Generally, most work on RNA localization in Xenopus oocytes has focused on mRNAs that localize to the vegetal pole. This study describes an mRNA that is transported to the animal pole. This mRNA forms the same RNP complexes as the vegetal-directed mRNAs with the exception of the protein Staufen, which therefore may be necessary to determine directionality.
Keywords: PHAX, RNA localization, Staufen, Vg1 mRNA, Xenopus oocyte
Abstract
The 3′ untranslated region of mRNA encoding PHAX, a phosphoprotein required for nuclear export of U-type snRNAs, contains cis-acting sequence motifs E2 and VM1 that are required for localization of RNAs to the vegetal hemisphere of Xenopus oocytes. However, we have found that PHAX mRNA is transported to the opposite, animal, hemisphere. A set of proteins that cross-link to the localization elements of vegetally localized RNAs are also cross-linked to PHAX and An1 mRNAs, demonstrating that the composition of RNP complexes that form on these localization elements is highly conserved irrespective of the final destination of the RNA. The ability of RNAs to bind this core group of proteins is correlated with localization activity. Staufen1, which binds to Vg1 and VegT mRNAs, is not associated with RNAs localized to the animal hemisphere and may determine, at least in part, the direction of RNA movement in Xenopus oocytes.
INTRODUCTION
The localization of mRNA underlies the spatial regulation of gene expression that, for example, determines the body plan of developing embryos, directs the turning of axonal growth cones, or enables sustained cell movement (Holt and Bullock 2009; Lecuyer et al. 2009; Martin and Ephrussi 2009; Marchand et al. 2012). There are at least three mechanisms through which subcellular enrichment of mRNA can be achieved. These include the local stabilization of a transcript from degradation, entrapment, or active transport using the cell cytoskeleton. The latter appears to be the predominant means to position RNA and entails the association of trans-acting factors with specific cis-acting sequence elements in the RNA to form a ribonucleoprotein (RNP) complex that usually engages a motor protein.
In some instances, the number of localized transcripts is exceedingly large. High-throughput in situ hybridization experiments revealed that >70% of the mRNAs in early Drosophila embryos are asymmetrically distributed in a variety of patterns (Lecuyer et al. 2007). Given the large number of localized transcripts and the assortment of destinations, it is unlikely that an individual RNA is targeted by a unique set of proteins. A more plausible alternative is that small differences in the composition of the localization RNP complex determine the pathway and final destination of the RNA. The notion that there is a limited number of localization factors, shared among a greater diversity of localization RNP complexes, is supported by the striking conservation of these proteins across species and disparate cell types. Several of the factors that comprise the localization RNP complex that forms on Vg1 mRNA in Xenopus oocytes have been identified, and nearly all have homologs that participate in the transport of RNA not only in oocytes and embryos, but also in differentiated somatic cells, ranging from Drosophila to mammals (Deshler et al. 1998; Havin et al. 1998; Cote et al. 1999; Zhao et al. 2001; Kroll et al. 2002, 2009; Allison et al. 2004; Yoon and Mowry 2004; Czaplinski et al. 2005; Arthur et al. 2009; Loeber et al. 2010).
The vegetal localization element (VLE) in Xenopus Vg1 mRNA is a 340-nucleotide (nt) sequence in the 3′ untranslated region (3′ UTR) that is composed of multiple repeats of E2 and VM1 motifs (Bubunenko et al. 2002) that are required for proper localization of the RNA to the vegetal cortex through the late pathway that operates during stages III and IV of oogenesis (Deshler et al. 1998; Cote et al. 1999). VegT mRNA is also transported to the vegetal cortex, and, like Vg1 mRNA, its localization element also contains multiple repeats of E2 and VM1 motifs (Bubunenko et al. 2002). Additional E2 and VM1 sequences occur in the 3′ UTR outside of the localization elements of both RNAs, and it has been proposed that it is the clustering of these motifs that defines a functional cis-acting element (Bubunenko et al. 2002). The significance of multiple, repeated sequences is not clear. It is possible that the clustering is a steric requirement that allows for protein–protein interactions within the RNP complex and/or the repeated sequences create a higher-order RNA structure required for productive factor binding.
We took note that the 3′ UTR of the mRNA encoding PHAX, a protein required for nuclear export of U-type snRNAs and intranuclear transport of snoRNAs (Ohno et al. 2000; Boulon et al. 2004; Watkins et al. 2004), contains a cluster of multiple E2 and VM1 motifs as well as a binding site for VgRBP71, another trans-acting factor implicated in vegetal localization (Kroll et al. 2002; Kolev and Huber 2003). Surprisingly, however, we have determined that this mRNA is localized to the animal hemisphere of Xenopus oocytes. Inspection of other RNAs that localize to the animal hemisphere revealed that they also contain multiple repeats of these sequence motifs. Using cross-linking experiments, we find that RNAs transported to either hemisphere associate with the exact same complement of six proteins, indicating that there is a core group that binds to localized RNAs irrespective of destination. However, the protein Staufen1, which binds to Vg1 and VegT mRNAs and mediates attachment of the resulting RNP complexes to the microtubule motor protein kinesin-1 (Yoon and Mowry 2004), is not associated with mRNAs that are localized to the animal hemisphere, indicating that this factor potentially plays a role in determining the direction of RNA movement in Xenopus oocytes.
RESULTS AND DISCUSSION
PHAX mRNA is localized to the animal hemisphere of Xenopus oocytes
VgRBP71 binds to the VLE of Vg1 mRNA (Kroll et al. 2002; Kolev and Huber 2003); orthologs of this protein bind to the localization elements of β-actin mRNA in fibroblasts and neurons (Gu et al. 2002; Snee et al. 2002) and MAP2 mRNA also in neurons (Rehbein et al. 2002). We have presented evidence that VgRBP71 may influence polyadenylation at a site proximal to the VLE in Vg1 mRNA (Kolev and Huber 2003). During the course of experiments to map polyadenylation sites in Vg1 mRNA, a PCR product was generated from another RNA that also has a consensus binding site for VgRBP71 immediately upstream of a polyadenylation signal (Kolev and Huber 2003). The sequence of the 137-bp fragment matched that of a Xenopus expressed sequence tag (EST) (AW766532) (Fig. 1A). We used this sequence as the starting point to search for overlapping sequences in EST databases and succeeded in constructing a hypothetical transcript of 3674 nt from five ESTs (Supplemental Table S1). Northern blot analysis confirmed the presence of a 3600-nt transcript that remains at a nearly constant level throughout all stages of oogenesis (Fig. 1B). A sequence derived from a Xenopus cDNA library (accession NM_001096115.1) was subsequently deposited that corresponds exactly to the hypothetical sequence constructed from ESTs. The latter cDNA, obtained from somatic tissue, ends at a polyadenylation signal 1811 nt downstream from the ORF; the oocyte transcript extends an additional 551 nt to a second polyadenylation site (Supplemental Fig. S1). The coding sequence of the Xenopus ORF has 70% identity to rat PHAX at the nucleotide level. PHAX is a component of the protein complex that mediates nuclear export of U snRNAs (Ohno et al. 2000); it also appears to participate in the intranuclear movement of snoRNA to Cajal bodies (Boulon et al. 2004; Watkins et al. 2004).
FIGURE 1.
PHAX mRNA is localized to the animal hemisphere of Xenopus oocytes. (A) A portion of the Xenopus EST AW766532 is shown, highlighting the binding site for VgRBP71 (red) and a downstream polyadenylation signal (green). The sequence at the binding site for VgRBP71 (bold, underlined) in the EST is compared with that in Vg1 mRNA. (B) Oocytes were separated according to developmental stage (I–VI) and total RNA isolated for Northern blot analysis. Each lane contains 10 oocyte equivalents of RNA. Complementary probes for PHAX and GAPDH (loading control) mRNAs were used simultaneously. The positions of size standards (nucleotides, nt) are indicated. (C) Scheme showing the relative positions of E2 and VM1 motifs, VgRBP71 binding sites, and polyadenylation signals in the 3′ UTRs of Vg1, VegT, and PHAX mRNAs. (D) Immunohistochemical staining of stage V/VI oocytes following in situ hybridization with digoxigenin-labeled probes for Vg1 mRNA (left panel) and PHAX mRNA (right panel). The discernible indentation on the oocyte surface acts as a marker for the animal hemisphere.
In addition to a binding site for VgRBP71, the 3′ UTR of PHAX mRNA also contains a cluster of E2 and VM1 motifs, similar to the organization seen in the localization elements of Vg1 and VegT mRNAs (Fig. 1C). This suggested that this mRNA is also localized to the vegetal hemisphere of Xenopus oocytes. Whole-mount in situ hybridization reveals that PHAX mRNA is indeed localized, but to the animal hemisphere (Fig. 1D). This result prompted us to examine the 3′ UTRs of other mRNAs that are localized to the animal hemisphere of Xenopus oocytes. Remarkably, most of them have multiple repeats of E2 and VM1 motifs that heretofore have been considered hallmarks of RNAs localized to the vegetal hemisphere (Supplemental Table S2). Many of these RNAs also contain putative binding sites for VgRBP71 upstream of polyadenylation signals. These results indicate that E2 and VM1 motifs constitute cis-acting localization elements for RNAs moving to either hemisphere of the oocyte.
Localized RNAs in Xenopus oocytes bind a core group of proteins irrespective of destination
E2 and VM1 sequences are the binding sites for Vg1RBP/Vera (Deshler et al. 1998) and VgRBP60/hnRNP I (Cote et al. 1999), respectively, which are two of six proteins that become UV-cross-linked to Vg1 and VegT mRNAs in oocyte extract (Mowry 1996). Subsets of these proteins can also be cross-linked to other RNAs localized to the vegetal cortex (Bubunenko et al. 2002; Chang et al. 2004; Claussen and Pieler 2004; Claussen et al. 2004; Horvay et al. 2006). Figure 2 shows that the same set of proteins is also cross-linked to PHAX and An1 mRNAs. Competition with unlabeled cognate RNAs establishes that these are the same proteins associated with Vg1 mRNA and that binding in each case is specific. A similar-sized RNA transcribed from pBluescript KS cannot compete for binding and exhibited no corresponding pattern of cross-linked proteins. A doublet of masking proteins, p54/p56, overlaps the band (p60) that corresponds to VgRBP60 and is not effectively displaced by competitor RNA (Mowry 1996). Vg1RBP/Vera corresponds to p69, while 40Love and at least one additional protein contribute to the p40 band. 40LoVe, specifically cross-linked to VLE RNA, can be immunoprecipitated from oocyte extract (Czaplinski et al. 2005; Kroll et al. 2009); however, a 40-kDa protein cross-linked to the VLE persists in extract depleted of 40LoVe (Czaplinski et al. 2005). Elr-type proteins have recently been shown to bind to the localization elements of several different vegetal mRNAs; one of these likely accounts for the additional p40 protein and possibly for p36 (Arthur et al. 2009). These results demonstrate that a core group of proteins directly binds to RNAs that are transported to either the animal or vegetal hemisphere of Xenopus oocytes.
FIGURE 2.
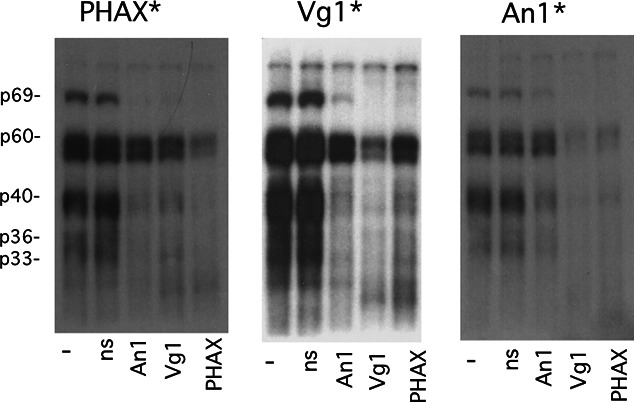
RNAs localized to either the vegetal or animal hemispheres are cross-linked to the same set of proteins. Internally radiolabeled PHAX (left), Vg1 (center), or An1 (right) RNA was added to whole-cell extract prepared from mixed-stage oocytes. Following UV irradiation, samples were treated with ribonuclease and then analyzed by SDS electrophoresis followed by autoradiography. A 20-fold excess of unlabeled, competitor RNA was added to individual samples, as indicated below each lane. Nonspecific RNA (ns) is a 356-nt transcript from the vector pBluescript KS.
Highly repeated E2 and VM1 sequences are not a universal signature of localization elements in Xenopus oocytes. Xdead end, Xvelo, fatvg, and Xbic-C mRNAs are all localized through the late pathway to the vegetal hemisphere, yet lack any appreciable combination of E2 and VM1 motifs (Chan et al. 1999; Betley et al. 2002; Claussen and Pieler 2004; Horvay et al. 2006). Importantly, however, Xdead end and Xvelo can be cross-linked to nearly the same group of proteins that bind to the VLE of Vg1 mRNA (Claussen and Pieler 2004; Horvay et al. 2006). Thus, it appears that the localization RNP complex that forms on these RNAs is similar, despite the differences in the composition of the cis-acting localization sequences. In vivo, RNAs that are transported through the late and early vegetal pathways can compete with each other, indicating that the two pathways share one or more common factors (Choo et al. 2005). Indeed, Xcat2 RNA, which uses the early pathway, is cross-linked to the same set of six proteins as Vg1 mRNA (Chang et al. 2004); another early pathway RNA, XNIF, binds many of these proteins as well (Claussen et al. 2004). The discovery that these same factors also bind to RNAs traveling to the animal hemisphere demonstrates that localization RNP complexes have a fundamental composition that is apparently found in most, if not all, localized RNAs in Xenopus oocytes.
A functional localization element extends throughout the length of An1 mRNA
The question that arises is whether the E2 and VM1 motifs that define a vegetal localization signal are also required for localization to the animal hemisphere. Since there are 12 E2 and 17 VM1 motifs in the 3′ UTR of PHAX and only three and five of these motifs, respectively, in the 3′ UTR of An1, we chose the latter for an analysis that included mutating individual or multiple repeats of these sequences. Based on previous work by others that successfully eliminated binding of Vg1RBP/Vera or VgRBP60/hnRNP I, we changed E2 motifs (WYCAC) to either AUUGC or UUUGC and VM1 motifs (YYUCU) to either GUGCG or CUGCG (Betley et al. 2002; Kwon et al. 2002; Lewis et al. 2008). Elimination of all three E2 motifs or four of the five VM1 motifs had no impact on the localization of An1 mRNA (Fig. 3A). Cross-linking of these mutant RNAs to the core group of six proteins was reduced, but not eliminated (results not shown). The transcripts used in these assays do not contain the 5′ UTR or the first 1289 nt of the An1 ORF, which indicated that a minimal localization element must reside in the 3′ half (nucleotides 1290–2106) of the ORF.
FIGURE 3.
Functional localization signals extend throughout the length of An1 mRNA. (A) Scheme showing the location of E2 and VM1 motifs in the An1 3′ UTR targeted for mutagenesis (left). Empty symbols represent sites of mutation. Localization phenotypes of injected RNAs (right). Following immunohistochemical staining of oocytes injected with DIG-labeled RNA, each was visually judged for localization. Representative oocytes injected with each RNA are shown along the side of the table oriented with the animal hemisphere upward. (B) UV cross-linking and localization assays of Vg1 and An1 mRNAs. Full-length Vg1 mRNA (lane 1) is cross-linked to the six core group of proteins. Deletion of the Vg1 3′ UTR, which removes the VLE, eliminates cross-linking of the binding proteins (lane 2) and abolishes localization of the RNA. Deletion of the An1 3′ UTR (lane 3) or the 3′ UTR and the last 850 nt of the ORF (lane 4) do not eliminate cross-linking activity or localization of these mutant RNAs.
Inspection of the An1 sequence revealed that clusters of E2 and VM1 sequences occur throughout the entire length of the transcript. We progressively truncated the RNA from the 3′ end of the full-length An1 transcript, beyond the start site of the constructs used for site-directed mutagenesis of E2 and VM1, in order to test the entire length of the RNA for localization activity. Deletion from the 3′ end up to nucleotide position 1256 had no effect on localization (Fig. 3B). These results indicate that the localization signal in An1 is highly redundant and that essentially any subregion of the transcript is sufficient to direct proper transport to the animal hemisphere.
While each fragment of An1 RNA tested has some cluster of E2 and VM1 motifs, we asked whether these were sufficient to engage the full complement of proteins that can be cross-linked to the Vg1 VLE. We compared deletion mutants of An1 that lack either the 3′ UTR or the 3′ UTR plus the last 850 nt of the ORF to both full-length Vg1 mRNA or Vg1 mRNA lacking the VLE, the removal of which is sufficient to prevent localization of the latter (Mowry and Melton 1992). Both truncated versions of An1 mRNA still cross-link to all six proteins; whereas, removal of the VLE from Vg1 mRNA results in elimination of all cross-linking (Fig. 3B). Thus, these assays demonstrate a correlation between localization activity and the ability of RNAs to associate with a core group of protein factors.
The localization of fragments of An1 reveals that the signals that confer this activity can be highly redundant and that the number of elements does not determine destination or even the apparent efficiency of transport. The occurrence of these sequences throughout the length of An1 mRNA is similar to yeast Ash1 mRNA, which has three localization elements in its ORF and a fourth extending into the 3′ UTR; each of these elements individually can direct localization of a reporter RNA demonstrating redundancy in this case as well (Chartrand et al. 1999; Gonzalez et al. 1999).
Staufen1 is associated only with RNA localized to the vegetal hemisphere
The late pathway used by Vg1 and VegT for transport to the vegetal hemisphere operates contemporaneously with one for movement of RNAs to the animal hemisphere (Perry-O’Keefe et al. 1990). The cross-linking experiments reveal that a core group of proteins binds to these localizing RNAs irrespective of destination, raising the question of how proper sorting is achieved. Cross-linking cannot be used to take account of the complete protein composition of localization RNP complexes. In our hands, the VLE-binding protein Prrp cannot be UV-cross-linked to Vg1 mRNA. The protein designated p78, which exhibits weak and variable cross-linking to the Vg1 VLE, could correspond to one of two Xenopus isoforms of Staufen (Stau1 79 kDa or Stau2 75 kDa) or to VgRBP71 (∼71 kDa) (Kroll et al. 2002; Allison et al. 2004). Using oocyte extract for immunoprecipitation assays, we previously showed that Prrp and VgRBP71 are associated not only with Vg1 and VegT, but also An1 and An3 mRNAs (Zhao et al. 2001; Kroll et al. 2002). Thus, these two proteins also appear to be part of the group of factors common to most, if not all, RNAs that are being transported through pathways that operate during the mid-stages of oogenesis and, furthermore, that cross-linked p78 likely corresponds to VgRBP71.
In Xenopus oocytes, both isoforms of Staufen become enriched at the vegetal cortex beginning at stage II of oogenesis (Allison et al. 2004). Staufen1 is associated with Vg1 and VegT mRNAs and is required for efficient localization (Yoon and Mowry 2004). Staufen1 also interacts with at least one of the two kinesin motors that direct transport of the Vg1 RNP complex, which tracks on a subpopulation of microtubules with their plus ends oriented toward the vegetal cortex (Yoon and Mowry 2004; Messitt et al. 2008). In light of its putative role as an adaptor to kinesin, we questioned whether Staufen1 plays a role in determining the direction of RNA movement and tested its association with other localized RNAs using immunoprecipitation assays (Fig. 4). We confirm that Staufen1 is bound to Vg1 and VegT mRNAs but did not detect any interaction with RNAs localized to the animal hemisphere (An1, An2, An3, or PHAX) or RNAs that use the early vegetal pathway (XWnt11 or Xcat-2). This result indicates that Staufen1 is restricted to RNP complexes transported to the vegetal hemisphere and, by extension, is a candidate for contributing to the directionality of RNA movement.
FIGURE 4.

Staufen1 is associated only with RNA localized to the vegetal hemisphere. Staufen1 was immunoprecipitated from whole-cell extract prepared from stage III/IV oocytes. The associated RNA was reverse-transcribed into cDNA and amplified by PCR using gene-specific primers (indicated above each panel). (Lanes 1) PCR standards generated using total oocyte RNA as a template; (lanes 2) control precipitation with protein A-Sepharose beads in the absence of antibody; (lanes 3) precipitation from extract incubated with Staufen1 antibody. Ribosomal protein L5 and actin are not localized mRNAs and serve as negative controls.
Staufen’s role in RNA localization extends from Drosophila oocytes, embryos, and neuroblasts to mammalian neurons (Roegiers and Jan 2000). However, it is unlikely that its function is identical across this wide variety of cell types and species. Staufen is associated with RNAs that localize to the anterior (bic) and posterior (osk) of Drosophila oocytes, seemingly eliminating any role in determining the final destination of RNA in these cells (St Johnston et al. 1991). It has been shown that in satu– mutants, oskar RNP complexes are trapped at the anterior end of Drosophila oocytes (Zimyanin et al. 2008). However, in these oocytes, a small number of particles escape and travel to the posterior at a velocity comparable to wild-type cells, adding additional evidence that Staufen does not influence directionality in these cells.
CONCLUSIONS
Our experiments uphold the proposal that the organization, namely clustering, of short sequences, represented by E2 and VM1 motifs, creates a functional localization element (Bubunenko et al. 2002). This is made apparent by comparing the sequences of Vg1 and An1 mRNAs. In the case of the former, these short sequences occur periodically throughout the length of the mRNA, including the ORF. However, the clustering of these two motifs is limited to the 340-nt localization element in the 3′ UTR, which is the only region of Vg1 mRNA that cross-links to the six core localization factors. On the other hand, E2 and VM1 motifs are clustered throughout the entire length of An1 mRNA, accounting for the localization activity of fragments derived from the 3′ UTR or the ORF of this mRNA, as well as the ability of each of these fragments to bind the entire set of cognate proteins in cross-linking assays. While it is not difficult to understand that the occurrences of these sequences enable the productive binding and organization of factors that result in the formation of an active localization RNP complex, it is not clear how, or whether, subtle differences in the distribution of these cis-acting elements engender features in the core RNP that might determine the destination of the RNA.
Our experiments demonstrate that a group of common factors binds to the localization element of RNAs that become localized during stage III/IV of Xenopus oogenesis irrespective of destination. This observation is not entirely surprising. Using immunoprecipitation assays from whole-cell extract, we reported earlier that three proteins that bind to the Vg1 VLE (Prrp, VgRBP71, and 40LoVe) are also found associated with An1 and An3 (Zhao et al. 2001; Kroll et al. 2002, 2009). In the case of VgRBP71, direct binding to An1 was also demonstrated in mobility shift assays (Kroll et al. 2002). The indication that RNP complexes that direct RNA movement are largely composed of the same group of proteins is satisfying in light of increasing evidence that many RNAs are localized in a variety of cell types (Marchand et al. 2012). In Drosophila early embryos, an estimated 70% of mRNAs are organized into distinct patterns (Lecuyer et al. 2007). It seems unlikely, if not impossible, that each RNA in this case would bind a unique or significantly dissimilar set of trans-acting factors. The more likely alternative is that the same core group of proteins assembles on the majority of localized RNAs with the inclusion of a smaller number of proteins that determine the destination of the transported RNA.
This model raises some challenging questions. What differences in the core RNP structure mediate the recruitment of factors that determine direction? Is the recruitment of these latter factors determined by the overall organization of the RNP, by direct contact to specific sequences in the RNA, or by a combination of both? In addition, is the destination of the RNA established in the nucleus or the cytoplasm? As more is learned about the composition of different localization RNP complexes, these key points can be addressed.
MATERIALS AND METHODS
Several of the experimental procedures have been described previously, including RNA isolation, Northern blot analysis, in vitro transcription, site-directed mutagenesis, isolation and culturing of oocytes, and UV-cross-linking assays (Zhao et al. 2001; Kolev and Huber 2003; Kroll et al. 2009).
Plasmids and nucleic acids
Individual RNAs and the plasmids used for their synthesis are described in Supplemental Table S3. All nucleotide positions for PHAX refer to accession number NM_001096115.1. This is the longest version of Xenopus PHAX mRNA presently in the nucleotide database, but it is missing the 3′ terminus of the oocyte transcript. The complete sequence of the oocyte mRNA was assembled by overlapping NM_001096115.1 with EST sequences AW767185 and AW766532 (Supplemental Fig. S1). The PHAX ORF is between nucleotides 80 and 1267. The parent plasmid for site-directed mutagenesis (QuickChange; Stratagene) of E2 and VM1 motifs in the An1 3′ UTR was pBSAn1PORF, which carries the 3′ half of An1 mRNA from nucleotides 1456–2836; the An1 ORF ends at nucleotide 2106.
In situ hybridization
Combined in situ hybridization and immunocytochemistry followed published procedures (Cote et al. 1999; Kloc and Etkin 1999). DIG-labeled antisense probes for Vg1 mRNA and PHAX mRNA were detected using anti-DIG-AP Fab fragments (Roche) in combination with BM Purple alkaline phosphatase substrate containing 5 mM levamisole.
Localization assays
Capped, DIG-labeled transcripts (∼50 pg) were injected into stage III/IV oocytes that were then cultured for 4 d at 18°C with daily changes of media containing purified vitellogenin (Kloc and Etkin 1999). Immunohistochemical staining of fixed oocytes followed the procedure described by Kloc and Etkin (1999).
UV cross-linking assay
Internally radiolabeled RNAs were prepared by run-off transcription using [α-32P]UTP. All RNAs were renatured in 50 mM HEPES (pH 7.4), 0.3 M KCl, and 10 mM MgCl2 by heating for 10 min to 55°C and cooling slowly to room temperature. In competition assays, a 20-fold excess of unlabeled RNA was added to S100 extract (∼40 μg of protein) and incubated for 10 min prior to the addition of radiolabeled RNA (80 ng). All other steps have been described (Kroll et al. 2009).
In vivo RNA binding assays (immunoprecipitation/RT-PCR)
Affinity-purified human Stau1 antibody (Ribonomics, Inc., RN012P) is certified for RNP immunoprecipitation (RIP). The antibody, directed against a 19-amino-acid sequence conserved between the human and Xenopus proteins, detected a single band in Western blot assays of whole-cell extract from mixed-stage oocytes (Supplemental Fig. S2). The RIP assays were performed as described previously, but with the following modifications (Huber and Zhao 2010): Immunoprecipitation of Staufen1 complexes required the addition of a cross-linking step. Oocytes were incubated in 0.2% formaldehyde in PBS for 1 h at room temperature, then quenched with 0.25 M glycine for 15 min prior to homogenization. Whole-cell extract was pre-cleared by incubation with protein A-Sepharose beads for 4 h at 4°C. After removal of the beads, 5 μg of Stau1 antibody was added to the extract and incubated overnight at 4°C with gentle rocking. Protein A beads (7.5 mg) were added to the extract and incubated for 1 h at 4°C to recover the antibody complexes. Protein–RNA complexes were eluted from the beads by incubation in 50 μL of TES buffer (50 mM Tris at pH 8.0, 10 mM EDTA, 1% SDS) for 10 min at 65°C. Cross-linking was reversed by the addition of Proteinase K (2 μg/μL) and an additional incubation for 1 h at 65°C. RNA was isolated by extraction twice with an equal volume of phenol (pH 4.5) and twice with an equal volume of chloroform:isoamylalcohol (24:1), and the procedure continued as before.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by the American Heart Association (0151445Z) and the College of Science and Office of Research, University of Notre Dame. This paper is dedicated to the memory of mentor and friend, Dr. Ira G. Wool.
REFERENCES
- Allison R, Czaplinski K, Git A, Adegbenro E, Stennard F, Houliston E, Standart N 2004. Two distinct Staufen isoforms in Xenopus are vegetally localized during oogenesis. RNA 10: 1751–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur PK, Claussen M, Koch S, Tarbashevich K, Jahn O, Pieler T 2009. Participation of Xenopus Elr-type proteins in vegetal mRNA localization during oogenesis. J Biol Chem 284: 19982–19992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betley JN, Frith MC, Graber JH, Choo S, Deshler JO 2002. A ubiquitous and conserved signal for RNA localization in chordates. Curr Biol 12: 1756–1761 [DOI] [PubMed] [Google Scholar]
- Boulon S, Verheggen C, Jady BE, Girard C, Pescia C, Paul C, Ospina JK, Kiss T, Matera AG, Bordonne R, et al. 2004. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell 16: 777–787 [DOI] [PubMed] [Google Scholar]
- Bubunenko M, Kress TL, Vempati UD, Mowry KL, King ML 2002. A consensus RNA signal that directs germ layer determinants to the vegetal cortex of Xenopus oocytes. Dev Biol 248: 82–92 [DOI] [PubMed] [Google Scholar]
- Chan AP, Kloc M, Etkin LD 1999. fatvg encodes a new localized RNA that uses a 25-nucleotide element (FVLE1) to localize to the vegetal cortex of Xenopus oocytes. Development 126: 4943–4953 [DOI] [PubMed] [Google Scholar]
- Chang P, Torres J, Lewis RA, Mowry KL, Houliston E, King ML 2004. Localization of RNAs to the mitochondrial cloud in Xenopus oocytes through entrapment and association with endoplasmic reticulum. Mol Biol Cell 15: 4669–4681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartrand P, Meng XH, Singer RH, Long RM 1999. Structural elements required for the localization of ASH1 mRNA and of a green fluorescent protein reporter particle in vivo. Curr Biol 9: 333–338 [DOI] [PubMed] [Google Scholar]
- Choo S, Heinrich B, Betley JN, Chen Z, Deshler JO 2005. Evidence for common machinery utilized by the early and late RNA localization pathways in Xenopus oocytes. Dev Biol 278: 103–117 [DOI] [PubMed] [Google Scholar]
- Claussen M, Pieler T 2004. Xvelo1 uses a novel 75-nucleotide signal sequence that drives vegetal localization along the late pathway in Xenopus oocytes. Dev Biol 266: 270–284 [DOI] [PubMed] [Google Scholar]
- Claussen M, Horvay K, Pieler T 2004. Evidence for overlapping, but not identical, protein machineries operating in vegetal RNA localization along early and late pathways in Xenopus oocytes. Development 131: 4263–4273 [DOI] [PubMed] [Google Scholar]
- Cote CA, Gautreau D, Denegre JM, Kress TL, Terry NA, Mowry KL 1999. A Xenopus protein related to hnRNP I has a role in cytoplasmic RNA localization. Mol Cell 4: 431–437 [DOI] [PubMed] [Google Scholar]
- Czaplinski K, Kocher T, Schelder M, Segref A, Wilm M, Mattaj IW 2005. Identification of 40LoVe, a Xenopus hnRNP D family protein involved in localizing a TGF-β-related mRNA during oogenesis. Dev Cell 8: 505–515 [DOI] [PubMed] [Google Scholar]
- Deshler JO, Highett MI, Abramson T, Schnapp BJ 1998. A highly conserved RNA-binding protein for cytoplasmic mRNA localization in vertebrates. Curr Biol 8: 489–496 [DOI] [PubMed] [Google Scholar]
- Gonzalez I, Buonomo SBC, Nasmyth K, von Ahsen U 1999. ASH1 mRNA localization in yeast involves multiple secondary structural elements and Ash1 protein translation. Curr Biol 9: 337–340 [DOI] [PubMed] [Google Scholar]
- Gu W, Pan F, Zhang H, Bassell GJ, Singer RH 2002. A predominantly nuclear protein affecting cytoplasmic localization of β-actin mRNA in fibroblasts and neurons. J Cell Biol 156: 41–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Havin L, Git A, Elisha Z, Oberman F, Yaniv K, Schwartz SP, Standart N, Yisraeli JK 1998. RNA-binding protein conserved in both microtubule- and microfilament-based RNA localization. Genes Dev 12: 1593–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt CE, Bullock SL 2009. Subcellular mRNA localization in animal cells and why it matters. Science 326: 1212–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvay K, Claussen M, Katzer M, Landgrebe J, Pieler T 2006. Xenopus Dead end mRNA is a localized maternal determinant that serves a conserved function in germ cell development. Dev Biol 291: 1–11 [DOI] [PubMed] [Google Scholar]
- Huber PW, Zhao WM 2010. Detection of protein–RNA complexes in Xenopus oocytes. Methods 51: 82–86 [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD 1999. Analysis of localized RNAs in Xenopus oocytes. In A comparative methods approach to the study of oocytes and embryos (ed. Richter JD), pp. 256–278 Oxford University Press, New York [Google Scholar]
- Kolev NG, Huber PW 2003. VgRBP71 stimulates cleavage at a polyadenylation signal in Vg1 mRNA, resulting in the removal of a cis-acting element that represses translation. Mol Cell 11: 745–755 [DOI] [PubMed] [Google Scholar]
- Kroll TT, Zhao W-M, Jiang C, Huber PW 2002. A homolog of FBP2/KSRP binds to localized mRNAs in Xenopus oocytes. Development 129: 5609–5619 [DOI] [PubMed] [Google Scholar]
- Kroll TT, Swenson LB, Hartland EI, Snedden DD, Goodson HV, Huber PW 2009. Interactions of 40LoVe within the ribonucleoprotein complex that forms on the localization element of Xenopus Vg1 mRNA. Mech Dev 126: 523–538 [DOI] [PubMed] [Google Scholar]
- Kwon S, Abramson T, Munro TP, John CM, Kohrmann M, Schnapp BJ 2002. UUCAC- and Vera-dependent localization of VegT RNA in Xenopus oocytes. Curr Biol 12: 558–564 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131: 174–187 [DOI] [PubMed] [Google Scholar]
- Lecuyer E, Yoshida H, Krause HM 2009. Global implications of mRNA localization pathways in cellular organization. Curr Opin Cell Biol 21: 409–415 [DOI] [PubMed] [Google Scholar]
- Lewis RA, Gagnon JA, Mowry KL 2008. PTB/hnRNP I is required for RNP remodeling during RNA localization in Xenopus oocytes. Mol Cell Biol 28: 678–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeber J, Claussen M, Jahn O, Pieler T 2010. Interaction of 42Sp50 with the vegetal RNA localization machinery in Xenopus laevis oocytes. FEBS J 277: 4722–4731 [DOI] [PubMed] [Google Scholar]
- Marchand V, Gaspar I, Ephrussi A 2012. An intracellular transmission control protocol: Assembly and transport of ribonucleoprotein complexes. Curr Opin Cell Biol 24: 202–210 [DOI] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A 2009. mRNA localization: Gene expression in the spatial dimension. Cell 136: 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messitt TJ, Gagnon JA, Kreiling JA, Pratt CA, Yoon YJ, Mowry KL 2008. Multiple kinesin motors coordinate cytoplasmic RNA transport on a subpopulation of microtubules in Xenopus oocytes. Dev Cell 15: 426–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL 1996. Complex formation between stage-specific oocyte factors and a Xenopus mRNA localization element. Proc Natl Acad Sci 93: 14608–14613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Melton DA 1992. Vegetal messenger RNA localization directed by a 340-nt RNA sequence element in Xenopus oocytes. Science 255: 991–994 [DOI] [PubMed] [Google Scholar]
- Ohno M, Segref A, Bachi A, Wilm M, Mattaj IW 2000. PHAX, a mediator of U snRNA nuclear export whose activity is regulated by phosphorylation. Cell 101: 187–198 [DOI] [PubMed] [Google Scholar]
- Perry-O’Keefe H, Kinter CR, Yisraeli J, Melton DA 1990. The use of in situ hybridisation to study the localisation of maternal mRNAs during Xenopus oogenesis. In In situ hybridisation: Application to developmental biology and medicine (ed. Harris N, Wilkinson DG), Vol. 40, pp. 115–130 Cambridge University Press, Cambridge [Google Scholar]
- Rehbein M, Wege K, Buck F, Schweizer M, Richter D, Kindler S 2002. Molecular characterization of MARTA1, a protein interacting with the dendritic targeting element of MAP2 mRNAs. J Neurochem 82: 1039–1046 [DOI] [PubMed] [Google Scholar]
- Roegiers F, Jan YN 2000. Staufen: A common component of mRNA transport in oocytes and neurons? Trends Cell Biol 10: 220–224 [DOI] [PubMed] [Google Scholar]
- Snee M, Kidd GJ, Munro TP, Smith R 2002. RNA trafficking and stabilization elements associate with multiple brain proteins. J Cell Sci 115: 4661–4669 [DOI] [PubMed] [Google Scholar]
- St Johnston D, Beuchle D, Nüsslein-Volhard C 1991. staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66: 51–63 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Lemm I, Ingelfinger D, Schneider C, Hossbach M, Urlaub H, Luhrmann R 2004. Assembly and maturation of the U3 snoRNP in the nucleoplasm in a large dynamic multiprotein complex. Mol Cell 16: 789–798 [DOI] [PubMed] [Google Scholar]
- Yoon YJ, Mowry KL 2004. Xenopus Staufen is a component of a ribonucleoprotein complex containing Vg1 RNA and kinesin. Development 131: 3035–3045 [DOI] [PubMed] [Google Scholar]
- Zhao WM, Jiang C, Kroll TT, Huber PW 2001. A proline-rich protein binds to the localization element of Xenopus Vg1 mRNA and to ligands involved in actin polymerization. EMBO J 20: 2315–2325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimyanin VL, Belaya K, Pecreaux J, Gilchrist MJ, Clark A, Davis I, St Johnston D 2008. In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134: 843–853 [DOI] [PMC free article] [PubMed] [Google Scholar]




