Abstract
The ability to sense metabolic stress is critical for successful cellular adaptation. In eukaryotes, the AMP-activated protein kinase (AMPK), a highly conserved serine/threonine kinase, functions as a critical metabolic sensor. AMPK is activated by the rising ADP/ATP and AMP/ATP ratios during conditions of energy depletion and also by increasing intracellular Ca2+. In response to metabolic stress, AMPK maintains energy homeostasis by phosphorylating and regulating proteins that are involved in many physiological processes including glucose and fatty acid metabolism, transcription, cell growth, mitochondrial biogenesis, and autophagy. Evidence is mounting that AMPK also plays a role in a number of pathways unrelated to energy metabolism. Here, we identify the recombination-activating gene 1 protein (RAG1) as a substrate of AMPK. The RAG1/RAG2 complex is a lymphoid-specific endonuclease that catalyzes specific DNA cleavage during V(D)J recombination, which is required for the assembly of the Ig and T-cell receptor genes of the immune system. AMPK directly phosphorylates RAG1 at serine 528, and the phosphorylation enhances the catalytic activity of the RAG complex, resulting in increased cleavage of oligonucleotide substrates in vitro, or increased recombination of an extrachromosomal substrate in a cellular assay. Our results suggest that V(D)J recombination can be regulated by AMPK activation, providing a potential new link between metabolic stress and development of B and T lymphocytes.
Keywords: energy coupling, immune diversity
The AMP-activated protein kinase (AMPK) is a highly conserved heterotrimeric serine/threonine kinase composed of a catalytic (α) subunit and two regulatory (β and γ) subunits, which plays a key role in sensing and responding to metabolic stress at both the cellular and the whole-body levels (1). Recently, it has become clear that AMPK also plays important roles in areas unrelated to metabolism such as lymphocyte survival. For example, triggering of the T-cell antigen receptor activates AMPK, and AMPK is required for survival of CD8 cytotoxic T lymphocytes following withdrawal of immune stimulation (2, 3). AMPK is activated by conditions that increase AMP/ATP and ADP/ATP ratios such as physical exercise, ischemia, or glucose deprivation (reviewed in ref. 4). An upstream kinase, the LKB1–STRAD–MO25 complex, phosphorylates threonine (T172) in the activation loop of AMPK and increases AMPK activity in response to an increase in AMP and ADP (5–7). AMPK activation is also triggered by Ca2+/calmodulin-activated protein kinase kinase β, CaMKKβ, under conditions that increase intracellular Ca2+ (8, 9). Activated AMPK directly phosphorylates a number of substrates that regulate glucose, lipid, and protein metabolism, as well as mitochondrial biogenesis and autophagy, to maintain energy homeostasis (reviewed in ref. 4). AMPK phosphorylates downstream targets at conserved Ser/Thr residues within a consensus sequence motif, which has hydrophobic residues at the −5 and +4 positions and basic residues at −4 or −3, or both positions (10, 11). In an effort to identify novel pathways that may be regulated by AMPK, we screened protein databases for proteins with AMPK consensus motif sequences by using the bioinformatics tool Scansite (http://scansite.mit.edu). One of the candidates we identified was recombination-activating gene 1 (RAG1), which is a crucial factor for V(D)J recombination during lymphocyte development.
Functional Ig or T-cell receptor (TCR) genes are assembled from germline variable (V), diversity (D), and joining (J) gene segments in developing B or T lymphocytes (12, 13). Each coding gene segment is flanked by a recombination signal sequence (RSS) consisting of conserved heptamer and nonamer sequences separated by nonconserved 12 or 23 bp of DNA (12-RSS or 23-RSS) (14, 15). V(D)J recombination occurs in two distinct steps: Double-stranded DNA cleavage mediated by the lymphoid-specific RAG1/RAG2 protein complex, followed by appropriate joining of pairs of coding ends and separately by joining of pairs of signal ends, mediated by nonhomologous DNA end joining, which requires DNA-dependent protein kinase, Ku70/80, Artemis, XRCC4, and DNA ligase 4 (16). The RAG1/RAG2 complex recognizes the RSS and catalyzes double-strand DNA cleavage in two enzymatic steps. A nick is introduced at the 5′end of the RSS, leaving a 3′ hydroxyl group on the coding end and a 5′ phosphate group on the signal end. In the second step, the RAG complex uses the 3′ hydroxyl group to attack the phosphodiester bond of the antiparallel strand by a nucleophilic reaction, resulting in the formation of a hairpin intermediate (17, 18). The purified RAG1/RAG2 complex is able to carry out these reactions in vitro. The majority of biochemical studies on RAG proteins have used truncated forms of RAG1 (amino acids 384–1,008) and RAG2 (amino acids 1–387), referred to as core RAG1 and RAG2, which have been easier to obtain in soluble form than the full-length proteins (19). Although the activity of the core RAG1/RAG2 complex is sufficient for DNA cleavage both in vivo and in vitro, noncore regions of RAG1 and RAG2 are crucial for efficient V(D)J recombination and its proper regulation (20–22). Normally, V(D)J recombination is kept under tight control by the expression of RAG1 and RAG2 only during the early stages of lymphocyte development and by the limited access of the RAG complex to particular antigen receptor loci, which is controlled by chromatin remodeling (23, 24). Here, we report that AMPK directly phosphorylates RAG1 at serine 528 (S528). Phosphomimetic substitutions of S528 enhance the catalytic ability of the RAG complex to induce nick and hairpin formation of recombination substrates, and also enhance recombination in cells, as does stimulation of AMPK by N1-(β-d-ribofuranosyl)-5-aminoimidazole-4-carboxamide (AICAR) (25) and metformin (26). These findings suggest that AMPK may link V(D)J recombination to metabolic stress.
Results
AMPK Directly Phosphorylates Mammalian RAG1 Protein in Vitro.
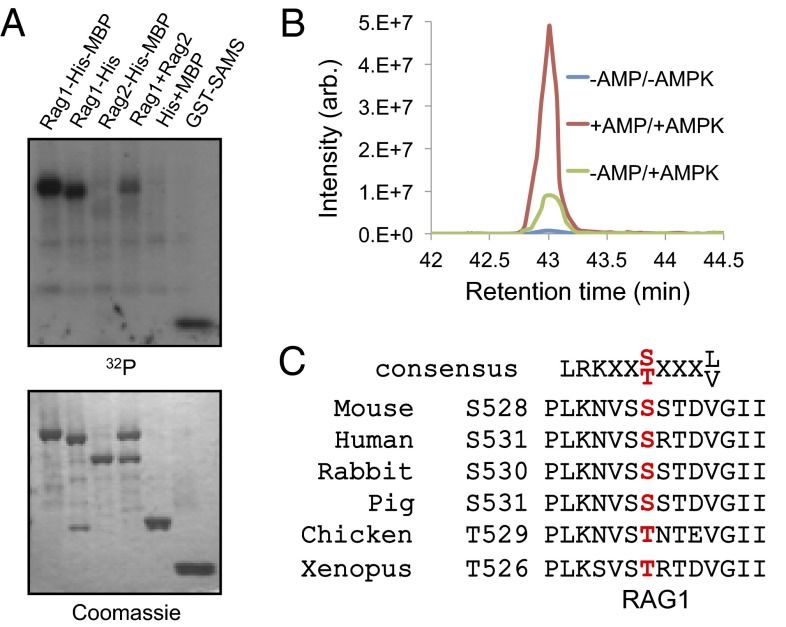
To test the possibility that RAG1 is an AMPK substrate, an in vitro phosphorylation reaction was performed by incubating AMPK in the presence of AMP and [γ-32P]ATP with recombinant maltose-binding protein (MBP)-His fusion RAG1 and RAG2. We found that RAG1 protein was directly phosphorylated by AMPK, but RAG2 protein, or MBP-His alone, were not (Fig. 1A). To identify the residues of RAG1 that are phosphorylated by AMPK, we analyzed the phosphopeptides of phosphorylated RAG1 by performing liquid chromatography followed by mass spectrometry (LC-MS). This LC-MS approach confirmed that a RAG1 peptide was singly phosphorylated either at S527 or S528. Extracted ion chromatograms at m/z 1,086.87 showed that phosphorylation of S527 or S528 is increased by more than threefold in the presence of AMP (Fig. 1B and Fig. S1). The consensus AMPK phosphorylation sequence, which is shown in Fig. 1C (Top), contains hydrophobic residues at the −5 and +4 positions and basic residues at −4 or −3, or both positions (27). Although both S527 and S528 are highly conserved in vertebrates, only S528 has a nonpolar residue at position +4 (Fig. 1C).
Fig. 1.
Phosphorylation of RAG1 protein by AMPK. (A) Recombinant RAG1 (amino acids 384–1,008) and RAG2 (amino acids 1–387) proteins, both tagged with His6 and MBP, were incubated with AMPK in the presence of [γ-32P]ATP and AMP. The optimized AMPK substrate GST-SAMS peptide (HMRSAMSGLHLVKRR) was used as the positive control. (B) Extracted ion chromatograms at m/z 1,086.87 showed that AMP plus AMPK significantly increased phosphorylation. Phosphorylation by AMPK increased more than threefold in the presence of AMP. (C) AMPK consensus target amino acid sequence in RAG1. Ser-528 is conserved in many vertebrate RAG1 proteins.
AMPK Phosphorylates Ser-528 of RAG1 in Vivo.
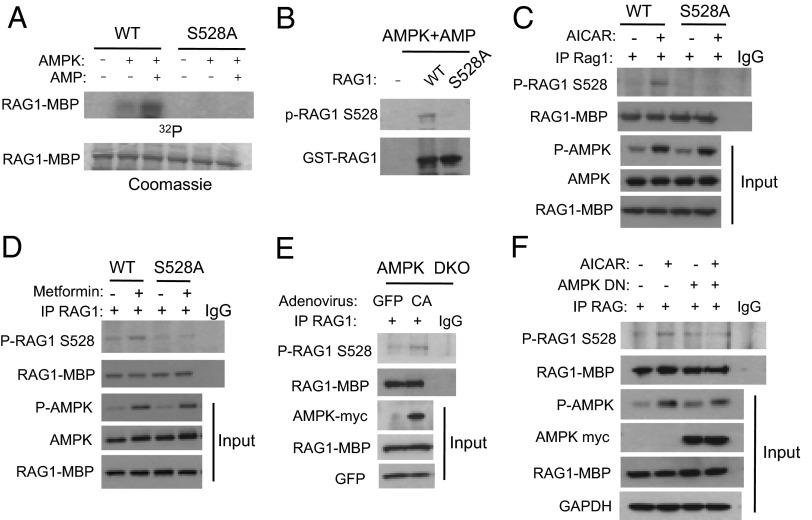
To further test whether S528 is the AMPK phosphorylation site, we mutated S528 to Ala (S528A) and used S528A RAG1 as an AMPK substrate in a kinase reaction. The S528A mutation completely abolished AMPK phosphorylation (Fig. 2A), suggesting that in vitro S528 is the AMPK phosphorylation site in RAG1. To assess the phosphorylation of RAG1 by AMPK in vivo, we generated a phosphospecific antibody to S528 (P-S528) of RAG1. The phosphospecific S528 antibody was tested by Western blotting after incubating RAG1 wild-type (WT) or S528A mutant in the presence of AMP and AMPK in vitro. We found that this antibody recognized WT RAG1 that was phosphorylated by AMPK, but did not recognize the nonphosphorylated form of either WT RAG1 or the S528A mutant (Fig. 2 A and B), suggesting that the P-S528 antibody is highly specific for the phosphorylated form of RAG1. As recombinant RAG1 was purified from 293T cells, we cannot formally rule out the possibility that S528 was not directly phosphorylated by AMPK, but by a kinase that copurified with RAG1 and is regulated by AMPK. In addition, we demonstrated AMPK-mediated phosphorylation of RAG1 expressed in 293T cells rather than endogenous RAG1 expressed in lymphocytes. Low sensitivity of the P-S528 antibody and low expression level of endogenous RAG1 in lymphocytes made it technically difficult for us to detect endogenous RAG1 phosphorylation.
Fig. 2.
Phosphorylation of Ser-528 of RAG1 by AMPK. (A and B) In vitro AMPK kinase assay using GST-tagged core WT RAG1 and S528A RAG1 mutants. Phosphorylated RAG1 was detected by radiography (A) or by immunoblot, using an antibody against phospho-S528 of RAG1 (B). (C and D) 293T cells were transiently transfected with either MBP fusion WT RAG1 or S528A mutant and treated with 2 mM AICAR or 1 mM metformin for 3 h. Phosphorylation of Ser-528 of RAG1 by endogenous AMPK was detected by immunoblotting following immunoprecipitation of MBP-RAG1. (E) AMPK double-knockout MEF (AMPK DKO) cells were transfected with MBP-RAG1 and infected with adenovirus expressing constitutively active (CA) AMPK or green fluorescent protein (GFP). MBP-RAG1 was immunoprecipitated and immunoblotted with antibody against phospho-S528 of RAG1. (F) NIH 3T3 cells were transiently transfected with MBP-RAG1 with or without AMPK dominant-negative (DN) plasmids and treated with 2 mM AICAR for 3 h. Phosphorylation of S528 was detected by immunoprecipitating MBP-RAG1 and immunoblotting with phosphospecific antibody against S528.
To determine whether AMPK phosphorylates RAG1 in vivo, MBP-WT or S528A mutant RAG1 were transiently expressed in 293T cells, which were treated with the AMPK activators AICAR or metformin. We found that AICAR (Fig. 2C) or metformin (Fig. 2D) induced phosphorylation of S528 but that phosphorylation was completely absent with the S528A mutant RAG1, indicating that S528 is phosphorylated in vivo by AMPK. To confirm that AMPK phosphorylates RAG1 in vivo, we expressed a constitutively active (CA) form of AMPKα2 in AMPKα1/α2 double-knockout (AMPK DKO) mouse embryo fibroblasts (MEFs) by infecting them with adenovirus expressing CA-AMPK. As shown in Fig. 2E, S528 phosphorylation was induced by CA-AMPK adenovirus in AMPK DKO cells. In contrast, AICAR-induced phosphorylation of RAG1 was inhibited if a dominant-negative (DN) form of AMPKα2 was overexpressed (Fig. 2F). Taken together, these results demonstrate that AMPK phosphorylates S528 in vivo.
Phosphorylation of RAG1 by AMPK Increases RAG1/2 Cleavage Activity in Vitro.
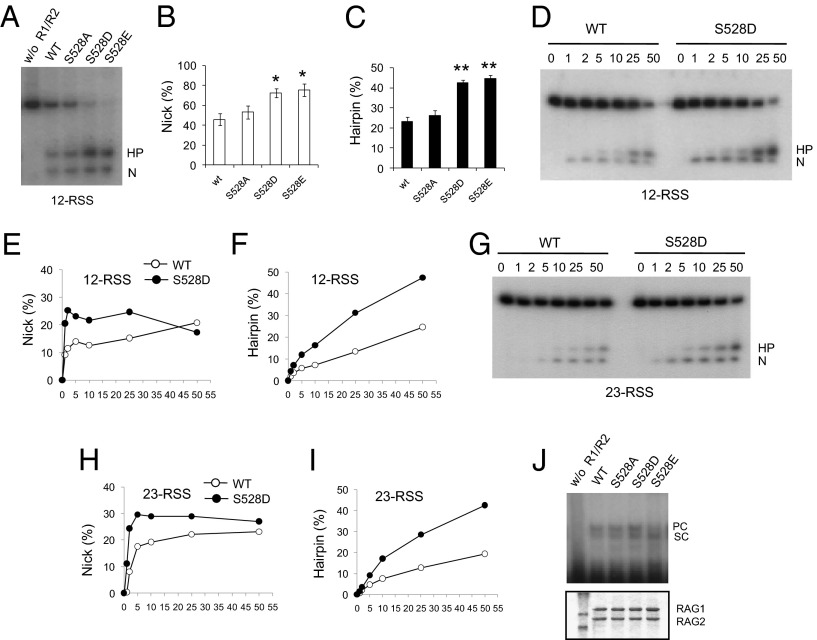
The RAG1/RAG2 complex initiates V(D)J recombination by recognizing and cleaving the RSSs that flank each coding segment, producing double-strand breaks. Cleavage by the RAG complex occurs in two steps. First, a nick is induced at the 5′ end of the RSS, followed by DNA hairpin formation by transesterification (14, 15, 19, 28). Incubation of purified RAG complex with 12-RSS and 23-RSS oligonucleotides in the presence of a divalent metal cation such as Mg2+ or Mn2+ can induce nick and hairpin formation (19, 29, 30). To examine the functional effect of the S528 phosphorylation, we generated phosphoserine mimetic mutants of RAG1 by mutating S528 to Asp (S528D) or Glu (S528E). A standard in vitro cleavage assay was performed in the presence of Mn2+ and either WT RAG1 or mutant RAG1 proteins that were copurified with RAG2 from 293T cells. The WT RAG protein complex efficiently generated nicks (N) and hairpins (HP) from the 12-RSS substrate (Fig. 3 A–C), increasing with the amount of the RAG1/RAG2 complex (Fig. S2E). S528A RAG1 generated similar levels of nicks and hairpins to WT RAG1, but the levels of both nicks and hairpins generated by S528D and S528E mutants were increased. In more detail, the time course of the reaction (1, 2, 5 min) showed that S528D RAG1 generated nicks at a rate ∼2.3-fold higher than WT RAG1. At later time points, the rates of nick generation of S528D and WT RAG1 converged, because nicked products were being converted into hairpin products. S528D RAG1 also generated hairpins at a rate approximately twofold higher than that of WT RAG1 (Fig. 3 D–F). A similar result was obtained with 23-RSS as substrate. Once again, S528D RAG1 generated nicks and hairpins significantly faster than WT RAG1 (Fig. 3 G–I). Similarly, coupled cleavage of both 12RSS and 23RSS substrates (in an assay containing Mg2+ and HMGB1 protein) was also increased with the S528D or S528E mutants compared with WT or S528A RAG1 (Fig. S2F).
Fig. 3.
Increased RAG cleavage activity by phosphomimetic mutants of RAG1 S528 in vitro. (A) RAG cleavage activity in vitro was measured. S528A, S528D, S528E, or WT RAG1 and WT RAG2 were incubated with radiolabeled 12-RSS and MnCl2 in reaction buffer. The nicked (N) and hairpin (HP) products from the cleavage reactions were analyzed by TBE-urea gel electrophoresis. (B and C) Relative percentage of nicked and hairpin products from total substrate from three independent experiments. Results are mean ± SE. *P < 0.05, **P < 0.001 between WT and S528 mutants. (D) Time course (in min) of nick and hairpin formation from a radiolabeled 12-RSS oligonucleotide substrate by WT RAG1/RAG2 or S528D RAG1/RAG2 protein. (E and F) Amount of nicked or hairpin product from D was plotted. (G) Time course (in min) of nick and hairpin formation with 23-RSS oligonucleotide substrate. (H and I) Time course of nicked (H) or hairpin (I) product obtained from G was plotted. (J) DNA binding assay was performed with S528A, S528D, S528E, or WT RAG1 and WT RAG2 in the presence of 5 nM radiolabeled 12-RSS and unlabeled 23-RSS. RAG–DNA complexes were separated on a 4% native polyacrylamide gel and were detected by autoradiography. PC, paired complex containing 12-RSS/23-RSS; SC, single substrate complex containing 12-RSS.
Because a binding complex is formed between the signal sequence DNA and RAG proteins before cleavage, we examined the DNA binding properties of WT, S528A, S528D, or S528E RAG1 in the RAG1/RAG2 complex by electrophoretic mobility shift assay (EMSA) using end-labeled 12-RSS and unlabeled 23-RSS substrates. As shown in Fig. 3J, WT, S528A, and S528D RAG1 formed comparable levels of 12-RSS complex [single substrate complex (SC)] and 12/23-RSS complex [paired complex (12/23RSS-PC)], suggesting that S528 mutations do not affect the interaction with RAG2 or with RSS.
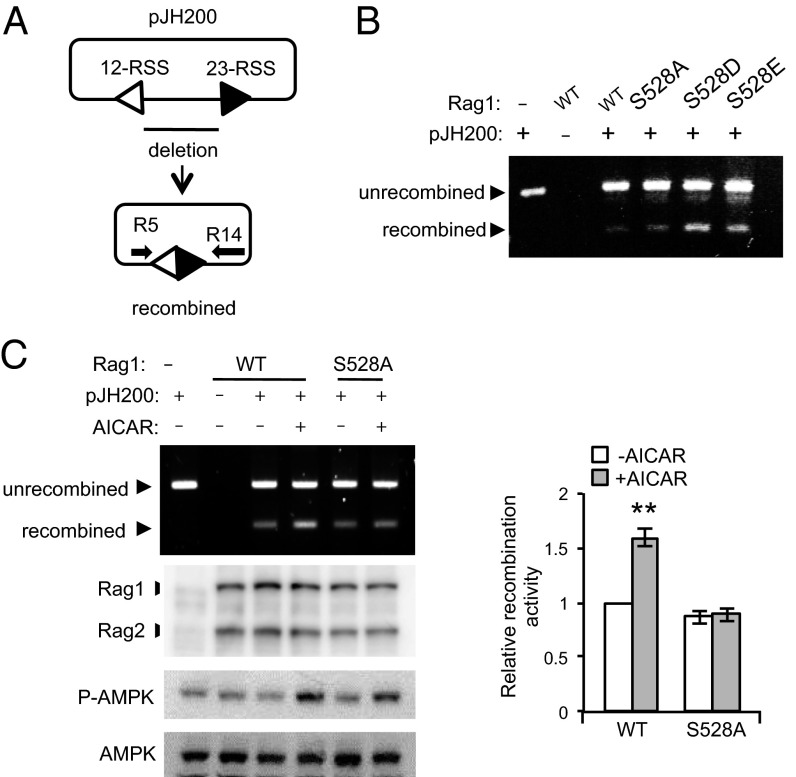
Phosphorylation of S528 of RAG1 by AMPK Increases Extrachromosomal V(D)J Recombination Activity in Cells.
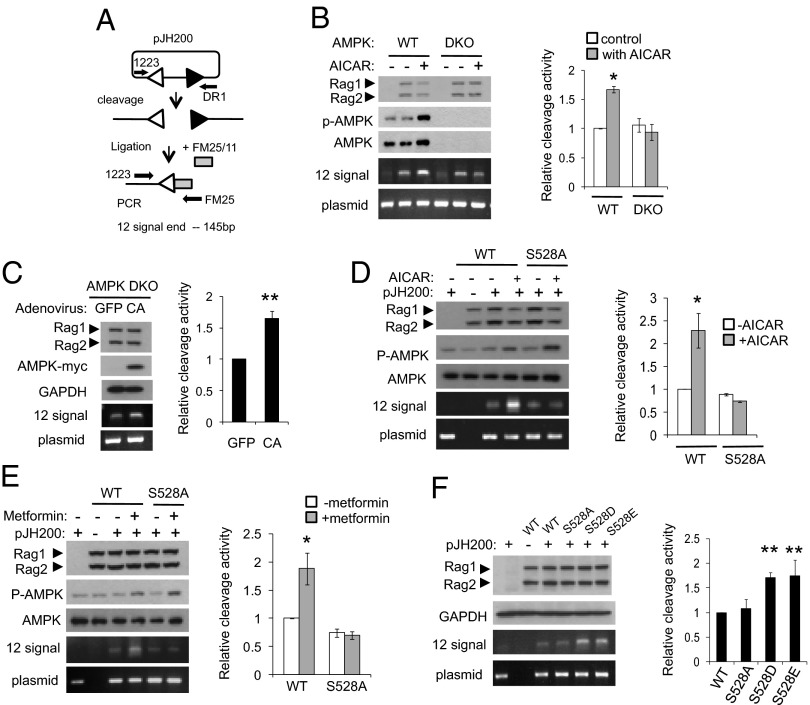
Previous studies showed that transient expression of the RAG complex (RAG1 and RAG2) in nonlymphoid cells can cleave the RSS of a plasmid recombination substrate (31). To investigate whether phosphorylation of RAG1 by AMPK could regulate RAG1/RAG2 activity in vivo, we tested cleavage of an extrachromosomal substrate in AMPK DKO cells and in 293T cells. RAG1, RAG2, and the extrachromosomal V(D)J recombination substrate pJH200 (see Fig. S2 legend) were cotransfected into either WT or AMPK DKO MEFs, which were then treated with 2 mM AICAR for 3 h before harvesting. We directly estimated the amount of protein expression and AMPK activation by Western blotting, and detected cleaved signal ends by ligation-mediated PCR (LM-PCR) (Fig. 4A). Although the basal cleavage activity of RAG proteins was similar between WT MEFs and AMPK DKO MEFs, AICAR treatment enhanced cleavage activity in WT MEFs, but not in AMPK DKO MEFs (Fig. 4B). Consistent with this result, RAG cleavage activity was induced by expressing a constitutively active form of AMPK but not by expressing a control protein in AMPK DKO MEF cells (Fig. 4C), suggesting that AMPK regulates RAG activity in cells. Similarly treating 293T cells, which had been transfected as above, with AICAR resulted in increased cleavage of substrate by ∼2.3-fold in the presence of WT RAG1, but did not affect cleavage of substrate in the presence of the S528A RAG1 mutant (Fig. 4D). Metformin also increased cleavage in the presence of WT RAG1 but not in the presence of S528A RAG1, again indicating that phosphorylation of S528 by AMPK augments cleavage in vivo (Fig. 4E). To determine whether cleavage in vivo would also respond to phosphomimetics of S528, we cotransfected S528A, S528D, S528E, or WT RAG1, WT RAG2, and the extrachromosomal substrate into 293T cells and analyzed cleavage by LM-PCR. As shown in Fig. 4F, cleavage by S528D and S528E RAG1 mutants was about twofold higher than that by either WT RAG1 or S528A RAG1. The Western blots of Fig. 4 show that protein expression was the same in all cases. Finally, we examined the role of S528 phosphorylation in V(D)J recombination by using the assay shown in Fig. 5A. The phosphomimetic mutants of full length of RAG1 increased recombination (Fig. 5B), and treatment with AICAR increased recombination with full-length WT RAG1, but not with the S528A mutant (Fig. 5C). Taken together, these findings indicate that AMPK increases RAG1 activity on extrachromosomal substrates. As the RAG1 function assays were performed in 293T cells, it is possible that some aspects of RAG1 activity may be different from that in a lymphoid cell line.
Fig. 4.
Increased cleavage of an extrachromosomal substrate by AMPK activation in vivo. (A) Diagram of the ligation-mediated PCR (LM-PCR) cleavage assay using the extrachromosomal V(D)J substrate pJH200. The linker (FM25/11) is shown as a shaded box. PCR primers 1223 and FM25 were used to amplify the cleaved fragment. To control for recovery of the recombination substrate, primers 1223 and DR1 were used to amplify a portion of the backbone of the plasmid (39, 40). (B) MBP-RAG1, MBP-RAG2, and pJH200 were transiently transfected in WT MEF cells or in AMPK DKO MEF cells. Cells were treated with 2 mM AICAR for 3 h. Cleavage was detected by LM-PCR. A representative experiment is shown (Left). Relative activities were quantified from at least three independent experiments (Right). Results are mean ± SE. *P < 0.05 between control and AICAR-treated cells. (C) Signal ends were detected in AMPK DKO cells expressing RAG1, RAG2, and an adenovirally expressed constitutively active form of AMPK (CA) or GFP. The level of the signal end was normalized to the level of the plasmid backbone and quantified from three independent experiments. Results are mean ± SE. **P < 0.001 between GFP and CA. (D and E) WT or S528A RAG1, RAG2, and pJH200 were cotransfected in 293T cells and treated with either 2 mM AICAR for 3 h (D) or 1 mM metformin for 3 h (E). Signal ends were detected by LM-PCR (Left). Relative amount of signal end was normalized with that of the unrecombined plasmid and quantified from three independent experiments (Right). Results are mean ± SE. *P < 0.05. (F) 293T cells were cotransfected with MBP-WT, S528A, S528D, or S528E RAG1, MBP-RAG2, and pJH200. Signal ends were detected by LM-PCR (Left). Relative activities were quantified from at least three independent experiments (Right). Results are mean ± SE. **P < 0.001 between WT and S528 mutants.
Fig. 5.
Increased V(D)J recombination of an extrachromosomal substrate by AMPK activation in vivo. (A) Diagram of the PCR assay for recombination. The recombined and unrecombined plasmids are shown. The PCR primers, R5 and R14, are indicated by arrows. (B) 293T cells were cotransfected with full-length MBP-WT, S528A, S528D, or S528E RAG1, full-length MBP-RAG2, and pJH200. The recombined and unrecombined products were detected by PCR. (C) Full-length WT or S528A RAG1, full-length RAG2, and pJH200 were cotransfected in 293T cells and treated with 2 mM AICAR for 3 h. Recombined product was detected by PCR (Left). Relative amount of recombined product was normalized with that of unrecombined plasmid and quantified from four independent experiments (Right). Results are mean ± SE. **P < 0.001.
Discussion
In this study, we identify the RAG1 protein as a substrate of AMPK. Our evidence suggests that S528 phosphorylation of RAG1 by AMPK increases the catalytic activity of the RAG complex (RAG1/RAG2) in the DNA cleavage reaction that initiates V(D)J recombination. Our results show that RAG1 with the phosphomimetic S528D mutation increases both nick and hairpin formation with artificial 12- and 23-RSS oligonucleotides in vitro. In addition, the ability of RAG proteins to induce cleavage of an extrachromosomal V(D)J substrate in cells was increased with either RAG1 phosphorylation of S528 by AMPK or with the use of S528D or S528E RAG1 mutants. Because mutating S528 to Ala did not affect RAG1’s ability to generate nicks and hairpins, we propose that this residue and its phosphorylation have a regulatory, but not catalytic, role in the function of RAG1 in V(D)J recombination.
Little is known about the biochemical pathways by which extracellular factors may regulate V(D)J recombination. A second messenger that has been shown to increase V(D)J recombination is cAMP (32). This effect has been attributed to increased expression of RAG1 and RAG2, because elevated intracellular cAMP induces an increase in the level of RAG1 and RAG2 mRNA. However, because increasing intracellular cAMP level also leads to AMPK activation (33–35), cAMP may additionally increase V(D)J recombination by increasing AMPK-mediated RAG1 phosphorylation and activity.
It is well known that immune function is linked to nutritional status (reviewed in ref. 36). Obesity, which decreases AMPK activity, is associated with lymphocyte subpopulations that have decreased proliferative capacity to mitogen stimulation, higher rates of infection, delayed wound healing, and lower production of antibodies after hepatitis B vaccination. However, the majority of studies indicate that energy restriction, which increases AMPK activity, increases lymphocyte count and proliferative responses to mitogens in obese individuals (reviewed in ref. 36). Although the potential effect of the energy status on V(D)J recombination has not been demonstrated at the organismal level or in a physiological context, it is tempting to speculate that AMPK-mediated phosphorylation of RAG1 may be involved in regulating lymphocyte development in response to changes in the energy status.
Materials and Methods
Proteins and DNA.
Previously described oligonucleotides were synthesized and gel purified to generate 12-RSS and 23-RSS substrates (DAR39/40 and DAR61/62) (19, 37). Oligonucleotides were radiolabeled with 32P by using polynucleotide kinase (Promega) before annealing. The pLEXm vector encoding MBP fused to either core RAG1 (384–1,008) or RAG2 (1–387) was used for expression. Core RAG1 and core RAG2 are referred to as RAG1 and RAG2 in this paper unless they are mentioned as full-length RAG1 and RAG2 (Fig. 5 B and C). RAG1 S528A, S528D, and S528E mutations were produced by QuikChange site-directed mutagenesis (Stratagene). The expression vectors were transiently cotransfected into 293T cells by using polyethylenimine (PEI), at a final concentration of 1 mg/L of each DNA and 4 mg/L PEI. Singly or coexpressed RAG1 and RAG2 were purified by polyhistidine or MBP affinity tags as described in ref. 19. Mutations were verified by DNA sequencing.
In Vitro AMPK Phosphorylation Assay.
Recombinant MBP or His6 fusions of RAG1 and RAG2, or GST-RAG1 and S528A mutant RAG1 proteins were purified from 293T cells that had been transiently transfected with expression vectors for the corresponding protein. Kinase reactions were performed in kinase buffer [20 mM Hepes-NaOH, pH 7.0, 0.4 mM DTT, 0.01% (vol/vol) Brij-35] containing 10 ng of AMPK produced in Escherichia coli (Millipore), 2 μg of substrates, 15 μCi of [γ-32P]ATP, with or without 300 μM AMP. The mixture was incubated at 30 °C for 30 min and was analyzed by SDS/PAGE. In the kinase reactions used for mass spectrometry analysis, nonradioactive ATP was used.
LC-MS.
A Coomassie-stained band corresponding to MBP-RAG1 phosphorylated by AMPK was excised from a SDS-polyacrylamide gel, reduced with DTT, and alkylated with iodoacetamide. Proteins were then digested with LysC protease overnight. Phosphopeptides were enriched with TiO2 columns, and then analyzed with an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific) equipped with a nanoLC system (Eksigent). Peptide IDs and phosphorylation sites were assigned by using Mascot 2.3 (Matrix Science). Peptide peak areas were calculated by using Proteome Discoverer 1.3 (Thermo Fisher Scientific).
Cell Culture and Western Blots.
293T cells, AMPKα1/a2 double-knockout (AMPK DKO) MEF cells and WT MEF cells were maintained in DMEM supplemented with 10% (vol/vol) FBS. Cells were lysed in RIPA buffer and subjected to immunoblotting. Phosphorylation of AMPKα was determined by using anti-AMPKα (T172) antibody from Cell Signaling. The antibody to phospho-RAG1 S528 was generated by injection of a phosphopeptide (PPLKNVSpSRTDVGI) into rabbits, and the phosphoantibody was purified by using an affinity column (21st Century Biochemicals). Antibody to MBP was obtained from Millipore. Myc-tagged AMPK (AMPK-myc) was detected by immunoblotting with anti-myc antibody (Cell Signaling).
DNA Cleavage Assay and EMSA.
Assays carried out in reaction buffer [25 mM Mops, pH 7.0, 32 mM KCl, 4 mM DTT, 0.1 mg/mL BSA, 1% (vol/vol) glycerol] containing 32P-labeled 2 nM 12-RSS, 100 ng/μL RAG1 (core)/RAG2 (core) with or without 12.5 nM 23-RSS and including 1 mM MnCl2 for 30 min at 37 °C as described (38). The reactions were stopped by using 1 vol (90% formamide-TBE), heated for 5 min at 95 °C, and separated on 15% Tris/borate/EDTA (TBE)-urea gels. Cleavage products were quantified and expressed as a percentage of the total signal in each lane by using ImageJ software. EMSA was performed in cleavage assay reaction buffer without MnCl2 with 100 ng of RAG1/RAG2 complex and 32P-labeled 12-RSS and unlabeled 23-RSS. Reaction mixtures were incubated for 30 min at 37 °C and fractionated by 4% TBE polyacrylamide gel electrophoresis.
Assay for DNA Cleavage Activity in Vivo.
The MBP-fusion S528A, S528D, S528E, and WT RAG1 (core) expression plasmids, MBP-fusion RAG2 (core) expression plasmid, and extrachromosomal recombination substrate pJH200 were cotransfected into 293T cells. Cells were incubated for 24 h and were treated with 2 mM AICAR for 3 h or with 1 mM metformin for 3 h. The signal end of the recombination substrate was assayed as described in ref. 39. Signal ends at the 12-RSS were assayed by LM-PCR by using the oligonucleotide linker FM25/11; amplification was performed with primers 1223 and FM25. To control for recovery of the recombination substrate, primers 1223 and DR1 were used to amplify a portion of the backbone of the plasmid (39, 40).
V(D)J Recombination Assay in Vivo.
Full-length versions of MBP-fusion RAG1 S528A and WT RAG1 expression plasmids, together with full-length MBP-fusion RAG2 plasmid and extrachromosomal recombination substrate pJH200, were cotransfected into 293T cells. Cells were incubated for 24 h and were then treated with AICAR. Total DNA was purified 3 h later by using a DNA purification kit (Promega). Recombined product was assayed by PCR as described previously (41).
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research Program, National Heart, Lung, and Blood Institute, National Institute of Diabetes and Digestive and Kidney Diseases, and National Cancer Institute, National Institutes of Health, and by National Institutes of Health/National Institute of Allergy and Infectious Diseases Grants R01 AI047829 and R01AI074953.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1307928110/-/DCSupplemental.
References
- 1.Kahn BB, Alquier T, Carling D, Hardie DG. AMP-activated protein kinase: Ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 2005;1(1):15–25. doi: 10.1016/j.cmet.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 2.Rolf J, et al. AMPKα1: A glucose sensor that controls CD8 T-cell memory. Eur J Immunol. 2013;43(4):889–896. doi: 10.1002/eji.201243008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamás P, et al. Regulation of the energy sensor AMP-activated protein kinase by antigen receptor and Ca2+ in T lymphocytes. J Exp Med. 2006;203(7):1665–1670. doi: 10.1084/jem.20052469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hardie DG, Ross FA, Hawley SA. AMPK: A nutrient and energy sensor that maintains energy homeostasis. Nat Rev Mol Cell Biol. 2012;13(4):251–262. doi: 10.1038/nrm3311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hawley SA, et al. Complexes between the LKB1 tumor suppressor, STRAD alpha/beta and MO25 alpha/beta are upstream kinases in the AMP-activated protein kinase cascade. J Biol. 2003;2(4):28. doi: 10.1186/1475-4924-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woods A, et al. LKB1 is the upstream kinase in the AMP-activated protein kinase cascade. Curr Biol. 2003;13(22):2004–2008. doi: 10.1016/j.cub.2003.10.031. [DOI] [PubMed] [Google Scholar]
- 7.Xiao B, et al. Structure of mammalian AMPK and its regulation by ADP. Nature. 2011;472(7342):230–233. doi: 10.1038/nature09932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hawley SA, et al. Calmodulin-dependent protein kinase kinase-beta is an alternative upstream kinase for AMP-activated protein kinase. Cell Metab. 2005;2(1):9–19. doi: 10.1016/j.cmet.2005.05.009. [DOI] [PubMed] [Google Scholar]
- 9.Woods A, et al. Ca2+/calmodulin-dependent protein kinase kinase-beta acts upstream of AMP-activated protein kinase in mammalian cells. Cell Metab. 2005;2(1):21–33. doi: 10.1016/j.cmet.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Dale S, Wilson WA, Edelman AM, Hardie DG. Similar substrate recognition motifs for mammalian AMP-activated protein kinase, higher plant HMG-CoA reductase kinase-A, yeast SNF1, and mammalian calmodulin-dependent protein kinase I. FEBS Lett. 1995;361(2-3):191–195. doi: 10.1016/0014-5793(95)00172-6. [DOI] [PubMed] [Google Scholar]
- 11.Gwinn DM, et al. AMPK phosphorylation of raptor mediates a metabolic checkpoint. Mol Cell. 2008;30(2):214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tonegawa S. Somatic generation of antibody diversity. Nature. 1983;302(5909):575–581. doi: 10.1038/302575a0. [DOI] [PubMed] [Google Scholar]
- 13.Hesslein DG, Schatz DG. Factors and forces controlling V(D)J recombination. Adv Immunol. 2001;78:169–232. doi: 10.1016/s0065-2776(01)78004-2. [DOI] [PubMed] [Google Scholar]
- 14.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: Complexes, ends, and transposition. Annu Rev Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 15.Gellert M. V(D)J recombination: RAG proteins, repair factors, and regulation. Annu Rev Biochem. 2002;71:101–132. doi: 10.1146/annurev.biochem.71.090501.150203. [DOI] [PubMed] [Google Scholar]
- 16.Lieber MR, Ma Y, Pannicke U, Schwarz K. Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol. 2003;4(9):712–720. doi: 10.1038/nrm1202. [DOI] [PubMed] [Google Scholar]
- 17.Roth DB, Zhu C, Gellert M. Characterization of broken DNA molecules associated with V(D)J recombination. Proc Natl Acad Sci USA. 1993;90(22):10788–10792. doi: 10.1073/pnas.90.22.10788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schlissel M, Constantinescu A, Morrow T, Baxter M, Peng A. Double-strand signal sequence breaks in V(D)J recombination are blunt, 5′-phosphorylated, RAG-dependent, and cell cycle regulated. Genes Dev. 1993;7(12B):2520–2532. doi: 10.1101/gad.7.12b.2520. [DOI] [PubMed] [Google Scholar]
- 19.McBlane JF, et al. Cleavage at a V(D)J recombination signal requires only RAG1 and RAG2 proteins and occurs in two steps. Cell. 1995;83(3):387–395. doi: 10.1016/0092-8674(95)90116-7. [DOI] [PubMed] [Google Scholar]
- 20.Steen SB, Han JO, Mundy C, Oettinger MA, Roth DB. Roles of the “dispensable” portions of RAG-1 and RAG-2 in V(D)J recombination. Mol Cell Biol. 1999;19(4):3010–3017. doi: 10.1128/mcb.19.4.3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Curry JD, Schlissel MS. RAG2’s non-core domain contributes to the ordered regulation of V(D)J recombination. Nucleic Acids Res. 2008;36(18):5750–5762. doi: 10.1093/nar/gkn553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akamatsu Y, et al. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc Natl Acad Sci USA. 2003;100(3):1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung D, Giallourakis C, Mostoslavsky R, Alt FW. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu Rev Immunol. 2006;24:541–570. doi: 10.1146/annurev.immunol.23.021704.115830. [DOI] [PubMed] [Google Scholar]
- 24.Roth DB, Roth SY. Unequal access: Regulating V(D)J recombination through chromatin remodeling. Cell. 2000;103(5):699–702. doi: 10.1016/s0092-8674(00)00173-2. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan JE, et al. Inhibition of lipolysis and lipogenesis in isolated rat adipocytes with AICAR, a cell-permeable activator of AMP-activated protein kinase. FEBS Lett. 1994;353(1):33–36. doi: 10.1016/0014-5793(94)01006-4. [DOI] [PubMed] [Google Scholar]
- 26.Zhou G, et al. Role of AMP-activated protein kinase in mechanism of metformin action. J Clin Invest. 2001;108(8):1167–1174. doi: 10.1172/JCI13505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hardie DG. AMP-activated/SNF1 protein kinases: Conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8(10):774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- 28.Swanson PC. The bounty of RAGs: Recombination signal complexes and reaction outcomes. Immunol Rev. 2004;200:90–114. doi: 10.1111/j.0105-2896.2004.00159.x. [DOI] [PubMed] [Google Scholar]
- 29.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13(23):3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13(23):3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Steen SB, Gomelsky L, Roth DB. The 12/23 rule is enforced at the cleavage step of V(D)J recombination in vivo. Genes Cells. 1996;1(6):543–553. doi: 10.1046/j.1365-2443.1996.d01-259.x. [DOI] [PubMed] [Google Scholar]
- 32.Menetski JP, Gellert M. V(D)J recombination activity in lymphoid cell lines is increased by agents that elevate cAMP. Proc Natl Acad Sci USA. 1990;87(23):9324–9328. doi: 10.1073/pnas.87.23.9324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Omar B, Zmuda-Trzebiatowska E, Manganiello V, Göransson O, Degerman E. Regulation of AMP-activated protein kinase by cAMP in adipocytes: Roles for phosphodiesterases, protein kinase B, protein kinase A, Epac and lipolysis. Cell Signal. 2009;21(5):760–766. doi: 10.1016/j.cellsig.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yin W, Mu J, Birnbaum MJ. Role of AMP-activated protein kinase in cyclic AMP-dependent lipolysis in 3T3-L1 adipocytes. J Biol Chem. 2003;278(44):43074–43080. doi: 10.1074/jbc.M308484200. [DOI] [PubMed] [Google Scholar]
- 35.Park SJ, et al. Resveratrol ameliorates aging-related metabolic phenotypes by inhibiting cAMP phosphodiesterases. Cell. 2012;148(3):421–433. doi: 10.1016/j.cell.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karlsson EA, Beck MA. The burden of obesity on infectious disease. Exp Biol Med (Maywood) 2010;235(12):1412–1424. doi: 10.1258/ebm.2010.010227. [DOI] [PubMed] [Google Scholar]
- 37.Grundy GJ, Hesse JE, Gellert M. Requirements for DNA hairpin formation by RAG1/2. Proc Natl Acad Sci USA. 2007;104(9):3078–3083. doi: 10.1073/pnas.0611293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones JM, Gellert M. Ordered assembly of the V(D)J synaptic complex ensures accurate recombination. EMBO J. 2002;21(15):4162–4171. doi: 10.1093/emboj/cdf394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hesse JE, Lieber MR, Gellert M, Mizuuchi K. Extrachromosomal DNA substrates in pre-B cells undergo inversion or deletion at immunoglobulin V-(D)-J joining signals. Cell. 1987;49(6):775–783. doi: 10.1016/0092-8674(87)90615-5. [DOI] [PubMed] [Google Scholar]
- 40.van Gent DC, et al. Initiation of V(D)J recombination in a cell-free system. Cell. 1995;81(6):925–934. doi: 10.1016/0092-8674(95)90012-8. [DOI] [PubMed] [Google Scholar]
- 41.Cortes P, et al. In vitro V(D)J recombination: Signal joint formation. Proc Natl Acad Sci USA. 1996;93(24):14008–14013. doi: 10.1073/pnas.93.24.14008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.