Abstract
Plants and green algae have a low pH-inducible mechanism in photosystem II (PSII) that dissipates excess light energy, measured as the nonphotochemical quenching of chlorophyll fluorescence (qE). Recently, nonphotochemical quenching 4 (npq4), a mutant strain of the green alga Chlamydomonas reinhardtii that is qE-deficient and lacks the light-harvesting complex stress-related protein 3 (LHCSR3), was reported [Peers G, et al. (2009) Nature 462(7272):518–521]. Here, applying a newly established procedure, we isolated the PSII supercomplex and its associated light-harvesting proteins from both WT C. reinhardtii and the npq4 mutant grown in either low light (LL) or high light (HL). LHCSR3 was present in the PSII supercomplex from the HL-grown WT, but not in the supercomplex from the LL-grown WT or mutant. The purified PSII supercomplex containing LHCSR3 exhibited a normal fluorescence lifetime at a neutral pH (7.5) by single-photon counting analysis, but a significantly shorter lifetime at pH 5.5, which mimics the acidified lumen of the thylakoid membranes in HL-exposed chloroplasts. The switch from light-harvesting mode to energy-dissipating mode observed in the LHCSR3-containing PSII supercomplex was sensitive to dicyclohexylcarbodiimide, a protein-modifying agent specific to protonatable amino acid residues. We conclude that the PSII-LHCII-LHCSR3 supercomplex formed in the HL-grown C. reinhardtii cells is capable of energy dissipation on protonation of LHCSR3.
Keywords: high light stress, light acclimation, photosynthesis, time-resolved fluorescence
Photosynthetic reactions in plants and algae fix CO2 by converting solar energy into electrochemical energy. In nature, unexpected changes in light intensity could lead to overexcitation of the photosystems, resulting in the accumulation of harmful reactive oxygen species (1). Plants and green algae have developed protective nonphotochemical quenching (NPQ) mechanisms that alleviate such photo-oxidative stress. Among these mechanisms, quenching of chlorophyll (Chl) fluorescence (qE)—a feedback process of photosystem II (PSII) regulation—dissipates excess light energy captured by PSII as heat on luminal acidification of the thylakoid membranes, which occurs along with elevated electron flow.
Numerous previous studies have focused on elucidating the molecular mechanism of qE quenching (see refs. 1–3 for reviews). In higher plants, qE induction depends on activation of the xanthophyll cycle (4–6) and the sensing of luminal acidification by PsbS, a protein homologous to light harvesting complex (LHC) proteins (7, 8). Protonation of PsbS induces a change in the macro-organization of the thylakoid membranes (3, 9, 10) that results in the aggregation of LHCII proteins (11) and/or induces conformational change(s) in LHCII (12), allowing for the formation of energy-quenching sites (13). Although both PsbS protein and zeaxanthin are thought to have crucial roles in qE quenching in higher plants, such molecular effectors have not been studied in depth in other photosynthetic organisms. The green alga Chlamydomonas reinhardtii does not express the PsbS protein (14), even though the PsbS gene is present (15), and a mutant deficient in violaxanthin deepoxidase activity still exhibits qE quenching (6, 16). Moreover, qE is inducible in C. reinhardtii. In contrast to higher plants, where qE quenching is activated immediately on exposure to high light (HL), the activation of qE quenching in C. reinhardtii requires prolonged exposure to HL (16) or low CO2 (17), suggesting that green algae have a distinct mechanism for qE induction and activation.
Niyogi et al. (18) recently reported that a C. reinhardtii mutant called nonphotochemical quenching 4 (npq4), which is deficient in the light-harvesting complex stress-related protein 3 (LHCSR3), induces little qE quenching. The genes for LHCSR3 (Lhcsr3.1 and Lhcsr3.2), formerly known as LI818 (19), encode a 25–26 –kDa integral membrane protein whose expression is induced under HL (20), low CO2 (21), or low iron (22) conditions and that can bind both Chl and xanthophylls (23). Furthermore, a recombinant LHCSR3 polypeptide reconstituted with Chl and xanthophylls is capable of dissipating excitation energy in a low-pH buffer, suggesting that this protein controls both the pH-sensing and energy-quenching functions in C. reinhardtii (23). Where this protein is localized in the thylakoid membranes, and whether it dissipates energy captured by PSII, remain unclear, however.
In this study, using both WT C. reinhardtii and its npq4 mutant grown in low light (LL) or HL and a newly established procedure (24), we isolated and characterized the PSII supercomplex associated with light-harvesting proteins.
Results
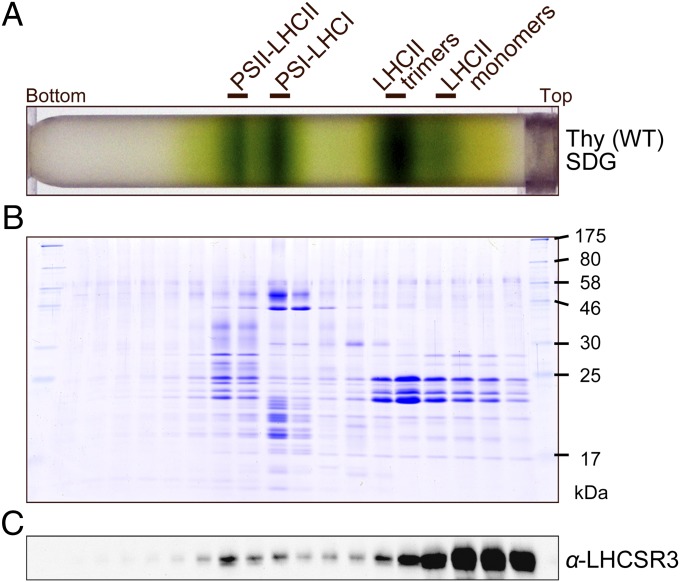
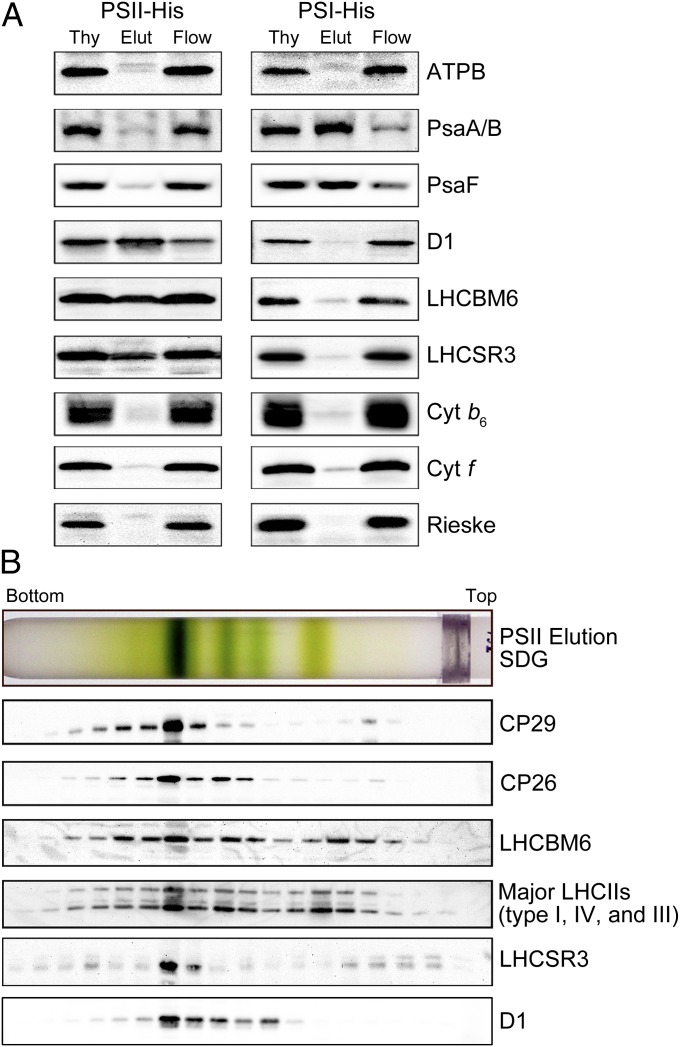
Sucrose density gradient (SDG) ultracentrifugation of the solubilized protein complexes from HL-grown WT C. reinhardtii cells resulted in four green bands—LHCII monomers, LHCII trimers, the photosystem I (PSI)-LHCI supercomplex, and the PSII-LHCII supercomplex (Fig. 1 A and B)—as reported previously (24). LHCSR3 signals were detected in the LHCII monomers and trimers (the fractions containing the free LHCII proteins) and the high molecular weight fraction of the PSII-LHCII supercomplex, but were barely discernible in the fraction containing the PSI-LHCI supercomplex (Fig. 1C). Immunoblot analysis of samples eluted from a nickel column indicated that the His-tagged PSII preparation, but not the His-tagged PSI preparation, included a significant amount of LHCSR3 (Fig. 2A), suggesting that LHCSR3 was associated almost exclusively with PSII. SDG ultracentrifugation of the His-tagged PSII preparation (Fig. 2B) yielded, as expected, a thick green band in the PSII-LHCII supercomplex position, and most of the LHCSR3 was localized to this fraction (Fig. 2B). The top three bands are likely the products of partial disassembly of the PSII-LHCII supercomplex including LHCII, the PSII core particle, and the PSII core particle plus a few LHCIIs, respectively (Fig. 2B). Thus, we concluded that LHCSR3 was expressed in C. reinhardtii under HL conditions, and that it associated predominantly with the PSII-LHCII supercomplex to form the PSII-LHCII-LHCSR3 supercomplex.
Fig. 1.
Purification of the PSII-LHCII-LHCSR3 supercomplex from WT C. reinhardtii. (A) Thylakoids from WT (137c) cells grown under HL conditions (500 μE/m2/s) were solubilized with α-DM and subjected to SDG centrifugation. The solubilized membranes (200 μg Chl) were loaded on a gradient. The four green bands were identified as dissociated LHCIIs (monomers and trimers), the PSI-LHCI supercomplex, and the PSII-LHCII supercomplex (24). (B) Polypeptides in the SDG fractions shown in A were analyzed by SDS/PAGE and staining with Coomassie brilliant blue R-250. (C) Polypeptides in the SDG fractions from the α-DM–solubilized WT thylakoids were subjected to immunoblotting with an antibody against LHCSR3.
Fig. 2.
Polypeptide composition of affinity-purified photosystems. (A) Immunoblotting analyses with antibodies against each photosynthetic protein were performed as labeled. The PSII and PSI supercomplexes isolated by nickel-affinity chromatography were obtained from PsbH-His and PsaA-His strains, respectively; 1 μg of Chl was loaded in each lane. Thy, thylakoids; Elut, elution fraction; Flow, flow-through fraction. (B) The affinity-purified PSII-LHCII-LHCSR3 supercomplexes were subjected to SDG fractionation and immunologically characterized using the specific antibodies as labeled.
To determine the stoichiometry of LHCSR3 with the PSII-LHCII-LHCSR3 supercomplex, we immunologically quantitated the amounts of a minor monomeric LHCII protein CP26 and LHCSR3 using a recombinant CP26 polypeptide and LHCSR3 polypeptide, respectively, as standards (Fig. S1). The ratio of LHCSR3/CP26 thus estimated was 0.14 (Table S1), suggesting that a substoichiometric amount of LHCSR3 is associated with the supercomplex, with one supercomplex binding 0.28 LHCSR3.
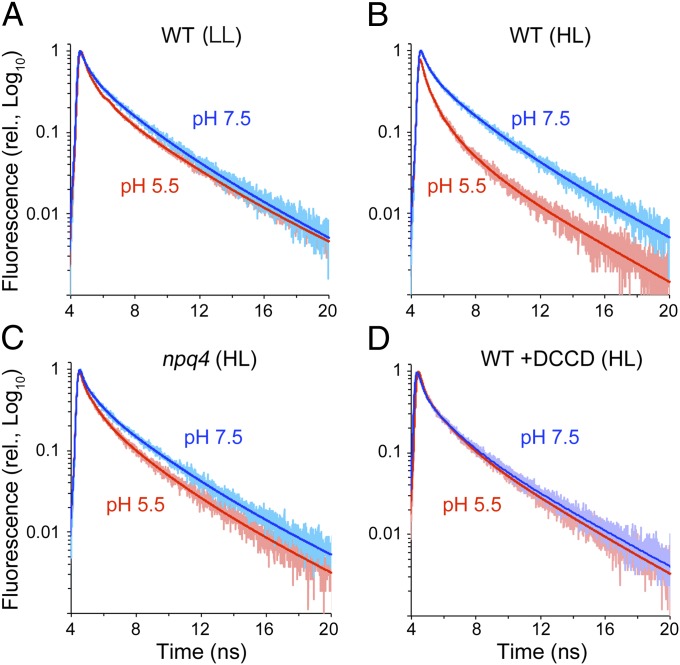
We subjected the PSII-LHCII and PSII-LHCII-LHCSR3 supercomplexes from the LL-grown and HL-grown WT alga to time-correlated single-photon counting experiments to examine whether association with LHCSR3 affects the fluorescence quenching capacity of the PSII-LHCII supercomplex (Fig. 3). When measured at pH 7.5 (which mimics the pH in the lumen of the chloroplasts in the LL-grown cells), the PSII-LHCII supercomplex from the LL-grown WT cells fluoresced with an average lifetime (τAVE) of 2.6 ns and had three components, one component each at 300 ps (12.8%), 1.6 ns (41.6%), and 4.1 ns (45.6%) (Fig. 3 and Table 1). Because lumen acidification is required for qE activation (4, 5, 25), we examined the fluorescence lifetime of the PSII-LHCII supercomplex at pH 5.5 (Fig. 3A); this lifetime did not differ significantly from that at pH 7.5, indicating that in a neutral or acidic buffer in the absence of LHCSR3, the PSII-LHCII supercomplex is in a high fluorescence state. We observed a similarly long lifetime fluorescence (τAVE = 2.6 ns) in the PSII-LHCII-LHCSR3 supercomplex from the HL-grown cells when measured at pH 7.5 (Fig. 3B). At pH 5.5, however, we detected a significantly shorter lifetime fluorescence (τAVE = 1.8 ns) (Fig. 3B), with exponential components of 200 ps (21.1%), 900 ps (40.1%), and 3.5 ns (38.8%) (Table 1). We ascribed the increased quenching capacity at pH 5.5 primarily to the amplitude increase (to 21.1% from 12.7%) and the lifetime decrease (to 200 ps from 300 ps) of the fastest component.
Fig. 3.
Time-resolved fluorescence analysis of the PSII supercomplexes. Time-correlated single-photon counting of fluorescence for the PSII supercomplexes from the LL-grown WT (A), HL-grown WT (B), HL-grown npq4 (C), and DCCD-treated HL-grown WT (D) were recorded at 682 nm (slit = 4 nm) at pH 5.5 and 7.5. Decay curves were fitted with three exponential functions. The faint colored dots are the actual data points, and the thick colored lines are the fitted curves.
Table 1.
Average Chl fluorescence lifetimes (τAVE) and the three lifetime components of the PSII-LHCII-LHCSR3 supercomplex at pH 5.5 and 7.5 in the presence or absence of DCCD after fitting
| PSII supercomplex (pH) | DCCD | τAve, ns | A1, % | τ1, ps | A2, % | τ2, ns | A3, % | τ3, ns |
| LL: WT (pH 7.5) | — | 2.6 | 12.8 | 300 | 41.6 | 1.6 | 45.6 | 4.1 |
| LL: WT (pH 5.5) | — | 2.4 | 20.8 | 200 | 29.0 | 1.3 | 50.2 | 4.0 |
| HL: WT (pH 7.5) | — | 2.6 | 12.7 | 300 | 31.9 | 1.5 | 55.4 | 3.8 |
| HL: WT (pH 5.5) | — | 1.8 | 21.1 | 200 | 40.1 | 0.9 | 38.8 | 3.5 |
| HL: WT (pH 7.5) | + | 2.6 | 12.8 | 300 | 41.6 | 1.6 | 45.6 | 4.1 |
| HL: WT (pH 5.5) | + | 2.5 | 17.3 | 300 | 32.0 | 1.4 | 50.7 | 3.9 |
| HL: npq4 (pH 7.5) | — | 2.7 | 12.8 | 200 | 29.0 | 1.3 | 58.1 | 3.8 |
| HL: npq4 (pH 5.5) | — | 2.3 | 14.3 | 200 | 34.0 | 1.0 | 51.7 | 3.6 |
A, relative amplitude; τ, lifetime; +, present; —, absent.
The activation of qE observed in the PSII-LHCII-LHCSR3 supercomplex after the decrease in pH was likely mediated by protonation of its acidic residues, as proposed in earlier work with recombinant LHCSR3 (23). Fig. 3D shows the inhibitory effect of dicyclohexylcarbodiimide (DCCD) on qE activation. A long lifetime fluorescence (τAVE = 2.5 ns) at pH 5.5 was evident after the supercomplex was treated with DCCD (Table 1), indicating that protonation of the PSII-LHCII-LHCSR3 supercomplex is necessary for qE activation. We further performed a binding assay of [14C]-DCCD to the supercomplex polypeptides to determine the potential targets of DCCD. After the PSII-LHCII-LHCSR3 supercomplex from the HL-grown WT and the PSII-LHCII supercomplex from the HL-grown npq4 mutant were treated with radioactive DCCD under the same conditions as those under which it inhibited qE activation, the decorated polypeptides were visualized by autoradiography after separation by SDS/PAGE. Fig. S2 shows the four DCCD-labeled bands corresponding to CP26, a minor monomeric LHCII protein CP29, major LHCII type I (LhcbM3/4/6/8/9)/LHCSR1/LHCSR3, and major LHCII type III (LhcbM2/7). The intensity of the third band from the PSII-LHCII supercomplex was less than that of the PSII-LHCII-LHCSR3 supercomplex (77%), suggesting that LHCSR3 is one of the targets of DCCD.
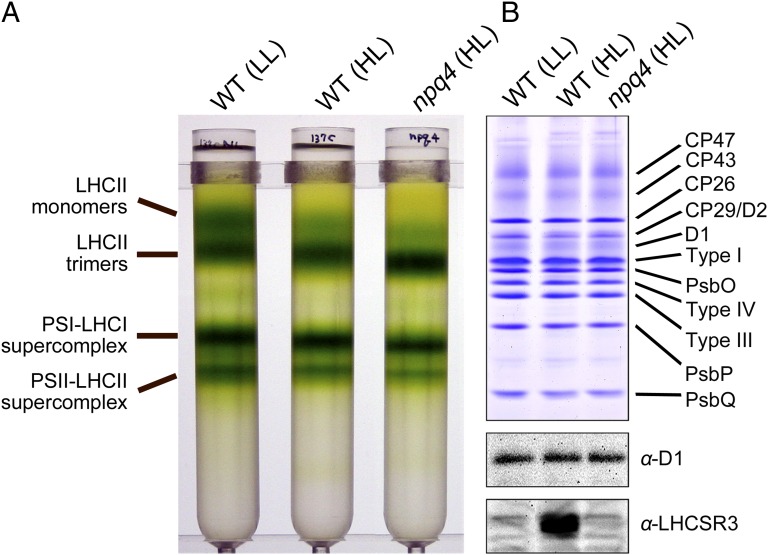
Examination of the photosynthetic supercomplexes in the HL-grown npq4 mutant revealed stable formation of the PSII-LHCII supercomplex in the absence of LHCSR3 (Fig. 4), suggesting that LHCSR3 could bind the periphery of the supercomplex. The supercomplex from the npq4 mutant exhibited fluorescence with an average lifetime comparable to that of the supercomplex from the LL- or HL-grown WT at pH 7.5 (τAVE = 2.7 ns) (Fig. 3C and Table 1). At pH 5.5, the supercomplex from the npq4 mutant exhibited fluorescence with an average lifetime of 2.3 ns (Fig. 3C and Table 1), much longer than that of the HL-grown WT supercomplex but still shorter than that measured at pH 7.5. These results indicate that LHCSR3 is necessary for the PSII-LHCII supercomplex to exhibit a large quenching capacity. Moreover, because the supercomplex prepared from the npq4 mutant exhibited fluorescence with an intermediate lifetime, it is likely that the supercomplex in the npq4 mutant retained additional quenching effector(s).
Fig. 4.
Purification of the PSII-LHCII-LHCSR3 supercomplex from C. reinhardtii npq4 strain. (A) Thylakoids from WT cells grown under LL conditions (40 μE m−2 s−1) and from the npq4 mutant grown under HL conditions (500 μE m−2 s−1) were solubilized with α-DM and subjected to SDG centrifugation. Solubilized membranes with 200 μg of Chl were loaded onto the gradients. (B) Polypeptides in the SDG fractions shown in A were analyzed by SDS/PAGE, stained with Coomassie brilliant blue R-250, and subjected to immunoblot analysis as indicated. Samples were normalized to the amount of D1 protein. Type I, major LHCII type I (LhcbM3/4/6/8/9); type III, major LHCII type III (LhcbM2/7); type IV, major LHCII type IV (LhcbM1).
Examiniation of the fluorescence lifetime of the free LHCII fractions to examine whether LHCSR3 exhibited quenching capacity for itself (Table S2) showed that the fraction from the HL-grown cells, which includes LHCSR3 (as in Fig. 1B), exhibited a long lifetime fluorescence (τAVE = 3.7 ns) at pH 7.5, similar to the isolated LHCII proteins (26). This result was not affected by a pH shift to 5.5 (τAVE = 3.7 ns). The free LHCII fractions prepared from the LL culture in which no LHCSR3 was present, also exhibited a long lifetime fluorescence (τAVE = 3.8 ns), demonstrating that the overall fluorescence lifetime was not affected by the presence of free LHCSR3.
Discussion
Plants and green algae alleviate HL stress via qE, a regulatory feedback process that safely dissipates excess energy as heat and in turn decreases the radiation decay yield (i.e, fluorescence) of Chl-excited states. Thus, active qE is manifested by the quenching of Chl fluorescence (NPQ). Because qE is so induced, at least two effectors are required: a pH sensor, which senses the luminal proton concentration, and a quenching site, in which the Chl-excited states decay primarily as heat. In vascular plants, the qE effector PsbS senses the luminal pH (1) and modifies the thylakoid membrane macrostructure accordingly (3, 9, 10); however, PsbS likely is not a quenching site, because it does not bind pigments (27). The altered thylakoid structure leads to conformational change(s) within LHCIIs (12), generating energy-quenching site(s) in the LHCII aggregates (11) and/or minor monomeric LHCII(s) (13), and establishing a low-fluorescence state that is measurable as NPQ. In contrast, in green algae, the qE effector LHCSR3 could bind Chl a and b, violaxanthin, lutein, and zeaxanthin, presumably acting as an energy-quenching site as well as a sensor for luminal acidification, as was demonstrated recently using a recombinant LHCSR3 that was reconstituted with the pigments (23).
In the present work, we have demonstrated that LHCSR3 is associated primarily with PSII (Fig. 1), and have isolated a PSII supercomplex containing LHCSR3 as well as the antenna proteins LHCII (PSII-LHCII-LHCSR3 supercomplex) from C. reinhardtii. A trace amount of LHCSR3 in the PSI-LHCI supercomplex (Fig. 1) possibly represents an LHCSR3 population associated with the PSI-LHCI supercomplex during state 2, as was reported in a recent study of HL-treated C. reinhardtii (28). Interestingly, the PSII-LHCII supercomplex was in an energy-dissipative state only in the presence of LHCSR3 and only at pH 5.5, not at pH 7.5. Our analysis of the pigment compositions of the PSII-LHCII and PSII-LHCII-LHCSR3 supercomplexes from the LL-grown and HL-grown WT and the npq4 mutant indicated only trace amounts of zeaxanthin in the samples (Fig. S3). Thus, the observed energy dissipation in the supercomplex was not related to the accumulation of zeaxanthin (5). Because the energy dissipation was inhibited by DCCD (Fig. 3D), it could be related to a protonation-induced conformational change within the PSII-LHCII-LHCSR3 supercomplex. The DCCD-binding amino acid residue(s) is more likely the site of the “low-pH switch.” The binding assay using radioactive DCCD revealed that DCCD could bind to LHCSR3, as well as to some other LHCII proteins (Fig. S2). Although further experiments are needed to pinpoint this low-pH switch by, for instance, site-directed mutagenesis of the possible DCCD-binding sites, we speculate that protonation(s) within LHCSR3 would switch the PSII-LHCII supercomplex into the energy-dissipative mode in C. reinhardtii.
Because qE is a part of the NPQ that is dependent on luminal acidification, we can estimate the extent of qE within the NPQ by canceling ΔpH across the thylakoid membranes using the ionophore nigericin (5). The HL-grown C. reinhardtii cells exposed to HL exhibited a nigericin-sensitive NPQ value at 1.77 (NPQWT – NPQnigericin) (Fig. S4). From the photon counts at pH 7.5 and 5.5 obtained through the fluorescence lifetime measurements, we can estimate how many photons were radiated at pH 7.5 but quenched at pH 5.5 (Table S3). After normalizing the quenched photon counts with all of the photon counts measured at pH 5.5, we estimated the quenching capacity (NPQcalc) of the protonated PSII-LHCII-LHCSR3 supercomplex as 1.06 and the protonated PSII-LHCII supercomplex as 0.28 according to the Stern–Volmer equation (Table S3). The difference between the two NPQcalc values (1.06 − 0.28 = 0.78), representing the LHCSR3-dependent NPQ of the supercomplex, was smaller than the LHCSR3-dependent NPQ observed in vivo (NPQWT − NPQnpq4 = 1.32; Fig. S4), which is tentatively accounted for by the substoichiometric amount of LHCSR3 in the isolated supercomplex (Fig. S1 and Table S1). The part of qE retained in the npq4 mutant (NPQnpq4 − NPQnigericin = 0.45) indicates that some NPQ components are independent of LHCSR3, which is possibly dependent on LHCSR1, another copy of the LHCSR protein, and/or xanthophyll cycle pigments located outside of the PSII-LHCII-LHCSR3 supercomplex.
Regarding the molecular basis of the energy dissipation observed in this study, we note that the increased amplitude and shortened lifetime of the fastest fluorescence lifetime component (Table 1) were most likely the primary cause of the induction of quenching by the shift to low pH. Recently reported fluorescence snapshots obtained during the HL treatment of living C. reinhardtii cells show an increase in the amplitudes of the 65-ps and 305-ps lifetime components (29), and the authors tentatively suggested that the changes in the 65-ps component were related to charge-transfer quenching in the minor LHCII and the changes in the 305-ps component were related to aggregated LHCII trimers. Because the major fluorescence lifetime change in our study was detected in the same range as that of the 305-ps component, the two studies could have observed the same quenching process; however, we suggest that the changes in the 200- to 300-ps component were not related to the aggregated LHCII trimers, because we did not observe such lifetime components in the free LHCII fractions (Fig. S5 and Table S2). Additional supporting evidence was obtained when we measured 77 K fluorescence emission spectra of the PSII-LHCII-LHCSR3 supercomplex (Fig. S6), where the (quenched) − (unquenched) difference fluorescence spectra (i.e., the NPQ spectra) showed no positive bands of fluorescence in the far red region, but rather exhibited spectra typical of PSII core particles (30). Such emerging far-red fluorescence has been considered a signature feature of LHCII aggregates (31, 32). Thus, we rather tentatively suggest that the changes seen in the 200- to 300-ps lifetime component were related to the formation of a charge-transfer center, presumably induced by the protonation of LHCSR3 within the supercomplex. Owing to the limited resolution of our single photon counting device, we are not able to address further details of the 65-ps component here.
In this study, we have demonstrated that free LHCSR3 did not exhibit fluorescence quenching using the free LHCII/LHCSR3 fraction (Table S2). Because LHCSR3 is a minor component in this fraction, excluding the possibility that the overall fluorescence lifetime of this fraction was not affected by the presence of LHCSR3 is difficult. In fact, a previous report indicated that a reconstituted LHCSR3 protein exhibited significant energy-quenching capability (23). However, we tentatively conclude that the association of LHCSR3 with the PSII-LHCII supercomplex is essential for inducing qE quenching in C. reinhardtii, because it is at least clear that a smaller amount of LHCSR3, which was associated with the PSII-LHCII supercomplex, caused a large quenching, whereas a larger amount of free LHCSR3 did not (Figs. 1 and 3 and Fig. S5). The Lhcsr3 genes are distributed not only in green algae, but also in moss (33) and in other algae, including diatoms (34, 35). Diatom studies demonstrate that low accumulations of LHCSR proteins (known as LHCX in diatoms) result in a low qE amplitude (34). The moss Physcomitrella pattens contains both PsbS and LHCSR, and disruption of the Lhcsr gene(s) leads to a dramatic decrease in qE (36). Thus, LHCSR3-based energy dissipation could be a widely adopted mechanism of coping with HL stress that preceded the establishment of terrestrial habitation by photosynthetic organisms.
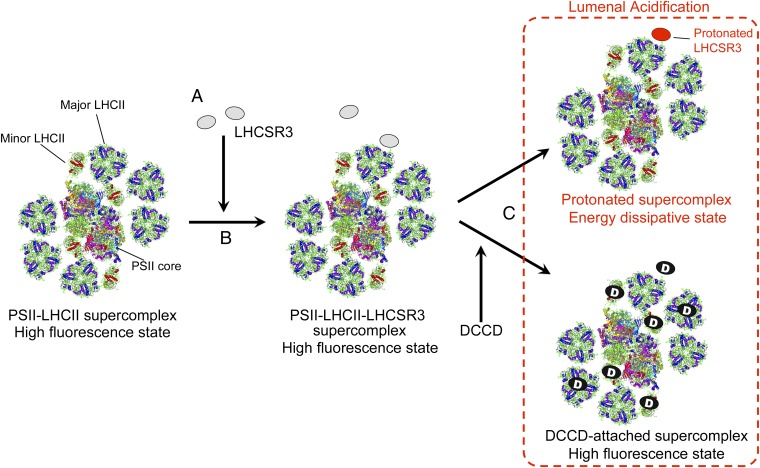
Based on our results and the literature, we propose the following molecular steps for the induction of qE in C. reinhardtii (Fig. 5): (i) LHCSR3 is expressed during HL illumination; (ii) LHCSR3 is bound to the PSII-LHCII supercomplex to form a PSII-LHCII-LHCSR3 supercomplex in the membranes; and (iii) the protonation of LHCSR3, which occurs on luminal acidification by HL illumination, directly modifies the antenna conformation within the supercomplex to form a quenching center.
Fig. 5.
A model for the induction of qE in C. reinhardtii. (A) LHCSR3 expression under HL conditions. (B) Binding of LHCSR3 to the PSII-LHCII supercomplex (24). (C) Acidification of the thylakoid lumen; energy dissipation inhibited by DCCD-binding to the PSII-LHCII-LHCSR3 supercomplex. The crystal coordinates were obtained from the Protein Data Bank (ID codes: PSII core dimer, 2AXT; LHCII, 2NHW; PSI-LHCI supercomplex, 2O01).
Materials and Methods
C.reinhardtii Strains and Growth Conditions.
C. reinhardtii WT strain 137c was obtained from the Chlamydomonas Center (http://www.chlamy.org). The npq4 (18), PsaA-His (37), and PsbH-His (38) mutants were as described previously. All strains were grown photoautotrophically at 23 °C in a high-salt minimal medium (39) with air bubbling under either LL (40 μE m−2 s−1) or HL (500 μE m−2 s−1) conditions.
Isolation of Photosystem Supercomplexes.
Thylakoid membranes from C. reinhardtii cells were prepared as described previously (24), suspended in a 25 mM Mes buffer (pH 6.5) at 0.4 mg Chl/mL, and solubilized with 1.0% dodecyl-α-maltoside (α-DM; Affymetrix) (24) for 5 min on ice. The photosynthetic supercomplexes were purified by SDG centrifugation as reported previously (24). To determine whether LHSCR3 was specifically bound to the PSII-LHCII supercomplex, we purified the PSI-LHCI and PSII-LHCII supercomplexes by nickel column chromatography using the thylakoids from a mutant strain carrying a His-tagged PsaA protein (PSI-LHCI supercomplex) (37) or a His-tagged PsbH protein (PSII-LHCII supercomplex) (38). Nickel-affinity chromatography of the His-tagged PSI and PSII supercomplexes was carried out as described previously (40, 41), but with 0.02% α-DM included in all buffers rather than other detergents.
Purification of Recombinant Proteins in Escherichia coli.
Lhcsr3.1 cDNA was cloned in the pQE52 vector (Qiagen). The resultant LHCSR3-His vector (pQE52-Lhcsr3.1-His) and the CP26-His vector (pQE52-lhcb5-His) (42) were used to transform E. coli host strains M15 [pREP4] and SG13009 [pREP4], respectively. His-tagged proteins were induced by isopropyl β-d-thiogalactopyranoside as described previously (42) and purified in a nickel-nitrilotriacetic acid spin column (Qiagen).
Pigment Analysis.
Pigments were extracted from the PSII supercomplexes with methanol/acetone (50:50 vol/vol) solution. The samples were incubated at 4 °C for 5 min in the dark before analysis. Pigments were separated by ultra-performance liquid chromatography (UPLC) using a Waters H-class system equipped with a 2.1 mm × 150 mm ACQUITY UPLC HSS C18 column (Waters). Gradients were run from 70:10:20 (vol/vol) acetonitrile/isopropanol/water to 85:10:20 (vol/vol) acetonitrile/isopropanol/water at 6.5 min and 50:50:0 (vol/vol) acetonitrile/isopropanol/water at 10 min before returning to 70:10:20 (vol/vol) acetonitrile/isopropanol/water at 15 min. The flow rate was 0.4 mL/min, and the column temperature was 40 °C. Pigment concentrations were calculated with Empower3 software (Waters) using commercial standards (DHI).
SDS/PAGE and Immunoblot Analysis.
SDS/PAGE and immunoblot analyses were conducted as described previously (24). The antibodies used have been described previously (40, 41). Densitometric analyses of the detected images were performed using Image Lab software (Bio-Rad).
Autoradiography.
For autoradiography, [14C]-DCCD (American Radiolabeled Chemicals) in toluene solution was dried with nitrogen gas and resolved in ethanol as reported previously (43), yielding a 2-mM stock solution. The [14C]-DCCD solution was added at a final concentration of 20 μM to the PSII supercomplexes. The [14C]-DCCD -labeled PSII supercomplexes (3 μg Chl) were separated using SDS/PAGE and exposed to an imaging plate (BAS-TR2025; Fuji Photo Film) for 62 h. Autoradiographs were processed on a BAS 5000 instrument (Fuji Photo Film).
Time-Resolved Fluorescence Lifetime Measurement.
Fluorescence lifetimes of the photosynthetic supercomplexes were determined by time-correlated single-photon counting of fluorescence on a FluoroCube instrument (HORIBA Jobin-Yvon). A light-emitting diode (NanoLED; HORIBA Jobin-Yvon) operating at 440 nm was used for excitation, and fluorescence was detected at 682 nm through a monochromator (slit = 4 nm). The supercomplex was treated with 20 μM DCCD, a known inhibitor of ∆pH-dependent qE quenching in higher plants (44). All experiments were performed at 23 °C. Fluorescence decays were analyzed using the reconvolution method with DAS6 software (HORIBA Jobin-Yvon).
Low-Temperature Fluorescence Emission Spectra.
Low-temperature fluorescence emission spectra were measured at 77 K using a FluoroMax4 spectrofluorometer (HORIBA Jobin-Yvon). Protein samples were excited at 440 nm (slit = 20 nm), and fluorescence was monitored between 600 and 800 nm (slit = 2 nm). Areas under the spectra were normalized to single photon counts of fluorescence from the same samples at 23 °C as described above.
Supplementary Material
Acknowledgments
We thank Dr. Michael Hippler (University of Münster) and Dr. Makio Yokono (Kobe University) for valuable discussions and Dr. Toru Hisabori (Tokyo Institute of Technology) and Dr. Yoshikatsu Matsubayashi [National Institute for Basic Biology (NIBB)] for technical advice. This work was supported in part by the Japan Society for the Promotion of Science, through a Research Fellowship for Young Scientists (21001384, to R.T.); the Cabinet Office, through the Funding Program for Next Generation World-Leading Researchers (NEXT Program) (GS026, to J.M.); the New Energy and Industrial Technology Development Organization, through a project for the strategic development of next-generation bioenergy utilization technology (P07015, to NIBB); and the Ministry of Education, Culture, Sports, Science and Technology, through the Network of Centers of Carbon Dioxide Resource Studies in Plants (to NIBB).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1222606110/-/DCSupplemental.
References
- 1.Li Z, Wakao S, Fischer BB, Niyogi KK. Sensing and responding to excess light. Annu Rev Plant Biol. 2009;60:239–260. doi: 10.1146/annurev.arplant.58.032806.103844. [DOI] [PubMed] [Google Scholar]
- 2.de Bianchi S, Ballottari M, Dall’osto L, Bassi R. Regulation of plant light harvesting by thermal dissipation of excess energy. Biochem Soc Trans. 2010;38(2):651–660. doi: 10.1042/BST0380651. [DOI] [PubMed] [Google Scholar]
- 3.Horton P, Johnson MP, Perez-Bueno ML, Kiss AZ, Ruban AV. Photosynthetic acclimation: Does the dynamic structure and macro-organisation of photosystem II in higher plant grana membranes regulate light harvesting states? FEBS J. 2008;275(6):1069–1079. doi: 10.1111/j.1742-4658.2008.06263.x. [DOI] [PubMed] [Google Scholar]
- 4.Demmig-Adams B, Adams WW., 3rd The role of xanthophyll cycle carotenoids in the protection of photosynthesis. Trends Plant Sci. 1996;1(1):21–26. [Google Scholar]
- 5.Horton P, Ruban AV, Walters RG. Regulation of light harvesting in green plants. Annu Rev Plant Physiol Plant Mol Biol. 1996;47:655–684. doi: 10.1146/annurev.arplant.47.1.655. [DOI] [PubMed] [Google Scholar]
- 6.Niyogi KK, Björkman O, Grossman AR. The roles of specific xanthophylls in photoprotection. Proc Natl Acad Sci USA. 1997;94(25):14162–14167. doi: 10.1073/pnas.94.25.14162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li XP, et al. A pigment-binding protein essential for regulation of photosynthetic light harvesting. Nature. 2000;403(6768):391–395. doi: 10.1038/35000131. [DOI] [PubMed] [Google Scholar]
- 8.Li XP, et al. Regulation of photosynthetic light harvesting involves intrathylakoid lumen pH sensing by the PsbS protein. J Biol Chem. 2004;279(22):22866–22874. doi: 10.1074/jbc.M402461200. [DOI] [PubMed] [Google Scholar]
- 9.Kiss AZ, Ruban AV, Horton P. The PsbS protein controls the organization of the photosystem II antenna in higher plant thylakoid membranes. J Biol Chem. 2008;283(7):3972–3978. doi: 10.1074/jbc.M707410200. [DOI] [PubMed] [Google Scholar]
- 10.Kereïche S, Kiss AZ, Kouřil R, Boekema EJ, Horton P. The PsbS protein controls the macro-organisation of photosystem II complexes in the grana membranes of higher plant chloroplasts. FEBS Lett. 2010;584(4):759–764. doi: 10.1016/j.febslet.2009.12.031. [DOI] [PubMed] [Google Scholar]
- 11.Johnson MP, et al. Photoprotective energy dissipation involves the reorganization of photosystem II light-harvesting complexes in the grana membranes of spinach chloroplasts. Plant Cell. 2011;23(4):1468–1479. doi: 10.1105/tpc.110.081646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonente G, Howes BD, Caffarri S, Smulevich G, Bassi R. Interactions between the photosystem II subunit PsbS and xanthophylls studied in vivo and in vitro. J Biol Chem. 2008;283(13):8434–8445. doi: 10.1074/jbc.M708291200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahn TK, et al. Architecture of a charge-transfer state regulating light harvesting in a plant antenna protein. Science. 2008;320(5877):794–797. doi: 10.1126/science.1154800. [DOI] [PubMed] [Google Scholar]
- 14.Bonente G, et al. The occurrence of the psbS gene product in Chlamydomonas reinhardtii and in other photosynthetic organisms and its correlation with energy quenching. Photochem Photobiol. 2008;84(6):1359–1370. doi: 10.1111/j.1751-1097.2008.00456.x. [DOI] [PubMed] [Google Scholar]
- 15.Anwaruzzaman M, et al. Genomic analysis of mutants affecting xanthophyll biosynthesis and regulation of photosynthetic light harvesting in Chlamydomonas reinhardtii. Photosynth Res. 2004;82(3):265–276. doi: 10.1007/s11120-004-2439-y. [DOI] [PubMed] [Google Scholar]
- 16.Niyogi KK, Bjorkman O, Grossman AR. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell. 1997;9(8):1369–1380. doi: 10.1105/tpc.9.8.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Förster B, Barry Osmond C, Boynton JE. Very high light-resistant mutants of Chlamydomonas reinhardtii: Responses of photosystem II, nonphotochemical quenching and xanthophyll pigments to light and CO2. Photosynth Res. 2001;67(1-2):5–15. doi: 10.1023/A:1010611509209. [DOI] [PubMed] [Google Scholar]
- 18.Peers G, et al. An ancient light-harvesting protein is critical for the regulation of algal photosynthesis. Nature. 2009;462(7272):518–521. doi: 10.1038/nature08587. [DOI] [PubMed] [Google Scholar]
- 19.Gagné G, Guertin M. The early genetic response to light in the green unicellular alga Chlamydomonas eugametos grown under light/dark cycles involves genes that represent direct responses to light and photosynthesis. Plant Mol Biol. 1992;18(3):429–445. doi: 10.1007/BF00040659. [DOI] [PubMed] [Google Scholar]
- 20.Richard C, Ouellet H, Guertin M. Characterization of the LI818 polypeptide from the green unicellular alga Chlamydomonas reinhardtii. Plant Mol Biol. 2000;42(2):303–316. doi: 10.1023/a:1006340308077. [DOI] [PubMed] [Google Scholar]
- 21.Miura K, et al. Expression profiling-based identification of CO2-responsive genes regulated by CCM1 controlling a carbon-concentrating mechanism in Chlamydomonas reinhardtii. Plant Physiol. 2004;135(3):1595–1607. doi: 10.1104/pp.104.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naumann B, et al. Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics. 2007;7(21):3964–3979. doi: 10.1002/pmic.200700407. [DOI] [PubMed] [Google Scholar]
- 23.Bonente G, et al. Analysis of LhcSR3, a protein essential for feedback de-excitation in the green alga Chlamydomonas reinhardtii. PLoS Biol. 2011;9(1):e1000577. doi: 10.1371/journal.pbio.1000577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tokutsu R, Kato N, Bui KH, Ishikawa T, Minagawa J. Revisiting the supramolecular organization of photosystem II in Chlamydomonas reinhardtii. J Biol Chem. 2012;287(37):31574–31581. doi: 10.1074/jbc.M111.331991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niyogi KK. Photoprotectioin revisited: Genetic and molecular approaches. Annu Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 26.Moya I, Silvestri M, Vallon O, Cinque G, Bassi R. Time-resolved fluorescence analysis of the photosystem II antenna proteins in detergent micelles and liposomes. Biochemistry. 2001;40(42):12552–12561. doi: 10.1021/bi010342x. [DOI] [PubMed] [Google Scholar]
- 27.Dominici P, et al. Biochemical properties of the PsbS subunit of photosystem II either purified from chloroplast or recombinant. J Biol Chem. 2002;277(25):22750–22758. doi: 10.1074/jbc.M200604200. [DOI] [PubMed] [Google Scholar]
- 28.Allorent G, et al. A dual strategy to cope with high light in Chlamydomonas reinhardtii. Plant Cell. 2013;25(2):545–557. doi: 10.1105/tpc.112.108274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amarnath K, Zaks J, Park SD, Niyogi KK, Fleming GR. Fluorescence lifetime snapshots reveal two rapidly reversible mechanisms of photoprotection in live cells of Chlamydomonas reinhardtii. Proc Natl Acad Sci USA. 2012;109(22):8405–8410. doi: 10.1073/pnas.1205303109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sugiura M, Minagawa J, Inoue Y. Properties of Chlamydomonas photosystem II core complex with a His-tag at the C-terminus of the D2 protein. Plant Cell Physiol. 1999;40(3):311–318. [Google Scholar]
- 31.Miloslavina Y, et al. Ultrafast fluorescence study on the location and mechanism of non-photochemical quenching in diatoms. Biochim Biophys Acta. 2009;1787(10):1189–1197. doi: 10.1016/j.bbabio.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Ruban AV, Horton P. Mechanism of ΔpH-dependent dissipation of absorbed excitation energy by photosynthetic membranes, I: Spectroscopic analysis of isolated light-harvesting complexes. Biochim Biophys Acta. 1992;1102(1):30–38. [Google Scholar]
- 33.Alboresi A, Caffarri S, Nogue F, Bassi R, Morosinotto T. In silico and biochemical analysis of Physcomitrella patens photosynthetic antenna: Identification of subunits which evolved upon land adaptation. PLoS ONE. 2008;3(4):e2033. doi: 10.1371/journal.pone.0002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailleul B, et al. An atypical member of the light-harvesting complex stress-related protein family modulates diatom responses to light. Proc Natl Acad Sci USA. 2010;107(42):18214–18219. doi: 10.1073/pnas.1007703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhu SH, Green BR. Photoprotection in the diatom Thalassiosira pseudonana: Role of LI818-like proteins in response to high light stress. Biochim Biophys Acta. 2010;1797(8):1449–1457. doi: 10.1016/j.bbabio.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 36.Alboresi A, Gerotto C, Giacometti GM, Bassi R, Morosinotto T. Physcomitrella patens mutants affected on heat dissipation clarify the evolution of photoprotection mechanisms upon land colonization. Proc Natl Acad Sci USA. 2010;107(24):11128–11133. doi: 10.1073/pnas.1002873107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gulis G, Narasimhulu KV, Fox LN, Redding KE. Purification of His6-tagged photosystem I from Chlamydomonas reinhardtii. Photosynth Res. 2008;96(1):51–60. doi: 10.1007/s11120-007-9283-9. [DOI] [PubMed] [Google Scholar]
- 38.Cullen M, et al. A highly active histidine-tagged Chlamydomonas reinhardtii photosystem II preparation for structural and biophysical analysis. Photochem Photobiol Sci. 2007;6(11):1177–1183. doi: 10.1039/b708611n. [DOI] [PubMed] [Google Scholar]
- 39.Sueoka N. Mitotic replication of deoxyribonucleic acid in Chlamydomonas reinhardi. Proc Natl Acad Sci USA. 1960;46(1):83–91. doi: 10.1073/pnas.46.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Iwai M, et al. Isolation of the elusive supercomplex that drives cyclic electron flow in photosynthesis. Nature. 2010;464(7292):1210–1213. doi: 10.1038/nature08885. [DOI] [PubMed] [Google Scholar]
- 41.Iwai M, Takahashi Y, Minagawa J. Molecular remodeling of photosystem II during state transitions in Chlamydomonas reinhardtii. Plant Cell. 2008;20(8):2177–2189. doi: 10.1105/tpc.108.059352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Minagawa J, Han KC, Dohmae N, Takio K, Inoue Y. Molecular characterization and gene expression of lhcb5 gene encoding CP26 in the light-harvesting complex II of Chlamydomonas reinhardtii. Plant Mol Biol. 2001;46(3):277–287. doi: 10.1023/a:1010643408100. [DOI] [PubMed] [Google Scholar]
- 43.Jahns P, Junge W. Dicyclohexylcarbodiimide-binding proteins related to the short circuit of the proton-pumping activity of photosystem II: Identified as light-harvesting chlorophyll-a/b-binding proteins. Eur J Biochem. 1990;193(3):731–736. doi: 10.1111/j.1432-1033.1990.tb19393.x. [DOI] [PubMed] [Google Scholar]
- 44.Ruban AV, Walters RG, Horton P. The molecular mechanism of the control of excitation energy dissipation in chloroplast membranes: Inhibition of delta pH-dependent quenching of chlorophyll fluorescence by dicyclohexylcarbodiimide. FEBS Lett. 1992;309(2):175–179. doi: 10.1016/0014-5793(92)81089-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.