Abstract
Base excision repair (BER) corrects DNA damage from oxidation, deamination and alkylation. Such base lesions cause little distortion to the DNA helix structure. BER is initiated by a DNA glycosylase that recognizes and removes the damaged base, leaving an abasic site that is further processed by short-patch repair or long-patch repair that largely uses different proteins to complete BER. At least 11 distinct mammalian DNA glycosylases are known, each recognizing a few related lesions, frequently with some overlap in specificities. Impressively, the damaged bases are rapidly identified in a vast excess of normal bases, without a supply of energy. BER protects against cancer, aging, and neurodegeneration and takes place both in nuclei and mitochondria. More recently, an important role of uracil-DNA glycosylase UNG2 in adaptive immunity was revealed. Furthermore, other DNA glycosylases may have important roles in epigenetics, thus expanding the repertoire of BER proteins.
Base excision repair corrects small DNA lesions (e.g., those caused by oxidation). A DNA glycosylase removes the damaged base; short-patch or long-patch repair then fills the gap.
Base excision repair (BER) corrects small base lesions that do not significantly distort the DNA helix structure. Such damage typically results from deamination, oxidation, or methylation (Fig. 1). Much of the damage is the result of spontaneous decay of DNA (Lindahl 1993), although similar damage may also be caused by environmental chemicals, radiation, or treatment with cytostatic drugs. BER takes place in nuclei, as well as in mitochondria, largely using different isoforms of proteins or genetically distant proteins. The identification of Escherichia coli uracil-DNA glycosylase (Ung) in 1974 by Tomas Lindahl marks the discovery of BER. Lindahl searched for an enzyme activity that would act on genomic uracil resulting from cytosine deamination. Such an activity was found, but rather unexpectedly, it was not a nuclease. Instead, Lindahl identified an enzyme that cleaved the bond between uracil and deoxyribose. The resulting abasic site (AP-site) was suggested to be further processed by an AP-endonuclease, an exonuclease, a DNA polymerase, and a ligase. Thus, the fundamental steps in the BER pathway were outlined already in the very first paper (Lindahl 1974). Enzymes that cleave the bond between deoxyribose and a modified or mismatched DNA base are now called DNA glycosylases. Collectively these enzymes initiate base excision repair of a large number of base lesions, each recognized by one or a few DNA glycosylases with overlapping specificities.
Figure 1.
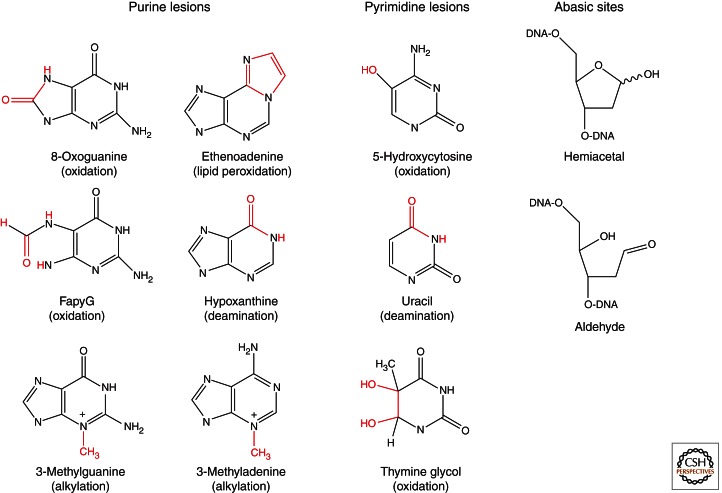
Chemistry of common base lesions and abasic sites.
This relatively brief review focuses on recent advances in the mechanism and function of BER with a focus on mammalian proteins. The current view is that BER is important in relation to cancer, neurodegeneration, and aging (Jeppesen et al. 2011; Wallace et al. 2012). Because of limited space, we have referred to reviews for the majority of results published more than 6–7 years ago. Also, for more detailed analyses of different aspects of BER, the reader is referred to excellent reviews on BER proteins and pathways published in Huffman et al. (2005), Beard and Wilson (2006), Berti and McCann (2006), Cortázar et al. (2007), Kavli et al. (2007), Sousa et al. (2007), Tubbs et al. (2007), Berger et al. (2008), Robertson et al. (2009), Friedman and Stivers (2010), Wilson et al. (2010), Svilar et al. (2011), and Jacobs and Schar (2012).
OVERVIEW OF THE BER PATHWAY
The BER pathway is initiated by one of at least 11 distinct DNA glycosylases, depending on the type of lesion (Table 1). The subsequent steps, incision, end processing, repair synthesis, and ligation are usually referred to as “common steps” but in reality take place by different mechanisms, depending on the type of glycosylase and physiological state of the cell. To what extent damage signaling is required before base excision is still a matter of debate.
Table 1.
Mammalian DNA glycosylases
| Enzyme | Subcellular localization | Mono-/ bifunct. | Substrates and (minor substrates) | Mouse knockout | Human diseasea |
|---|---|---|---|---|---|
| UNG2 | Nuclei | M | U, 5-FU in ss and dsDNA, U:A and U:G context (alloxan, 5-hydroxyuracil, isodialuric acid) | Partial defect in CSR, skewed SHM, B-cell lymphomas | Complete defect in CSR, HIGM syndrome, infections, lymphoid hyperplasia |
| UNG1 | Mitochondria | M | Like UNG2 | Unknown | Unknown |
| SMUG1 | Nucleus | M | 5-hmU, U:G > U:A > ssU, 5-FU, εC in ss and dsDNA | Viable and fertile, SMUG1/UNG/MSH triple k.o. reduced longevity | Unknown |
| TDG | Nucleus | M | U:G > T:G (5-hmU in dsDNA, 5-FU) | Embryonic lethal, epigenetic role in development | Unknown |
| MBD4 (MED1) | Nucleus | M | U:G and T:G, 5-hmU in CpG context (εC, 5-FU in dsDNA) | Viable and fertile, C to T transitions, intestinal neoplasia | Mutated in carcinomas with microsatellite instability |
| MPG (AAG) | Nucleus | M | 3meA, 7meG, 3meG, Hx, εA | Viable and fertile, triple knockouts in MPG/ AlkBH2/AlkBH3 hypersensitive to inflammatory bowel disease | Unknown |
| OGG1 | Nucleus | M/B | 8-oxoG:C, Fapy:C | Viable and fertile, OGG1/MUTYH double knockouts cancer prone | OGG1 activity associated with CAG repeat expansion in Huntington’s disease |
| MUTYH | Nucleus | M | A opposite 8-oxoG/C/G | OGG1/MUTYH double knockouts cancer prone | MUTYH variants associated with colon polyposis |
| NTHL1 | Nucleus | B | Tg, FapyG, 5-hC, 5-hU in dsDNA | Viable and fertile, NTHL1/NEIL1 double knockouts cancer prone | Unknown |
| NEIL1 | Nucleus | B | Tg, FapyG, FapyA, 8-oxoG, 5-hU, DHU, Sp and Gh in ss and dsDNA | Viable and normal at birth, obese after 7 months, NTHL1/NEIL1 double knockouts cancer prone | Unknown |
| NEIL2 | Nucleus | B | Similar to NEIL1 | Unknown | Unknown |
| NEIL3 | Nucleus | M/B | FapyG, FapyA, Sp and Gh in ssDNA | Viable and fertile, memory and learning deficit | Unknown |
aAssociation of germline alterations in DNA glycosylases NTHL1, NEIL1, NEIL2, MPG, TDG, UNG, and SMUG1 with colorectal cancer has been reported, but significance uncertain (Broderick et al. 2006). bifunct., bifunctional; k.o., knockout.
Base Removal
A damage-specific DNA glycosylase removes the flipped out damaged base, leaving an abasic site (AP-site) (Figs. 2 and 3A). DNA glycosylases are generally well conserved in evolution, but there are striking exceptions to this (e.g., in alkylation repair), where no prokaryote homolog of mammalian methyl purine DNA glycosylase (MPG, also called AAG or ANPG) has been identified. Furthermore, the evolutionary conservation of glycosylases is largely limited to the enzymatic core domain. Mammalian DNA glycosylases, in addition, have amino- and/or carboxy-terminal extensions that are not found in prokaryotic counterparts (Hegde et al. 2010). These extensions are usually disordered and are required for subcellular targeting, interaction with other proteins (Nilsen et al. 1997; Otterlei et al. 1999), and even target DNA recognition (Hegde et al. 2010).
Figure 2.
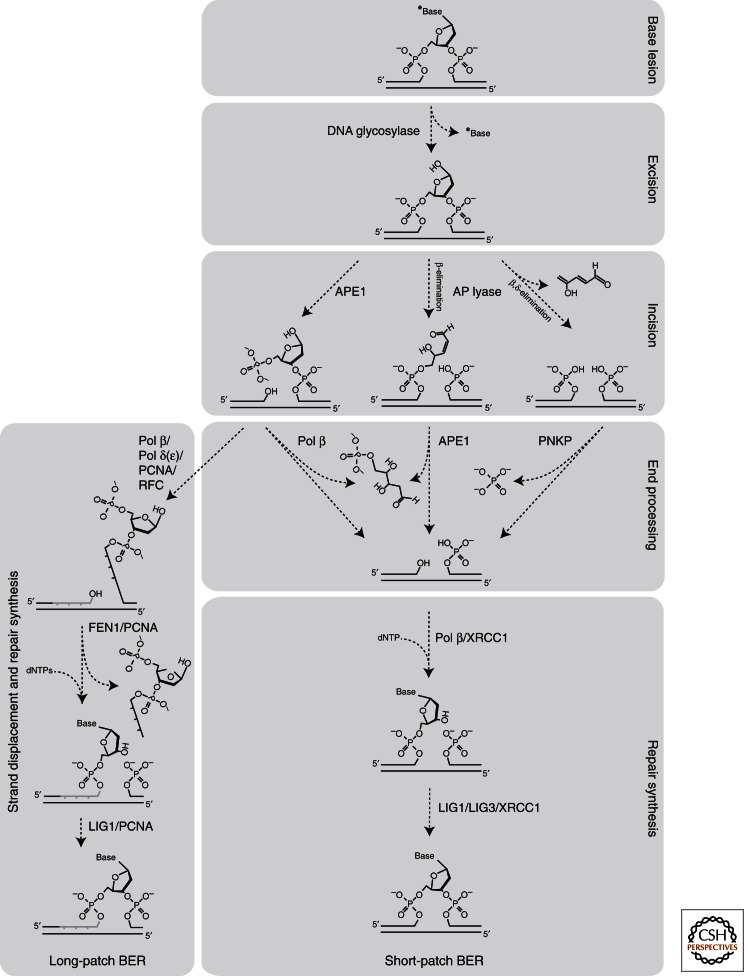
Subpathways in base excision repair (BER). BER takes place by short-patch repair or long-patch repair that largely use different proteins downstream of the base excision. The repair process takes place in five core steps: (1) excision of the base, (2) incision, (3) end processing, and (4) repair synthesis, including gap filling and ligation.
Figure 3.
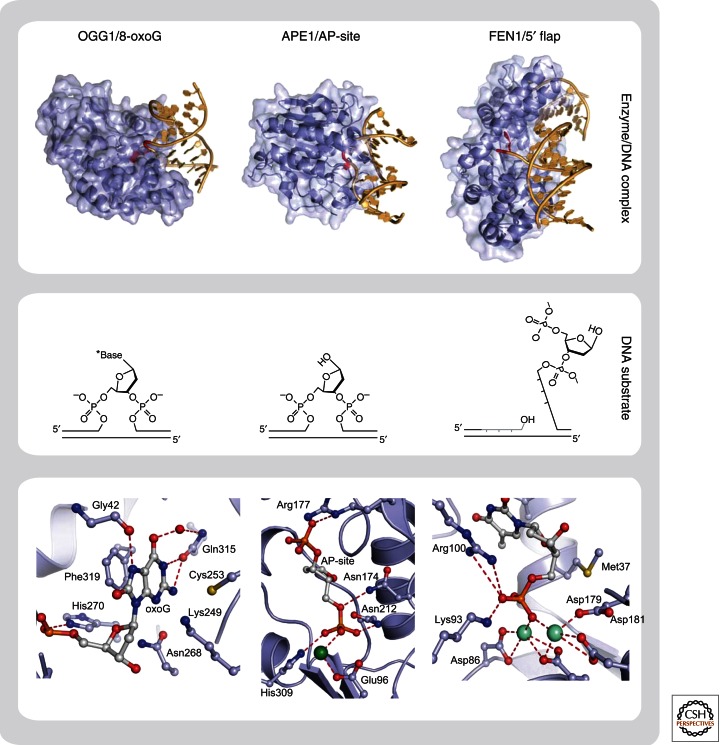
Structural basis for interaction of BER enzymes with their DNA substrates. Overall structure (upper panel) and close-up of active site (lower panel) of three different repair proteins in complex with their DNA substrates. Recognition and extrahelical flipping of damaged base (8-oxoG) by DNA glycosylase (OGG1) (Radom et al. 2007), PDB code 2NOZ; binding of AP-site by AP-endonuclease (APE1) (Mol et al. 2000), PDB code 1DE8; and recognition of 5′ flap by structure specific endonuclease (FEN1) (Tsutakawa et al. 2011), PDB code 3Q8K. The chemical structures of DNA substrates are shown in the middle panel.
Cleavage of the AP-Site, Gap Filling, and Ligation
An essential role of BER has been documented by inactivating functions of proteins in the common steps of BER. Thus, the major AP-endonuclease in mammalian cells, APE1, is essential for survival, as shown using knockout mice (Friedberg and Meira 2006). APE1 (also called HAP1 and Apex) carries both an AP-endonuclease activity and a redox function required for activation of several transcription factors. It also protects against oxidative stress and it is not clear which function is the most important one for survival (Tell et al. 2009). However, Pol-β null mutants are hypersensitive to alkylating agents producing substrates for MPG (Sobol and Wilson 2001) and disruption of the Pol β gene is embryonic lethal in mice (Gu et al. 1994). This supports the view that the BER pathway is an essential DNA repair mechanism because this polymerase has no known role in DNA replication. Recent evidence has indicated that DNA ligase I, rather than DNA ligase 3, may be the major nuclear DNA ligase both in short-patch BER and long-patch BER, whereas DNA ligase 3 is essential in mitochondria (Gao et al. 2011; Simsek et al. 2011). This surprising finding is discussed in more detail below.
Overlapping Functions of BER Proteins and DNA Repair Pathways
Mice with targeted disruptions of a DNA glycosylase (“knockout mice”) are generally viable and fertile with only moderately increased mutation frequencies and no overt early disease phenotype (Table 1) (Friedberg and Meira 2006; Vartanian et al. 2006). As an exception, thymine DNA glycosylase (TDG) knockouts are embryonic lethal, probably because TDG also has an epigenetic function in regulation of DNA methylations (Cortázar et al. 2011; Cortellino et al. 2011). The limited effect of single-glycosylase knockout is probably best explained by overlapping substrate recognition of DNA glycosylases, as well as overlap between BER and other repair pathways. As could be expected, the phenotypes are enhanced in double or triple knockouts affecting backup functions. Thus, double-knockout mice deficient in OGG1 (8-oxoguanine DNA glycosylase) and MUTYH (MutY homolog) are strongly cancer prone and have shortened life spans (Xie et al. 2004), a finding that makes sense because MUTYH removes adenines that have been misincorporated opposite 8-oxoG. NTHL1 (E. coli nth endonuclease III-like 1) and NEIL1/2 (nei endonuclease VIII-like 1/2) that mainly repair oxidized pyrimidines and ring-opened purines have overlapping substrate specificities and serve as backups for each other. Double knockouts develop lung and liver tumors (see also Table 1) (Chan et al. 2009). Surprisingly, double knockouts in UNG/SMUG1 (single-strand selective monofunctional uracil glycosylase) are viable without an overt early phenotype. However, triple knockout mice deficient in uracil glycosylases UNG and SMUG1 and mismatch repair protein MSH2 have substantially shortened life spans and die mostly from lymphomas (Kemmerich et al. 2012). Most likely, MMR serves as a last major option in repair of genomic uracil. Several translesion bypass polymerases function as backup for the single mammalian DNA glycosylase removing 3-methyladenine, methylpurine DNA glycosylase (MPG, also called AAG) (Johnson et al. 2007; Monti et al. 2008). These results show extensive backup within and between pathways processing base damage.
Does Poly(ADP)-Ribose Polymerase 1 (PARP1) Have a Role in BER?
PARP1 is the founding member of a large family of proteins involved in cellular signaling (Gibson and Kraus 2012). PARP1 poly(ADP)-ribosylates numerous proteins, including itself, and has a role in DNA damage sensing and recruitment of DNA repair proteins (De Vos et al. 2012; Gibson and Kraus 2012). Although mice deficient in PARP1 have no overt phenotype and are fertile, they are sensitive to DNA-damaging alkylating agents and γ-irradiation that cause a variety of lesions, including damage that is recognized by BER (Wang et al. 1995; de Murcia et al. 1997). Furthermore, PARP1-deficient cells are sensitive to methyl methanesulfonate (MMS) that causes DNA damage recognized by BER (Horton and Wilson 2007). PARP1 has a distinct role as a DNA damage sensor in single-strand break repair (SSBR) (Fisher et al. 2007) and protects against generation of excessive SSBs during BER (Parsons et al. 2005a; Woodhouse et al. 2008). However, its role in BER signaling and recruitment of repair proteins is not settled (De Vos et al. 2012). Recent evidence indicates that PARP1 may not be required for BER of alkylation lesions. In these experiments, PARP1-depleted cells were exposed to the alkylating agent dimethyl sulfate. It was confirmed that SSBR was affected, but BER was not (Strom et al. 2011). Thus, PARP1 is present at SSBs and is important for SSBR, but its role in BER remains unclear. One explanation for the divergent results may be that PARP1 is only required for some of the base lesions repaired by BER. Furthermore, it is not obvious that SSBs formed directly by genotoxic agents are biologically equivalent to those formed as intermediates in BER, although they may be chemically similar at the DNA level. Furthermore, it remains unclear whether specific DNA damage signaling is required to recruit BER proteins or whether the damage is simply identified by a DNA glycosylase scanning DNA.
Chromatin Remodeling in BER
Chromatin remodeling is possibly important for most DNA transactions, including DNA repair. As one example, ATP-dependent chromatin remodeling by the classical SWI/SNF complex was found to be required for in vitro repair of 8-oxoG. This was caused by strong inhibition of OGG1 and APE1 by histone octamers (Menoni et al. 2007). The emerging picture is that chromatin remodeling is important in BER, perhaps requiring different complexes in early and late stages (Nakanishi et al. 2007; Odell et al. 2013).
DNA GLYCOSYLASES—GENERAL PROPERTIES AND SUBSTRATE RECOGNITION
Presently, at least 11 different mammalian DNA glycosylases are known, not including mitochondrial isoforms usually resulting from alternative splicing (Svilar et al. 2011; Jacobs and Schar 2012). They are generally relatively small, positively charged, and most frequently single-domain proteins. Each DNA glycosylase recognizes a small number of related base lesions, although the degree of selectivity varies (Table 1). Some DNA glycosylases recognize a damaged or inappropriate base both in a base pair, in a mismatch, and even in single-stranded DNA (e.g., UNG, SMUG1, and NEIL1). Others are highly specific for base lesions in double-stranded DNA and are inactive on single-stranded DNA (e.g., TDG, MBD4 [methyl binding domain 4 protein], OGG1, and MUTYH) (Robertson et al. 2009; Svilar et al. 2011; Jacobs and Schar 2012). For some types of mismatched bases (e.g., T:G, 5-FU:G, and possibly U:G) mismatch repair can be an alternative to BER (Fischer et al. 2007; Pettersen et al. 2011). As an example, repair of U:G mismatches is predominantly by BER, but the mismatch repair system may serve as a backup, at least if a DNA strand break is present not too far from the U:G mismatch, as shown in in vitro experiments with cell extracts (Schanz et al. 2009) and indicated from studies on knockout mice (Kemmerich et al. 2012).
DNA glycosylases are remarkable by being able to find and remove bases carrying minor lesions hidden in undistorted DNA and in the presence of a very large excess of normal bases. The active site of DNA glycosylases can only accommodate an extrahelical base (Fig. 3A). Such binding of a flipped out base applies to several other types of DNA repair proteins, including AlkB type oxidative demethylases (Sundheim et al. 2008; Yang et al. 2008) and O6-guanine alkyl transferase (Tubbs et al. 2007), as well as DNA-modifying enzymes, such as cytosine 5-methyltransferase (Bashtrykov et al. 2012). DNA glycosylases bind to the minor groove, kink DNA at the site of damage and flip the damaged base out of the major groove (Huffman et al. 2005). It remains controversial whether the glycosylase performs active base flipping (Slupphaug et al. 1996; Huffman et al. 2005) or merely captures the base when it is temporarily in an extrahelical state (Cao et al. 2004; Stivers 2008).
DNA glycosylases are called monofunctional if they have glycosylase activity only (e.g., UNG), thus producing an AP-site. Bifunctional glycosylases, in addition to the glycosylase activity, have a β-lyase activity (e.g., NTHL1) or a β,δ-lyase activity (e.g., NEIL1) that cleaves the DNA strand by β-elimination and β/δ-elimination, respectively, leaving ends that require tailoring before BER can proceed (Fig. 2) (Svilar et al. 2011; Jacobs and Schar 2012). All mammalian DNA glycosylases that remove uracil are monofunctional. Other monofunctional glycosylases include MPG and MUTYH, that removes A mispaired to 8-oxoG, G, or C. In contrast, most glycosylases that are specialized for removal of oxidized bases (NTHL1, NEIL1-3) are bifunctional.
SIX DIFFERENT PROTEIN FOLDS HAVE ACQUIRED DNA GLYCOSYLASE ACTIVITIES
Through convergent evolution, six structural superfamilies have acquired DNA glycosylase activities (for review see Dalhus et al. 2009). The HhH DNA glycosylase superfamily is widespread in all kingdoms of life and a number of subfamilies are well characterized (Denver et al. 2003). Some of these have additional domains or contain characteristic cofactors, in particular, an [Fe4S4]2+ cluster. The archetypical H2TH DNA glycosylase contains an amino-terminal domain with an α-helix carrying the catalytic residue Pro2 and a two-layered β-sandwich structure. The carboxy-terminal domain is built from α-helices, in addition to two β-strands forming a zinc finger (Sugahara et al. 2000). The third and fourth classes of DNA glycosylases are the alkyladenine DNA glycosylase (MPG/AAG) and uracil DNA glycosylase (UDG) superfamilies, named after two well-characterized human DNA glycosylases. Both MPG and UDG DNA glycosylases are compact single-domain enzymes. A comprises an antiparallel β-sheet surrounded by α-helices (Lau et al. 1998). This superfamily was originally believed to be limited to mammals, but has now been characterized also in prokaryotes (Aamodt et al. 2004). The members of the UDG superfamily contain a core of a four-stranded parallel twisted β-sheet flanked by α-helices (Mol et al. 1995). The most recently discovered DNA glycosylase superfamily (Alseth et al. 2006) comprises enzymes built entirely from α-helices in a solenoid-like superhelix fold and has been termed the HEAT-like repeat (HLR) DNA glycosylase superfamily (Dalhus et al. 2007). The HLR DNA glycosylases are widespread in prokaryotes, but have not yet been found in multicellular organisms. Finally, a sixth DNA glycosylase superfamily that is present in bacteriophages hydrolyzes the N-glycosylic bond of the 5′ base in cyclobutane pyrimidine dimers. The 3D structures of enzymes from all six DNA glycosylase superfamilies have been solved during the last 20 years. These structures show that although these enzymes have evolved from different ancestors and target a wide range of base lesions, there are some common mechanisms for the action of all DNA glycosylases.
SHORT-PATCH AND LONG-PATCH BER
After initiation of BER by a DNA glycosylase, further processing may take place by “short-patch” BER (also called “single-nucleotide BER”), in which a single nucleotide gap is generated and subsequently filled and ligated, or by long-patch BER in which a gap of 2–10 nucleotides is generated and filled (Fig. 2A) (as reviewed in Lindahl 2001; Almeida and Sobol 2007; Fortini and Dogliotti 2007; Robertson et al. 2009; Svilar et al. 2011; Wallace et al. 2012). Although short-patch BER is generally the dominant pathway, long-patch BER may be the dominant mechanism of postreplicative BER initiated by, for example, UNG2 (Otterlei et al. 1999) or NEIL1 (Hegde et al. 2008b), both expressed at highest levels during S phase.
Short-patch BER requires several repair-specific proteins that do not participate in replication and is equally efficient in proliferating and nonproliferating cells. The major core proteins required in the different steps in short-patch repair are an initiating DNA glycosylase, AP-endonuclease APE1, DNA polymerase β (Pol β), and DNA ligase I or III (LIG1/3), (Figs. 2 and 3). In addition, poly(ADPribose) polymerase 1 (PARP1) and XRCC1 participate, but apparently not in all types of short-patch BER (Hanssen-Bauer et al. 2011). Many other proteins also have roles in BER, including damage signaling, tailoring of ends in the gap, posttranscriptional modifications and chromatin remodeling.
Long-patch repair is mainly taking place in proliferating cells and uses, to a large extent, replication proteins for processing subsequent to the glycosylase action and strand cleavage by APE1 (Figs. 2 and 3). These include DNA polymerase δ/ε, PCNA, FEN1, and LIG1 (Svilar et al. 2011). However, incorporation of the second nucleotide during repair is apparently independent on proliferation status of the cell and uses Pol β in nonproliferating cells, but preferentially Pol δ/ε in proliferating cells (Akbari et al. 2009). The choice of pathway is also depending on the specificity of the initiating glycosylase (Fortini et al. 1999), as well as the cell type and availability of BER factors (Narciso et al. 2007; Bauer et al. 2011; Tichy et al. 2011). The abundant nonhistone protein high mobility group box 1 (HMGB1) is a chromatin protein that binds to DNA and regulates chromatin structure, gene expression, and several DNA repair pathways, including BER (Liu et al. 2010b). HMGB1 stimulates the rate of BER through interaction with BER proteins APE1 and FEN1 and appears to direct repair to the long-patch pathway (Prasad et al. 2007; Liu et al. 2010b).
The enzymology of the downstream steps in BER depends on whether the DNA glycosylase is monofunctional, bifunctional with β-lyase activity, or bifunctional with β/δ lyase activity (Fig. 2). If the glycosylase is monofunctional, the AP-endonuclease APE1 cleaves the DNA strand 5′ of the abasic site, generating 3′OH and 5′deoxyribose phosphate (5′dRP) ends. Then Pol β inserts the correct nucleotide while its phosphodiesterase activity (dRPase) removes 5′dRP, generating the 5′phosphate end required for ligation (Beard and Wilson 2006; Hegde et al. 2008a; Robertson et al. 2009; Svilar et al. 2011) (Fig. 2). Although the polymerase and phosphodiesterase activities are in separate domains (Beard and Wilson 2006), the downstream strand containing the 5′phosphate end increases nucleotide incorporation several-fold both for Pol β and Pol λ, a closely related X-family member. Interestingly, this is because of an increased incorporation rate rather than increased nucleotide binding affinity (Duym et al. 2006).
If the damaged base is removed by a DNA glycosylase with associated β-lyase activity (OGG1 or NTHL1), β-elimination by the lyase generates an unsaturated hydroxyaldehyde linked to the 3′ end (3′dRP) and a phosphate at the 5′ end (Svilar et al. 2011). The 3′dRP is efficiently removed by APE1 that generates a 3′OH end, preparing the intermediate for the polymerase step. A glycosylase with β,δ-lyase activity cleaves at both sides of the AP-site, releasing the unsaturated deoxyribose as trans-4-hydroxy-2,4-pentadienal. This generates a one-nucleotide gap flanked by 3′-phosphate and 5′-phosphate ends (Svilar et al. 2011). The 3′ phosphate can be removed by the bifunctional enzyme polynucleotide kinase/phosphatase (PNKP) that has 5′kinase and 3′phosphatase activities (Fig. 2). Although β,δ-elimination and gap trimming by PNKP circumvents the requirement for APE1, the latter does have a weak 3′ phosphatase activity that may remove the phosphate (Beard et al. 2006).
Pol β IS A MAJOR FAMILY X POLYMERASE IN BER AND IS MUTATED IN A LARGE NUMBER OF EPITHELIAL CANCERS
The major gap-filling polymerase in short-patch BER is Pol β. Two other members of the polymerase family X, Pol µ and TdT, contribute to double-strand break repair but apparently have no role in BER (Nick McElhinny and Ramsden 2004). Pol λ, the closest relative of Pol β, contributes in BER of oxidative DNA damage and is recruited to the site of damage (Braithwaite et al. 2005). However, Pol β was also the major contributor in an 8-oxoG:C context although a smaller contribution of Pol λ was detectable as well (Braithwaite et al. 2010). These results were supported by cell survival studies using single- and double-polymerase knockout mouse embryonic fibroblasts (MEFs). Here a significant role of Pol λ for survival was observed after exposure to H2O2. In contrast, the significance of Pol λ for survival after alkylation damage or incorporation of 5-hydroxymethyldeoxyuridine (hmdUrd) was very minor (Braithwaite et al. 2010). BER of U:G in cell extracts required Pol β and the marginal contribution of Pol λ could only be revealed in a Pol β-knockout background. In conclusion, the current view is that Pol β is the major short-patch BER polymerase. Pol λ has a significant, but modest role in repair of oxidative base lesions. Interestingly, the effects appeared to be additive, indicating that their functions are independent.
Importantly, mutations in the Pol β reading frame have been identified in approximately 30% of human solid cancers (Starcevic et al. 2004), as first observed in colorectal cancers (Wang et al. 1992). Mutations in the catalytic domain, e.g., Ile260Met, may reduce rate, DNA binding, and fidelity, leading to genomic instability and cellular transformation (Nemec et al. 2012). Furthermore, Pol β variants may act as dominant negative and sequence-specific mutators (Lang et al. 2007; Murphy et al. 2012).
LIG1 IS IMPORTANT IN NUCLEAR SHORT-PATCH BER, WHEREAS LIG3 IS ESSENTIAL IN MITOCHONDRIA
Mechanistically, the last step in BER is ligation of the nick after gap filling (Fig. 3). Mammalian cells have three DNA ligases (DNA ligase I, III, and IV). DNA ligase I (LIG1) interacts with PCNA and is required for long-patch repair, and in the joining of Okazaki fragments in replication, but its role in short-patch repair has been unclear. The main function of DNA ligase IV (LIG4) is in repair of double-strand breaks by nonhomologous end joining (NHEJ) and V(D)J recombination, the latter restricted to immature B-cells. LIG4 is not thought to have a role in BER. LIG3 is present both in nuclei and mitochondria, in contrast to LIG1 and LIG4 that are only found in nuclei. Inactivation of murine LIG3 is known to be embryonic lethal, which has made functional studies on intact organisms difficult. In addition, LIG3 is apparently restricted to vertebrates so simpler eukaryotic model systems have not been available. For a long time LIG3 was thought to be the major ligase in short-patch nuclear BER. This was mostly based on the interaction of LIG3 with the scaffolding protein XRCC1, which stabilizes LIG3 and enhances ligation. Furthermore, mammalian cells lacking XRCC1 are hypersensitive to DNA-alkylating agents that produce base lesions that are repaired by BER (Friedberg et al. 2006). However, LIG1 was found to efficiently substitute for the XRCC1–LIG3 complex in short-patch repair in vitro assays, whereas LIG3 could not substitute for LIG1 in long-patch repair (Sleeth et al. 2004). Furthermore, it was recently reported that LIG1 is the major short-patch BER polymerase for nuclear DNA repair, whereas LIG3 is essential for mtDNA repair and cell survival (Gao et al. 2011; Simsek et al. 2011). Using a mouse model in which Lig3 was conditionally inactivated either in the nervous system (Lig3Nes-cre) or heart and skeletal muscle (Lig3Ckmm-cre), mice were born alive and were indistinguishable from littermates. However, they displayed growth retardation, disrupted neurogenesis, and ataxia already after 2 weeks and died within 3 weeks. Morphological abnormalities in mitochondria, as well as dramatically reduced content of mtDNA were observed (Gao et al. 2011). Similarly, Lig3Ckmm-cre mice displayed abnormal heart and muscle morphology and heart failure with death within 3.5 and 4.5 weeks of age (Gao et al. 2011). Thus, these mice displayed massive mitochondrial dysfunction as a consequence of LIG3 deficiency, likely affecting mtDNA replication and/or repair. By complementing conditional Lig3 knockouts by different forms of LIG3, the significance of the mitochondrial form of LIG3 could be confirmed (Simsek et al. 2011). In addition, the mitochondrial ligase function could also be performed by LIG1 tailored to enter mitochondria, as well as a viral ligase, suggesting that it is the ligase activity that is essential, not the type of ligase (Simsek et al. 2011). This is in accordance with the finding that the ligation step in in vitro BER in mitochondrial extracts from human cells is stimulated by the addition of bacteriophage T4 DNA ligase (Akbari et al. 2008). Furthermore, whereas nuclear BER apparently takes place in large protein complexes (Akbari et al. 2004, 2007; Hanssen-Bauer et al. 2011), there is so far no evidence for the existence of mitochondrial BER complexes (Akbari et al. 2007), making it plausible that the type of DNA ligase activity is not critical. Surprisingly, nuclear DNA repair, as assessed with alkaline comet analysis, was not affected in cultured quiescent astrocytes from Lig3Ckmm-cre mice after challenge with H2O2 or ionizing radiation or methyl methane sulfonate (MMS). Knockdown of Lig3 in MEFs with shRNA gave similar results. In contrast, Lig1 knockdown seriously affected nuclear DNA repair, indicating that Lig1 is the major BER ligase. Lig1/Lig3 double knockdown was even more deleterious, indicating that LIG3 also contributes, but in a minor way. These results were probably not caused by a switch from short-patch to long-patch repair, as inhibition of the presumed long-patch polymerases (Pol δ or ε) by aphidicolin had no effect. These results show the crucial role of LIG3 in mtDNA maintenance and cell survival and further suggest that LIG1, rather than LIG3, is the major ligase in nuclear BER. However, there is also ample evidence for a role of LIG3 in nuclear BER. More research on repair of different types of base lesions, other cell types, as well as other mammalian species, is therefore warranted before a firm conclusion on the roles of LIG1 and LIG3 in nuclear BER can be made.
BER IN MITOCHONDRIA TAKES PLACE BY SHORT-PATCH REPAIR AND LONG-PATCH REPAIR
Although it has been known for a long time that mitochondria can perform BER, it was previously thought that mitochondria can only perform short-patch repair (Stierum et al. 1999). This repair would use a DNA glycosylase, APE1, Pol γ, and LIG3 (Liu and Demple 2010). However, three papers later, long-patch repair in mitochondria independently documented (Akbari et al. 2008; Liu et al. 2008; Szczesny et al. 2008). Whereas the possible role of FEN1 was initially controversial, the current view is that FEN1 has a role by acting synergistically with human structure-specific nuclease/helicase DNA2, which in human cells is a mitochondrial protein (Zheng et al. 2008; Liu and Demple 2010).
URACIL IN DNA FROM CYTOSINE DEAMINATION AND dUTP MISINCORPORATION
Genomic Uracil from dUTP Misincorporation and Chemical Deamination of Cytosine
Small amounts of uracil in DNA result from cytosine deamination and incorporation of dUTP instead of dTTP during replication, giving rise to U:A pairs. Spontaneous cytosine deamination results from the inherent instability of the base and introduces highly mutagenic U:G mismatches. Uracil in DNA is not a replication-blocking lesion and is easily copied by replicative DNA polymerases, as well as RNA polymerases. U:G mismatches therefore give rise to C:G to T:A transition mutations unless repaired before replication (Friedberg et al. 2006). Unfortunately, we still lack information on the number of spontaneous cytosine deamination events per mammalian genome per day. This is because deamination is 2–3 orders of magnitude higher in single-stranded than in double-stranded DNA and we do not know which fraction of DNA is single stranded at any one time (Kavli et al. 2007). Furthermore, we do not know whether histones and other chromatin proteins may affect deamination rates. dUTP is an intermediate in pyrimidine metabolism, but is normally kept at very low levels by a highly efficient dUTPase (Friedberg et al. 2006). If incorporated instead of dTTP, this results in U:A pairs that are not directly mutagenic, but possibly cytotoxic because some transcription factors cannot bind to target sequences containing U instead of T. U:A pairs are rapidly and efficiently repaired in mammalian cells (Otterlei et al. 1999; Nilsen et al. 2000). However, abasic sites that are intermediates in repair are weakly mutagenic. Therefore, a contribution of A:U pairs to mutagenesis would depend on the number of dUMP residues incorporated relative to the fidelity of repair. Unfortunately, we do not know how many dUMP residues are incorporated per round of replication. First, we do not have methods to directly quantify dUTP incorporation. Second, the incorporation cannot presently be accurately estimated by indirect methods. This is because we do not know how dUTP is distributed between different subcellular and subnuclear compartments, and in addition, information on average pool sizes in cells is limited. However, genomic uracil was threefold lower in nonproliferating hepatocytes, as compared with MEFs from Ung−/− mice, indicating that the largest fraction of genomic uracil may come from dUTP incorporation during replication, rather than from deamination (Andersen et al. 2005). Furthermore, in yeast, abasic sites largely result from uracil-removal by UNG, with misincorporation of dUTP representing the major source of genomic uracil (Guillet and Boiteux 2003). Whereas only weakly mutagenic in wild-type yeast cells, abasic sites from dUTP-incorporation and excision are toxic and a major cause of spontaneous mutations in yeast cells deficient in BER and RAD17-Mec3-Ddc1 (9-1-1) damage checkpoint clamp (Collura et al. 2012). In sum, these results indicate that genomic uracil, including U:G mismatches and U:A pairs, are quantitatively important lesions that must be efficiently and accurately processed.
REPAIR OF URACIL IN DNA
Processing of uracil in DNA is initiated by a DNA glycosylase that generates an abasic site and completed by BER proteins performing the common steps in BER. In mammalian cells, four distinct DNA glycosylases, UNG2, SMUG1, TDG, and MBD4, are known to remove uracil from nuclear DNA. Their properties have been extensively reviewed (Stivers and Drohat 2001; Krokan et al. 2002; Berti and McCann 2006; Kavli et al. 2007; Sousa et al. 2007; Stivers 2008; Robertson et al. 2009; Friedman and Stivers 2010; Svilar et al. 2011; Jacobs and Schar 2012; Wallace et al. 2012). UNG1 is the only mitochondrial uracil-DNA glycosylase known. Mitochondrial UNG1 and nuclear UNG2 are both encoded by the UNG-gene using alternative splicing and alternative promoters. This results in alternative amino-terminal sequences that are required for subcellular localization and protein–protein interactions, principally PCNA and RPA for UNG2 (Kavli et al. 2007). Although UNG2 and SMUG1 have very limited sequence homology, there are clear similarities in the active site, as well as in function and overall structure (Pettersen et al. 2007, 2011. Whereas SMUG1 was originally thought to be found only in mammalian cells, subsequent work has also identified it in some eubacteria (Pettersen et al. 2007; Lucas-Lledo et al. 2011).
Role of Different Uracil-DNA Glycosylases
UNG proteins are found in eukaryotes, eubacteria, and some viruses and are by far the most efficient in terms of catalytic turnover number (Krokan et al. 2002). Nuclear UNG2 is most highly expressed in S phase and has an important function in postreplicative repair of U:A pairs. However, it also appears to the major contributor to U:G repair (Hagen et al. 2008; Visnes et al. 2008). UNG2 is sequentially phosphorylated during S phase and the triple-phosphorylated form is also ubiquitylated and removed, but apparently not by proteasomal degradation. It is present in lower amounts, but not absent in G2, M, and G1 phases (Hagen et al. 2008). The phosphorylation status affects enzyme activity and possibly protein–protein interactions. Interestingly, a germline variant of UNG2, Arg88Cys, has essentially abolished binding of RPA and reduced recruitment to single-stranded DNA, but normal binding to PCNA. However, mutations that disrupted UNG2–PCNA interaction, impaired recruitment of UNG2 to replication foci (Torseth et al. 2012). These results indicate that UNG2 interaction partners regulate subcellular localization and possibly the function of the glycosylase. Strikingly, TDG is oppositely regulated when compared with UNG2 and has the highest level of expression in G1 phase, indicating separate roles regulated by expression pattern (Hardeland et al. 2007). SMUG1 is not cell cycle regulated and, contrary to expectations, appears to contribute less to U:G repair than does TDG, at least in nuclear extracts (Visnes et al. 2008). Interestingly, SMUG1 knockout mice were phenotypically normal, fertile, and had normal life span. Nearly all 5-hmU activity was ablated in SMUG1-deficient mice, confirming its major role as a general 5-hmU-DNA glycosylase. UNG-SMUG1 double knockouts were also viable, whereas UNG-SMUG1-MSH2 triple knockouts had strongly reduced life span and died from lymphomas (Kemmerich et al. 2012). These results also show that the 5-hmU DNA glycosylase activity of MBD4 is likely to be insignificant in most sequence contexts. It may, however, be important in CpG contexts where it may have a role in 5-meC demethylation in a scheme involving hydroxylation of 5-meC and subsequent deamination to 5-hmU and removal by MBD4. Alternatively, 5-hmU could be removed by TDG (Hashimoto et al. 2012a,b). Although these schemes require further exploration, these results and previous work strongly suggest that TDG and MBD4 may have epigenetic functions that may be an integrated part of their function as DNA glycosylases (Cortázar et al. 2011; Cortellino et al. 2011). Interestingly, MBD4 is mutated in a large fraction of human colorectal cancers that display microsatellite instability (MSI) (Riccio et al. 1999).
ADAPTIVE IMMUNITY BY MUTAGENIC PROCESSING OF U:G MISMATCHES—THE ROLE OF BER PROTEINS
Intriguingly, U:G mismatches are also used to create immunoglobulin diversity in adaptive immunity. Here, antigen-activated B lymphocytes express the enzyme activation-induced cytidine deaminase (AID or AICD), a seminal discovery from T. Honjo’s group (Muramatsu et al. 1999). Originally thought to be an RNA cytosine deaminase, convincing evidence now indicates that it is a DNA deaminase (Conticello et al. 2007). Uracil-DNA glycosylase UNG2 has a unique role in the processing of the U:G mismatches generated by AID, and is required for both normal somatic hypermutation (SHM) and class switch recombination (CSR) (Rada et al. 2002; Imai et al. 2003). SHM and CSR also require mismatch repair proteins and AP-endonucleases APE1 and APE2. In contrast, nucleotide excision repair proteins are apparently not required in adaptive immunity (Guikema et al. 2007; Schrader et al. 2007). Furthermore, it is well established that double-strand break repair (DSBR) proteins are essential in late steps of CSR (Soulas-Sprauel et al. 2007). Thus, there is extensive functional overlap between two of the most important defense mechanisms, DNA repair and immunity. AID creates mutagenic U:G mismatches in Ig variable (V) regions and switch (S) regions. If replicated across, U:G mismatches cause G:C to T:A transition mutations. If uracil is removed by UNG2, the abasic site is subject to translesion synthesis, principally by Pol η (eta), giving rise to a wider spectrum of mutations. The properties of UNG2 are uniquely suited for the purpose and cannot be substituted by SMUG1 under physiological conditions (Doseth et al. 2012; Perez-Duran et al. 2012). The role of UNG2, AID, mismatch repair proteins, and NHEJ proteins in adaptive immunity has been extensively reviewed and shall not be further elaborated here (Di Noia and Neuberger 2007; Maul and Gearhart 2010a,b; Stavnezer 2011).
OXIDATIVE DNA BASE LESIONS
Reactive oxygen species (ROS) are generated in all aerobic organisms as by-products of the normal cellular metabolism, and are also formed by exposure to exogenous sources such as pollution, UV-, and ionizing radiation. ROS are highly reactive and can chemically modify many components of the cell via oxidation, including DNA bases. Oxidative DNA base lesions that cause minor structural changes to DNA are mainly repaired by the BER pathway (David et al. 2007; Hegde et al. 2008a; Dalhus et al. 2009). Purines undergo oxidation of the ring atoms leading to various chemical modifications. Most notably, the highly mutagenic guanine derivative 7,8-dihydro-8-oxoguanine (8-oxoG) is formed in large quantities, and the formamidopyrimidine lesions FapyA and FapyG, with opened imidazole rings, are also abundant (Fig. 1). The 8-oxoG lesion has strong miscoding properties because of Hoogsteen pairing and both bacterial and eukaryotic DNA polymerases insert adenine opposite 8-oxoG with high frequencies. Other important oxidation lesions include several premutagenic oxidized pyrimidines such as thymine glycol (Tg), 5-hydroxycytosine (5-hC), 5-hydroxyuracil (5hU), dihydrothymine (DHT), and dihydrouracil (DHU).
REMOVAL OF OXIDATIVE DNA BASE LESIONS—ESSENTIAL BUT ALSO ASSOCIATED WITH TRINUCLEOTIDE EXPANSION
Endonuclease III (Nth), endonuclease VIII (Nei), and formamidopyrimidine DNA glycosylase (Fpg) are DNA glycosylases for removal of oxidized base lesions in E. coli. Nth and Nei mainly recognize and excise oxidized pyrimidine lesions whereas Fpg primarily functions on oxidized purines (Dalhus et al. 2009). Human cells express several DNA glycosylases for removal of oxidized bases, including OGG1 for removal of 8-oxoG and the corresponding ring-fragmented purine formamidopyrimidine derivative FapyG (Aburatani et al. 1997; Bjoras et al. 1997; Lu et al. 1997; Radicella et al. 1997; Roldan-Arjona et al. 1997; Rosenquist et al. 1997), NTH1 for removal of FapyA/FapyG and oxidized pyrimidines (Aspinwall et al. 1997), and NEIL (Nei-like) DNA glycosylase for removal of oxidized pyrimidines and purines. In total, three NEIL glycosylases, termed NEIL1-3, have been identified in human cells (Hazra et al. 2002a,b; Morland et al. 2002; Takao et al. 2002; Dou et al. 2003). NEIL1 has a broad substrate specificity, removing both pyrimidine and purine derived lesions from DNA, whereas NEIL2 is reported to be rather specific for 5-OHU and oxidized derivatives of cytosine, particularly in mismatched double-stranded DNA (DNA bubbles) (Bandaru et al. 2002; Hazra et al. 2002a,b; Morland et al. 2002; Takao et al. 2002; Dou et al. 2003; Parsons et al. 2005b, 2007; Grin et al. 2010). More specifically, NEIL1 primarily recognizes and removes oxidized pyrimidines like 5-OHU, 5-OHC, and TG as well as the ring-saturated DHT and DHU (Bandaru et al. 2002; Hazra et al. 2002a; Morland et al. 2002; Dou et al. 2003). However, it has also been shown that NEIL1 removes 8-oxoG in different contexts such as in double-stranded DNA (dsDNA), (Morland et al. 2002; Forsbring et al. 2009) or close to the 3′ end of DNA single-strand breaks (Parsons et al. 2005b), but not in single-stranded DNA (Dou et al. 2003). NEIL1 and NEIL2 also remove the oxidized guanine lesions guanidinohydantoin (Gh) and spiroiminodihydantoin (Sp) from single- and double-stranded DNA (Hailer et al. 2005; Zhao et al. 2010), 8-oxoadenine (8-oxoA) from duplex DNA when paired with a cytosine (Grin et al. 2010), as well as FapyA and FapyG (Hazra et al. 2002a). Neil3 recognizes the oxidized purines, spiroiminodihydantoin (Sp), guanidinohydantoin (Gh), FapyG, and FapyA, but not 8-oxo-7,8-dihydroguanine (8-oxoG). Neil3 prefers lesions in single-stranded DNA and in bubble structures (Liu et al. 2010a). NEIL1 is also reported to be involved in nucleotide excision repair of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosine (Jaruga et al. 2010).
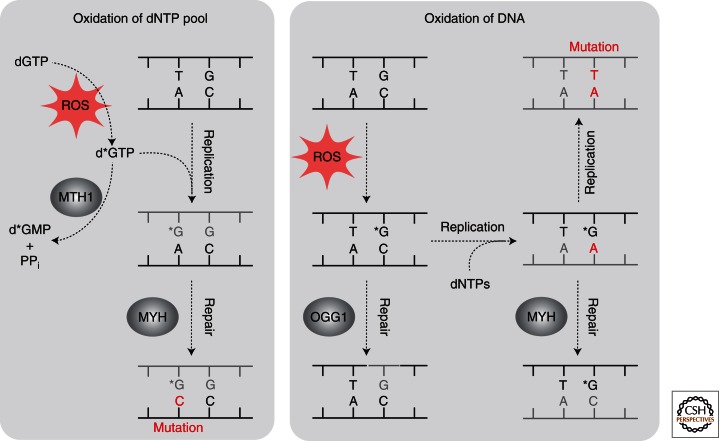
MUTYH is also involved in protection against oxidative damage (Williams and David 1998). It does not remove oxidatively damaged bases as such, but is specific for removal of adenine base-paired with 8-oxoG, which is frequently found as the result of erroneous replication of DNA containing 8-oxoG that has escaped repair by OGG1 (Fig. 4). Inactivation of both MUTYH and OGG1 in mice strongly increased the incidence of lung and small intestinal cancer (Russo et al. 2004), supporting a casual role for unrepaired oxidized bases in cancer development. Inherited variants in the human MUTYH are associated with somatic mutations in colorectal tumors or adenomas (Jones et al. 2002, 2004). The Nth1−/− Neil1−/− mice developed pulmonary and hepatocellular tumors at a higher incidence whereas the single knockouts, Nth1−/− and Neil1−/−, showed no significant increase in tumor formation (Chan et al. 2009).
Figure 4.
Repair of 8-oxoG (*G) resulting from incorporation of 8-oxo-dGTP (d*GTP) in the dNTP pool (left) and from oxidation of G in DNA (right) require different mechanisms. Most of the 8-oxo-dGTPs in the dNTP pool are hydrolysed by MTH, thus avoiding misincorporation during DNA replication. If 8-oxo-dGTP is misincorporated opposite an A in the template strand and the A is removed by MYH, this repair process would result in a mutation. Alternatively, repair by MMR may restore the correct basepair. Repair of 8-oxoG in DNA (right panel): BER initiated by OGG1 removes the majority of 8-oxoG oxidized in DNA. If 8-oxoG is not repaired before DNA replication, replicative DNA polymerases δ and ε incorporate dAMP opposite 8-oxoG with a high frequency. The resulting 8-oxoG:A is repaired by MYH, restoring the correct basepair. However, if the 8-oxoG:A basepair is not repaired before a second round of DNA replication, it leads to an A:T transversion mutation.
The progressive cognitive decline associated with age-dependent neurodegeneration is proposed to be caused by accumulation of oxidative damage to macromolecules (Barja 2004). High metabolic activity and low levels of antioxidant enzymes make neurons particularly prone to damage by reactive oxygen species. Thus, repair of oxidative DNA damage is essential for normal brain function. NEIL1, NEIL2, OGG1, and NTHL1 expressions are widespread at all ages, whereas NEIL3 expression is restricted to discrete regions harboring stem cell populations (i.e., the subventricular zone, the rostral migratory stream, and the subgranular zone) (Rolseth et al. 2008). UNG, OGG1, and Neil-deficient mice show major increases in infarct size after reversible middle cerebral artery occlusion and reperfusion. NEIL3, in contrast to UNG (Endres et al. 2004), OGG1 (Liu et al. 2011), and NEIL1 (Canugovi et al. 2012), has no significant function in postmitotic cells but rather plays a role in regeneration (Sejersted et al. 2011). It appears that NEIL3 is involved in repair of oxidative damage in proliferating cells in brain such as neuronal progenitors, reactive astrocytes, and activated microglia.
Intriguingly, 8-oxoG processing by OGG1 in trinucleotide repeats of the CAG type is associated with trinucleotide expansion in Huntington’s disease and loss of OGG1 suppresses trinucleotide expansion in a mouse model for Huntington’s disease (Kovtun et al. 2007). A recent report shows that Neil1 DNA glycosylase also contributes to germline and somatic HD CAG repeat expansion (Mollersen et al. 2012). These rather surprising findings show that damage processing by a normal repair protein may be harmful in some sequence contexts in a particular genetic background.
REMOVAL OF ALKYLATED BASES AND ETHENOADDUCTS IN DNA
The primary DNA base lesions arising from endogenous methyl donors and from environmental toxins and chemotherapeutic drugs with alkylating properties comprise a range of N- and O-alkylated DNA bases, including N3- and N7-substituted alkylpurines and O2-subsituted alkylpyrimidines, which are recognized and removed by DNA glycosylases. Of these, 3-methyladenine (3-mA), which blocks replication, is the major cytotoxic alkylating damage. Other common alkylated base products, including N1-substituted purines and O6-methylated guanine, are repaired by direct damage reversal processes. Moreover, several alkylbase DNA glycosylases remove deaminated adenine (inosine) and cyclic ethenoadducts such as 1,N6-ethenoadenine and 3,N4-ethenocytosine.
Five different classes of alkyl DNA glycosylases have been identified, but only one is present in mammalian cells (Dalhus et al. 2009). The 3-mA DNA glycosylase I (E. coli Tag), 3-mA DNA glycosylase II (E. coli AlkA), and the Helicobacter pylori 3-mA DNA glycosylase III (MagIII) all belong to the HhH superfamily described above (Labahn et al. 1996; Hollis et al. 2000; Drohat et al. 2002; Cao et al. 2003; Eichman et al. 2003; O’Brien and Ellenberger 2004; Metz et al. 2007). The human Aag (Lau et al. 1998; Lau et al. 2000) and the prokaryotic HLR 3-mA DNA glycosylases (B. cereus AlkC and AlkD) (Dalhus et al. 2007; Rubinson et al. 2008) represent the two other known folds that have evolved to recognize and remove alkylated bases in DNA.
Chronic inflammation is associated with an increased risk of cancer and generates large quantities of reactive oxygen and nitrogen species (RONS) that leads to oxidation and deamination of DNA bases. In addition, RONS indirectly induce etheno bases and other exocyclic DNA adducts via lipid peroxidation. The AAG glycosylase, which removes ethenoadducts, protects against inflammation-associated colon cancer in mice (Meira et al. 2008). In a chemically induced colitis and colon cancer model in mice, AAG display apparent epistasis with the oxidative demethylases, ALKBH2 and ALKBH3, which repair ethenoadducts by direct reversal (Calvo et al. 2012).
CONCLUDING REMARKS
Our understanding of the mechanism and significance of BER and BER proteins has advanced substantially in the last decade. BER is performed by a set of related pathways that have roles in prevention of cancer, neurodegeneration, and aging. Animal models, particularly knockout mice, combined with biochemical studies, have revealed extensive backup and complementary functions of enzymes involved. These explain the modest effects of single-DNA glycosylase deficiencies. Disease association has been shown by double knockouts, as well as with some gene variants in DNA glycosylases or proteins in the common steps of BER. However, unlike the distinct syndromes associated with defects in nucleotide excision repair, mismatch repair, or double-strand break repair, the disease association in the case of BER is usually more subtle and phenotypically blends in with disease from causes other than DNA repair. Furthermore, BER proteins have been shown to have important roles beyond DNA repair, particularly in adaptive immunity and epigenetics. The interaction BER and other DNA transactions, overall regulation of BER, as well as the regulation of individual proteins remain topics that warrant further work.
ACKNOWLEDGMENTS
This work is supported by The Research Council of Norway, The Norwegian Cancer Association, The Svanhild and Arne Must Fund for Medical Research, and the Cancer Fund at St. Olav’s Hospital, Oslo University Hospital, and Norwegian University of Science and Technology. We also thank Yngve Sejersted and Paul Backe for help in preparing figures, and other members of our laboratories for critical reading of the manuscript.
Footnotes
Editors: Errol C. Friedberg, Stephen J. Elledge, Alan R. Lehmann, Tomas Lindahl, and Marco Muzi-Falconi
Additional Perspectives on DNA Repair, Mutagenesis, and Other Responses to DNA Damage available at www.cshperspectives.org
REFERENCES
- Aamodt RM, Falnes PO, Johansen RF, Seeberg E, Bjoras M 2004. The Bacillus subtilis counterpart of the mammalian 3-methyladenine DNA glycosylase has hypoxanthine and 1,N6-ethenoadenine as preferred substrates. J Biol Chem 279: 13601–13606 [DOI] [PubMed] [Google Scholar]
- Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M 1997. Cloning and characterization of mammalian 8-hydroxyguanine-specific DNA glycosylase/apurinic, apyrimidinic lyase, a functional mutM homologue. Cancer Res 57: 2151–2156 [PubMed] [Google Scholar]
- Akbari M, Otterlei M, Pena-Diaz J, Aas PA, Kavli B, Liabakk NB, Hagen L, Imai K, Durandy A, Slupphaug G, et al. 2004. Repair of U/G and U/A in DNA by UNG2-associated repair complexes takes place predominantly by short-patch repair both in proliferating and growth-arrested cells. Nucleic Acids Res 32: 5486–5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbari M, Otterlei M, Pena-Diaz J, Krokan HE 2007. Different organization of base excision repair of uracil in DNA in nuclei and mitochondria and selective upregulation of mitochondrial uracil-DNA glycosylase after oxidative stress. Neuroscience 145: 1201–1212 [DOI] [PubMed] [Google Scholar]
- Akbari M, Visnes T, Krokan HE, Otterlei M 2008. Mitochondrial base excision repair of uracil and AP sites takes place by single-nucleotide insertion and long-patch DNA synthesis. DNA Repair (Amst) 7: 605–616 [DOI] [PubMed] [Google Scholar]
- Akbari M, Pena-Diaz J, Andersen S, Liabakk NB, Otterlei M, Krokan HE 2009. Extracts of proliferating and non-proliferating human cells display different base excision pathways and repair fidelity. DNA Repair (Amst) 8: 834–843 [DOI] [PubMed] [Google Scholar]
- Almeida KH, Sobol RW 2007. A unified view of base excision repair: Lesion-dependent protein complexes regulated by post-translational modification. DNA Repair (Amst) 6: 695–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I, Rognes T, Lindback T, Solberg I, Robertsen K, Kristiansen KI, Mainieri D, Lillehagen L, Kolsto AB, Bjoras M 2006. A new protein superfamily includes two novel 3-methyladenine DNA glycosylases from Bacillus cereus, AlkC and AlkD. Mol Microbiol 59: 1602–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen S, Heine T, Sneve R, Konig I, Krokan HE, Epe B, Nilsen H 2005. Incorporation of dUMP into DNA is a major source of spontaneous DNA damage, while excision of uracil is not required for cytotoxicity of fluoropyrimidines in mouse embryonic fibroblasts. Carcinogenesis 26: 547–555 [DOI] [PubMed] [Google Scholar]
- Aspinwall R, Rothwell DG, Roldan-Arjona T, Anselmino C, Ward CJ, Cheadle JP, Sampson JR, Lindahl T, Harris PC, Hickson ID 1997. Cloning and characterization of a functional human homolog of Escherichia coli endonuclease III. Proc Natl Acad Sci 94: 109–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandaru V, Sunkara S, Wallace SS, Bond JP 2002. A novel human DNA glycosylase that removes oxidative DNA damage and is homologous to Escherichia coli endonuclease VIII. DNA Repair (Amst) 1: 517–529 [DOI] [PubMed] [Google Scholar]
- Barja G 2004. Free radicals and aging. Trends Neurosci 27: 595–600 [DOI] [PubMed] [Google Scholar]
- Bashtrykov P, Ragozin S, Jeltsch A 2012. Mechanistic details of the DNA recognition by the Dnmt1 DNA methyltransferase. FEBS Lett 586: 1821–1823 [DOI] [PubMed] [Google Scholar]
- Bauer M, Goldstein M, Christmann M, Becker H, Heylmann D, Kaina B 2011. Human monocytes are severely impaired in base and DNA double-strand break repair that renders them vulnerable to oxidative stress. Proc Natl Acad Sci 108: 21105–21110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Wilson SH 2006. Structure and mechanism of DNA polymerase β. Chem Rev 106: 361–382 [DOI] [PubMed] [Google Scholar]
- Beard WA, Prasad R, Wilson SH 2006. Activities and mechanism of DNA polymerase β. Methods Enzymol 408: 91–107 [DOI] [PubMed] [Google Scholar]
- Berger SH, Pittman DL, Wyatt MD 2008. Uracil in DNA: Consequences for carcinogenesis and chemotherapy. Biochem Pharmacol 76: 697–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berti PJ, McCann JA 2006. Toward a detailed understanding of base excision repair enzymes: Transition state and mechanistic analyses of N-glycoside hydrolysis and N-glycoside transfer. Chem Rev 106: 506–555 [DOI] [PubMed] [Google Scholar]
- Bjoras M, Luna L, Johnsen B, Hoff E, Haug T, Rognes T, Seeberg E 1997. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. EMBO J 16: 6314–6322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braithwaite EK, Kedar PS, Lan L, Polosina YY, Asagoshi K, Poltoratsky VP, Horton JK, Miller H, Teebor GW, Yasui A, et al. 2005. DNA polymerase λ protects mouse fibroblasts against oxidative DNA damage and is recruited to sites of DNA damage/repair. J Biol Chem 280: 31641–31647 [DOI] [PubMed] [Google Scholar]
- Braithwaite EK, Kedar PS, Stumpo DJ, Bertocci B, Freedman JH, Samson LD, Wilson SH 2010. DNA polymerases β and λ mediate overlapping and independent roles in base excision repair in mouse embryonic fibroblasts. PLoS ONE 5: e12229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broderick P, Bagratuni T, Vijayakrishnan J, Lubbe S, Chandler I, Houlston RS 2006. Evaluation of NTHL1, NEIL1, NEIL2, MPG, TDG, UNG and SMUG1 genes in familial colorectal cancer predisposition. BMC Cancer 6: 243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calvo JA, Meira LB, Lee CY, Moroski-Erkul CA, Abolhassani N, Taghizadeh K, Eichinger LW, Muthupalani S, Nordstrand LM, Klungland A, et al. 2012. DNA repair is indispensable for survival after acute inflammation. J Clin Invest 122: 2680–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canugovi C, Yoon JS, Feldman NH, Croteau DL, Mattson MP, Bohr VA 2012. Endonuclease VIII-like 1 (NEIL1) promotes short-term spatial memory retention and protects from ischemic stroke-induced brain dysfunction and death in mice. Proc Natl Acad Sci 109: 14948–14953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao C, Kwon K, Jiang YL, Drohat AC, Stivers JT 2003. Solution structure and base perturbation studies reveal a novel mode of alkylated base recognition by 3-methyladenine DNA glycosylase I. J Biol Chem 278: 48012–48020 [DOI] [PubMed] [Google Scholar]
- Cao C, Jiang YL, Stivers JT, Song F 2004. Dynamic opening of DNA during the enzymatic search for a damaged base. Nat Struct Mol Biol 11: 1230–1236 [DOI] [PubMed] [Google Scholar]
- Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW 2009. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 8: 786–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collura A, Kemp PA, Boiteux S 2012. Abasic sites linked to dUTP incorporation in DNA are a major cause of spontaneous mutations in absence of base excision repair and Rad17-Mec3-Ddc1 (9–1-1) DNA damage checkpoint clamp in Saccharomyces cerevisiae. DNA Repair (Amst) 11: 294–303 [DOI] [PubMed] [Google Scholar]
- Conticello SG, Langlois MA, Yang Z, Neuberger MS 2007. DNA deamination in immunity: AID in the context of its APOBEC relatives. Adv Immunol 94: 37–73 [DOI] [PubMed] [Google Scholar]
- Cortázar D, Kunz C, Saito Y, Steinacher R, Schär P 2007. The enigmatic thymine DNA glycosylase. DNA Repair (Amst). 6: 489–504 [DOI] [PubMed] [Google Scholar]
- Cortázar D, Kunz C, Selfridge J, Lettieri T, Saito Y, MacDougall E, Wirz A, Schuermann D, Jacobs AL, Siegrist F, et al. 2011. Embryonic lethal phenotype reveals a function of TDG in maintaining epigenetic stability. Nature 470: 419–423 [DOI] [PubMed] [Google Scholar]
- Cortellino S, Xu J, Sannai M, Moore R, Caretti E, Cigliano A, Le Coz M, Devarajan K, Wessels A, Soprano D, et al. 2011. Thymine DNA glycosylase is essential for active DNA demethylation by linked deamination-base excision repair. Cell 146: 67–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhus B, Helle IH, Backe PH, Alseth I, Rognes T, Bjoras M, Laerdahl JK 2007. Structural insight into repair of alkylated DNA by a new superfamily of DNA glycosylases comprising HEAT-like repeats. Nucleic Acids Res 35: 2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalhus B, Laerdahl JK, Backe PH, Bjoras M 2009. DNA base repair–recognition and initiation of catalysis. FEMS Microbiol Rev 33: 1044–1078 [DOI] [PubMed] [Google Scholar]
- David SS, O’Shea VL, Kundu S 2007. Base-excision repair of oxidative DNA damage. Nature 447: 941–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. 1997. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci 94: 7303–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denver DR, Swenson SL, Lynch M 2003. An evolutionary analysis of the helix-hairpin-helix superfamily of DNA repair glycosylases. Mol Biol Evol 20: 1603–1611 [DOI] [PubMed] [Google Scholar]
- De Vos M, Schreiber V, Dantzer F 2012. The diverse roles and clinical relevance of PARPs in DNA damage repair: Current state of the art. Biochem Pharmacol 84: 137–146 [DOI] [PubMed] [Google Scholar]
- Di Noia JM, Neuberger MS 2007. Molecular mechanisms of antibody somatic hypermutation. Annu Rev Biochem 76: 1–22 [DOI] [PubMed] [Google Scholar]
- Doseth B, Ekre C, Slupphaug G, Krokan HE, Kavli B 2012. Strikingly different properties of uracil-DNA glycosylases UNG2 and SMUG1 may explain divergent roles in processing of genomic uracil. DNA Repair (Amst) 11: 587–593 [DOI] [PubMed] [Google Scholar]
- Dou H, Mitra S, Hazra TK 2003. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J Biol Chem 278: 49679–49684 [DOI] [PubMed] [Google Scholar]
- Drohat AC, Kwon K, Krosky DJ, Stivers JT 2002. 3-Methyladenine DNA glycosylase I is an unexpected helix-hairpin-helix superfamily member. Nat Struct Biol 9: 659–664 [DOI] [PubMed] [Google Scholar]
- Duym WW, Fiala KA, Bhatt N, Suo Z 2006. Kinetic effect of a downstream strand and its 5′-terminal moieties on single nucleotide gap-filling synthesis catalyzed by human DNA polymerase λ. J Biol Chem 281: 35649–35655 [DOI] [PubMed] [Google Scholar]
- Eichman BF, O’Rourke EJ, Radicella JP, Ellenberger T 2003. Crystal structures of 3-methyladenine DNA glycosylase MagIII and the recognition of alkylated bases. EMBO J 22: 4898–4909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endres M, Biniszkiewicz D, Sobol RW, Harms C, Ahmadi M, Lipski A, Katchanov J, Mergenthaler P, Dirnagl U, Wilson SH, et al. 2004. Increased postischemic brain injury in mice deficient in uracil-DNA glycosylase. J Clin Invest 113: 1711–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer F, Baerenfaller K, Jiricny J 2007. 5-Fluorouracil is efficiently removed from DNA by the base excision and mismatch repair systems. Gastroenterology 133: 1858–1868 [DOI] [PubMed] [Google Scholar]
- Fisher AE, Hochegger H, Takeda S, Caldecott KW 2007. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol 27: 5597–5605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsbring M, Vik ES, Dalhus B, Karlsen TH, Bergquist A, Schrumpf E, Bjoras M, Boberg KM, Alseth I 2009. Catalytically impaired hMYH and NEIL1 mutant proteins identified in patients with primary sclerosing cholangitis and cholangiocarcinoma. Carcinogenesis 30: 1147–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortini P, Dogliotti E 2007. Base damage and single-strand break repair: Mechanisms and functional significance of short- and long-patch repair subpathways. DNA Repair (Amst) 6: 398–409 [DOI] [PubMed] [Google Scholar]
- Fortini P, Parlanti E, Sidorkina OM, Laval J, Dogliotti E 1999. The type of DNA glycosylase determines the base excision repair pathway in mammalian cells. J Biol Chem 274: 15230–15236 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Meira LB 2006. Database of mouse strains carrying targeted mutations in genes affecting biological responses to DNA damage Version 7. DNA Repair (Amst) 5: 189–209 [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T 2006. DNA repair and mutagenesis. ASM Press, Washington, DC [Google Scholar]
- Friedman JI, Stivers JT 2010. Detection of damaged DNA bases by DNA glycosylase enzymes. Biochemistry 49: 4957–4967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Katyal S, Lee Y, Zhao J, Rehg JE, Russell HR, McKinnon PJ 2011. DNA ligase III is critical for mtDNA integrity but not Xrcc1-mediated nuclear DNA repair. Nature 471: 240–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BA, Kraus WL 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13: 411–424 [DOI] [PubMed] [Google Scholar]
- Grin IR, Dianov GL, Zharkov DO 2010. The role of mammalian NEIL1 protein in the repair of 8-oxo-7,8-dihydroadenine in DNA. FEBS Lett 584: 1553–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Marth JD, Orban PC, Mossmann H, Rajewsky K 1994. Deletion of a DNA polymerase β gene segment in T cells using cell type-specific gene targeting. Science 265: 103–106 [DOI] [PubMed] [Google Scholar]
- Guikema JE, Linehan EK, Tsuchimoto D, Nakabeppu Y, Strauss PR, Stavnezer J, Schrader CE 2007. APE1- and APE2-dependent DNA breaks in immunoglobulin class switch recombination. J Exp Med 204: 3017–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillet M, Boiteux S 2003. Origin of endogenous DNA abasic sites in Saccharomyces cerevisiae. Mol Cell Biol 23: 8386–8394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen L, Kavli B, Sousa MM, Torseth K, Liabakk NB, Sundheim O, Pena-Diaz J, Otterlei M, Horning O, Jensen ON, et al. 2008. Cell cycle-specific UNG2 phosphorylations regulate protein turnover, activity and association with RPA. EMBO J 27: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD 2005. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 4: 41–50 [DOI] [PubMed] [Google Scholar]
- Hanssen-Bauer A, Solvang-Garten K, Sundheim O, Pena-Diaz J, Andersen S, Slupphaug G, Krokan HE, Wilson DM 3rd, Akbari M, Otterlei M 2011. XRCC1 coordinates disparate responses and multiprotein repair complexes depending on the nature and context of the DNA damage. Environ Mol Mutagen 52: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardeland U, Kunz C, Focke F, Szadkowski M, Schar P 2007. Cell cycle regulation as a mechanism for functional separation of the apparently redundant uracil DNA glycosylases TDG and UNG2. Nucleic Acids Res 35: 3859–3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Liu Y, Upadhyay AK, Chang Y, Howerton SB, Vertino PM, Zhang X, Cheng X 2012a. Recognition and potential mechanisms for replication and erasure of cytosine hydroxymethylation. Nucleic Acids Res 40: 4841–4849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H, Zhang X, Cheng X 2012b. Excision of thymine and 5-hydroxymethyluracil by the MBD4 DNA glycosylase domain: Structural basis and implications for active DNA demethylation. Nucleic Acids Res 40: 8276–8284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S 2002a. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci 99: 3523–3528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra TK, Kow YW, Hatahet Z, Imhoff B, Boldogh I, Mokkapati SK, Mitra S, Izumi T 2002b. Identification and characterization of a novel human DNA glycosylase for repair of cytosine-derived lesions. J Biol Chem 277: 30417–30420 [DOI] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S 2008a. Early steps in the DNA base excision/single-strand interruption repair pathway in mammalian cells. Cell Res 18: 27–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Theriot CA, Das A, Hegde PM, Guo Z, Gary RK, Hazra TK, Shen B, Mitra S 2008b. Physical and functional interaction between human oxidized base-specific DNA glycosylase NEIL1 and flap endonuclease 1. J Biol Chem 283: 27028–27037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde ML, Hazra TK, Mitra S 2010. Functions of disordered regions in mammalian early base excision repair proteins. Cell Mol Life Sci 67: 3573–3587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollis T, Ichikawa Y, Ellenberger T 2000. DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA. EMBO J 19: 758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JK, Wilson SH 2007. Hypersensitivity phenotypes associated with genetic and synthetic inhibitor-induced base excision repair deficiency. DNA Repair (Amst) 6: 530–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffman JL, Sundheim O, Tainer JA 2005. DNA base damage recognition and removal: New twists and grooves. Mutat Res 577: 55–76 [DOI] [PubMed] [Google Scholar]
- Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, et al. 2003. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat Immunol 4: 1023–1028 [DOI] [PubMed] [Google Scholar]
- Jacobs AL, Schar P 2012. DNA glycosylases: In DNA repair and beyond. Chromosoma 121: 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaruga P, Xiao Y, Vartanian V, Lloyd RS, Dizdaroglu M 2010. Evidence for the involvement of DNA repair enzyme NEIL1 in nucleotide excision repair of (5′R)- and (5′S)-8,5′-cyclo-2′-deoxyadenosines. Biochemistry 49: 1053–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeppesen DK, Bohr VA, Stevnsner T 2011. DNA repair deficiency in neurodegeneration. Prog Neurobiol 94: 166–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Yu SL, Prakash S, Prakash L 2007. A role for yeast and human translesion synthesis DNA polymerases in promoting replication through 3-methyl adenine. Mol Cell Biol 27: 7198–7205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S, Emmerson P, Maynard J, Best JM, Jordan S, Williams GT, Sampson JR, Cheadle JP 2002. Biallelic germline mutations in MYH predispose to multiple colorectal adenoma and somatic G:C–>T:A mutations. Hum Mol Genet 11: 2961–2967 [DOI] [PubMed] [Google Scholar]
- Jones S, Lambert S, Williams GT, Best JM, Sampson JR, Cheadle JP 2004. Increased frequency of the k-ras G12C mutation in MYH polyposis colorectal adenomas. Br J Cancer 90: 1591–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kavli B, Otterlei M, Slupphaug G, Krokan HE 2007. Uracil in DNA–general mutagen, but normal intermediate in acquired immunity. DNA Repair (Amst) 6: 505–516 [DOI] [PubMed] [Google Scholar]
- Kemmerich K, Dingler FA, Rada C, Neuberger MS 2012. Germline ablation of SMUG1 DNA glycosylase causes loss of 5-hydroxymethyluracil- and UNG-backup uracil-excision activities and increases cancer predisposition of Ung-/-Msh2-/- mice. Nucleic Acids Res 40: 6016–6025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kovtun IV, Liu Y, Bjoras M, Klungland A, Wilson SH, McMurray CT 2007. OGG1 initiates age-dependent CAG trinucleotide expansion in somatic cells. Nature 447: 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krokan HE, Drablos F, Slupphaug G 2002. Uracil in DNA—occurrence, consequences and repair. Oncogene 21: 8935–8948 [DOI] [PubMed] [Google Scholar]
- Labahn J, Scharer OD, Long A, Ezaz-Nikpay K, Verdine GL, Ellenberger TE 1996. Structural basis for the excision repair of alkylation-damaged DNA. Cell 86: 321–329 [DOI] [PubMed] [Google Scholar]
- Lang T, Dalal S, Chikova A, DiMaio D, Sweasy JB 2007. The E295K DNA polymerase β gastric cancer-associated variant interferes with base excision repair and induces cellular transformation. Mol Cell Biol 27: 5587–5596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau AY, Scharer OD, Samson L, Verdine GL, Ellenberger T 1998. Crystal structure of a human alkylbase-DNA repair enzyme complexed to DNA: Mechanisms for nucleotide flipping and base excision. Cell 95: 249–258 [DOI] [PubMed] [Google Scholar]
- Lau AY, Wyatt MD, Glassner BJ, Samson LD, Ellenberger T 2000. Molecular basis for discriminating between normal and damaged bases by the human alkyladenine glycosylase, AAG. Proc Natl Acad Sci 97: 13573–13578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T 1974. An N-glycosidase from Escherichia coli that releases free uracil from DNA containing deaminated cytosine residues. Proc Natl Acad Sci 71: 3649–3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindahl T 1993. Instability and decay of the primary structure of DNA. Nature 362: 709–715 [DOI] [PubMed] [Google Scholar]
- Lindahl T 2001. Keynote: Past, present, and future aspects of base excision repair. Prog Nucleic Acid Res Mol Biol 68: xvii–xxx [DOI] [PubMed] [Google Scholar]
- Liu P, Demple B 2010. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen 51: 417–426 [DOI] [PubMed] [Google Scholar]
- Liu P, Qian L, Sung JS, de Souza-Pinto NC, Zheng L, Bogenhagen DF, Bohr VA, Wilson DM 3rd, Shen B, Demple B 2008. Removal of oxidative DNA damage via FEN1-dependent long-patch base excision repair in human cell mitochondria. Mol Cell Biol 28: 4975–4987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS 2010a. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc Natl Acad Sci 107: 4925–4930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Prasad R, Wilson SH 2010b. HMGB1: Roles in base excision repair and related function. Biochim Biophys Acta 1799: 119–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Croteau DL, Souza-Pinto N, Pitta M, Tian J, Wu C, Jiang H, Mustafa K, Keijzers G, Bohr VA, et al. 2011. Evidence that OGG1 glycosylase protects neurons against oxidative DNA damage and cell death under ischemic conditions. J Cereb Blood Flow Metab 31: 680–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R, Nash HM, Verdine GL 1997. A mammalian DNA repair enzyme that excises oxidatively damaged guanines maps to a locus frequently lost in lung cancer. Curr Biol 7: 397–407 [DOI] [PubMed] [Google Scholar]
- Lucas-Lledo JI, Maddamsetti R, Lynch M 2011. Phylogenomic analysis of the uracil-DNA glycosylase superfamily. Mol Biol Evol 28: 1307–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Gearhart PJ 2010a. AID and somatic hypermutation. Adv Immunol 105: 159–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul RW, Gearhart PJ 2010b. Controlling somatic hypermutation in immunoglobulin variable and switch regions. Immunol Res 47: 113–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meira LB, Bugni JM, Green SL, Lee CW, Pang B, Borenshtein D, Rickman BH, Rogers AB, Moroski-Erkul CA, McFaline JL, et al. 2008. DNA damage induced by chronic inflammation contributes to colon carcinogenesis in mice. J Clin Invest 118: 2516–2525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menoni H, Gasparutto D, Hamiche A, Cadet J, Dimitrov S, Bouvet P, Angelov D 2007. ATP-dependent chromatin remodeling is required for base excision repair in conventional but not in variant H2A.Bbd nucleosomes. Mol Cell Biol 27: 5949–5956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metz AH, Hollis T, Eichman BF 2007. DNA damage recognition and repair by 3-methyladenine DNA glycosylase I (TAG). EMBO J 26: 2411–2420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mol CD, Arvai AS, Slupphaug G, Kavli B, Alseth I, Krokan HE, Tainer JA 1995. Crystal structure and mutational analysis of human uracil-DNA glycosylase: Structural basis for specificity and catalysis. Cell 80: 869–878 [DOI] [PubMed] [Google Scholar]
- Mol CD, Izumi T, Mitra S, Tainer JA 2000. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected]. Nature 403: 451–456 [DOI] [PubMed] [Google Scholar]
- Mollersen L, Rowe AD, Illuzzi JL, Hildrestrand GA, Gerhold KJ, Tveteras L, Bjolgerud A, Wilson DM 3rd, Bjoras M, Klungland A 2012. Neil1 is a genetic modifier of somatic and germline CAG trinucleotide repeat instability in R6/1 mice. Hum Mol Genet 21: 4939–4947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monti P, Ciribilli Y, Russo D, Bisio A, Perfumo C, Andreotti V, Menichini P, Inga A, Huang X, Gold B, et al. 2008. Rev1 and Polzeta influence toxicity and mutagenicity of Me-lex, a sequence selective N3-adenine methylating agent. DNA Repair (Amst) 7: 431–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morland I, Rolseth V, Luna L, Rognes T, Bjoras M, Seeberg E 2002. Human DNA glycosylases of the bacterial Fpg/MutM superfamily: An alternative pathway for the repair of 8-oxoguanine and other oxidation products in DNA. Nucleic Acids Res 30: 4926–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T 1999. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. J Biol Chem 274: 18470–18476 [DOI] [PubMed] [Google Scholar]
- Murphy DL, Donigan KA, Jaeger J, Sweasy JB 2012. The E288K colon tumor variant of DNA polymerase β is a sequence specific mutator. Biochemistry 51: 5269–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi S, Prasad R, Wilson SH, Smerdon M 2007. Different structural states in oligonucleosomes are required for early versus late steps of base excision repair. Nucleic Acids Res 35: 4313–4321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narciso L, Fortini P, Pajalunga D, Franchitto A, Liu P, Degan P, Frechet M, Demple B, Crescenzi M, Dogliotti E 2007. Terminally differentiated muscle cells are defective in base excision DNA repair and hypersensitive to oxygen injury. Proc Natl Acad Sci 104: 17010–17015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemec AA, Donigan KA, Murphy DL, Jaeger J, Sweasy JB 2012. Colon cancer-associated DNA polymerase β variant induces genomic instability and cellular transformation. J Biol Chem 287: 23840–23849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick McElhinny SA, Ramsden DA 2004. Sibling rivalry: Competition between Pol X family members in V(D)J recombination and general double strand break repair. Immunol Rev 200: 156–164 [DOI] [PubMed] [Google Scholar]
- Nilsen H, Otterlei M, Haug T, Solum K, Nagelhus TA, Skorpen F, Krokan HE 1997. Nuclear and mitochondrial uracil-DNA glycosylases are generated by alternative splicing and transcription from different positions in the UNG gene. Nucleic Acids Res 25: 750–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE 2000. Uracil-DNA glycosylase (UNG)-deficient mice reveal a primary role of the enzyme during DNA replication. Mol Cell 5: 1059–1065 [DOI] [PubMed] [Google Scholar]
- O’Brien PJ, Ellenberger T 2004. The Escherichia coli 3-methyladenine DNA glycosylase AlkA has a remarkably versatile active site. J Biol Chem 279: 26876–26884 [DOI] [PubMed] [Google Scholar]
- Odell ID, Wallace SS, Pederson DS 2013. Rules of engagement for base excision repair in chromatin. J Cell Physiol 228: 258–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterlei M, Warbrick E, Nagelhus TA, Haug T, Slupphaug G, Akbari M, Aas PA, Steinsbekk K, Bakke O, Krokan HE 1999. Post-replicative base excision repair in replication foci. EMBO J 18: 3834–3844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Dianova, II, Allinson SL, Dianov GL 2005a. Poly(ADP-ribose) polymerase-1 protects excessive DNA strand breaks from deterioration during repair in human cell extracts. FEBS J 272: 2012–2021 [DOI] [PubMed] [Google Scholar]
- Parsons JL, Zharkov DO, Dianov GL 2005b. NEIL1 excises 3′ end proximal oxidative DNA lesions resistant to cleavage by NTH1 and OGG1. Nucleic Acids Res 33: 4849–4856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons JL, Kavli B, Slupphaug G, Dianov GL 2007. NEIL1 is the major DNA glycosylase that processes 5-hydroxyuracil in the proximity of a DNA single-strand break. Biochemistry 46: 4158–4163 [DOI] [PubMed] [Google Scholar]
- Perez-Duran P, Belver L, de Yebenes VG, Delgado P, Pisano DG, Ramiro AR 2012. UNG shapes the specificity of AID-induced somatic hypermutation. J Exp Med 209: 1379–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B 2007. Uracil-DNA glycosylases SMUG1 and UNG2 coordinate the initial steps of base excision repair by distinct mechanisms. Nucleic Acids Res 35: 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen HS, Visnes T, Vagbo CB, Svaasand EK, Doseth B, Slupphaug G, Kavli B, Krokan HE 2011. UNG-initiated base excision repair is the major repair route for 5-fluorouracil in DNA, but 5-fluorouracil cytotoxicity depends mainly on RNA incorporation. Nucleic Acids Res 39: 8430–8444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad R, Liu Y, Deterding LJ, Poltoratsky VP, Kedar PS, Horton JK, Kanno S, Asagoshi K, Hou EW, Khodyreva SN, et al. 2007. HMGB1 is a cofactor in mammalian base excision repair. Mol Cell 27: 829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS 2002. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr Biol 12: 1748–1755 [DOI] [PubMed] [Google Scholar]
- Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S 1997. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc Natl Acad Sci 94: 8010–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radom CT, Banerjee A, Verdine GL 2007. Structural characterization of human 8-oxoguanine DNA glycosylase variants bearing active site mutations. J Biol Chem 282: 9182–9194 [DOI] [PubMed] [Google Scholar]
- Riccio A, Aaltonen LA, Godwin AK, Loukola A, Percesepe A, Salovaara R, Masciullo V, Genuardi M, Paravatou-Petsotas M, Bassi DE, et al. 1999. The DNA repair gene MBD4 (MED1) is mutated in human carcinomas with microsatellite instability. Nat Genet 23: 266–268 [DOI] [PubMed] [Google Scholar]
- Robertson AB, Klungland A, Rognes T, Leiros I 2009. DNA repair in mammalian cells: Base excision repair: The long and short of it. Cell Mol Life Sci 66: 981–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roldan-Arjona T, Wei YF, Carter KC, Klungland A, Anselmino C, Wang RP, Augustus M, Lindahl T 1997. Molecular cloning and functional expression of a human cDNA encoding the antimutator enzyme 8-hydroxyguanine-DNA glycosylase. Proc Natl Acad Sci 94: 8016–8020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolseth V, Runden-Pran E, Luna L, McMurray C, Bjoras M, Ottersen OP 2008. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair (Amst) 7: 1578–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenquist TA, Zharkov DO, Grollman AP 1997. Cloning and characterization of a mammalian 8-oxoguanine DNA glycosylase. Proc Natl Acad Sci 94: 7429–7434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinson EH, Metz AH, O’Quin J, Eichman BF 2008. A new protein architecture for processing alkylation damaged DNA: The crystal structure of DNA glycosylase AlkD. J Mol Biol 381: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo MT, De Luca G, Degan P, Parlanti E, Dogliotti E, Barnes DE, Lindahl T, Yang H, Miller JH, Bignami M 2004. Accumulation of the oxidative base lesion 8-hydroxyguanine in DNA of tumor-prone mice defective in both the Myh and Ogg1 DNA glycosylases. Cancer Res 64: 4411–4414 [DOI] [PubMed] [Google Scholar]
- Schanz S, Castor D, Fischer F, Jiricny J 2009. Interference of mismatch and base excision repair during the processing of adjacent U/G mispairs may play a key role in somatic hypermutation. Proc Natl Acad Sci 106: 5593–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader CE, Guikema JE, Linehan EK, Selsing E, Stavnezer J 2007. Activation-induced cytidine deaminase-dependent DNA breaks in class switch recombination occur during G1 phase of the cell cycle and depend upon mismatch repair. J Immunol 179: 6064–6071 [DOI] [PubMed] [Google Scholar]
- Sejersted Y, Hildrestrand GA, Kunke D, Rolseth V, Krokeide SZ, Neurauter CG, Suganthan R, Atneosen-Asegg M, Fleming AM, Saugstad OD, et al. 2011. Endonuclease VIII-like 3 (Neil3) DNA glycosylase promotes neurogenesis induced by hypoxia-ischemia. Proc Natl Acad Sci 108: 18802–18807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek D, Furda A, Gao Y, Artus J, Brunet E, Hadjantonakis AK, Van Houten B, Shuman S, McKinnon PJ, Jasin M 2011. Crucial role for DNA ligase III in mitochondria but not in Xrcc1-dependent repair. Nature 471: 245–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleeth KM, Robson RL, Dianov GL 2004. Exchangeability of mammalian DNA ligases between base excision repair pathways. Biochemistry 43: 12924–12930 [DOI] [PubMed] [Google Scholar]
- Slupphaug G, Mol CD, Kavli B, Arvai AS, Krokan HE, Tainer JA 1996. A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA. Nature 384: 87–92 [DOI] [PubMed] [Google Scholar]
- Sobol RW, Wilson SH 2001. Mammalian DNA β-polymerase in base excision repair of alkylation damage. Prog Nucleic Acid Res Mol Biol 68: 57–74 [DOI] [PubMed] [Google Scholar]
- Soulas-Sprauel P, Rivera-Munoz P, Malivert L, Le Guyader G, Abramowski V, Revy P, de Villartay JP 2007. V(D)J and immunoglobulin class switch recombinations: A paradigm to study the regulation of DNA end-joining. Oncogene 26: 7780–7791 [DOI] [PubMed] [Google Scholar]
- Sousa MM, Krokan HE, Slupphaug G 2007. DNA-uracil and human pathology. Mol Aspects Med 28: 276–306 [DOI] [PubMed] [Google Scholar]
- Starcevic D, Dalal S, Sweasy JB 2004. Is there a link between DNA polymerase β and cancer? Cell Cycle 3: 998–1001 [PubMed] [Google Scholar]
- Stavnezer J 2011. Complex regulation and function of activation-induced cytidine deaminase. Trends Immunol 32: 194–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierum RH, Dianov GL, Bohr VA 1999. Single-nucleotide patch base excision repair of uracil in DNA by mitochondrial protein extracts. Nucleic Acids Res 27: 3712–3719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers JT 2008. Extrahelical damaged base recognition by DNA glycosylase enzymes. Chemistry 14: 786–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stivers JT, Drohat AC 2001. Uracil DNA glycosylase: Insights from a master catalyst. Arch Biochem Biophys 396: 1–9 [DOI] [PubMed] [Google Scholar]
- Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T 2011. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res 39: 3166–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugahara M, Mikawa T, Kumasaka T, Yamamoto M, Kato R, Fukuyama K, Inoue Y, Kuramitsu S 2000. Crystal structure of a repair enzyme of oxidatively damaged DNA, MutM (Fpg), from an extreme thermophile, Thermus thermophilus HB8. EMBO J 19: 3857–3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundheim O, Talstad VA, Vagbo CB, Slupphaug G, Krokan HE 2008. AlkB demethylases flip out in different ways. DNA Repair (Amst) 7: 1916–1923 [DOI] [PubMed] [Google Scholar]
- Svilar D, Goellner EM, Almeida KH, Sobol RW 2011. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxid Redox Signal 14: 2491–2507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczesny B, Tann AW, Longley MJ, Copeland WC, Mitra S 2008. Long patch base excision repair in mammalian mitochondrial genomes. J Biol Chem 283: 26349–26356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Kanno S, Kobayashi K, Zhang QM, Yonei S, van der Horst GT, Yasui A 2002. A back-up glycosylase in Nth1 knock-out mice is a functional Nei (endonuclease VIII) homologue. J Biol Chem 277: 42205–42213 [DOI] [PubMed] [Google Scholar]
- Tell G, Quadrifoglio F, Tiribelli C, Kelley MR 2009. The many functions of APE1/Ref-1: Not only a DNA repair enzyme. Antioxid Redox Signal 11: 601–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichy ED, Liang L, Deng L, Tischfield J, Schwemberger S, Babcock G, Stambrook PJ 2011. Mismatch and base excision repair proficiency in murine embryonic stem cells. DNA Repair (Amst) 10: 445–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torseth K, Doseth B, Hagen L, Olaisen C, Liabakk NB, Graesmann H, Durandy A, Otterlei M, Krokan HE, Kavli B, et al. 2012. The UNG2 Arg88Cys variant abrogates RPA-mediated recruitment of UNG2 to single-stranded DNA. DNA Repair (Amst) 11: 559–569 [DOI] [PubMed] [Google Scholar]
- Tsutakawa SE, Classen S, Chapados BR, Arvai AS, Finger LD, Guenther G, Tomlinson CG, Thompson P, Sarker AH, Shen B, et al. 2011. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell 145: 198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubbs JL, Pegg AE, Tainer JA 2007. DNA binding, nucleotide flipping, and the helix-turn-helix motif in base repair by O6-alkylguanine-DNA alkyltransferase and its implications for cancer chemotherapy. DNA Repair (Amst) 6: 1100–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS 2006. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci 103: 1864–1869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visnes T, Akbari M, Hagen L, Slupphaug G, Krokan HE 2008. The rate of base excision repair of uracil is controlled by the initiating glycosylase. DNA Repair (Amst) 7: 1869–1881 [DOI] [PubMed] [Google Scholar]
- Wallace SS, Murphy DL, Sweasy JB 2012. Base excision repair and cancer. Cancer Lett 327: 73–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Patel U, Ghosh L, Banerjee S 1992. DNA polymerase beta mutations in human colorectal cancer. Cancer Res 52: 4824–4827 [PubMed] [Google Scholar]
- Wang ZQ, Auer B, Stingl L, Berghammer H, Haidacher D, Schweiger M, Wagner EF 1995. Mice lacking ADPRT and poly(ADP-ribosyl)ation develop normally but are susceptible to skin disease. Genes Dev 9: 509–520 [DOI] [PubMed] [Google Scholar]
- Williams SD, David SS 1998. Evidence that MutY is a monofunctional glycosylase capable of forming a covalent Schiff base intermediate with substrate DNA. Nucleic Acids Res 26: 5123–5133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson SH, Beard WA, Shock DD, Batra VK, Cavanaugh NA, Prasad R, Hou EW, Liu Y, Asagoshi K, Horton JK, et al. 2010. Base excision repair and design of small molecule inhibitors of human DNA polymerase β. Cell Mol Life Sci 67: 3633–3647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhouse BC, Dianova II, Parsons JL, Dianov GL 2008. Poly(ADP-ribose) polymerase-1 modulates DNA repair capacity and prevents formation of DNA double strand breaks. DNA Repair (Amst) 7: 932–940 [DOI] [PubMed] [Google Scholar]
- Xie Y, Yang H, Cunanan C, Okamoto K, Shibata D, Pan J, Barnes DE, Lindahl T, McIlhatton M, Fishel R, et al. 2004. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res 64: 3096–3102 [DOI] [PubMed] [Google Scholar]
- Yang CG, Yi C, Duguid EM, Sullivan CT, Jian X, Rice PA, He C 2008. Crystal structures of DNA/RNA repair enzymes AlkB and ABH2 bound to dsDNA. Nature 452: 961–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Krishnamurthy N, Burrows CJ, David SS 2010. Mutation versus repair: NEIL1 removal of hydantoin lesions in single-stranded, bulge, bubble, and duplex DNA contexts. Biochemistry 49: 1658–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng L, Zhou M, Guo Z, Lu H, Qian L, Dai H, Qiu J, Yakubovskaya E, Bogenhagen DF, Demple B, et al. 2008. Human DNA2 is a mitochondrial nuclease/helicase for efficient processing of DNA replication and repair intermediates. Mol Cell 32: 325–336 [DOI] [PMC free article] [PubMed] [Google Scholar]