Abstract
Background
A blood-based test that could be used as a screen for Alzheimer disease (AD) may enable early intervention and better access to treatment.
Objective
To apply a multiplex immunoassay panel to identify plasma biomarkers of AD using plasma samples from the Alzheimer’s Disease Neuroimaging Initiative cohort.
Design
Cohort study.
Setting
The Biomarkers Consortium Alzheimer’s Disease Plasma Proteomics Project.
Participants
Plasma samples at baseline and at 1 year were analyzed from 396 (345 at 1 year) patients with mild cognitive impairment, 112 (97 at 1 year) patients with AD, and 58 (54 at 1 year) healthy control subjects.
Main Outcome Measures
Multivariate and univariate statistical analyses were used to examine differences across diagnostic groups and relative to the apolipoprotein E (ApoE) genotype.
Results
Increased levels of eotaxin 3, pancreatic polypeptide, and N-terminal protein B–type brain natriuretic peptide were observed in patients, confirming similar changes reported in cerebrospinal fluid samples of patients with AD and MCI. Increases in tenascin C levels and decreases in IgM and ApoE levels were also observed. All participants with Apo ε3/ε4 or ε4/ε4 alleles showed a distinct biochemical profile characterized by low C-reactive protein and ApoE levels and by high Cortisol, interleukin 13, apolipoprotein B, and gamma interferon levels. The use of plasma biomarkers improved specificity in differentiating patients with AD from controls, and ApoE plasma levels were lowest in patients whose mild cognitive impairment had progressed to dementia.
Conclusions
Plasma biomarker results confirm cerebrospinal fluid studies reporting increased levels of pancreatic polypeptide and N-terminal protein B–type brain natriuretic peptide in patients with AD and mild cognitive impairment. Incorporation of plasma biomarkers yielded high sensitivity with improved specificity, supporting their usefulness as a screening tool. The ApoE genotype was associated with a unique biochemical profile irrespective of diagnosis, highlighting the importance of genotype on blood protein profiles.
Alzheimer disease (AD) is a progressive neurodegenerative disorder estimated to affect 27 million individuals worldwide, with numbers doubling every 20 years.1 Although current diagnostic standards require the manifestation of dementia before diagnosis, neuropathologic features of AD occur well before the onset of dementia.2 Studies3–5 examining patients with prodromal AD that has progressed to dementia have identified a symptomatic predementia stage characterized by deficits in episodic memory and executive function. In addition, results of longitudinal natural history studies3,4,6 examining biomarker changes have led to a general recognition that predementia stages of AD can he identified using cerebrospinal fluid (CSF) Aβ42 and t-tau/p-tau profiles, positron emission tomography (PET) amyloid imaging, and volumetric magnetic resonance imaging biomarker data. Furthermore, CSF studies and PET can help to confirm AD pathologic changes in patients diagnosed as having dementia of the Alzheimer type. As a result, revisions to the current diagnostic criteria are under consideration that incorporate CSF testing and imaging as aids in the diagnosis of and risk assessment for AD.7–9
Although CSF and PET studies are proving to be useful tools to confirm AD pathologic changes and to assess the risk of progression, there is a widespread interest in noninvasive, cost-effective tests that can be used as screening tools to identify patients who would be eligible for more detailed confirmatory AD testing. Several studies10–21 have described blood-based biomarkers of AD. Of note, recent studies17, 22, 21 have used a multiplex immunoassay panel to examine changes in CSF and blood of patients with AD and mild cognitive impairment (MCI). Similar expression patterns in CSF and plasma have been described, including elevations in eotaxin 3 and pancreatic polypeptide (PP) levels. Data suggest that multiplex immunoassay panels using CSF and blood samples may have some usefulness in differentiating patients with AD from healthy control subjects (HCs) and perhaps in identifying patients with early stages of the disease.
To further explore the potential of multiplex immunoassay panels to identify plasma biomarkers of AD, the present study uses a 190-analyte version of the panel applied in prior plasma and CSF studies17, 22,23 to examine changes in patients with AD and MCI using a subset of baseline and 1-year plasma samples from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) cohort. Multivariate analysis and analysis of covariance (ANCOVA) were conducted to examine differences across diagnostic groups. Finally, the influence of the apolipoprotein E (ApoE) genotype on plasma protein expression was explored. The study focused on whether some of the literature findings for putative blood-based biomarkers of AD were replicated in the ADNI cohort.
METHODS
ADNI COHORT AND SELECTION OF PLASMA SAMPLES
The ADNI study (http://adni.loni.ucla.edu) was launched by the National Institute on Aging, the Foundation for the National Institutes of Health, and by a group of private and public partners as a precompetitive AD biomarker consortium. The enrollment target for the ADNI study was 400 patients with amnestic MCI, 200 patients with mild AD, and 200 HCs. The ADNI protocol was approved by the human studies committees at 58 institutions in the United States and Canada. Written and verbal informed consents were obtained from participants at screening and enrollment. At the time of plasma sample collection, participants were 60 years or older and in good general health, having no other neurological, psychiatric, or major medical diagnoses that could contribute substantially to dementia. Further details about the ADNI study, including participant selection procedures and complete study protocols, are available online (http://www.alzheimers.org/clinicaltrials/fullrec.asp?PrimaryKey=208). Only a subset of the ADNI plasma sample set was included in the present study. All the baseline and 1-year plasma samples from the MCI arm were analyzed. Baseline (n = 112) and matched 1-year (n = 97) plasma samples from the AD arm were selected based on the availability of concurrent CSF or PET data. For the HC arm, plasma samples were selected for analysis if the subjects had concurrent CSF biomarker data and if CSF Aβ42 levels were above the above median value of the total HC Aβ42 distribution.
LABORATORY TESTS
ApoE Genotyping
ApoE genotyping was performed for all study participants using EDTA blood samples. Quantitative polymerase chain reaction assays (TaqMan; Qiagen) were used for genotyping ApoE nucleotides 334 T/C (rs 429358) and 472 C/T (rs 7412) with a realtime thermocycler (ABI 7900; Applied Biosystems) using DNA freshly prepared from EDTA whole blood.
Multiplex Immunoassay Panel
Plasma samples were evaluated by an independent laboratory (Myriad RBM) for levels of 190 analytes using a multianalyte profile panel (Human Discovery MAP version 1.0; Myriad RBM) and a commercially available platform (Luminex 100; Luminex Corporation). A complete list of the analytes is given in eTable 1 (http://www.archneurol.com). In total, 146 of 190 analytes met quality control criteria for subsequent statistical analysis. Details of the methods can be found in the eAppendix, and validation reports are available from Myriad RBM (www.myriadrbm.com). Myriad RBM provided 3 levels of quality control for each analyte, and details of the quality control results, detection limits for assays, and dynamic ranges for plasma are given in the data primer (http://adni.loni.ucla.edu/wp-content/uploads/2010/11/BC_Plasma_Proteomics_Data_Primer.pdf) and in eTable 2.
Plasma Sample Collection
Patients had fasted overnight (approximately 8 hours) before plasma sample collection. All EDTA plasma samples were obtained in the morning and were processed per ADNI laboratory standard operating procedures (http://adni.loni.ucla.edu). Additional details are provided in the eAppendix.
STATISTICAL ANALYSIS
Analytes that had more than 10% missing values or values listed as low were excluded from the analysis. For analytes that had more than 90% of data available, missing or low values were imputed. Each analyte was checked for normal distribution and multidimensional scaling, and Mahalanobis distances (or generalized squared interpoint distance for its squared value) were used to detect outliers. The statistical management of outliers and false discovery rates for multiple comparisons are described in detail in the data primer. Positive false discovery corrections were applied to P values to account for the multiple comparisons. ANCOVA models, including diagnosis, age, sex, and ApoE allelic status, were used to examine the contributions of these variables to differences across diagnostic groups. Detailed results of the ANCOVA are given in eTable 3. In addition, individual group comparisons were made between the AD vs HC arms and between the MCI vs HC arms at baseline and at 1 year according to diagnosis at the time of blood sample collection. Preliminary group comparisons were also made between (1) patients whose MCI had progressed to dementia (MCIp group) with follow-up periods of up to 48 months vs HCs and (2) MCIp group vs patients whose MCI had not yet progressed to dementia (MCInp group). Details of the statistical analysis plan for the project are given in the data primer.
Multivariate flexible linear discriminant analysis, logistic regression, partial least squares, random forests, nearest shrunken centroids, and support vector machine algorithms were conducted to compare the AD vs HC arms. Within die AD and HC data set, 170 participants were placed into a training set (n=96) and a test set (n=74). Assignment to the AD and HC arms was based on diagnosis at 1 year. Details of the multivariate approaches are provided in the eAppendix.
RESULTS
The demographics were calculated for the following 2 diagnostic states: (1) clinical diagnosis at the time of enrollment (referred to as baseline demographics) and (2) clinical diagnosis at the time of data download (referred to as progression demographics). According to baseline demographics, Table 1 summarizes the characteristics of 1062 analyzed plasma samples, comprising those at baseline and at 1 year from 396 (345 at 1 year) patients with mild cognitive impairment, 112 (97 at 1 year) patients with AD, and 58 (54 at 1 year) healthy control subjects. No significant group differences in age were observed, and men outnumbered women in all the groups. Among the AD arm, the ApoE allele was present in 67.9%, whereas the prevalence among the HC arm was significantly lower than population norms, which was expected based on the use of CSF Aβ42 values above the group median as part of the selection criteria for the HC group.
Table 1.
Baseline Demographics of the Alzheimer’s Disease Neuroimaging Initiative Cohorta
| Variable | Study Arm
|
||
|---|---|---|---|
| Healthy Control Subjects | Mild Cognitive Impairment | Alzheimer Disease | |
| No. at baseline (No. at 1y) | 58 (54) | 396 (345) | 112 (97) |
| Age. mean (SD), y | 75 (6) | 75 (8) | 75 (8) |
| Male-female ratio | 30:28 | 256:140 | 65:47 |
| ApoE allele carrier, % | 8.6 | 53.3 | 67.9 |
| Mini-Mental State Examination score, mean (SD) | 28.9 (1.2) | 27.0 (1.8) | 23.6 (1.9) |
| Cognitive Dementia Rating Scale global score | 0.0 | 0.5 | 0.5 (n = 59), 1.0 (no = 53) |
The clinical diagnosis at the time of study enrollment is referred to as baseline. The clinical status at baseline determined the visit and procedure schedule for participants enrolled in the study.
According to progression demographics, Table 2 summarizes the numbers of participants whose diagnosis changed from enrollment (baseline) based on a January 2011 version of the ADNI database. The demographic information includes CSF Aβ42 levels and PET indexes of amyloid burden based on follow-up clinical diagnostic status. Follow-up periods from baseline ranged from 6 to 48 months. Within the MCI arm, 164 patients progressed to dementia of the Alzheimer type (MCIp group), leaving 218 patients with MCI who did not progress to dementia (MCInp group) (some members of the MCI arm regressed back to normal and were not counted). No significant differences in age were observed across groups. Among the MCIp group, the ApoE allele was present in 67.7%, which was higher than the 44.0% prevalence among the MCInp group. The Aβ burden (as evidenced by low CSF Aβ42 levels between 142 and 147 pg/mL and by increased standard uptake value ratios for the PET indexes of amyloid binding between 1.71 and 1.79) was greatest in patients with AD and those in the MCIp group compared with HCs and patients in the MCInp group who remained classified as having MCI.
Table 2.
Follow-up Demographics of the Alzheimer’s Disease Neuroimaging Initiative Cohorta
| Variable | Diagnosis as of January 2011
|
|||||
|---|---|---|---|---|---|---|
| Healthy Control Subject | Healthy Control Subject to MCI | MCI to Healthy Control Subject | MCInp | MCIp | Alzheimer Disease | |
| No. at baseline (No. at 1 y) | 54(50) | 4 (4) | 14 (14) | 218 (175) | 164 (156) | 112 (97) |
| Age, mean (SD), y | 75(6) | 73 (9) | 72 (8) | 75 (8) | 75 (7) | 75 (8) |
| Male-female ratio | 27:27 | 3:1 | 10:4 | 146:72 | 100:64 | 65:47 |
| ApoE allele carrier, % | 7.4 | 25.0 | 28.6 | 44.0 | 67.7 | 67.9 |
| Mini-Mental State Examination score, mean (SD) | 28.9 (1.2) | 29.0 (0.8) | 27.9 (1.6) | 27.2 (1.8) | 26.6 (1.7) | 23.6 (1.9) |
| Cerebrospinal fluid Aβ42 level at baseline, mean (SD), pg/mL | 253 (55) | 238 (4) | 233 (6) | 174 (106) | 147 (86) | 142 (102) |
| PET index of amyloid burden at 1 y, SUVR | 1.23 | No data | 1.57 | 1.56 | 1.79 | 1.71 |
Abbreviations: MCI, mild cognitive impairment; MCInp, patients whose MCI had not yet progressed to dementia; MCIp, patients whose MCI had progressed to dementia; PET, positron emission tomography; SUVR, standard uptake value ratio.
The clinical diagnosis at the time of data download (January 2011) was used to calculate demographics for the preliminary progression analysis. Patients may have progressed anywhere between 6 months and 48 months.
MULTIVARIATE ANALYSIS
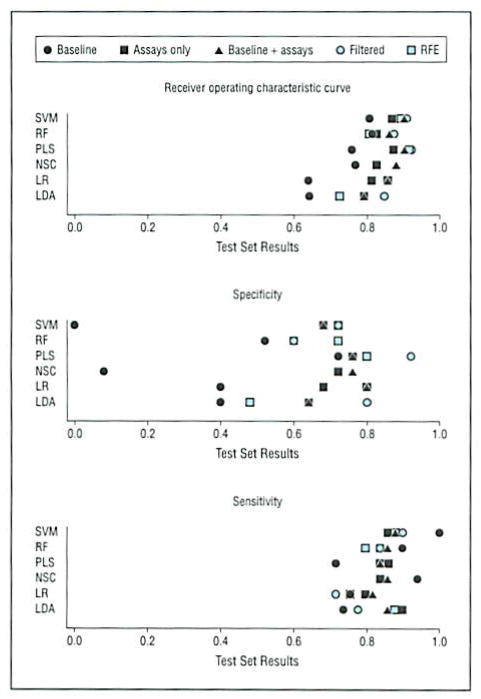
Six algorithms (linear discriminant analysis, logistic regression, partial least squares, random forests, nearest shrunken centroids, and support vector machine) were applied to the ADNI cohort to identify, in an unbiased fashion, the top plasma analytes of AD. Sensitivity, specificity, and receiver operating characteristic curves from the multivariate analysis were generated on the training set, and results are shown for the test set in Figure 1. Performance across algorithms was similar. Inclusion of plasma analytes improved specificity across the models. For example, in the logistic regression model, specificity improved from 40% in baseline models (ie, age, sex, and ApoE allelic status alone) to 80% in models that included plasma biomarkers. Using a comparison method by Hanley and McNeil,24 the improvement in the logistic regression model from 40% to 80% was statistically significant (P = .005). The ApoE genotype and N-terminal protein B–type brain natriuretic peptide (NT-proBNP) levels were identified as 2 primary contributors to improved specificity across logistic regression, partial least squares, random forests, nearest shrunken centroids, and support vector machine models. In summary, inclusion of a subset of blood-based biomarkers improved specificity over the baseline model alone.
Figure 1.
Results of the multivariate analysis. The Alzheimer disease and healthy control subject arms were split into a training set (n = 96) and a test set (n = 74). Multivariate approaches included support vector machine (SVM). random forests (RF), partial least squares (PLS), nearest shrunken centroids (NSC), logistic regression (LR), and flexible linear discriminant analysis (LDA). Baseline models included age, sex, and ApoE allelic status (ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4) only. Baseline + assays included baseline covariates and all the analytes. In addition, 2 feature selection schemes were used, including a filtered approach (top analytes by univariate analysis with P < .05) and recursive feature selection (RFE) to reduce the number of analytes in the predictor sets before modeling. Inclusion of plasma analytes improved specificity over the baseline model for the SVM, RF, NSC, LR, and LDA models. Additional details are given in the eTables and eAppendix.
UNIVARIATE ANALYSIS
Figure 2 shows a Venn diagram highlighting analytes that were significant across group comparisons and demonstrated consistent directional changes in both baseline and 1-year comparisons. ANCOVAs were conducted to examine differences between the AD vs HC arms and between the MCI vs HC arms at baseline and at 1 year based on diagnostic classification at the time of plasma sample collection, with age, sex, and ApoE allelic status included as covariates. Analytes showing inconsistencies in directional changes were excluded. ANCOVA details for all the analytes are given in eTable 3. The IgM and ApoE plasma protein levels were decreased, while the PP, eotaxin 3, NT-proBNP, tenascin C (TN-C), and matrix metalloproteinase 1 (MMP-1) levels were increased relative to HCs in all AD and MCI comparisons at baseline and at 1 year. In addition, increases were noted in alpha-2-macroglobulin, Cortisol, apolipoprotein A-IV, adiponectin, interleukin 3 (IL-3), vascular endothelial growth factor, and macrophage inflammatory protein 1α levels, and decreases were noted in apolipoprotein D and Tamm-Horsfall protein levels in a subset of comparisons.
Figure 2.
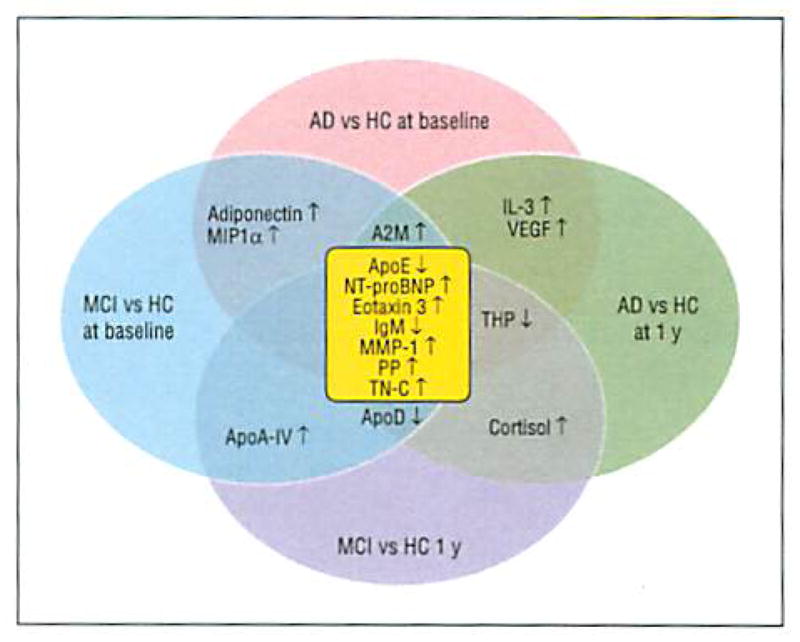
Venn diagram of analytes with P < .05 based on analyses of covariance across groups. Diagnostic categorization was based on diagnosis at the time of plasma sample collection. Analytes highlighted in center (yellow) were common across group comparisons. AD indicates Alzheimer disease; A2M, alpha-2-macroglobulin: ApoA-IV, apolipoprotein A-IV; ApoD, apolipoprotein D; ApoE. apolipoprotein E; HC, healthy control subject; IL-3, interleukin 3; MCI, mild cognitive impairment; MIP1α, macrophage inflammatory protein 1; MMP-1, matrix metalloproteinase 1; NT-proBNP, N-terminal protein B–type brain natriuretic peptide; PP, pancreatic polypeptide; TN-C, tenascin C; THP, Tamm-Horsfall protein; VEGF, vascular endothelial growth factor; ↑, increase; and ↓, decrease.
EXPRESSION OF ApoE PROTEIN LEVELS
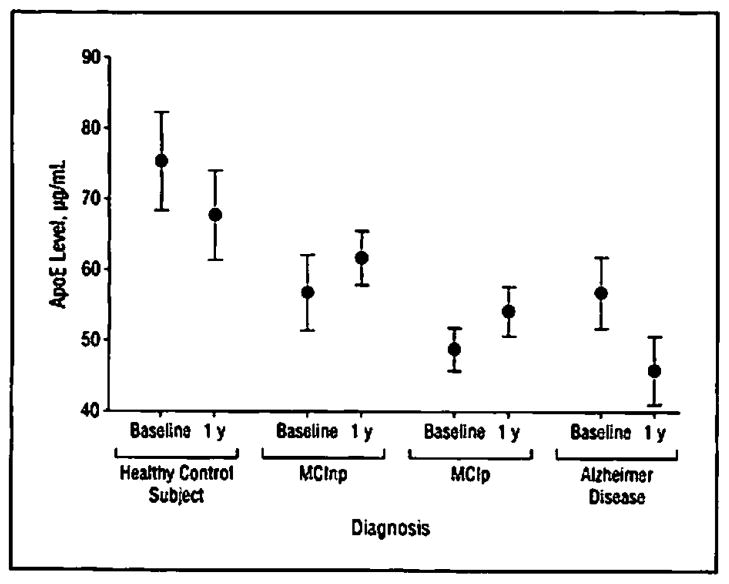
Expression patterns of the top analytes from the multivariate and univariate ANCOVAs were examined to determine if levels in patients with AD were similar to levels among patients in the MCIp group. No differences in expression of the top plasma analytes expression were observed between the MCInp vs MCIp groups when comparisons were corrected for age, sex, and ApoE allelic status (data not shown). However, ApoE protein levels were lower in the MCIp group compared with the MCInp group and compared with HCs. Figure 3 shows ApoE plasma expression levels based on 48-month follow-up diagnostic classification.
Figure 3.
Plasma protein expression levels of plasma apolipoprotein E (ApoE) at baseline and at 1 year. Diagnosis was based on that at up to 48 months’ follow-up. Levels of ApoE were lowest in patients with Alzheimer disease and in patients with mild cognitive impairment (MCI) who progressed to dementia compared with healthy control subjects and patients with MCI who remained dementia free at 48 months’ follow-up. Data are expressed as mean values (nonimputed raw data) and 95% CIs.
PLASMA PHENOTYPE ASSOCIATED WITH THE ApoE GENOTYPE
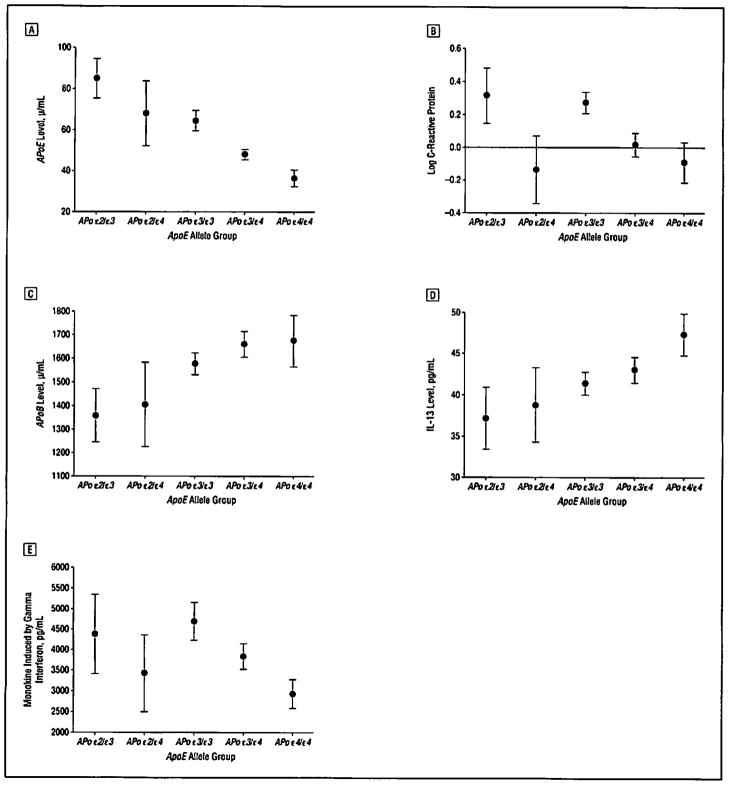
Expression patterns of the other analytes were examined based on ApoE allelic status rather than on diagnostic status (Figure 4). Significant differences in ApoE plasma concentrations as a function of ApoE allelic status were noted (eg, ε2/ε2, ε2/ε3, ε2/ε4, ε3/ε3, ε3/ε4, and ε4/ε4), with ε3/ε4 or ε4/ε4 carriers expressing the lowest ApoE plasma protein levels irrespective of diagnosis, while those with an ε2 allele expressed the highest levels of ApoE protein even in the presence of an ε4 allele (Figure 4A). The ApoE allelic status was associated with differential concentrations of other proteins that were identified in the univariate analysis for some group comparisons. For example, C-reactive protein was low in participants with 1 or more ε4 alleles and was highest in ε2/ε3 carriers (Figure 4B). Monokine induced by gamma interferon (also known as CXCL9) exhibited lowest levels in participants with 1 or more ε4 alleles (Figure 4E), while IL-13 (Figure 4D) showed elevations. Notably, apolipoprotein B levels were elevated in participants with an ε4 allele (Figure 4C). Other top biomarker analytes identified in the ADNI cohort showed some associations with ApoE genotype, including Cortisol, Tamm-Horsfall protein, some of the interleukins, and several of the microglobulins. Additional analytes associated with AD (including tissue inhibitor of metalloproteinases, cystatin C, chromogranin A, α-1 antichymotrypsin, and pregnancy-associated plasma protein CA 19-9) showed some associations with the ApoE genotype (eTable 3 ANCOVA results). A complete list of the analytes that demonstrated a significant association with ApoE is given in eTable 3, and expression patterns by diagnostic groups are also provided. Similar patterns were observed when the AD, MCI, and HC arms were analyzed separately (data not shown).
Figure 4.
Baseline plasma expression patterns based on ApoE allelic status in all participants irrespective of disease stage (healthy control subject, mild cognitive impairment, or Alzheimer disease). A, Apolipoprotein E (ApoE). B, C-reactive protein. C, Apolipoprotein B (ApoB). D, Interleukin 13 (IL-3). E, Monokine induced by gamma interferon. Data are expressed as mean (95% CI) values (nonimputed raw data, with missing values excluded).
COMMENT
The present study examined changes in plasma biomarkers in the ADNI cohort. Statistical analysis showed on average 80% to 90% sensitivity across models, and incorporation of plasma analytes improved specificity from 40% up to 70% and 80% in differentiating patients with AD from HCs, suggesting some usefulness of the multiplex immunoassay panel as a screening tool. Increases in PP, TN-C, MMP-1, eotaxin 3, and NT-proBNP levels and decreases in IgM and ApoE levels were consistently observed across all group comparisons in the ADNI cohort. Notably, ApoE protein levels were lowest in patients with MCI who progressed to dementia. Perhaps the most intriguing observation was the identification of a biochemical profile based on ApoE genotype in the AD, MCI, and HC arms.
Previous studies13,14,17,18,21,25,26 have described blood-based biomarkers in patients having AD, with little consistency across studies. When the same platform is applied across cohorts, similarities in the biomarkers begin to emerge. Specifically, increases in PP, eotaxin 3, and plasma NT-proBNP levels, observed in the present ADNI cohort, have been previously described in CSF samples obtained from patients with AD.22,23 In addition, evidence from the Australian Imaging, Biomarkers and Lifestyle Flagship Study of Aging group (H.D.S., written communication, June 2012) and from the Texas Alzheimer’s Research Consortium17 shows increased levels of PP, TN-C, and eotaxin 3 in plasma samples, suggesting that changes in these analytes are robust across cohorts when tested on the same platform. It should be noted that the Texas Alzheimer’s Research Consortium used an earlier version of the panel (which did not include ApoE or NT-proBNP) than that used in the present study.
Perhaps the most notable finding from the ADNI cohort was the identification of a protein profile associated with ApoE allelic status, a known risk factor for AD. For example, ApoE protein, C-reactive protein, and gamma interferon plasma protein levels were lowest in Apo ε4 carriers, while IL-13 levels were elevated. Many of the plasma-based biomarkers previously described in the literature13,15,17,26–30 (eg, Cortisol, cystatin C, chromogranin A, tissue inhibitor of metalloproteinases, alpha-1 antichymotrypsin, and pregnancy-associated plasma protein CA 19-9) also showed some associations with the ApoE genotype (eTables 1–3 and eAppendix), suggesting that differences may in part be driven by genotypic status in addition to dementia status. The present data support a phenotypic plasma signature associated with the ApoE genotype and could provide some explanation for the biological variability of blood-based biomarkers of AD described in the literature.13,15,18 Clearly, examination in much larger cohorts is needed.
Three common biological themes seemed to be associated with the top plasma biomarker changes in the ADNI cohort. The first biological theme appeared to be associated with metabolic markers that might be altered by cholinergic tone but most likely were driven by concomitant medication use (eg, PP). In fact, evidence indicates that acetylcholinesterase inhibitors can alter PP release,31 suggesting that alterations in PP observed in the ADNI cohort may be a function of acetylcholinesterase use. Alternatively, abnormal cholinergic tone may be driving abnormal expression of PP in patients with AD. The second biological theme appeared to be linked to vascular pathologic conditions (eg, TN-C, MMP-1, and NT-proBNP). For example, TN-C has been associated with vascular remodeling, and NT-proBNP has long been studied in the context of cardiovascular disease.32,33 A recent study34 examining 464 individuals 75 years or older without dementia demonstrated a significant association of NT-proBNP levels with cognitive decline, and NT-proBNP has been reported to be elevated in patients with subcortical vascular dementia.35 Notably, increases in natriuretic peptide levels in patients with AD have previously been described by Ewers et al11,12 and by Buerger and colleagues,36 with speculation that endothelial remodeling during dementia might be detectable by peripheral vascular markers. The third biological theme seemed to revolve around the phenotypic signature of an ApoE genotype, perhaps best characterized by ApoE protein levels, the acute-phase C-reactive protein, and a subset of interleukins, and is probably the most notable of the categories. It has been hypothesized that ApoE is critical for Aβ clearance, although it remains less clear whether low levels of ApoE actually contribute to AD pathologic changes.37,38 The ApoE genotype also seems to be loosely associated with increases in several inflammatory cytokines. Indeed, eotaxin 3, one of the top biomarkers in the ADNI plasma data set, can be released following stimulation with interleukins such as IL-3 and IL-4,39 and levels might be driven by modulation of interleukin levels. Further longitudinal studies relating changes in plasma and CSF ApoE and cytokine concentrations over time to changes in cognition and AD brain pathologic conditions in at-risk older individuals will likely further clarify the biological relevance of these associations.
Significant care should be taken in the interpretation of the present data set. For example, many of the observed changes may be a function of concurrent medication use (eg, PP). Furthermore, several common comorbidities exist in patients with AD, including cardiovascular disease and diabetes mellitus, that may be more reflective of vascular pathologic conditions (eg, TN-C, MMP-1, and NT-proBNP) and are not that specific to AD. Finally, changes in one biomarker may drive changes in others within the panel (eg, interleukin influence on eotaxin 3), suggesting that many of the top biomarkers may be redundant and part of a larger common signaling cascade.
In summary, the ADNI plasma biomarker data confirm CSF studies reporting increased levels of PP, eotaxin 3, tenascin C, and NT-proBNP in patients with AD and MCI. When plasma biomarkers were combined with baseline demographics, the combined models showed high sensitivity and improved specificity, suggesting their usefulness as a potential screening tool. Further studies to confirm these findings are merited. Finally, the ADNI plasma biomarker data suggest a distinct phenotypic profile associated with ApoE allelic status. Additional research will help to delineate whether a blood-based multiplex immunoassay panel that includes these analytes can deliver as a screening tool for AD.
Acknowledgments
Funding/Support: The ADNI study is funded by grant U01 AG024904 from the National Institute on Aging and the National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health and through generous contributions from the following: Abbott, Alzheimer’s Association, Alzheimer’s Drug Discovery Foundation, Amorfix Life Sciences Ltd, AstraZeneca, Bayer HealthCare, BioClinica Inc. Biogen Idec Inc, Bristol-Myers Squibb, Eisai Inc, Elan Pharmaceuticals Inc, Eli Lilly and Company, F. Hoffmann-La Roche Ltd and its affiliated company Genentech Inc, GE Healthcare, Innogenetics, NV, Janssen Alzheimer Immunotherapy Research & Development, LLC, Johnson & Johnson Pharmaceutical Research & Development LLC, Medpace Inc, Merck & Co Inc, Meso Scale Diagnostics LLC, Novartis Pharmaceuticals Corporation, Pfizer Inc, Servier, Syn-arc Inc, and Takeda Pharmaceutical Company. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. The ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of California, Los Angeles. This research was also supported by grants P30 AG010129 and KOI AG030514 from the National Institutes of Health and by The Dana Foundation.
Footnotes
Group Information: The Biomarkers Consortium Alzheimer’s Disease Plasma Proteomics Project members are listed at http://adni.loni.ucla.edu/wp-content/uploads/how_to_apply/ADNl_Acknowledgement_List.pdf.
Online-Only Material: The eTables and eAppendix and are available at http://www.archneurol.com.
Author Contributions: Data used in the preparation of this article were obtained from the ADNI database (adni.loni.ucla.edu). As such, the investigators within the ADNI study contributed to the design and implementation of ADNI or provided data but did not participate in the analysis or writing of the manuscript. Study concept and design: Soares, Potter, Pickering, Shera, Ferm, Dean, Simon, Siuciak, Koroshetz, Wan, Trojanowski, and Shaw. Acquisition of data: Soares, Potter, Trojanowski, and Shaw. Analysis and interpretation of data: Soares, Potter, Pickering, Kuhn, Immermann, Ferm, Dean, Swenson, Kaplow, Thambisetty, Zagouras, Koroshetz, Wan, Trojanowski, and Shaw. Drafting of the manuscript: Soares, Potter, Pickering, Kuhn, Ferm, Dean, Swenson, Siuciak, Thambisetty, Koroshetz, and Trojanowski. Critical revision of the manuscript for important intellectual content: Soares, Potter, Pickering, Kuhn, Immermann, Shera, Ferm, Simon, Swenson, Siuciak, Kaplow, Zagouras, Koroshetz, Wan, Trojanowski, and Shaw. Statistical analysts: Soares, Pickering, Kuhn, Immermann, Thambisetty, and Trojanowski. Obtained funding: Soares, Potter, Dean, Simon, Siuciak, and Wan. Administrative, technical, and material support: Potter, Simon, Swenson, Siuciak, and Trojanowski. Study supervision: Soares, Wan, and Trojanowski.
Financial Disclosure: Dr Soares reported owning shares in Bristol-Myers Squibb. Mr Immermann and Dr Wan reported owning shares in Pfizer Inc. Dr Simon reported serving as an independent biomarker consultant. Data collection and sharing for this study represent the work of the Biomarkers Consortium Project “Use of Targeted Multiplex Proteomic Strategies to Identify Plasma-Based Biomarkers in Alzheimer’s Disease.” This project was submitted to the Biomarkers Consortium Neuroscience Steering Committee by a subgroup of the ADNI Private Partner Scientific Board for execution and was managed by a Biomarkers Consortium Project Team that includes members from academia, government, and the pharmaceutical industry. Funding for this project was provided through an overage of funds raised by the Foundation for the National Institutes of Health for the ADNI partnership. The entire data set described in the present study, along with the Biomarkers Consortium plasma proteomics project team data primer and statistical analysis plan, are available on the ADNI website (http://adni.loni.ucla.edu).
References
- 1.Thies W, Bleiler L Alzheimer’s Association. 2011 Alzheimer’s disease facts and figures. Alzheimers Dement. 2011;7(2):208–244. doi: 10.1016/j.jalz.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 2.Savva GM, Wharton SB, Ince PG, Forster G, Matthews FE, Brayne C Medical Research Council Cognitive Function and Ageing Study. Age, neuropathology, and dementia. N Engl J Med. 2009;360(22):2302–2309. doi: 10.1056/NEJMoa0806142. [DOI] [PubMed] [Google Scholar]
- 3.Dubois B, Feldman HH, Jacova C, et al. Revising the definition of Alzheimer’s disease: a new lexicon. Lancet Neurol. 2010;9(11):1118–1127. doi: 10.1016/S1474-4422(10)70223-4. [DOI] [PubMed] [Google Scholar]
- 4.Dubois B, Feldman HH, Jacova C, et al. Research criteria for the diagnosis of Alzheimer’s disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 2007;6(8):734–746. doi: 10.1016/S1474-4422(07)70178-3. [DOI] [PubMed] [Google Scholar]
- 5.Petersen RC, Negash S. Mild cognitive impairment: an overview. CNS Spectr. 2008;13(1):45–53. doi: 10.1017/s1092852900016151. [DOI] [PubMed] [Google Scholar]
- 6.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albert MS, DeKosky ST, Dickson D, et al. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):270–279. doi: 10.1016/j.jalz.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):263–269. doi: 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 2011;7(3):280–292. doi: 10.1016/j.jalz.2011.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Booij BB, Lindahl T, Wetterberg P, et al. A gene expression pattern in blood for the early detection of Alzheimer’s disease. J Alzheimers Dis. 2011;23(1):109–119. doi: 10.3233/JAD-2010-101518. [DOI] [PubMed] [Google Scholar]
- 11.Ewers M, Mielke MM, Hampel H. Blood-based biomarkers of microvascular pathology in Alzheimer’s disease. Exp Gerontol. 2010;45(1):75–79. doi: 10.1016/j.exger.2009.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewers M, Walsh C, Trojanowski JO, et al. North American Alzheimer’s Disease Neuroimaging Initiative (ADNI). Prediction of conversion from mild cognitive impairment to Alzheimer’s disease dementia based upon biomarkers and neuropsychological test performance. Neurobiol Aging. 2012;33(7):1203–1214. e2. doi: 10.1016/j.neurobiolaging.2010.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hye A, Lynham S, Thambisetty M, et al. Proteome-based plasma biomarkers for Alzheimer’s disease. Brain. 2006;129(Pt 11):3042–3050. doi: 10.1093/brain/awl279. [DOI] [PubMed] [Google Scholar]
- 14.Lovestone S, Francis P, Kloszewska I, et al. Add Neuro Med Consortium. Add-NeuroMed—the European collaboration for the discovery of novel biomarkers for Alzheimer’s disease. Ann N Y Acad Sci. 2009;1180:36–46. doi: 10.1111/j.1749-6632.2009.05064.x. [DOI] [PubMed] [Google Scholar]
- 15.Marksteiner J, Kemmler G, Weiss EM, et al. Five out of 16 plasma signaling proteins are enhanced in plasma of patients with mild cognitive impairment and Alzheimer’s disease. Neurobiol Aging. 2011;32(3):539–540. doi: 10.1016/j.neurobiolaging.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagele E, Han M, Demarshall C, Belinka B, Nagele R. Diagnosis of Alzheimer’s disease based on disease-specific autoantibody profiles In human sera. [Accessed May 26, 2012];PLoS One. 2011 6(8):e23112. doi: 10.1371/journal.pone.0023112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Bryant SE, Xiao G, Barber R, et al. Texas Alzheimer’s Research Consortium. A serum protein-based algorithm for the detection of Alzheimer disease. Arch Neurol. 2010;67(9):1077–1081. doi: 10.1001/archneurol.2010.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ray S, Britschgi M, Herbert C, et al. Classification and prediction of clinical Alzheimer’s diagnosis based on plasma signaling proteins. Nat Med. 2007;13 (11):1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 19.Reddy MM, Wilson R, Wilson J, et al. Identification of candidate IgG biomarkers for Alzheimer’s disease via combinatorial library screening. Cell. 2011;144 (1):132–142. doi: 10.1016/j.cell.2010.11.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rye PD, Booij BB, Grave G, et al. A novel blood test for the early detection of Alzheimer’s disease. J Alzheimers Dis. 2011;23(1):121–129. doi: 10.3233/JAD-2010-101521. [DOI] [PubMed] [Google Scholar]
- 21.Soares HD, Chen Y, Sabbagh M, Roher A, Schrijvers E, Breteler M. Identifying early markers of Alzheimer’s disease using quantitative multiplex proteomic immunoassay panels [published correction appears in Ann N Y Acad Sci 2010 Feb;1188:231] Ann N Y Acad Sci. 2009;1180:56–67. doi: 10.1111/j.1749-6632.2009.05066.x. [DOI] [PubMed] [Google Scholar]
- 22.Craig-Schapiro R, Kuhn M, Xiong C, et al. Multiplexed Immunoassay panel identifies novel CSF biomarkers for Alzheimer’s disease diagnosis and prognosis. [Accessed May 26, 2012];PLoS One. 2011 6(4):e18850. doi: 10.1371/journal.pone.0018850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu WT, Chen-Plotkin A, Arnold SE, et al. Novel CSF biomarkers for Alzheimer’s disease and mild cognitive impairment. Acta Neuropathol. 2010;119(6):669–678. doi: 10.1007/s00401-010-0667-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(3):839–843. doi: 10.1148/radiology.148.3.6878708. [DOI] [PubMed] [Google Scholar]
- 25.Zhang R, Barker L, Pinchev D, et al. Mining biomarkers in human sera using proteomic tools. Proteomics. 2004;4(1):244–256. doi: 10.1002/pmic.200300495. [DOI] [PubMed] [Google Scholar]
- 26.Laske C, Leyhe T, Stransky E, Hoffmann N, Fallgatter AJ, Dietzsch J. Identification of a blood-based biomarker panel for classification of Alzheimer’s disease. Int J Neuropsychopharmacol. 2011;14(9):1147–1155. doi: 10.1017/S1461145711000459. [DOI] [PubMed] [Google Scholar]
- 27.Csemansky JG, Dong H, Fagan AM, et al. Plasma Cortisol and progression of dementia in subjects with Alzheimer-type dementia. Am J Psychiatry. 2006;163(12):2164–2169. doi: 10.1176/appi.ajp.163.12.2164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gupta VB, Laws SM, Villemagne VL, et al. AIBL Research Group. Plasma apolipoprotein E and Alzheimer disease risk: the AIBL study of aging. Neurology. 2011;76(12):1091–1098. doi: 10.1212/WNL.0b013e318211c352. [DOI] [PubMed] [Google Scholar]
- 29.Laske C, Stransky E, Fritsche A, Eschweiler GW, Leyhe T. Inverse association of Cortisol serum levels with T-tau, P-tau 181 and P-tau 231 peptide levels and T-tau/Abeta 1-42 ratios in CSF in patients with mild Alzheimer’s disease dementia. Eur Arch Psychiatry Clin Neurosci. 2009;259(2):80–85. doi: 10.1007/s00406-008-0838-3. [DOI] [PubMed] [Google Scholar]
- 30.Bruno D, Nierenberg JJ, Ritchie JC, Lutz MW, Pomara N. Cerebrospinal fluid Cortisol concentrations in healthy elderly are affected by both APOE and TOMM40 variants. Psychoneuroendocrinology. 2012;37(3):366–371. doi: 10.1016/j.psyneuen.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.degli Uberti EC, Bondanelli M, Margutti A, et al. Acute administration of human galanin in normal subjects reduces the potentiating effect of pyridostigmine-induced cholinergic enhancement on release of norepinephrine and pancreatic polypeptide. Neuroendocrinology. 1996;64(5):398–404. doi: 10.1159/000127143. [DOI] [PubMed] [Google Scholar]
- 32.Nishikimi T, Kuwahara K, Nakao K. Current biochemistry, molecular biology, and clinical relevance of natriuretic peptides. J Cardiol. 2011;57(2):131–140. doi: 10.1016/j.jjcc.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 33.Prado J, Baltrons MA, Pifarré P, García A. Glial cells as sources and targets of natriuretic peptides. Neurochem Int. 2010;57(4):367–374. doi: 10.1016/j.neuint.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 34.Kerola T, Nieminen T, Hartikainen S, Sulkava R, Vuolteenaho O, Kettunen R. B-type natriuretic peptide as a predictor of declining cognitive function and dementia—a cohort study of an elderly general population with a 5-year follow-up. Ann Med. 2010;42(3):207–215. doi: 10.3109/07853891003652542. [DOI] [PubMed] [Google Scholar]
- 35.Kondziella D, Göthlin M, Fu M, Zetterberg H, Wallin A. B-type natriuretic peptide plasma levels are elevated in subcortical vascular dementia. Neuroreport. 2009;20(9):825–827. doi: 10.1097/WNR.0b013e328326f82f. [DOI] [PubMed] [Google Scholar]
- 36.Buerger K, Uspenskaya O, Hartmann O, et al. Prediction of Alzheimer’s disease using midregional proadrenomedullin and midregional proatrial natriuretic peptide: a retrospective analysis of 134 patients with mild cognitive impairment. J Clin Psychiatry. 2011;72(4):556–563. doi: 10.4088/JCP.09m05872oli. [DOI] [PubMed] [Google Scholar]
- 37.Riddell DR, Zhou H, Atchison K, et al. Impact of apolipoprotein E (ApoE) polymorphism on brain ApoE levels. J Neurosci. 2008;28(45):11445–11453. doi: 10.1523/JNEUROSCI.1972-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim J, Jiang H, Park S, et al. Haploinsufflciency of human APOE reduces amyloid deposition in a mouse model of amyloid-β amyloidosis. J Neurosci. 2011;31(49):18007–18012. doi: 10.1523/JNEUROSCI.3773-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Banwell ME, Tolley NS, Williams TJ, Mitchell TJ. Regulation of human eotaxin-3/CCL26 expression: modulation by cytokines and glucocorticoids. Cytokine. 2002;17(6):317–323. doi: 10.1006/cyto.2002.1021. [DOI] [PubMed] [Google Scholar]