Pre-mRNA splicing occurs in two steps. This paper reports studies of protein factor requirements for step 2 catalysis. The studies reveal that the only remodeling of the spliceosome between step 1 and step 2 is mediated by the DEAH-box ATPase Prp16.
Keywords: spliceosome, step 2 catalysis, step 2 factors/Prp16, Slu7 Prp18, Prp22
Abstract
Step 2 catalysis of pre-mRNA splicing entails the excision of the intron and ligation of the 5′ and 3′ exons. The tasks of the splicing factors Prp16, Slu7, Prp18, and Prp22 in the formation of the step 2 active site of the spliceosome and in exon ligation, and the timing of their recruitment, remain poorly understood. Using a purified yeast in vitro splicing system, we show that only the DEAH-box ATPase Prp16 is required for formation of a functional step 2 active site and for exon ligation. Efficient docking of the 3′ splice site (3′SS) to the active site requires only Slu7/Prp18 but not Prp22. Spliceosome remodeling by Prp16 appears to be subtle as only the step 1 factor Cwc25 is dissociated prior to step 2 catalysis, with its release dependent on docking of the 3′SS to the active site and Prp16 action. We show by fluorescence cross-correlation spectroscopy that Slu7/Prp18 and Prp16 bind early to distinct, low-affinity binding sites on the step-1-activated B* spliceosome, which are subsequently converted into high-affinity sites. Our results shed new light on the factor requirements for step 2 catalysis and the dynamics of step 1 and 2 factors during the catalytic steps of splicing.
INTRODUCTION
Splicing catalysis proceeds by way of two consecutive phosphoester transfer reactions that involve three chemically reactive sites in the pre-mRNA: the 5′ splice site (5′SS), the 3′ splice site (3′SS), and the branch site (BS). In the first step, the BS adenosine attacks the 5′SS nucleophilically, yielding the free 5′ exon and the lariat-3′ exon intermediates. In the second step, the 3′ hydroxyl group of the 5′ exon attacks the 3′SS; the exons are ligated and the intron lariat is excised (Will and Lührmann 2011).
The spliceosome is a dynamic molecular machine which undergoes several remodeling steps during its functional cycle; these are driven by at least eight evolutionarily conserved DExH/D-box ATPases or RNA helicases (Cordin et al. 2012). Initially, U1 and U2 snRNPs recognize (by base-pairing) the 5′SS and BS, respectively. The preformed U4/U6.U5 tri-snRNP is then recruited, yielding the B complex, which however does not yet have an active catalytic center. Subsequent activation of the spliceosome (yielding the Bact complex) requires the action of the DExH/D-box ATPases Prp28 and Brr2 and involves a major rearrangement of the spliceosome’s RNA–RNA interaction network. This includes the displacement of the U1 and U4 snRNAs and the formation of new base-pair interactions of U6 snRNA with U2 snRNA and the 5′SS. The resulting RNA structure forms the core of the catalytic center of the spliceosome. In addition, the conserved loop I of the U5 snRNA is also part of the catalytic center and helps to correctly position the 3′ and 5′ ends of the exons for both catalytic steps (Sontheimer and Steitz 1993; Staley and Guthrie 1998; Horowitz 2012).
During the transition from the B to the Bact complex, not only is the spliceosome’s RNA network rearranged, but also its protein composition changes radically. In the yeast spliceosome, all of the U1 and U4/U6 proteins are released, while approximately 20 proteins are stably recruited during this transition. These include eight proteins of the “Nineteen Complex” (NTC) and 12 additional proteins, which are henceforth termed NTC-related proteins (Chen et al. 2002; Fabrizio et al. 2009; Warkocki et al. 2009). Although the Bact complex now contains a catalytic center, it is catalytically inactive. In yeast, step 1 catalysis of pre-mRNA splicing still requires three proteins: Prp2, Spp2, and Cwc25 (Roy et al. 1995; Kim and Lin 1996; Silverman et al. 2004; Chiu et al. 2009; Warkocki et al. 2009). The DEAH-box ATPase Prp2 and its coactivator Spp2 are needed for the ATP-dependent creation of the active site of the catalytic center for step 1 (Kim and Lin 1996; Warkocki et al. 2009; Lardelli et al. 2010). During this step, the 45S Bact complex undergoes a substantial structural change that leads to the 40S B* complex and involves, among others, the displacement of at least two NTC-related proteins, Cwc24 and Cwc27, as well as the destabilization of the binding of the U2 SF3a/SF3b and retention and splicing (RES) complex proteins. As a result, the BS adenosine becomes accessible, allowing nucleophilic attack by its 2′OH group on the phosphodiester bond at the 5′SS (Warkocki et al. 2009; Lardelli et al. 2010). Interestingly, the Prp2-mediated remodeling of the spliceosome also creates a high-affinity binding site for Cwc25 on the B* complex. Cwc25 remains firmly bound to the spliceosome after the completion of step 1 catalysis, which generates the spliceosomal C complex (Ohrt et al. 2012). It has been suggested that Cwc25 may stabilize the step 1 conformation of the catalytic center, thereby facilitating step 1 catalysis (Chiu et al. 2009; Warkocki et al. 2009).
As a prerequisite for step 2 catalysis, complex C also must be remodeled. This is achieved by the activity of the DEAH-box ATPase Prp16 (Schwer and Guthrie 1992). Further participants in the efficient catalysis of step 2 are Slu7, Prp18, and Prp22, which like Prp16, are not already present in the Bact complex (Warkocki et al. 2009; Horowitz 2012). A known task of Slu7 and Prp18, which form a heterodimeric complex in the spliceosome, is to dock the 3′SS into the active site of the catalytic center (James et al. 2002; Horowitz 2012), a process that does not require ATP. Consistent with this, Slu7 can be crosslinked to the 3′SS (Umen and Guthrie 1995; McPheeters et al. 2000). Prp22 is a DEAH-box ATPase that is required for the dissociation of the mature mRNA from the intron-lariat spliceosome after step 2 catalysis (Schwer 2008). In whole-cell extracts in vitro, Slu7 and Prp18 cooperate with Prp22, which also has an ATP-independent function before step 2 catalysis (Schwer and Gross 1998). Depletion/reconstitution studies performed with whole-cell splicing extracts showed that the requirement of step 2 factors for step 2 catalysis is less strict when the distance between the BS and the 3′SS is relatively short (Ansari and Schwer 1995; Brys and Schwer 1996; Zhang and Schwer 1997; Schwer and Gross 1998; James et al. 2002). However, little more is known about the mechanism of action of these factors in step 2 catalysis because investigation of the catalytic mechanisms of the spliceosome was until very recently only possible in whole-cell extracts, making it necessary to use immunodepletion and immunoprecipitation techniques with their accompanying limitations.
The extent to which complex C is restructured during Prp16-mediated catalytic activation preceding step 2 is also poorly understood. It has been hypothesized that the spliceosome requires two different conformations during its catalytic phase (Konarska and Query 2005). However, at the same time, there is evidence for a single active site in the catalytic center of the spliceosome (Steitz and Steitz 1993). Thus, one possible structural change catalyzed by the ATPase Prp16 would be the removal of the branch structure from the step 1 active site to allow accommodation of the 3′SS (Konarska and Query 2005). It is not known, however, whether this is accompanied by remodeling events as extensive as those observed during Prp2-mediated catalytic activation preceding step 1 (Ohrt et al. 2012). On the basis of immunoprecipitation experiments, it has been shown that at least two proteins, Cwc25 and Yju2, are displaced during the action of Prp16 (Tseng et al. 2011), but it is not clear whether the action of additional step 2 factors is also needed. Moreover, it is not clear whether the creation of the step 2 active site requires only Prp16 or whether one or more of the other step 2 factors are also required for this process—either by helping to create the active site or stabilizing the step 2 conformation. Alternatively, or in addition, the docking of the 3′SS into the step 2 active site may also have an important effect on stabilizing the catalytic center in a conformation suitable for step 2 catalysis. Further open questions concern the timing of the recruitment of the step 2 factors to the spliceosome during the catalytic phase and the locations of their binding sites.
Here we have studied the role of the four transacting step 2 factors (i.e., that are not present in the Bact spliceosome) in step 2 catalysis of yeast pre-mRNA splicing, employing a recently described purified splicing system (Warkocki et al. 2009). We used purified and well-characterized yeast spliceosomal complexes stalled before catalytic activation at the Bact stage and complemented them with recombinantly expressed step 1 and step 2 splicing factors. We show that only the ATPase Prp16 is required to efficiently transform the spliceosome after step 1 (the C complex) into the catalytically activated C* complex, whose step 2 active site conformation is sufficiently stable to allow its isolation. The C* complex catalyzes exon ligation in the absence of Slu7, Prp18, and Prp22 when the BS-3′SS distance is short. Surprisingly, efficient splicing of a pre-mRNA with a longer BS-3′SS distance requires only Slu7 and Prp18 (in addition to Prp16) for exon ligation but not Prp22. Our results indicate that the main function of Slu7 and Prp18 is to provide efficient docking of the 3′SS into the step 2 active site, and Prp22 is not needed for this step. We show via mass spectrometry and dual-color fluorescence cross-correlation spectroscopy (dcFCCS) that the only protein displaced from the spliceosome as a prerequisite for step 2 catalysis is the step 1 factor Cwc25. Only the stable docking of the 3′SS—and not the productive step 2 catalysis—is the prerequisite for the dissociation of Cwc25, in addition to the action of Prp16. Thus, the Prp16-mediated remodeling of the spliceosome appears to be subtle. Finally, we show by dcFCCS that Prp16 and Slu7 each have a specific, low-affinity binding site already on the B* spliceosome; these sites are distinct from one another and from the binding site of the step 1 factor Cwc25. The low-affinity entry sites for Prp16 and Slu7 in the B* complex are sequentially converted into high-affinity binding sites during or after completion of step 1 catalysis (for Prp16) and of C* complex formation (for Slu7/Prp18). Thus, our results suggest that all the factors investigated here bind simultaneously to the catalytically activated spliceosome, and each factor’s binding site is “toggled” between a low-affinity and a high-affinity state.
RESULTS
Prp16 and Slu7/Prp18, but not Prp22, are required for efficient step 2 catalysis of wild-type actin pre-mRNA
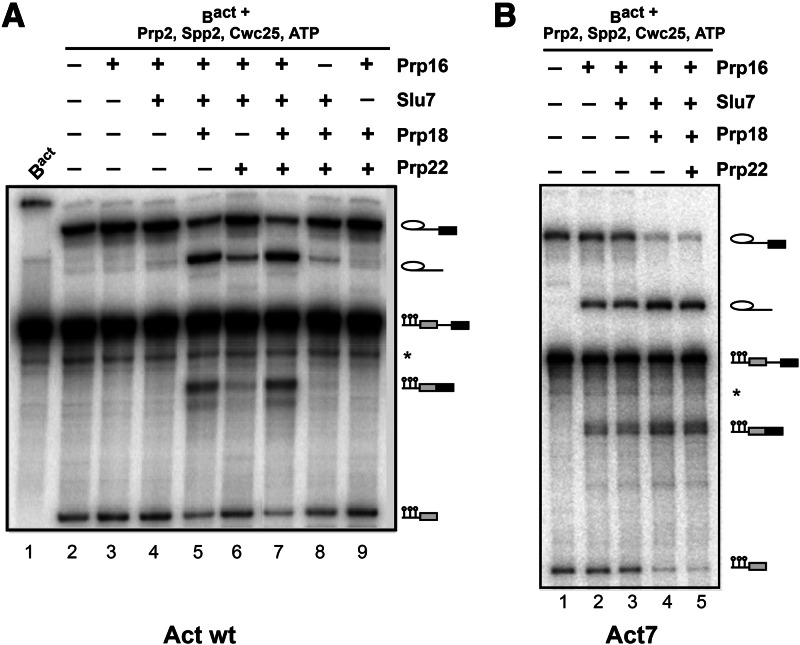
First we investigated the requirements for step 2 catalysis in our purified splicing system using wild-type actin pre-mRNA, which contains 38 nucleotides between the branch consensus sequence (i.e., UACUAACA) and the 3′SS consensus sequence (i.e., UAG). For this purpose, we purified activated B complexes that had been assembled on a wild-type actin pre-mRNA to which three MS2 stem–loops had been added at its 5′ end (these loops have no effect on splicing; the pre-mRNA is referred hereafter as Actwt and the corresponding spliceosome as Actwt-Bact). Spliceosomes were assembled in heat-inactivated splicing extract from yeast cells expressing a temperature-sensitive Prp2 helicase (prp2-1) and isolated by a three-step affinity purification method described previously (Fabrizio et al. 2009; Warkocki et al. 2009). The purified Bact complexes lacking Prp2 were eluted with maltose from the affinity matrix and fractionated by centrifugation in a glycerol gradient.
Fractions containing the purified Bact complex were combined, and aliquots containing 10 fmol of this complex were mock-treated (Fig. 1A, lane 1), or complemented in solution with Prp2, Spp2, Cwc25, and ATP (generating a reconstituted C complex, lanes 2–9) plus various combinations of recombinant Prp16, Slu7, Prp18, and Prp22 (lanes 3–9). All were then incubated for 45 min at 23°C, and splicing catalysis was subsequently analysed by denaturing gel electrophoresis. As reported previously (Warkocki et al. 2009), step 1 catalysis occurred in the presence of Prp2, Spp2, Cwc25, and ATP (lane 2). Interestingly, efficient step 2 catalysis was observed upon further addition of Prp16, Slu7, and Prp18 (lane 5). Prp22 was not required, as its addition to the other step 2 factors did not increase step 2 splicing efficiency (cf. lanes 5,7). Omission of Prp18 (lane 4), Prp16 (lane 8), or Slu7 (lane 9) reduced step 2 catalysis to near background levels. Prp22 appeared to compensate to a minor extent for the absence of Prp18, yielding a step 2 splicing efficiency of 1%–2% (cf. lane 6 with lane 7), but Prp22 could not compensate at all for the absence of Slu7 from the reaction mixture (lane 9). We conclude that Prp16, Slu7, and Prp18 are sufficient for step 2 catalysis of actin pre-mRNA, and Prp22 is not required to increase the efficiency of exon ligation in the extract-free splicing system.
FIGURE 1.
Spliceosomal factor requirements for step 2 catalysis of Bact spliceosomes assembled on actin pre-mRNAs. (A) Extent of splicing in vitro catalyzed by affinity-purified spliceosomes assembled on the Actwt pre-mRNA substrate. The Bact complex (lane 1) was affinity-purified by a three-step procedure (see Supplemental Methods). Peak fractions of the second glycerol gradient were used for complementation assays with recombinant proteins at a 30-fold molar excess over the Bact complex (10 fmol) as indicated (lanes 2–9). Reaction mixtures were incubated (45 min at 23°C) in the presence of ATP. RNAs were extracted, separated by denaturing polyacrylamide gel electrophoresis (PAGE), and visualized by autoradiography. (B) Bact complexes assembled on Act7 pre-mRNA were purified and eluted from the amylose matrix. Subsequently, the complexes were complemented with the recombinant proteins indicated and were further incubated in the presence of ATP. Symbols for pre-mRNA, splicing intermediates, and products are indicated on the right: the 5′ exon (with the three MS2 loops shown) in gray, intron as a thin black line, 3′ exon in black. (*) Uncharacterized pre-mRNA bands.
Prp16 is sufficient for step 2 catalysis of Act7 pre-mRNA
Next we investigated step 2 catalysis and its factor requirements using a pre-mRNA (designated Act7) with a much shorter distance between the BS and the 3′SS consensus sequences (seven versus 38 nucleotides in Actwt) (Brys and Schwer 1996). As a control, we first biochemically characterized the purified Bact complexes assembled on Act7 (Supplemental Fig. S1). As observed for Bact complexes assembled on Actwt substrate, complexes formed on Act7 contained only unspliced pre-mRNA and the U2, U5, and U6 snRNAs. Mass-spectrometric (MS) analysis of Bact complexes assembled on Act7 (Act7-Bact) (see Fourmann et al. [2013] for MS analysis of Bact complexes assembled on Act7 substrate) revealed that their proteome was essentially the same as that of Bact complexes assembled on the wild-type substrate and, like Actwt-Bact, lacked Prp2, Spp2, Cwc25, and most importantly, step 2 factors as well as disassembly factors (see Supplemental Table S1 for MS analysis of Bact complexes assembled on the wild-type substrate, column 1).
Affinity-purified Act7-Bact complexes were eluted from the matrix and then used for in vitro reconstitution of both catalytic steps of splicing (Fig. 1B). Act7-Bact complexes yielded efficient step 1 intermediates upon incubation with Prp2, Spp2, Cwc25, and ATP (lane 1). Interestingly, the addition of Prp16 alone resulted in the formation of significant amounts of step 2 splicing products (i.e., mRNA and intron lariat) (Fig. 1B, lane 2). However, further addition of both Slu7 and Prp18 increased the amounts of these products (Fig. 1B, cf. lane 4 with lanes 2,3), whereas the additional inclusion of Prp22 had no further stimulatory effect (lane 5).
As Slu7/Prp18 significantly stimulated the efficiency of Act7 pre-mRNA splicing, we investigated this effect in more detail by performing a time course of the Act7 splicing reaction in the presence and absence of Slu7/Prp18. Supplemental Figure S2 shows that there was a twofold difference in the yield of mRNA product. Although ∼40% of step 1 splicing intermediates were transformed into products in the absence of Slu7/Prp18 after ∼40 min incubation (Supplemental Fig. S2B), a yield of ∼80% was obtained in the presence of Slu7/Prp18 (Supplemental Fig. S2A). In summary, these data show that Slu7/Prp18 is not essential for the generation of the step 2 active site; and thus, the 3′SS can interact productively with the step 2 active site in its absence, provided that the distance between the BS and 3′SS consensus sequences is short (here, 7 nucleotides). On the other hand, the data also show that the Slu7/Prp18 heterodimer enhances significantly the efficiency of step 2 catalysis of Act7 pre-mRNA splicing (Supplemental Fig. S2C).
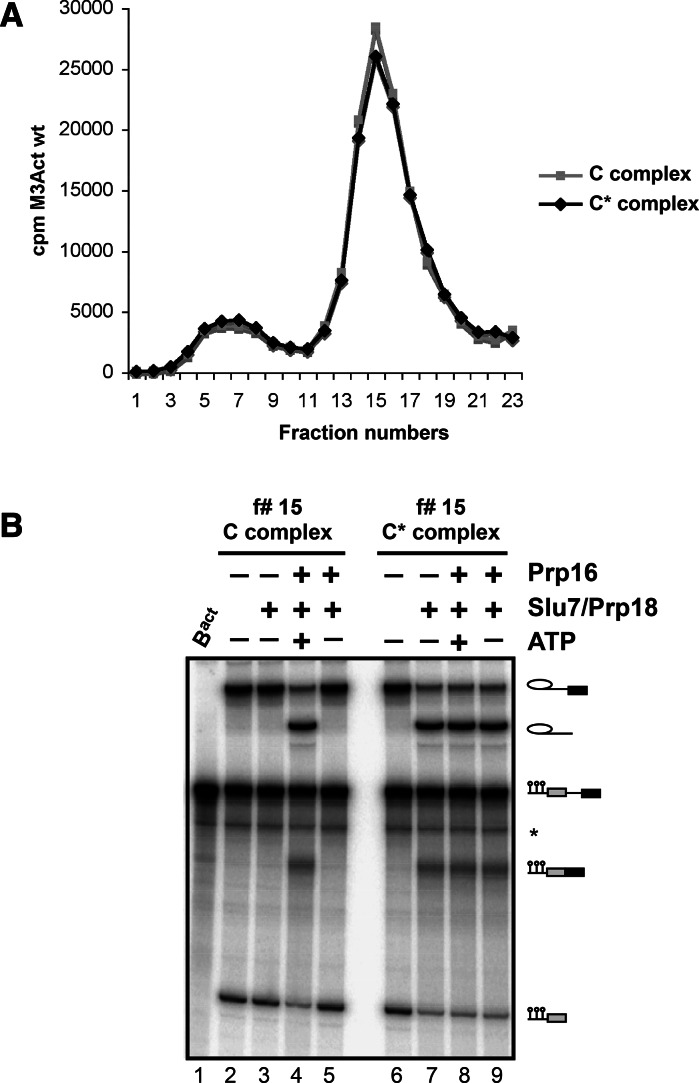
Prp16 is sufficient to create a stable C* spliceosome, which contains an active step 2 catalytic site
One possible explanation for the observed lower efficiency of step 2 catalysis of Act7 pre-mRNA in the absence of Slu7/Prp18 could be that the Prp16-catalyzed transition from the step 1 to step 2 conformation of the spliceosome’s catalytic center is inefficient in the absence of these two proteins. Alternatively, or in addition, the conformation of the step 2 catalytic center might be less stable in the absence of Slu7/Prp18. To distinguish between these possibilities, we carried out the following experiment. First, we generated a C complex by supplementing affinity-purified, amylose-matrix-bound Actwt-Bact complexes with Prp2, Spp2, Cwc25, and ATP (Fig. 2A). After incubation for 45 min at 23°C, the matrix-bound spliceosomes were divided into equal portions; one portion was supplemented with buffer, the other with Prp16 and ATP. The two reaction mixtures were incubated for 45 min at 23°C and then depleted of residual ATP by treatment with hexokinase and glucose. The amylose matrices were then washed extensively with GK75 buffer (containing 75 mM KCl; see Materials and Methods for details), and the spliceosomal complexes were eluted with maltose and subjected to glycerol gradient centrifugation in GK75 buffer. Figure 2A shows that the sedimentation profiles of the spliceosomes from the two reactions were essentially the same.
FIGURE 2.
ATP-dependent Prp16 activity is sufficient to generate stable, catalytically activated C* complexes in the absence of Slu7, Prp18, and Prp22. (A) Bact complexes were purified and complemented with Prp2, Spp2, and Cwc25 on the amylose matrix to generate C complexes. One-half of the reaction mixture was supplemented with buffer, the other half with Prp16 and ATP to generate C* complexes. After incubation, complexes were washed with GK75 buffer and eluted with maltose. Complexes (specific activity 100 cpm/fmol) were sedimented on glycerol gradients containing 75 mM KCl. Fractions were analyzed by Cherenkov counting. (B) Fraction 15 of both complexes was analyzed directly (lanes 2,6) or was additionally complemented with Slu7/Prp18 and Prp16 +/− ATP as indicated. After incubation, RNAs were extracted, separated by denaturing PAGE, and visualized by autoradiography. RNA species (right), see Figure 1.
Peak fraction 15 from each gradient was divided into aliquots (each containing 10 fmol spliceosomes), which were then incubated for 45 min at 23°C in the absence or presence of Prp16 +/− ATP and/or Slu7/Prp18. The yields of splicing intermediates and products were subsequently determined by denaturing gel electrophoresis. Figure 2B (lanes 2,3) shows that the purified C complex, which comprises the Actwt pre-mRNA splicing intermediates (lane 2), did not yield any step 2 products upon addition of Slu7/Prp18 (lane 3). Only when Prp16 and ATP were included together with Slu7/Prp18 was efficient step 2 catalysis observed (lane 4). In contrast, with purified C complexes that had been preincubated with Prp16 and ATP before glycerol-gradient centrifugation, significant step 2 catalysis was observed already after addition of Slu7/Prp18 (Fig. 2B, lane 7), and the yield of step 2 splicing products was not enhanced when Prp16 +/− ATP was included in addition to Slu7/Prp18 (lanes 8,9). These data therefore demonstrate that Prp16 and ATP together generate an active step 2 conformation of the spliceosome’s catalytic center, which is stable in the absence of Slu7/Prp18 (and Prp22) and remains active during glycerol gradient centrifugation. As this purified spliceosome is catalytically activated for step 2 catalysis, we term it the “C* complex.” These data also suggest that the significant stimulation of Act7 step 2 catalysis by Slu7/Prp18 described above (Fig. 1B; Supplemental Fig. S2), is unlikely due to less efficient C* formation by Prp16 in the absence of Slu7/Prp18 (see also below).
Prp16-mediated remodeling of the spliceosome does not per se lead to dissociation of proteins from the C* complex
The ability to isolate stable C* complexes allowed us to investigate whether the Prp16-mediated activation of the spliceosome for step 2 catalysis leads to the dissociation of proteins. For this purpose we first compared the protein composition of the isolated C* complex with that of the C complex. In addition, we determined the protein composition of spliceosomes that had catalyzed step 2, by the addition of Slu7/Prp18 to the C* complex, and thus contained mRNA product. All complexes were assembled on Actwt pre-mRNA and isolated by glycerol gradient centrifugation at 75 mM salt (Fabrizio et al. 2009; Warkocki et al. 2009). Supplemental Table S1 shows that the peptide numbers for the U2, U5, NTC, and NTC-related proteins, as well as for the proteins of the RES complex in C and C* complexes, were essentially identical. In both complexes, only trace quantities of Cwc24 and Cwc27 were detected; these proteins are known to dissociate already during the Prp2-mediated transformation of the Bact to B* spliceosome (Warkocki et al. 2009; Ohrt et al. 2012). These data show that the Prp16-mediated spliceosome remodeling step does not involve the dissociation of any spliceosomal protein (see also Ilagan et al. 2013). However, when one compares the peptide numbers of the proteins from complexes C and C* with those from the post-catalytic complex—that is, after addition of Slu7/Prp18—it becomes clear that Cwc25 is under-represented in the post-catalytic complex. Similar amounts of all other proteins, including Yju2, were found. This suggests that Slu7/Prp18 may be directly or indirectly involved in the dissociation of Cwc25. Our results differ in this respect from those published recently by Tseng et al. (2011), who suggested that Cwc25 is released together with Yju2 from the spliceosome solely by the action of Prp16. However, they conducted immunoprecipitations with antibodies against Cwc25 and Yju2 under high-salt conditions, where many spliceosomal proteins likely are destabilized or dissociate (Ohrt et al. 2012). Moreover, a role of Slu7/Prp18 in Cwc25 release could not be excluded under their experimental conditions.
Slu7/Prp18 is required in addition to Prp16 to displace Cwc25 from the spliceosome assembled on Actwt pre-mRNA as determined by dcFCCS
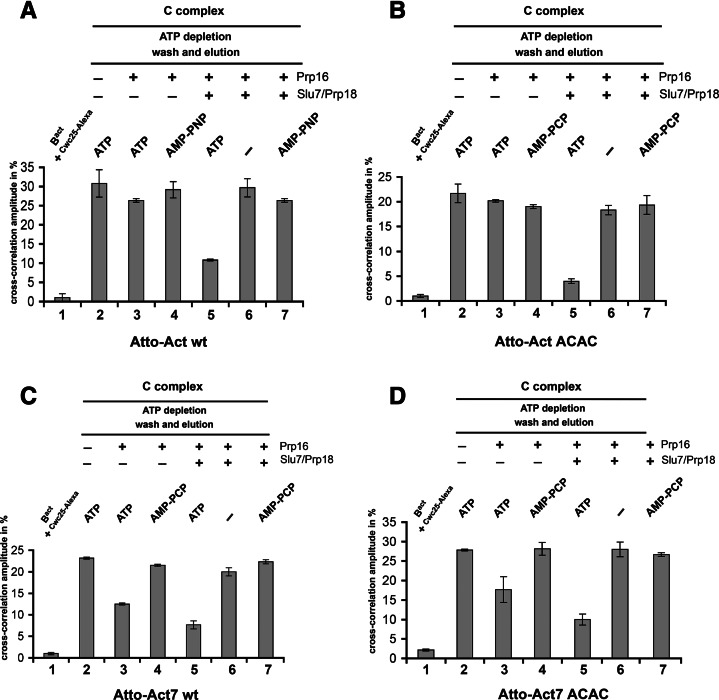
To clarify the requirements for the dissociation of Cwc25 from the spliceosome, we used dcFCCS, which allows the quantitative analysis of the recruitment and dissociation of a protein (Ohrt et al. 2008, 2011, 2012). The experimental set-up for the dcFCCS measurements was described in detail previously (Ohrt et al. 2012). In brief, we first purified Bact spliceosomes that were assembled on Actwt pre-mRNA containing a red fluorescent dye, Atto647N-maleimide (Atto), at its 5′ end. While the spliceosomes were stably bound to the amylose affinity matrix, they were incubated with recombinant Prp2, Spp2, ATP, and a modified version of Cwc25, which carried the green fluorescent dye, Alexa488, at its C terminus (Cwc25-Alexa) (Ohrt et al. 2012). After removal of ATP by incubation with hexokinase and glucose, the column was extensively washed with GK75 buffer. The C complexes carrying simultaneously a red dye (at their 5′ exon) and a green dye (on Cwc25) were eluted from the affinity matrix with maltose and supplemented in solution with Prp16+/−Slu7/Prp18 in the presence or absence of ATP or in the presence of the nonhydrolysable ATP analog AMP-PNP or AMP-PCP. The mixtures were incubated for 45 min at 23°C and then subjected to dcFCCS measurement.
The resulting cross-correlation amplitudes are shown in Figure 3A. Consistent with our previous findings (Ohrt et al. 2012), the purified C complex showed high cross-correlation amplitude of ∼30% (column 2), indicating that Cwc25-Alexa is stably bound to the C complex. Consistent with its recruitment first during B* complex formation, Cwc25-Alexa did not bind to purified Actwt-Bact complexes (column 1). Importantly, incubation of the C complex with Prp16 and ATP, yielding the C* complex, did not decrease significantly the cross-correlation amplitude compared with the control reaction of the C complex with Prp16 and AMP-PNP (columns 3 and 4). This indicates that Prp16 and its ATP-dependent catalytic activation of the spliceosome for step 2 catalysis are not sufficient to displace Cwc25 under near-physiological conditions. However, when C* complexes were further supplemented with Slu7/Prp18, a substantial decrease in the cross-correlation amplitude (by ∼65%) was observed (column 5). This was not observed when C complexes were incubated with Prp16 and Slu7/Prp18 in the absence of ATP or in the presence of AMP-PNP, i.e., when C* complex formation was inhibited (columns 6 and 7). As a control, we also analyzed the products of the splicing reaction by denaturing gel electrophoresis, using aliquots from the same reactions that had been used for the dcFCCS studies in Figure 3A. This analysis confirmed that step 2 products were obtained only when C complexes were incubated with Prp16, Slu7, and Prp18 in the presence of ATP (Supplemental Fig. S3, lane 4, corresponding to the reaction conditions in Fig. 3A, column 5). Consistent with the MS results (Supplemental Table S1), our dcFCCS results show that the release of Cwc25 requires Slu7/Prp18, in addition to the ATPase activity of Prp16, to displace Cwc25 from the spliceosome.
FIGURE 3.
Displacement of the step 1 factor Cwc25 from the spliceosome by Slu7/Prp18 as analyzed by dcFCCS. (A) Affinity-purified Bact complexes assembled on Atto-Actwt pre-mRNA were complemented on the matrix with Prp2, Spp2, ATP, and Cwc25-Alexa. The reaction mixture was incubated and then depleted of ATP. After washing of the matrix, reconstituted C complexes were eluted with maltose and complemented in solution as indicated above each column, and dcFCCS measurements were performed. (B) Affinity-purified Bact complexes assembled on Atto-ActACAC 3′SS mutant (see text) were complemented on the matrix as described in A. The reaction mixture was incubated, C complexes were eluted and complemented in solution as indicated above each column, and dcFCCS measurements were performed. Affinity-purified Bact complexes assembled on Atto-Act7wt (C) and Atto-Act7ACAC 3′SS mutant (D) were complemented on the matrix to generate C complexes. Eluted C complexes were then complemented in solution as indicated above each column, and dcFCCS measurements were performed. Cross-correlation amplitudes derived from two independent experiments are shown for each complex. Error bars indicate the standard deviation from two independent measurements.
To determine whether step 2 catalysis is required per se for the dissociation of Cwc25 from the spliceosome, we carried out the same series of dcFCCS experiments as described in Figure 3A but using spliceosomes assembled onto an actin pre-mRNA that carried a mutated 3′SS (ACAC instead of AGAG) and that were therefore unable to undergo step 2 catalysis. Figure 3B shows that Cwc25-Alexa was also efficiently released from the spliceosome when C complexes were incubated with Prp16, Slu7/Prp18 in the presence of ATP (column 5), but not under any other of the conditions tested (columns 2–7). As a control, we verified that under the experimental conditions of column 5 in Figure 3B, no step 2 catalysis occurred (Supplemental Fig. S4). These data indicate that Slu7/Prp18 is required for the displacement of Cwc25 from the spliceosome irrespective of whether step 2 catalysis occurs, at least when the spliceosome is assembled onto a pre-mRNA with a long distance between the BS and 3′SS.
We also used dcFCCS to study the binding behavior of Yju2-EGFP, which has also been reported to dissociate during the catalytic phase (Liu et al. 2007; Tseng et al. 2011). Under low-salt conditions, we observed a similarly high cross-correlation amplitudes (i.e., for the red Atto dye on the actin pre-mRNA and the Yju2-EGFP, tagged with the Enhanced Green Fluorescent Protein) in the spliceosomal complexes Bact, B*, C* and also in post-catalytic spliceosomes (Supplemental Fig. S5). This result is consistent with our MS analyses of the various spliceosomal complexes (Supplemental Table S1) and shows that Yju2 remains associated with the spliceosome during the entire catalytic phase.
Evidence that docking of the 3′SS to the step 2 active site triggers the dissociation of Cwc25 from the spliceosome
For pre-mRNAs with a long distance between the BS and the 3′SS—such as Actwt pre-mRNA—Slu7/Prp18 is essential to bind the 3′SS efficiently into the step 2 active site (Umen and Guthrie 1995). Slu7/Prp18 may also facilitate docking of the 3′SS ACAC mutant of actin pre-mRNA, as crosslinking revealed that Slu7 directly contacts mutant 3′SS (Umen and Guthrie 1995). Thus, the observed requirements of Slu7/Prp18 for the efficient release of Cwc25 from spliceosomes assembled on Actwt pre-mRNA could also indicate that it is the actual docking of the 3′SS of Actwt pre-mRNA to the step 2 active site that induces the release of Cwc25 from the spliceosome and not the recruitment of Slu7/Prp18 to the spliceosome per se.
Since spliceosomes assembled on Act7 pre-mRNA catalyze step 2 catalysis to a significant extent in the absence of Slu7/Prp18 but twice as efficiently in their presence (Fig. 1B; Supplemental Fig. S2), we investigated by dcFCCS whether Cwc25 is released, at least in part, from these spliceosomes before the recruitment of Slu7/Prp18. Interestingly, when purified C complexes were incubated solely with Prp16 and ATP (but not AMP-PCP), Cwc25-Alexa was indeed displaced from ∼35% of the spliceosomes (Fig. 3C, columns 2–4). Incubation of C complexes with Slu7/Prp18 in addition to Prp16 and ATP, resulted in the displacement of Cwc25 from a further ∼25% of the spliceosomes, again in an ATP-dependent manner (Fig. 3C, columns 5–7). The relative amounts of spliceosomes from which Cwc25 is released in the absence and presence of Slu7/Prp18 corresponds roughly to the yields of step 2 products obtained in the absence and presence of Slu7/Prp18 (Fig. 1B; Supplemental Fig. S2). Finally, the release of Cwc25-Alexa from spliceosomes was equally efficient in the presence of the Act7 3′SS ACAC mutant pre-mRNA (Fig. 3D), demonstrating that also with Act7 pre-mRNA step 2 catalysis is not a prerequisite for the dissociation of Cwc25.
In summary, these results are consistent with the idea that the docking of the 3′SS to its binding site in the step 2 active site is indeed sufficient for the release of Cwc25 from the spliceosome when the distance between the BS and the 3′SS is short. The requirement of Slu7/Prp18 to displace Cwc25 from an additional fraction of Act7-C* spliceosomes (Fig. 1B; Supplemental Fig. S2) would indicate that even when the distance between the BS and 3′SS is short, as in Act7 pre-mRNA, the 3′SS accesses the step 2 active site only inefficiently, and Slu7/Prp18 facilitates this docking step.
A low-affinity entry site that is distinct from the Cwc25 binding site is already created for Slu7/Prp18 during catalytic activation of the spliceosome by Prp2
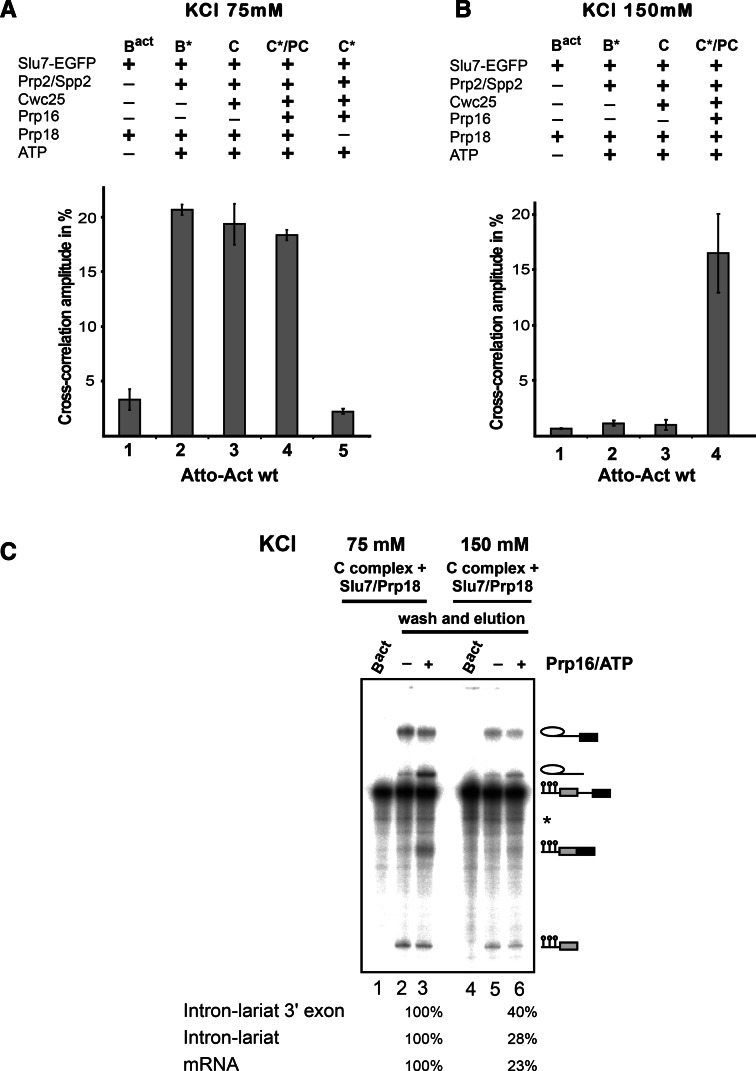
The timing of the recruitment of the step 2 factors Slu7/Prp18 and Prp16 to the spliceosome during the catalytic phase and the location of their binding sites remain poorly understood. To address this question, we first investigated the recruitment of Slu7 to the spliceosome in our purified splicing system using dcFCCS. To obtain a fluorescent version of Slu7, we expressed a fusion protein of Slu7 with EGFP in yeast cells. Purified Slu7-EGFP was functional in an in vitro splicing assay (Supplemental Fig. S6). Although previous immunoprecipitation studies from splicing extracts suggested that Slu7 binds stably to the spliceosome after the Prp16-mediated remodeling of the spliceosome (Ansari and Schwer 1995; Umen and Guthrie 1995; Horowitz 2012), it remains unclear whether Slu7 already has a binding site on the spliceosome at an earlier stage of the spliceosomal cycle. We therefore tested whether Slu7-EGFP can bind specifically Bact, B*, C, or C* complexes under low-salt (75 mM) conditions.
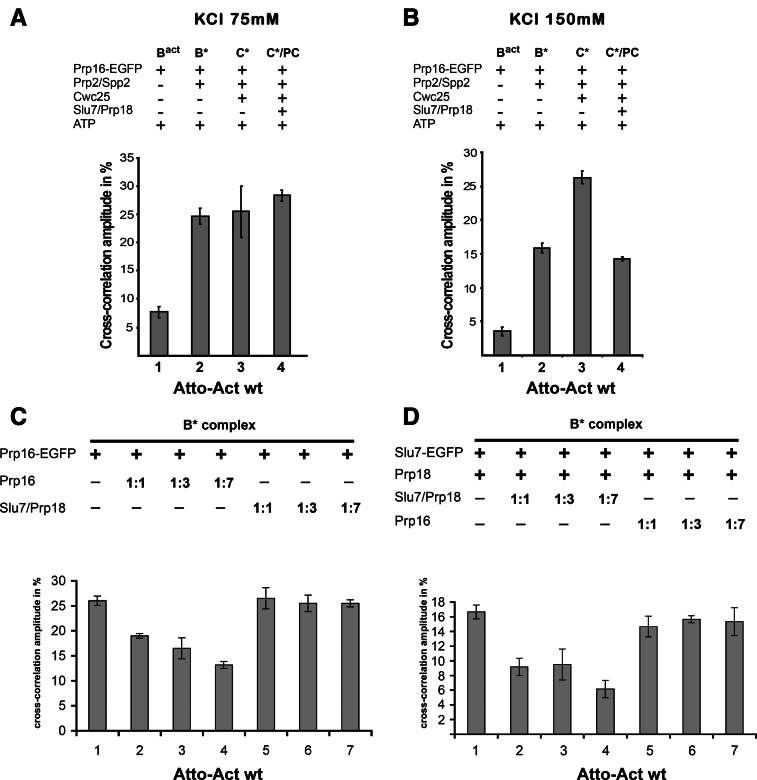
Bact spliceosomes were assembled on Atto-Actwt pre-mRNA as described above. Affinity-purified Bact spliceosomes were supplemented on the amylose affinity matrix with recombinant step 1 factors and Prp16, then incubated with Slu7-EGFP in the presence or absence of Prp18 and (Fig. 4A), washed with GK75 buffer, and then eluted from the matrices with maltose and directly subjected to dcFCCS. The resulting cross-correlation amplitudes between Slu7-EGFP and the Atto-pre-mRNA are shown in Figure 4A. Although Bact spliceosomes did not bind Slu7-EGFP in the presence of Prp18 (as indicated by the low cross-correlation amplitude of ∼3%, column 1), B* exhibited a high cross-correlation amplitude (∼22%), indicating that Slu7/Prp18 bound efficiently to B* complexes at 75 mM salt (column 2). A similar binding efficiency of Slu7-EGFP was observed with C and C* complexes (columns 3 and 4; note that addition of Slu7-EGFP and Prp18 to C* will result in a post-catalytic complex). These data suggest that a binding site for Slu7/Prp18 is already created on the spliceosome at the stage when it is catalytically activated by Prp2, Spp2, and ATP. As binding of Slu7-EGFP to the B* and C* complexes is only observed in the presence of Prp18 (Fig. 4A, column 5), this is consistent with the idea that Slu7 and Prp18 bind as a heterodimer to the spliceosome.
FIGURE 4.
A low-affinity binding site is created for Slu7-EGFP during catalytic activation of the spliceosome by Prp2 as analyzed by dcFCCS. (A) Affinity-purified Bact complexes assembled on Atto-Actwt were complemented on the matrix with recombinant Slu7-EGFP plus Prp18 (column 1, Bact) and additionally with Prp2 and Spp2 (column 2, B*), Cwc25 (column 3, C), Prp16 (column 4, C*/PC), or without Prp18 (column 5, C*). C*/PC indicates that upon Slu7/Prp18 binding to the C* complex, the latter is transformed into the post-catalytic complex (PC). After incubation, complexes were washed with GK75 buffer or GK150 buffer (B) and then eluted from the matrix with maltose. dcFCCS measurements were then performed at complex concentrations of 1.0 nM. Cross-correlation amplitudes derived from two independent experiments are shown for each complex. Error bars indicate the standard deviation from two independent measurements. (C) Reconstituted C complexes were incubated on the matrix with Slu7/Prp18 at 75 or 150 mM KCl, respectively. Complexes were washed with buffers containing 75 or 150 mM KCl, eluted from the matrix in GK75 buffer and mock-treated (lanes 2,5) or incubated in solution with Prp16 plus ATP (lanes 3,6). After incubation, RNAs were extracted, separated by denaturing PAGE, and visualized by autoradiography. Symbols for RNA species (as in Fig. 1) are shown on the right. The intensities of the intermediate and mRNA product signals were quantified using the ImageQuant software (Molecular Dynamics). The signals in lane 3 were used as 100%.
To gain more insight into the strength of Slu7/Prp18 binding to the spliceosome at various functional stages, we carried out the same set of experiments as described in Figure 4A except that the final washing step following incubation of the spliceosomes with Slu7-EGFP was performed with GK150 buffer, containing 150 mM KCl instead of 75 mM. Slu7-EGFP showed a high cross-correlation amplitude only when it was added to the C* complex (Fig. 4B, column 4), whereas the signals observed with B* and C complexes were only slightly above the background value observed with Bact complexes (columns 1–3).
The stable binding of Slu7-EGFP to the C* complex was only observed in the presence of Prp16 and ATP (and not AMP-PCP) (Supplemental Fig. S7), demonstrating that the stable binding of Slu7/Prp18 takes place only concomitantly with, or after, the generation of the step 2 conformation of the catalytic center by the ATPase Prp16. In summary, we conclude that a low-affinity spliceosomal binding site is created for Slu7/Prp18 during Prp2-mediated catalytic activation, and this is converted into a high-affinity binding site by the Prp16 ATP-dependent remodeling of the C complex that yields the C* complex. Our observation that Slu7/Prp18 binds equally well to the B* and C complex, despite the presence of Cwc25 in the C complex, suggests that the binding sites of Cwc25 and Slu7/Prp18 on the C complex do not overlap. Thus, Slu7/Prp18 do not appear to aid displacement of Cwc25 by competing for its binding site.
To determine whether the low-affinity Slu7/Prp18 binding site in the C complex is a functional site, i.e., whether Slu7/Prp18 can facilitate step 2 catalysis when bound to this site, we carried out the following experiment. Affinity-purified matrix-bound Actwt-Bact spliceosomes were incubated with Prp2, Spp2, ATP, and Cwc25 to yield C complexes at 75 mM KCl. The latter were then washed with GK75 buffer and subsequently incubated with Slu7 and Prp18 for 45 min at 23°C, followed by extensive washing (10 column volumes) with GK75 buffer. Under these conditions, excess free Slu7 and Prp18 are removed while Slu7/Prp18 remain bound to the low-affinity binding site of the C complex (see Fig. 4A, column 3). The C complexes were then eluted with maltose and complemented with buffer or Prp16 and ATP. As a control, a similar experiment was carried out as described above except that incubation of the C complex with Slu7/Prp18 and washing step of the affinity matrix were performed with buffer containing 150 mM KCl. Elution with maltose was then performed at 75 mM KCl. Under these conditions, not only is the excess of free Slu7/Prp18 removed but also most of Slu7/Prp18 bound to the low-affinity binding site (see Fig. 4B, column 3). Figure 4C shows that C complexes that have retained Slu7/Prp18 at the low-affinity binding site (i.e., those washed with GK75 buffer) catalyze step 2 of Actwt pre-mRNA efficiently but only in the presence of Prp16 and ATP (lane 3). In contrast, C complexes washed with GK150 buffer yielded only comparatively low amounts of mRNA product (lane 6). These data suggest that Slu7/Prp18, which are bound to their low-affinity binding site in the C complex, can directly facilitate step 2 catalysis following Prp16/ATP action; thus, the low-affinity binding site for Slu7/Prp18 is a functionally relevant site. We propose that the low-affinity binding site be termed the “entry site” of Slu7/Prp18 on the spliceosome.
Prp16's entry site on the B* spliceosome is distinct from the Slu7/Prp18 entry site
It was previously shown that Prp16 may act already during step 1 of splicing (Burgess et al. 1990; Burgess and Guthrie 1993). To test whether Prp16, like Slu7/Prp18, is recruited to the spliceosome at the B* complex stage, we carried out dcFCCS experiments similar to those described above for Slu7-EGFP, using purified Prp16-EGFP that had been expressed in yeast cells. Figure 5A shows that at 75 mM salt, Prp16-EGFP indeed binds efficiently to the B* complex, as indicated by the high cross-correlation amplitude of ∼25% (column 2), whereas with the Bact complex, a cross-correlation amplitude of only 7% was observed (column 1). A high binding efficiency, similar to that with B*, was also observed when Prp16-EGFP was added to the C complex (column 3), which was converted into C* complex by the action of Prp16-EGFP plus ATP, and also after the further addition of Slu7/Prp18, which yields post-catalytic spliceosomes (column 4).
FIGURE 5.
A low-affinity binding site is created for Prp16-EGFP during catalytic activation of the spliceosome by Prp2 as analyzed by dcFCCS. Affinity-purified Bact complexes assembled on Atto-Actwt were complemented on the matrix with recombinant Prp16-EGFP plus ATP (column 1, Bact) and additionally with the proteins indicated on the left. After incubation, complexes were washed with GK75 buffer (A) or GK150 buffer (B) and then eluted from the matrix with maltose. dcFCCS measurements were then performed at complex concentrations of 1.0 nM. C*/PC indicates that upon Slu7/Prp18 binding to the C* complex, the latter is transformed into the post-catalytic complex (PC). (C) The B* complex was complemented on the matrix with Prp16-EGFP and incubated at 23°C, the matrix was washed with GK75 buffer, and supplemented with increasing amount of untagged Prp16 (columns 2–4) or Slu7/Prp18 (columns 5–7). The matrix was washed, complexes were eluted with maltose, and cross-correlation amplitudes were measured by dcFCCS. (D) The B* complex was complemented on the matrix with Slu7-EGFP and Prp18 and incubated at 23°C. The matrix was then washed with GK75 buffer, and the complexes were supplemented with an increasing amount of untagged Slu7 and Prp18 (columns 2–4) or Prp16 (columns 5–7). After washing, complexes were eluted with maltose and cross-correlation amplitudes were measured by dcFCCS. Cross-correlation amplitudes derived from two independent experiments are shown for each complex. Error bars indicate the standard deviation from two independent measurements.
When we carried out the same binding experiments as described above at 150 mM salt, Prp16-EGFP showed a cross-correlation amplitude of ∼15% (Fig. 5B, column 2), which is significantly lower than the 25% measured at 75 mM salt (Fig. 5A, column 2), but still clearly above background (<4%; cf. columns 1 and 2 of Fig. 5B). The binding of Prp16-EGFP is most efficient to the C* complex, i.e., in the presence of ATP (column 3), and the cross-correlation amplitude is reduced to <15% in the post-catalytic spliceosome (column 4). These data show clearly that Prp16 has a binding site on the catalytically activated B* spliceosome, which is somewhat stronger, i.e., more salt-resistant, than the one for Slu7/Prp18. Surprisingly, they also indicate that Prp16 remains bound after activation for step 2 catalysis and is destabilized from the spliceosome only after step 2 has occurred.
The observation that both Slu7/Prp18 and Prp16 have binding sites on the B* complex raised the question of whether these sites are distinct or partially overlap. Therefore, we used dcFCCS to monitor the interaction of Prp16-EGFP and Slu7-EGFP with catalytically activated B* complexes in the presence of excess unlabeled Prp16 or excess unlabeled Slu7/Prp18. Figure 5C shows that the binding of Prp16-EGFP to the B* complex is reduced by increasing amounts of untagged Prp16 but not by Slu7/Prp18. In the complementary experiment, the binding of Slu7-EGFP to the B* complex was reduced by addition of unlabeled Slu7/Prp18 but not by Prp16 (Fig. 5D). These data indicate that the entry sites for Slu7/Prp18 and Prp16 on the catalytically activated B* spliceosome are distinct from one another, as well as from the binding site of Cwc25, as suggested above.
DISCUSSION
ATP-dependent Prp16 activity is sufficient to create a functional and stable step 2 conformation of the spliceosome
In this work, we used a purified yeast splicing system to investigate the requirement of step 2 catalysis of the pre-mRNA splicing for the proteins known as step 2 splicing factors: Prp16, Prp22, and the Slu7/Prp18 heterodimer. The first surprising observation was that, in the purified splicing system, efficient step 2 catalysis of Actwt pre-mRNA requires only Slu7/Prp18, and not Prp22 (Fig. 1). This indicates that Slu7 and Prp18 together are sufficient for the docking of the 3′SS in the spliceosome’s step 2 active site. It is not clear why in previous studies, the removal of Prp22 from whole-cell extracts led to the inhibition of step 2 catalysis of Actwt pre-mRNA splicing (Schwer and Gross 1998), as our results exclude a helper function of Prp22 in step 2 catalysis in the purified system. It is possible that in whole-cell extracts Prp22 supports the binding of Slu7/Prp18 to the spliceosome and thus indirectly increases the efficiency of step 2 catalysis. We note, however, that our results are consistent with the findings of others (Company et al. 1991)—heat inactivation of Prp22 in whole-cell extracts still permits step 2 catalysis to occur, inhibiting only the dissociation of the spliced mRNA from the intron-lariat spliceosome. The conclusion that Prp22 is not required for step 2 is based on the lack of an effect of adding recombinant Prp22. However, although recombinant Prp22 is not required for step 2, it allows efficient Prp22-mediated dissociation of post-catalytic spliceosomes into intron-lariat spliceosome and mRNA/RNP, thus demonstrating that our recombinant Prp22 is functional (Fourmann et al. 2013). In addition, these results do not exclude a role for Prp22 in proofreading the 3′SS during step 2, and a role for Prp22 in step 2 catalysis could therefore depend on the particular RNA substrate used.
A second important observation using our purified system was that splicing of Act7 pre-mRNA, in which the distance between the BS and the 3′SS consensus sequences is short, only requires Prp16 and ATP for exon ligation and not Slu7, Prp18, or Prp22. In earlier studies it was shown that single or double depletions from splicing extracts of Slu7, Prp18, or Prp22 allow step 2 catalysis to take place; however, these results did not exclude the possibility that the step 2 factors still present made a contribution to this step (Ansari and Schwer 1995; Brys and Schwer 1996; Zhang and Schwer 1997; Schwer and Gross 1998; James et al. 2002). Here, we show unequivocally that the ATP-dependent action of Prp16 is sufficient for the formation of a functional step 2 active site, and exon ligation takes place without the help of the step 2 factors, Slu7/Prp18 and Prp22.
These results shed new light on the mechanism of step 2 catalysis and on the role of step 2 factors. First, they show that the ATP-dependent action of Prp16 is sufficient for the generation of a functional step 2 active site in the spliceosomal catalytic center, and Slu7/Prp18 and Prp22 per se are not essential for the formation of the step 2 active site (Fig. 1B). Second, they show that Slu7/Prp18 and Prp22 are not needed for the maintenance or the stabilization of this site. This is revealed by the fact that we were able to purify a functionally active C* complex by gradient centrifugation in the absence of Slu7, Prp18, and Prp22, implying that the step 2 conformation, once induced by Prp16 and ATP, is stable enough not to require maintenance by continual action of Prp16′s ATPase activity (Fig. 2). Furthermore, the requirement for ATP hydrolysis implies that the step 1 and step 2 conformations of the catalytic center are not in equilibrium with one another (Konarska and Query 2005; Konarska et al. 2006). Third, our results support the idea that, in the case of pre-mRNAs with a short distance between the BS and the 3′SS, the latter reaches its docking position in the catalytic center by diffusion through space without the need for scanning by Slu7/Prp18 (see also Horowitz 2012 for discussion and references). Why then does the addition of Slu7/Prp18 to the Act7 C* complex stimulate step 2 catalysis if the reaction can in principle proceed without these proteins? (Fig. 1; Supplemental Fig. S2). The most plausible explanation seems to be that the docking of the 3′SS into the step 2 active site is more efficient in the presence of Slu7/Prp18, even when the 3′SS–BS distance is short. Mechanistically, this may also suggest that Slu7/Prp18 acts by altering the conformation of the spliceosome. The docking of the 3′SS by three-dimensional diffusion alone becomes very inefficient and may even be prevented by secondary structure with longer 3′SS–BS distances (Meyer et al. 2011), so that in such cases Slu7/Prp18 is essential for step 2 catalysis.
Finally, our results with the Act7 pre-mRNA show that the step 2 active site recognizes the 3′SS and binds it stably, even in the absence of the step 2 factors Slu7, Prp18, and Prp22. Moreover, in the absence of these step 2 factors, even the spliced Act7 mRNA remains stably bound to the step 2 spliceosome upon density-gradient centrifugation and only dissociates from the spliceosome through the action of the RNA helicase Prp22 (Fourmann et al. 2013). The components of the spliceosome known to be involved in the recognition and binding of the 3′SS are loop 1 of the U5 snRNA and Prp8 (Frank et al. 1992; Teigelkamp et al. 1995; Collins and Guthrie 1999; Grainger and Beggs 2005; Aronova et al. 2007). Slu7/Prp18 may also help to stabilize the interaction of the 3′SS and the active site, either directly or indirectly through Prp8, especially if the interaction of the 3′ SS is weak (Crotti et al. 2007).
Remodeling of the spliceosome by Prp16 ATPase is subtle
The catalytic activation of the spliceosome requires three helicases, and the action of each of these leads to significant structural changes in the spliceosome. For example, the action of Prp28 displaces the U1 snRNP (and other components) from the spliceosome. Brr2-mediated activation leads to the displacement of U4 snRNA from the spliceosome and a dramatic exchange of more than 20 proteins, as well as the reorganization of the spliceosome’s RNA–RNA interaction network (Staley and Guthrie 1998; Fabrizio et al. 2009). The Prp2-mediated transformation of Bact into the catalytically active B* complex also involves the displacement of several proteins, even at low salt concentrations, whereas the binding of others is destabilized (Ohrt et al. 2012). This process is accompanied by a substantial change in hydrodynamic behavior of the spliceosome (Kim and Lin 1996; Warkocki et al. 2009). On the other hand, the transition from the C complex to the step-2-activated C* complex, induced by Prp16, is considerably more subtle. The C and C* complexes have identical Svedberg values (Fig. 2), making a major structural change unlikely. Likewise, the protein composition of C* remains largely unchanged, as shown by the results of MS analyses (Supplemental Table S1). At the same time, we show that the C* complex contains a stable step 2 catalytic center, the maintenance of which does not require either continual Prp16 ATPase activity or Slu7/Prp18 (Fig. 2). It is therefore likely that the Prp16-mediated remodeling is relatively local in nature and affects primarily the catalytic core structure. As a minimum structural change, the branch structure must be removed from the step 1 catalytic center to accomodate the 3′SS (Konarska et al. 2006). Nevertheless, as discussed in more detail below, the action of the ATPase Prp16 alters the affinities of several proteins for the spliceosome, such as reducing Cwc25′s binding affinity and increasing that of Slu7/Prp18.
Prp16 ATPase activity is necessary but not sufficient for displacement of the step 1 factor Cwc25 from the spliceosome
In earlier work, we demonstrated that although the catalytically activated B* complex generates small amounts of step 1 intermediates, efficient step 1 catalysis only occurs in the presence of Cwc25 (Warkocki et al. 2009). The exact task of Cwc25 is at present unknown, but it is possible that Cwc25 stabilizes the step 1 conformation of the catalytic center or induces a more closed conformation of the step 1 active site. Consistent with these two possibilities, Cwc25 was suggested to bind to the BS (Tseng et al. 2011). It is therefore likely that the site of interaction between Cwc25 and the step 1 spliceosome must be remodeled as a prerequisite for efficient catalysis of step 2.
Our MS and dcFCCS results show that Prp16 ATPase activity is necessary for the dissociation of Cwc25 from the spliceosome, but not sufficient, as a second signal is needed. The finding that Cwc25 dissociates from the C* complex assembled on Actwt pre-mRNA when Slu7/Prp18 is added initially led us to assume that the binding of Slu7/Prp18 to the spliceosome directly displaces Cwc25. However, our experiments with C* complexes assembled on Act7 pre-mRNA suggest a different mechanism; in this case, Cwc25 dissociates in the absence of Slu7/Prp18, and the dissociated fraction correlates with the proportion of ligated exons (Fig. 3; discussion above). We therefore favor the following model: First, the ATPase activity of Prp16 induces a local rearrangement of the catalytic center from the step 1 to the step 2 conformation. The effects of this structural change include a reduction in the binding affinity of Cwc25 for the catalytic center. The docking of the 3′SS into the newly formed step 2 catalytic center then leads—directly or, more probably, indirectly—to the displacement of Cwc25 from the spliceosome via an additional conformational change at the catalytic center. Only the stable docking of the 3′SS—and not productive step 2 catalysis—is the prerequisite for the dissociation of Cwc25, as shown by the results with Actwt and Act7 3′SS mutants pre-mRNAs (Fig. 3). The results of earlier work (Umen and Guthrie 1995; Horowitz 2012; and references therein)—in which mutations in the pre-mRNA’s 3′SS inhibit step 2 catalysis, but these mutant 3′SS are nevertheless bound by Slu7 and thus potentially also at the spliceosome’s catalytic center—are consistent with this model.
In this model, the recruitment of Slu7/Prp18 in the case of the Actwt C* spliceosomes leads indirectly to the dissociation of Cwc25 in that its primary task is to dock the 3′SS efficiently into the step 2 catalytic center. We cannot at present exclude an additional role for Slu7/Prp18. For example, these proteins might trigger (directly or indirectly, such as through Prp8) a conformational change in the catalytic center that contributes to the stabilization of the step 2 conformation and thus additionally destabilizes the association of Cwc25 with the catalytic center. Our results obtained with Act7-C* complexes indicate that this possible function of Slu7/Prp18 is not a critical one.
Our results contrast with the findings of Cheng and coworkers, who observed that Cwc25 dissociates from the spliceosome already after ATP hydrolysis by Prp16 (Tseng et al. 2011). However, their experiments were conducted at high salt concentrations (i.e., 200 mM KCl), at which Cwc25 is more easily displaced from the spliceosome. Furthermore, under their experimental conditions, Slu7/Prp18 was probably present. In contrast to a previous report (Liu et al. 2007), we do not observe any dissociation of Yju2 from the spliceosome. Unlike Cwc25, Yju2, which is already recruited in the Bact complex, is an integral component of the spliceosome during its catalytic phase.
Prp16 and Slu7/Prp18 bind to nonoverlapping entry sites on the B* spliceosome well in advance of their functional involvement
Our dcFCCS binding studies led to the surprising result that the conversion of the activated spliceosome to a step 1 catalytically activated B* complex is accompanied by the generation of binding sites not only for the step 1 factor, Cwc25 (Ohrt et al. 2012), but also—in advance of their function in step 2 catalysis—for the step 2 factors, Prp16 and Slu7/Prp18. Although the binding of Prp16 and Slu7/Prp18 to the B* complex is salt-sensitive (i.e., found only to be stable at low salt concentration), there are several observations that suggest that this binding is specific. First, the binding of the fluorescent proteins Slu7-EGFP and Prp16-EGFP is subject to competition by unlabeled Slu7 and Prp16, respectively; but Slu7 does not compete with Prp16 binding or vice versa. The later indicates that the binding sites for Slu7/Prp18 and Prp16 on B* do not overlap. Furthermore, the fact that Slu7/Prp18 is also equally efficiently bound to the B* complex and to the Cwc25-containing C complex suggests that the binding site of Slu7/Prp18 likewise does not overlap with that of Cwc25. Second, binding of Slu7-EGFP to the B* spliceosome was only observed in the presence of Prp18, which is consistent with earlier findings that Slu7 and Prp18 are only functionally active as a heterodimer in the spliceosome (Zhang and Schwer 1997; James et al. 2002; Aronova et al. 2007). Third, we show that C complexes to which Slu7/Prp18 were bound under low-salt conditions were able, after the addition of Prp16 and ATP, to carry out step 2 catalysis; this indicates that the initial low-affinity binding of Slu7/Prp18 on the C complex is functionally relevant (Fig. 4C).
Our observation that Prp16 can be recruited to the spliceosome as early as in the B* stage is consistent with observations that Prp16 can, under conditions where the splicing reaction is compromised by mutations in the BS or the U6 snRNA, act during or before step 1 catalysis, thereby exercising a proofreading function (Burgess et al. 1990; Koodathingal et al. 2010; Horowitz 2011; Tseng et al. 2011). As Prp16 is not required for step 1 catalysis, its involvement at such an early stage would be consistent with Prp16 having such a proofreading function. The creation of binding sites for the step 2 factors Prp16 and Slu7/Prp18 at the B* stage could also offer an advantage with respect to the kinetics of the splicing reaction, as they would already be present in the spliceosome before their catalytic function is required at a later stage.
Binding and activity of step 1 and step 2 spliceosomal factors
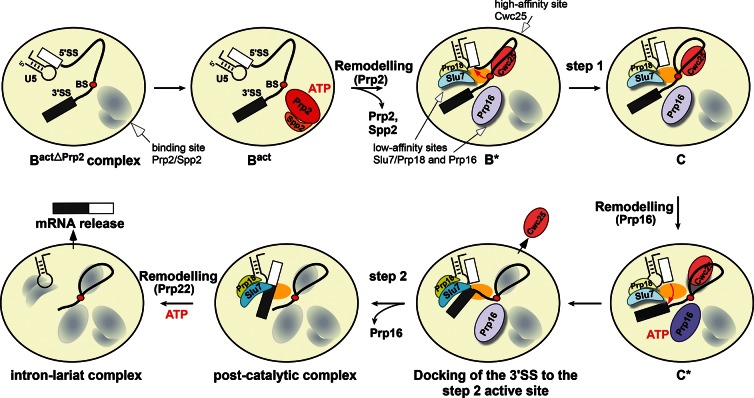
A summary of the catalytic steps of splicing is illustrated schematically in Figure 6. First, the Prp2-mediated catalytic activation of the Bact complex generates a high-affinity binding site for the step 1 factor, Cwc25, and low-affinity sites for Prp16 and Slu7/Prp18. After the binding of Cwc25 to the B* complex, step 1 catalysis takes place, leading to formation of the C complex, in which Cwc25 is still stably bound. During this event, it is likely that a conformational change occurs, transforming the low-affinity binding site of Prp16 to a high-affinity binding site (i.e., which withstands 150 mM salt) (Fig. 5B). ATP hydrolysis by Prp16 occurs with concomitant transformation of the catalytic center from the step 1 to the step 2 conformation. This rearrangement leads in turn to a reduction in the spliceosome’s affinity for Cwc25, although the latter still remains bound. A further consequence of Prp16 ATPase activity is the transformation of the low-affinity binding site for Slu7/Prp18 (the entry site) into a high-affinity one, defined operationally by the increased salt-resistance of Slu7/Prp18 binding (Fig. 4A,B), and efficient docking of the 3′SS into the step 2 active site. It is likely that the docking of the 3′SS induces a further conformational change at the catalytic center, which ultimately leads to the dissociation of Cwc25, thus opening the way for step 2 catalysis. It is interesting to note that our dcFCCS results indicate that Prp16 remains associated with the C* complex after hydrolyzing ATP and dissociates only when Slu7/Prp18 is stably bound/present (Fig. 5B).
FIGURE 6.
Binding and activity of step 1 and step 2 factors during the catalytic phase of the spliceosome. Schematic representation of the substantial remodeling that the precatalytic Bact spliceosome undergoes during the transition from step 1 to step 2 catalysis. The ATPases Prp2 and Prp16 are required for activating the spliceosome for the first and second catalytic steps, respectively. The ATPase Prp2 plus ATP creates a high-affinity binding site for Cwc25, which stabilizes the step 1 conformation of the catalytic center, thereby facilitating step 1 catalysis. Prp2 also creates low-affinity binding sites for Prp16 and Slu7; the latter binds to the spliceosome only together with Prp18. The ATPase Prp16 plus ATP is required for remodeling the spliceosome for step 2 catalysis and to stabilize the binding of Slu7/Prp18, which is needed to dock efficiently the 3′SS into the step 2 active site, leading to displacement of Cwc25 before exon ligation. The exons are then ligated and the post-catalytic spliceosome is generated. Proteins are oval-shaped, shadowed, and highlighted with light colors (low binding affinity) or with dark colors without a shadow (high binding affinity). The yellow oval in the middle of the spliceosome represents the active site. The conserved U5 stem–loop I is also shown.
Until now, it was generally assumed that the spliceosome’s binding sites for the step 1 and step 2 factors were generated consecutively. However, our results suggest that all of these factors bind simultaneously to the catalytically activated spliceosome, and each factor’s binding site is converted or “toggled” from a low-affinity, to a high-affinity, and again to a low-affinity state. It will be interesting to see, in future work, at which stages of spliceosome assembly and function the binding sites for the other helicases, such as Prp22 and Prp43, are formed.
MATERIALS AND METHODS
Splicing reconstitution assays
Ten femtomoles of BactΔPrp2 complexes eluted from the amylose matrix or from peak fraction(s) of the second gradient were supplemented with 100 fmol (10-fold molar excess or 30-fold molar excess for the experiment shown in Fig. 1A) of recombinant proteins in a 40-µL reaction. After incubating the reconstitution reaction for 10 min on ice, one-tenth volume of a 10-fold “rescue” solution (200 mM phosphate buffer, KPO4 [pH 7.3], 10 mM MgCl2, 20 mM ATP, 14% PEG 8000) was added to the reaction. The reactions were incubated for 45 min at 23°C. After Proteinase K digestion, the RNAs were extracted by phenol–chloroform–isoamyl alcohol (PCI) extraction and precipitated by ethanol and then analyzed by 8% denaturing PAGE followed by autoradiography. For reconstitution assays on the amylose matrix, complex concentration was determined by Cherenkov counting, and recombinant proteins were added in 10-fold molar excess. A typical reconstitution reaction was performed with 250–300 µL amylose beads in a final volume of 500–600 µL (in GK75 buffer; 20 mM HEPES-KOH [pH 7.9], 1.5 mM MgCl2, 75 mM KCl, 5% glycerol, 0.01% NP40, 0.5 mM DTT, and 0.5 mM PMSF) containing 50–60 µL 10-fold “rescue” solution and were incubated at 23°C for 45 min. For ATP depletion, the reaction was supplemented with 2 mM glucose and 2 units of recombinant hexokinase and incubated for 10 min at 23°C. The matrix was then washed thoroughly with 100 column volumes of GK75 buffer and the complexes were eluted with 12–20 mM maltose. For dual color FCCS measurements, reactions were carried out in solution in a final volume of 60 µL with a final complex concentration of 1.0 nM. Pre-mRNA was fluorescently labeled as previously described (Ohrt et al. 2012). The purified, labeled BactΔPrp2 spliceosomes were then complemented with a 10-fold molar excess of recombinant proteins, and the extent of binding of Prp16 and Slu7 fused to EGFP or Cwc25-Alexa488 to the spliceosome was measured. Recombinant Cwc25 was chemically labeled with Atto488 as described (Ohrt et al. 2012). Instead of adding the 10-fold rescue mix, the reaction conditions were only adjusted to final concentrations of 2.5 mM MgCl2 and 2 mM ATP (or ATP analogs), as PEG8000 increased the background of the dcFCCS measurements. Complexes were measured in a volume of 25 µL at a concentration of 1.0 nM in a reaction chamber consisting of two glass slides separated by a 1-mm spacer.
Dual-color fluorescence cross correlation spectroscopy (dcFCCS)
dcFCCS allows for the analysis of molecular interactions such as binding or release of proteins in a complex system (Kettling et al. 1998; Földes-Papp and Rigler 2001; Ohrt et al. 2011). dcFCCS requires particles labeled with two spectrally separated colors (e.g., green and red). The fluorescence of the labeled molecules is spectrally separated and detected in two channels (see Supplemental Methods). The signals are analyzed by computing the auto-correlation of each channel and their cross-correlation. If molecules bearing different labels are not part of the same complex, then they will diffuse independently through the confocal volume. In that case, there will be no cross-correlation between both signals. However, if the molecules are part of the same complex, they will form doubly labeled entities. Codiffusion of the two labels will generate correlated signals in both detection channels and therefore a cross-correlation. The amplitude of the cross-correlation is proportional to the number of doubly labeled molecules and can therefore be used to monitor binding and release reactions (Ricka and Binkert 1989; Schwille et al. 1997; Földes-Papp 2005; Mütze et al. 2011).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
We thank Elke Penka, Thomas Conrad, and Wiebke Behrens-Kranz for excellent technical assistance. We thank Henning Urlaub and Monika Raabe for MS analysis. This work was supported by a grant from the Deutsche Forschungsgemeinschaft SFB 860 to R.L. and J.E.
REFERENCES
- Ansari A, Schwer B 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J 14: 4001–4009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronova A, Bacikova D, Crotti LB, Horowitz DS, Schwer B 2007. Functional interactions between Prp8, Prp18, Slu7, and U5 snRNA during the second step of pre-mRNA splicing. RNA 13: 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brys A, Schwer B 1996. Requirement for SLU7 in yeast pre-mRNA splicing is dictated by the distance between the branchpoint and the 3′ splice site. RNA 2: 707–717 [PMC free article] [PubMed] [Google Scholar]
- Burgess SM, Guthrie C 1993. A mechanism to enhance mRNA splicing fidelity: The RNA-dependent ATPase Prp16 governs usage of a discard pathway for aberrant lariat intermediates. Cell 73: 1377–1391 [DOI] [PubMed] [Google Scholar]
- Burgess S, Couto JR, Guthrie C 1990. A putative ATP binding protein influences the fidelity of branchpoint recognition in yeast splicing. Cell 60: 705–717 [DOI] [PubMed] [Google Scholar]
- Chen C-H, Yu W-C, Tsao TY, Wang L-Y, Chen H-R, Lin J-Y, Tsai W-Y, Cheng S-C 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res 30: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu YF, Liu YC, Chiang TW, Yeh TC, Tseng CK, Wu NY, Cheng SC 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol Cell Biol 29: 5671–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins CA, Guthrie C 1999. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev 13: 1970–1982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Company M, Arenas J, Abelson J 1991. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature 349: 487–493 [DOI] [PubMed] [Google Scholar]
- Cordin O, Hahn D, Beggs JD 2012. Structure, function and regulation of spliceosomal RNA helicases. Curr Opin Cell Biol 24: 431–438 [DOI] [PubMed] [Google Scholar]
- Crotti LB, Bacikova D, Horowitz DS 2007. The Prp18 protein stabilizes the interaction of both exons with the U5 snRNA during the second step of pre-mRNA splicing. Genes Dev 21: 1204–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabrizio P, Dannenberg J, Dube P, Kastner B, Stark H, Urlaub H, Lührmann R 2009. The evolutionarily conserved core design of the catalytic activation step of the yeast spliceosome. Mol Cell 36: 593–608 [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z 2005. How the molecule number is correctly quantified in two-color fluorescence cross-correlation spectroscopy: Corrections for cross-talk and quenching in experiments. Curr Pharm Biotechnol 6: 437–444 [DOI] [PubMed] [Google Scholar]
- Földes-Papp Z, Rigler R 2001. Quantitative two-color fluorescence cross-correlation spectroscopy in the analysis of polymerase chain reaction. Biol Chem 382: 473–478 [DOI] [PubMed] [Google Scholar]
- Fourmann JB, Schmitzová J, Christian H, Urlaub H, Ficner R, Boon KL, Fabrizio P, Lührmann R 2013. Dissection of the factor requirements for spliceosome disassembly and the elucidation of its dissociation products using a purified splicing system. Genes Dev 27: 413–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D, Patterson B, Guthrie C 1992. Synthetic lethal mutations suggest interactions between U5 small nuclear RNA and four proteins required for the second step of splicing. Mol Cell Biol 12: 5197–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD 2005. Prp8 protein: At the heart of the spliceosome. RNA 11: 533–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS 2011. The splice is right: Guarantors of fidelity in pre-mRNA splicing. RNA 17: 551–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horowitz DS 2012. The mechanism of the second step of pre-mRNA splicing. Wiley Interdiscip Rev RNA 3: 331–350 [DOI] [PubMed] [Google Scholar]
- Ilagan JO, Chalkley RJ, Burlingame AL, Jurica MS 2013. Rearrangements within human spliceosomes captured after exon ligation. RNA 19: 400–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James SA, Turner W, Schwer B 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8: 1068–1077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettling U, Koltermann A, Schwille P, Eigen M 1998. Real-time enzyme kinetics monitored by dual-color fluorescence cross-correlation spectroscopy. Proc Natl Acad Sci 95: 1416–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Lin RJ 1996. Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol Cell Biol 16: 6810–6819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Query CC 2005. Insights into the mechanisms of splicing: More lessons from the ribosome. Genes Dev 19: 2255–2260 [DOI] [PubMed] [Google Scholar]
- Konarska MM, Vilardell J, Query CC 2006. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell 21: 543–553 [DOI] [PubMed] [Google Scholar]
- Koodathingal P, Novak T, Piccirilli JA, Staley JP 2010. The DEAH box ATPases Prp16 and Prp43 cooperate to proofread 5′ splice site cleavage during pre-mRNA splicing. Mol Cell 39: 385–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lardelli RM, Thompson JX, Yates JR III, Stevens SW 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YC, Chen HC, Wu NY, Cheng SC 2007. A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol Cell Biol 27: 5403–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPheeters DS, Schwer B, Muhlenkamp P 2000. Interaction of the yeast DExH-box RNA helicase prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res 28: 1313–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Plass M, Pérez-Valle J, Eyras E, Vilardell J 2011. Deciphering 3′ss selection in the yeast genome reveals an RNA thermosensor that mediates alternative splicing. Mol Cell 43: 1033–1039 [DOI] [PubMed] [Google Scholar]
- Mütze J, Ohrt T, Schwille P 2011. Fluorescence correlation spectroscopy in vivo. Laser Photon Rev 5: 52–67 [Google Scholar]
- Ohrt T, Mütze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P 2008. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res 36: 6439–6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Staroske W, Mütze J, Crell K, Landthaler M, Schwille P 2011. Fluorescence cross-correlation spectroscopy reveals mechanistic insights into the effect of 2′-O-methyl modified siRNAs in living cells. Biophys J 100: 2981–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohrt T, Prior M, Dannenberg J, Odenwälder P, Dybkov O, Rasche N, Schmitzová J, Gregor I, Fabrizio P, Enderlein J, et al. 2012. Prp2-mediated protein rearrangements at the catalytic core of the spliceosome as revealed by dcFCCS. RNA 18: 1244–1256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricka J, Binkert T 1989. Direct measurement of a distinct correlation-function by fluorescence cross-correlation. Phys Rev A 39: 2646–2652 [DOI] [PubMed] [Google Scholar]
- Roy J, Kim K, Maddock JR, Anthony JG, Woolford JL Jr 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1: 375–390 [PMC free article] [PubMed] [Google Scholar]
- Schwer B 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol Cell 30: 743–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Gross CH 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre- mRNA splicing. EMBO J 17: 2086–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Guthrie C 1992. A conformational rearrangement in the spliceosome is dependent on PRP16 and ATP hydrolysis. EMBO J 11: 5033–5039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwille P, Meyer-Almes FJ, Rigler R 1997. Dual-color fluorescence cross-correlation spectroscopy for multicomponent diffusional analysis in solution. Biophys J 72: 1878–1886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman EJ, Maeda A, Wei J, Smith P, Beggs JD, Lin RJ 2004. Interaction between a G-patch protein and a spliceosomal DEXD/H-box ATPase that is critical for splicing. Mol Cell Biol 24: 10101–10110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA 1993. The U5 and U6 small nuclear RNAs as active site components of the spliceosome. Science 262: 1989–1996 [DOI] [PubMed] [Google Scholar]
- Staley JP, Guthrie C 1998. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell 92: 315–326 [DOI] [PubMed] [Google Scholar]
- Steitz TA, Steitz JA 1993. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci 90: 6498–6502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teigelkamp S, Newman AJ, Beggs JD 1995. Extensive interactions of PRP8 protein with the 5′ and 3′ splice sites during splicing suggest a role in stabilization of exon alignment by U5 snRNA. EMBO J 14: 2602–2612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CK, Liu HL, Cheng SC 2011. DEAH-box ATPase Prp16 has dual roles in remodeling of the spliceosome in catalytic steps. RNA 17: 145–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C 1995. Prp16p, Slu7p, and Prp8p interact with the 3′ splice site in two distinct stages during the second catalytic step of pre-mRNA splicing. RNA 1: 584–597 [PMC free article] [PubMed] [Google Scholar]
- Warkocki Z, Odenwälder P, Schmitzová J, Platzmann F, Stark H, Urlaub H, Ficner R, Fabrizio P, Lührmann R 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat Struct Mol Biol 16: 1237–1243 [DOI] [PubMed] [Google Scholar]
- Will CL, Lührmann R 2011. Spliceosome structure and function. In RNA Worlds (ed. Gesteland RF, Cech TR, Atkins JF), pp. 181–203 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Schwer B 1997. Functional and physical interaction between the yeast splicing factors Slu7 and Prp18. Nucleic Acids Res 25: 2146–2152 [DOI] [PMC free article] [PubMed] [Google Scholar]