reduced to levels comparable to those in Nkx3. heterozygous mice (Asatiani et al., 2005). NKX3.1 is located centrally in the 1Mb region of human chromosome 8p21 that is deleted in prostate cancer (Swalwell et al., 2002). Loss of this region or loss of one allele of NKX3.1 is found in the majority of early stage human prostate cancer (Vocke et al., 1996; Asatiani et al., 2005). In human prostate cancer the 5′ UTR of the gene is subjected to non-classical DNA methylation that nevertheless correlates with loss of protein expression (Asatiani et al., 2005). A number of genetic polymorphisms have been found in both the coding and noncoding regions of NKX3.1 (Zheng et al., 2006). Only one of these, a single nucleotide polymorphism at nucleotide 154 of the coding region, is associated with prostate cancer risk (Gelmann et al., 2002).
Furthermore, the role of NKX3.1 as a tumor suppressor has been reinforced by the discovery of a kindred with a germ-line mutation in the homeodomain that co-segregates with early onset of prostate cancer (Zheng et al., 2006). The mutation changes the threonine at homeodomain position 41, to an alanine residue, which affects α-helical capping. Interestingly, a threonine to methionine amino acid substitution at the precisely analogous position of NKX2.5, the human homolog of the tinman homeodomain protein, determines hereditary cardiac abnormalities in a family (Kasahara and Benson, 2004). Mutation of the α-helical capping amino acid destabilizes the helix and decreases DNA binding affinity by 95% (Zheng et al., 2006).
Cooperativity of Nkx3.1 with Pten in suppression of prostate cancer
Although Nkx3.1 loss-of-function is not sufficient for prostate cancer, Nkx3.1 can cooperate with Pten, in suppression of prostate carcinogenesis. In particular, the combined loss-of-function of Nkx3.1 and the Pten tumor suppressor gene in mice with compound mutations (Nkx3.1−/−; Pten+/−) display striking cooperativity, as evidenced by the development of high-grade PIN lesions by 6 months of age (Kim et al., 2002b). Ultimately these Nkx3.1; Pten compound mutant mice develop adenocarcinoma, in some cases with metastases to the lymph nodes as well as distant organs (Abate-Shen et al., 2003). Histologically, these lesions are multifocal and poorly differentiated with prominent and multiple nucleoli, increased nuclear:cytoplasmic ratio, and numerous mitoses (Park et al., 2002). Aside from these malignant lesions, the compound mutants display no additional phenotypes compared with the single mutants, which emphasizes the prostate-specificity of the cooperativity between Nkx3.1 and Pten.
The cooperation of Nkx3.1 and Pten loss in prostate carcinogenesis is mediated, at least in part, by the PI3-Kinase–Akt signaling pathway, whose activation is a primary consequence of Pten loss-of-function (reviewed in Cantley and Neel, 1999; Di Cristofano and Pandolfi, 2000; Cantley, 2002; Vivanco and Sawyers, 2002). Indeed, the activation of Akt by phosphorylation is increased considerably in protein lysates from Nkx3.1−/−; Pten+/− prostates, while p-Akt is immunohistochemically localized to the carcinomas (Kim et al., 2002b). Notably, p-Akt staining is also observed in the prostatic epithelium of Nkx3.1 single mutants, suggesting that loss-of-function of Nkx3.1 can affect Akt activation in the context of wild-type Pten activity (Kim et al., 2002b). It has been proposed that Nkx3.1 regulates phosphorylation of Akt via an androgen-receptor dependent signaling pathway (Lei et al., 2006).
The mouse as a model for human prostate cancer progression
Cancer progression in these Nkx3.1; Pten mutant mice display many features in common with human prostate cancer, particularly at the gene expression level (Ouyang et al., 2008), which makes them highly advantageous to investigating the molecular mechanisms of disease progression as relevant to human prostate cancer. In particular, the p27kip1 cell cycle regulator is frequently down-regulated in human prostate cancer and correlated with poor clinical outcome, yet rarely undergoes mutational inactivation (Cote et al., 1998; Yang et al., 1998; Erdamar et al., 1999; Kuczyk et al., 1999). Mouse models lacking the Nkx3.1 homeobox gene and the Pten tumor suppressor revealed that triple mutant mice heterozygous for a p27kip1 null allele (Nkx3.1+/−or−/−; Pten+/−; p27+/−) displayed enhanced prostate carcinogenesis, whereas mice that are homozygous null for p27kip1 (Nkx3.1+/−or−/−; Pten+/−; p27−/−) show inhibition of cancer progression, which was dependent on expression of Cyclin D1 (Gao et al., 2004). These findings suggest that p27kip1 possesses dosage-sensitive positive as well as negative modulatory roles in prostate cancer progression. Notably, this function of p27kip1 is apparently dependent on inactivation of both Nkx3.1 and Pten, because p27kip1 does not display dosage-sensitivity in combination with loss of function of either gene alone (Di Cristofano et al., 2001; Gary et al., 2004).
Furthermore, analyses of the Nkx3.1; Pten compound mutant have also provided new insights regarding the mechanisms underlying androgen independent prostate cancer, which is the final stage of disease progression in patients. In particular, the Nkx3.1; Pten mutant mice develop androgen-independent prostate cancer following androgen ablation, which is critically dependent on activation of Akt and Erk MAP kinase signaling pathways (Abate-Shen et al., 2003; Gao et al., 2006a, 2006b). The Akt and Erk MAP kinase signaling act in concert to promote tumorigenicity and androgen-independence in the context of the prostate microenvironment, but not in cell culture (Gao et al., 2006a). This has led to the proposal that androgen-dependence in vivo involves an epithelial-stromal interaction wherein Akt and Erk MAP kinase signaling in epithelial cells confer androgen-independence by overcoming the antagonistic effects of AR signaling in the stroma (Gao et al., 2006a).
Investigations of the evolution of androgen-independence in Nkx3.1; Pten mutant mice have also provided new insights regarding the emergence of the disease. Such analyses have shown that the prostate epithelial cells from these Nkx3.1; Pten mutant mice are capable of surviving and proliferating in the absence of androgens well before they display overt PIN or cancer phenotypes (Gao et al., 2006b). Furthermore, experimentally manipulating serum levels of testosterone in these mice for up to 7 months, has demonstrated that prolonged exposure of Nkx3.1; Pten mutant mice to androgen levels that are one tenth of normal results in a marked acceleration of prostate tumorigenesis compared with those exposed to androgen levels within the normal range, which suggests that exposure to reduced androgens may promote prostate tumorigenesis by selecting for molecular events that promote more aggressive, hormone-refractory tumors (Banach-Petrosky et al., 2007). These findings have important implications for treatment of prostate cancer patients as well as evaluating the risk of individuals predisposed to prostate cancer and demonstrate the value of the Nkx3.1; Pten mutant mouse model for studying androgen independence.
Nkx3.1 haploinsufficiency and epigenetic inactivation in mouse and human prostate cancer
Loss of heterozygosity of NKX3.1 is not accompanied by mutation of the residual contralateral allele, thus diverging from the suppressor gene paradigm of biallelic inactivation (Voeller et al., 1997; Ornstein et al., 2001). An analogous situation is observed in Nkx3.1+/−; Pten+/− compound heterozygotes, in which the Nkx3.1 wild-type allele remains intact, indicating that Nkx3.1 does not behave as a classical tumor suppressor that requires two hits for inactivation (Kim et al., 2002b). Furthermore, expression profiling of prostate tissues from Nkx3.1 mutants reveals altered gene expression in Nkx3.1 heterozygotes (Magee et al., 2003). Among genes that are down-regulated in Nkx3.1 homozygotes, several are poorly or not expressed in heterozygous mice; interestingly, however, up-regulated genes are generally not affected in heterozygotes compared with wild-type. These findings are consistent with previous observations of altered secretory protein expression in Nkx3.1 heterozygotes (Bhatia-Gaur et al., 1999), and indicate that Nkx3.1 heterozygosity results in significantly altered gene expression patterns in the prostate.
Residual levels of NKX3.1 protein expression in early stage prostate cancer are likely to exert a continued effect on cell growth, whereas during progression to metastatic disease there may be selective pressure for protein loss because >80% of metastatic foci have lost all NKX3.1 expression (Bowen et al., 2000). Levels of NKX3.1 expression in primary prostate cancer are lower when NKX3.1 undergoes allelic loss and methylation of the contralateral allele. In cancers that undergo either genetic loss or methylation NKX3.1 protein expression tends to be above the median of protein expression levels in cancer (Asatiani et al., 2005). Lower levels of NKX3.1 expression in primary prostate cancer is associated with higher Gleason grade. The mechanisms of NKX3.1 protein down-regulation during progression to metastatic disease have not been elucidated.
Loss-of-function of Nkx3.1 in mutant mice recapitulates this scenario for inactivation of the human gene in prostate cancer. In particular, Nkx3.1 heterozygous mutant mice display a similar, albeit less severe, prostatic epithelial phenotype compared with homozygotes, because Nkx3.1 heterozygotes (Nkx3.1+/−) develop PIN, and the Nkx3.1; Pten compound heterozygotes (Nkx3.1+/−; Pten+/−) develop high-grade PIN and carcinoma in situ (Bhatia-Gaur et al., 1999; Schneider et al., 2000; Tanaka et al., 2000; Kim et al., 2002b, 2002a). Notably, the PIN and carcinoma in situ lesions arising in these heterozygotes display a loss of Nkx3.1 protein expression (Abdulkadir et al., 2002; Kim et al., 2002b), analogous to the situation in human prostate cancer. Interestingly, in these mutant mouse models, this loss of Nkx3.1 protein expression even occurs in PIN and carcinoma lesions of mice having two wild-type Nkx3.1 alleles (Kim et al., 2002b). More generally, loss of Nkx3.1 protein expression occurs in various other mouse models of prostate cancer including those dependent on loss of function of Pten (Wang et al., 2003) or on gain of function of c-Myc or SV40 T antigen (Ellwood-Yen et al., 2003; Bethel and Bieberich, 2007). In combination, these findings indicate that Nkx3.1 haploinsufficiency may be due initially to altered gene expression patterns in heterozygotes, and is greatly enhanced during aging by the epigenetic inactivation of the wild-type Nkx3.1 allele during PIN formation.
Perspectives
In summary, NKX3.1 is a classic example of a developmental regulatory protein that is required for tissue differentiation and whose loss of function leads to carcinogenesis (Fig. 1). Indeed, analyses of NKX3.1 have provided insights into the mechanisms by which differentiation factors are linked to cancer mechanisms. Thus, in the normal prostate, NKX3.1 is expressed from the earliest stages of prostate formation. In adults, NKX3.1 controls normal differentiation and protects against oxidative damage by regulating gene expression in conjunction with other transcription factors. In cancer, NKX3.1 protein is reduced by a number of mechanisms, such as disruption of the gene or by affecting the degradation of the protein. Down-regulation of NKX3.1 protein in turn has profound effects on prostate epithelial cells, affecting their proliferation, differentiation, and polarity. In addition, loss of NKX3.1 increases susceptibility to DNA damage, thereby furthering oncogenic insult, which is a consequence of perturbed expression of proteins that affect oxidative damage as well as by activation of topoisomerase I.
However, despite the fact that loss of function of Nkx3.1 predisposes to prostate cancer, it is not sufficient for tumorigenesis. Therefore, the activities of Nkx3.1 in prostate cancer cells are not consistent with activities of “classic” tumor suppressor genes. In this regard, the role of Nkx3.1 as a tumor suppressor highlights the broader functions of homeobox genes in development and cancer. Indeed, homeobox genes are often deregulated in cancer, as reflected by their frequent “gain” or “loss” of expression, which is associated with oncogene-like or tumor suppressor-like activities, respectively (Cillo et al., 2001; Abate-Shen, 2002). However, their tumorigenic activities have been attributed to their aberrant usage in inappropriate cellular contexts, rather than the acquisition of “new” functions.
Accordingly, these tumorigenic activities tend to be more subtle than “classic” oncogenes or tumor suppressors; rather than being the driving force for tumorigenesis, deregulated homeobox genes appear to tip the balance in favor of the cancer phenotype, for example by promoting cellular proliferation versus quiescence. It is intriguing to note that another NK family member, NKX2.1, was recently found to play the role of dominant oncogene by gene amplification and over expression in a subset of non-small cell carcinoma of the lung (Weir et al., 2007). These features, combined with their known functions as regulators of differentiation, have led to the idea that homeobox genes may contribute to the tissue-specific features of cancer phenotypes (Abate-Shen, 2002). Thus, NKX3.1 provides an excellent example of how loss of the normal functions of a homeobox gene can predispose to cancer in a tissue-specific manner.
Acknowledgments
Work in the authors’ laboratories is supported by grants from the NCI, NIDDK, and the Department of Defense Prostate Cancer Research Program.
Contributor Information
Cory Abate-Shen, Department of Urology Columbia University College of Physicians and Surgeons Herbert Irving Comprehensive Cancer Center New York, NY, USA.
Michael M. Shen, Department of Genetics and Development Columbia University College of Physicians and Surgeons Herbert Irving Comprehensive Cancer Center New York, NY, USA; Department of Medicine Columbia University College of Physicians and Surgeons Herbert Irving Comprehensive Cancer Center New York, NY, USA
Edward Gelmann, Department of Medicine Columbia University College of Physicians and Surgeons Herbert Irving Comprehensive Cancer Center New York, NY, USA; Department of Pathology Columbia University, College of Physicians and Surgeons Herbert Irving Comprehensive Cancer Center New York, NY, USA.
References
- Abate-Shen C. Deregulated homeobox gene expression in cancer: cause or consequence? Nat Rev Cancer. 2002;2:777–785. doi: 10.1038/nrc907. [DOI] [PubMed] [Google Scholar]
- Abate-Shen C, Banach-Petrosky WA, Sun X, Economides KD, Desai N, Gregg JP, Borowsky AD, Cardiff RD, Shen MM. Nkx3.1; Pten mutant mice develop invasive prostate adenocarcinoma and lymph node metastases. Cancer Res. 2003;63:3886–3890. [PubMed] [Google Scholar]
- Abdulkadir SA, Magee JA, Peters TJ, Kaleem Z, Naughton CK, Humphrey PA, Milbrandt J. Conditional loss of Nkx3.1 in adult mice induces prostatic intraepithelial neoplasia. Mol Cell Biol. 2002;22:1495–1503. doi: 10.1128/mcb.22.5.1495-1503.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azpiazu N, Frasch M. Tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- Asatiani E, Huang WX, Wang A, Rodriguez Ortner E, Cavalli LR, Haddad BR, Gelmann EP. Deletion, methylation, and expression of the NKX3.1 suppressor gene in primary human prostate cancer. Cancer Res. 2005;65:1164–1173. doi: 10.1158/0008-5472.CAN-04-2688. [DOI] [PubMed] [Google Scholar]
- Banach-Petrosky W, Jessen WJ, Ouyang X, Gao H, Rao J, Quinn J, Aronow BJ, Abate-Shen C. Prolonged exposure to reduced levels of androgen accelerates prostate cancer progression in Nkx3.1; Pten mutant mice. Cancer Res. 2007;67:9089–9096. doi: 10.1158/0008-5472.CAN-07-2887. [DOI] [PubMed] [Google Scholar]
- Bethel CR, Bieberich CJ. Loss of Nkx3.1 expression in the transgenic adenocarcinoma of mouse prostate model. Prostate. 2007;67:1740–1750. doi: 10.1002/pros.20579. [DOI] [PubMed] [Google Scholar]
- Bethel CR, Faith D, Li X, Guan B, Hicks JL, Lan F, Jenkins RB, Bieberich CJ, De Marzo AM. Decreased NKX3.1 protein expression in focal prostatic atrophy, prostatic intraepithelial neoplasia, and adenocarcinoma: association with gleason score and chromosome 8p deletion. Cancer Res. 2006;66:10683–10690. doi: 10.1158/0008-5472.CAN-06-0963. [DOI] [PubMed] [Google Scholar]
- Bhatia-Gaur R, Donjacour AA, Sciavolino PJ, Kim M, Desai N, Young P, Norton CR, Gridley T, Cardiff RD, Cunha GR, Abate-Shen C, Shen MM. Roles for Nkx3.1 in prostate development and cancer. Genes Dev. 1999;13:966–977. doi: 10.1101/gad.13.8.966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieberich CJ, Fujita K, He WW, Jay G. Prostate-specific and androgen-dependent expression of a novel homeobox gene. J Biol Chem. 1996;271:31779–31782. doi: 10.1074/jbc.271.50.31779. [DOI] [PubMed] [Google Scholar]
- Bowen C, Bubendorf L, Voeller HJ, Slack R, Willi N, Sauter G, Gasser TC, Koivisto P, Lack EE, Kononen J, Kallioniemi OP, Gelmann EP. Loss of NKX3.1 expression in human prostate cancers correlates with tumor progression. Cancer Res. 2000;60:6111–6115. [PubMed] [Google Scholar]
- Bowen C, Stuart A, Ju JH, Tuan J, Blonder J, Conrads TP, Veenstra TD, Gelmann EP. NKX3.1 homeodomain protein binds to topoisomerase I and enhances its activity. Cancer Res. 2007;67:455–464. doi: 10.1158/0008-5472.CAN-06-1591. [DOI] [PubMed] [Google Scholar]
- Cantley LC. The phosphoinositide 3-kinase pathway. Science. 2002;296:1655–1657. doi: 10.1126/science.296.5573.1655. [DOI] [PubMed] [Google Scholar]
- Cantley LC, Neel BG. New insights into tumor suppression: PTEN suppresses tumor formation by restraining the phosphoinositide 3-kinase/AKT pathway. Proc Natl Acad Sci USA. 1999;96:4240–4245. doi: 10.1073/pnas.96.8.4240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson JA, Fillmore RA, Schwartz RJ, Zimmer WE. The smooth muscle gamma-actin gene promoter is a molecular target for the mouse bagpipe homologue, mnkx3-1, and serum response factor. J Biol Chem. 2000;275:39061–39072. doi: 10.1074/jbc.M006532200. [DOI] [PubMed] [Google Scholar]
- Chen CY, Schwartz RJ. Identification of novel DNA binding targets and regulatory domains of a murine tinman homeodomain factor, nkx-2.5. J Biol Chem. 1995;270:15628–15633. doi: 10.1074/jbc.270.26.15628. [DOI] [PubMed] [Google Scholar]
- Chen H, Bieberich CJ. Structural and functional analysis of domains mediating interaction between NKX-3.1 and PDEF. J Cell Biochem. 2005;94:168–177. doi: 10.1002/jcb.20297. [DOI] [PubMed] [Google Scholar]
- Chen H, Mutton LN, Prins GS, Bieberich CJ. Distinct regulatory elements mediate the dynamic expression pattern of Nkx3.1. Dev Dyn. 2005;234:961–973. doi: 10.1002/dvdy.20596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Nandi AK, Li X, Bieberich CJ. NKX-3.1 interacts with prostate-derived Ets factor and regulates the activity of the PSA promoter. Cancer Res. 2002;62:338–340. [PubMed] [Google Scholar]
- Choi CY, Kim YH, Kwon HJ, Kim Y. The homeodomain protein NK-3 recruits Groucho and a histone deacetylase complex to repress transcription. J Biol Chem. 1999;274:33194–33197. doi: 10.1074/jbc.274.47.33194. [DOI] [PubMed] [Google Scholar]
- Cillo C, Cantile M, Faiella A, Boncinelli E. Homeobox genes in normal and malignant cells. J Cell Physiol. 2001;188:161–169. doi: 10.1002/jcp.1115. [DOI] [PubMed] [Google Scholar]
- Cote RJ, Shi Y, Groshen S, Feng AC, Cordon-Cardo C, Skinner D, Lieskovosky G. Association of p27Kip1 levels with recurrence and survival in patients with stage C prostate carcinoma. J Natl Cancer Inst. 1998;90:916–920. doi: 10.1093/jnci/90.12.916. [DOI] [PubMed] [Google Scholar]
- Cunha GR, Donjacour AA, Cooke PS, Mee S, Bigsby RM, Higgins SJ, Sugimura Y. The endocrinology and developmental biology of the prostate. Endocrine Rev. 1987;8:338–362. doi: 10.1210/edrv-8-3-338. [DOI] [PubMed] [Google Scholar]
- Damante G, Di Lauro R. Several regions of Antennapedia and thyroid transcription factor 1 homeodomains contribute to DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:5388–5392. doi: 10.1073/pnas.88.12.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damante G, Fabbro D, Pellizzari L, Civitareale D, Guazzi S, Polycarpou-Schwartz M, Cauci S, Quadrifoglio F, Formisano S, Di Lauro R. Sequence-specific DNA recognition by the thyroid transcription factor-1 homeodomain. Nucleic Acids Res. 1994;22:3075–3083. doi: 10.1093/nar/22.15.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27kip1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–224. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–390. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- Ellwood-Yen K, Graeber TG, Wongvipat J, Iruela-Arispe ML, Zhang J, Matusik R, Thomas GV, Sawyers CL. Myc-driven murine prostate cancer shares molecular features with human prostate tumors. Cancer Cell. 2003;4:223–238. doi: 10.1016/s1535-6108(03)00197-1. [DOI] [PubMed] [Google Scholar]
- Erdamar S, Yang G, Harper JW, Lu X, Kattan MW, Thompson TC, Wheeler TM. Levels of expression of p27kip1 protein in human prostate and prostate cancer: an immunohistochemical analysis. Mod Pathol. 1999;12:751–755. [PubMed] [Google Scholar]
- Gao H, Ouyang X, Banach-Petrosky W, Borowsky AD, Lin Y, Kim M, Lee H, Shih WJ, Cardiff RD, Shen MM, Abate-Shen C. A critical role for p27kip1 gene dosage in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2004;101:17204–17209. doi: 10.1073/pnas.0407693101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Ouyang X, Banach-Petrosky WA, Gerald WL, Shen MM, Abate-Shen C. Combinatorial activities of Akt and B-Raf/Erk signaling in a mouse model of androgen-independent prostate cancer. Proc Natl Acad Sci USA. 2006a;103:14477–14482. doi: 10.1073/pnas.0606836103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Ouyang X, Banach-Petrosky WA, Shen MM, Abate-Shen C. Emergence of androgen independence at early stages of prostate cancer progression in Nkx3.1; Pten mice. Cancer Res. 2006b;66:7929–7933. doi: 10.1158/0008-5472.CAN-06-1637. [DOI] [PubMed] [Google Scholar]
- Gary B, Azuero R, Mohanty GS, Bell WC, Eltoum IE, Abdulkadir SA. Interaction of Nkx3.1 and p27kip1 in prostate tumor initiation. Am J Pathol. 2004;164:1607–1614. doi: 10.1016/S0002-9440(10)63719-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmann EP, Steadman DJ, Ma J, Ahronovitz N, Voeller HJ, Swope S, Abbaszadegan M, Brown KM, Strand K, Hayes RB, Stampfer MJ. Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function. Cancer Res. 2002;62:2654–2659. [PubMed] [Google Scholar]
- Gruschus JM, Tsao DH, Wang LH, Nirenberg M, Ferretti JA. Interactions of the vnd/NK-2 homeodomain with DNA by nuclear magnetic resonance spectroscopy: basis of binding specificity. Biochemistry. 1997;36:5372–5380. doi: 10.1021/bi9620060. [DOI] [PubMed] [Google Scholar]
- Guan B, Pungaliya P, Li X, Uquillas C, Mutton LN, Rubin EH, Bieberich CJ. Ubiquitination by TOPORS Regulates the Prostate Tumor Suppressor NKX3.1. J Biol Chem. 2008;283:4834–4840. doi: 10.1074/jbc.M708630200. [DOI] [PubMed] [Google Scholar]
- Harvey RP. NK-2 homeobox genes and heart development. Dev Biol. 1996;178:203–216. doi: 10.1006/dbio.1996.0212. [DOI] [PubMed] [Google Scholar]
- Hayashi N, Sugimura Y, Kawamura J, Donjacour AA, Cunha GR. Morphological and functional heterogeneity in the rat prostatic gland. Biol Reprod. 1991;45:308–321. doi: 10.1095/biolreprod45.2.308. [DOI] [PubMed] [Google Scholar]
- Hayward SW, Rosen MA, Cunha GR. Stromal-epithelial interactions in the normal and neoplastic prostate. Br J Urol. 1997;79(Suppl 2):18–26. doi: 10.1111/j.1464-410x.1997.tb16917.x. [DOI] [PubMed] [Google Scholar]
- He WW, Sciavolino PJ, Wing J, Augustus M, Hudson P, Meissner PS, Curtis RT, Shell BK, Bostwick DG, Tindall DJ, Gelmann EP, Abate-Shen C, Carter KC. A novel human prostate-specific, androgen-regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997;43:69–77. doi: 10.1006/geno.1997.4715. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Paroush Z, Ish-Horowicz D. Groucho acts as a corepressor for a subset of negative regulators, including Hairy and Engrailed. Genes Dev. 1997;11:3072–3082. doi: 10.1101/gad.11.22.3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju JH, Maeng JS, Zemedkun M, Ahronovitz N, Mack JW, Ferretti JA, Gelmann EP, Gruschus JM. Physical and functional interactions between the prostate suppressor homeoprotein NKX3.1 and serum response factor. J Mol Biol. 2006;360:989–999. doi: 10.1016/j.jmb.2006.05.064. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Benson DW. Biochemical analyses of eight NKX2.5 homeodomain missense mutations causing atrioventricular block and cardiac anomalies. Cardiovasc Res. 2004;64:40–51. doi: 10.1016/j.cardiores.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Kim MJ, Bhatia-Gaur R, Banach-Petrosky WA, Desai N, Wang Y, Hayward SW, Cunha GR, Cardiff RD, Shen MM, Abate-Shen C. Nkx3.1 mutant mice recapitulate early stages of prostate carcinogenesis. Cancer Res. 2002a;62:2999–3004. [PubMed] [Google Scholar]
- Kim MJ, Cardiff RD, Desai N, Banach-Petrosky WA, Parsons R, Shen MM, Abate-Shen C. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci USA. 2002b;99:2884–2889. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Nirenberg M. Drosophila NK-homeobox genes. Proc Natl Acad Sci USA. 1989;86:7716–7720. doi: 10.1073/pnas.86.20.7716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Choi CY, Lee SJ, Conti MA, Kim Y. Homeodomain-interacting protein kinases, a novel family of corepressors for homeodomain transcription factors. J Biol Chem. 1998;273:25875–25879. doi: 10.1074/jbc.273.40.25875. [DOI] [PubMed] [Google Scholar]
- Korkmaz KS, Korkmaz CG, Ragnhildstveit E, Kizildag S, Pretlow TG, Saatcioglu F. Full-length cDNA sequence and genomic organization of human NKX3A—alternative forms and regulation by both androgens and estrogens. Gene. 2000;260:25–36. doi: 10.1016/s0378-1119(00)00453-4. [DOI] [PubMed] [Google Scholar]
- Kos L, Chiang C, Mahon KA. Mediolateral patterning of somites: multiple axial signals, including Sonic hedgehog, regulate Nkx-3.1 expression. Mech Dev. 1998;70:25–34. doi: 10.1016/s0925-4773(97)00168-8. [DOI] [PubMed] [Google Scholar]
- Kuczyk M, Machtens S, Hradil K, Schubach J, Christian W, Knuchel R, Hartmann J, Bokemeyer C, Jonas U, Serth J. Predictive value of decreased p27Kip1 protein expression for the recurrence-free and long-term survival of prostate cancer patients. Br J Cancer. 1999;81:1052–1058. doi: 10.1038/sj.bjc.6690806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Q, Jiao J, Xin L, Chang CJ, Wang S, Gao J, Gleave ME, Witte ON, Liu X, Wu H. NKX3.1 stabilizes p53, inhibits AKT activation, and blocks prostate cancer initiation caused by PTEN loss. Cancer Cell. 2006;9:367–378. doi: 10.1016/j.ccr.2006.03.031. [DOI] [PubMed] [Google Scholar]
- Li X, Guan B, Maghami S, Bieberich CJ. NKX3.1 is regulated by protein kinase CK2 in prostate tumor cells. Mol Cell Biol. 2006;26:3008–3017. doi: 10.1128/MCB.26.8.3008-3017.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lints TJ, Parsons LM, Hartley L, Lyons I, Harvey RP. Nkx-2.5: a novel murine homeobox gene expressed in early heart progenitor cells and their myogenic descendants. Development. 1993;119:419–431. doi: 10.1242/dev.119.2.419. [DOI] [PubMed] [Google Scholar]
- Magee JA, Abdulkadir SA, Milbrandt J. Haploinsufficiency at the Nkx3.1 locus. A paradigm for stochastic, dosage-sensitive gene regulation during tumor initiation. Cancer Cell. 2003;3:273–283. doi: 10.1016/s1535-6108(03)00047-3. [DOI] [PubMed] [Google Scholar]
- Marker PC, Dahiya R, Cunha GR. Spontaneous mutation in mice provides new insight into the genetic mechanisms that pattern the seminal vesicles and prostate gland. Dev Dyn. 2003;226:643–653. doi: 10.1002/dvdy.10276. [DOI] [PubMed] [Google Scholar]
- Mogal AP, van der Meer R, Crooke PS, Abdulkadir SA. Haploinsufficient prostate tumor suppression by Nkx3.1: a role for chromatin accessibility in dosage-sensitive gene regulation. J Biol Chem. 2007;282:25790–25800. doi: 10.1074/jbc.M702438200. [DOI] [PubMed] [Google Scholar]
- Muhr J, Andersson E, Persson M, Jessell TM, Ericson J. Groucho-mediated transcriptional repression establishes progenitor cell pattern and neuronal fate in the ventral neural tube. Cell. 2001;104:861–873. doi: 10.1016/s0092-8674(01)00283-5. [DOI] [PubMed] [Google Scholar]
- Murtaugh LC, Zeng L, Chyung JH, Lassar AB. The chick transcriptional repressor Nkx3.2 acts downstream of Shh to promote BMP-dependent axial chondrogenesis. Dev Cell. 2001;1:411–422. doi: 10.1016/s1534-5807(01)00039-9. [DOI] [PubMed] [Google Scholar]
- Oettgen P, Finger E, Sun Z, Akbarali Y, Thamrongsak U, Boltax J, Grall F, Dube A, Weiss A, Brown L, Quinn G, Kas K, Endress G, Kunsch C, Libermann TA. PDEF, a novel prostate epithelium-specific ETS transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J Biol Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- Ornstein DK, Cinquanta M, Weiler S, Duray PH, Emmert-Buck MR, Vocke CD, Linehan WM, Ferretti JA. Expression studies and mutational analysis of the androgen regulated homeobox gene NKX3.1 in benign and malignant prostate epithelium. J Urol. 2001;165:1329–1334. [PubMed] [Google Scholar]
- Ouyang X, DeWeese TL, Nelson WG, Abate-Shen C. Loss-of-function of Nkx3.1 promotes increased oxidative damage in prostate carcinogenesis. Cancer Res. 2005;65:6773–6779. doi: 10.1158/0008-5472.CAN-05-1948. [DOI] [PubMed] [Google Scholar]
- Ouyang X, Jessen W, Al-Ahmadie H, Serio A, Lin Y, Shih W, Reuter V, Scardino P, Shen M, Aronow B, Vickers A, Gerald W, Abate-Shen C. AP-1 transcription factors are associated with progression and recurrence of prostate cancer. Cancer Res. 2008;68:2133–2144. doi: 10.1158/0008-5472.CAN-07-6055. [DOI] [PubMed] [Google Scholar]
- Park JH, Walls JE, Galvez JJ, Kim M, Abate-Shen C, Shen MM, Cardiff RD. Prostatic intraepithelial neoplasia in genetically engineered mice. Am J Pathol. 2002;161:727–735. doi: 10.1016/S0002-9440(10)64228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott JL, Blok L, Tindall DJ. Isolation and androgen regulation of the human homeobox cDNA, NKX3.1. Prostate. 1998;35:71–80. doi: 10.1002/(sici)1097-0045(19980401)35:1<71::aid-pros10>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Schneider A, Brand T, Zweigerdt R, Arnold H. Targeted disruption of the Nkx3.1 gene in mice results in morphogenetic defects of minor salivary glands: parallels to glandular duct morphogenesis in prostate. Mech Dev. 2000;95:163–174. doi: 10.1016/s0925-4773(00)00355-5. [DOI] [PubMed] [Google Scholar]
- Sciavolino PJ, Abrams EW, Yang L, Austenberg LP, Shen MM, Abate-Shen C. Tissue-specific expression of murine Nkx3.1 in the male urogenital system. Dev Dyn. 1997;209:127–138. doi: 10.1002/(SICI)1097-0177(199705)209:1<127::AID-AJA12>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- Simmons SO, Horowitz JM. Nkx3.1 binds and negatively regulates the transcriptional activity of Sp-family members in prostate-derived cells. Biochem J. 2006;393:397–409. doi: 10.1042/BJ20051030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ST, Jaynes JB. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steadman DJ, Giuffrida D, Gelmann EP. DNA-binding sequence of the human prostate-specific homeodomain protein NKX3.1. Nucleic Acids Res. 2000;28:2389–2395. doi: 10.1093/nar/28.12.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura Y, Cunha GR, Donjacour AA. Morphogenesis of ductal networks in the mouse prostate. Biol Reprod. 1986;34:961–971. doi: 10.1095/biolreprod34.5.961. [DOI] [PubMed] [Google Scholar]
- Swalwell JI, Vocke CD, Yang Y, Walker JR, Grouse L, Myers SH, Gillespie JW, Bostwick DG, Duray PH, Linehan WM, Emmert-Buck MR. Determination of a minimal deletion interval on chromosome band 8p21 in sporadic prostate cancer. Genes Chromosomes Cancer. 2002;33:201–205. doi: 10.1002/gcc.10015. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Komuro I, Inagaki H, Jenkins NA, Copeland NG, Izumo S. Nkx3.1, a murine homolog of Drosophila bagpipe, regulates epithelial ductal branching and proliferation of the prostate and palatine glands. Dev Dyn. 2000;219:248–260. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1054>3.3.co;2-5. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Lyons GE, Izumo S. Expression of the Nkx3.1 homobox gene during pre and postnatal development. Mech Dev. 1999;85:179–182. doi: 10.1016/s0925-4773(99)00084-2. [DOI] [PubMed] [Google Scholar]
- Timms BG, Mohs TJ, Didio LJA. Ductal budding and branching patterns in the developing prostate. J Urol. 1994;151:1427–1432. doi: 10.1016/s0022-5347(17)35273-4. [DOI] [PubMed] [Google Scholar]
- Treier M, Gleiberman AS, O’Connell SM, Szeto DP, McMahon JA, McMahon AP, Rosenfeld MG. Multistep signaling requirements for pituitary organogenesis in vivo. Genes Dev. 1998;12:1691–1704. doi: 10.1101/gad.12.11.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribioli C, Frasch M, Lufkin T. Bapx1: an evolutionary conserved homologue of the Drosophila bagpipe homeobox gene is expressed in splanchnic mesoderm and the embryonic skeleton. Mech Dev. 1997;65:145–162. doi: 10.1016/s0925-4773(97)00067-1. [DOI] [PubMed] [Google Scholar]
- Tribioli C, Lufkin T. The murine Bapx1 homeobox gene plays a critical role in embryonic development of the axial skeleton and spleen. Development. 1999;126:5699–5711. doi: 10.1242/dev.126.24.5699. [DOI] [PubMed] [Google Scholar]
- Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase AKT pathway in human cancer. Nat Rev Cancer. 2002;2:489–501. doi: 10.1038/nrc839. [DOI] [PubMed] [Google Scholar]
- Vocke CD, Pozzatti RO, Bostwick DG, Florence CD, Jennings SB, Strup SE, Duray PH, Liotta LA, Emmert-Buck MR, Linehan WM. Analysis of 99 microdissected prostate carcinomas reveals a high frequency of allelic loss on chromosome 8p12-21. Cancer Res. 1996;56:2411–2416. [PubMed] [Google Scholar]
- Voeller HJ, Augustus M, Madike V, Bova GS, Carter KC, Gelmann EP. Coding region of NKX3.1, a prostate-specific homeobox gene on 8p21, is not mutated in human prostate cancers. Cancer Res. 1997;57:4455–4459. [PubMed] [Google Scholar]
- Wang S, Gao J, Lei Q, Rozengurt N, Pritchard C, Jiao J, Thomas GV, Li G, Roy-Burman P, Nelson PS, Liu X, Wu H. Prostate-specific deletion of the murine Pten tumor suppressor gene leads to metastatic prostate cancer. Cancer Cell. 2003;4:209–221. doi: 10.1016/s1535-6108(03)00215-0. [DOI] [PubMed] [Google Scholar]
- Weir BA, Woo MS, Getz G, Perner S, Ding L, Beroukhim R, Lin WM, Province MA, Kraja A, Johnson LA, Shah K, Sato M, Thomas RK, Barletta JA, Borecki IB, Broderick S, Chang AC, Chiang DY, Chirieac LR, Cho J, Fujii Y, Gazdar AF, Giordano T, Greulich H, Hanna M, Johnson BE, Kris MG, Lash A, Lin L, Lindeman N, Mardis ER, McPherson JD, Minna JD, Morgan MB, Nadel M, Orringer MB, Osborne JR, Ozenberger B, Ramos AH, Robinson J, Roth JA, Rusch V, Sasaki H, Shepherd F, Sougnez C, Spitz MR, Tsao MS, Twomey D, Verhaak RG, Weinstock GM, Wheeler DA, Winckler W, Yoshizawa A, Yu S, Zakowski MF, Zhang Q, Beer DG, Wistuba LL, Watson MA, Garraway LA, Ladanyi M, Travis WD, Pao W, Rubin MA, Gabriel SB, Gibbs RA, Varmus HE, Wilson RK, Lander ES, Meyerson M. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang RM, Naitoh J, Murphy M, Wang HJ, Phillipson J, deKernion JB, Loda M, Reiter RE. Low p27 expression predicts poor disease-free survival in patients with prostate cancer. J Urol. 1998;159:941–945. [PubMed] [Google Scholar]
- Zheng SL, Ju JH, Chang BL, Ortner E, Sun J, Isaacs SD, Wiley KE, Liu W, Zemedkun M, Walsh PC, Ferretti J, Gruschus J, Isaacs WB, Gelmann EP, Xu J. Germ-line mutation of NKX3.1 cosegregates with hereditary prostate cancer and alters the homeodomain structure and function. Cancer Res. 2006;66:69–77. doi: 10.1158/0008-5472.CAN-05-1550. [DOI] [PubMed] [Google Scholar]