Abstract
Formins promote processive elongation of actin filaments for cytokinetic contractile rings and other cellular structures. In vivo, these structures are exposed to tension, but the effect of tension on these processes was unknown. Here we used single-molecule imaging to investigate the effects of tension on actin polymerization mediated by yeast formin Bni1p. Small forces on the filaments dramatically slowed formin-mediated polymerization in the absence of profilin, but resulted in faster polymerization in the presence of profilin. We propose that force shifts the conformational equilibrium of the end of a filament associated with formin homology 2 domains toward the closed state that precludes polymerization, but that profilin–actin associated with formin homology 1 domains reverses this effect. Thus, physical forces strongly influence actin assembly by formin Bni1p.
A host of proteins regulate the actin cytoskeleton by controlling filament nucleation, elongation, capping, branching, and bundling. Members of the formin family of proteins nucleate new filaments and remain processively attached to barbed ends while promoting the elongation of unbranched filaments (1, 2). Formin homology (FH)2 domains form a donut-shaped head-to-tail homodimer that encircles the fast-growing barbed end of actin filaments and promotes nucleation and polymerization (1, 3). When an actin monomer binds to the barbed end of a filament, one FH2 domain steps onto the new subunit, allowing the formin to remain attached to the filament through thousands of cycles of subunit addition (Fig. 1A) (2, 4). The FH2-bound end of the filament binds incoming actin monomers when in an “open” conformation but not in the “closed” conformation. As a consequence, FH2 domains slow barbed-end elongation compared with free barbed ends, a phenomenon termed “gating” (1, 5). Despite gating, FH2 domains can promote rapid filament elongation when coupled to FH1 domains (6), which are located N-terminal to the FH2 domain (7). Multiple polyproline tracks in FH1 domains bind the small actin-binding protein profilin, which mediates association of several profilin–actin complexes in close proximity to the end of a filament. Diffusive motions of the FH1 domain transfer actin rapidly to the filament barbed end (5), allowing elongation at rates faster than addition of subunits from the bulk phase.
Fig. 1.
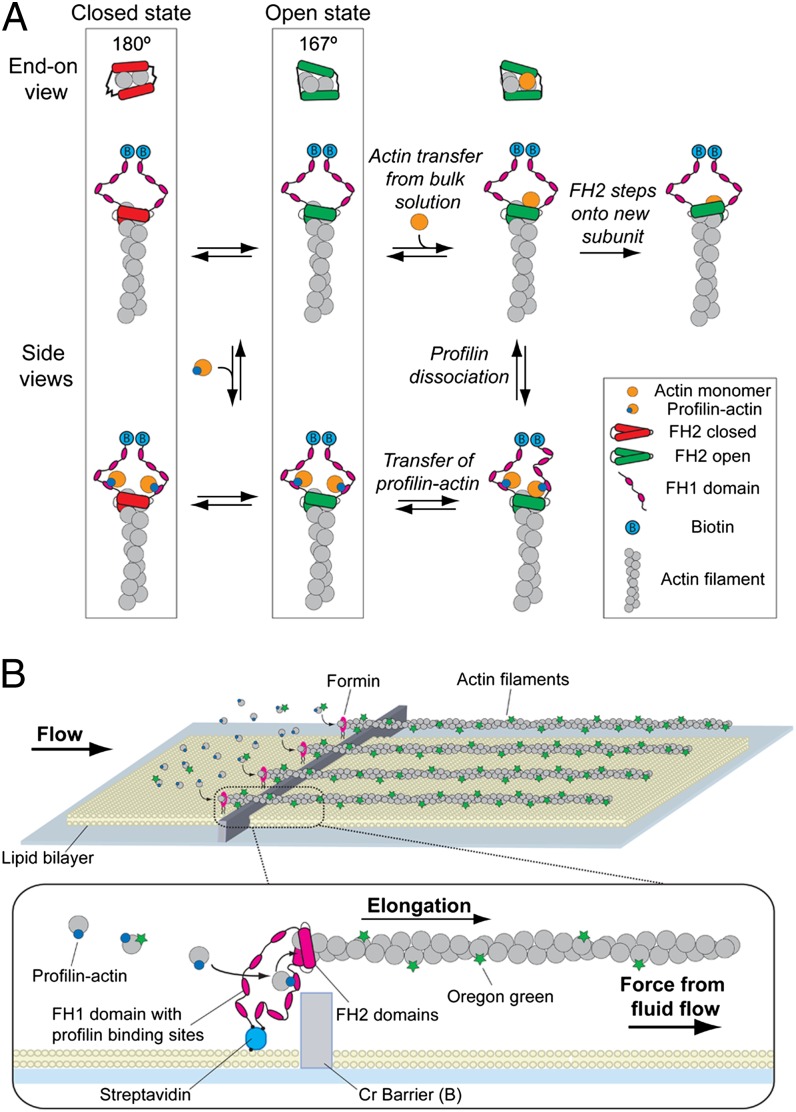
Formin-mediated actin filament polymerization in actin curtains. (A) Schematic of the reaction pathways for actin filament elongation by formin. FH2 dimers are shown in red for the closed conformation and green for the open conformation. End-on views of the filament illustrate the hypothesis that the closed conformation corresponds to a 180° pitch of the filament, whereas the open conformation has a 167° pitch (25). (B) Schematic of actin curtains. Biotinylated formin (Bni1p) is anchored via streptavidin to a lipid bilayer and polymerizes actin filaments that are aligned along nanofabricated barriers by solvent flow, allowing visualization by total internal reflection fluorescence microscopy.
Actin filaments are subject to tension in cells, yet the influence of tension on formin-mediated polymerization was unknown, and theories predicted different outcomes in the absence and presence of profilin. Kozlov and colleagues proposed that the elasticity of the FH2 domain and the formin–barbed end binding energy govern the polymerization rate (8, 9). Their simulations suggested that tension increases the rate of polymerization by lowering the activation barrier for subunit addition and energetically favoring FH2 domain stepping onto the incoming subunit (8). Vavylonis et al. (10) postulated that force-induced stretching of FH1 domains slows the transfer of profilin–actin to the growing filament, so tension would inhibit polymerization in the presence of profilin. Here we describe experiments to test these predictions. We find that the effects of force are opposite both predictions for budding yeast formin Bni1p.
Results
To assess the effect of tension on actin filament elongation mediated by formin, we visualized actin filaments that were anchored to the surface of a microfluidic chamber coated with a lipid bilayer (Fig. 1B) (11–15). We used a construct consisting of the FH1 and FH2 domains (residues 1228–1766) of Saccharomyces cerevisiae formin Bni1p fused to a biotinylated N-terminal tag. Formin was coupled to biotinylated lipids through a streptavidin linkage (Fig. 1B, Inset), so that the N terminus of the protein was anchored to the bilayer in a configuration similar to what is expected in living cells. One or both of the FH1 domains of a single formin dimer might be anchored by streptavidin to the bilayer. Anchored formins initiated actin filament elongation, and application of buffer flow aligned formin-bound actin filaments into “actin curtains” along nanofabricated barriers for imaging by total internal reflection fluorescence microscopy (TIRFM) (Figs. 1B and 2A). Flow confined the filaments within the ∼224-nm depth of the evanescent field, and they disappeared from view when buffer flow stopped, confirming they were linked to the bilayer only through the biotinylated formin (Fig. 2 A and B and Movie S1).
Fig. 2.
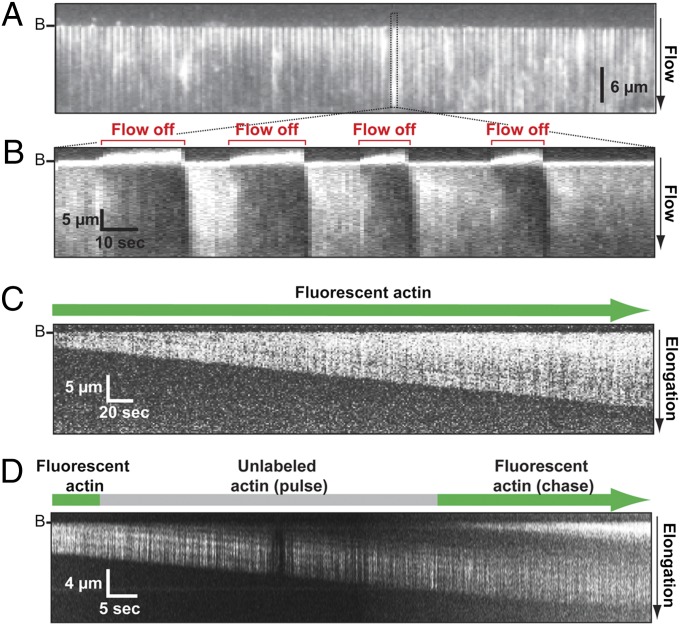
Lipid-tethered formins polymerize actin filament curtains. Buffer conditions: 1.5 μM actin (33% Oregon Green-actin) in microscopy buffer (10 mM imidazole, pH 7.0, 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 0.02 mM CaCl2, 15 mM glucose, 0.02 mg/mL catalase, 0.1 mg/mL glucose oxidase). (A) Image of an actin curtain with flow on. The dotted box highlights an individual actin filament. B, location of barriers. Buffer flow is from top to bottom. (B) Kymograph showing alternating examples of extension and diffusion of a single actin filament that result from applying and stopping buffer flow over the actin curtain shown in A. (C) Kymograph showing a single formin-anchored actin filament growing over time. (D) Pulse-chase experiment demonstrating that polymer growth occurs only at the barbed end of the filament associated with the formin anchored at the barrier. Polymerization was initiated with fluorescent actin monomers, followed by a pulse of unlabeled actin monomers and finally chased with additional fluorescent actin monomers. The dark section in the middle of the actin filament at the right of the kymograph is due to the incorporation of unlabeled actin monomers.
Actin filaments increased in length over time at an initial rate of ∼12 subunits per s in buffer lacking methylcellulose, always growing in the direction of the buffer flow (Fig. 2C). Pulse-chase experiments, involving alternate injections of fluorescent and unlabeled actin, confirmed that polymerization occurred only at formin-bound barbed ends (Fig. 2D). Tethered filaments could grow for >1 h without dissociating from formin, reaching lengths >40 μm (>14,000 actin subunits).
Influence of Buffer Viscosity on Actin Polymerization.
Initial experiments illustrated in Fig. 2 used a low-viscosity polymerization buffer [η = 0.9 centipoise (cP)], which required bulk flow rates of 0.3 mL/min to confine the filaments within the evanescent field. The filaments could not be visualized by TIRFM at lower flow rates with this buffer, and thus each experimental run consumed large quantities of fluorescently tagged actin. To minimize protein consumption, we included 0.25% methylcellulose [η = 1.4 cP, molecular weight ∼14 kDa (16, 17)] to increase the solution viscosity of the polymerization buffer and allow the filaments to be observed by TIRFM at lower flow rates. Buffers containing low concentrations of 14-kDa methylcellulose are expected to behave as newtonian fluids, for which the viscosity should not depend on the shear rate (17–19).
In the absence of profilin, the rate of formin-mediated actin assembly for the filaments aligned on bilayer-coated surfaces (13.2 ± 0.9 subunits per s at 0.05 mL/min bulk flow) was the same as the rate observed for filaments attached to myosin-coated slides (13.6 ± 1.6 subunits per s without bulk flow) (see Fig. 4A; Table S1). In the same buffer, free actin filament barbed ends elongated on myosin-coated slides at 17.6 ± 0.7 subunits per s in the absence of formin. The ratio of the polymerization rates measured in the presence and absence of formin is a means of quantifying how much the FH2 domain slows subunit addition. This gating factor is viewed as the fraction of time that the end is available for adding subunits. In this case, the gating factor of ∼0.75 indicates that the actin curtains accurately reflect the known attributes of formin-mediated actin polymerization.
Fig. 4.
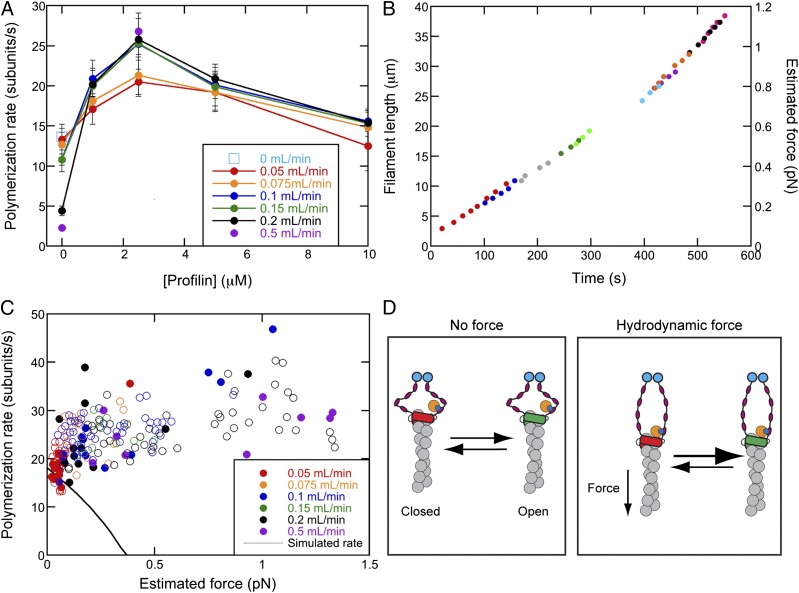
Tension increases the rate of formin-mediated polymerization in the presence of profilin. Buffer conditions were as in Fig. 3 except for the presence of profilin. (A) Dependence of polymerization rates on the concentration of profilin and the flow rate: open cyan square, no flow; red circles, 0.05 mL/min; orange circles, 0.075 mL/min; blue circles, 0.1 mL/min; green circles, 0.15 mL/min; black circles, 0.2 mL/min; purple circles, 0.5 mL/min. Error bars are SEMs. (B) Elongation profiles of 10 filaments in the presence of 2.5 μM profilin and 0.2 mL/min bulk flow, aligned by length. Each filament is represented by a different color, and every third data point is shown. (C) Force-dependence of polymerization rates of filaments growing in the presence of 2.5 μM profilin with bulk flow rates of 0.05 (red circles), 0.075 (orange circles), 0.1 (blue circles), 0.15 (green circles), 0.2 (black circles), and 0.5 (purple circles) mL/min to produce drag force on each filament. Data were collected using either 10-nm barriers (filled circles) or 25-nm barriers (open circles). The solid black line represents data simulated using a thermodynamic model of formin-mediated polymerization in the presence of 2.5 μM profilin with the same tension-sensitive gating factor as in Fig. 3B. (D) Cartoon depiction of the anticipated effect of tension on the equilibrium between the open (green) and closed (red) conformations of the filament (gray)-bound formin in the presence of profilin (dark blue). FH1 domains are depicted in black and magenta, streptavidin-bound biotin labels are shown in cyan, and the incoming actin subunit is shown in orange.
Estimation of Surface Proximity and Applied Drag Force.
The force experienced by an anchored formin varies in proportion to the length of the actin filament, flow rate, and solution viscosity (20), and the relative force on the tethered formin always increases as the filament grows. We used the Batchelor equation to estimate the drag force on a cylindrical filament (20), including a term to account for the proximity of the filament to the surface (Fig. S1A) (21, 22). This formula relates the drag force (Fd) on a filament of length L to the flow velocity (v), which when corrected for the height of the filament from the surface (h) is given as (20–22)
 |
where η is solution viscosity (1.4 cP at 0.25% methylcellulose) and r is the radius of the filament (8 nm for an actin filament). We determined the flow rate at the height of the filament by tracking the trajectories of actin filaments that broke from anchored filaments and passed through the field of view in the plane of the tethered filaments (Table S2). We estimated the height of filaments at all flow rates used by calculating and comparing the flow profile of the sample chamber [with dimensions of ∼200 µm × ∼4.5 mm (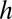 ×
× 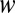 )] with the linear velocities of the broken filament fragments measured at the imaging plane (Fig. S1B). The flow velocity profile
)] with the linear velocities of the broken filament fragments measured at the imaging plane (Fig. S1B). The flow velocity profile 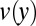 of a channel whose height (
of a channel whose height (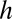 ) is much less than its width
) is much less than its width 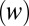 can be calculated using the Navier–Stokes equation in the form
can be calculated using the Navier–Stokes equation in the form
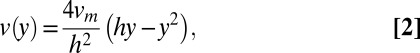 |
where 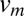 in the maximum velocity at the center of the channel (23). The maximum velocity can be calculated by recognizing that the bulk flow rate can be calculated as previously described (24) using the following equation:
in the maximum velocity at the center of the channel (23). The maximum velocity can be calculated by recognizing that the bulk flow rate can be calculated as previously described (24) using the following equation:
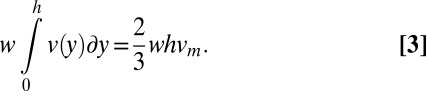 |
Comparison of the linear velocity data (Table S2) with the flow profiles at each of the different bulk flow rates revealed that the broken actin filaments passed through the chamber at a height of approximately ∼100–130 nm from the surface (Fig. S1B and Table S2). We used the heights calculated at each flow rate to estimate the drag on the anchored filaments using Eq. 1.
Our analysis yielded an estimated force of ∼0.14 pN/μm per mL/min of bulk flow (Table S2). These force estimates may be subject to error if the filament fragments do not remain in the same plane after breaking from anchored filaments. However, the magnitude of these errors is likely to be small (Fig. S1A), and does not affect our conclusion that the relative force is proportional to filament length, nor does it influence our conclusions regarding the effects of tension on formin-mediated polymerization (see below).
Tension Slows Bni1p-Mediated Polymerization in the Absence of Profilin.
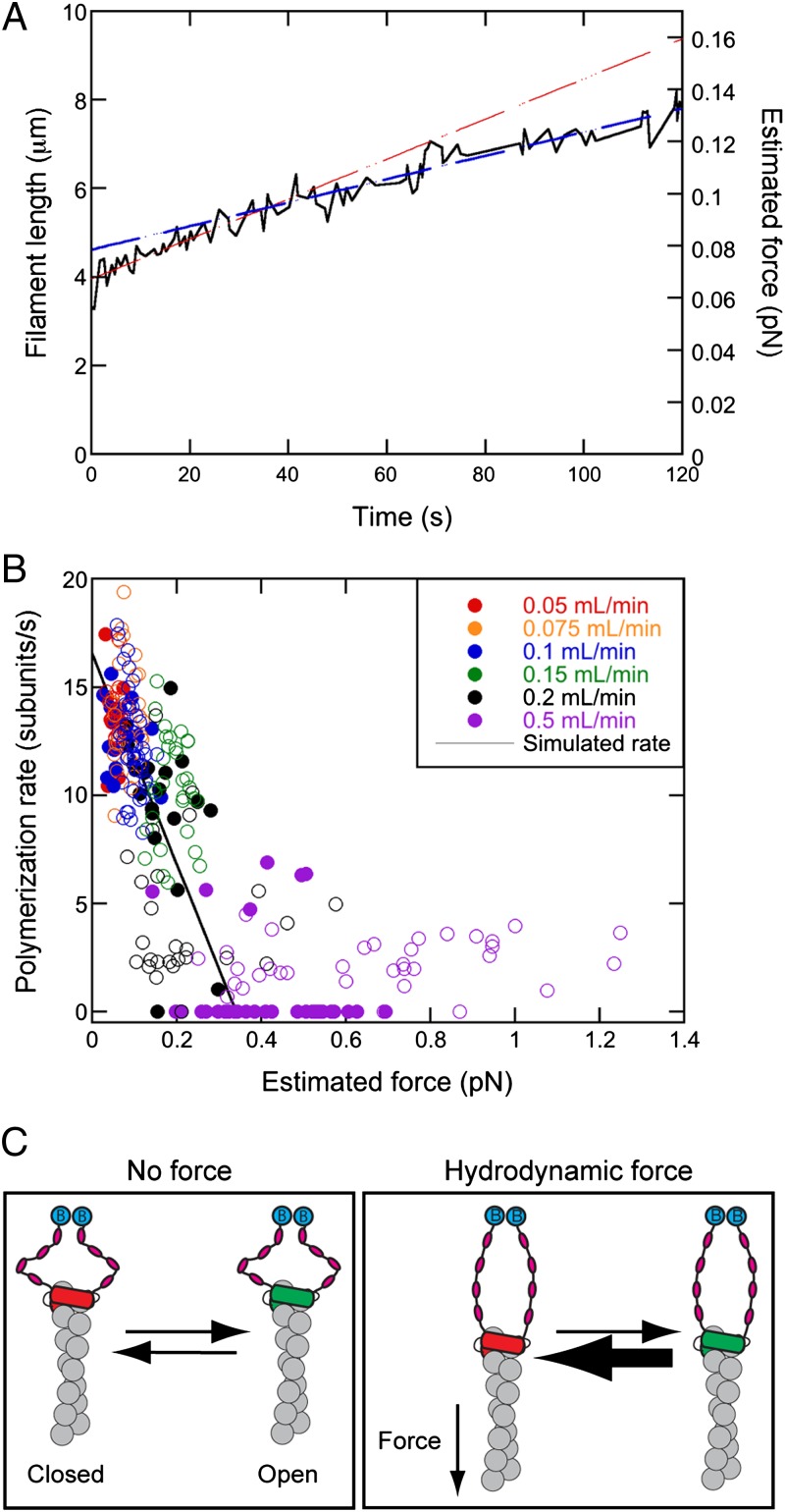
In the absence of profilin, the FH2 domain alone mediates actin monomer addition, allowing us to ask whether FH2-mediated polymerization is sensitive to tension. We observed under these conditions that elongation rates of individual filaments declined as the relative force increased with the length of the filament (Fig. 3A). The elongation rate decreased linearly from ∼13 subunits per s in the absence of force to zero at forces greater than ∼0.3 pN when using 10-nm barriers (Figs. 3 A and B and 4A and Table S1), where the entire formin can clear the barrier. When using 25-nm barriers, some filaments elongated at ∼2 subunits per s at forces exceeding ∼0.2 pN (Fig. 3B, open circles), suggesting that tall barriers may insulate formins from the effects of force. Decreasing the applied tension via spontaneous filament breakage or a change in the flow rate resulted in a corresponding increase in the polymerization rate (Fig. S2), demonstrating that the effect of force is reversible. Thus, tension unexpectedly inhibits formin-mediated actin filament elongation in the absence of profilin (8, 9).
Fig. 3.
Tension inhibits formin-mediated elongation of actin filaments in the absence of profilin. Buffer conditions: 1.5 μM actin (33% Oregon Green-actin) in microscopy buffer with 0.25% methylcellulose. (A) Time course of elongation of a single filament in the absence of profilin with a bulk flow of 0.1 mL/min. The black line is the length. The red and blue dashed lines are tangents to the beginning and the end of the elongation curve to emphasize the decline in the rate of elongation as the filament grew longer. The polymerization rate changes smoothly with filament length (Fig. 3B), so the tangents are meant simply as guides and not to suggest that the polymerization rate changes at a discrete length. The estimated hydrodynamic force applied to the filament is shown on the right axis and is proportional to the length of the filament. (B) Force-dependence of polymerization rate of filaments growing with bulk flow rates of 0.05 (red circles), 0.075 (orange circles) 0.1 (blue circles), 0.15 (green circles), 0.2 (black circles), and 0.5 (purple circles) mL/min. Data were collected using 10-nm barriers (filled circles) or 25-nm barriers (open circles). The solid line represents data simulated using a thermodynamic model of formin-mediated polymerization with a force-sensitive gating factor (5). (C) Cartoon depiction of the effect of tension on the equilibrium between the open (green) and closed (red) conformations of the filament (gray)-bound formin in the absence of profilin. FH1 domains are depicted in black and magenta and streptavidin-bound biotin labels are shown in cyan.
Tension Promotes Bni1p-Mediated Polymerization in the Presence of Profilin.
We next examined the influence of profilin on formin-mediated polymerization. Profilin stimulates polymerization, with maximum rates observed at intermediate profilin concentrations (Fig. 4A), owing to competition between profilin and profilin–actin for FH1 domains (5, 6, 25). Formin-mediated polymerization increases with profilin concentration up to the Kd of profilin–actin [∼2.9 μM for S. cerevisiae profilin and chicken muscle actin (26)] but decreases at higher profilin concentrations, because the FH1 domains become saturated with free profilin (5, 6, 25). These effects of profilin on formin-mediated actin polymerization are recapitulated in our experiments, confirming that Bni1p binds and delivers profilin–actin to the barbed end of the filaments in the actin curtains.
We initially anticipated that higher forces might stretch the natively disordered FH1 domain and compromise delivery of profilin–actin (10, 27). Surprisingly, the application of tension increased the rate of formin-mediated actin polymerization at all profilin concentrations tested (Fig. 4 B and C and Fig. S3). This outcome was in striking contrast to the inhibition of polymerization seen when tension was applied to anchored formin in the absence of profilin. Decreases in the applied tension via filament breakage or a change in the flow rate resulted in corresponding decreases in the polymerization rate. Formin-mediated elongation rates increased by up to ∼20% at the highest flow rates tested (Fig. 4C and Fig. S3), and tension produced the largest increase in average polymerization rates at 2.5 µM profilin (Fig. 4A). Tension-enhanced polymerization was less pronounced at high profilin concentrations, where free profilin occupies some of the sites on the FH1 domains. This biphasic effect of profilin concentration on elongation indicates that the binding of profilin–actin to the FH1 domain enables rapid formin-mediated polymerization under tension, and argues that profilin alone is insufficient to overcome the inhibitory influence of applied tension.
Discussion
We demonstrate that actin filaments can be tethered to lipid-bound biotinylated Bni1(FH1FH2)p and that applying flow aligns formin-anchored barbed ends at nanofabricated barriers resulting in the formation of actin curtains. Several groups have used buffer flow to apply force to surface-tethered molecules (28, 29). However, the actin curtain approach facilitates data collection and analysis by greatly increasing the number of simultaneously imaged, formin-bound filaments and eliminating unbound filaments altogether. In addition, individual actin filaments are all aligned in the direction of the applied hydrodynamic force, minimizing filament overlap. The lipid bilayer also presents a more biologically relevant medium for polymerization than a glass surface. Using this approach, we observed that the tension on tethered formin Bni1p has unanticipated effects on the elongation of actin filaments in both the absence and presence of profilin.
Prior theory suggested that tension would increase FH2 stepping rates, leading to more rapid polymerization rates (8). Our finding that tension slows formin-mediated elongation in the absence of profilin suggests that tension influences another aspect of the process. In one model, FH2 dimers on the barbed end of a filament are thought to transition between a closed state with a conformation similar to the 180° orientation of the subunits observed in crystal structures (1) and a 167° open state, which favors binding of actin monomers to the barbed end of the growing filament (Fig. 1A) (25). At any moment in time, a fraction of the population is in the closed conformation that is incapable of adding subunits (1, 5, 25). Thus, the overall polymerization rate is governed by the equilibrium between the FH2 closed and open states.
A simple extension of this hypothesis is that tension slows the rate of polymerization by shifting the FH2–barbed end equilibrium toward the closed conformation (Fig. 3C). This hypothesis is consistent with a thermodynamic model for formin-mediated polymerization (5, 30, 31), and simulations with a force-sensitive gating mechanism reproduce the effect of tension on the formin-mediated actin polymerization rate in the absence of profilin (theoretical line in Fig. 3B). We suggest that even if tension were to increase the rate at which the FH2 domain steps onto the newly added actin subunit as predicted (8), this higher stepping rate is offset by a shift toward the closed conformation (Fig. 3C).
We considered two mechanisms whereby force might promote actin elongation in the presence of profilin. Tension on the FH1 domains might stimulate transfer of profilin–actin from the FH1 domains to the barbed end of the filament. However, simulations of actin polymerization using the force-sensitive FH2 gating mechanism (Fig. 3C) showed that even extremely fast transfer of profilin–actin from the FH1 domain cannot overcome the negative effect of force on the gating factor (Fig. 4C and Fig. S4). Alternatively, profilin–actin bound to the FH1 domain might allosterically shift the FH2 conformational equilibrium toward the open configuration that allows actin polymerization (Fig. 4D). This mechanism can readily explain how profilin enables rapid actin filament polymerization by surface-tethered formin subjected to tension.
The formin-bound actin filaments that make up the contractile ring during cytokinesis are subjected to a minimum force of ∼6 pN (10). Our finding that profilin overcomes the inhibitory effect of applied tension on formin-mediated elongation of actin filaments explains why profilin binding to both actin- and polyproline-containing FH1 domains is required for cytokinesis in fission yeast (32). Detailed structural and mechanistic studies will be crucial for understanding how formins function under tension in vivo. This work highlights that even small forces have dramatic and unanticipated effects on biological reactions.
Materials and Methods
Buffers.
Lipid buffer (10 mM Tris⋅HCl, pH 7.8, 100 mM NaCl) was used for bilayer deposition on the slide surface, and BSA buffer [40 mM Tris⋅HCl, pH 8.0, 50 mM NaCl, 1 mM MgCl2, 20% (wt/vol) BSA] was used for surface passivation (blocking surfaces). Monomeric actin was stored in G buffer (2 mM Tris⋅HCl, pH 8.0, 0.2 mM ATP, 0.5 mM DTT, 0.1 mM CaCl2, 1 mM sodium azide). Formin construct Bni1(FH1FH2)p and S. cerevisiae profilin were stored in KMEI buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM imidazole, pH 7.0). Polymerization conditions were 10 mM imidazole (pH 7.0), 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 50 mM DTT, 0.2 mM ATP, 0.02 mM CaCl2, 15 mM glucose, 0.02 mg/mL catalase, and 0.1 mg/mL glucose oxidase with or without 0.25% methylcellulose. The 0.25% methylcellulose buffer was prepared from a 2% (wt/vol) methylcellulose stock solution (η = 15 cP, molecular weight ∼14 kDa; M7140; Sigma-Aldrich).
Purification of Actin, Formin, and Profilin.
Chicken skeletal muscle actin was purified from an acetone powder of chicken breast muscle by one cycle of polymerization and depolymerization followed by gel filtration (25). A fraction of the actin was labeled at cysteine 374 with Oregon Green 488 iodoacetamide, followed by depolymerization and purification by ion exchange and gel filtration (33).
Residues 1228–1766 of the S. cerevisiae formin Bni1p were cloned into a pQE70 vector using primers that encoded an N-terminal biotinylation sequence (31). This construct, which also contained a C-terminal 6-His tag, was transformed into BL21 DE3 RP CodonPlus cells (Agilent Technologies). Expression was induced with 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) in the presence of 50 μM biotin and overnight incubation at 16 °C. Cells were lysed by sonication in 500 mM NaCl, 50 mM Tris⋅HCl (pH 8.0) and clarified. The formin was purified by affinity chromatography on a 5-mL column of Ni-NTA resin (Qiagen) eluted with 250 mM imidazole (pH 8.0) in lysis buffer. The protein was then incubated with rotation for 1 h at 4 °C with 2 mL avidin resin (Promega) in the elution buffer with 1 mM DTT. The resin was poured into a column and washed with lysis buffer, and biotinylated Bni1(FH1FH2)p was eluted with 5 mM biotin in lysis buffer. Eluted protein was concentrated using spin columns (Millipore) and dialyzed into KMEI buffer with 1 mM DTT. The protein was flash-frozen in dialysis buffer and stored at −80 °C.
S. cerevisiae profilin was expressed from plasmid pMW172-SpPRF in BL21 DE3 cells and induced with 0.4 mM IPTG at 37 °C for 4 h (25, 26). Cells were suspended in 150 mM KCl, 20 mM Tris⋅HCl (pH 7.5) and lysed by sonication. After the lysate was spun, the supernatant was applied to a poly-l-proline-agarose column, washed with 2 M urea in lysis buffer, and eluted with 7 M urea in lysis buffer (25). After dialyzing into KMEI buffer, eluted protein was concentrated using spin columns and stored at 4 °C.
Nanofabrication of Diffusion Barrier Slides.
Diffusion barriers were fabricated on fused-silica slides (Finkenbeiner) as described (12–14). In brief, slides were cleaned in Nano-Strip solution (Cyantek), rinsed with acetone and isopropanol (IPA), and dried with N2. A double layer of polymethylmethacrylate (PMMA) (MicroChem) was spin-coated on the slides: first 3% PMMA of 25K (molecular weight) in anisole and then 1.5% PMMA of 495K in anisole. The PMMA was coated with a layer of AquaSAVE (Mitsubishi Rayon). Each layer was spun at 4,000 rpm for 45 s with 300 rpm/s acceleration. Barrier patterns were made using electron-beam lithography. After patterning, the AquaSAVE was removed with a water wash and the patterns were developed using a 3:1 solution of IPA to methylisobutylketone (MicroChem) for 1 min with bath sonication at 4 °C. Chromium (Cr) was deposited to the desired thickness using an electron-beam evaporator. Barrier height was verified by tapping-mode atomic force microscopy (Dimension Icon; Veeco) (Fig. S5). PMMA layers and Cr except patterned regions were peeled off by heating and sonicating the slide in acetone for 30 min at 65 °C and for 5 min at room temperature, respectively, leaving behind the Cr barriers on the slide surface.
Flow-Cell Preparation.
Patterned slides were cleaned by successive treatments with 99% acetone, 2% Hellmanex detergent (Hellma Analytics), 99% ethanol, 1 M NaOH, and water. Sample chambers with volumes of about 10 µL were formed using double-side tape (3M) between the diffusion barrier slide and a 50 × 22-mm coverslip (12-548-5E; Fisher Scientific). Nanoports (Upchurch Scientific) were glued on each hole and connected to 1/16-inch tubing (Upchurch Scientific) for injection of solutions from 1- or 3-mL Luer lock syringes. The chamber was then rinsed with 3 mL of water followed by 3 mL of lipid buffer. Biotinylated liposome solution, composed of 10 mg/mL DOPC (1,2-dioleoyl-sn-glycerophosphocholine), 0.8 mg/mL mPEG 2000-DOPE (1,2-dioleoyl-sn-glycero-3-phosphoethanolamine-N-[methoxy(polyethylene glycol)-2000]), and 0.05 mg/mL biotinylated-DPPE [1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(cap biotinyl)], was prepared in lipid buffer (11–13). The liposome solution was diluted 25-fold in lipid buffer, and 1 mL was introduced into the sample chamber and incubated for 15 min. Excess lipids were washed out and the chamber was incubated for 20 min to allow vesicle fusion and bilayer formation on the slide surface. BSA buffer (1 mL) was injected to block exposed surfaces, followed by 1 mL of 25 µg/mL streptavidin in BSA buffer. Free streptavidin was flushed out with 3 mL of BSA buffer, and 1 mL of 0.5–5 nM biotinylated Bni1(FH1FH2)p in KMEI buffer was injected and incubated for ∼10 min. Unbound formin was washed out with 1 mL of BSA buffer. Flow cells were connected to the flow system and placed on a microscope for TIRF imaging. Actin monomers (1.5 μM, 33% Oregon Green-labeled) and other solutions were introduced into the sample chamber with a syringe pump (KD Scientific) at flow rates specified in each figure caption.
Microscopy and Data Analysis.
An inverted microscope (TE-2000U; Nikon) was modified for TIRFM using a 488-nm solid-state laser (Sapphire 488 CDHR; Coherent) for illumination. The flow cell was placed on a motorized microscope stage (Ludl Electronic Products) to control the x-y-z position of the flow cell. The laser was directed through a fused-silica dove prism (ZCMI125012; ESCO) at θi = 67° angle of incidence (critical angle: θc = 66°) to generate an evanescent field on the sample side of the slide with a penetration depth of 224 nm. Fluorescence was collected by a water-immersion objective lens (PlanApo, 60×, NA 1.2; Nikon) and imaged by an EM-CCD (Cascade 512B; Photometrics) after passing through a notch filter (NF01-488U-25; Semrock) to block the 488-nm excitation laser. Exposure times were 200 ms unless otherwise specified.
Time-lapse movies of actin curtains were digitized using commercial software (NIS-Elements; Nikon) and analyzed using ImageJ software (http://rsb.info.nih.gov/ij). We typically measured elongation for 300 s. Individual filament lengths were measured over time using ImageJ software and the Measure Smooth Length plugin (Pollard laboratory plugin). Polymerization rates were determined by applying linear fits to short spans of elongation data (30–60 s), thereby yielding multiple polymerization rates for each filament as its length increased. Polymerization rates were determined for 10–40 filaments that were separated by an average distance of 13 μm at each profilin concentration and flow rate. Analyzing only actin filaments that were well-separated from their nearest neighbors ensured that we were visualizing single filaments and also minimized potential effects of local heterogeneities in buffer flow arising from closely juxtaposed filaments.
Simulations of Formin-Mediated Polymerization.
We adapted a model using Virtual Cell software (National Resource for Cell Analysis and Modeling and the National Center for Research Resources; http://nrcam.uchc.edu) to simulate the effects of force-sensitive gating on formin-mediated polymerization in the absence and presence of profilin (5, 30, 31). This public model considers profilin, actin, FH2-associated barbed ends, and FH1 domains as distinct species and the minimal components required for polymerization. Table S3 summarizes the rate constants for the formation of intermediate species (FH1–profilin, FH1–profilin–actin, FH1–profilin–actin–barbed end, barbed end–profilin, and filamentous actin).
To calculate the effect of force on the gating factor, we applied a linear fit to the polymerization data for drag forces <0.4 pN in Fig. 3C, and divided the polymerization rates at each drag force by the polymerization rate of free (not formin-bound) filaments (measured in the absence of force to be 17.6 subunits per s). We used this relationship between drag force and gating factor in our simulations of formin-mediated polymerization in the absence and presence of 2.5 μM profilin.
Supplementary Material
Acknowledgments
We thank Yongli Zhang for helpful discussions and critical reading of the manuscript. This work was supported by National Institutes of Health Research Grants GM026338 (to T.D.P.), and GM074739 (to E.C.G.), a National Science Foundation Presidential Early Career Award for Scientists and Engineers (to E.C.G.), and the Howard Hughes Medical Institute (E.C.G.). J.Y.L. was supported in part by Korean Research Foundation Grant KRF-2008-357-C00048. N.C. is a postdoctoral fellow of the Leukemia and Lymphoma Society.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1308257110/-/DCSupplemental.
References
- 1.Otomo T, et al. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature. 2005;433(7025):488–494. doi: 10.1038/nature03251. [DOI] [PubMed] [Google Scholar]
- 2.Paul AS, Pollard TD. Review of the mechanism of processive actin filament elongation by formins. Cell Motil Cytoskeleton. 2009;66(8):606–617. doi: 10.1002/cm.20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu Y, et al. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 2004;116(5):711–723. doi: 10.1016/s0092-8674(04)00210-7. [DOI] [PubMed] [Google Scholar]
- 4.Kovar DR, Pollard TD. Insertional assembly of actin filament barbed ends in association with formins produces piconewton forces. Proc Natl Acad Sci USA. 2004;101(41):14725–14730. doi: 10.1073/pnas.0405902101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vavylonis D, Kovar DR, O’Shaughnessy B, Pollard TD. Model of formin-associated actin filament elongation. Mol Cell. 2006;21(4):455–466. doi: 10.1016/j.molcel.2006.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kovar DR, Harris ES, Mahaffy R, Higgs HN, Pollard TD. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124(2):423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- 7.Higgs HN. Formin proteins: A domain-based approach. Trends Biochem Sci. 2005;30(6):342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- 8.Kozlov MM, Bershadsky AD. Processive capping by formin suggests a force-driven mechanism of actin polymerization. J Cell Biol. 2004;167(6):1011–1017. doi: 10.1083/jcb.200410017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shemesh T, Kozlov MM. Actin polymerization upon processive capping by formin: A model for slowing and acceleration. Biophys J. 2007;92(5):1512–1521. doi: 10.1529/biophysj.106.098459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vavylonis D, Wu JQ, Hao S, O’Shaughnessy B, Pollard TD. Assembly mechanism of the contractile ring for cytokinesis by fission yeast. Science. 2008;319(5859):97–100. doi: 10.1126/science.1151086. [DOI] [PubMed] [Google Scholar]
- 11.Granéli A, Yeykal CC, Prasad TK, Greene EC. Organized arrays of individual DNA molecules tethered to supported lipid bilayers. Langmuir. 2006;22(1):292–299. doi: 10.1021/la051944a. [DOI] [PubMed] [Google Scholar]
- 12.Greene EC, Wind S, Fazio T, Gorman J, Visnapuu ML. DNA curtains for high-throughput single-molecule optical imaging. Methods Enzymol. 2010;472:293–315. doi: 10.1016/S0076-6879(10)72006-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visnapuu ML, Fazio T, Wind S, Greene EC. Parallel arrays of geometric nanowells for assembling curtains of DNA with controlled lateral dispersion. Langmuir. 2008;24(19):11293–11299. doi: 10.1021/la8017634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groves JT, Ulman N, Boxer SG. Micropatterning fluid lipid bilayers on solid supports. Science. 1997;275(5300):651–653. doi: 10.1126/science.275.5300.651. [DOI] [PubMed] [Google Scholar]
- 15.Mossman KD, Campi G, Groves JT, Dustin ML. Altered TCR signaling from geometrically repatterned immunological synapses. Science. 2005;310(5751):1191–1193. doi: 10.1126/science.1119238. [DOI] [PubMed] [Google Scholar]
- 16.Whistler RL, BeMiller JN, editors. Industrial Gums: Polysaccharides and Their Derivatives. 3rd Ed. San Diego: Academic; 1993. [Google Scholar]
- 17.Togrul H, Arslan N. Flow properties of sugar beet pulp cellulose and intrinsic viscosity-molecular weight relationship. Carbohydr Polym. 2003;54(1):63–71. [Google Scholar]
- 18.Herraez-Dominguez JV, Gil Garcia de Leon F, Diez-Sales O, Herraez-Dominguez M. Rheological characterization of two viscosity grades of methylcellulose: An approach to the modeling of the thixotropic behaviour. Colloid Polym Sci. 2005;284(1):86–91. [Google Scholar]
- 19.Kirk DE, Othmer DF. Encyclopedia of Chemical Technology. 2nd Ed. New York: Wiley; 2004. [Google Scholar]
- 20.Batchelor GK. Slender-body theory for particles of arbitrary cross-section in Stokes flow. J Fluid Mech. 1970;44(3):419–440. [Google Scholar]
- 21.Brennen C, Winet H. Fluid mechanics of propulsion by cilia and flagella. Annu Rev Fluid Mech. 1977;9:339–398. [Google Scholar]
- 22.Gittes F, Mickey B, Nettleton J, Howard J. Flexural rigidity of microtubules and actin filaments measured from thermal fluctuations in shape. J Cell Biol. 1993;120(4):923–934. doi: 10.1083/jcb.120.4.923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berg HC. Random Walks in Biology. Princeton, NJ: Princeton Univ Press; 1993. [Google Scholar]
- 24.Gorman J, Plys AJ, Visnapuu ML, Alani E, Greene EC. Visualizing one-dimensional diffusion of eukaryotic DNA repair factors along a chromatin lattice. Nat Struct Mol Biol. 2010;17(8):932–938. doi: 10.1038/nsmb.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Paul AS, Pollard TD. The role of the FH1 domain and profilin in formin-mediated actin-filament elongation and nucleation. Curr Biol. 2008;18(1):9–19. doi: 10.1016/j.cub.2007.11.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eads JC, et al. Structure determination and characterization of Saccharomyces cerevisiae profilin. Biochemistry. 1998;37(32):11171–11181. doi: 10.1021/bi9720033. [DOI] [PubMed] [Google Scholar]
- 27.Pollard TD. Progress towards understanding the mechanism of cytokinesis in fission yeast. Biochem Soc Trans. 2008;36(Pt 3):425–430. doi: 10.1042/BST0360425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jégou A, et al. Individual actin filaments in a microfluidic flow reveal the mechanism of ATP hydrolysis and give insight into the properties of profilin. PLoS Biol. 2011;9(9):e1001161. doi: 10.1371/journal.pbio.1001161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Oijen AM, et al. Single-molecule kinetics of lambda exonuclease reveal base dependence and dynamic disorder. Science. 2003;301(5637):1235–1238. doi: 10.1126/science.1084387. [DOI] [PubMed] [Google Scholar]
- 30.Courtemanche N, Pollard TD. Determinants of Formin Homology 1 (FH1) domain function in actin filament elongation by formins. J Biol Chem. 2012;287(10):7812–7820. doi: 10.1074/jbc.M111.322958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paul AS, Pollard TD. Energetic requirements for processive elongation of actin filaments by FH1FH2-formins. J Biol Chem. 2009;284(18):12533–12540. doi: 10.1074/jbc.M808587200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu J, Pollard TD. Profilin binding to poly-L-proline and actin monomers along with ability to catalyze actin nucleotide exchange is required for viability of fission yeast. Mol Biol Cell. 2001;12(4):1161–1175. doi: 10.1091/mbc.12.4.1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuhn JR, Pollard TD. Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys J. 2005;88(2):1387–1402. doi: 10.1529/biophysj.104.047399. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.