Abstract
It was previously shown that enhanced nisin resistance in some mutants was associated with increased expression of three genes, pbp2229, hpk1021, and lmo2487, encoding a penicillin-binding protein, a histidine kinase, and a protein of unknown function, respectively. In the present work, we determined the direct role of the three genes in nisin resistance. Interruption of pbp2229 and hpk1021 eliminated the nisin resistance phenotype. Interruption of hpk1021 additionally abolished the increase in pbp2229 expression. The results indicate that this nisin resistance mechanism is caused directly by the increase in pbp2229 expression, which in turn is brought about by the increase in hpk1021 expression. We also found a degree of cross-protection between nisin and class IIa bacteriocins and investigated possible mechanisms. The expression of virulence genes in one nisin-resistant mutant and two class IIa bacteriocin-resistant mutants of the same wild-type strain was analyzed, and each mutant consistently showed either an increase or a decrease in the expression of virulence genes (prfA-regulated as well as prfA-independent genes). Although the changes mostly were moderate, the consistency indicates that a mutant-specific change in virulence may occur concomitantly with bacteriocin resistance development.
Nisin and the class IIa bacteriocins (also called pediocin-like bacteriocins) are antimicrobial peptides that are produced by lactic acid bacteria and that have the greatest potential as biopreservatives for food. One of the main target organisms in this context is Listeria monocytogenes, a food-borne pathogen that causes severe human illness as well as economic losses for the food industry.
Nisin exerts its antimicrobial action by forming pores in the cytoplasmic membrane through an interaction with the peptidoglycan precursor lipid II (for a recent review, see reference 17). Enhanced nisin resistance in L. monocytogenes generally constitutes less than a 10-fold increase in the MIC. Nisin resistance in several, but not all, spontaneous mutants of L. monocytogenes was associated with increased expression of three genes, encoding a putative penicillin-binding protein (pbp2229), a histidine protein kinase (hpk1021), and a protein of unknown function (lmo2487) (15). Inactivation of the two-component regulatory system encoded by lisRK resulted in decreased nisin sensitivity (10). Although various cell envelope changes have been observed in different nisin-resistant mutants, the molecular mechanisms involved remain poorly understood.
High-level resistance to class IIa bacteriocins (at least a 103-fold increase in the MIC) in L. monocytogenes and some other gram-positive bacteria is conveyed by one general mechanism, which presumably involves eradication of the docking molecule (14). The putative docking molecule is a mannose-specific phosphoenolpyruvate-dependent phosphotransferase system (PTS) encoded by mptACD, which is transcribed by the alternative sigma factor σ54 (encoded by rpoN) in conjunction with the activator ManR (12). The mechanism concomitantly gives rise to a substantial increase in the expression of two β-glucoside-specific PTS genes, encoding EIIBgl and a phospho-β-glucosidase (14).
A preventative measure that has frequently been suggested for avoiding bacteriocin resistance problems is the simultaneous use of two bacteriocins with different specific pore-forming mechanisms, e.g., nisin and a class IIa bacteriocin. This strategy raises the question of whether cross-resistance between the compounds could occur. Cross-resistance between nisin and IIa bacteriocins is not apparent in wild-type L. monocytogenes isolates (34). There are, however, contradictory reports regarding cross-resistance following the spontaneous development of resistance to one of the two types of bacteriocins (5, 11, 35).
Bacteriocins are cationic antimicrobial peptides and are related to the defensin-type components of the innate immune system (28). Nonspecific cross-protection between cationic peptides could conceivably render bacteriocin-resistant L. monocytogenes mutants less sensitive to defensins. Additionally, high-level class IIa bacteriocin resistance involves changes in the expression of several PTSs (14), including β-glucoside-specific PTSs, which are known to repress prfA-dependent virulence gene induction (7). All in all, it is therefore important to consider whether the development of bacteriocin resistance could influence the virulence of L. monocytogenes.
The aims of the present work were to provide a mechanistic understanding of nisin resistance and to assess whether the development of bacteriocin resistance could concomitantly confer reduced sensitivity to other bacteriocins or changes in virulence. We determined the role in the nisin resistance mechanism of the three genes that showed increased expression in certain nisin-resistant mutants. Furthermore, we investigated the occurrence of and mechanisms for cross-resistance between nisin and class IIa bacteriocins. Finally, possible changes in virulence were assessed by analysis of virulence gene expression in representative nisin-resistant and class IIa bacteriocin-resistant mutants.
MATERIALS AND METHODS
Strains and growth conditions.
The wild-type L. monocytogenes strains and derived spontaneous bacteriocin-resistant mutants used here are described in Table 1. Cultures were grown in brain heart infusion (BHI) broth (Difco, Detroit, Mich.) at 30°C without agitation or on tryptone soya agar (TSA; Oxoid, Basingstoke, United Kingdom), unless stated otherwise.
TABLE 1.
Bacteriocin phenotypes of bacteriocin-resistant mutants and corresponding wild-type L. monocytogenes strains
| Straina | Phenotypeb for:
|
Reference or source | |||
|---|---|---|---|---|---|
| Nisin A | Pediocin PA-1 | Leucocin A | Carnobacteriocin B2 | ||
| 412 | ES | ES | ES | S | 16 |
| 412N | I | S | S | S | 15 |
| 412C | S | I | I | I | This workc |
| 412P | S | HR | HR | HR | 16 |
| 412C2 | S | HR | HR | HR | 14 |
| 358 | ES | ES | S | S | 15 |
| 358N | I | S | I | S | 15 |
| 358C | S | HR | HR | HR | This workc |
| 409 | S | ES | ES | S | 15 |
| 409N | I | S | I | S | 15 |
| B73 | ES | ES | ES | ES | 13a |
| B73-V1 | ES | I | I | S | 43 |
| B73-V2 | ES | I | I | S | 43 |
| B73-MR1 | ES | HR | HR | HR | 33 |
| B73P | ES | HR | HR | HR | This workd |
| 31 | S | ES | ES | S | 16a |
| 31P | S | I | I | I | 16a |
| O57 | ES | S | S | S | 1a |
| 3.33A | ES | HR | HR | HR | 14 |
| O57C | ES | HR | HR | HR | This workc |
| EGDe | ES | ES | ES | ES | 13b |
| EGY2 | ES | HR | HR | HR | 12 |
Spontaneous mutants were isolated from wild-type strains 412, 358, 409, B73, 31, and O57. Mutants were isolated by selection as follows: N, with nisin A; P, with pediocin PA-1; C, C2, and A, with carnobacteriocin B2; and MR1, V1, and V2, with leucocin A. EGY2 was made from wild-type EGDe by in-frame deletion in the mptD gene.
ES, S, I, and HR indicate extremely sensitive, sensitive, intemediately resistant, and highly resistant, respectively.
Isolated with 100 C-AU of carnobacteriocin B2 ml−1 and 0.1% Tween 80 on TSA plates incubated at 5°C for 24 h and then at 30°C.
Isolated with 2,560 P-AU of pediocin PA-1 ml−1 on TSA plates (pH 6.5) incubated at 30°C.
Bacteriocin sensitivity assay.
The class IIa bacteriocins, pediocin PA-1, leucocin A, and carnobacteriocin B2, were prepared as fermentation products of the producer organisms Pediococcus acidilactici PA-2 (C. Hansen A/S, Hørsholm, Denmark), Leuconostoc gelidum UAL 187-22 (29), and Carnobacterium piscicola A9b (26), respectively, as described previously (14). The activities of the pediocin and leucocin fermentates were determined by a turbidimetric screening assay (modified from that described in reference 25) with L. monocytogenes 412 as an indicator organism and were expressed as arbitrary units (AU) specific for each compound (P-AU and L-AU, respectively). Carnobacteriocin B2 was kindly supplied by Lilian Nilsson (Danish Institute for Fisheries Research, Lyngby, Denmark); its activity (C-AU) was determined by an agar well diffusion assay with L. monocytogenes O57 as an indicator as described previously (27). Pure nisin A was kindly supplied by Danisco Beaminster Ltd. (Beaminster, United Kingdom) and prepared as a stock solution of 500,000 IU/ml in 0.02 M HCl.
Bacteriocin sensitivity was assessed as growth inhibition by a low, an intermediate, or a high level of bacteriocin in an agar spot assay. Overnight cultures (BHI broth with the pH adjusted to 6.5) were diluted to an optical density at 600 nm (OD600) of 0.5 and serially diluted to 10−2 in physiological saline with peptone, and two 10-μl samples were spotted on TSA plates (pH 6.5) supplemented with 0.1% Tween 80 and the appropriate amount of bacteriocin. For each bacteriocin, concentrations were chosen from preliminary experiments such that the low concentration permitted uninhibited growth of most wild-type strains, the intermediate concentration inhibited the growth of most wild-type strains and some mutants, and the high concentration prevented the growth of all wild-type strains and inhibited most mutants. The levels used were 40, 200, and 750 IU of nisin A ml−1; 50, 400, and 2,000 C-AU of carnobacteriocin B2 ml−1; 20, 40, and 3,072 P-AU of pediocin PA-1 ml−1; and 20, 40, and 768 L-AU of leucocin A ml−1. The plates were incubated at 5°C for 24 h and subsequently at 25°C for 24 h, and growth was monitored visually. The assay allowed the designation of four bacteriocin phenotypes: extremely sensitive strains could not grow even at the lowest concentration, sensitive strains were inhibited by the intermediate concentration, intermediately resistant strains were inhibited by the highest concentration, and highly resistant strains were not affected by the highest concentration of bacteriocin.
Construction of insertion mutants.
Genes pbp2229, hpk1021, and lmo2487 were interrupted in L. monocytogenes 412 and 412N. The three restriction fragment differential display-PCR products (16) were cloned in temperature-sensitive integration vector pAUL-A (9), resulting in plasmids pLG10, pBK26, and pLG11. These plasmids contained a 241-bp internal fragment of pbp2229, encoding putative penicillin-binding protein PBP2229 (annotated as lmo2229 in the L. monocytogenes genome sequence; http://genolist.pasteur.fr/ListiList/); 316 bp of hpk1021, encoding the histidine kinase homologue HPK1021 (lmo1021); and 199 bp of a gene encoding a protein of unknown function (lmo2487), respectively. Each of the plasmids was transformed into the two strains by electroporation (30) and integrated into the chromosome by propagation at 42°C in the presence of 5 μg of erythromycin ml−1 (9). Integration of pLG10, pBK26, and pLG11 into L. monocytogenes 412 resulted in strains AG100, AG111, and AG106, and integration of these plasmids into L. monocytogenes 412N resulted in strains AG105, AG114, and AG104, respectively (Table 2). Correct insertion was verified by PCR with standard primers for the vector sequence and a primer complementary to chromosomal DNA adjacent to the segment used for insertion. The multiple cloning site in pAUL-A is flanked by terminators, preventing transcription out of and into the inserted fragment (37).
TABLE 2.
MICs of nisin A and pediocin PA-1 of L. monocytogenes strains
| Strain | Descriptiona | MIC of:
|
|
|---|---|---|---|
| Nisin (IU/ml) | Pediocin (P-AU/ml) | ||
| 412 | Wild type | 1,000 | 80 |
| AG100 | Inactivation of pbp2229 in 412 | 750 | 80 |
| AG111 | Inactivation of hpk1021 in 412 | 1,000 | 80 |
| AG106 | Inactivation of lmo2487 in 412 | 1,000 | 80 |
| 412N | Nisin-resistant mutant of 412 | 1,500 | 160 |
| AG105 | Inactivation of pbp2229 in 412N | 750 | 80 |
| AG114 | Inactivation of hpk1021 in 412N | 25 | 80 |
| AG104 | Inactivation of lmo2487 in 412N | 1,500 | 160 |
Genes were inactivated by plasmid insertion (see Materials and Methods).
The MICs of nisin and pediocin for the six insertion mutants and strains 412 and 412N were determined by an agar spot assay. Overnight cultures grown in BHI broth at 37°C without shaking were diluted to approximately 107 CFU ml−1 in physiological saline with peptone, and two 10-μl samples were spotted on TSA plates containing 1,500, 1,250, 1,000, 750, 500, 400, 300, 200, 100, 50, 25, and 10 IU of nisin ml−1 (TSA adjusted to pH 6.5) or 320, 160, 80, 40, 20, and 10 P-AU of pediocin ml−1. The plates were incubated at 37°C for 24 h, and the MIC was determined as the median minimal concentration preventing growth for three independent cultures for nisin and for two independent cultures for pediocin. The insertion in the spotted cultures was verified by parallel enumeration at 37 and 42°C on plates containing 5 μg of erythromycin ml−1.
Construction of pbp2229-lacZ and lmo2487-lacZ transcriptional fusions and β-galactosidase assays.
A PCR fragment of 345 bp containing the promoter region of pbp2229 was amplified with primers pbp2229-F (5′-GGGGAATTCGATCAGCTTTTGCGTCATATTCC) and pbp2229-R (5′-GGGGGATCCGTCCATGTAACTCTCCTATCTTC); to facilitate cloning of the PCR fragments, the primers contained EcoRI or BamHI restriction sites (underlined). A 332-bp fragment containing the promoter region of lmo2487 was generated with primers lmo2487-F (5′-GGGGGAATTCAAAGAACGCACGGAAGAACG) and lmo2487-R (5′-GGGGGGATCCCAAGTAGAGTAAGGGCTTCTTC). The PCR fragments were digested with EcoRI and BamHI and cloned into EcoRI/BamHI-digested pTCV-lac (32), thereby generating plasmids pPBP2229-lacZ and pLMO2487-lacZ. The fusions of the pbp2229 and lmo2487 promoter regions to lacZ in pPBP2229-lacZ and pLMO2487-lacZ, respectively, were verified by DNA sequencing. The plasmids were introduced into L. monocytogenes strains by electroporation (30).
For the β-galactosidase assay, the L. monocytogenes strains containing the lacZ fusion plasmids were grown in BHI medium at 37°C with agitation, and 1-ml samples of cells were collected at various times. Cells were permeabilized by treatment with 0.5% toluene and 4.5% ethanol, and β-galactosidase activity was determined as described previously (22). The specific activity of β-galactosidase was calculated as follows: (OD420 of the reaction mixture − OD550 of the reaction mixture)/(reaction time in minutes × OD600 of the cells used in the reaction mixture). Three independent experiments were carried out. The β-galactosidase activities presented are the averages of the three determinations. The observed variations in the three experiments did not exceed 10%.
RNA extraction for analysis of gene expression.
The expression of selected genes in exponentially growing cultures of L. monocytogenes 412, 412C, 412P, and 412N was analyzed by DNA microarray hybridization. Strains were grown in charcoal-activated BHI medium (0.2% vegetable charcoal; Merck) at 37°C with agitation, and cells were harvested in mid-exponential phase at an OD600 of approximately 1. RNA was isolated as described previously (14, 16) with an RNeasy kit supplemented with in-column DNase treatment (Qiagen, Hilden, Germany).
The expression of hpk1021 in L. monocytogenes 412 and 412N and the six insertion mutants was analyzed by Northern blot hybridization. The wild-type strain and insertion mutants were grown at 37°C without shaking, and RNA was extracted from mid-exponential-phase cells (OD600, approximately 0.6). The harvested cultures were enumerated in parallel at 37 and 42°C on plates containing 5 μg of erythromycin ml−1 to verify the insertion.
DNA microarray design and fabrication.
A DNA microarray was designed to comprise a total of 64 genes from L. monocytogenes covering, among other functions, genes involved in virulence and in putative bacteriocin resistance mechanisms. The microarray also contained a number of spots representing negative and ratio controls. The negative control spots were either samples without DNA, reporting possible carryover contamination, or nonrelevant genes, reporting nonspecific hybridization. The ratio control spots contained a gene sequence complementary to an in vitro-synthesized RNA molecule which was used in equal amounts to spike the cDNA labeling reactions of the mutant and wild-type total RNAs.
Internal gene fragments ranging from 500 to 1,000 bp were made by PCR with specific primers designed for each gene. Excess primers were removed from the PCR products by using GFX columns (Amersham Biosciences, Hørsholm, Denmark), and the DNA amplicons were concentrated to at least 100 ng μl−1. Purified and concentrated DNA amplicons were mixed 1:1 in dimethyl sulfoxide (Sigma, Gillingham, United Kingdom) and spotted in quadruplicate by using a QPix2 robot (Genetix Ltd., New Milton, United Kingdom) equipped with a microarray accessory unit. Spotting was conducted with ChipMaker CMP3 microspotting split pins (TeleChem International Inc., Sunnyvale, Calif.) and Corning CMT-Gaps-2 coated slides (Biotech Line AS, Slangerup, Denmark). After spotting was complete, the slides were baked at 80°C for 4 h, washed in 70% ethanol for 15 min, heat denatured at 95°C in double-distilled H2O for 2 min, and finally rinsed for 1 min in 96% ethanol. The slides were subsequently allowed to air dry in a clean and dust-free environment. The slides were kept sealed at room temperature in a humidity-free environment (e.g., in a dessicator) until used.
Preparation of ivt-RNA.
In vitro-transcribed control RNA (ivt-RNA) was synthesized by using a MEGAscript T7 RNA polymerase kit (Ambion) in accordance with the manufacturer's instructions. The RNA was transcribed from a cloned full-length human urokinase plasminogen activator receptor gene (36). This gene did not have any significant homology to the L. monocytogenes genome sequence.
Preparation of labeled cDNA.
Fluorescence-labeled cDNA copies of total RNA from L. monocytogenes were prepared by direct incorporation of Cy5- or Cy3-labeled dUTP analogues through a first-strand reverse transcription reaction with a CyScribe first-strand cDNA labeling kit (Amersham Biosciences) in accordance with kit instructions. For each reaction, 20 μg of total RNA and 1 ng of ivt-RNA were mixed with 5 pmol of random-nonamer oligonucleotides before labeling. To remove unincorporated fluorophore-labeled dUTP after the labeling procedure, a CyScribe GFX purification protocol was applied as recommended by the manufacturer (Amersham Biosciences), yielding a final volume of 30 μl of labeled cDNA. The efficiency of cDNA labeling was determined spectrophotometrically at 550 nm (Cy3) or 650 nm (Cy5) in combination with OD260 measurements.
Microarray hybridization.
Custom-made DNA microarray slides were prehybridized with 1 ml of prehybridization buffer, containing 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 10 mg of bovine serum albumin/ml, and 25% (vol/vol) formamide, at 42°C for 45 min without a cover glass. The slides were rinsed briefly with distilled H2O and dried with pressurized air. Subsequently, purified labeled cDNA fractions prepared from equal amounts of wild-type L. monocytogenes and mutant L. monocytogenes total RNAs were mixed 1:1, i.e., 30 μl of Cy3-labeled cDNA and 30 μl of Cy5-labeled cDNA. To this mixture was added 10 μl of salmon sperm DNA (2 μg/μl) and 70 μl of hybridization buffer, containing 10× SSC, 0.2% SDS, and 50% (vol/vol) formamide. The hybridization mixture was heated to 95°C for 5 min in a 1.5-ml Eppendorf tube, followed by centrifugation for 2 min at 6,000 × g. The entire volume of the hybridization mixture was dispensed on the surface of the prehybridized microarray and gently covered with a clean cover glass. The slides were allowed to hybridize in 80% relative humidity at 42°C overnight in the dark. After hybridization, the cover glass was gently removed by dipping the slides into 2× SSC-0.1% SDS at 42°C. The slides then were transferred to 0.1× SSC-0.1% SDS and left at room temperature for 10 min, followed by four washes in 0.1× SSC for 1 min each. The slides were rinsed in distilled H2O and briefly dipped in 99% ethanol before being air dried in the dark. At this point, the slides were ready for scanning or could be kept in the dark for later analysis.
Gene expression data analysis.
DNA microarray data were analyzed from digital pictures of the slide surface acquired by using an arrayWoRx white-light charge-coupled device-based scanner (Applied Precision, Issaquah, Washington) with a 0.5-s exposure time in both the Cy3 and the Cy5 channels and a 10-μm image resolution. The arrayWoRx software package was used to determine ratio and intensity values. A microarray grid defining a spot size and a region of interest for each spot was used to calculate spot total intensities and local background intensities in both the Cy3 and the Cy5 channels. Ratio values were normalized automatically according to the average intensity of the eight ratio control spots on each slide. Signal intensities of negative control spots defined the detection limit for expression analysis, and all genes with signal intensities lower than the average negative control spot intensity plus two times the standard deviation were considered not detectable.
To evaluate the statistical significance of changes in individual gene expression levels between the wild-type and the mutant strains, we performed a t test of whether the means of two different sets of ratio values, i.e., the ratio values for the control spots and the ratio values for a specific gene, were likely to be equal. The t test requires Gaussian data and, by doing a QQ-plot test, we found that the deviation of our data from normality was not large enough to warrant rejection of the t test as a method. For each wild-type-mutant combination, a total of 32 data points represented the ratio values for the controls and a total of 16 data points represented the ratio values for each sample gene in two independent experiments performed in duplicate. The t test returns a P value that predicts the smallest level of significance that leads to rejection of the null hypothesis, i.e.,: the average ratio value for the control equals the average ratio value for the sample gene. Since we performed multiple testing of 64 different genes at a time, a Bonferroni correction (2) of the P value was required. Thus, we considered only P values of less than 0.05/64, or 0.0008, to imply significant regulation, representing less than a 5% probability of false-positive regulation in any of the genes.
Northern blot analysis.
Northern hybridization with 3 μg of RNA was carried out as described previously (14, 16), and the signal intensity was quantified by using ImageQuant software, version 5.0 (Molecular Dynamics, Amersham Biosciences). The expression of hpk1021 in L. monocytogenes 412 and 412N and the six insertion mutants was analyzed with the same PCR product (545 bp, nucleotides 24 to 568) as that used on the DNA microarray as a probe. This fragment is situated upstream of and partially overlapping the restriction fragment differential display-PCR fragment used for insertional inactivation of the gene (nucleotides 335 to 650). The expression of selected virulence genes (prfA, inlA, inlB, inlC2, and inlD) in L. monocytogenes 412, 412N, 412C, and 412P was analyzed with the same internal gene fragments as those used on the DNA microarray as probes and hybridization to the RNA used in the DNA microarray experiment.
Protein extraction and 2-D gel electrophoresis.
The total cellular proteins of L. monocytogenes 412 and 412C were extracted, resolved by 2-D gel electrophoresis, stained with colloidal Coomassie brilliant blue, and analyzed as described previously (33), except that 18-cm isoelectric focusing strips were used for one-dimensional electrophoresis. Proteins were extracted from two independent cultures of each strain, and at least two 2-D gel electrophoresis analyses were conducted for each sample.
RESULTS
Bacteriocin cross-resistance in spontaneous mutants.
In order to assess whether resistance that developed to one bacteriocin concomitantly conferred resistance to other bacteriocins, the sensitivities to nisin A and three class IIa bacteriocins, pediocin PA-1, leucocin A, and carnobacteriocin B2, were determined for 14 spontaneous mutants (Table 1). The bacteriocin sensitivity assay was carried out with low, intermediate, and high levels of the four bacteriocins and allowed designation of the phenotypes extremely sensitive, sensitive, intermediately resistant, and highly resistant to the respective compound. Of the 14 spontaneous mutants, 5 mutants were isolated following exposure to carnobacteriocin, and 3 each were isolated on pediocin, leucocin, and nisin. Additionally, a mutant with an in-frame deletion in the mptD gene, L. monocytogenes EGY2 (12), was tested.
The three mutants isolated on nisin, 412N, 358N, and 409N, were all intermediately nisin resistant and also had reduced sensitivity to pediocin and leucocin (Table 1). The mutants isolated on class IIa bacteriocins showed complete cross-resistance among these bacteriocins: intermediately resistant mutants (strains 412C, 31P, B73-V1, and B73-V2) isolated on one class IIa bacteriocin also had reduced sensitivity to the other two compounds, and highly resistant mutants (strains 412P, 412C2, 358C, B73-MR1, B73P, 3.33A, O57C, and EGY2) had this phenotype for all three class IIa bacteriocins tested. There was, however, no consistent correlation between developed class IIa bacteriocin resistance and nisin sensitivity. The mutants of L. monocytogenes 412 that were intermediately and highly resistant to class IIa bacteriocins all had reduced sensitivity to nisin. This finding was also observed for the mutant of strain 358, which was highly resistant to class IIa bacteriocins. The remaining three mutants with an intermediate level of resistance and five mutants with a high level of resistance to class IIa bacteriocins, originating from wild-type strains 409, B73, 31, O57, and EGDe, had unaltered nisin sensitivity.
Interruption of genes associated with nisin resistance.
In order to investigate whether the increased expression of the pbp2229, hpk1021, and lmo2487 genes had a direct role in the nisin resistance mechanism, the genes were inactivated by plasmid integration. Gene inactivation in L. monocytogenes 412 and 412N resulted in various degrees of cell elongation and chain formation. This finding was particularly pronounced in strain AG105, the 412N derivative with an insertion in pbp2229, for which no normal rods were observed in an overnight liquid culture. The strain with the fewest chains was AG114, the 412N derivative with an insertion in hpk1021. The two strains with insertions in lmo2487 showed reduced growth (monitored as the OD600), and this finding was particularly pronounced for the 412 derivative (Fig. 1C and F).
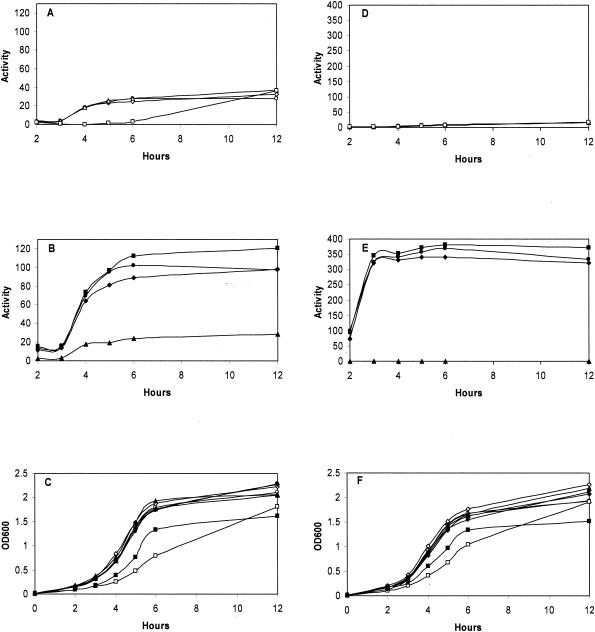
FIG. 1.
Analysis of promoter activity by lacZ fusions. Beta-galactosidase activity (A, B, D, and E) and growth measured as OD600 (C and F) of strains transformed with a plasmid containing a fusion of the lacZ reporter gene to the promoter region of pbp2229 (A, B, and C) or lmo2487 (D, E, and F). Open symbols: L. monocytogenes 412 and derived insertion mutants; closed symbols: strain 412N and derived insertion mutants; diamonds, 412 or 412N; circles, strain with insertion in pbp2229; triangles, strain with insertion in hpk1021; squares, strain with insertion in lmo2487.
The MICs of nisin and pediocin were determined for L. monocytogenes 412 and 412N and the six insertion mutants (Table 2). In nisin-resistant strain 412N, insertional inactivation of pbp2229 reduced the nisin MIC, an insertion in hpk1021 conferred extreme nisin sensitivity, and an insertion in lmo2487 had no effect on the nisin MIC. The nisin MICs for the three insertion mutants of strain 412 were unaltered, except for slightly increased nisin sensitivity in the pbp2229 mutant.
In accordance with the bacteriocin sensitivity assay results (Table 1), the pediocin MIC for L. monocytogenes 412N was twofold higher than that for the wild-type strain (Table 2). Inactivation of pbp2229 and hpk1021 but not of lmo2487 in 412N reduced the pediocin MIC to the wild-type level. Inactivation of the three genes in L. monocytogenes 412 had no effect on pediocin sensitivity.
Analysis of the expression of insertion mutants.
In order to elucidate whether there was a connection between the increased expression of pbp2229, hpk1021, and lmo2487 in L. monocytogenes 412N, the expression of each of the three genes in the insertion mutants was analyzed.
The expression of pbp2229 and lmo2487 was analyzed by fusion of the respective promoter regions to the lacZ reporter gene in plasmid pTCV-lac. The resulting plasmids were transformed into the six insertion mutants and strains 412 and 412N, and β-galactosidase specific activity was determined during growth (Fig. 1). The two genes were expressed at higher levels in nisin-resistant strain 412N (Fig. 1B and E) than in wild-type strain 412 (Fig. 1A and D), in accordance with previous observations (15), irrespective of the difference in growth conditions (37°C with aeration in the present study versus static culturing at 30°C in the previous work).
The expression of pbp2229 was not affected by an insertion in pbp2229 or hpk1021 in strain 412 (Fig. 1A). Expression was decreased in the lmo2487 insertion mutant, but the reduction conceivably reflected a general decrease in gene and protein expression due to the impeded growth of this strain (Fig. 1C). Strain 412N had an approximately threefold-higher level of pbp2229 expression than strain 412, and this expression was reduced to about the wild-type level following insertion in hpk1021 (Fig. 1B). An insertion in pbp2229 or lmo2487 in strain 412N resulted in a slight increase in pbp2229 expression (Fig. 1B).
The expression of lmo2487 was negligible in strain 412 and the three derived insertion mutants (Fig. 1D). Expression in strain 412N was increased about 30-fold relative to that in the wild-type strain (Fig. 1E). An insertion in hpk1021 in strain 412N abolished lmo2487 expression, and an insertion in pbp2229 or lmo2487 resulted in a minor increase in expression (Fig. 1E).
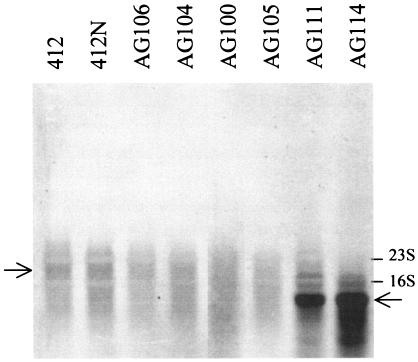
The promoter region for hpk1021 could not be unequivocally deduced from the genome sequence, and the expression of this gene therefore was analyzed by Northern blotting (Fig. 2). The blot showed a low level of expression of hpk1021 in strain 412 and a slight increase in strain 412N. There was a very high level of expression of a truncated transcript in the hpk1021 insertion mutants of strains 412 and 412N (strains AG111 and AG114, respectively). Expression was not appreciably affected in the pbp2229 and lmo2487 insertion mutants.
FIG. 2.
Northern blot analysis of hpk1021 expression in L. monocytogenes 412 and 412N and derived insertion mutants. L. monocytogenes AG100, AG111, and AG106 are pbp2229, hpk1021, and lmo2487 insertion mutants of strain 412, and L. monocytogenes AG105, AG114, and AG104 are the corresponding insertion mutants of strain 412N. The arrow at the left shows the wild-type hpk1021 transcript, the arrow at the right indicates the truncated hpk1021 transcript. The positions of the 16S and 23S rRNA bands are indicated.
DNA microarray analysis of genes associated with bacteriocin resistance.
In order to investigate the mechanisms of the observed cross-resistance, DNA microarray hybridization was performed for genes previously shown to be associated with bacteriocin resistance. L. monocytogenes 412 and spontaneous mutants 412C, 412P, and 412N were selected for the analysis, since these mutants represent the occurring bacteriocin phenotypes. RNA was isolated from cultures grown in charcoal-activated BHI medium at 37°C with aeration, in contrast to previous studies (15), where static cultures in untreated BHI medium at 30°C were used. The DNA microarray data are presented in Table 3.
TABLE 3.
DNA microarray data from genes associated with bacteriocin resistance mechanismsa
Ratios and P values in bold represent genes with statistically significant differences in expression (P < 0.008, corresponding to <5% probability of false-positive regulation; see Materials and Methods). E, exponent; n.d., not detected.
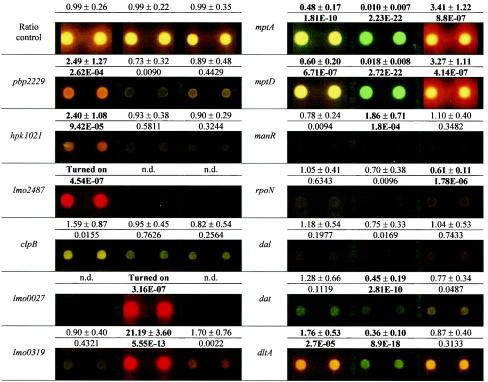
Analysis of genes associated with nisin resistance showed increased expression of pbp2229, hpk1021, and lmo2487 in L. monocytogenes 412N, in accordance with other results (see above and reference 15). The four genes had unaltered expression in class IIa bacteriocin-resistant mutants 412C and 412P.
The DNA microarray analysis of genes associated with resistance to class IIa bacteriocins (Table 3) showed that under the growth conditions used, L. monocytogenes 412P had substantial increases in EIIBgl (lmo0027) and phospho-β-glucosidase (lmo0319) expression and considerably reduced mptA and mptD expression. The strain additionally had a significant 1.9-fold increase in manR expression. L. monocytogenes 412C had a significant 1.6-fold decrease in rpoN expression, but this strain had an approximate 3-fold increase in mptA and mptD expression. The expression of mptA and mptD in nisin-resistant L. monocytogenes 412N was about half that in the wild-type strain.
A reduced d-alanine content in cell wall teichoic acids (TAs) was previously shown to confer nonspecific sensitivity to cationic antimicrobial peptides, probably through an increased negative charge of the cell wall (1, 31). Therefore, the expression of the dal, dat, and dltA genes, which mediate the synthesis of d-alanine and its incorporation into TAs in Listeria (1, 41), in the spontaneous bacteriocin-resistant mutants was analyzed (Table 3). L. monocytogenes 412N had a significant 1.8-fold increase in dltA expression. In L. monocytogenes 412P, dat and dltA expression was reduced to less than half that in the wild-type strain.
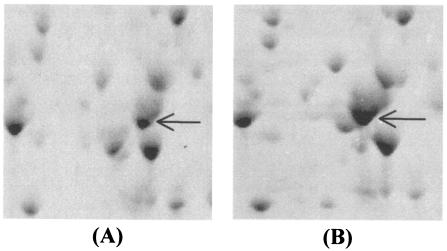
Verification of the increase in MptA expression in L. monocytogenes 412C.
The increase in mptA and mptD expression in intermediately resistant mutant L. monocytogenes 412C was in contrast to previous observations for spontaneous mutants with a high level of resistance to class IIa bacteriocins (14). The location of the MptA protein in the proteome is well known (14), and the intensities of the corresponding spots in the proteomes of L. monocytogenes 412 and 412C therefore could be compared easily (Fig. 3). The intensity of the MptA protein spot in L. monocytogenes 412C was on average sixfold higher than the intensity of this spot in wild-type strain 412.
FIG. 3.
2-D gel electrophoresis of L. monocytogenes 412 (A) and 412C (B). Enlargements of the region encompassing the MptA protein spot (arrows) are shown.
DNA microarray analysis of virulence gene expression.
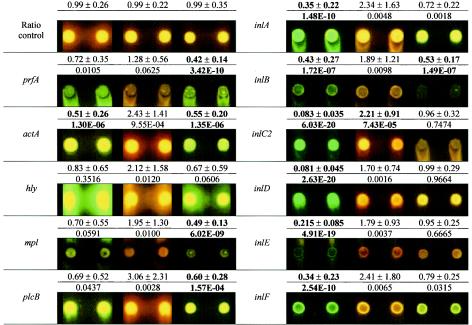
In order to assess whether the development of bacteriocin resistance may concomitantly alter virulence, the expression of 13 virulence-associated genes in L. monocytogenes 412N, 412C, and 412P was analyzed by DNA microarray hybridization (Table 4). The expression of inlC and lmaA was below the detection limit in all strains. The expression of prfA, encoding the main virulence gene regulator, was reduced 2.4-fold in strain 412C. The expression of PrfA-dependent genes actA, mpl, plcB, and inlB in strain 412C also was significantly reduced by about twofold. Strain 412N had a significant decrease in the expression of seven of the tested virulence genes, including a >10-fold reduction for inlC2 and inlD, an ∼5-fold reduction for inlE, an ∼3-fold reduction for inlA and inlF, and an ∼2-fold reduction for inlB and actA. The only significant change observed in strain 412P was increased expression of inlC2.
TABLE 4.
DNA microarray data from genes associated with virulencea
Ratios and P values in bold represent genes with statistically significant differences in expression (P < 0.008, corresponding to <5% probability of false-positive regulation; see Materials and Methods). E, exponent.
Verification of differences in virulence gene expression.
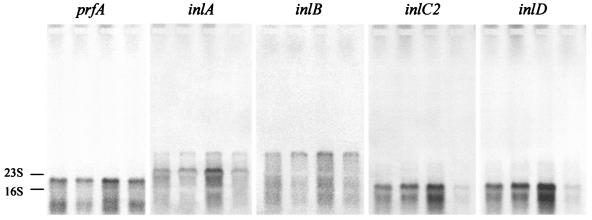
The changes in the expression of selected virulence genes were verified by Northern blotting (Fig. 4). The tested genes were prfA, encoding the central virulence gene regulator, two prfA-regulated genes (inlA and inlB), and two prfA-independent genes (inlC2 and inlD). Quantification of the hybridization signals confirmed the approximate levels of expression changes determined by DNA microarray hybridization. The up-regulation in strain 412P and the down-regulation of inlC2 and inlD in strain 412N were particularly apparent.
FIG. 4.
Northern blot analysis of virulence gene expression. RNAs from L. monocytogenes 412 (first lane from left), 412C (second lane), 412P (third lane), and 412N (fourth lane) were hybridized with probes for prfA, inlA, inlB, inlC2, and inlD. The positions of the 16S and 23S rRNA bands are indicated.
DISCUSSION
pbp2229-mediated nisin resistance mechanism.
In the present study, we analyzed the direct involvement of increased expression of pbp2229, hpk1021, and lmo2487 in the nisin resistance phenotype of L. monocytogenes 412N (15) by constructing insertion mutants for each of the three genes in strain 412N and wild-type strain 412. Nisin MIC determinations showed that the resistance phenotype was eradicated following pbp2229 and hpk1021 inactivation in strain 412N, indicating that PBP2229 and HPK1021 are directly involved in mediating nisin resistance. Furthermore, promoter fusion analysis established that hpk1021 inactivation in strain 412N reduced pbp2229 expression to the wild-type level, demonstrating that hpk1021 is required for pbp2229 induction. A similar involvement of transcriptional induction of pbpB was evident in intermediate vancomycin resistance in Staphylococcus aureus (6). The increased lmo2487 expression also required an active hpk1021 gene in strain 412N; however, inactivation of lmo2487 had no effect on the nisin sensitivity of strain 412N. Taken together, the results indicate that 412N-type nisin resistance is caused directly by enhanced PBP2229 expression, which in turn is caused by the increased expression of HPK1021.
Various phenotypes of insertion mutants could be brought about through polar effects on coexpressed downstream genes. The size of the pbp2229 transcript has been determined by Northern blot analysis to be about 2.1 kb (A. Gravesen, unpublished results), which corresponds very well to the 2,145-bp reading frame. Accordingly, the L. monocytogenes EGDe genome sequence (http://genolist.pasteur.fr/ListiList/) has a terminator sequence after the pbp2229 open reading frame. An insertion in hpk1021 presumably affects the expression of the downstream cognate response regulator, rrp1022, and possibly also the three following genes. None of the latter three genes has been suggested on the basis of homology to have a regulatory role. These observations strongly suggest that the phenotypes of the pbp2229 and hpk1021 insertion mutants are caused directly by eradication of PBP2229 and the HPK1021-RRP1022 two-component signal transduction system, respectively.
The two-component signal transduction system encoded by lisRK also affects nisin sensitivity (10); a lisK deletion mutant displayed enhanced nisin resistance but reduced expression of pbp2229, hpk1021, and lmo2487. The lisR gene was included in the DNA microarray analysis in the present study, and the hybridization data established that lisR expression was not substantially changed in L. monocytogenes 412N compared to wild-type 412 (average ratio, 0.87) (data not shown). It is currently not clear how the lisRK and hpk1021 response regulator systems interact in determining nisin sensitivity.
Nisin forms membrane pores through an interaction with lipid II (17), which is an essential molecule in cell wall synthesis. The fact that only intermediately resistant nisin mutants were isolated could be explained by the hypothesis that high-level resistance, like that seen with class IIa bacteriocins, is attainable only through one mechanism, namely, elimination of the docking molecule, and the elimination of lipid II is not possible. Intermediate-level resistance, on the other hand, can be obtained through several mechanisms, e.g., modifications in cell envelope charge or density that reduce the accessibility of the docking molecule. Accordingly, nisin sensitivity in various Listeria mutants has been related to membrane charge and fluidity (20, 24, 45), cell wall thickness (21), cell wall charge (1), or a combination of these factors (11). Intermediate class IIa bacteriocin resistance in two mutants was related to membrane fluidity (43).
Penicillin-binding proteins catalyze the incorporation of the disaccharide-pentapeptide moiety of lipid II into the growing peptidoglycan chain, and PBP2229 conceivably mediates enhanced nisin resistance by shielding lipid II and possibly also by reducing the extracellular lipid II concentration. The results presented here thus support previous suggestions that nisin sensitivity is affected by the accessibility of lipid II (8, 15).
Possible mechanisms for cross-protection between nisin and class IIa bacteriocins.
As mentioned above, previous reports on cross-resistance between nisin and class IIa bacteriocins have presented various results. Some Listeria mutants ostensibly did not exhibit cross-resistance (35, 38, 46), while others showed various degrees of reduced sensitivity to the other compound (11, 40). In one case, class IIa bacteriocin resistance development was reported to confer increased nisin sensitivity (5).
In the present work, we observed that the three mutants (originating from three different wild-type strains) that were intermediately nisin resistant all had reduced class IIa bacteriocin sensitivity, but high-level class IIa bacteriocin resistance did not occur. One of the mutants was analyzed by DNA microarray hybridization, and the results suggested two possible mechanisms for the observed cross-protection. L. monocytogenes 412N had an approximate twofold decrease in mptA and mptD expression, corresponding to the twofold increase in the pediocin MIC. A similar expression change occurred in class IIa bacteriocin-resistant mutants B73-V1 and B73-V2, which had comparable class IIa bacteriocin resistance levels (V. Vadyvaloo, personal communication). Additionally, strain 412N had a slight increase in dltA expression, which might lead to a more negative cell wall through increased d-alanine content in TAs presenting nonspecific protection against cationic antimicrobial peptides (1, 31).
Only a few of the tested class IIa bacteriocin-resistant mutants displayed cross-protection to nisin; three out of eight mutants that were highly resistant to class IIa bacteriocins and one out of four mutants that were intermediately resistant to class IIa bacteriocins had reduced nisin sensitivity. There was thus no correlation between the class IIa bacteriocin resistance level and the occurrence of cross-protection to nisin. DNA microarray analysis of one mutant that was intermediately resistant and one mutant that was highly resistant to class IIa bacteriocins showed that the nisin cross-protection was neither PBP2229 associated nor related to increased d-alanine content. Additionally, Northern blot analysis showed that none of the 12 class IIa bacteriocin-resistant mutants used in this study had an appreciable increase in pbp2229 expression (data not shown). It is therefore presently not possible to suggest a mechanism for the nisin cross-protection in the class IIa bacteriocin-resistant mutants.
Changed virulence gene expression in bacteriocin-resistant mutants—is virulence altered?
This report presents, to our knowledge, the first assessment of virulence in spontaneously bacteriocin-resistant L. monocytogenes. We analyzed the expression of 11 virulence genes under conditions that provide prfA-mediated induction. It is customary to use a twofold cutoff value when considering differential gene expression determined by DNA arrays (see, e.g., reference 23). However, differences in transcript abundance of less than 2-fold can be biologically significant (18), and a lower cutoff value of a 1.5-fold difference in expression therefore also has been used (39, 42). Several studies on the function of listerial virulence genes have used gene inactivation (3, 9, 13), which would yield a marked reduction in expression. We are not aware of reports on the effects of moderate changes in the expression of these genes.
L. monocytogenes 412C, which is intermediately resistant to class IIa bacteriocins, had a 2.4-fold reduction in prfA expression. In accordance with this result, a prfA deletion mutant (4) was intermediately resistant to pediocin (A. Gravesen, unpublished results). In addition to the statistically significant decreases in virulence gene expression described here, L. monocytogenes 412N and 412C showed nonsignificant decreases in the expression of the remaining prfA-dependent genes. L. monocytogenes 412P, on the other hand, showed nonsignificant increases (up to 2.4-fold) in the expression of all tested virulence genes. The tested prfA-dependent genes included genes involved in different steps of the infection cycle: inlA and inlB, which are essential for invasion of the intestinal epithelium; actA, which mediates intracellular motility; and plcB and mpl, which are required for escape from the phagosome (for reviews, see references 19 and 44). The consistent changes in the expression of the central virulence genes suggest that virulence may be decreased in L. monocytogenes 412N and 412C and increased in L. monocytogenes 412P. However, since nisin resistance can be acquired through several different mechanisms, it is not possible to extend the presumption to nisin-resistant mutants in general.
All in all, our results indicate that enhanced pbp2229 expression, brought about by an increased level of the signal transduction system encoded by hpk1021-rrp1022, is directly involved in the prevalent nisin resistance mechanism occurring, e.g., in L. monocytogenes 412N. We have also shown that intermediate cross-resistance occurs between nisin and class IIa bacteriocins. This observation is important when the simultaneous use of the two compounds is being considered. Furthermore, our results imply that mutant-specific changes in virulence may occur, and if this implication holds true, it may have considerable impact on assessment of the threat imposed by bacteriocin resistance. Supplementary studies with more mutants and virulence assays are required to investigate the validity of the results for bacteriocin-resistant mutants in general and to substantiate the biological significance of the observed changes in virulence gene expression.
Acknowledgments
We thank Lilian Nilsson for supplying carnobacteriocin B2 and Danisco Beaminster Ltd. for supplying pure nisin A. The expert technical assistance of Lene Gertman and Hanne Mordhorst is highly appreciated. We acknowledge Steen Knudsen for helpful comments on the statistical analysis of the array data.
This work was supported by the Food Biotechnology Program of the Danish Ministry for Food, Agriculture, and Fisheries (BIOT 99-DFU-8), by the Danish Research Council (9315013), and by a grant from The Danish Rectors' Conference and the National Research Foundation (South Africa) to M.R.
REFERENCES
- 1.Abachin, E., C. Poyart, E. Pellegrini, E. Milohanic, F. Fiedler, P. Berche, and P. Trieu-Cuot. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1-14. [DOI] [PubMed] [Google Scholar]
- 1a.Ben Embarek, P. K., and H. H. Huss. 1993. Heat resistance of Listeria monocytogenes in vacuum packaged pasteurized fish fillets. Int. J. Food. Microbiol. 20:85-95. [DOI] [PubMed] [Google Scholar]
- 2.Bender, R., and S. Lange. 2001. Adjusting for multiple testing—when and how? J. Clin. Epidemiol. 54:343-349. [DOI] [PubMed] [Google Scholar]
- 3.Bergmann, B., D. Raffelsbauer, M. Kuhn, M. Goetz, S. Hom, and W. Goebel. 2002. InlA- but not InlB-mediated internalization of Listeria monocytogenes by non-phagocytic mammalian cells needs the support of other internalins. Mol. Microbiol. 43:557-570. [DOI] [PubMed] [Google Scholar]
- 4.Bohne, J., H. Kestler, C. Uebele, Z. Sokolovic, and W. Goebel. 1996. Differential regulation of the virulence genes of Listeria monocytogenes by the transcriptional activator PrfA. Mol. Microbiol. 20:1189-1198. [DOI] [PubMed] [Google Scholar]
- 5.Bouttefroy, A., and J. B. Milliere. 2000. Nisin-curvaticin 13 combinations for avoiding the regrowth of bacteriocin resistant cells of Listeria monocytogenes ATCC 15313. Int. J. Food Microbiol. 62:65-75. [DOI] [PubMed] [Google Scholar]
- 6.Boyle-Vavra, S., S. Yin, M. Challapalli, and R. S. Daum. 2003. Transcriptional induction of the penicillin-binding protein 2 gene in Staphylococcus aureus by cell wall-active antibiotics oxacillin and vancomycin. Antimicrob. Agents Chemother. 47:1028-1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm, K., M. T. Ripio, J. Kreft, and J. A. Vazquez-Boland. 1999. The bvr locus of Listeria monocytogenes mediates virulence gene repression by beta-glucosides. J. Bacteriol. 181:5024-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breukink, E., I. Wiedemann, C. van Kraaij, O. P. Kuipers, H. Sahl, and B. de Kruijff. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361-2364. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cotter, P. D., C. M. Guinane, and C. Hill. 2002. The LisRK signal transduction system determines the sensitivity of Listeria monocytogenes to nisin and cephalosporins. Antimicrob. Agents Chemother. 46:2784-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crandall, A. D., and T. J. Montville. 1998. Nisin resistance in Listeria monocytogenes ATCC 700302 is a complex phenotype. Appl. Environ. Microbiol. 64:231-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalet, K., Y. Cenatiempo, P. Cossart, and Y. Héchard. 2001. A sigma(54)-dependent PTS permease of the mannose family is responsible for sensitivity of Listeria monocytogenes to mesentericin Y105. Microbiology 147:3263-3269. [DOI] [PubMed] [Google Scholar]
- 13.Dramsi, S., I. Biswas, E. Maguin, L. Braun, P. Mastroeni, and P. Cossart. 1995. Entry of Listeria monocytogenes into hepatocytes requires expression of InlB, a surface protein of the internalin multigene family. Mol. Microbiol. 16:251-261. [DOI] [PubMed] [Google Scholar]
- 13a.Dykes, G. A., and J. W. Hastings. 1998. Fitness costs associated with class IIa bacteriocin resistance in Listeria monocytogenes B73. Lett. Appl. Microbiol. 26:5-8. [DOI] [PubMed] [Google Scholar]
- 13b.Glaser, P., et al. 2001. Comparative genomics of Listeria species. Science 294:849-852. [DOI] [PubMed] [Google Scholar]
- 14.Gravesen, A., M. Ramnath, K. B. Rechinger, N. Andersen, L. Jansch, Y. Héchard, J. W. Hastings, and S. Knøchel. 2002. High-level resistance to class IIa bacteriocins is associated with one general mechanism in Listeria monocytogenes. Microbiology 148:2361-2369. [DOI] [PubMed] [Google Scholar]
- 15.Gravesen, A., K. Sørensen, F. M. Aarestrup, and S. Knøchel. 2001. Spontaneous nisin-resistant Listeria monocytogenes mutants with increased expression of a putative penicillin-binding protein and their sensitivity to various antibiotics. Microb. Drug Resist. 7:127-135. [DOI] [PubMed] [Google Scholar]
- 16.Gravesen, A., P. Warthoe, S. Knøchel, and K. Thirstrup. 2000. Restriction fragment differential display of pediocin-resistant Listeria monocytogenes 412 mutants shows consistent overexpression of a putative beta-glucoside-specific PTS system. Microbiology 146:1381-1389. [DOI] [PubMed] [Google Scholar]
- 16a.Gravesen, A., A. M. Jydegaard Axelsen, J. Mendes Da Silva, T. B. Hansen, and S. Knøchel. 2002. Frequency of bacteriocin resistance development and associated fitness costs in Listeria monocytogenes. Appl. Environ. Microbiol. 68:756-764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Héchard, Y., and H. G. Sahl. 2002. Mode of action of modified and unmodified bacteriocins from Gram-positive bacteria. Biochimie 84:545-557. [DOI] [PubMed] [Google Scholar]
- 18.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102:109-126. [DOI] [PubMed] [Google Scholar]
- 19.Kreft, J., and J. A. Vazquez-Boland. 2001. Regulation of virulence genes in Listeria. Int. J. Med. Microbiol. 291:145-157. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., M. L. Chikindas, R. D. Ludescher, and T. J. Montville. 2002. Temperature- and surfactant-induced membrane modifications that alter Listeria monocytogenes nisin sensitivity by different mechanisms. Appl. Environ. Microbiol. 68:5904-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maisnier-Patin, S., and J. Richard. 1996. Cell wall changes in nisin-resistant variants of Listeria innocua grown in the presence of high nisin concentrations. FEMS Microbiol. Lett. 140:29-35. [DOI] [PubMed] [Google Scholar]
- 22.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Milohanic, E., P. Glaser, J. Y. Coppee, L. Frangeul, Y. Vega, J. A. Vazquez-Boland, F. Kunst, P. Cossart, and C. Buchrieser. 2003. Transcriptome analysis of Listeria monocytogenes identifies three groups of genes differently regulated by PrfA. Mol. Microbiol. 47:1613-1625. [DOI] [PubMed] [Google Scholar]
- 24.Ming, X., and M. A. Daeschel. 1995. Correlation of cellular phospholipid content with nisin resistance of Listeria monocytogenes Scott A. J. Food Prot. 58:416-420. [DOI] [PubMed] [Google Scholar]
- 25.Mørtved, C. I., and I. Nes. 1990. Plasmid-associated bacteriocin production by a Lactococcus sake strain. J. Gen. Microbiol. 136:1601-1607. [Google Scholar]
- 26.Nilsson, L., L. Gram, and H. H. Huss. 1999. Growth control of Listeria monocytogenes on cold-smoked salmon using a competitive lactic acid bacteria flora. J. Food Prot. 62:336-342. [DOI] [PubMed] [Google Scholar]
- 27.Nilsson, L., M. K. Nielsen, Y. Ng, and L. Gram. 2002. Role of acetate in production of an autoinducible class IIa bacteriocin in Carnobacterium piscicola A9b. Appl. Environ. Microbiol. 68:2251-2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nissen-Meyer, J., and I. F. Nes. 1997. Ribosomally synthesized antimicrobial peptides: their function, structure, biogenesis, and mechanism of action. Arch. Microbiol. 167:67-77. [PubMed] [Google Scholar]
- 29.Papathanasopoulos, M. A., F. Krier, A. M. Revol-Junelles, G. Lefebvre, J. P. Le Caer, A. von Holy, and J. W. Hastings. 1997. Multiple bacteriocin production by Leuconostoc mesenteroides TA33a and other Leuconostoc/Weissella strains. Curr. Microbiol. 35:331-335. [DOI] [PubMed] [Google Scholar]
- 30.Park, S. F., and G. S. Stewart. 1990. High-efficiency transformation of Listeria monocytogenes by electroporation of penicillin-treated cells. Gene 94:129-132. [DOI] [PubMed] [Google Scholar]
- 31.Peschel, A., M. Otto, R. W. Jack, H. Kalbacher, G. Jung, and F. Gotz. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405-8410. [DOI] [PubMed] [Google Scholar]
- 32.Poyart, C., and P. Trieu-Cuot. 1997. A broad-host-range mobilizable shuttle vector for the construction of transcriptional fusions to beta-galactosidase in gram-positive bacteria. FEMS Microbiol. Lett. 156:193-198. [DOI] [PubMed] [Google Scholar]
- 33.Ramnath, M., K. B. Rechinger, L. Jänsch, J. W. Hastings, S. Knøchel, and A. Gravesen. 2003. Development of a Listeria monocytogenes EGDe partial proteome reference map and comparison with the protein profiles of food isolates. Appl. Environ. Microbiol. 69:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rasch, M., and S. Knøchel. 1998. Variations in tolerance of Listeria monocytogenes to nisin, pediocin PA-1 and bavaricin A. Lett. Appl. Microbiol. 27:275-278. [DOI] [PubMed] [Google Scholar]
- 35.Rekhif, N., A. Atrih, and G. Lefebvre. 1994. Selection and properties of spontaneous mutants of Listeria monocytogenes ATCC 15313 resistant to different bacteriocins produced by lactic acid bacteria strains. Curr. Microbiol. 28:237-241. [Google Scholar]
- 36.Roldan, A. L., M. V. Cubellis, M. T. Masucci, N. Behrendt, L. R. Lund, K. Dano, E. Appella, and F. Blasi. 1990. Cloning and expression of the receptor for human urokinase plasminogen activator, a central molecule in cell surface, plasmin dependent proteolysis. EMBO J. 9:467-474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schaferkordt, S., and T. Chakraborty. 1995. Vector plasmid for insertional mutagenesis and directional cloning in Listeria spp. BioTechniques 19:720-725. [PubMed] [Google Scholar]
- 38.Schillinger, U., H. S. Chung, K. Keppler, and W. H. Holzapfel. 1998. Use of bacteriocinogenic lactic acid bacteria to inhibit spontaneous nisin-resistant mutants of Listeria monocytogenes Scott A. J. Appl. Microbiol. 85:657-663. [DOI] [PubMed] [Google Scholar]
- 39.Smoot, L. M., J. C. Smoot, M. R. Graham, G. A. Somerville, D. E. Sturdevant, C. A. Migliaccio, G. L. Sylva, and J. M. Musser. 2001. Global differential gene expression in response to growth temperature alteration in group A Streptococcus. Proc. Natl. Acad. Sci. USA 98:10416-10421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Song, H. J., and J. Richard. 1997. Antilisterial activity of three bacteriocins used at sub minimal inhibitory concentrations and cross-resistance of the survivors. Int. J. Food Microbiol. 36:155-161. [DOI] [PubMed] [Google Scholar]
- 41.Thompson, R. J., H. G. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tusher, V. G., R. Tibshirani, and G. Chu. 2001. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98:5116-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vadyvaloo, V., J. W. Hastings, M. J. Van Der Merwe, and M. Rautenbach. 2002. Membranes of class IIa bacteriocin-resistant Listeria monocytogenes cells contain increased levels of desaturated and short-acyl-chain phosphatidylglycerols. Appl. Environ. Microbiol. 68:5223-5230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vazquez-Boland, J. A., M. Kuhn, P. Berche, T. Chakraborty, G. Dominguez-Bernal, W. Goebel, B. Gonzalez-Zorn, J. Wehland, and J. Kreft. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584-640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Verheul, A., N. J. Russell, R. Van't Hof, F. M. Rombouts, and T. Abee. 1997. Modifications of membrane phospholipid composition in nisin-resistant Listeria monocytogenes Scott A. Appl. Environ. Microbiol. 63:3451-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vignolo, G., J. Palacios, M. E. Farias, F. Sesma, U. Schillinger, W. Holzapfel, and G. Oliver. 2000. Combined effect of bacteriocins on the survival of various Listeria species in broth and meat system. Curr. Microbiol. 41:410-416. [DOI] [PubMed] [Google Scholar]