Abstract
To achieve the high degree of processivity required for DNA replication, DNA polymerases associate with ring-shaped sliding clamps that encircle the template DNA and slide freely along it. The closed circular structure of sliding clamps necessitates an enzyme-catalyzed mechanism, which not only opens them for assembly and closes them around DNA, but specifically targets them to sites where DNA synthesis is initiated and orients them correctly for replication. Such a feat is performed by multisubunit complexes known as clamp loaders, which use ATP to open sliding clamp rings and place them around the 3′ end of primer–template (PT) junctions. Here we discuss the structure and composition of sliding clamps and clamp loaders from the three domains of life as well as T4 bacteriophage, and provide our current understanding of the clamp-loading process.
Ring-shaped sliding clamps and DNA polymerases slide freely along the DNA template during replication. Clamp loaders utilize ATP to open the clamp rings and place them around the 3′ ends of primer–template junctions.
During each round of DNA replication, thousands to billions of nucleotides must be faithfully copied in a short period of time. However, by themselves, replicative DNA polymerases are distributive, synthesizing only ten or so nucleotides of complementary DNA before dissociating. To achieve the high degree of processivity required for efficient DNA replication, replicative DNA polymerases associate with ring-shaped sliding clamps that encircle the template DNA and slide freely along it. Such an association effectively tethers the polymerase to DNA, substantially increasing the amount of continuous replication. The closed circular structure of sliding clamps necessitates an enzyme-catalyzed mechanism, which not only opens them for assembly and closes them around DNA, but specifically targets them to sites where DNA synthesis is initiated and orients them correctly for interaction with DNA polymerases. Such a feat is performed by multisubunit complexes known as clamp loaders, which use ATP to open sliding clamp rings and place them around the 3′ end of primer–template (PT) junctions. Here we discuss the structure and composition of sliding clamps and clamp loaders from the three domains of life as well as T4 bacteriophage, and provide our current understanding of the clamp-loading process.
SLIDING CLAMPS
Crystal Structures
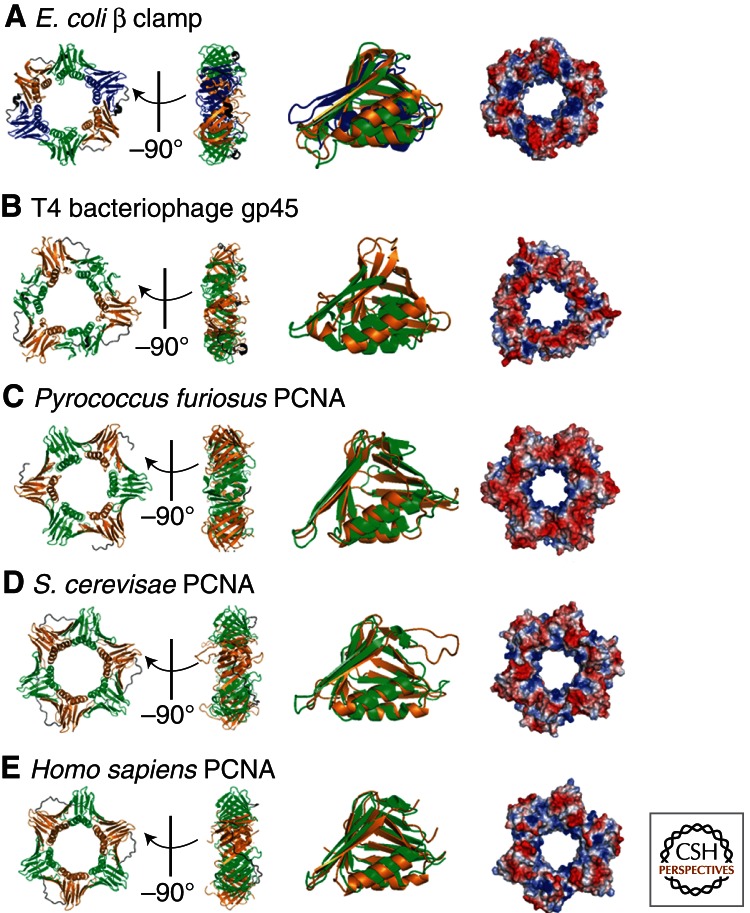
Based on early biochemical assays, it had been predicted that sliding clamps encircle DNA in a sequence-independent manner that allows them to slide freely along DNA (Stukenberg et al. 1991). In 1992, this hypothesis was confirmed when the first crystal structure of a sliding clamp, E. coli β clamp, was solved (Fig. 1A, left). In the structure, two semicircular β subunits are aligned head-to-tail, forming a ring with an inner diameter large enough to accommodate duplex DNA. As a result of the head-to-tail arrangement, the ring has two structurally distinct faces, one of which, the “carboxy-terminal face,” contains both carboxyl termini and serves as a platform for interaction with other proteins, including polymerases and the clamp loader, as discussed below. Each β subunit consists of three independent domains connected by a long loop, referred to as the interdomain connecting loop (IDCL), which protrudes from the carboxy-terminal face. Although the amino acid sequence of each domain is quite different, all fold into similar three-dimensional structures with identical chain topologies; two adjacent α-helices flanked by two, four-stranded antiparallel β sheets (Fig. 1A, middle). Thus, the β clamp has pseudo sixfold symmetry (Kong et al. 1992). The 12 α helices line the inner surface of the ring while the β sheets form a continuous outer surface. Although the β clamp has a net negative charge, a strong positive electrostatic potential lines the inner surface of the ring where water molecules mediate protein–DNA interactions, allowing the β clamp to move freely along DNA in a sequence-independent manner (Fig. 1A, right).
Figure 1.
Sliding clamps from different organisms. Crystal structures of (A) the Escherichia coli β clamp (Protein Data Bank [PDB] code 2POL), (B) gene 45 protein (gp45) from T4 bacteriophage (PDB code 1CZD), (C) Pyrococcus furiosus proliferating cell nuclear antigen (PCNA, PDB code 1GE8), (D) Saccharomyces cerevisiae PCNA (PDB code 1PLQ), and (E) Homo sapiens PCNA (PDB code 1AXC). For each organism, the top and side views of the three-dimensional crystal structure are shown on the left and alignment of the globular domains in the middle. The electrostatic surface potential for each is shown on the right with red and blue representing negative and positive electrostatic potentials, respectively. All images were constructed using PyMOL. The sliding clamps from all domains of life have a similar architecture comprised of six domains arranged in a circle. The chain-folding topologies of each domain are the same. The E. coli β clamp is homodimeric (three domains per monomer), whereas T4 gp45 and PCNA from eukaryotes and archaea are each homotrimeric (two domains per monomer). The monomers are arranged head-to-tail in all clamps, resulting in structurally distinct faces. The face from which the carboxyl termini protrude (“carboxyl-terminus face”) interacts with clamp loaders and polymerases. A strong positive electrostatic potential lines the inner surface of each ring where double-stranded DNA resides, favoring nonspecific interactions with the negatively charged phosphate backbone.
The functional equivalents of the β subunit in eukaryotes (proliferating cell nuclear antigen, PCNA) and T4 bacteriophage (gene 45 protein, gp45) do not share any significant sequence similarity (<10%) with the β subunit and each is approximately two-thirds its size (Kong et al. 1992; Krishna et al. 1994; Gulbis et al. 1996; Moarefi et al. 2000). Early biochemical analyses suggested that these proteins trimerize to form a six-domain ring like the β clamp (Bauer and Burgers 1988; Jarvis et al. 1989b; Yao et al. 1996). Subsequent crystallographic studies confirmed that each sliding clamp is a homotrimeric ring with each monomer contributing two nearly superimposable domains with chain topologies identical to that in the β subunit (Fig. 1B,D,E). The three monomers are aligned head-to-tail, forming a ring with two structurally distinct faces, one of which contains the carboxyl termini and the IDCLs. The negatively charged outer surface of each ring is composed of continuous β sheets, whereas α helices comprise the positively charged inner surface. Each ring has a diameter large enough to completely encircle duplex DNA (Krishna et al. 1994; Gulbis et al. 1996; Moarefi et al. 2000).
Most archaeal genomes, such as Pyrococcus furiosus (Pfu), encode a homotrimeric sliding clamp (PfuPCNA in Fig. 1C) similar to their evolutionary counterparts (Cann et al. 1999; Iwai et al. 2000; Matsumiya et al. 2001). However, in archaea of the crenarchaeal subdomain, three PCNA homologs have been identified (Cann and Ishino 1999; Iwai et al. 2000; Daimon et al. 2002; Dionne et al. 2003). In Sulfolobus solfataricus, the three PCNA homologs (ssoPCNAs 1–3) share only 8%–22% sequence identity and do not form homotrimers due to electrostatic and steric clashes (Pascal et al. 2006). Rather, they exist as monomers in solution and self-assemble into a functional heterotrimer (ssoPCNA123) in a defined order (Dionne et al. 2003; Williams et al. 2006; Hlinkova et al. 2008). Unlike its evolutionary counterparts, each interface within the ssPCNA123 heterotimer is unique, which has implications on the clamp-loading mechanism, as discussed below. However, the ring-shaped heterotrimer has an overall shape and dimensions similar to the homotrimeric sliding clamps in Figure 1 (Pascal et al. 2006; Williams et al. 2006; Hlinkova et al. 2008). Thus, despite the various subunit compositions and lack of any significant sequence homology, the three-dimensional structure of the sliding clamp has been highly conserved throughout evolution.
Solution Structures
Crystal structures are static representations of a particular conformation and may not accurately reflect the solution structure of a given protein. For sliding clamps, one or more of the subunit interfaces may dissociate because of thermal fluctuations and flexibility of the protein, causing the closed ring to transiently or permanently open in solution. Such behavior has profound implications in both the clamp loading and unloading pathways, as discussed in future sections.
The stability of a closed sliding clamp is largely governed by the intermolecular interactions at subunit interfaces. Although the subunits of the β clamp appear to be under “spring tension” caused by strenuous bending within the β monomers, continuation of the β-sheet structures across subunit interfaces contributes six intermolecular ion pairs and four strong hydrogen bonds to each interface, suggesting that the β-clamp ring remains closed in solution (Kong et al. 1992; Jeruzalmi et al. 2001b). Indeed, biochemical analyses have shown that the closed β-clamp ring remains stable in solution, with a Kd for subunit dissociation less than 60 pm, and only opens in the presence of the clamp loader (Yao et al. 1996; Paschall et al. 2011). Similar behavior is observed for PfuPCNA, where four hydrogen bonds and 10 intermolecular ion pairs are present at each subunit interface. Such enhancement of electrostatic interactions is thought to be responsible for the increased thermostability of the sliding clamp from P. furiosus, a hyperthermophilic archaeon that grows optimally at 100°C (Matsumiya et al. 2001, 2003). However, it is interesting to note that a recent report suggests that PCNA from Methanosarcina acetivorans (Mac), a mesophilic archaeon that grows best in moderate temperatures (25°C–40°C), may exist as a monomer in solution (Liu et al. 2011).
In contrast to β clamp and PfuPCNA, various experimental approaches have collectively shown that gp45 exists exclusively in an open state in solution, with one interface opened within the plane of the ring (Soumillion et al. 1998; Alley et al. 1999b; Millar et al. 2004). Such behavior may be ascribed to differences in the architecture of the gp45 sliding clamp that weakens the binding affinity at subunit interfaces. Indeed, contacts at the subunit interfaces are far less extensive; the hydrogen bonding networks are less regular, no intermolecular ion pairs are present, and the buried surface area is roughly half that for β clamp and PfuPCNA (Moarefi et al. 2000). Consequently, the Kd for subunit dissociation in gp45 (∼210 nm) is approximately 3500-fold greater than that for β clamp (Yao et al. 1996).
Eukaryotic PCNA seems to exist as a closed ring in solution (Schurtenberger et al. 1998; Zhuang et al. 2006b). However, recent molecular dynamics (MD) simulations suggest that the PCNA ring spontaneously relaxes in-plane to adopt more open conformations and may transiently sample out-of-plane, spiral conformations (Kazmirski et al. 2005; Adelman et al. 2010; Tainer et al. 2010). Importantly, the barrier to reclose is essentially nonexistent and occurs on a nanosecond timescale in the absence of clamp loader, preventing DNA from entering the PCNA ring. Although the total surface area buried at each interface of eukaryotic PCNA is comparable that for β clamp and PfuPCNA, only one ion pair is present at each interface compared with six in β clamp (Kong et al. 1992; Krishna et al. 1994; Gulbis et al. 1996). In agreement, the Kd for subunit dissociation in human PCNA (21 nm) is ∼350-fold greater than that for β clamp (Yao et al. 1996). Thus, the closed ring structure may be favored in eukaryotic PCNA but weakened intermolecular contacts may permit spontaneous and transient opening.
CLAMP LOADER COMPOSITION AND ARCHITECTURE
Bacterial Clamp Loaders
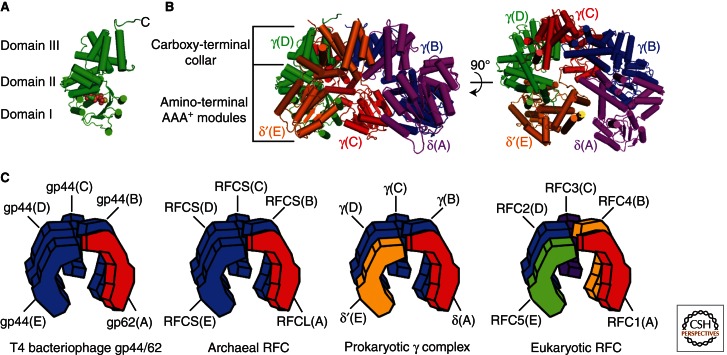
The E. coli clamp loader is comprised of three different subunits, δ, δ′, and γ/τ, which assemble into a heteropentamer with stoichiometry δδ′(γ/τ)3 (Onrust et al. 1995; Pritchard et al. 2000; Jeruzalmi et al. 2001a). Two other subunits, χ and ψ, are associated with the clamp loader but these subunits are not required for its assembly nor its clamp-loading activity and will not be discussed further (Onrust and O’Donnell 1993; Xiao et al. 1993; Naktinis et al. 1995; Olson et al. 1995; Glover and McHenry 1998; Kelman et al. 1998; Turner et al. 1999). The γ and τ subunits are encoded by the same gene. The τ subunit is the full-length product and, when present within the clamp loader, binds the DNA polymerase III core and helicase via its carboxyl terminus, effectively trimerizing the polymerase at the replication fork. However, the carboxyl terminus of the τ subunit is not required for clamp loading and its truncation by a translational frameshift produces the γ subunit (Leu et al. 2003; Reyes-Lamothe et al. 2010). Thus, clamp loader comprised of δδ′γ3 has been predominantly used in in vitro studies on the bacterial clamp loader and is referred to as the minimal clamp loader or simply the γ complex. Herein, our description of the bacterial clamp loader refers only to the γ complex. Each subunit of the γ complex has the same general architecture consisting of three distinct domains (Fig. 2A). The amino-terminal domain (Domain I) is a RecA-Type ATPase domain containing Walker A (P-Loop), Walker B, sensor 1, and Ser-Arg-Cys (SRC) motifs (Guenther et al. 1997; Jeruzalmi et al. 2001a; Kazmirski et al. 2004). Walker A and B motifs are common among NTPases and are important for nucleotide binding and hydrolysis (Walker et al. 1982). The sensor 1 motif functions to monitor and relay information regarding the state of bound ATP through structural rearrangements of the γ complex. The highly conserved arginine residue of the SRC motif functions as an “arginine finger” within an interfacial ATPase site, discussed further below (Davey et al. 2002). Domain II contains a sensor 2 motif that is analogous to sensor 1. Together, Domains I and II form an ATP-binding module, referred to as an AAA+ module, which is structurally conserved among members of the AAA+ superfamily of ATPases. The carboxy-terminal domains (Domain III) are unique to clamp loaders among AAA+ ATPases and pack together to form a ring-shaped “collar” that holds the complex together (Fig. 2B). The five subunits are arranged such that the γ subunits are adjacent to one another, in positions B–D, with the δ and δ′ subunits flanking each side, in the A and E positions, respectively. The amino-terminal domains are arranged in an open “horseshoe” below the carboxy-terminal collar with a gap present between the AAA+ modules of the δ(A) and δ′(E) subunits to allow entry of PT DNA into the complex, as discussed further below. This arrangement positions the SRC motif of a given subunit near the ATP-binding module of the next such that each ATPase site is located at a subunit interface and faces the inner surface of the complex (Fig. 2A). However, only the γ subunits bind ATP as the Walker A motifs of the δ and δ′ subunits are degenerate. The δ subunit also lacks an SRC motif (Xiao et al. 1995; Guenther et al. 1997; Jeruzalmi et al. 2001a; Kazmirski et al. 2004). Thus, only three functional ATPase sites are present: between subunits δ′(E) and γ(D), γ(D) and γ(C), and γ(C) and γ(B) (Tsuchihashi and Kornberg 1989; Hingorani et al. 1999; Johnson and O’Donnell 2003). The SRC motif of γ(B) does not abut a nucleotide in the δ(A) subunit.
Figure 2.
Clamp loader structure. (A) Close-up of the E. coli γD subunit (from PDB code 1XXH) bound to ATPγS (shown in space-filling format). The carboxyl terminus and the three domains (I, II, and III) are indicated. (B) Front (left) and carboxy-terminal/top (right) views of the γ3δδ′ minimal clamp loader structure (PDB code 1XXH). The “carboxy-terminal collar” and amino-terminal AAA+ ATPase modules are indicated. All images were generated using PyMOL. (C) Stylized images of the clamp loaders from bacteria (γ complex), eukaryotes (RFC), bacteriophage T4 (gp44/62), and archaea (RFC). The positions of the five subunits of each clamp loader complex are denoted by the letters A–E, with analogous subunits indicated by matching colors.
T4 Bacteriophage Clamp Loader
In T4, four gp44 subunits and one gp62 subunit associate to form the gp44/62 clamp loader complex (Spicer et al. 1984; Jarvis et al. 1989b). The gp44 subunits contain AAA+ modules and are homologous to the E. coli γ subunits (Spicer et al. 1984; Jarvis et al. 1989b; Zhuang et al. 2006a). However, gp62 lacks both SRC and Walker A motifs, analogous to the E. coli δ subunit (Spicer et al. 1984; O’Donnell et al. 1993). Thus, based on homology between the subunits of the γ complex and gp44/62, it was presumed that the subunit arrangement within gp44/62 is analogous to that in the γ complex, with the gp62 subunit occupying position A while the four gp44 subunits occupy positions B–E (Fig. 2C). This has recently been verified in a crystal structure of gp44/62 complexed to both gp45 and PT DNA. Such an arrangement provides for only three interfacial ATPase sites as the gp62 subunit in position A does not present an “arginine finger” to the gp44 subunit in position E (Kelch et al. 2011). However, it should be noted that several studies have each independently showed that gp44/62 hydrolyzes four ATP molecules during the clamp-loading process, suggesting that each gp44 subunit contains an equally active ATPase site (Jarvis et al. 1989a; Berdis and Benkovic 1996; Sexton et al. 1996; Young et al. 1996; Zhuang et al. 2006a; Pietroni and von Hippel 2008). In the aforementioned crystal structure, ADP bound in the E subunit packs closely against a carboxy-terminal extension of the gp62 (A) collar domain, referred to as the Á domain. This domain has the same fold topology as a corresponding domain in the A subunit of eukaryotic RFC (discussed below), tempting one to speculate that a novel interfacial ATPase site may be present between the A and E subunits of both T4 and eukaryotic clamp loaders (Bowman et al. 2004; Kelch et al. 2011).
Eukaryal Clamp Loaders
The eukaryotic clamp loader, referred to as replication factor C (RFC) or activator 1, is highly conserved from yeast to humans and is composed of five distinct subunits, RFCs 1–5, all of which share a similar three-domain core architecture reminiscent of their ancestral counterparts (Burbelo et al. 1993; Cullmann et al. 1995; Uhlmann et al. 1997b; Bowman et al. 2004). RFCs 2–4 are homologous to each other and to the ATP-hydrolyzing subunits of the T4 (gp44 subunits) and E. coli (γ subunits) clamp loaders. Thus, each contains an AAA+ module and an SRC motif (Chen et al. 1992a,b; O’Donnell et al. 1993; Li and Burgers 1994a,b; Noskov et al. 1994; Yao et al. 2003). The RFC5 subunit is analogous to the δ′ subunit of the E. coli γ complex in that it contains an SRC motif but lacks several key residues that are conserved at the active sites of functional ATPases (Gary and Burgers 1995; Cai et al. 1998; Podust et al. 1998a; Schmidt et al. 2001; Yao et al. 2003; Bowman et al. 2004). However, it is still capable of binding ATP (Bowman et al. 2004). Collectively, the RFC2–5 subunits are often referred to as the “small subunits” and occupy positions analogous to their bacterial counterparts in the five-subunit complex (Fig. 2C); RFC5 in position E followed by RFCs 2, 3, and 4 in positions D, C, and B, respectively (Pan et al. 1993; Uhlmann et al. 1996; Cai et al. 1997; Ellison and Stillman 1998; Yao et al. 2003; Bowman et al. 2004). The remaining subunit, RFC1, occupies position A and is similar to the gp62 subunit of T4 gp44/62 in that it lacks an SRC motif but contains an A′ domain, which abuts the nucleotide bound in position E (RFC5 [Li and Burgers 1994a; Cullmann et al. 1995; Bowman et al. 2004; Kelch et al. 2011]). However, unlike gp62, RFC1 contains a consensus Walker A motif (Bunz et al. 1993; Cullmann et al. 1995; Cai et al. 1998; Podust et al. 1998a; Schmidt et al. 2001; Yao et al. 2003). Thus, four interfacial ATPase sites are present within RFC; between RFC5 (E) and RFC2 (D), RFC2 (D) and RFC3 (C), RFC3 (C) and RFC4 (B), and RFC4 (B) and RFC1 (A) (Gomes et al. 2001; Yao et al. 2003). Furthermore, the RFC1 (A) subunit also contains unique amino- and carboxy-terminal extensions beyond the three-domain core. Hence, RFC1 is often referred to as the “large subunit.” The carboxy-terminal extensions are required for complex formation with the small RFC subunits whereas the amino-terminal extensions are dispensable for PCNA loading. In fact, their deletion actually increases the PCNA-loading activity of RFC and subsequent DNA synthesis by polymerase δ (Burbelo et al. 1993; Uhlmann et al. 1996, 1997a; Allen et al. 1998; Podust et al. 1998b; Gomes et al. 2000; Gomes and Burgers 2001). Apart from a minor role in DNA damage repair, the function of the amino-terminal extension remains largely unknown (Gomes et al. 2000). Interestingly, it shares homology with all known bacterial DNA ligases and several eukaryotic poly-ADP ribose polymerases (PARPs) although there is no evidence of either activity. Rather, this region of RFC1, designated the ligase homology domain, is most likely responsible for the nonspecific DNA-binding affinity of RFC, discussed further below (Bunz et al. 1993; Burbelo et al. 1993; Cullmann et al. 1995; Fotedar et al. 1996; Allen et al. 1998).
Archaeal Clamp Loaders
Similar to their eukaryotic counterparts, archaeal clamp loaders are heteropentameric complexes comprised of a single large subunit in position A and four smaller subunits in positions B–E, as shown in Figure 2C (Pisani et al. 2000; Cann et al. 2001; Seybert et al. 2002; Miyata et al. 2005; Chen et al. 2009b). Each subunit adopts the conserved three-domain core architecture (Mayanagi et al. 2001; Oyama et al. 2001; Miyata et al. 2004, 2005; Seybert et al. 2006). The large subunit, RFCL, is homologous to the carboxy-terminal portion of the large subunit of eukaryotic RFC (RFC1) and contains a conserved Walker A motif but lacks an SRC motif (Cann and Ishino 1999; Pisani et al. 2000; Cann et al. 2001; Oyama et al. 2001; Henneke et al. 2002; Chen et al. 2005). Based on low-resolution structures of RFC from Pyrococcus furiosus, RFCL may also contain an A′ domain, similar to yeast RFC1 and T4 gp62 (Miyata et al. 2005). In most archaea, a single small subunit (RFCS) is present, which contains both Walker A and SRC motifs, homologous to RFCs 2–4 of eukaryotic RFC (Cann and Ishino 1999; Kelman and Hurwitz 2000; Cann et al. 2001; Oyama et al. 2001; Henneke et al. 2002). However, in several archaea, such as Methanosarcina acetivorans (Mac), an additional small subunit is present, which contains an SRC motif and a degenerate Walker A motif. This subunit, referred to as RFCS2, is homologous to RFC5 from eukaryotic RFC and occupies an analogous position (E) in the five-subunit complex (Chen et al. 2005, 2009b). Thus, the five-subunit archaeal clamp loader contains four interfacial ATPase sites, similar to eukaryotic RFC (Seybert and Wigley 2004; Seybert et al. 2006). However, it should be noted that a unique heterohexameric clamp loader in which two large subunits (RFCL) are complexed to four small subunits (RFCS) has been reported for Methanobacterium thermoautotrophicum (Kelman and Hurwitz 2000).
CLAMP LOADERS ARE DNA-DEPENDENT ATPases
Amid the numerous nucleic acid structures within cells, clamp loaders must target their respective clamps to sites where DNA synthesis initiates. Thus, clamp loaders must have a higher affinity for PT junctions than any other nucleic acid structure. However, if the high affinity was static, clamp loaders would compete with replicative polymerases for binding to the same PT junctions. On the contrary, if their binding affinity were dynamic, switching reversibly between high- and low-affinity states, clamp loaders could associate with PT junctions to load their respective clamps and then dissociate to allow polymerase binding (Ason et al. 2000). Detailed analyses over the years have revealed that the affinity of clamp loaders for PT junctions is indeed dynamic and modulated by ATP. Recent crystal structures of the E. coli γ complex have provided the structural basis for such behavior.
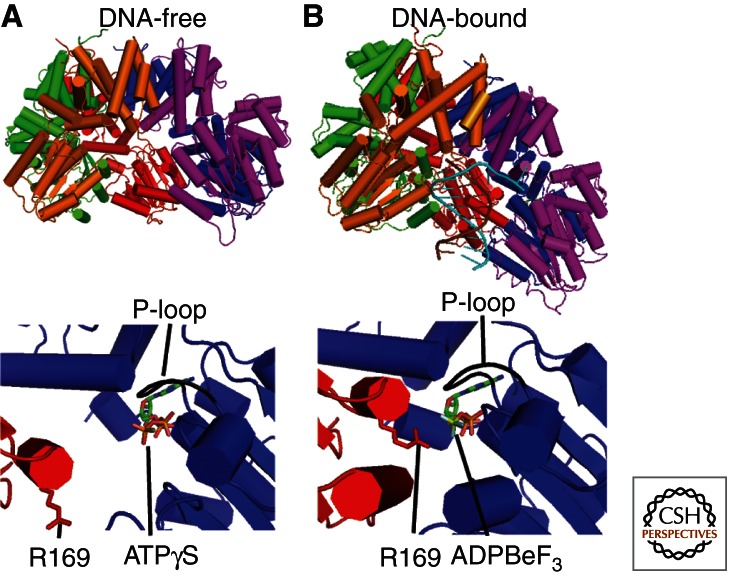
In the absence of DNA, the γ complex binds tightly to ATP but the “arginine fingers” of the SRC motifs are held out of reach (Fig. 3A) (Hingorani et al. 1999; Johnson and O’Donnell 2003; Kazmirski et al. 2004; Thompson et al. 2009). These motifs increase the rate of ATP hydrolysis by stabilizing the transition state (Ahmadian et al. 1997; Kazmirski et al. 2004). Consequently, ATPase activity of the γ complex alone is negligible (Onrust et al. 1991; Hingorani et al. 1999; Ason et al. 2000). This ATP-bound form of the γ complex has high affinity for PT junctions and binds rapidly (Reems et al. 1995; Bloom et al. 1996; Hingorani and O’Donnell 1998; Ason et al. 2000). The single-stranded portion of the template-strand is presumed to enter through the gap between the AAA+ modules of the δ (A) and δ′ (E) subunits, inducing a right-handed spiral conformation that closely mimics the helical geometry of the DNA duplex and “screws” onto the double-stranded portion. In this “notched screw cap” arrangement, the γ complex interacts intimately with only the template strand; conserved positively charged/polar residues within the AAA+ modules contact the duplex region and track along the minor groove while the 5′ single-stranded overhang exits the central chamber and binds to the exterior surface of the δ (A) subunit. In contrast, the primer strand is mostly surface-exposed. Apart from its 3′ terminal nucleotide, which runs into the interior wall of the carboxy-terminal collar, the primer strand is disengaged from the γ complex. This lack of intimate contact is vital to the clamp loader function as it allows recognition of both RNA and DNA primers during DNA replication and repair (Simonetta et al. 2009). The “notched screw cap” arrangement of the γ complex subunits realigns the interfacial ATPase sites such that the SRC motif from one subunit is presented to the ATP binding site of the next, promoting ATP hydrolysis (Fig. 3B) (Hingorani et al. 1999). The sensor 1 and sensor 2 motifs, “sensing” the absence of the γ phosphate of ATP, subsequently trigger a conformational change converting the γ complex to a low-affinity state and promoting rapid dissociation (Ason et al. 2003; Simonetta et al. 2009; Thompson et al. 2009). On release from DNA, the γ complex can exchange ADP for ATP, allowing the dynamic DNA binding cycle to continue (Ason et al. 2000, 2003; Thompson et al. 2009).
Figure 3.
DNA promotes ATP hydrolysis by triggering a structural rearrangement of the interfacial ATPase sites of the γ complex. The DNA-free (A, PDB code 1XXH) and DNA-bound (B, PDB code 3GLF) forms of the γ complex are shown. For each, the structure of the γ complex is shown on top with a close-up of the γC-γB interfacial ATPase site shown at the bottom. All images were generated using PyMOL. Nucleotide is bound in the AAA+ module of the γB subunit. The “P-loop” is indicated and shown in black. In the absence of DNA, the “arginine finger” (Arg169) from the γC subunit is too far away to interact with the γ phosphate of ATP in the γB subunit. On binding DNA, the γ complex adopts a right-handed spiral configuration with a uniform rise and rotation that closely mimics that of the bound DNA. Such a structural rearrangement presents the “arginine finger” (Arg169) from one subunit (γC) to the γ phosphate of ATP bound in the next subunit (γB), promoting ATP hydrolysis.
All clamp loaders must bind and hydrolyze multiple ATP molecules to power site-specific loading of their respective clamps to PT junctions. The DNA-dependent ATPase activity discussed above for the E. coli γ complex is conserved among clamp loaders from T4 (Piperno et al. 1978; Rush et al. 1989; Capson et al. 1991; Munn and Alberts 1991b; Zhuang et al. 2006a), eukaryotes (Lee and Hurwitz 1990; Lee et al. 1991; Tsurimoto and Stillman 1991; Yoder and Burgers 1991; Fien and Stillman 1992; Cai et al. 1996; Ellison and Stillman 1998; Hingorani and Coman 2002; Sakato et al. 2011a), and archaea (Kelman and Hurwitz 2000; Pisani et al. 2000; Cann et al. 2001; Seybert et al. 2002; Seybert and Wigley 2004; Chen et al. 2005). Historically, it had been assumed that clamp loaders use the energy from ATP hydrolysis solely to apply mechanical force to open sliding clamps. Although ATP hydrolysis may contribute to ring opening within T4 (discussed below), detailed studies over the last 20 years or so have revealed that ATP hydrolysis primarily serves to modulate the interaction between clamp loaders and PT DNA by allowing interconversion between high- and low-affinity DNA-binding states. Such a dynamic DNA-binding mechanism specifically targets clamp loaders to PT junctions yet triggers their departure to prevent competition with replicative polymerases, as discussed further below.
Clamp-Loading Mechanism
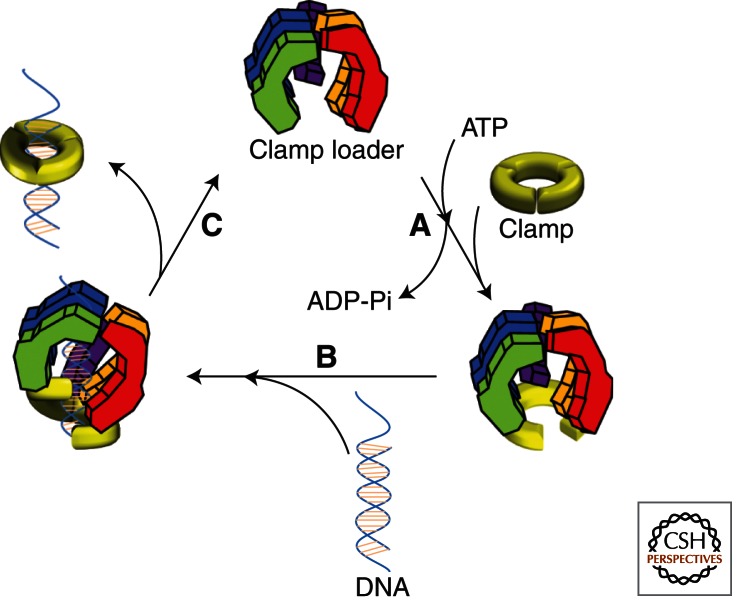
Detailed mechanistic studies on the clamp-loading process from all domains of life have revealed a similar sequential mechanism that is outlined in Figure 4. Given the conservation of the clamp and clamp loader structures depicted in Figures 1 and 2, this is no surprise. Nonetheless, subtle differences have emerged that distinguish each system. These are discussed below.
Figure 4.
The clamp-loading mechanism. The catalytic reaction cycle for the loading of sliding clamps onto DNA by clamp loaders is shown as a schematic diagram. (A) Opening of the sliding clamp ring. In the absence of ATP, clamp loaders bind their respective clamps very weakly. On binding ATP, clamp loaders undergo a conformational change, which permits optimal interaction with the carboxy-terminal face of their respective clamp and subsequent opening of the clamp ring. (B) PT junction binding. The open clamp–clamp loader complex together specifically recognizes and binds a PT junction, adopting a “notched screw cap arrangement,” which matches the helical geometry of the DNA duplex and properly aligns the interfacial ATPase sites for hydrolysis. (C) Closure of the clamp ring. On hydrolysis of ATP, clamp loaders revert back to a low-affinity DNA-binding state and eject, leaving the PT DNA positioned within an opened sliding clamp ring. Concurrent or subsequent to ejection, electrostatic interactions between the positively charged inner surface of the sliding clamp ring and the negatively charged DNA drives closure of the open sliding clamp around DNA.
Opening of the Sliding Clamp Ring
In the absence of ATP, clamp loaders interact with their respective clamps very weakly. On binding ATP, clamp loaders undergo a conformational change that permits optimal interaction with the carboxy-terminal face of their respective clamp and subsequent opening of the clamp ring (Fig. 4A). In E. coli, all subunits of the γ complex bind the β clamp but the δ subunit is the major contact point and is mainly responsible for ring opening (Naktinis et al. 1995; Turner et al. 1999; Leu et al. 2000; Jeruzalmi et al. 2001b; Leu and O’Donnell 2001; Johnson and O’Donnell 2003; Paschall et al. 2011). In the absence of ATP, the δ subunit is sequestered within the γ complex, largely because of the interaction with the nearby δ′ subunit, and the γ complex interacts with the β clamp very weakly (Onrust and O’Donnell 1993; Naktinis et al. 1995; Onrust et al. 1995; Leu and O’Donnell 2001; Indiani and O’Donnell 2003). On binding ATP, the interaction between δ and δ′ is disrupted, presenting δ for interaction with the carboxy-terminal face of a closed β clamp (Naktinis et al. 1995; Bloom et al. 1996; Goedken et al. 2004; Snyder et al. 2004; Thompson et al. 2009; Paschall et al. 2011). A “hydrophobic plug” of the δ subunit subsequently wedges into a hydrophobic pocket on either β monomer, destabilizing the adjacent dimer interface and prying open the β-clamp ring. This process is favored by the release of “spring tension” between domains within the β monomers and occurs in the absence of ATP hydrolysis (Onrust et al. 1991; Naktinis et al. 1995; Hingorani and O’Donnell 1998; Hingorani et al. 1999; Turner et al. 1999; Jeruzalmi et al. 2001b; Stewart et al. 2001; Goedken et al. 2004; Williams et al. 2004; Anderson et al. 2009; Paschall et al. 2011). Thus, ATP binding but not hydrolysis is required for γ-complex binding to and opening the β clamp. The same is true for RFC-catalyzed opening of PCNA rings in both eukaryotes (Lee and Hurwitz 1990; Shiomi et al. 2000; Gomes and Burgers 2001; Schmidt et al. 2001; Johnson et al. 2006; Yao et al. 2006; Zhuang et al. 2006b; Chen et al. 2009a; Sakato et al. 2011a,b; Thompson et al. 2011) and archaea (Henneke et al. 2002; Matsumiya et al. 2002; Seybert et al. 2002, 2006; Seybert and Wigley 2004), suggesting a similar mechanism applies to these domains of life. However, it remains to be seen whether ATP-bound RFC “actively” pries open the PCNA ring as in E. coli or “passively” waits for it open (Kazmirski et al. 2005; Thompson et al. 2009). Also, subtle variations in some archaea should be noted. As alluded to above, three unique interfaces are present within the ssoPCNA123 heterotrimer of Sulfolobus solfataricus and only the PCNA3–PCNA1 interface is opened by ssoRFC (Dionne et al. 2003, 2008). On the contrary, each subunit interface within a homopolymeric sliding clamp is identical and, thus, the one selected by the clamp loader for opening is random. Furthermore, a recent report suggests that, in Methanosarcina acetivorans (Mac), PCNA exists as monomers in solution and RFC functions to assemble an open PCNA homotrimer by binding individual monomers in an ATP-dependent manner (Liu et al. 2011). Regardless of these slight differences, it is tempting to speculate that sliding clamps within bacteria, eukaryotes, and archaea are opened predominantly in-plane in the clamp–clamp loader complexes as tandem out-of-plane movements are presumed to induce the right-handed spiral conformation of clamp loaders that is competent for ATP hydrolysis.
Like RFC and the γ complex, T4 gp44/62 also undergoes an ATP-dependent conformational change that permits optimal interaction with the carboxy-terminal face of an open gp45 sliding clamp (Latham et al. 1997; Pietroni et al. 1997, 2001; Zhuang et al. 2006a; Pietroni and von Hippel 2008). However, on interaction with ATP-bound gp44/62, the open gp45 sliding clamp opens further, stimulating ATP hydrolysis at two of the four ATPase sites within gp44/62 (Mace and Alberts 1984; Rush et al. 1989; Berdis and Benkovic 1996; Sexton et al. 1996, 1998; Young et al. 1996; Alley et al. 2000; Trakselis et al. 2003). Such partial ATP hydrolysis is required to open and stabilize gp45 in an “activated” conformation that is competent for optimal loading onto DNA, in stark contrast to that described above (Sexton et al. 1996; Pietroni et al. 1997; Alley et al. 2000; Trakselis et al. 2003). This implies that the gp45 ring opens out-of plane on interacting with ATP-bound gp44/62, permitting the gp45–gp44/62 complex to adopt an intermediate, right-handed spiral conformation in which only two of the four ATP sites of gp44/62 are aligned properly for hydrolysis. Because of the unique open structure of gp45 in solution, perhaps gp44/62 uses ATP hydrolysis to passively “catch” and stabilize gp45 rings that have spontaneously adopted this productive conformation. However, it is also possible that gp44/62 uses ATP hydrolysis to “actively” sculpt gp45 rings into this intermediate conformation.
PT Junction Binding
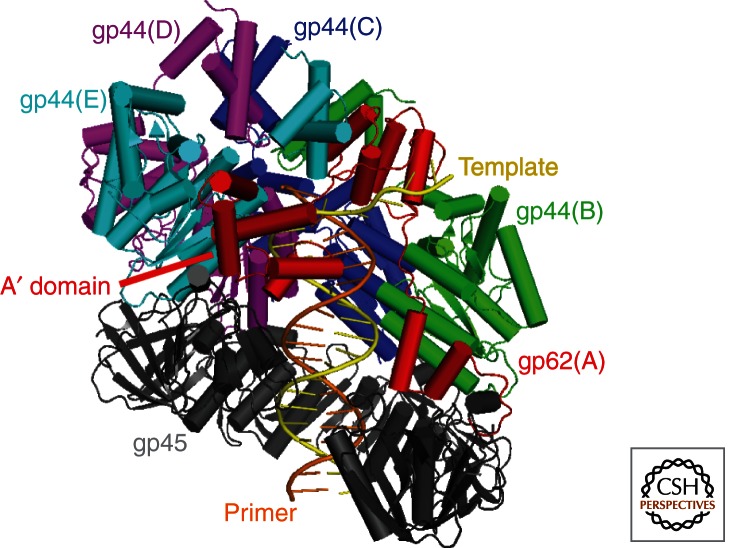
Once formed, the open clamp–clamp loader complex together specifically recognizes and binds a PT junction (Fig. 4B) undergoing another conformational change, which is illustrated in the recent crystal structure of the gp45–gp44/62–DNA complex (Jarvis et al. 1989a; Munn and Alberts 1991b; Hockensmith et al. 1993; Pietroni et al. 2001). The PT DNA is positioned such that the duplex region is threaded through the central pore of gp45 and is sequestered within the inner chamber of gp44/62 (Fig. 5). The gp45 ring is held open by extensive interactions between its carboxy-terminal face and the entire undersurface of gp44/62 and the gp45–gp44/62 complex together adopts a “notched screw cap arrangement” that matches the helical geometry of the DNA duplex. In this orientation, all four interfacial ATPase sites are aligned properly for optimal hydrolysis (Piperno et al. 1978; Mace and Alberts 1984; Young et al. 1996; Kelch et al. 2011). Furthermore, the gap between domains in the open clamp is not wide enough to allow passage of duplex DNA. Hence, it is presumed that the single-stranded portion of the template strand enters through the gaps in the gp45 clamp and between the A′ and AAA+ domains of the gp62 (A) subunit of gp44/62 and the complex “screws” onto the double-stranded portion of the DNA. Similar to that observed in the crystal structure of the E. coli clamp loader–DNA complex described above, the closest contacts between gp44/62 and the DNA involve the template strand (Jarvis et al. 1989a; Hockensmith et al. 1993; Kelch et al. 2011). Positively-charged/polar residues from the inner surface of the clamp and the AAA+ modules within the inner chamber of gp44/62 track the minor groove of the duplex region. The single-stranded template overhang protrudes through the gap between the A and A′ domains of the gp62 (A) subunit and binds to its collar domain. The 3′ end of the primer is blocked by the collar domain of the gp44B (B) subunit. This structure depicted in Figure 5 closely resembles a lower resolution, electron microscopy (EM) structure from the archaeon Pyrococcus furiosus in which PfuRFC is bound to DNA and an open PfuPCNA ring. Indeed, most of the T4 crystal structure fits well into the archaeal EM structure, showing that the open clamp–clamp loader–DNA complex is a common intermediate in the clamp-loading process for both T4 and archaea (Miyata et al. 2004, 2005; Kelch et al. 2011).
Figure 5.
The clamp–clamp loader–DNA complex from T4 bacteriophage. Structure of the T4 clamp loader (gp44/62) bound to an open clamp (gp45) and PT DNA (from PDB 3U60) generated using PyMOL. PT DNA, gp44/62 (multicolored), and gp45 (gray) are shown in cartoon form. The A′ domain (red) as well as the primer (orange) and template (yellow) strands of the duplex DNA are indicated.
For bacteria and eukaryotes, it is suggested that the ATP-bound clamp loader holds the clamp in an open, right-handed spiral around DNA as depicted in Figure 5 but such complexes have yet to be captured. However, substantial evidence supports their existence. The DNA-dependent ATPase activity of the E. coli γ complex (Onrust et al. 1991; Xiao et al. 1995; Williams et al. 2004) and eukaryotic RFC (Tsurimoto and Stillman 1990; Lee et al. 1991; Podust et al. 1992; Cai et al. 1996; Ellison and Stillman 1998; Zhang et al. 1999; McNally et al. 2010) is stimulated by their respective sliding clamp, suggesting that an open clamp stabilizes the “notched screw cap” arrangement of the clamp loader on DNA. Furthermore, the experimentally-measured distance between PCNA subunits in the PCNA–RFC–DNA complex from eukaryotes (34 Å) agrees fairly well with that determined from a MD-based model (39 Å) of the open PCNA–RFC–DNA complex (Kazmirski et al. 2005; Kelch et al. 2011; Sakato et al. 2011b). Finally, the asymmetric DNA footprint of the eukaryotic PCNA–RFC complex agrees with that predicted by the MD-based model (Tsurimoto and Stillman 1991). Thus, the clamp-loading process within all domains of life likely proceeds through a common open clamp–clamp loader–DNA intermediate.
Closure of the Clamp Ring
In the “notched screw cap” arrangement of the clamp–clamp loader complex on DNA, the interfacial ATPase sites are aligned properly for optimal hydrolysis. As described above, the clamp loaders revert back to a low-affinity DNA-binding state on hydrolysis of ATP and eject (Fig. 4C), leaving the PT DNA positioned within the opened sliding clamp ring (Tsurimoto and Stillman 1991; Kaboord and Benkovic 1995, 1996; Berdis and Benkovic 1996; Zhang et al. 1999; Gomes and Burgers 2001; Stewart et al. 2001; Trakselis et al. 2003; Seybert and Wigley 2004; Johnson et al. 2006; Seybert et al. 2006; Zhuang et al. 2006b; Chen et al. 2009a; Park and O’Donnell 2009). Concurrent or subsequent to ejection of the clamp loader, electrostatic interactions between the positively charged inner surface of the sliding clamp ring and the negatively charged DNA drive contraction of the open sliding clamp around DNA. For bacteria, eukaryotes, and presumably archaea, such a process results in complete closure of the sliding clamp ring (Tinker et al. 1994; Fukuda et al. 1995; Johnson et al. 2006; Zhuang et al. 2006b; Georgescu et al. 2008; Anderson et al. 2009; McNally et al. 2010). However, the T4 gp45 ring remains partially open and T4 gp43 DNA polymerase subsequently fills the void by inserting its carboxyl terminus into the open subunit interface and docks onto the carboxy-terminal face of gp45 (Pietroni et al. 1997; Alley et al. 1999a, 2000; Latham et al. 1999; Trakselis et al. 2001). Regardless, in all domains of life, ATP hydrolysis is required for ejection of the clamp loader from DNA and closure of the sliding clamp ring. Indeed, ATP hydrolysis is necessary for association of replicative polymerases with loaded sliding clamps, a process presumed to require closure of the clamp ring as well as displacement of the clamp loader as clamp loaders and polymerases share common binding sites on sliding clamps (Piperno and Alberts 1978; Tsurimoto and Stillman 1990; Burgers 1991; Lee et al. 1991; Munn and Alberts 1991a; Fotedar et al. 1996; Mossi et al. 1997; Oku et al. 1998; Zhang et al. 1999; Gomes et al. 2001; Seybert and Wigley 2004; Liu et al. 2011). However, it should be noted that numerous studies have reported that eukaryotic RFC remains engaged with DNA throughout DNA replication (Waga and Stillman 1998; Masuda et al. 2007; Kumar et al. 2010). Although such behavior may be ascribed to nonspecific binding of RFC either to DNA through the ligase homology domain of RFC1 (Podust et al. 1998b) or to replication protein A (RPA) through RFCs 1, 4, and 5 (Waga and Stillman 1998; Yuzhakov et al. 1999), this raises the possibility that RFC may remain in the vicinity of the PT junction after ATP hydrolysis to either “hold” or reload PCNA at the primer terminus on dissociation of polymerase δ (Masuda et al. 2007; Waga and Stillman 1998). Regardless, once clamp loaders exchange ADP for ATP, the catalytic cycle may continue (Bertram et al. 1998, 2000; Hingorani and O’Donnell 1998; Pietroni et al. 2001).
The aforementioned mechanism for assembly of sliding clamps onto DNA is most likely the predominant pathway in all domains of life (Gomes et al. 2001; Thompson et al. 2009; Sakato et al. 2011b). However, the unique open structure of free gp45 clamp may also provide alternative pathways. ATP-bound gp44/62 may first bind to a PT junction and then load gp45 (Smiley et al. 2006; Zhuang et al. 2006a). For other domains of life, the geometry of the clamp loader–DNA complex likely prevents the clamp loader from productively binding a closed clamp (Thompson et al. 2009). Although this pathway is productive in T4 bacteriophage, it is most likely insignificant as binding of clamp loaders to PT DNA triggers rapid ATP hydrolysis and their subsequent dissociation such that clamp loader–DNA complexes are very short-lived (Chen et al. 2009a; Thompson et al. 2009). Alternatively, the open gp45 ring may spontaneously load onto DNA in a nonproductive manner and subsequently be “corrected” by gp44/62 before entry of gp43 polymerase. However, this pathway is highly inefficient and requires high concentrations of gp45 and/or molecular crowding (Reddy et al. 1993; Sanders et al. 1994; Smiley et al. 2006).
CLAMP UNLOADING
In a replicating cell, Okazaki fragments are in excess of sliding clamps, necessitating an efficient unloading mechanism to recycle sliding clamps during DNA replication (Yao et al. 1996; Leu et al. 2000). For example, in an E. coli cell, there are only about 350 β clamps for 2000–4000 Okazaki fragments. With each round of replication taking only 40 or so min, this requires each β clamp to recycle every 3.5 to 7 min (Leu et al. 2000). Similar arguments apply to other domains of life (Yao et al. 1996). In T4, the open gp45 ring is not stable on DNA and rapidly dissociates on departure of gp43 polymerase. Such a process occurs with a half-life of approximately 1.4 min, well within the allotted time window for clamp recycling. Thus, enzyme-catalyzed clamp removal is not necessary (Kaboord and Benkovic 1996; Yao et al. 1996; Soumillion et al. 1998). However, as mentioned above, the closed circular structure of sliding clamps from bacteria, eukaryotes, and archaea is very stable. Consequently, spontaneous dissociation from DNA occurs much slower than that for gp45 with half-lives of 24 and 72 min reported for eukaryotic PCNA and E. coli β clamp, respectively (Podust et al. 1995; Yao et al. 1996; Leu et al. 2000). Thus, sliding clamps must be catalytically recycled within these domains of life to keep up with ongoing DNA replication (Lee and Hurwitz 1990; Yao et al. 2006). Surprisingly, clamp loaders seem to perform this feat. In addition to loading their respective clamps onto DNA, clamp loaders from bacteria (γ complex), archaea (PfuRFC), and eukaryotes (RFC) have been shown to remove loaded clamps from DNA in an ATP-dependent process (Naktinis et al. 1995; Cai et al. 1996; Yao et al. 1996; Shibahara and Stillman 1999; Gomes et al. 2000; Leu et al. 2000; Cann et al. 2001). Although the mechanistic details have yet to be revealed, this suggests that replicative polymerases prevent unwanted clamp unloading during ongoing replication by blocking the clamp loaders access to loaded sliding clamps. On completion of an Okazaki fragment, the polymerase dissociates, providing an opportunity for the clamp loader to recycle the sliding clamp (Naktinis et al. 1995; Oku et al. 1998).
CONCLUDING REMARKS
The twenty-first century has been an exciting time for the study of clamps and clamp loaders. Three-dimensional structures of sliding clamps and clamp loaders from all domains of life as well as T4 bacteriophage have now been reported, showing that their basic architecture and composition has been conserved throughout evolution. Also, thorough biochemical analyses along with ground-breaking crystal structures have revealed the primary role of ATP as a modulator of the interaction between clamp loaders and PT DNA and outlined similar sequential mechanisms for the clamp-loading process within the different life forms that are sprinkled with subtle differences. However, many important questions still remain unanswered. For instance, what is the nature of the ATP-dependent conformational change within archaeal, eukaryotic, and T4 clamps loaders that permit optimal interaction with their respective clamps? Do these clamp loaders “actively” pry open sliding clamp rings like the bacterial clamp loader or “passively” wait for them to open once bound? What is the timing of clamp loader ejection and ring closure? How is the unloading activity of clamp loaders controlled during DNA replication? It will be interesting to see what answers future studies will provide for these lingering questions.
ACKNOWLEDGMENTS
We thank Dr. Jarrod French of the Benkovic Laboratory for critically reading this chapter. This work is supported by National Institutes of Health (NIH) grant GM13306 (S.J.B.). M.H. is supported by the National Cancer Institute of the NIH under Award Number F32CA165471. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
Editors: Stephen D. Bell, Marcel Méchali, and Melvin L. DePamphilis
Additional Perspectives on DNA Replication available at www.cshperspectives.org
REFERENCES
- Adelman JL, Chodera JD, Kuo IF, Miller TF III, Barsky D 2010. The mechanical properties of PCNA: Implications for the loading and function of a DNA sliding clamp. Biophys J 98: 3062–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR, Stege P, Scheffzek K, Wittinghofer A 1997. Confirmation of the arginine-finger hypothesis for the GAP-stimulated GTP-hydrolysis reaction of Ras. Nat Struct Biol 4: 686–689 [DOI] [PubMed] [Google Scholar]
- Allen BL, Uhlmann F, Gaur LK, Mulder BA, Posey KL, Jones LB, Hardin SH 1998. DNA recognition properties of the N-terminal DNA binding domain within the large subunit of replication factor C. Nucleic Acids Res 26: 3877–3882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alley SC, Jones AD, Soumillion P, Benkovic SJ 1999a. The carboxyl terminus of the bacteriophage T4 DNA polymerase contacts its sliding clamp at the subunit interface. J Biol Chem 274: 24485–24489 [DOI] [PubMed] [Google Scholar]
- Alley SC, Shier VK, Abel-Santos E, Sexton DJ, Soumillion P, Benkovic SJ 1999b. Sliding clamp of the bacteriophage T4 polymerase has open and closed subunit interfaces in solution. Biochemistry 38: 7696–7709 [DOI] [PubMed] [Google Scholar]
- Alley SC, Abel-Santos E, Benkovic SJ 2000. Tracking sliding clamp opening and closing during bacteriophage T4 DNA polymerase holoenzyme assembly. Biochemistry 39: 3076–3090 [DOI] [PubMed] [Google Scholar]
- Anderson SG, Thompson JA, Paschall CO, O’Donnell M, Bloom LB 2009. Temporal correlation of DNA binding, ATP hydrolysis, and clamp release in the clamp loading reaction catalyzed by the Escherichia coli γ complex. Biochemistry 48: 8516–8527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ason B, Bertram JG, Hingorani MM, Beechem JM, O’Donnell M, Goodman MF, Bloom LB 2000. A model for Escherichia coli DNA polymerase III holoenzyme assembly at primer/template ends. DNA triggers a change in binding specificity of the γ complex clamp loader. J Biol Chem 275: 3006–3015 [DOI] [PubMed] [Google Scholar]
- Ason B, Handayani R, Williams CR, Bertram JG, Hingorani MM, O’Donnell M, Goodman MF, Bloom LB 2003. Mechanism of loading the Escherichia coli DNA polymerase III β sliding clamp on DNA. Bona fide primer/templates preferentially trigger the γ complex to hydrolyze ATP and load the clamp. J Biol Chem 278: 10033–10040 [DOI] [PubMed] [Google Scholar]
- Bauer GA, Burgers PM 1988. The yeast analog of mammalian cyclin/proliferating-cell nuclear antigen interacts with mammalian DNA polymerase δ. Proc Natl Acad Sci 85: 7506–7510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berdis AJ, Benkovic SJ 1996. Role of adenosine 5′-triphosphate hydrolysis in the assembly of the bacteriophage T4 DNA replication holoenzyme complex. Biochemistry 35: 9253–9265 [DOI] [PubMed] [Google Scholar]
- Bertram JG, Bloom LB, Turner J, O’Donnell M, Beechem JM, Goodman MF 1998. Pre-steady state analysis of the assembly of wild type and mutant circular clamps of Escherichia coli DNA polymerase III onto DNA. J Biol Chem 273: 24564–24574 [DOI] [PubMed] [Google Scholar]
- Bertram JG, Bloom LB, Hingorani MM, Beechem JM, O’Donnell M, Goodman MF 2000. Molecular mechanism and energetics of clamp assembly in Escherichia coli. The role of ATP hydrolysis when γ complex loads β on DNA. J Biol Chem 275: 28413–28420 [DOI] [PubMed] [Google Scholar]
- Bloom LB, Turner J, Kelman Z, Beechem JM, O’Donnell M, Goodman MF 1996. Dynamics of loading the β sliding clamp of DNA polymerase III onto DNA. J Biol Chem 271: 30699–30708 [DOI] [PubMed] [Google Scholar]
- Bowman GD, O’Donnell M, Kuriyan J 2004. Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex. Nature 429: 724–730 [DOI] [PubMed] [Google Scholar]
- Bunz F, Kobayashi R, Stillman B 1993. cDNAs encoding the large subunit of human replication factor C. Proc Natl Acad Sci 90: 11014–11018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbelo PD, Utani A, Pan ZQ, Yamada Y 1993. Cloning of the large subunit of activator 1 (replication factor C) reveals homology with bacterial DNA ligases. Proc Natl Acad Sci 90: 11543–11547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgers PM 1991. Saccharomyces cerevisiae replication factor C II formation and activity of complexes with the proliferating cell nuclear antigen and with DNA polymerases δ and ε. J Biol Chem 266: 22698–22706 [PubMed] [Google Scholar]
- Cai J, Uhlmann F, Gibbs E, Flores-Rozas H, Lee CG, Phillips B, Finkelstein J, Yao N, O’Donnell M, Hurwitz J 1996. Reconstitution of human replication factor C from its five subunits in baculovirus-infected insect cells. Proc Natl Acad Sci 93: 12896–12901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai J, Gibbs E, Uhlmann F, Phillips B, Yao N, O’Donnell M, Hurwitz J 1997. A complex consisting of human replication factor C p40, p37, and p36 subunits is a DNA-dependent ATPase and an intermediate in the assembly of the holoenzyme. J Biol Chem 272: 18974–18981 [DOI] [PubMed] [Google Scholar]
- Cai J, Yao N, Gibbs E, Finkelstein J, Phillips B, O’Donnell M, Hurwitz J 1998. ATP hydrolysis catalyzed by human replication factor C requires participation of multiple subunits. Proc Natl Acad Sci 95: 11607–11612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann IK, Ishino Y 1999. Archaeal DNA replication: Identifying the pieces to solve a puzzle. Genetics 152: 1249–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann IK, Ishino S, Hayashi I, Komori K, Toh H, Morihawa K, Ishino Y 1999. Functional interactions of a homolog of proliferating cell nuclear antigen with DNA polymerases in Archaea. J Bacteriol 181: 6591–6599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cann IK, Ishino S, Yuasa M, Daiyasu H, Toh H, Ishino Y 2001. Biochemical analysis of replication factor C from the hyperthermophilic archaeon Pyrococcus furiosus. J Bacteriol 183: 2614–2623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capson TL, Benkovic SJ, Nossal NG 1991. Protein-DNA cross-linking demonstrates stepwise ATP-dependent assembly of T4 DNA polymerase and its accessory proteins on the primer-template. Cell 65: 249–258 [DOI] [PubMed] [Google Scholar]
- Chen M, Pan ZQ, Hurwitz J 1992a. Sequence and expression in Escherichia coli of the 40-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci 89: 2516–2520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Pan ZQ, Hurwitz J 1992b. Studies of the cloned 37-kDa subunit of activator 1 (replication factor C) of HeLa cells. Proc Natl Acad Sci 89: 5211–5215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Kocherginskaya SA, Lin Y, Sriratana B, Lagunas AM, Robbins JB, Mackie RI, Cann IK 2005. Biochemical and mutational analyses of a unique clamp loader complex in the archaeon Methanosarcina acetivorans. J Biol Chem 280: 41852–41863 [DOI] [PubMed] [Google Scholar]
- Chen S, Levin MK, Sakato M, Zhou Y, Hingorani MM 2009a. Mechanism of ATP-driven PCNA clamp loading by S. cerevisiae RFC. J Mol Biol 388: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Lin Y, Yoshinaga A, Chhotani B, Lorenzini JL, Crofts AA, Mei S, Mackie RI, Ishinio Y, Cann IK 2009b. Molecular analyses of a three-subunit euryarchaeal clamp loader complex from Methanosarcina acetivorans. J Bacteriol 191: 6539–6549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullmann G, Fien K, Kobayashi R, Stillman B 1995. Characterization of the five replication factor C genes of Saccharomyces cerevisiae. Mol Cell Biol 15: 4661–4671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daimon K, Kawarabayasi Y, Kikuchi H, Sako Y, Ishino Y 2002. Three proliferating cell nuclear antigen-like proteins found in the hyperthermophilic archaeon Aeropyrum pernix: Interactions with the two DNA polymerases. J Bacteriol 184: 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davey MJ, Jeruzalmi D, Kuriyan J, O’Donnell M 2002. Motors and switches: AAA+ machines within the replisome. Nat Rev Mol Cell Biol 3: 826–835 [DOI] [PubMed] [Google Scholar]
- Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD 2003. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Mol Cell 11: 275–282 [DOI] [PubMed] [Google Scholar]
- Dionne I, Brown NJ, Woodgate R, Bell SD 2008. On the mechanism of loading the PCNA sliding clamp by RFC. Mol Microbiol 68: 216–222 [DOI] [PubMed] [Google Scholar]
- Ellison V, Stillman B 1998. Reconstitution of recombinant human replication factor C (RFC) and identification of an RFC subcomplex possessing DNA-dependent ATPase activity. J Biol Chem 273: 5979–5987 [DOI] [PubMed] [Google Scholar]
- Fien K, Stillman B 1992. Identification of replication factor C from Saccharomyces cerevisiae: A component of the leading-strand DNA replication complex. Mol Cell Biol 12: 155–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fotedar R, Mossi R, Fitzgerald P, Rousselle T, Maga G, Brickner H, Messier H, Kasibhatia S, Hübscher U, Fotedar A 1996. A conserved domain of the large subunit of replication factor C binds PCNA and acts like a dominant negative inhibitor of DNA replication in mammalian cells. EMBO J 15: 4423–4433 [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T 1995. Structure-function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. J Biol Chem 270: 22527–22534 [DOI] [PubMed] [Google Scholar]
- Gary SL, Burgers MJ 1995. Identification of the fifth subunit of Saccharomyces cerevisiae replication factor C. Nucleic Acids Res 23: 4986–4991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgescu RE, Kim SS, Yurieva O, Kuriyan J, Kong XP, O’Donnell M 2008. Structure of a sliding clamp on DNA. Cell 132: 43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover BP, McHenry CS 1998. The χ ψ subunits of DNA polymerase III holoenzyme bind to single-stranded DNA-binding protein (SSB) and facilitate replication of an SSB-coated template. J Biol Chem 273: 23476–23484 [DOI] [PubMed] [Google Scholar]
- Goedken ER, Levitus M, Johnson A, Bustamante C, O’Donnell M, Kuriyan J 2004. Fluorescence measurements on the E.coli DNA polymerase clamp loader: Implications for conformational changes during ATP and clamp binding. J Mol Biol 336: 1047–1059 [DOI] [PubMed] [Google Scholar]
- Gomes XV, Burgers PM 2001. ATP utilization by yeast replication factor C. I. ATP-mediated interaction with DNA and with proliferating cell nuclear antigen. J Biol Chem 276: 34768–34775 [DOI] [PubMed] [Google Scholar]
- Gomes XV, Gary SL, Burgers PM 2000. Overproduction in Escherichia coli and characterization of yeast replication factor C lacking the ligase homology domain. J Biol Chem 275: 14541–14549 [DOI] [PubMed] [Google Scholar]
- Gomes XV, Schmidt SL, Burgers PM 2001. ATP utilization by yeast replication factor C. II. Multiple stepwise ATP binding events are required to load proliferating cell nuclear antigen onto primed DNA. J Biol Chem 276: 34776–34783 [DOI] [PubMed] [Google Scholar]
- Guenther B, Onrust R, Sali A, O’Donnell M, Kuriyan J 1997. Crystal structure of the δ′ subunit of the clamp-loader complex of E. coli DNA polymerase III. Cell 91: 335–345 [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O’Donnell M, Kuriyan J 1996. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell 87: 297–306 [DOI] [PubMed] [Google Scholar]
- Henneke G, Gueguen Y, Flament D, Azam P, Querellou J, Dietrich H, Hübscher U, Raffin JP 2002. Replication factor C from the hyperthermophilic archaeon Pyrococcus abyssi does not need ATP hydrolysis for clamp-loading and contains a functionally conserved RFC PCNA-binding domain. J Mol Biol 323: 795–810 [DOI] [PubMed] [Google Scholar]
- Hingorani MM, Coman MM 2002. On the specificity of interaction between the Saccharomyces cerevisiae clamp loader replication factor C and primed DNA templates during DNA replication. J Biol Chem 277: 47213–47224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani MM, O’Donnell M 1998. ATP binding to the Escherichia coli clamp loader powers opening of the ring-shaped clamp of DNA polymerase III holoenzyme. J Biol Chem 273: 24550–24563 [DOI] [PubMed] [Google Scholar]
- Hingorani MM, Bloom LB, Goodman MF, O’Donnell M 1999. Division of labor—sequential ATP hydrolysis drives assembly of a DNA polymerase sliding clamp around DNA. EMBO J 18: 5131–5144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlinkova V, Xing G, Bauer J, Shin YJ, Dionne I, Rajashankar KR, Bell SD, Ling H 2008. Structures of monomeric, dimeric and trimeric PCNA: PCNA-ring assembly and opening. Acta Crystallogr D Biol Crystallogr 64: 941–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockensmith JW, Kubasek WL, Evertsz EM, Mesner LD, von Hippel PH 1993. Laser cross-linking of proteins to nucleic acids. II. Interactions of the bacteriophage T4 DNA replication polymerase accessory proteins complex with DNA. J Biol Chem 268: 15721–15730 [PubMed] [Google Scholar]
- Indiani C, O’Donnell M 2003. Mechanism of the δ wrench in opening the β sliding clamp. J Biol Chem 278: 40272–40281 [DOI] [PubMed] [Google Scholar]
- Iwai T, Kurosawa N, Itoh YH, Horiuchi T 2000. Phylogenetic analysis of archaeal PCNA homologues. Extremophiles 4: 357–364 [DOI] [PubMed] [Google Scholar]
- Jarvis TC, Paul LS, Hockensmith JW, von Hippel PH 1989a. Structural and enzymatic studies of the T4 DNA replication system. II. ATPase properties of the polymerase accessory protein complex. J Biol Chem 264: 12717–12729 [PubMed] [Google Scholar]
- Jarvis TC, Paul LS, von Hippel PH 1989b. Structural and enzymatic studies of the T4 DNA replication system. I. Physical characterization of the polymerase accessory protein complex. J Biol Chem 264: 12709–12716 [PubMed] [Google Scholar]
- Jeruzalmi D, O’Donnell M, Kuriyan J 2001a. Crystal structure of the processivity clamp loader gamma (γ) complex of E. coli DNA polymerase III. Cell 106: 429–441 [DOI] [PubMed] [Google Scholar]
- Jeruzalmi D, Yurieva O, Zhao Y, Young M, Stewart J, Hingorani M, O’Donnell M, Kuriyan J 2001b. Mechanism of processivity clamp opening by the δ subunit wrench of the clamp loader complex of E. coli DNA polymerase III. Cell 106: 417–428 [PubMed] [Google Scholar]
- Johnson A, O’Donnell M 2003. Ordered ATP hydrolysis in the γ complex clamp loader AAA+ machine. J Biol Chem 278: 14406–14413 [DOI] [PubMed] [Google Scholar]
- Johnson A, Yao NY, Bowman GD, Kuriyan J, O’Donnell M 2006. The replication factor C clamp loader requires arginine finger sensors to drive DNA binding and proliferating cell nuclear antigen loading. J Biol Chem 281: 35531–35543 [DOI] [PubMed] [Google Scholar]
- Kaboord BF, Benkovic SJ 1995. Accessory proteins function as matchmakers in the assembly of the T4 DNA polymerase holoenzyme. Curr Biol 5: 149–157 [DOI] [PubMed] [Google Scholar]
- Kaboord BF, Benkovic SJ 1996. Dual role of the 44/62 protein as a matchmaker protein and DNA polymerase chaperone during assembly of the bacteriophage T4 holoenzyme complex. Biochemistry 35: 1084–1092 [DOI] [PubMed] [Google Scholar]
- Kazmirski SL, Podobnik M, Weitze TF, O’Donnell M, Kuriyan J 2004. Structural analysis of the inactive state of the Escherichia coli DNA polymerase clamp-loader complex. Proc Natl Acad Sci 101: 16750–16755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmirski SL, Zhao Y, Bowman GD, O’Donnell M, Kuriyan J 2005. Out-of-plane motions in open sliding clamps: Molecular dynamics simulations of eukaryotic and archaeal proliferating cell nuclear antigen. Proc Natl Acad Sci 102: 13801–13806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelch BA, Makino DL, O’Donnell M, Kuriyan J 2011. How a DNA polymerase clamp loader opens a sliding clamp. Science 334: 1675–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z, Hurwitz J 2000. A unique organization of the protein subunits of the DNA polymerase clamp loader in the archaeon Methanobacterium thermoautotrophicum δH. J Biol Chem 275: 7327–7336 [DOI] [PubMed] [Google Scholar]
- Kelman Z, Yuzhakov A, Andjelkovic J, O’Donnell M 1998. Devoted to the lagging strand—the subunit of DNA polymerase III holoenzyme contacts SSB to promote processive elongation and sliding clamp assembly. EMBO J 17: 2436–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong XP, Onrust R, O’Donnell M, Kuriyan J 1992. Three-dimensional structure of the β subunit of E. coli DNA polymerase III holoenzyme: A sliding DNA clamp. Cell 69: 425–437 [DOI] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J 1994. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell 79: 1233–1243 [DOI] [PubMed] [Google Scholar]
- Kumar R, Nashine VC, Mishra PP, Benkovic SJ, Lee TH 2010. Stepwise loading of yeast clamp revealed by ensemble and single-molecule studies. Proc Natl Acad Sci 107: 19736–19741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham GJ, Bacheller DJ, Pietroni P, von Hippel PH 1997. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. II. The Gp44/62 clamp loader interacts with a single defined face of the sliding clamp ring. J Biol Chem 272: 31677–31684 [DOI] [PubMed] [Google Scholar]
- Latham GJ, Dong F, Pietroni P, Dozono JM, Bacheller DJ, von Hippel PH 1999. Opening of a monomer-monomer interface of the trimeric bacteriophage T4-coded GP45 sliding clamp is required for clamp loading onto DNA. Proc Natl Acad Sci 96: 12448–12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Hurwitz J 1990. Mechanism of elongation of primed DNA by DNA polymerase δ, proliferating cell nuclear antigen, and activator 1. Proc Natl Acad Sci 87: 5672–5676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Kwong AD, Pan ZQ, Hurwitz J 1991. Studies on the activator 1 protein complex, an accessory factor for proliferating cell nuclear antigen-dependent DNA polymerase δ. J Biol Chem 266: 594–602 [PubMed] [Google Scholar]
- Leu FP, O’Donnell M 2001. Interplay of clamp loader subunits in opening the β sliding clamp of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem 276: 47185–47194 [DOI] [PubMed] [Google Scholar]
- Leu FP, Hingorani MM, Turner J, O’Donnell M 2000. The δ subunit of DNA polymerase III holoenzyme serves as a sliding clamp unloader in Escherichia coli. J Biol Chem 275: 34609–34618 [DOI] [PubMed] [Google Scholar]
- Leu FP, Georgescu R, O’Donnell M 2003. Mechanism of the E. coli τ processivity switch during lagging-strand synthesis. Mol Cell 11: 315–327 [DOI] [PubMed] [Google Scholar]
- Li X, Burgers PM 1994a. Cloning and characterization of the essential Saccharomyces cerevisiae RFC4 gene encoding the 37-kDa subunit of replication factor C. J Biol Chem 269: 21880–21884 [PubMed] [Google Scholar]
- Li X, Burgers PM 1994b. Molecular cloning and expression of the Saccharomyces cerevisiae RFC3 gene, an essential component of replication factor C. Proc Natl Acad Sci 91: 868–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C, McKinney MC, Chen YH, Earnest TM, Shi X, Lin LJ, Ishino Y, Dahmen K, Cann IK, Ha T 2011. Reverse-chaperoning activity of an AAA+ protein. Biophys J 100: 1344–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace DC, Alberts BM 1984. The complex of T4 bacteriophage gene 44 and 62 replication proteins forms an ATPase that is stimulated by DNA and by T4 gene 45 protein. J Mol Biol 177: 279–293 [DOI] [PubMed] [Google Scholar]
- Masuda Y, Suzuki M, Piao J, Gu Y, Tsurimoto T, Kamiya K 2007. Dynamics of human replication factors in the elongation phase of DNA replication. Nucleic Acids Res 35: 6904–6916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumiya S, Ishino Y, Morikawa K 2001. Crystal structure of an archaeal DNA sliding clamp: Proliferating cell nuclear antigen from Pyrococcus furiosus. Protein Sci 10: 17–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumiya S, Ishino S, Ishino Y, Morikawa K 2002. Physical interaction between proliferating cell nuclear antigen and replication factor C from Pyrococcus furiosus. Genes Cells 7: 911–922 [DOI] [PubMed] [Google Scholar]
- Matsumiya S, Ishino S, Ishino Y, Morikawa K 2003. Intermolecular ion pairs maintain the toroidal structure of Pyrococcus furiosus PCNA. Protein Sci 12: 823–831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayanagi K, Miyata T, Oyama T, Ishino Y, Morikawa K 2001. Three-dimensional electron microscopy of the clamp loader small subunit from Pyrococcus furiosus. J Struct Biol 134: 35–45 [DOI] [PubMed] [Google Scholar]
- McNally R, Bowman GD, Goedken ER, O’Donnell M, Kuriyan J 2010. Analysis of the role of PCNA-DNA contacts during clamp loading. BMC Struct Biol 10: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar D, Trakselis MA, Benkovic SJ 2004. On the solution structure of the T4 sliding clamp (gp45). Biochemistry 43: 12723–12727 [DOI] [PubMed] [Google Scholar]
- Miyata T, Oyama T, Mayanagi K, Ishino S, Ishino Y, Morikawa K 2004. The clamp-loading complex for processive DNA replication. Nat Struct Mol Biol 11: 632–636 [DOI] [PubMed] [Google Scholar]
- Miyata T, Suzuki H, Oyama T, Mayanagi K, Ishino Y, Morikawa K 2005. Open clamp structure in the clamp-loading complex visualized by electron microscopic image analysis. Proc Natl Acad Sci 102: 13795–13800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moarefi I, Jeruzalmi D, Turner J, O’Donnell M, Kuriyan J 2000. Crystal structure of the DNA polymerase processivity factor of T4 bacteriophage. J Mol Biol 296: 1215–1223 [DOI] [PubMed] [Google Scholar]
- Mossi R, Jonsson ZO, Allen BL, Hardin SH, Hubscher U 1997. Replication factor C interacts with the C-terminal side of proliferating cell nuclear antigen. J Biol Chem 272: 1769–1776 [DOI] [PubMed] [Google Scholar]
- Munn MM, Alberts BM 1991a. DNA footprinting studies of the complex formed by the T4 DNA polymerase holoenzyme at a primer-template junction. J Biol Chem 266: 20034–20044 [PubMed] [Google Scholar]
- Munn MM, Alberts BM 1991b. The T4 DNA polymerase accessory proteins form an ATP-dependent complex on a primer-template junction. J Biol Chem 266: 20024–20033 [PubMed] [Google Scholar]
- Naktinis V, Onrust R, Fang L, O’Donnell M 1995. Assembly of a chromosomal replication machine: Two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. II. Intermediate complex between the clamp loader and its clamp. J Biol Chem 270: 13358–13365 [PubMed] [Google Scholar]
- Noskov V, Maki S, Kawasaki Y, Leem SH, Ono B, Araki H, Pavlov Y, Sugino A 1994. The RFC2 gene encoding a subunit of replication factor C of Saccharomyces cerevisiae. Nucleic Acids Res 22: 1527–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donnell M, Onrust R, Dean FB, Chen M, Hurwitz J 1993. Homology in accessory proteins of replicative polymerases—E coli to humans. Nucleic Acids Res 21: 1–3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oku T, Ikeda S, Sasaki H, Fukuda K, Morioka H, Ohtsuka E, Yoshikawa H, Tsurimoto T 1998. Functional sites of human PCNA which interact with p21 (Cip1/Waf1), DNA polymerase δ and replication factor C. Genes Cells 3: 357–369 [DOI] [PubMed] [Google Scholar]
- Olson MW, Dallmann HG, McHenry CS 1995. DnaX complex of Escherichia coli DNA polymerase III holoenzyme. The χ ψ complex functions by increasing the affinity of tau and γ for δ.δ′ to a physiologically relevant range. J Biol Chem 270: 29570–29577 [PubMed] [Google Scholar]
- Onrust R, O’Donnell M 1993. DNA polymerase III accessory proteins. II. Characterization of δ and δ′. J Biol Chem 268: 11766–11772 [PubMed] [Google Scholar]
- Onrust R, Stukenberg PT, O’Donnell M 1991. Analysis of the ATPase subassembly which initiates processive DNA synthesis by DNA polymerase III holoenzyme. J Biol Chem 266: 21681–21686 [PubMed] [Google Scholar]
- Onrust R, Finkelstein J, Naktinis V, Turner J, Fang L, O’Donnell M 1995. Assembly of a chromosomal replication machine: Two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. I. Organization of the clamp loader. J Biol Chem 270: 13348–13357 [DOI] [PubMed] [Google Scholar]
- Oyama T, Ishino Y, Cann IK, Ishino S, Morikawa K 2001. Atomic structure of the clamp loader small subunit from Pyrococcus furiosus. Mol Cell 8: 455–463 [DOI] [PubMed] [Google Scholar]
- Pan ZQ, Chen M, Hurwitz J 1993. The subunits of activator 1 (replication factor C) carry out multiple functions essential for proliferating-cell nuclear antigen-dependent DNA synthesis. Proc Natl Acad Sci 90: 6–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park MS, O’Donnell M 2009. The clamp loader assembles the β clamp onto either a 3′ or 5′ primer terminus: The underlying basis favoring 3′ loading. J Biol Chem 284: 31473–31483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascal JM, Tsodikov OV, Hura GL, Song W, Cotner EA, Classen S, Tomkinson AE, Tainer JA, Ellenberger T 2006. A flexible interface between DNA ligase and PCNA supports conformational switching and efficient ligation of DNA. Mol Cell 24: 279–291 [DOI] [PubMed] [Google Scholar]
- Paschall CO, Thompson JA, Marzahn MR, Chiraniya A, Hayner JN, O’Donnell M, Robbins AH, McKenna R, Bloom LB 2011. The E. coli clamp loader can actively pry open the β-sliding clamp. J Biol Chem 286: 42704–42714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietroni P, von Hippel PH 2008. Multiple ATP binding is required to stabilize the “activated” (clamp open) clamp loader of the T4 DNA replication complex. J Biol Chem 283: 28338–28353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietroni P, Young MC, Latham GJ, von Hippel PH 1997. Structural analyses of gp45 sliding clamp interactions during assembly of the bacteriophage T4 DNA polymerase holoenzyme. I. Conformational changes within the gp44/62-gp45-ATP complex during clamp loading. J Biol Chem 272: 31666–31676 [DOI] [PubMed] [Google Scholar]
- Pietroni P, Young MC, Latham GJ, von Hippel PH 2001. Dissection of the ATP-driven reaction cycle of the bacteriophage T4 DNA replication processivity clamp loading system. J Mol Biol 309: 869–891 [DOI] [PubMed] [Google Scholar]
- Piperno JR, Alberts BM 1978. An ATP stimulation of T4 DNA polymerase mediated via T4 gene 44/62 and 45 proteins. The requirement for ATP hydrolysis. J Biol Chem 253: 5174–5179 [PubMed] [Google Scholar]
- Piperno JR, Kallen RG, Alberta BM 1978. Analysis of a T4 DNA replication protein complex. Studies of the DNA recognition site for T4 gene 44/62 and 45 protein-catalyzed ATP hydrolysis. J Biol Chem 253: 5180–5185 [PubMed] [Google Scholar]
- Pisani FM, De Felice M, Carpentieri F, Rossi M 2000. Biochemical characterization of a clamp-loader complex homologous to eukaryotic replication factor C from the hyperthermophilic archaeon Sulfolobus solfataricus. J Mol Biol 301: 61–73 [DOI] [PubMed] [Google Scholar]
- Podust VN, Georgaki A, Strack B, Hubscher U 1992. Calf thymus RF-C as an essential component for DNA polymerase δ and ε holoenzymes function. Nucleic Acids Res 20: 4159–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust LM, Podust VN, Sogo JM, Hubscher U 1995. Mammalian DNA polymerase auxiliary proteins: Analysis of replication factor C-catalyzed proliferating cell nuclear antigen loading onto circular double-stranded DNA. Mol Cell Biol 15: 3072–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podust VN, Tiwari N, Ott R, Fanning E 1998a. Functional interactions among the subunits of replication factor C potentiate and modulate its ATPase activity. J Biol Chem 273: 12935–12942 [DOI] [PubMed] [Google Scholar]
- Podust VN, Tiwari N, Stephan S, Fanning E 1998b. Replication factor C disengages from proliferating cell nuclear antigen (PCNA) upon sliding clamp formation, and PCNA itself tethers DNA polymerase δ to DNA. J Biol Chem 273: 31992–31999 [DOI] [PubMed] [Google Scholar]
- Pritchard AE, Dallmann HG, Glover BP, McHenry CS 2000. A novel assembly mechanism for the DNA polymerase III holoenzyme DnaX complex: Association of δδ′ with DnaX(4) forms DnaX(3)δδ′. EMBO J 19: 6536–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy MK, Weitzel SE, von Hippel PH 1993. Assembly of a functional replication complex without ATP hydrolysis: A direct interaction of bacteriophage T4 gp45 with T4 DNA polymerase. Proc Natl Acad Sci 90: 3211–3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reems JA, Wood S, McHenry CS 1995. Escherichia coli DNA polymerase III holoenzyme subunits α, β, and γ directly contact the primer-template. J Biol Chem 270: 5606–5613 [DOI] [PubMed] [Google Scholar]
- Reyes-Lamothe R, Sherratt DJ, Leake MC 2010. Stoichiometry and architecture of active DNA replication machinery in Escherichia coli. Science 328: 498–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rush J, Lin TC, Quinones M, Spicer EK, Douglas I, Williams KR, Konigsberg WH 1989. The 44P subunit of the T4 DNA polymerase accessory protein complex catalyzes ATP hydrolysis. J Biol Chem 264: 10943–10953 [PubMed] [Google Scholar]
- Sakato M, O’Donnell M, Hingorani MM 2011a. A central swivel point in the RFC clamp loader controls PCNA opening and loading on DNA. J Mol Biol 416: 163–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakato M, Zhou Y, Hingorani MM 2011b. ATP-binding- and hydrolysis-driven rate-determining events in the RFC-catalyzed PCNA clamp loading reaction. J Mol Biol 416: 176–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders GM, Kassavetis GA, Geiduschek EP 1994. Use of a macromolecular crowding agent to dissect interactions and define functions in transcriptional activation by a DNA-tracking protein: Bacteriophage T4 gene 45 protein and late transcription. Proc Natl Acad Sci 91: 7703–7707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt SL, Gomes XV, Burgers PM 2001. ATP utilization by yeast replication factor C. III. The ATP-binding domains of Rfc2, Rfc3, and Rfc4 are essential for DNA recognition and clamp loading. J Biol Chem 276: 34784–34791 [DOI] [PubMed] [Google Scholar]
- Schurtenberger P, Egelhaaf SU, Hindges R, Maga G, Jónsson ZO, May RP, Glatter O, Hübscher U 1998. The solution structure of functionally active human proliferating cell nuclear antigen determined by small-angle neutron scattering. J Mol Biol 275: 123–132 [DOI] [PubMed] [Google Scholar]
- Sexton DJ, Carver TE, Berdis AJ, Benkovic SJ 1996. Protein–protein and protein-DNA interactions at the bacteriophage T4 DNA replication fork. Characterization of a fluorescently labeled DNA polymerase sliding clamp. J Biol Chem 271: 28045–28051 [DOI] [PubMed] [Google Scholar]
- Sexton DJ, Kaboord BF, Berdis AJ, Carver TE, Benkovic SJ 1998. Dissecting the order of bacteriophage T4 DNA polymerase holoenzyme assembly. Biochemistry 37: 7749– 7756 [DOI] [PubMed] [Google Scholar]
- Seybert A, Wigley DB 2004. Distinct roles for ATP binding and hydrolysis at individual subunits of an archaeal clamp loader. EMBO J 23: 1360–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A, Scott DJ, Scaife S, Singleton MR, Wigley DB 2002. Biochemical characterisation of the clamp/clamp loader proteins from the euryarchaeon Archaeoglobus fulgidus. Nucleic Acids Res 30: 4329–4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seybert A, Singleton MR, Cook N, Hall DR, Wigley DB 2006. Communication between subunits within an archaeal clamp-loader complex. EMBO J 25: 2209–2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibahara K, Stillman B 1999. Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585 [DOI] [PubMed] [Google Scholar]
- Shiomi Y, Usukura J, Masamura Y, Takeyasu K, Nakayama Y, Obuse C, Yoshikawa H, Tsurimoto T 2000. ATP-dependent structural change of the eukaryotic clamp-loader protein, replication factor C. Proc Natl Acad Sci 97: 14127–14132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonetta KR, Kazmirski SL, Goedken ER, Cantor AJ, Kelch BA, McNally R, Seyedin SN, Makino DL, O’Donnell M, Kuriyan J 2009. The mechanism of ATP-dependent primer-template recognition by a clamp loader complex. Cell 137: 659–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley RD, Zhuang Z, Benkovic SJ, Hammes GG 2006. Single-molecule investigation of the T4 bacteriophage DNA polymerase holoenzyme: Multiple pathways of holoenzyme formation. Biochemistry 45: 7990–7997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snyder AK, Williams CR, Johnson A, O’Donnell M, Bloom LB 2004. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: II. Uncoupling the β and DNA binding activities of the γ complex. J Biol Chem 279: 4386–4393 [DOI] [PubMed] [Google Scholar]
- Soumillion P, Sexton DJ, Benkovic SJ 1998. Clamp subunit dissociation dictates bacteriophage T4 DNA polymerase holoenzyme disassembly. Biochemistry 37: 1819–1827 [DOI] [PubMed] [Google Scholar]
- Spicer EK, Nossal NG, Williams KR 1984. Bacteriophage T4 gene 44 DNA polymerase accessory protein. Sequences of gene 44 and its protein product. J Biol Chem 259: 15425–15432 [PubMed] [Google Scholar]
- Stewart J, Hingorani MM, Kelman Z, O’Donnell M 2001. Mechanism of β clamp opening by the δ subunit of Escherichia coli DNA polymerase III holoenzyme. J Biol Chem 276: 19182–19189 [DOI] [PubMed] [Google Scholar]
- Stukenberg PT, Studwell-Vaughan PS, O’Donnell M 1991. Mechanism of the sliding β-clamp of DNA polymerase III holoenzyme. J Biol Chem 266: 11328–11334 [PubMed] [Google Scholar]
- Tainer JA, McCammon JA, Ivanov I 2010. Recognition of the ring-opened state of proliferating cell nuclear antigen by replication factor C promotes eukaryotic clamp-loading. J Am Chem Soc 132: 7372–7378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Paschall CO, O’Donnell M, Bloom LB 2009. A slow ATP-induced conformational change limits the rate of DNA binding but not the rate of β clamp binding by the Escherichia coli γ complex clamp loader. J Biol Chem 284: 32147–32157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JA, Marzahn MR, O’Donnell M, Bloom LB 2011. Replication factor C is a more effective PCNA opener than the checkpoint clamp loader, RAD24-RFC. J Biol Chem 287: 2203–2209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker RL, Kassavetis GA, Geiduschek EP 1994. Detecting the ability of viral, bacterial and eukaryotic replication proteins to track along DNA. EMBO J 13: 5330–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakselis MA, Alley SC, Abel-Santos E, Benkovic SJ 2001. Creating a dynamic picture of the sliding clamp during T4 DNA polymerase holoenzyme assembly by using fluorescence resonance energy transfer. Proc Natl Acad Sci 98: 8368–8375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trakselis MA, Berdis AJ, Benkovic SJ 2003. Examination of the role of the clamp-loader and ATP hydrolysis in the formation of the bacteriophage T4 polymerase holoenzyme. J Mol Biol 326: 435–451 [DOI] [PubMed] [Google Scholar]
- Tsuchihashi Z, Kornberg A 1989. ATP interactions of the tau and γ subunits of DNA polymerase III holoenzyme of Escherichia coli. J Biol Chem 264: 17790–17795 [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B 1990. Functions of replication factor C and proliferating-cell nuclear antigen: Functional similarity of DNA polymerase accessory proteins from human cells and bacteriophage T4. Proc Natl Acad Sci 87: 1023–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurimoto T, Stillman B 1991. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem 266: 1950–1960 [PubMed] [Google Scholar]
- Turner J, Hingorani MM, Kelman Z, O’Donnell M 1999. The internal workings of a DNA polymerase clamp-loading machine. EMBO J 18: 771–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Cai J, Flores-Rozas H, Dean FB, Finkelstein J, O’Donnell M, Hurwitz J 1996. In vitro reconstitution of human replication factor C from its five subunits. Proc Natl Acad Sci 93: 6521–6526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann F, Cai J, Gibbs E, O’Donnell M, Hurwitz J 1997a. Deletion analysis of the large subunit p140 in human replication factor C reveals regions required for complex formation and replication activities. J Biol Chem 272: 10058–10064 [DOI] [PubMed] [Google Scholar]
- Uhlmann F, Gibbs E, Cai J, O’Donnell M, Hurwitz J 1997b. Identification of regions within the four small subunits of human replication factor C required for complex formation and DNA replication. J Biol Chem 272: 10065–10071 [DOI] [PubMed] [Google Scholar]
- Waga S, Stillman B 1998. Cyclin-dependent kinase inhibitor p21 modulates the DNA primer-template recognition complex. Mol Cell Biol 18: 4177–4187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker JE, Saraste M, Runswick MJ, Gay NJ 1982. Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J 1: 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams CR, Snyder AK, Kuzmic P, O’Donnell M, Bloom LB 2004. Mechanism of loading the Escherichia coli DNA polymerase III sliding clamp: I. Two distinct activities for individual ATP sites in the γ complex. J Biol Chem 279: 4376–4385 [DOI] [PubMed] [Google Scholar]
- Williams GJ, Johnson K, Rudolf J, McMahon SA, Carter L, Oke M, Liu H, Taylor GL, White MF, Naismith JH 2006. Structure of the heterotrimeric PCNA from Sulfolobus solfataricus. Acta Crystallogr Sect F Struct Biol Cryst Commun 62: 944–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao H, Dong Z, O’Donnell M 1993. DNA polymerase III accessory proteins. IV. Characterization of χ and ψ. J Biol Chem 268: 11779–11784 [PubMed] [Google Scholar]
- Xiao H, Naktinis V, O’Donnell M 1995. Assembly of a chromosomal replication machine: Two DNA polymerases, a clamp loader, and sliding clamps in one holoenzyme particle. IV. ATP-binding site mutants identify the clamp loader. J Biol Chem 270: 13378–13383 [DOI] [PubMed] [Google Scholar]
- Yao N, Turner J, Kelman Z, Stukenberg PT, Dean F, Shechter D, Pan ZQ, Hurwitz J, O’Donnell M 1996. Clamp loading, unloading and intrinsic stability of the PCNA, β and gp45 sliding clamps of human. E coli and T4 replicases. Genes Cells 1: 101–113 [DOI] [PubMed] [Google Scholar]
- Yao N, Coryell L, Zhang D, Georgescu RE, Finkelstein J, Coman MM, Hingorani MM, O’Donnell M 2003. Replication factor C clamp loader subunit arrangement within the circular pentamer and its attachment points to proliferating cell nuclear antigen. J Biol Chem 278: 50744–50753 [DOI] [PubMed] [Google Scholar]
- Yao NY, Johnson A, Bowman GD, Kuriyan J, O’Donnell M 2006. Mechanism of proliferating cell nuclear antigen clamp opening by replication factor C. J Biol Chem 281: 17528–17539 [DOI] [PubMed] [Google Scholar]
- Yoder BL, Burgers PM 1991. Saccharomyces cerevisiae replication factor C. I. Purification and characterization of its ATPase activity. J Biol Chem 266: 22689–22697 [PubMed] [Google Scholar]
- Young MC, Weitzel SE, von Hippel PH 1996. The kinetic mechanism of formation of the bacteriophage T4 DNA polymerase sliding clamp. J Mol Biol 264: 440–452 [DOI] [PubMed] [Google Scholar]
- Yuzhakov A, Kelman Z, Hurwitz J, O’Donnell M 1999. Multiple competition reactions for RPA order the assembly of the DNA polymerase δ holoenzyme. EMBO J 18: 6189–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Gibbs E, Kelman Z, O’Donnell M, Hurwitz J 1999. Studies on the interactions between human replication factor C and human proliferating cell nuclear antigen. Proc Natl Acad Sci 96: 1869–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang Z, Berdis AJ, Benkovic SJ 2006a. An alternative clamp loading pathway via the T4 clamp loader gp44/62-DNA complex. Biochemistry 45: 7976–7989 [DOI] [PubMed] [Google Scholar]
- Zhuang Z, Yoder BL, Burgers PM, Benkovic SJ 2006b. The structure of a ring-opened proliferating cell nuclear antigen-replication factor C complex revealed by fluorescence energy transfer. Proc Natl Acad Sci 103: 2546–2551 [DOI] [PMC free article] [PubMed] [Google Scholar]