MILI (mouse Piwi-like), one of the three mouse PIWI family members, is an enzyme participating in the biogenesis of piRNA (PIWI-interacting RNA), which is male germ cell–specific small RNA required for gene expression regulation. The authors’ novel strategy using germline stem (GS) cells revealed that glycerol-3-phosphate acyltransferase 2 (GPAT2), a mitochondrial outer membrane protein, bound to MILI and was indispensable for the piRNA biogenesis. GPAT2 presumably functions as an anchor to construct piRNA-producing machinery, including MILI, because its catalytic activity is not necessary for the function. Thus, this study clearly demonstrates the utility of GS cells and the importance of GPAT2 for studying the biogenesis of piRNAs.
Keywords: MILI, piRNA, GPAT2, GS cells, mitochondria
Abstract
piRNA (PIWI-interacting RNA) is a germ cell–specific small RNA in which biogenesis PIWI (P-element wimpy testis) family proteins play crucial roles. MILI (mouse Piwi-like), one of the three mouse PIWI family members, is indispensable for piRNA production, DNA methylation of retrotransposons presumably through the piRNA, and spermatogenesis. The biogenesis of piRNA has been divided into primary and secondary processing pathways; in both of these MILI is involved in mice. To analyze the molecular function of MILI in piRNA biogenesis, we utilized germline stem (GS) cells, which are derived from testicular stem cells and possess a spermatogonial phenotype. We established MILI-null GS cell lines and their revertant, MILI-rescued GS cells, by introducing the Mili gene with Sendai virus vector. Comparison of wild-type, MILI-null, and MILI-rescued GS cells revealed that GS cells were quite useful for analyzing the molecular mechanisms of piRNA production, especially the primary processing pathway. We found that glycerol-3-phosphate acyltransferase 2 (GPAT2), a mitochondrial outer membrane protein for lysophosphatidic acid, bound to MILI using the cells and that gene knockdown of GPAT2 brought about impaired piRNA production in GS cells. GPAT2 is not only one of the MILI bound proteins but also a protein essential for primary piRNA biogenesis.
INTRODUCTION
The P-element wimpy testis (PIWI) family is a gene family in animals that plays a pivotal role in the production of PIWI-interacting RNA (piRNA), a germ cell–specific subset of small RNAs (Kim et al. 2009). In mice, there are three Piwi family members: MIWI (mouse Piwi), MILI (mouse Piwi-like), and MIWI2, all of which are essential for spermatogenesis at different developmental stages (Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007). PIWI proteins not only bind to piRNAs but also participate in their biogenesis (Kim et al. 2009).
It has been generally believed that mammalian piRNA biogenesis takes place in nuage, that is, cytoplasmic nonmembranous structures also known as germline granules (Eddy 1975; Chuma et al. 2009). Recent studies have shown that at least two kinds of nuage are involved in piRNA processing: the pi-body, which is also known as inter-mitochondrial cement (IMC) because of its amorphous structure and cytoplasmic localization (as observed by electron microscopy); and the piP-body (Aravin et al. 2009). Based on biochemical analysis of piRNA production in Drosophila and large-scale sequencing of piRNAs in various species, the biogenesis of piRNA has been divided into primary and secondary processing pathways (Brennecke et al. 2007; Gunawardane et al. 2007; Aravin et al. 2008). Although the primary pathway is only vaguely understood, long single-strand precursor RNAs transcribed from genomic regions termed “piRNA clusters” are believed to be catalyzed into MILI-bound primary piRNAs, which typically contain uracil at their 5′ ends (1st U) (Aravin et al. 2006; Girard et al. 2006; Grivna et al. 2006; Lau et al. 2006; Watanabe et al. 2008). Pi-bodies could be the organelle in which primary processing takes place.
The secondary pathway of piRNA production is the so-called “ping-pong amplification cycle,” in which PIWI proteins and other proteins such as tudor domain–containing proteins (TDRDs) and mouse vasa homolog (MVH) play pivotal roles (Reuter et al. 2009; Shoji et al. 2009; Kuramochi-Miyagawa et al. 2010). In the initial step of this process, complementary transcripts annealed to MILI-bound piRNAs are cleaved at the 10th nucleotide from the 5′ end by the slicer activity of MILI, which takes in the pi-body (De Fazio et al. 2011). The resultant secondary piRNAs are complementary to the primary piRNAs with an adenine base at the 10th position (10th A), which corresponds to the 1st U of the primary piRNAs. In the next step of the cycle, the secondary piRNAs are incorporated into MIWI2, which is colocalized in piP-body with the proteins involved in the cycle (Aravin et al. 2009; Shoji et al. 2009).
In male germ cells, de novo DNA methylation of retrotransposons such as Line-1 and intracisternal A particle (IAP) is introduced during embryonic days 15.5–18.5 (La Salle et al. 2004), when both MILI and MIWI2 are expressed, to prevent retrotransposon-induced mutagenesis. The sequences of the majority of embryonic piRNAs during this phase correspond to retrotransposon genes (Aravin et al. 2008). Various gene-targeted mice in which embryonic piRNA production is severely damaged show the impairment of de novo DNA methylation in retrotransposons (Aravin et al. 2007; Carmell et al. 2007; Kuramochi-Miyagawa et al. 2008, 2010). Taking these data into account, although there is a lack of direct evidence, it is quite likely that piRNAs have critical roles in the de novo DNA methylation of retrotransposons in the embryonic testis. Both MILI- and MIWI2-null mice show severe impairment of piRNA production, as well as reduced DNA methylation and enhanced expression of retrotransposons in male germ cells.
Cultured cells are quite useful for analyzing molecular events because they can easily be obtained in good numbers. The only mammalian cell lines possessing germ cell characteristics are germline stem (GS) cells, which are established from the testes of neonates and bear features of spermatogonial stem cells (Kanatsu-Shinohara et al. 2003). We explored the GS cells for studying the functional piRNA pathway in this study. First, we analyzed GS cell lines established from control and MILI-null mice and compared them to the MILI-null GS cells in which MILI expression had been restored. GS cells turned out to be quite useful for analyzing the molecular mechanisms of piRNA production, especially the primary processing pathway. Moreover, using the GS cells, we showed by coimmunoprecipitation and mass analysis that glycerol-3-phosphate acyltransferase 2 (GPAT2), a mitochondrial outer membrane protein that has a catalytic domain for the synthesis of lysophosphatidic acid from glycerol-3-phosphate and long-chain acyl-CoA (Wang et al. 2007), is one of MILI-binding proteins. We further showed in gene knockdown experiments that GPAT2 plays a critical role in piRNA production.
RESULTS AND DISCUSSION
Characterization of MILI-null GS cells and revertant cells
We first examined whether GS cell lines derived from the testes of neonatal MILI-heterozygous and MILI-deficient mice were useful for the study of piRNA production and subsequent DNA methylation. The morphology and proliferation rates of four MILI-null GS cell lines were almost the same as those of four MILI-heterozygous GS cells (Supplemental Fig. S1A,B). Because the expression of 1Δ1-type IAP (hereafter, just IAP) and Line-1 retrotransposons was repressed in normal testes, but not MILI-null testes, we analyzed their expression in MILI-heterozygous and MILI-deficient GS cells (Fig. 1A). While expression of Line-1 was not affected, that of the IAP retrotransposon was significantly enhanced by the absence of MILI (P < 0.005; Student’s t-test). The enhancement was presumably due to defective DNA methylation in the promoter region of IAP (Supplemental Fig. S2A). Meanwhile, IAP expression level in the MILI-null GS cells was higher than that in the day 14 MILI-null germ cells (Supplemental Fig. S3).
FIGURE 1.
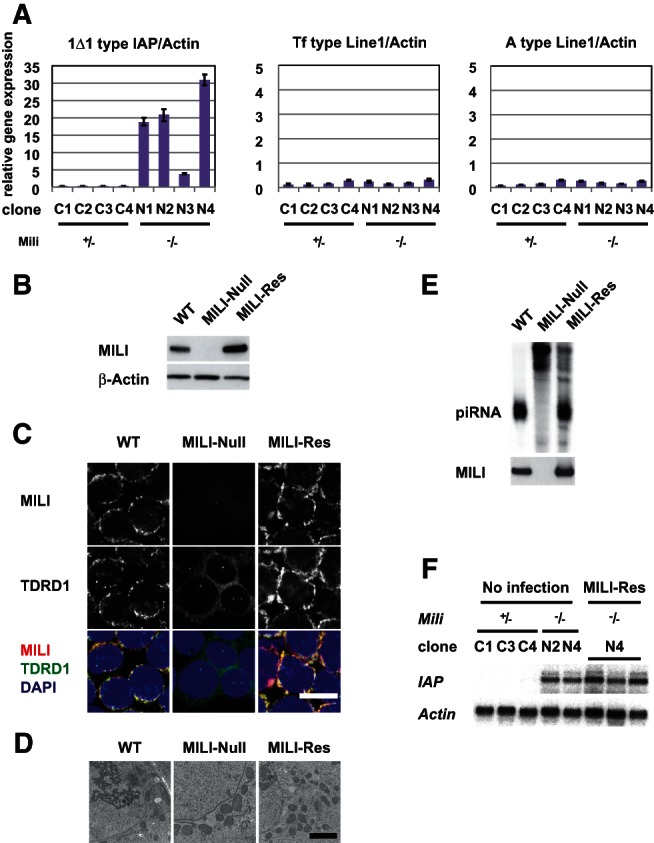
Characterization of MILI-null and MILI-rescued GS cells. (A) Expression of retrotransposons (intracisternal A particle [IAP] and Line-1) in MILI-null GS cells. Quantitative RT-PCR analysis of four independent MILI-heterozygous (+/−) GS cell lines and four MILI-null (−/−) GS cell lines. (B) Western blot analysis of MILI in wild-type, MILI-null, and MILI-rescued GS cells. (C) Immunohistochemical staining of MILI and TDRD1 in wild-type, MILI-null, and MILI-rescued GS cells. MILI and TDRD1 were stained with Alexa Fluor 568 (red) and Alexa Fluor 647 (green), respectively. DNA was stained with DAPI (blue). Scale bar, 20 μm. (D) Electron microscopic images of wild-type, MILI-null, and MILI-rescued GS cells. Scale bar, 2 μm. (E) MILI-bound piRNA in wild-type, MILI-null, and MILI-rescued GS cells. RNA coimmunoprecipitated using anti-MILI antibody was labeled with 32P and subjected to gel electrophoresis. (F) Expression of IAP (5.4 kb) in heterozygous (C1, C3, C4), MILI-null (N2, N4), and MILI-rescued GS cells, as assessed by Northern blotting. The three samples of MILI-rescued GS cells were collected from different cultures.
DNA methylation of Line-1 promoters in MILI-null GS cells was not significantly different than that in MILI-heterozygous GS cells. Therefore, the lack of a significant increase in Line-1 transcript levels in MILI-null GS cells was attributable to the DNA methylation states (Supplemental Fig. S2B,C). These data show that GS cells had a phenotype reminiscent of that of male germ cells in vivo. We next decided to focus on the IAP retrotransposon and to use clone N4 mainly as a representative clone of MILI-null GS cells, because it showed the highest expression of IAP by RT-PCR.
We expressed MILI in MILI-deficient GS cells, thereby producing “MILI-rescued” GS cells, to further analyze the function of MILI in GS cell lines. We used a Sendai virus vector (Nishimura et al. 2011) because of its high gene transfer ability and its high and stable expression after infection (Supplemental Fig. S4). Western blot analysis showed that the level of expression of MILI in the MILI-rescued GS cells was higher than that in wild-type GS cells (Fig. 1B). Immunohistochemistry confirmed the expression of MILI and its perinuclear granular localization in MILI-rescued cells (Fig. 1C). TDRD1, another protein essential for piRNA production and normal spermatogenesis, is known to bind to MILI and to localize in pi-bodies (Chuma et al. 2006; Kojima et al. 2009; Wang et al. 2009). Although TDRD1 showed a perinuclear granular staining pattern and colocalized with MILI in wild-type GS cells, it was diffusely distributed in MILI-null GS cells. However, MILI expression restored its perinuclear granular staining pattern. Electron microscopic analysis revealed that there was no nuage structure in GS cells and, therefore, that no significant alterations were present in wild-type, MILI-KO, or MILI-rescued GS cells (Fig. 1D).
Although piRNA expression was restored by the expression of MILI (Fig. 1E), piRNAs in the MILI-rescued GS cells did not induce post-transcriptional gene silencing (PTGS) of IAP retrotransposons. This deficient PTGS was due to a deficiency in the piRNA-dependent PTGS machinery and/or the low abundance of antisense piRNAs (discussed below). In addition, expression of IAP and DNA methylation of IAP promoters was not restored (Fig. 1F; Supplemental Fig. S5, S6), which might have been due to the lack of DNMT3L and MIWI2 (Takashima et al. 2009), both of which are necessary for de novo DNA methylation (Bourc’his and Bestor 2004; Carmell et al. 2007). However, expression of DNMT3L and MIWI2 did not bring about the IAP DNA methylation, suggesting that some other factors would be necessary for the methylation. Although MILI could not rescue DNA methylation, efficient regeneration of MILI-bound piRNAs by MILI expression prompted us to analyze the piRNAs in the cells comprehensively.
Comprehensive analysis of piRNAs in MILI-rescued GS cells
The size distribution of small RNAs in wild-type and rescued GS cells was determined by deep sequencing analysis (Fig. 2A). Small RNAs of 22–23 and 25–31 nucleotides (nt) in length represent microRNAs (miRNAs) and piRNAs, respectively. Numbers of piRNAs per miRNA calculated from the data in Figure 2B showed that about twice as piRNAs were present in MILI-rescued GS cells as in wild-type GS cells. Numbers of piRNAs relative to miRNAs can be used as a good index of the amount of piRNAs, because the biogenesis of miRNA is independent of that of piRNA. No alteration of miRNA expression by gene knockout and gene introduction experiments is shown in Supplemental Figure S7. Therefore, piRNAs were significantly more abundant in MILI-rescued GS cells than in wild-type GS cells, which suggests that the amount of piRNA would be dependent on the amount of MILI. This phenomenon is essentially the same as the previous data that Argonaute protein abundance controls the amount of miRNA in somatic cells (Diederichs and Haber 2007).
FIGURE 2.
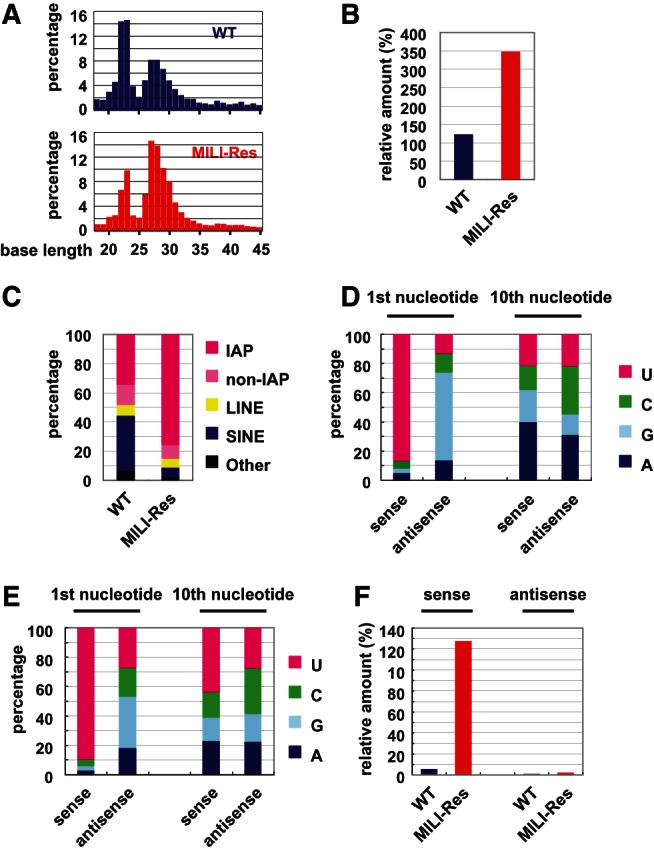
piRNA expression analysis in wild-type and MILI-rescued GS cells by deep sequencing. (A) The size distribution of small RNAs. Total numbers of small RNAs (18–45 nt) analyzed in wild-type and MILI-rescued GS cell libraries were 9,683,216 and 7,420,549, respectively. (B) Ratio of total piRNA to miRNA. (C) Genomic annotation of the piRNAs corresponding to retrotransposons in wild-type and MILI-rescued GS cells. (D,E) Ratio of the first-sense nucleotide (left) and 10th antisense nucleotide (right) of IAP piRNAs in wild-type and MILI-null GS cells. (F) Comparison of the relative amounts of sense and antisense IAP piRNAs (per miRNA).
The genomic annotations of the piRNAs in the wild-type and MILI-rescued GS cells are shown in Figure 2C and Supplemental Figure S8. The ratios of IAP and non-IAP LTR, Line, and Sine retrotransposons in wild-type GS cells were similar to those for prepachytene piRNAs reported previously (Aravin et al. 2007). In addition, both prepachytene piRNAs and GS cell piRNAs showed relatively high amount of piRNAs corresponding to mRNA. In the MILI-rescued GS cells, the percentage of IAP LTR was significantly higher and the number of IAP piRNA molecules per miRNA was about 13 times higher than that in wild-type GS cells. Next, a more detailed analysis of IAP piRNAs was carried out.
The first and 10th nucleotides of piRNAs can be used as “signatures” of the primary and the secondary processing pathways, respectively, as described in the Introduction. The data for IAP piRNAs in wild-type and MILI-rescued GS cells are shown in Figure 2, D and E, respectively. As for wild-type GS cells, the percentage of 1st U sense piRNAs was >85%. In contrast, the percentage of 1st U antisense piRNAs was only 15%. A previous report on prepachytene male germ cells showed that the percentages of sense and antisense piRNAs with 1st U was ∼80% and 70%, respectively (Aravin et al. 2007). As we reported, the same trend was also observed in embryonic male germ cells (Kuramochi-Miyagawa et al. 2008). Meanwhile, the percentages of 10th A sense and antisense piRNAs were <40% in the wild-type GS cells. Statistical analysis of piRNAs in wild-type GS cells using z-score showed that the percentage of 1st U piRNA was significantly higher than the average, but 10th A piRNAs exhibited no preference (Supplemental Fig. S9). These data demonstrated that the secondary processing does rarely proceed in the cells.
The sequence data for MILI-rescued GS cells were essentially the same as those for wild-type GS cells (Fig. 2E; Supplemental Fig. S9). They showed that the production of piRNAs in wild-type and MILI-rescued GS cells almost exclusively occurs via the primary processing pathway and that GS cells are a superb resource for the analysis of this pathway. The numbers of piRNAs corresponding to IAP per miRNA (Fig. 2F) showed that IAP sense piRNAs were much more abundant in MILI-rescued GS cells than in wild-type GS cells. This increase can be explained by the enhanced primary processing of IAP, higher amount of IAP transcript, and/or the increased amount of MILI protein level in the MILI-rescued GS cells.
GPAT2 as a MILI-binding protein and its function in piRNA production
Based on the possible function of MILI in primary piRNA production, we attempted to identify the proteins that bound to MILI in MILI-rescued GS cells. Proteins that were precipitated by the anti-MILI antibody were subjected to SDS-PAGE and silver staining. Comparison of the data for MILI-rescued GS cells and MILI-null GS cells revealed several discrete bands only in the MILI-rescued GS cells (Supplemental Fig. S10A). Mass spectrometric analysis of these bands identified proteins of approximately 135, 110, and 79 kD as Mov10l1, MILI, and GPAT2, which is one of four GPAT enzymes preferentially expressed in testis, respectively. The numbers of peptides and coverage were shown in Supplemental Figure S10B. Mov10l1 has been reported to be a binding partner of MILI and to function in the production of piRNA (Frost et al. 2010; Zheng et al. 2010), which verifies the feasibility of this method. Binding of MILI to GPAT2 was confirmed by coimmunoprecipitation analysis of testis samples (Fig. 3A) and HEK 293T cells in which MILI and GPAT2 were overexpressed (Supplemental Fig. S10C). Based on these data, we entertained the idea that GPAT2 has some biological relevance in piRNA production, especially the primary processing pathway.
FIGURE 3.
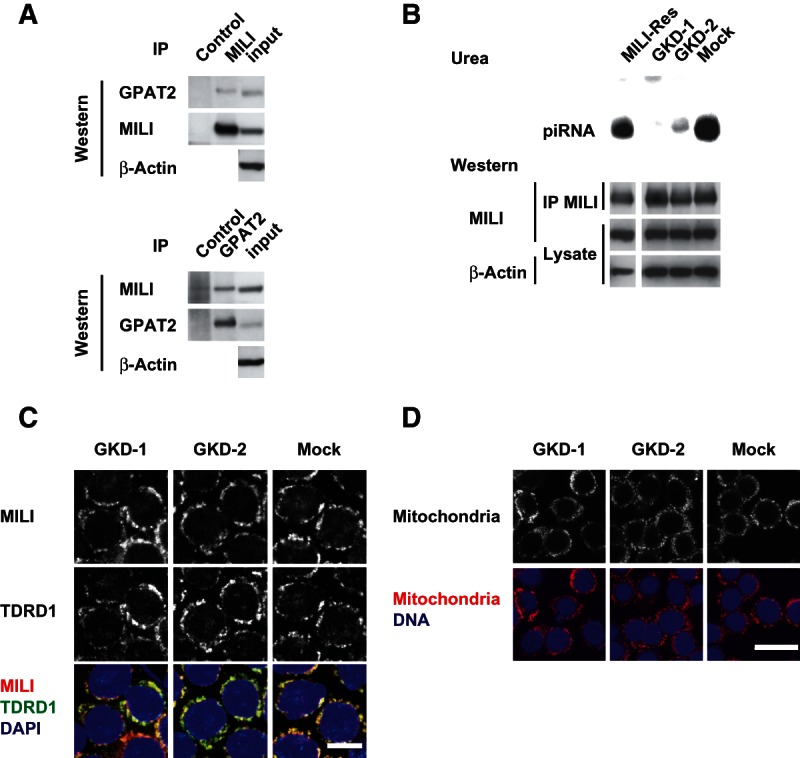
GPAT2 as a MILI-binding protein and effects of its knockdown in GS cells. (A) Binding of GPAT2 and MILI in the testis. Adult (day 12) testes were immunoprecipitated with anti-MILI and anti-GPAT2 antibodies, and the precipitates were subjected to Western blotting with anti-GPAT2 and anti-MILI antibodies, respectively. (B) Impairment of piRNA production as a result of GPAT knockdown. RNA coimmunoprecipitated with anti-MILI antibody was labeled with 32P and subjected to urea PAGE. Amounts of MILI were confirmed by Western blotting of immunoprecipitated materials (IPs) and cellular lysates, as described. (C) Immunohistochemical staining of MILI and TDRD1 in mock-infected control and GPAT2-knockdown MILI-rescued GS cells. MILI and TDRD1 were stained with Alexa Fluor 568 (red) and Alexa Fluor 647 (green), respectively. DNA was stained with DAPI (blue). Scale bar, 20 μm. (D) Staining of mitochondria in GPAT2-knockdown GS cells.
To examine the possible role of GPAT2 in piRNA production, we reduced the expression of GPAT2 in MILI-rescued GS cells using a lentiviral gene knockdown vector (Wiznerowicz and Trono 2003). Two knockdown vectors for GPAT2 (GKD-1 and GKD-2) efficiently reduced GPAT2 expression in MILI-rescued GS cells at the mRNA and protein levels (Supplemental Fig. S11A,B, respectively). We first analyzed amounts of piRNAs in GPAT2-knockdown MILI-rescued cells (Fig. 3B). Compared with MILI-rescued GS cells (Fig. 2A), the amounts of piRNAs that bound to MILI were significantly reduced. These data clearly show that GPAT2 is essential for the biogenesis of piRNA. As shown in Figure 3B (lower panel), Western blot analysis of lysates and immunoprecipitated materials confirmed that the level of expression of MILI was not affected by the knockdown of GPAT2 expression. These data indicated that the stability of MILI was not affected by the presence or absence of piRNA, which is unlike the situation of Drosophila Piwi (Haase et al. 2010). Expression levels of Miwi2, Mov10L1, and MItoPLD were not altered either by GPTA2 knockdown (Supplemental Fig. S11C).
As shown in Figure 1C, a null mutation in MILI caused the subcellular localization of TDRD1 to be abnormal. We next analyzed whether the knockdown of GPAT2 would affect the localization of MILI and TDRD1. In knockdown cells, both MILI and TDRD1 showed perinuclear granular staining patterns similar to those in MILI-rescued GS cells (Fig. 3C). In addition, there were no significant differences in the gross distribution (Fig. 3D) or electron microscopic features (Supplemental Fig. S12) of mitochondria between mock-infected and knockdown GS cells. To determine the function of GPAT2 in piRNA production, we performed deep sequencing analysis. piRNA production was severely impaired in knockdown cells, as shown in Figure 4A. Moreover, it was more severely impaired in GS cells expressing GKD-1 shRNA than in those expressing GKD-2 shRNA, which presumably reflects a difference in the amount of GPAT2 protein (Fig. 3B). The size distribution of small RNAs in knockdown GS cells was essentially the same as that in MILI-null GS cells (Fig. 4B). These data clearly show that GPAT2 plays a critical role in primary processing during piRNA production.
FIGURE 4.
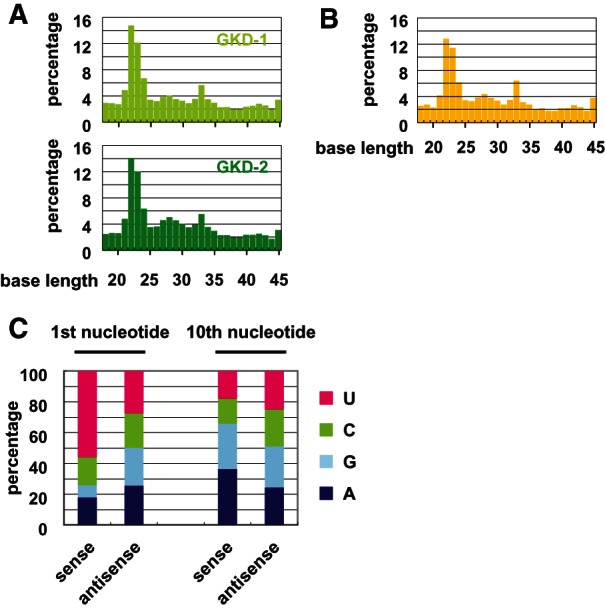
piRNA expression analysis in GPAT2-knockdown GS cells by deep sequencing. (A,B) Size distribution of small RNAs. Total numbers of small RNAs (18–45 nt) analyzed in MILI-rescued GS cells transfected with the GKD-1- and GKD-2-knockdown vectors (A), and in MILI-null GS cells (B), were 5,400,278, 5,485,436, and 5,122,719, respectively. (C) Ratio of the first and 10th nucleotide of sense and antisense IAP piRNAs, respectively, in MILI-rescued GS cells expressing the GKD-1-knockdown vector.
Our comprehensive analysis of knockdown GS cells and MILI-null GS cells revealed additional features. The percentage of 1st U IAP small RNAs for the same size of piRNA (25–31 nt) was ∼50% in MILI-null GS cells. Considering the high preponderance of 1st U in piRNAs, a molecular mechanism independent of MILI may give rise to 1st U small RNAs in GS cells (Supplemental Fig. S13A,C). This observation suggests that a molecular mechanism independent of MILI may give rise to 1st U small RNAs.
GS cells as a resource for analyzing the primary processing of piRNA
To elucidate the molecular mechanisms of piRNA biogenesis, it is necessary to obtain a large number of cells expressing piRNAs. The Bombyx ovary germ cell line BmN4 and Drosophila ovarian somatic cell line OSC have been quite useful for analyzing the production and function of piRNAs (Kawaoka et al. 2009; Saito et al. 2009). However, no piRNA-producing mammalian cell line has previously been reported, and it is rather difficult to obtain cells containing piRNAs because of their low abundance in vivo. Deep sequencing analysis showed that the majority of piRNA clusters of wild-type GS cells was same as those of prepachytene piRNA clusters (Supplemental Table 1). Taking these data into account, the utility of GS cells is a big advance for analyzing several aspects of mammalian piRNAs and is especially valuable for studying their primary processing. The biggest advantage of using GS cell lines to study piRNAs is the utility of null mutant cell lines and their revertants. In addition, the highly efficient introduction of genes using Sendai virus vectors, as well as gene knockdown and gene transfer using lentiviral vectors, are also quite useful for testing the functions of genes involved in primary piRNA production. We also would like to emphasize that this novel method is quite useful to search for unknown factors involved in piRNA biogenesis as shown in this paper.
Deep sequencing of piRNAs in GS cells showed a characteristic feature: the strong preference for the production of sense primary piRNAs. In contrast, male germ cells synthesize both sense and antisense primary piRNAs in vivo. Moreover, the vast majority of IAP piRNAs in GS cells corresponded to sense transcript, but the ratio of sense and antisense piRNAs in male germ cells was almost equal. These contradictory data suggest that the orientation of primary piRNAs merely reflects the abundance of the substrate transcripts. Meanwhile, GS cells lack the distinct structure of germ cell granules observed as intermitochondrial cement (IMC) by electron microscope (Supplemental Fig. S12), and a ping-pong amplification cycle does not operate in GS cells. This finding suggests that germ cell granules are required for the amplification of piRNAs, but not for their primary production. On the other hand, a ping-pong amplification cycle does not operate in GS cells. Considering the report showing that MILI was able to carry out the ping-pong cycle in male germ cells without MIWI2 to some extent (Aravin et al. 2007), some molecules essential for the process are presumably lacking in GS cells. It may be possible to reconstitute the full piRNA production pathway by replenishing those factors and antisense transcripts.
piRNA production in mitochondria
pi-bodies are found in small numbers in embryonic male germ cells, prospermatogonia, and A-type spermatogonia. They become undetectable after the formation of B-type spermatogonia, but they reappear in pachytene-stage spermatocytes. We recently reported that the pi-body would be the site of the initial phase of ping-pong amplification cycle, which involves MILI and TDRD1 (Kuramochi-Miyagawa et al. 2010). Many proteins, including MILI, TDRD1, and MVH, have been reported to be necessary for integrating the structure of pi-bodies (Chuma et al. 2006; Kuramochi-Miyagawa et al. 2008, 2010). However, it is unknown whether these proteins themselves are essential or whether the piRNAs produced by their action are necessary for the formation of IMC. Although the close proximity between mitochondria and the pi-bodies, which are also known as IMC, strongly suggests the possible function of mitochondria in piRNA production, there was, until recently, no evidence for this.
Two groups have independently reported that the mitochondrial outer membrane enzyme, mitochondrial phospholipase D (MitoPLD), which contains the domain hydrolyzing cardiolipin to generate phosphatidic acid, is necessary for piRNA generation. Genetic analysis of its Drosophila homolog, Zucchini, showed that it was required for piRNA production. It has been postulated that lipid metabolism on the mitochondrial membrane would play a role in piRNA production. However, very recently it was shown that MitoPLD does not show phospholipase activity but plays critical roles in piRNA production through its single-strand RNA cleavage activity.
GPAT2 is an enzyme of the mitochondrial outer membrane that contains a catalytic domain for the synthesis of lysophosphatidic acid, a precursor of phosphatidic acid, from fatty acids and acyl CoA. Therefore, we designed and examined the function of mutant GPAT2, in which the essential HXXXXD motif of the GPAT domain was disrupted. These mutants were able to rescue the function of GPAT2 in piRNA production in GPAT2 knockdown GS cells (Fig. 5). These data suggest that the acyltransferase activity of GPAT2 is dispensable for the biogenesis of piRNA. Currently, it remains unknown as to how GPAT2 participates in the production of piRNA independently of its possible enzymatic activities. One explanation is that the protein functions as a scaffold on the mitochondrial outer membrane to which many factors essential for piRNA production are tethered, including MILI. GPAT2 would thus be a key molecule for understanding the involvement of mitochondria in piRNA biogenesis.
FIGURE 5.
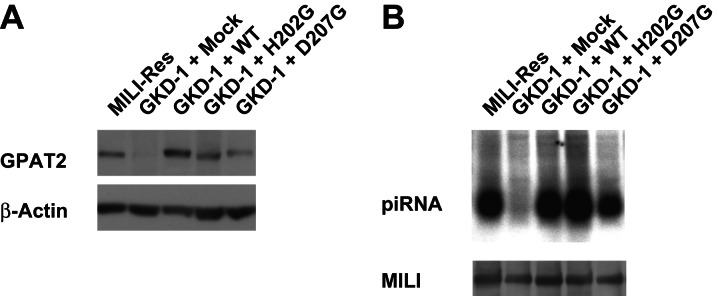
Rescue of piRNA production by GPAT2 mutants. (A) Expression of GPAT2 and its mutants in the GPAT knocked-down GS cells. Western blot analysis was carried out after introducing the GPAT2 and its mutant genes in the GPAT2 knocked-down GKD-1 GS cells. (B) Analysis of piRNA production in the GKD-1 GS cells after the introduction of GPAT2 and its mutant genes.
MATERIALS AND METHODS
Cell culture
MILI-deficient testis cells were collected from newborn (1- to 3-d-old) mice that had been crossed to the DBA/2 strain more than three generations. The establishment and culture of GS cells were performed according to previously published methods (Kanatsu-Shinohara et al. 2003, 2004).
Preparation of Sendai virus vectors and establishment of MILI-rescued GS cells
The Bsr (blasticidin S deaminase), EGFP, and Mili genes were cloned into the replication-defective and persistent Sendai virus (SeVdp) vector (Supplemental Fig. S4) according to the method described in a previously published paper (Nishimura et al. 2011). MILI-deficient GS cells were infected with Sendai virus and incubated for 2 h at room temperature. After washing with phospate-buffered saline, the GS cells were cultured on mouse embryonic fibroblasts (MEFs). To select infected cells, the cells were incubated in the presence of blasticidin S (20 μg/mL).
Quantitative PCR and Northern blotting
Total RNAs prepared from GS cells were treated with DNase I and subjected to RT-PCR using the ThermoScript RT-PCR system (Invitrogen) and random hexamers. Primers specific for IΔI-type IAP (forward, AACGCTGCTGCTTTAACTCC; reverse, ATTGTTCCCTCACTGGCAAA), Tf-type Line-1 (forward, CTGTACCACCTGGGAACTGC; reverse, TGCTGGCAAGCTCTCTTACA), and A-type Line-1 (forward, CAGCTGAGTCGCCTGACAC; reverse, CTCTCCTTAGTTTCAGTGG) were used in conjunction with an ABI PRISM 7900HT Sequence Detection System (Applied Biosystems). Northern blotting was performed according to the method described as previously published (Kuramochi-Miyagawa et al. 2008).
Antibodies
The anti-MILI antibodies used were PM044 (MBL) for immunoprecipitation and ab36763 (Abcam) for Western blotting and immunostaining. The anti-TDRD1 antibody used for immunostaining has been described previously (Chuma et al. 2006). Affinity-purified anti-GPAT2 polyclonal antibody was generated by immunizing rabbits with mixed peptides from GPAT2 (amino acid 1–17 [MLKSNPQTQQRSNHNGQC] and 414–428 [PDTEKEQEWTPPTGLC]), obtained by MBL.
Electron microscopy
GS cells were fixed with 2% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2), postfixed with 1% OsO4 and 0.1 M sucrose in 0.1 M phosphate buffer, dehydrated with a graded ethanol series, and embedded in epoxy resin. Semi-thin sections (1 μm thick) were stained with 0.1% toluidine blue for light microscopic analysis. Sections (70–90 nm thick) were placed on 150-mesh copper grids, stained with uranyl acetate followed by lead citrate, and examined under an H7000 electron microscope (Hitachi).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. T. Shinohara for providing wild-type GS cells and for valuable advice concerning the GS cell culture. We also thank Dr. M. Wiznerowicz and Dr. D. Trono for providing the pLVTH lentivirus vector, Dr. H. Miyoshi for providing the CSII-EF-IRES-Venus lentivirus vector, and M. Imaizumi for secretarial work. This work was supported in part by grants from the Ministry of Education, Science, Sports and Culture, and Core Research for Evolutional Science and Technology (CREST). S.K.-M. was a Japan Science and Technology Agency Precursory Research for Embryonic Science and Technology (PREST) researcher.
REFERENCES
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourc’his D, Bestor TH 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431: 96–99 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N 2006. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci 103: 15894–15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N 2009. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol 306: 17–23 [DOI] [PubMed] [Google Scholar]
- De Fazio S, Bartonicek N, Di Giacomo M, Abreu-Goodger C, Sankar A, Funaya C, Antony C, Moreira PN, Enright AJ, O’Carroll D 2011. The endonuclease activity of Mili fuels piRNA amplification that silences LINE1 elements. Nature 480: 259–263 [DOI] [PubMed] [Google Scholar]
- Deng W, Lin H 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- Diederichs S, Haber DA 2007. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell 131: 1097–1108 [DOI] [PubMed] [Google Scholar]
- Eddy EM 1975. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol 43: 229–280 [DOI] [PubMed] [Google Scholar]
- Frost RJ, Hamra FK, Richardson JA, Qi X, Bassel-Duby R, Olson EN 2010. MOV10L1 is necessary for protection of spermatocytes against retrotransposons by Piwi-interacting RNAs. Proc Natl Acad Sci 107: 11847–11852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442: 199–202 [DOI] [PubMed] [Google Scholar]
- Grivna ST, Beyret E, Wang Z, Lin H 2006. A novel class of small RNAs in mouse spermatogenic cells. Genes Dev 20: 1709–1714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane LS, Saito K, Nishida KM, Miyoshi K, Kawamura Y, Nagami T, Siomi H, Siomi MC 2007. A slicer-mediated mechanism for repeat-associated siRNA 5′ end formation in Drosophila. Science 315: 1587–1590 [DOI] [PubMed] [Google Scholar]
- Haase AD, Fenoglio S, Muerdter F, Guzzardo PM, Czech B, Pappin DJ, Chen C, Gordon A, Hannon GJ 2010. Probing the initiation and effector phases of the somatic piRNA pathway in Drosophila. Genes Dev 24: 2499–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Ogonuki N, Inoue K, Miki H, Ogura A, Toyokuni S, Shinohara T 2003. Long-term proliferation in culture and germline transmission of mouse male germline stem cells. Biol Reprod 69: 612–616 [DOI] [PubMed] [Google Scholar]
- Kanatsu-Shinohara M, Inoue K, Lee J, Yoshimoto M, Ogonuki N, Miki H, Baba S, Kato T, Kazuki Y, Toyokuni S, et al. 2004. Generation of pluripotent stem cells from neonatal mouse testis. Cell 119: 1001–1012 [DOI] [PubMed] [Google Scholar]
- Kawaoka S, Hayashi N, Suzuki Y, Abe H, Sugano S, Tomari Y, Shimada T, Katsuma S 2009. The Bombyx ovary-derived cell line endogenously expresses PIWI/PIWI-interacting RNA complexes. RNA 15: 1258–1264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Han J, Siomi MC 2009. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol 10: 126–139 [DOI] [PubMed] [Google Scholar]
- Kojima K, Kuramochi-Miyagawa S, Chuma S, Tanaka T, Nakatsuji N, Kimura T, Nakano T 2009. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 14: 1155–1165 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Totoki Y, Toyoda A, Ikawa M, Asada N, Kojima K, Yamaguchi Y, Ijiri TW, et al. 2008. DNA methylation of retrotransposon genes is regulated by Piwi family members MILI and MIWI2 in murine fetal testes. Genes Dev 22: 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. 2010. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 24: 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Salle S, Mertineit C, Taketo T, Moens PB, Bestor TH, Trasler JM 2004. Windows for sex-specific methylation marked by DNA methyltransferase expression profiles in mouse germ cells. Dev Biol 268: 403–415 [DOI] [PubMed] [Google Scholar]
- Lau NC, Seto AG, Kim J, Kuramochi-Miyagawa S, Nakano T, Bartel DP, Kingston RE 2006. Characterization of the piRNA complex from rat testes. Science 313: 363–367 [DOI] [PubMed] [Google Scholar]
- Nishimura K, Sano M, Ohtaka M, Furuta B, Umemura Y, Nakajima Y, Ikehara Y, Kobayashi T, Segawa H, Takayasu S, et al. 2011. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem 286: 4760–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Saito K, Inagaki S, Mituyama T, Kawamura Y, Ono Y, Sakota E, Kotani H, Asai K, Siomi H, Siomi MC 2009. A regulatory circuit for piwi by the large Maf gene traffic jam in Drosophila. Nature 461: 1296–1299 [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Takashima S, Takehashi M, Lee J, Chuma S, Okano M, Hata K, Suetake I, Nakatsuji N, Miyoshi H, Tajima S, et al. 2009. Abnormal DNA methyltransferase expression in mouse germline stem cells results in spermatogenic defects. Biol Reprod 81: 155–164 [DOI] [PubMed] [Google Scholar]
- Wang S, Lee DP, Gong N, Schwerbrock NM, Mashek DG, Gonzalez-Baro MR, Stapleton C, Li LO, Lewin TM, Coleman RA 2007. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch Biochem Biophys 465: 347–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H 2009. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 19: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D 2003. Conditional suppression of cellular genes: Lentivirus vector-mediated drug-inducible RNA interference. J Virol 77: 8957–8961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Xiol J, Reuter M, Eckardt S, Leu NA, McLaughlin KJ, Stark A, Sachidanandam R, Pillai RS, Wang PJ 2010. Mouse MOV10L1 associates with Piwi proteins and is an essential component of the Piwi-interacting RNA (piRNA) pathway. Proc Natl Acad Sci 107: 11841–11846 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


