Significance
Mosquitoes transmit some of the most dangerous human diseases. Microarray analysis of developmental gene dynamics in the female Aedes aegypti fat body has demonstrated that 6,146 genes are differentially expressed during juvenile hormone-dependent posteclosion development. These genes show striking temporal and functional separation. A RNAi microarray screen revealed a differential action of Met in the down- and up-regulation of genes expressed during early and late PE, respectively. Thus, this study provides substantial evidence of a central role for insect-specific JH and its receptor Met in the regulation of female mosquito reproductive biology.
Keywords: hormone action, insect disease vector, insect-specific hormone
Abstract
Juvenile hormone III (JH) plays a key role in regulating the reproduction of female mosquitoes. Microarray time-course analysis revealed dynamic changes in gene expression during posteclosion (PE) development in the fat body of female Aedes aegypti. Hierarchical clustering identified three major gene clusters: 1,843 early-PE (EPE) genes maximally expressed at 6 h PE, 457 mid-PE (MPE) genes at 24 h PE, and 1,815 late-PE (LPE) genes at 66 h PE. The RNAi microarray screen for the JH receptor Methoprene-tolerant (Met) showed that 27% of EPE and 40% of MPE genes were up-regulated whereas 36% of LPE genes were down-regulated in the absence of this receptor. Met repression of EPE and MPE and activation of LPE genes were validated by an in vitro fat-body culture experiment using Met RNAi. Sequence motif analysis revealed the consensus for a 9-mer Met-binding motif, CACGC/TGA/GT/AG. Met-binding motif variants were overrepresented within the first 300 bases of the promoters of Met RNAi–down-regulated (LPE) genes but not in Met RNAi–up-regulated (EPE) genes. EMSAs using a combination of mutational and anti-Met antibody supershift analyses confirmed the binding properties of the Met consensus motif variants. There was a striking temporal separation of expression profiles among major functional gene groups, with carbohydrate, lipid, and xenobiotics metabolism belonging to the EPE and MPE clusters and transcription and translation to the LPE cluster. This study represents a significant advancement in the understanding of the regulation of gene expression by JH and its receptor Met during female mosquito reproduction.
Mosquitoes transmit some of the most dangerous human diseases. Despite significant efforts, controlling these disease vectors remains challenging. Mosquitoes are disease vectors because they require vertebrate blood for their egg development; disease pathogens have evolved to use this link between a mosquito and a vertebrate host for their life cycles (1, 2). Therefore, a clear understanding of mechanisms governing mosquito egg production is essential for developing novel approaches to vector control (1, 2).
An insect-specific sesquiterpenoid hormone, juvenile hormone III (JH), and a steroid hormone, 20-hydroxyecdysone (20E), are the two major hormones that govern reproduction of female mosquitoes. Vitellogenesis is the central process of egg maturation, which involves the production of massive amounts of yolk protein precursors in the fat body and their accumulation by developing oocytes. This process is activated by blood feeding and is controlled by 20E. The molecular basis of this hormone's action in mosquito vitellogenesis has been studied in great detail (3–7).
A unique aspect of female mosquito reproduction is its JH-dependent posteclosion (PE) development, a period critical for a female to become responsive to 20E and digest blood, subsequently resulting in egg maturation. The JH hemolymph titer increases after adult eclosion, reaching a peak at 48–50 h PE and declining slowly over the next 5 d in females not fed a blood meal (8). Several aspects of the JH-dependent PE development have been characterized: Ovarian primary follicles grow double in length, their nurse cells increase in ploidy, ovaries develop an endocytic complex of coated vesicles and receptors, and the follicular epithelium undergoes differentiation (9–11). Likewise, extensive JH-dependent cellular remodeling occurs in the fat body, an insect analog of the vertebrate liver and adipose tissue. This remodeling involves dramatic enlargement of nucleoli, accumulation of ribosomes, and the development of both Golgi complexes and extensive invaginations of the plasma membrane (12, 13). JH also controls translation of the nuclear receptor betaFTZ-F1 (fushi tarazu binding factor 1), known as the competence factor (14). However, our understanding of molecular mechanisms underlying JH regulation of female mosquito PE development has been limited.
Recent studies have established that Methoprene-tolerant (Met), a member of the family of basic helix–loop–helix (bHLH)-Per-Arnt-Sim (PAS) transcription factors, is the JH receptor. In Drosophila melanogaster, null mutants of the Met gene convey potent resistance to JH and the insecticide methoprene, one of the JH analogs (15, 16). Met binds to JH with a high-affinity–inducing reporter gene transcription (17–19). RNAi-mediated depletion of Met in the beetle Tribolium castaneum leads to precocious pupation, suggesting an anti-metamorphic action of Met (20). In addition, Parthasarathy et al. (21) showed that Met depletion also results in precocious formation of adult features in pupae. Met belongs to the family of bHLH-PAS transcription factors, members of which require the formation of homo- or heterodimers for DNA binding and transcriptional regulation (22). Studies in Aedes aegypti, Tribolium castaneum, and the silkworm Bombyx mori have shown that Met interacts with a bHLH-PAS domain-containing steroid receptor coactivator, SRC [also known as betaFTZ-F1 interacting steroid receptor coactivator (“FISC”) and “Taiman”] (19, 23, 24). In addition to SRC/FISC, Aedes Met has been shown to interact with another bHLH-PAS transcription factor, Cycle (25). Moreover, Met and the circadian proteins Cycle and Clock are required for JH regulation of the photoperiod-dependent switch from diapause to reproduction in Pyrrhocoris apterus (26). Several studies have reported identification of the Met-binding response motifs; however, no unified consensus has been reached (19, 23, 25, 27).
In this work, we have characterized the developmental dynamics of genes expressed during JH-regulated PE development of the female A. aegypti fat body, a tissue central to female reproduction. The microarray analysis revealed an unexpectedly high level of gene activity, with 6,146 genes expressed in temporally and functionally separated cohorts. Met RNAi microarray screens showed a differential action of Met, which down-regulated genes that were maximally expressed early during PE and up-regulated those that showed maximal expression late during PE. An in vitro, fat-body culture experiment using Met RNAi validated the role of Met in mediating JH action. Bioinformatics analysis and EMSA delineated a consensus for a 9-mer Met-binding motif, CACGC/TGA/GT/AG, which was present in the promoters of a number of Met–up-regulated genes. We thus have provided substantial evidence for the central role of JH and its receptor Met in the regulation of female mosquito reproductive biology.
Results
Expression Dynamics of Fat Body Genes During JH-Dependent PE Development of Female A. aegypti Mosquitoes.
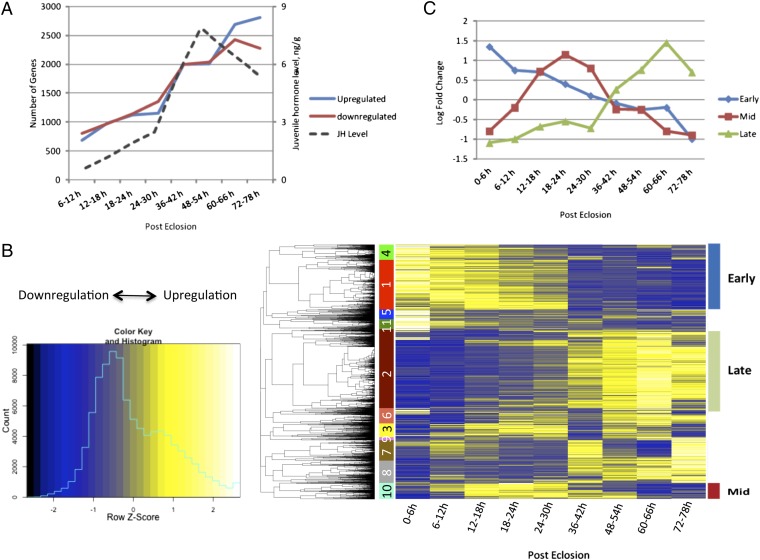
We used custom-made Agilent microarray chips that contained probe sets corresponding to 15,321 A. aegypti genes (28) and examined fat-body samples collected at nine time points spanning the entire PE development of female mosquitoes, from 0–6 h until 72–78 h (Fig. 1A). Differentially expressed gene (DEG) sets were established by comparing transcripts from each of the eight PE time points with that from 0–6 h PE using a minimum fold change of ≥1.75 (0.8 in a log2 scale) as the confidence threshold and a false-discovery rate (P value) of ≤0.01, because it has been used previously (28). There was an unexpectedly high increase of gene expression activity during the PE developmental stage in the fat body of female Aedes mosquitoes (Fig. 1A and Dataset S1). The number of DEGs increased dramatically over this PE time period, reaching a maximum at 60–66 h PE, with 6,146 genes being either up- or down-regulated (Fig. 1A).
Fig. 1.
The microarray analysis of the fat-body transcriptome during PE development of A. aegypti female mosquitoes. (A) Number of significantly up-regulated and down-regulated genes in the microarray experiment [fold change ≥1.75 (0.8 in log2 scale) and a false-discovery rate (P value) of ≤0.01] referred in a chronological time order (h PE). JH titers (dashed line), labeled on the right y-axis, are from Shapiro et al. (8). (B) Hierarchical clustering result of DEGs. The heatmap shows the expression values (mean among three replicates) for the nine different time points. Each row represents data for one probe, and each column corresponds to a time point between 0–6 h and 72–78 h PE, with 6-h intervals. Cutting the gene dendrogram identified 11 discrete clusters, which are color coded on the left. Clusters 1 and 4 were designated as EPE, cluster 10 as the MPE, and cluster 2 as the LPE gene cohorts. (C) Expression profiles of EPE, MPE, and LPE gene cohorts during the 78-h PE period.
To analyze the gene-expression dynamics during PE development, we performed hierarchical clustering of the DEGs identified in the previous step (Fig. 1B). Partitioning of the resulting gene dendrogram identified 11 clusters. Three major clusters with expression trends relevant for fat-body PE development were identified: early posteclosion (EPE), mid posteclosion (MPE), and late posteclosion (LPE) (Fig. 1 B and C and Dataset S1). The 1,843 genes of the EPE cluster (clusters 1 and 4) have the highest expression around 0–6 h PE, followed by a gradual decline throughout PE development (Fig. 1C). The MPE cluster (cluster 10) contained 457 genes. Their expression levels peaked at 18–24 h PE and declined thereafter. The LPE cluster (cluster 2) included 1,815 genes; transcript levels of these genes were low at 0–6 h PE, rose throughout PE to reach peak expression at 60–66 h, and declined moderately at 72–78 h PE (Fig. 1C and Dataset S1). Quantitative RT-PCR (qPCR) analysis of selected EPE, MPE, and LPE genes showed correlation with microarray data (Fig. S1 and Dataset S2).
Overall, our data suggest that EPE genes are maximally expressed at a low JH titer and MPE genes are maximally expressed at an intermediate JH level; expression levels within both of these gene groups is inhibited at a high JH titer. The expression of LPE genes, however, exhibits an opposite trend, requiring a high JH titer for their maximal expression.
PE Gene Expression in the Fat Body of Female Mosquitoes Deprived of JH or Met.
To establish further that the JH signaling pathway is the major determinant of gene regulation in PE development, we used preparations of isolated mosquito abdomens deprived of JH. The specimens were prepared as described in Materials and Methods. Total RNA was isolated 24 h after preparing JH-deprived mosquitoes, and transcript levels were measured by qPCR for seven EPE and nine LPE genes (Dataset S2). Transcript levels of tested EPE genes increased in the JH-deprived mosquitoes with the topical application of solvent in the absence of JH but were inhibited after JH was topically applied (Fig. 2A, Fig. S2A and Dataset S2). In contrast, LPE genes showed an opposite trend; their transcript levels were suppressed in the absence of JH and were elevated with the application of the hormone (Fig. 2B, Fig. S2B and Dataset S2). Thus, we confirmed that JH has an inhibitory effect on a subset of the EPE gene cluster and an activating effect on a subset of the LPE gene cluster.
Fig. 2.
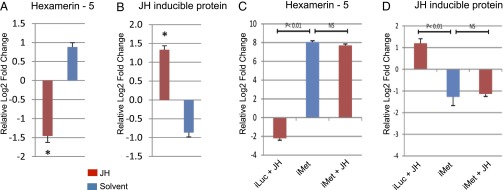
(A, B) Analysis of JH target genes in JH-deprived female mosquitoes. These mosquitoes were prepared by severing their body between meso- and meta-thorax at 6 h PE. Then, meta-thorax/abdomen-containing bodies were sealed by paraffin and topically treated with JH III or with solvent only. Total RNA was extracted 24 h post-treatment and analyzed by means of quantitative RT-PCR. Target genes were selected from the cohorts of gene time-series PE transcriptome. The EPE gene encoding Hexamerin-5 was repressed (A), while the LPE gene encoding for JH-inducible protein was induced by JH (B). Data were normalized using S7 as an internal control. Relative log2-fold changes were established by comparing transcript levels of each sample to 0–6 h PE mosquito sample. Asterisk (*) represents P < 0.01 (t test). (C, D) Met RNAi renders JH target genes insensitive to JH. Fat bodies from 72 h after injection of Met dsRNA were dissected and incubated with or without JH (10 μg/ml JH III) for 3 h at 27°C. Transcript levels of the same JH target genes as in (A, B) were characterized by means of quantitative RT-PCR. Data were normalized using S7 as an internal control. Relative log2-fold changes were established by comparing transcript levels of dsMet (iMet) sample to dsLuc (iLuc) control mosquito sample. Statistically significant differences (two sample t-test; P value ≤0.01) between samples are labeled.
To test whether these JH gene regulation patterns were mediated by Met, we performed in vitro fat-body culture experiments. In preparation for the Met RNAi experiments, we conducted preliminary tests to evaluate the effectiveness of the Met RNAi (iMet) knockdown (Dataset S2). At 6 h PE female mosquitoes were injected with either Met dsRNA or with luciferase (Luc) dsRNA as a control, and the Met transcript level and ovarian follicle growth were examined 72 h postinjection. Mosquitoes with Met RNAi depletion also exhibited retardation of ovarian follicle growth, similar to that of female mosquitoes deprived of JH, reported earlier (Fig. S3 A and B) (9, 10). Compared with the iLuc control, the Met transcript levels, visualized using RT-PCR, was very low in the female mosquito fat body after the injection of Met dsRNA (Fig. S3C).
Fat bodies from Met dsRNA- and Luc dsRNA-injected mosquitoes were incubated in a complete culture medium supplemented with either JH or solvent (acetone) for 8 h. The same set of genes as for JH-deprived mosquitoes was analyzed by means of qPCR (Fig. 2 C and D and Fig. S2 C and D). In fat bodies from iLuc control mosquitoes, these genes showed a significant response to the presence of JH in the culture medium; transcript levels of selected EPE genes decreased (Fig. 2C and Fig. S2C) and those of LPE genes were elevated (Fig. 2D and Fig. S2D), correlating with the effect of JH application on these genes in JH-deprived mosquitoes. Transcript levels of the same EPE genes were elevated and those of LPE genes were down-regulated in fat bodies of Met-depleted mosquitoes incubated with solvent only, in a fashion similar to that in JH-deprived mosquitoes treated with solvent (Fig. 2 C and D and Fig. S2 C and D). No change was observed in transcript levels of either EPE or LPE genes in fat bodies of Met-depleted mosquitoes incubated in the presence of JH as compared with those incubated in solvent only (Fig. 2 C and D and Fig. S2 C and D).
Effect of Met RNAi Depletion on the Fat-Body Transcriptome in Aedes Female Mosquitoes PE.
Met RNAi depletion was conducted as described above, and fat-body RNA was isolated from both Met dsRNA-injected and Luc dsRNA-injected (control) mosquitoes at 72 h postinjection and was subjected to microarray analysis. Transcripts with differential expression of ≥1.75 folds (0.8 in a log2 scale) and a false-discovery rate (P value) of ≤0.01 were selected for further examination. An impressively large number of genes were affected by iMet. In total, 1,385 genes were up-regulated and 1,169 were down-regulated in the iMet transcriptome (Dataset S3). The qPCR analysis of 11 selected genes (seven down-regulated and four up-regulated) from iMet-depleted fat bodies demonstrated levels of these transcripts similar to those from the iMet microarray transcriptome, confirming the validity of the latter screen (Fig. S3 D and E).
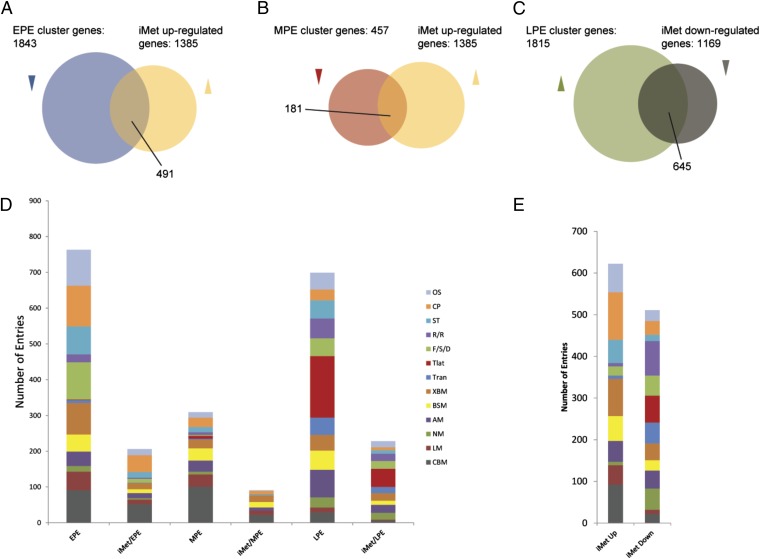
Comparison of the fat-body transcriptome from Met-depleted mosquitoes with that of the PE developmental time course revealed that iMet resulted in up-regulation of 491 (27%) and 181 (40%) of EPE and MPE genes, respectively (Fig. 3 A and B and Dataset S4) The two-tailed P values of <0.001 (Fisher’s exact test) suggests that the overlaps (associations) in both cases are highly statistically significant. In contrast, 645 (36%) (Fisher’s exact test two-tailed P value <0.001) of LPE genes were down-regulated by iMet (Fig. 3C and Dataset S4). Only 5% of LPE genes were up-regulated by iMet. A mere 1% of EPE and 4% of MPE genes were down-regulated in the iMet transcriptome. Taken together, these experiments clearly established that Met mediates JH action downregulating EPE and MPE genes and up-regulating LPE genes in the fat body of Aedes female mosquitoes.
Fig. 3.
(A and B) Venn diagrams comparing the up-regulated genes from the Met RNAi-depleted transcriptome with (A) the EPE gene cohort and (B) the MPE gene cohort. (C) Venn diagram comparing the down-regulated genes from the iMet RNAi-depleted transcriptome with the LPE gene cohort. (D) Gene ontology analysis of gene cohorts expressed in the female fat body PE. EPE, early posteclosion gene cohort; MPE, mid-posteclosion gene cohort; LPE, late posteclosion gene cohort. iMet/EPE, iMet/MPE, and iMet/LPE, comparison of the Met RNAi-depleted transcriptome with respective clusters. (E) Distribution of gene functional groups within up- and down-regulated Met RNAi-depleted transcriptome. Functional group abbreviations are OS, organismal systems; CP, cellular process; ST, signal transduction; R/R, replication and repair; F/S/D, folding sorting and degradation; Tlat, translation; Tran, transcription; XBM, xenobiotics biodegradation and metabolism; BSM, biosynthesis of other secondary metabolites; AM, amino acid metabolism; NM, nucleotide metabolism; LM, lipid metabolism; CBM, carbohydrate metabolism.
Kyoto Encyclopedia of Genes and Genomes Functional Group Analysis of Met-Regulated Fat Body Genes in Female Mosquitoes PE.
Using the Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis, we examined the ontology of the genes belonging to the EPE, MPE, and LPE clusters (28). We found that the genes could be categorized into 13 functional groups based on the biological processes and molecular functions (Fig. 3D). There was a remarkable temporal separation of major functional gene groups in the fat-body transcriptome. A large proportion of genes from the EPE and MPE clusters encoded enzymes involved in carbohydrate and lipid metabolism. Other notable functional gene groups were xenobiotics, organismal systems, cellular processes, and folding, sorting and degradation (Fig. 3D). The functional identity of the LPE cluster genes was markedly different from those of the EPE and MPE clusters (Fig. 3D). In particular, genes encoding the translation machinery were the largest functional group in the LPE cluster, followed by genes encoding amino acid metabolism. Other noteworthy differences with EPE and MPE genes included gene groups encoding factors involved in transcription and replication. In addition, in each of these clusters, a large proportion of genes belonged to diverse or unknown functional classes, defined as having either insufficient information or no significant matches to other organisms.
The KEGG analysis revealed that the functional distributions of iMet–up-regulated and –down-regulated genes were significantly different. The functional groups that showed significant overrepresentation (P < 0.01 in a hypergeometric distribution) among the iMet–up-regulated genes but not among the iMet–down-regulated genes were carbohydrate and lipid metabolism, cellular processes, xenobiotics biodegradation and metabolism, organismal systems, biosynthesis of other secondary metabolites and signal transduction (Fig. 3E). On the other hand, functional groups of replication and repair, translation, transcription, nucleotide metabolism, and folding sorting and degradation were enriched (P < 0.01 in a hypergeometric distribution) among the iMet–down-regulated genes but not among the iMet–up-regulated genes (Fig. 3E). Amino acid metabolism was the only functional group that was overrepresented within both gene sets. Large overlaps between iMet–up-regulated and EPE genes were observed in the functional categories cellular processes and carbohydrate metabolism; the two-tailed P values of <0.001 (Fisher’s exact test) confirms the very high statistical significance of the overlaps. Significant overlaps (P < 0.01) were observed in case of carbohydrate metabolism, xenobiotics, biodegradation and metabolism, and biosynthesis of other secondary metabolites when the iMet–up-regulated and the MPE gene sets were considered (Fig. 3D). Genes down-regulated in the Met-depleted transcriptome largely overlapped with those of the LPE cluster, especially those encoding for the transcription and translational machinery (P < 0.001) (Fig. 3D). Additionally, the genes encoding for the DNA replication system were found to be down-regulated by Met knockdown (Fig. 3D).
Identification of the Consensus for the Met-Binding Motif in Met-Regulated Genes from Aedes Mosquitoes.
We searched for putative Met-binding motifs in upstream regulatory regions of Met-dependent genes identified by the Met RNAi screen. Previously published data provided experimental evidence that two sequences—CCACACGCGAAG and CACGCGGTG—in the promoters of A. aegypti midgut-specific early trypsin (ET) and kruppel homolog 1 (Kr-h1) genes, respectively, bind Met (23, 25). Both of these Met-binding sequences shared the subpattern CACGCG, which is similar to but not identical with the palindromic canonical E-box motif, CACGTG, with thymine replaced by cytosine at the fifth position. E-box is a characteristic signature of recognition for the family of transcription factors containing the bHLH-PAS protein structural motif (24). The JH response element from the B. mori Kr-h1 gene (kJHRE)—GGCTCCACGTG—contains the canonical E-box sequence (19). The responsiveness of the kJHRE to Met has been confirmed experimentally in a cell-culture transfection assay (19). However, a putative JH binding motif sequence reported for Drosophila melanogaster and the honey bee Apis mellifera was suggested to be significantly different and contained neither the E-box nor any E-box–like motifs. Moreover, direct interaction of this motif with Met has not been confirmed (27).
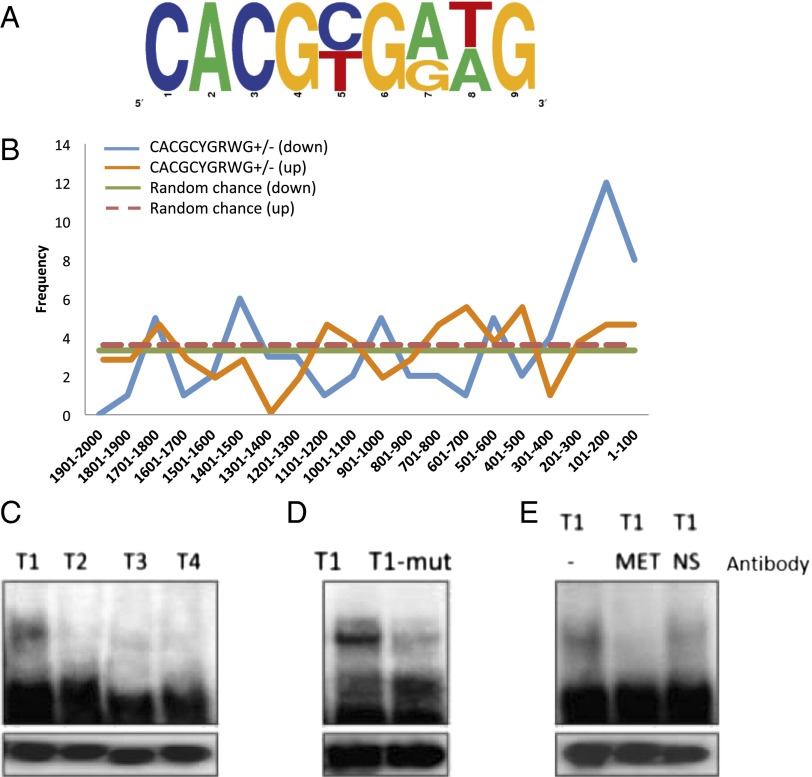
We used a pattern-search approach to identify the presence of putative Met-binding sites in Met-coregulated gene cohorts. We extracted 2-kbp regions upstream of the translation start sites (TSSs) for 1,154 of the 1,169 iMet–down-regulated (up-regulated in presence of Met) genes from the Vectorbase database. These regions were divided into twenty 100-base windows and were searched for the presence of the known mosquito Met-binding motifs CACGCGGTG and CACGCGAAG. There were only 8 and 16 exact matches for these two Met-binding motifs, respectively (Dataset S5), within the search space of 1,154 genes. This result suggested that the consensus for the Met-binding site might be more degenerate. Therefore, we searched the same regions allowing one-base degeneracy, N(A/T/C/G) (Dataset S5). Since the palindromic canonical E-box has a thymine in the fifth position, we replaced cytosine with thymine before repeating the same analysis (Dataset S5). Based on the bioinformatics analysis and prior experimental evidence, we propose, that the consensus for the Met-binding motif in A. aegypti is CACGC/TGA/GT/AG (CACGYGRWG (Fig. 4A). This definition is inclusive of the experimentally proven Met-binding motifs in A. aegypti, the JH response element in B. mori, and the palindromic canonical E-box motif. Of the 1,154 Met-depleted genes that were down-regulated by a fold change of ≥1.75 (0.8 in log2 scale), 68 were found to carry the putative Met-binding motif CACGYGRWG, in either the “+” or “−” strand. The motif appeared twice within the 2-kb upstream regions of five of these sequences. The GC content for the 1,154 sequences searched was ∼40%. Therefore, the probability of finding the consensus within each of the 100-base windows for all 1,154 sequences by random chance is approximately 3. It was observed that the occurrence of the putative Met-binding motif CACGYGRWG within the first 300 bp from the TSSs was three (1–100 bases; 201–300 bases) to four (101–200 bases) times more, which is significantly higher (P = < 0.1 and < 0.05 respectively; chi square test) than what one would expect by random chance (Fig. 4B) and suggests a positional bias for this region. For the 1,334 of 1,385 iMet genes that were up-regulated by a fold change of ≥1.75 (0.8 in log2 scale), no such pronounced peaks were observed when 2 kbp upstream of the TSS of these genes was searched (Fig. 4B). Two of the experimentally proven Met-binding sites have five and three bases, respectively, upstream of the E-box–like 6-mer (19, 23), we therefore aligned each of the 73 CACGYGRWG motifs detected, along with 10 bases upstream of their start sites, but this alignment did not yield any meaningful consensus.
Fig. 4.
(A and B) Computational analysis of gene promoters from the Met-depleted transcriptome. (A) Characterization of a Met-binding motif. The putative Met-binding motif was defined as CACGYGRWG. The nine-base consensus sequence has three degenerate positions, with cytosine/thymine at position five, adenine/guanine at position seven, and thymine/adenine at position eight. (B) Distribution of putative Met-binding sites within the upstream regions of up- and down-regulated genes from the Met RNAi-depleted transcriptome. The occurrence of the putative Met-binding motifs was enriched within the first 300 bp from the TSSs of iMet–down-regulated genes. (C–E) Validation of the Met-binding consensus. (C) Four motifs from three iMet–down-regulated genes designated as T1 (Ribosomal protein S28), T2 (RNA m5u methyltransferase), T3, and T4 (DNA-directed RNA polymerase E1) were tested using EMSA with fat-body nuclear extract from female mosquitoes 48 h PE. (D) The six-base, E-box–like motif CACGCGGAG of T1 was mutated to the T1 mutant, TCAATAGAG, and was tested using EMSA with the fat-body nuclear extract. (E) The presence of Met in the DNA–protein complex formed by T1 in EMSA with the fat-body nuclear extract was confirmed by adding polyclonal antibody against Met. NS, nonspecific antibody against actin. Lower panels in C–E represent loading controls of unbound probes.
Validation of Met-Binding Consensus Using EMSA.
EMSA was used to evaluate the consensus characterized by the computational analysis. We used 35–38 bases from three Aedes gene promoters within the iMet–down-regulated gene repertoire (Dataset S6), which carried nine base motifs that either matched or were very close to the consensus defined above. The promoter of the gene encoding ribosomal protein S28 [RPS28, AAEL006860 (Vectorbase, https://www.vectorbase.org/organisms/aedes-aegypti)] carried the motif CACGCGGAG (T1) 158 bases upstream of the TSS that matched the consensus. That of RNA m5u methyltransferase (AAEL000674) had CACGTGTCA (T2) 1,477 bases upstream, and the DNA-directed RNA polymerase E1 (AAEL007568) promoter carried CACTCGGTG (T3) and CACTTGGAG (T4) 401 and 1,501 bases, respectively, upstream of the TSS. EMSA showed that T1 formed a dense band for the DNA–protein complex in nuclear extracts isolated from the fat body of mosquitoes 48 h PE (Fig. 4C). Binding was greatly reduced for T3 and T4 (Fig. 4C), each of which had one base mismatch to the consensus in the fourth position of the six-base, E-box-like motif. In the case of T2, in which there were three mismatches at the 3′ end of the six-base, E-box like motif, binding was almost nonexistent (Fig. 4C). We then mutated the entire six-base, E-box-like motif of T1, leaving the three 3′-end bases intact, and observed that binding also was greatly diminished in this case (Fig. 4D). This result showed that all nine bases are important for proper binding. The presence of Met in the DNA-binding complex for T1 was tested using polyclonal antibodies against A. aegypti Met (AaMet AB) in EMSA with nuclear extracts from fat bodies of female mosquitoes 48 h PE. There was interference in the formation of the DNA–protein complex when the AaMet AB was added to the EMSA reaction mixture (Fig. 4E). The formation of the complex was not disrupted by nonspecific (NS) antibody. This result proved the presence of Met within the complex formed by the T1 sequence.
The putative Met-binding consensus has three degenerate bases at positions 5, 7, and 8 (Fig. 4A). Therefore, EMSA was used further to study the binding properties of all other possible variations of the Met-binding consensus, three with cytosine in the fifth position and four with thymine in the same position. To examine the formation of DNA-binding complexes to possible sequences with cytosine in the fifth position, the T1 sequence was mutated in the seventh and eighth positions, resulting in three sequences designated B1, B2, and B3 (Fig. S4 A and B). In all three cases, dense bands were observed in EMSA using fat-body nuclear extracts (Fig. S4 A and B). The binding specificity was confirmed by competition with a 50× concentration of their respective unlabeled specific probes (Fig. S4A). The presence of Met in DNA–protein complexes was proven by interference of the DNA–protein complex formation when the reactions were performed with the addition of AaMet AB (Fig. S4B). Next, we checked binding properties of the four possible combinations with thymine in the fifth position. Within the promoters of four genes from the iMet–down-regulated gene cohort, which were designated S21–24 (Fig. S4C), 37 bases were used as probes. We found that the bands formed by these sequences differed from each other in EMSA with the fat-body nuclear extract (Fig. S4D). The S22 and S23 sequences formed dense bands similar to that of the T1 motif. Addition of AaMet AB but not NS serum to the EMSA reaction mixture interfered with the binding of both the S22 and S23 motifs (Fig. S4E). Formation of the DNA–protein complex was reduced substantially in EMSA with the S21 sequence, in which adenine and thymine were present in the seventh and eighth positions, whereas guanine and adenine in these same positions in the S24 sequence diminished binding to an undetectable level. Taken together, our results suggest that the first six bases of the nine-base motif to which Met can bind are similar to the E-box or E-box–like sequences with a degenerate fifth position (either cytosine or thymine). The three bases downstream of the E-box–like motif are important, and two of these bases can be degenerate with invariable guanine in the ninth position. The binding properties are similar for all tested sequence combinations with cytosine in the fifth position but not for those with thymine in that position.
Discussion
Our transcriptome study combined with the Met RNAi screen has revealed important biological features related to the fat body of female A. aegypti mosquitoes. Very high transcriptional activity was observed in this tissue during the JH-dependent PE development. Although we anticipated finding putative JH gene targets using microarray analysis during PE development, such a spectacular rise in transcription was unexpected and reflects the great importance of JH-regulated periods in governing gonadotrophic cycles in mosquitoes. The time-course microarray analysis has identified temporal distribution of major peaks of transcriptional activity with a large number of genes being maximally up-regulated at the beginning (1,843 genes of the EPE cluster), the middle (457 genes of the MPE cluster), or the end (1,815 genes of the LPE cluster) of PE development. Using selected EPE cluster genes, we obtained experimental proof that these genes require a low level of JH for their maximal expression and are inhibited by a high level of JH; this result correlates well with the data on the JH titers in female mosquitoes PE (8). An inverse relationship exists between selected LPE genes and JH; these genes need a high JH titer for their maximal expression.
In our study, we have identified the significant role of the JH receptor Met in regulating transcriptional activity in the fat body of female mosquitoes PE. The RNAi transcriptome screen has shown that 1,385 genes were up-regulated and 1,169 genes down-regulated as a result of Met depletion; 27% of EPE and 40% of MPE genes were up-regulated and 36% of LPE genes were down-regulated by Met RNAi. RNAi experiments using an in vitro fat-body culture demonstrated that Met mediates the action of JH. In these experiments, selected EPE genes were activated and selected LPE genes were repressed in the Met-depleted tissue. Met RNAi depletion rendered repressive and activating actions of JH on either of these genes ineffective. It is important to stress that a large percentage of EPE, MPE, and LPE genes appear not to be controlled by Met. Future research should investigate the regulation of expression of these genes.
A 9-mer consensus for the Met-binding motif CACGC/TGA/GT/AG was identified using pattern-search and EMSA analyses. The core of this motif is conserved between the mosquito A. aegypti and the moth B. mori. However, further analysis is needed to establish conservation of this Met consensus sequence among other insects. Along with conservation, the binding sites of many transcription factors show bias for a certain position within the upstream regions, because these transcription factors can act most effectively when at a certain distance from the TSS (29, 30). The effect of positional bias for transcription factor binding sites has been observed in insects (31), vertebrates (32, 33), and plants (34). In our case, the in silico analysis determined that Met-binding motif variants showed a strong positional bias for the first 300 bases of the promoters of iMet–down-regulated genes. This bias suggests that Met might be directly involved in the up-regulation of some LPE genes. The fact that no such bias was detected in the iMet–up-regulated genes indicates that the EPE and MPE genes might be controlled indirectly by Met in accordance with some additional transcription factors. Two genes encoding the transcription factors hairy and Kr-h1 have been shown to be activated by Met, but their targets have not been characterized (19, 26). Overall, the complexity of JH/Met transcriptional regulation requires additional investigation.
The insect fat body is a storage depot of energy reserves and is a center of metabolism, immunity, detoxification, and production of yolk protein precursors during reproduction. It responds to changing the physiological needs of an insect organism by altering its functions. Gene ontology analysis has provided insight into the biological significance of the fat body in female mosquitoes and identified large and diverse sets of differentially expressed genes during PE development. We have found that the expressions of major functional groups are temporally separated, with carbohydrate, lipid, and xenobiotics metabolism being overrepresented in the EPE and MPE gene clusters and translation and transcription overrepresented in the LPE gene cluster. Met controls a large portion of these genes; it down-regulates EPE/MPE genes and up-regulates LPE genes. This unique partition of major functional gene groups reflects the complexity of fat-body activities that accommodate the requirements of reproducing female mosquitoes. A newly eclosed female mosquito needs a rapid increase in carbohydrate and lipid metabolism to support nectar feeding and energy-consuming host seeking. By the end of PE development, a female mosquito prepares for the intense demands of post–blood-feeding reproductive events. The high expression level of the genes involved in translation, transcription, and amino acid metabolism in the fat body at the end of PE development reflects a distinctive adaptation of mosquitoes as hematophagous insects. This study advances our understanding of JH regulation of mosquito reproduction and provides important insight into the control of gene expression by JH and its receptor Met.
Materials and Methods
Experimental Animals.
The mosquito A. aegypti UGAL/Rockefeller strain was raised as described previously (35). Adult mosquitoes were fed continuously on water and 10% (wt/vol) sucrose solution. All dissections were performed in Aedes physiological solution at room temperature (35).
Microarray Assays.
Single-color hybridizations of custom-made Agilent microarrays with 15,321 A. aegypti genes were conducted at the University of Chicago Core Instrument Facility following standard protocols (28). Three independent biological replicates were performed for the each treatment. The raw expression data were processed using Agilent Feature Extraction Software (36). Subsequent analysis steps were performed in the statistical programming environment R using Bioconductor packages (37). The expression data were background corrected and quantile normalized with functions from the Agi4 × 44 PreProcess and Limma libraries (38). Statistical analysis of DEGs was performed with the Limma library. When filtering for DEGs, we used a fold change of ≥1.75 (0.8 in log2 scale) and an adjusted P value of ≤0.01, in accordance with our previously used filtering criterion (28). The latter was adjusted for multiple testing with the Benjamin–Hochberg method to determine false-discovery rates (39). Hierarchical clustering with complete linkage was performed with the hclust function in R. The distance matrix required for this method was obtained by computing distance-transformed Spearman correlation coefficients among the expression profiles of all DEGs. Discrete clusters were obtained by cutting the resulting cluster dendrogram with the cutree function using a visually determined height value.
Preparation of JH-Deprived Mosquitoes.
We used a previously described approach, in which the head and the JH-producing endocrine gland, the corpora allata, were isolated from the rest of a female mosquito body by severing the metathorax and abdomen (10). The wound was sealed with paraffin, and the abdomens then were attached with paraffin to a microscopic slide. Slides with adhered abdomens were placed in Petri dishes with a wet filter paper and kept at 27 °C. These JH-deprived mosquito abdomens were prepared at 6 h PE, a time of very low JH titer (8). The abdomens were treated topically with three applications of 500 pg JH III (Sigma-Aldrich) in acetone (solvent) or with acetone alone (10). JH response was characterized 24 h after hormonal treatment. The effectiveness of eliminating JH was evaluated by measuring the length of ovarian follicles. In JH-deprived mosquitoes treated with acetone (control solvent) only, follicles remained small (50–60 μm), similar to those early in PE, indicating a low level of JH in these preparations. Application of JH III resulted in growth of ovarian follicles to about 80–90 μm, clearly showing that this treatment was effective in rescuing the arrested follicular growth phenotype. Total RNA was extracted from fat bodies 24 h after hormonal treatment. The experiment was repeated three times under the same conditions.
In Vitro Fat-Body Culture.
Female mosquitoes were injected with Met dsRNA or Luc dsRNA within 0–6 h PE. Fat bodies were dissected from mosquitoes 72 h after injection and were incubated in a complete culture medium supplemented with amino acids for 8 h, as described previously (34). JH (10 μg/mL JH III) or solvent (acetone) was added to the culture medium. Total RNA was isolated and analyzed using qPCR. The experiment was repeated three times under same conditions.
RNA Extraction and cDNA Preparation.
For the microarray transcriptome analysis, RNA samples collected at nine time points at 6-h intervals after eclosion of female mosquitoes were selected. For Met RNAi analysis, fat body samples were collected 72 h after injection. RNA was extracted from fat bodies of 10 female mosquitoes using the TRIzol method (Invitrogen) according to the manufacturer’s protocol. It was concentrated using the RNeasy MiniElute cleanup kit (Qiagen) for further processing.
Met RNAi.
To synthesize Met dsRNA, we followed a method described previously (35). In brief, dsRNA of a specific gene template was synthesized using the MEGAscript kit (Ambion). The luciferase gene was used to generate control iLuc dsRNA. After dsRNA synthesis, samples were subjected to phenol/chloroform extraction and ethanol precipitation. dsRNA then was suspended in RNase-free water to a final concentration of 5 µg/µL. At 24 h PE, female mosquitoes were injected with 300 nL dsRNA. The Picospritzer II (General Valve) was used to introduce corresponding dsRNAs into the thorax of CO2-anesthetized female mosquitoes.
qRT-PCR Analysis.
cDNAs were synthesized from 2 µg total RNA using the Omniscript Reverse Transcriptase kit (Qiagen). RNA was treated with DNase I (Invitrogen) before cDNA synthesis. PCR was performed using the Platinum High Fidelity Supermix (Invitrogen). qRT-PCR was performed using the iCycler iQ system (Bio-Rad) and an IQ SYBR Green Supermix (Bio-Rad). Quantitative measurements were performed in triplicate and normalized to the internal control of S7 ribosomal protein mRNA for each sample. Real-time data were collected from the software iCycler v3.0. Raw data were exported to Excel (Microsoft) for analysis.
Computational Analyses of Regulatory Gene Regions of Met-Depletion Transcriptome.
The upstream regions of the Met-depleted genes were extracted from the database for the A. aegypti genome (http://aaegypti.vectorbase.org/), and pattern searches were performed with an in-house PerlScript for the different 9-mers. The 2-kb regions were divided into twenty 100-base windows to check if the patterns show a bias for a certain region within the entire search space. The actual GC content of the Met-depleted up- and down-regulated genes were calculated with the help of another in-house PerlScript. These values were used to calculate the probability of finding the 9-mers by random chance. A pattern search allowing one base mismatch, N (A/T/C/G), for each of the 9-mers was used to check for the degeneracy. The reverse complements for the 9-mers were used to search the “+” strand to account for the “−” strand, and the frequencies of hits from both strands were added. The frequencies of observation by random chance within both stands were calculated when results were plotted.
EMSA Analysis.
The annealed deoxyoligonucleotide of each motif was purified from 15% TBE Criterion Precast Gel (Bio-Rad) and labeled with γ-32P ATP. EMSA was performed using a gel-shift assay system (Promega) with the fat-body nuclear extract of female mosquitoes 48 h PE. Nuclear extracts were prepared as described previously (40). The DNA–protein complex was separated on 5% TBE Criterion Precast Gel (Bio-Rad). After electrophoresis, the gel was dried and incubated with phosphor imaging screen in the cassette overnight to reach highest exposure and was visualized in the Personal Molecular Imager (Bio-Rad) by auto-radiography. For the competition assay, 50-fold unlabeled E-box–like motif or unlabeled AP-2 motif (nonspecific competitor oligonucleotides; Promega) was incubated with nuclear extract for 10 min and then was incubated further with the labeled probe for 20 min. Identity of a complex was verified by directly adding polyclonal antibodies against A. aegypti Met.
Supplementary Material
Acknowledgments
We thank Marc H. Domanus and the technical staff of the Joint Institute for Genomics and Systems Biology, Argonne National Laboratory for conducting the microarray assays and Dr. J. Zhu for providing with antibodies against A. aegypti Met. This work was supported by National Institutes of Health/National Institute of Allergy and Infectious Diseases Award 5R01 AI036959.
Footnotes
The authors declare no conflict of interest.
See Commentary on page 9623.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305293110/-/DCSupplemental.
References
- 1.Attardo GM, Hansen IA, Raikhel AS. Nutritional regulation of vitellogenesis in mosquitoes: Implications for anautogeny. Insect Biochem Mol Biol. 2005;35(7):661–675. doi: 10.1016/j.ibmb.2005.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Clements AN. 1992. The Biology of Mosquitoes (Chapman & Hall, London) Vol 1. Development, Nutrition and Reproduction, p. 551.
- 3.Raikhel AS, et al. Molecular biology of mosquito vitellogenesis: From basic studies to genetic engineering of antipathogen immunity. Insect Biochem Mol Biol. 2002;32(10):1275–1286. doi: 10.1016/s0965-1748(02)00090-5. [DOI] [PubMed] [Google Scholar]
- 4.Sun G, Zhu J, Chen L, Raikhel AS. Synergistic action of E74B and ecdysteroid receptor in activating a 20-hydroxyecdysone effector gene. Proc Natl Acad Sci USA. 2005;102(43):15506–15511. doi: 10.1073/pnas.0503501102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu J, Chen L, Sun G, Raikhel AS. The competence factor β Ftz-F1 potentiates ecdysone receptor activity via recruiting a p160/SRC coactivator. Mol Cell Biol. 2006;26(24):9402–9412. doi: 10.1128/MCB.01318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mane-Padros D, Cruz J, Cheng A, Raikhel AS. A critical role of the nuclear receptor HR3 in regulation of gonadotrophic cycles of the mosquito Aedes aegypti. PLoS ONE. 2012;7(9):e45019. doi: 10.1371/journal.pone.0045019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz J, Mane-Padros D, Zou Z, Raikhel AS. Distinct roles of heme-liganted nuclear receptor E75, an insect orthologue of vertebrate Rev-erb, in mosquito vitellogenesis. Mol Cell Endocrinol. 2012;349:262–271. doi: 10.1016/j.mce.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiro AB, et al. Juvenile hormone and juvenile esterase in adult females in Aedes aegypti. J Insect Physiol. 1986;32:867–877. [Google Scholar]
- 9.Gwadz RW, Spielman A. Corpus allatum control of ovarian development in Aedes aegypti. J Insect Physiol. 1973;19(7):1441–1448. doi: 10.1016/0022-1910(73)90174-1. [DOI] [PubMed] [Google Scholar]
- 10.Raikhel AS, Lea AO. Hormone-mediated formation of the endocytic complex in mosquito oocytes. Gen Comp Endocrinol. 1985;57(3):422–433. doi: 10.1016/0016-6480(85)90224-2. [DOI] [PubMed] [Google Scholar]
- 11.Raikhel AS, Lea AO. Control of vitelline envelope formation in the mosquito follicle; role of juvenile hormone and 20-hydroxyecdysone. Tissue Cell. 1991;23:577–591. doi: 10.1016/0040-8166(91)90015-l. [DOI] [PubMed] [Google Scholar]
- 12.Raikhel AS, Lea AO. Previtellogenic development and vitellogenin synthesis in the fat body of a mosquito: An ultrastructural and immunocytochemical study. Tissue Cell. 1983;15(2):281–299. doi: 10.1016/0040-8166(83)90023-x. [DOI] [PubMed] [Google Scholar]
- 13.Raikhel AS, Lea AO. Juvenile hormone controls previtellogenic proliferation of ribosomal RNA in the mosquito fat body. Gen Comp Endocrinol. 1990;77(3):423–434. doi: 10.1016/0016-6480(90)90233-c. [DOI] [PubMed] [Google Scholar]
- 14.Zhu J, Chen L, Raikhel AS. Posttranscriptional control of the competence factor betaFTZ-F1 by juvenile hormone in the mosquito Aedes aegypti. Proc Natl Acad Sci USA. 2003;100(23):13338–13343. doi: 10.1073/pnas.2234416100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson TG, Ashok M. Insecticide resistance resulting from an absence of target-site gene product. Proc Natl Acad Sci USA. 1998;95(24):14040–14044. doi: 10.1073/pnas.95.24.14040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashok M, Turner C, Wilson TG. Insect juvenile hormone resistance gene homology with the bHLH-PAS family of transcriptional regulators. Proc Natl Acad Sci USA. 1998;95(6):2761–2766. doi: 10.1073/pnas.95.6.2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miura K, Oda M, Makita S, Chinzei Y. Characterization of the Drosophila Methoprene -tolerant gene product. Juvenile hormone binding and ligand-dependent gene regulation. FEBS J. 2005;272(5):1169–1178. doi: 10.1111/j.1742-4658.2005.04552.x. [DOI] [PubMed] [Google Scholar]
- 18.Charles JP, et al. Ligand-binding properties of a juvenile hormone receptor, Methoprene-tolerant. Proc Natl Acad Sci USA. 2011;108(52):21128–21133. doi: 10.1073/pnas.1116123109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kayukawa T, et al. Transcriptional regulation of juvenile hormone-mediated induction of Krüppel homolog 1, a repressor of insect metamorphosis. Proc Natl Acad Sci USA. 2012;109(29):11729–11734. doi: 10.1073/pnas.1204951109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Konopova B, Jindra M. Juvenile hormone resistance gene Methoprene-tolerant controls entry into metamorphosis in the beetle Tribolium castaneum. Proc Natl Acad Sci USA. 2007;104(25):10488–10493. doi: 10.1073/pnas.0703719104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parthasarathy R, Tan A, Palli SR. bHLH-PAS family transcription factor methoprene-tolerant plays a key role in JH action in preventing the premature development of adult structures during larval-pupal metamorphosis. Mech Dev. 2008;125(7):601–616. doi: 10.1016/j.mod.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kewley RJ, Whitelaw ML, Chapman-Smith A. The mammalian basic helix-loop-helix/PAS family of transcriptional regulators. Int J Biochem Cell Biol. 2004;36(2):189–204. doi: 10.1016/s1357-2725(03)00211-5. [DOI] [PubMed] [Google Scholar]
- 23.Li M, Mead EA, Zhu J. Heterodimer of two bHLH-PAS proteins mediates juvenile hormone-induced gene expression. Proc Natl Acad Sci USA. 2011;108(2):638–643. doi: 10.1073/pnas.1013914108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Xu J, Sheng Z, Sui Y, Palli SR. Steroid receptor co-activator is required for juvenile hormone signal transduction through a bHLH-PAS transcription factor, methoprene tolerant. J Biol Chem. 2011;286(10):8437–8447. doi: 10.1074/jbc.M110.191684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shin SW, Zou Z, Saha TT, Raikhel AS. bHLH-PAS heterodimer of methoprene-tolerant and Cycle mediates circadian expression of juvenile hormone-induced mosquito genes. Proc Natl Acad Sci USA. 2012;109(41):16576–16581. doi: 10.1073/pnas.1214209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajgar A, Jindra M, Dolezel D. Autonomous regulation of the insect gut by circadian genes acting downstream of juvenile hormone signaling. Proc Natl Acad Sci USA. 2013;(Feb):26. doi: 10.1073/pnas.1217060110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Y, Zhang Z, Robinson GE, Palli SR. Identification and characterization of a juvenile hormone response element and its binding proteins. J Biol Chem. 2007;282(52):37605–37617. doi: 10.1074/jbc.M704595200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zou Z, et al. Transcriptome analysis of Aedes aegypti transgenic mosquitoes with altered immunity. PLoS Pathog. 2011;7(11):e1002394. doi: 10.1371/journal.ppat.1002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, et al. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bellora N, Farré D, Albà MM. Positional bias of general and tissue-specific regulatory motifs in mouse gene promoters. BMC Genomics. 2007;8:459. doi: 10.1186/1471-2164-8-459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Mol Cell. 2007;28(2):337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Down TA, Bergman CM, Su J, Hubbard TJ. Large-scale discovery of promoter motifs in Drosophila melanogaster. PLOS Comput Biol. 2007;3(1):e7. doi: 10.1371/journal.pcbi.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie X, et al. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434(7031):338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ma S, et al. Discovery of stress responsive DNA regulatory motifs in Arabidopsis. PLoS ONE. 2012;7(8):e43198. doi: 10.1371/journal.pone.0043198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roy SG, Hansen IA, Raikhel AS. Effect of insulin and 20-hydroxyecdysone in the fat body of the yellow fever mosquito, Aedes aegypti. Insect Biochem Mol Biol. 2007;37(12):1317–1326. doi: 10.1016/j.ibmb.2007.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zahurak M, et al. Pre-processing Agilent microarray data. BMC Bioinformatics. 2007;8:142. doi: 10.1186/1471-2105-8-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gentleman RC, et al. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004;5(10):R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smyth GK. 2004. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 3(1):Article3.
- 39.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practicaland powerful approach to multiple testing. J R Stat Soc, B. 1995;57:289–300. [Google Scholar]
- 40.Miura K, Wang SF, Raikhel AS. Two distinct subpopulations of ecdysone receptor complex in the female mosquito during vitellogenesis. Mol Cell Endocrinol. 1999;156(1-2):111–120. doi: 10.1016/s0303-7207(99)00136-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.