Abstract
Transgenic animals are an important source of protein and nutrition for most humans and will play key roles in satisfying the increasing demand for food in an ever-increasing world population. The past decade has experienced a revolution in the development of methods that permit the introduction of specific alterations to complex genomes. This precision will enhance genome-based improvement of farm animals for food production. Precision genetics also will enhance the development of therapeutic biomaterials and models of human disease as resources for the development of advanced patient therapies.
1. INTRODUCTION
1.1. The Need for Genetically Modified Large Animals
Hunger worldwide is increasing; approximately 1 billion people are already chronically malnourished (Godfray et al., 2010). Contemporary efforts to meet demand are degrading an already taxed environment (Foley et al., 2011; Tilman, Balzer, Hill, & Befort, 2011). Improvements in the efficiency of production and safety are becoming even more important considerations for protection of the environment and reduction in land usage (Clark & Whitelaw, 2003). Global climate change will only exacerbate the lack of animal protein production (McMichael, 2012; Schmidhuber & Tubiello, 2007; Wolkovich et al., 2012). The green revolution has practically peaked according to its father, Borlaug (2000), who asserted that farm animals are critical to nutrition and that genetic engineering of foodstuffs will be required to feed the world. Both genetic- and management-based increases in sustainable productivity will be a key to satisfying global protein needs (Fahrenkrug et al., 2010).
Genetically engineered animals have a larger role than just as food (Fig. 1). They contribute to our health by serving as model systems for treatment of diseases and disorders as well as a source of biomaterials used for rebuilding tissues and organs (Kues & Niemann, 2004; Snaith & Törnell, 2002). Mice have historically been the prime medical models for finding disease-causing genes and testing drugs. Owing to their large numbers and the availability of in-bred lines that improve the reproducibility of experimental results, molecular and cellular investigations generally are first conducted in mice. Moreover, powerful selection protocols in cultured mouse embryonic stem cells allow identification and incorporation into genomes of genetic alterations that occur at very low frequencies, i.e. 10−5–10−8 (Mansour, Thomas, & Capecchi, 1988; Smithies, Gregg, Boggs, Koralewski, & Kucherlapati, 1985). As a result, specific mutants can be made that mimic human mutations, e.g. cystic fibrosis (Snouwaert et al., 1992). However, the complete panoply of symptoms in humans does not always manifest in mice with the same genetic defects [e.g. the cystic fibrosis mouse does not have the same range of problems that humans encounter with the same mutant genes (Rogers et al., 2008)]. Moreover, many of the advantages for academic studies are disadvantages for translation to human studies. For example, in-bred strains of mice provide highly reproducible experimental results because important alleles that control physiological pathways are homozygous at every locus and identical in every individual (Erickson, 1996), a situation that does not apply to the heterogeneous human population. Likewise, mice that have major differences in overall physiology have been selected for high-density, low-activity living, which results in abnormal metabolic characteristics that interferes with translation to humans (Martin, Ji, Maudsley, & Mattson, 2010).
Figure 1.
The multiple applications of genetically modified large animals. The pig is shown as an example. The first application is to improve traits in the farm animal. Examples of the potential improved traits include (1) resistance to diseases, (2) improved nutrition such as introducing a gene to produce the healthier omega-3 fatty acids to replace the normal omega-6 fatty acids (Lai et al., 2006), and (3) reducing the environmental impact of major pig production facilities by reducing phosphorous in manure (Golovan et al., 2001). The second application of genetically modified pigs is for biomedical products such as organ transplantation (http://web.archive.org/web/20071210031618/http://www.fda.gov/fdac/features/596_xeno.html) or specific functional organ parts such as heart valves and subcellular structures. Examples include inactivating genes such as α-1,3-galactose that produce powerful immune responses when introduced into humans and eliminating the potential spread of porcine endogenous retroviruses. The third application of genetically modified pigs is the creation of animals that closely mimic human diseases such as cystic fibrosis (Rogers et al., 2008), cardiovascular disease, and cancer. For color version of this figure, the reader is referred to the online version of this book.
Unfortunately, the selection techniques that are so powerful in conjunction with mouse embryonic stem cells have not been translated to other animals. For human applications where safety is paramount, larger animals are desirable as model systems for testing therapeutic procedures. Deleterious mutations that are similar to those in humans have been identified in certain breeds of cats and dogs because of the close relationship to their owners (Ellinwood, Vite, & Haskins, 2004; Haskins, Desnick, DiFerrante, Jezyk, & Patterson, 1984; Koeberl, Pinto, Brown, & Chen, 2009; Ponder et al., 2006; Wolfe, 2009), but the spontaneous appearance of these animals in veterinary clinics does not provide for on-demand and replicable lines for scientific studies. Generally, the range of spontaneous disease models in large animals is highly limited compared to the number of genetic disorders in humans.
That will change. Precision genetics, developed in the first decade of the twenty-first century, will be a key player for the challenges ahead. Specific genetic alterations in the genomes of the pig, which is similar in size, physiology, organ development, and disease progression (Kuzmuk & Schook, 2011; Lunney, 2007), will provide subjects that significantly accelerate the development of new medical devices, pharmaceuticals, therapeutic protocols, and tissue-based products from humanized transgenic lines. In this review, we summarize the game-changing genetic methods that are under development that will support unprecedented progress in adapting the genomes of farm animals to support their multiple roles in human societies. The implications of the new genetic technologies can be appreciated by acknowledging problems and issues that arose during the early years of genetic engineering.
1.2. Genetic Engineering of Animals Pre-2000
Transgenic animal technology is entering its fourth decade. The first recombinant DNAs were designed to express specific genes in bacteria (Cohen et al., 1973). Almost immediately, there was concern by some that reshaping genetic systems might be hazardous in some unknown way, which led to a self-imposed moratorium on recombinant eukaryotic genetic material (Berg et al., 1974). As a consequence, elucidation of the gene expression machinery in animals was slowed until it became evident that the fears were based on fears of the unknown rather than any scientific evidence (Berg & Singer, 1995). The moratorium served as an unfortunate precedent for ignorance and unspecified fears impeding progress in animal genetics.
1.2.1. Classical Methods for Genetic Engineering of Animals
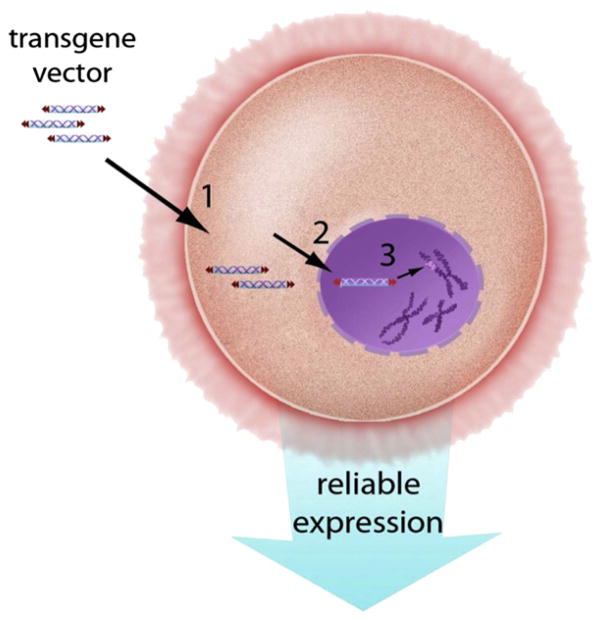
Once anxieties of cloning eukaryotic genes were addressed, plasmid-based recombinant DNA technology supported the rapid characterization of the molecular genetic mechanisms by which genes are expressed in complex animals and plants. Introduction of genetic material into an animal’s genome requires overcoming the elaborate cellular mechanisms that minimize DNA modification and keep out foreign DNA. These mechanisms have evolved to maintain the integrity of the information in genomes and to prevent the subversion or destruction of cellular activities. In animals, transgenic DNA faces three barriers to its introduction into genomes—the cell membrane, the nuclear membrane, and the structure of chromosomes (Fig. 2).
Figure 2.
The three barriers to the introduction of foreign DNA into genomes: (1) the cell membrane, (2) the nuclear membrane, and (3) the chromosomal DNA in the chromosomes. For effective transgenesis, the foreign DNA must overcome the three barriers and then be able to withstand protective measures such as methylation that are employed to reduce expression of transgenic DNA that has inserted into the chromatin. For color version of this figure, the reader is referred to the online version of this book.
There are two fundamental ways of delivering genetic material into an animal genome (Fig. 3). Plasmid-based gene delivery has been the most common because these vectors can be made and isolated in abundance in most laboratories using simple procedures. Plasmids nearly always contain an antibiotic resistance gene to raise the concentration of the recombinant plasmid in host Escherichia coli cells. However, organisms containing a transgenic antibiotic gene, often referred to as a selection marker, generally are not advised for release outside laboratories, even though there is not any evidence whatsoever that such transgenes will have any effect on the environment. Although plasmids can be easily produced and purified, their introduction into genomes is difficult. The astonishing integrity of the boundaries is best appreciated by realizing that the average human consumes more than 1000 trillion genes per day, all of which are kept from the chromosomes of his/her cells. Hence, chemical treatments of the cells or direct injections generally are required for delivery of plasmids to cells. Of the hundreds of plasmids that actually enter the cell, only a few are incorporated into a chromosome. The outcome of plasmid delivery is uncertain in two ways. First, the transgenic DNA can integrate into any of billions of sites in a mammalian genome and second, the actual sequence that integrates into any site can vary. Consequently, these uncontrollable features can result in undefined sequences integrating into resident genes, which can lead to unwanted genetic effects. This is called insertional mutagenesis. Most concerns with genetically engineered organisms derive from the potential collateral effects that are hard to predict. An important, relatively recent modification of the plasmid delivery involves the use of transposons to carry the transgene into genome. DNA transposons insert a rigorously defined sequence into a genome with much higher efficiency than occurs by random recombination. Transposons are described in more detail in Section 2.5.1.
Figure 3.
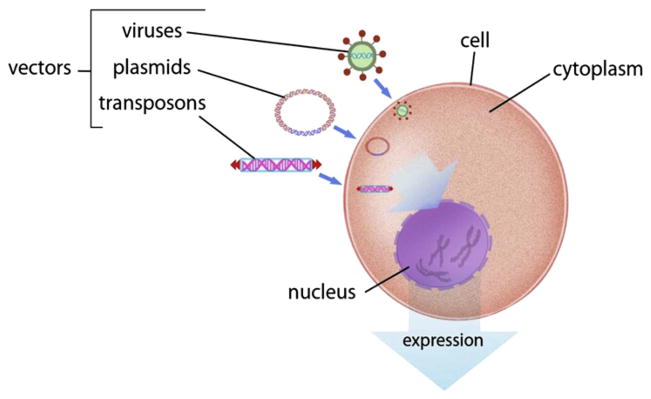
The three vectors for introduction of foreign DNA into genomes: (1) plasmids, (2) viruses, and (3) transposons. For color version of this figure, the reader is referred to the online version of this book.
Viruses comprise the second generic method used for gene delivery into animal cells. Their activities and properties have been studied for decades. There are several hurdles with the use of viruses (Hackett, Largaespada, & Cooper, 2010). The first is cost of manufacture and purification in amounts required for effective delivery to cells, which prohibits their use in most laboratories. Second, viruses often direct integration into and/or proximal to resident genes and thereby influence normal cellular function. Third, cells have evolved elaborate defenses against viruses. Fourth, for commercial animals, there has always been anxiety about undefined virus effects.
A major issue in genetic engineering animals is controlling expression of the new genetic material so that the protein it encodes is made at the appropriate level in the right tissues (Jaenisch, 1988). Genetic elements called enhancers and promoters regulate the expression of a gene. The combination of an appropriate promoter with a transgene is called an expression cassette. For an expression cassette to be useful in commercial animals, it must be reliably expressed as it is inherited from one generation to the next. Regardless of whether the transgenic material is introduced as a plasmid, transposon, or viral genome, the site of its integration may affect the spatial and temporal features of its expression.
1.2.2. Early Genetic Engineering in Mice, Chickens, and Fish
The first transgenic animals were produced more than 30 years ago (Brinster et al., 1981; Cline et al., 1980; Gordon, Scangos, Plotkin, Barbosa, & Ruddle, 1980) and stable lines of animals were produced soon after (Gordon & Ruddle, 1981, 1982). The expression cassettes for the transgenes generally had viral promoters and were delivered on plasmids that integrated fairly randomly. As a result, they lacked tissue-specific expression of the transgenes (Lacy, Roberts, Evans, Burtenshaw, & Costantini, 1983). The dramatic demonstration of growth enhancement in mice, a phenotype with clear relevance to food animals, following delivery of transgenic growth hormone genes (Palmiter et al., 1982; Palmiter, Norstedt, Gelinas, Hammer, & Brinster, 1983), led to predictions that recombinant DNAs would be introduced into food crops and animals (Bauman, McCutcheon, Steinhour, Eppard, & Sechen, 1985; Seidel, 1985; Wagner & Murray, 1985). However, in some cases random integration led to adverse effects, including death (ref). These observations led many to appreciate the delicate balance between introducing new desirable traits without incurring unwanted genetic effects. Insertional mutagenesis also rekindled the lingering fears of genetic tampering in animals (Rollin, 1985).
Two of the earliest genetic engineering projects in agricultural animals involved chickens and fish. Chickens are a major agricultural product and their susceptibility to viral infections stimulated interest in genetically engineering resistance to diseases. Moreover, transforming chicken eggs into bioreactors for the production of therapeutic proteins of high value appeared to be significantly better than transforming mammalian mammary glands to secrete the biological milk (Ivarie, 2003). The earliest experiments in avian transgenesis utilized retroviruses. Retroviral infections of poultry can cause sarcomas (Rous, 1910) and leukemias (Beard, Sharp, Eckert, Beard, & Mommaerts, 1952). However, cells that express viral envelope (env) proteins are resistant to infection. This observation led investigators to engineer lines of chickens that would be immune to infection by avian viruses by using modified avian viruses as vectors to deliver env genes to chicken genomes (Crittenden & Salter, 1985, 1986). Transgenic lines of chickens were achieved (Bosselman et al., 1989; Mizuarai et al., 2001; Salter, Smith, Hughes, Wright, & Crittenden, 1987; Thoraval et al., 1995); however, the efficiencies using retroviral vectors were low, the cargo capacity of retroviruses was limited, and some of the transgenic birds shed replicating virus. Other viral vectors, including lentiviruses, and transposons have been used to introduce transgenes into the chicken germline (Macdonald et al., 2012; Sang, 2004), but the efficiencies remain low, expression of the transgenes may be subject to epigenetic effects (Hofmann et al., 2006), and use of viral vectors to engineer food remains unsettling to the public. No transgenic poultry have been commercialized.
Genetic engineering in fish has a very long history because fish comprise a major source of protein and produce large numbers of eggs whose nuclei are easy to genetically manipulate (Yan, 1998). A further stimulus to genetic engineering of fish is the worldwide over-exploitation of fisheries that has led to a declining marine capture since its peak in 1996 (Smith, Asche, Guttormsen, & Wiener, 2010; Worm et al., 2009). Genetic engineering in fish is as simple as it gets. Microinjection of plasmids into eggs is easy but the efficiency of actually obtaining fish that will pass on the gene in an expressible state is quite low (Hackett, 1993). Nevertheless, owing to the large numbers of eggs and the ability to inject hundreds of fertilized embryos per hour, even inefficient random recombination of transgenic DNA into genomes with subsequent, reliable expression through multiple generations can be achieved. Consequently, following the isolation of vertebrate growth hormone genes, several groups throughout the world initiated programs to engineer fish with accelerated growth and development (Hackett & Alvarez, 2000). The most visible product from these endeavors was the Aqua-Advantage salmon (Salmo salar), fish that contained a single expression cassette comprising a Chinook salmon (Oncorhynchus tshawytscha) growth hormone gene transcriptionally controlled by a promoter from the ocean pout (Zoarces americanus) antifreeze protein gene. A critical achievement was the specific introduction of defined eukaryotic genetic sequences without attendant genes of either bacterial origin or known antibiotic activity that are commonly used for cloning of transgenic DNA sequences. Nevertheless, the genetically engineered salmon encountered intense opposition by a variety of groups concerned with food safety, environmental impact, and other assorted issues, despite the finding that the fish were essentially equivalent to domesticated salmon (Devlin, Sakhrani, Tymchuk, Rise, & Goh, 2009; Smith et al., 2010; Van Eenennaam & Muir, 2011).
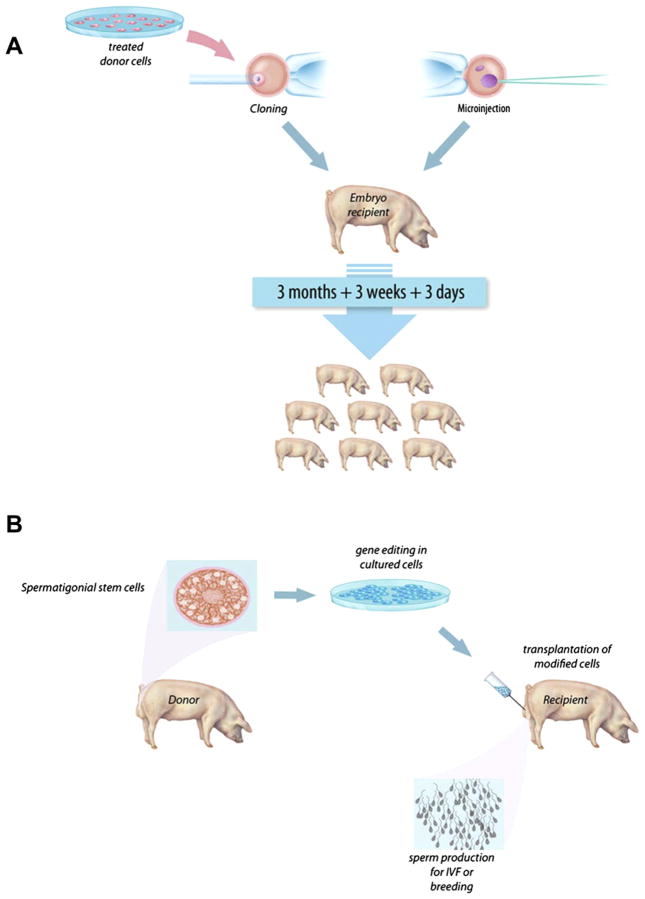
A large number of genes encoding both markers and proteins of commercial interest have been introduced into animal germlines using plasmids, naked DNA sequences, and viruses (Tables 1–4). Several effective methods of introduction of recombinant genomes into embryos have been developed. The most common are illustrated in Figure 4—somatic cell nuclear transfer (SCNT), microinjection, and sperm-mediated gene transfer (SMGT) (Carlson, Garbe, et al., 2011; Clark & Whitelaw, 2003). The studies reported in Tables 1–4 show that all three of the applications of transgenic technologies in large animals shown in Figure 1 have been initiated— improvement of intrinsic traits, improved medical products, and creation of better models of human disease. In all of these cases, the integration sites of the DNA sequences were uncontrolled and the efficiencies of producing germ-line transgenic animals were invariably low.
Table 1.
Transgenic animals for enhanced production or with marker genes
| Cassette* | Delivery† | F0 Exp‡ | F1 Exp‡ | Reference |
|---|---|---|---|---|
| Animal production | ||||
|
| ||||
| Pigs | ||||
|
| ||||
| mMT/hGH | PNI | 11/18 | Yes | (Brem, 1985; Hammer et al., 1985; Miller et al., 1989; Pursel et al., 1987) |
| mMT/hGRF | PNI | 2/7 | Yes | (Pinkert, 1987; Pursel et al., 1989) |
| mMT/bGH | PNI | 8/11 | Yes | (Pursel et al., 1987) |
| hMT/pGH | PNI | 1/6, 5/22 | Yes | (Nottle, 1999; Vize et al., 1988) |
| MLV/rGH | PNI | 1/1 | ND | (Ebert et al., 1988) |
| mMT/hGRF | PNI | ND | NA | (Brem and Winnacker, 1988) |
| bPRL/bGH | PNI | 2/4 | ND | (Polge et al., 1989) |
| hALB/hGRF | PNI | 3/3 | ND | (Pursel et al., 1989) |
| mMT/hIGF-1 | PNI | 1/4 | ND | (Miller et al., 1989; Pursel et al., 1989) |
| rPEPCK/bGH | PNI | 5/7 | Yes | (Wieghart et al., 1990) |
| CMV/pGH | PNI | 3/31 | ND | (Ebert et al., 1990) |
| MLV/pGH | PNI | 1/1 | ND | (Ebert et al., 1990) |
| MSV/cc-ski | PNI | 10/29 | ND | (Pursel et al., 1992) |
| oMT/oGH | PNI | 6/15 | ND | (Pursel et al., 1997) |
| ba-LA/ba-LA | PNI | ND | Yes | (Bleck et al., 1998) |
| cASK/hIGF-1 | PNI | NA | Yes | (Pursel et al., 1999; Pursel et al., 2004) |
| bCsn/hGH | PNI | 1/1 | ND | (Hirabayashi et al., 2001) |
| mPSP/APPA | PNI | 29/33 | Yes | (Golovan et al., 2001) |
| maP2/FAD2 | PNI | 2/3 | Yes | (Saeki et al., 2004) |
| bα-LA/hIGF-1 | PNI | NA | Yes | (Monaco et al., 2005) |
| CAG/hfat-1 | SCNT | 3/6, 12/13 | ND | (Lai et al., 2006; Pan et al., 2010) |
| bCsn/hLz | SCNT | 1/2 | Yes | (Tong et al., 2011) |
|
| ||||
| Cattle | ||||
|
| ||||
| MMTV/bGH | PNI | ND | ND | (Roshlau and Zackel, 1989) |
| cASK/hER | PNI | ≤1/1 | ND | (Hill, 1992; Massey, 1990) |
| bCsn/hLF | PNI | ND | ND | (Krimpenfort et al., 1991) |
| cASK/hlGF-1 | PNI | ND | Yes | (Hill, 1992) |
| MMTV/hlGF-1 | PNI | ND | ND | (Hill, 1992) |
| MSV/cc-ski | PNI | 1/1 | ND | (Bowen et al., 1994) |
| bβCsn/bβCsn & bκ-Csn | SCNT | 9/11 | ND | (Brophy et al., 2003) |
| bCsn/hGH | SCNT | 1/15 | Yes | (Salamone et al., 2006) |
| hα-LA/hα-LA | SCNT | 3/3 | Yes | (Wang et al., 2008) |
| hLF/hLF | SCNT | 2/2 | ND | (Yang et al., 2008) |
| bCsn/hLz | SCNT | 17/30 | ND | (Yang et al., 2011) |
| mTF/bGH | PNI | NA | NA | Bondioli, Hammer (unpubl.) |
| EF1α/anti-GDF8 shRNA | LV-MI | 5/5 | ND | (Tessanne et al., 2012) |
|
| ||||
| Goats | ||||
|
| ||||
| bCsn/hLz | PNI | Yes | Yes | (Maga et al., 2003) |
| oCsn/hGH | PNI | NA | NA | (Lee et al., 2006) |
| oCsn/hLF | PNI | NA | Yes | (Zhang et al., 2008) |
|
| ||||
| Sheep | ||||
|
| ||||
| mMT/hGH | PNI | ND, 0/1 | ND | (Hammer et al., 1985; Pursel et al., 1987) |
| mMT/bGH | PNI, MI | 2/2, 2/2 | ND, No | (Pursel et al., 1987; Rexroad et al., 1989) |
| oMT/oGH | PNI | 3/3 | ND | (Murray et al., 1989) |
| mMT/hGRF | MI | 1/7 | No | (Rexroad et al., 1989) |
| RSV/CE, CK, oMT/CE, CK | PNI | NA | NA | (Rogers, 1990; Ward, 1991) |
| mTF/bGH, mAlb/hGRF | PNI | 3/11 | NA | (Rexroad et al., 1991) |
| mKER/oIGF-I | PNI | 2/5 | Yes | (Damak et al., 1996a) |
|
| ||||
| Marker genes | ||||
|
| ||||
| Pigs | ||||
|
| ||||
| CMV/EGFP | RV, SCNT, EIAV, SCNT, SMGT, LV | 1/2, 1/1, 34/37, 4/4, 4/4, 6/7, ND | Yes | (Cabot et al., 2001; Garcia-Vazquez et al., 2010; Lai et al., 2002b; Liu et al., 2008; Whitelaw et al., 2004; Whyte et al., 2011; Zhang et al., 2012) |
| SV40/hSEAP | SMGT | 35/57 | Yes | (Chang et al., 2002) |
| K14/GFP, PGK/GFP | LV, SCNT | 32/34, 10/10 | ND | (Hofmann et al., 2003; Kurome et al., 2008) |
| CMV/EBFP, EGFP, DsRed2 | SMGT | 7/7 triple TG | ND | (Webster et al., 2005) |
| pCMV/huKO | RV-WCI | 18/18 | ND | (Matsunari et al., 2008) |
| CAG/EGFP | SCNT | 9/9 | Yes | (Whitworth et al., 2009) |
| mOCT4/EGFP, hOCT4/EGFP | SCNT | 6/11 | Yes, no | (Nowak-Imialek et al., 2011) |
| CAG/VenusFP | SB-CPI | 2/5 | Yes | (Garrels et al., 2011) |
| CAG/YFP, CAG/TFP | SCNT | 7/7 | ND | (Deng et al., 2011) |
| Ub/GFP | SB-SCNT | 4/5 | ND | (Jakobsen et al., 2011) |
| PGK/YFP | SB-SCNT | 6/6 | ND | (Carlson, Garbe, et al., 2011) |
| mStra8/EYFP-mito | SCNT | ND | ND | (Sommer et al., 2012) |
|
| ||||
| Cattle | ||||
|
| ||||
| RV/Neo | RV-MI | NA | No | (Haskell and Bowen, 1995) |
| CMV/βGEO | SCNT | 3/3 | ND | (Cibelli et al., 1998) |
| PGK/EGFP | LV | 4/4 | ND | (Hofmann et al., 2004) |
|
| ||||
| Sheep | ||||
|
| ||||
| mKER/CAT | PNI | 1/4 | Yes | (Damak et al., 1996b) |
| PGK/GFP | LV-MI | 3/9 | No | (Ritchie et al., 2009) |
Species of origin are given by lower case letters: m, mouse; b, bovine; c, chicken; h, human; o, ovine; p, porcine; r, rat.
Transgenic expression cassettes show the transcriptional regulatory motifs/transgene. Promoters: ALB, albumin; aP2, adipocyte lipid-binding protein P2; ASK, α-skeletal actin; BLG, β-lactoglobulin; CAG (also called CAGG/CAGGS), human CMV early enhancer fused to β-actin promoter; CMV, cyto-megalovirus; Csn, casein; EF1α, Elongation Factor 1α; H1, pol III-dependent RNA promoter, human RNase P; H-2Kb, major histocompatibility complex H-2Kb; ICAM2, intercellular adhesion molecule 2; IgSV, immunoglobulin heavy chain enhancer; INV, suprabasal keratinocyte-specific involucrin; K14, keratin K14; KER, keratin; LA, lactalbumin; mAb, mouse monoclonal antibody; MCP, membrane cofactor protein; mIgA, mouse immunoglobulin A; MLV, mouse leukemia virus LTR; MMTV, mouse mammary tumor virus LTR; MSV, mouse sarcoma virus LTR; MT, metallothionein; MTla, Metallothionein la; MX, interferon-induced GTP-binding protein Mx1; NSE, neuron-specific enolase; NTA-RCA, auto-regulative tetracycline-responsive bicistronic expression cassette regulator of complement activation; OCT4, Octamer-binding Transcription factor 4; PEPCK, phosphoenolpyr-uvate carboxykinase; PGK, phophoglycerol kinase; PRL, prolactin; PSP, parotid secretory protein; RHO, human Rhodopsin; Rho, rhodopsin; Rho4.4, Rhodopsin promoter 4.4; RSV, Rous sarcoma virus LTR; β-Lac, β-Lactoglobulin; Stra8, Stimulated by Retinoic Acid 8; SV40, simian virus 40; TF, transferin; Tie2, Tyrosine kinase with immunoglobulin-like and EGF-like domains 1; Ub, ubiquitin; Visna virus LTR, Visna virus LTR; WAP, whey acidic protein; κP, kappa protein. Transgenes: α1AT, α1 antitrypsin; α-1,3GT, α-1,3 = GGTA1, galactosyltransferase; A20, tumor necrosis factor-α-induced protein 3 (TNFaip3); anti-GDF8 shRNA, anti-Myostatin short hairpin RNA; anti-PERV shRNA, anti-porcine endogenous retrovirus short hairpin RNA; anti-PrP shRNA, anti-major prion protein or CD230 short hairpin RNA; ApoBEC3G, apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3G; APPA, E. coli Phytase gene; AT, antithrombin III; BChE, Butyr-ylcholinesterase; bi-scFV r28M, bispecific single-chain variable fragment (bi-scFV) molecule with anti-human CD28 anti-human melanoma specificity; BLVenv, Bovine Leukemia Virus Envelope; BSSL, bile salt-stimulated lipase; CAT, Chloramphenicol Acetyl Transferase; CD46, CD46 complement regulatory protein or Membrane Cofactor Protein; CD55, Decay-accelerating Factor; CD59, Pro-tectin, a complement regulatory protein; CE, E. coli cysE; CFTR, Cystic fibrosis transmembrane conductance regulator; CK, E. coli cysK; COL, Collagen; COL1A1, α1(I) procollagen; Cre, Cre recombinase; CTLA4-Ig, fusion gene between Cytotoxic T-Lymphocyte Antigen 4 and human IgG1; Cκ, immunoglobulin light chain; ELOVL4-5bpdel, elongation of very long chain fatty acids-4 with 5 bp deletions; ELOVL4-Y270ter, elongation of very long chain fatty acids-4 with 270 stop mutation; eNOS, nitric oxide synthase; EPO, Erythropoietin; ER, Estrogen Receptor; EYFP-mito, mito-chondria localized EYFP; FAD2, spinach Delta-12 fatty acid desaturase; FIX, coagulation Factor IX; FVIII, coagulation factor VIII; G-CSF, granulocyte colony stimulating factor; GH, growth hormone; GnT-III, N-Acetylglucosaminyltransferase III; GRF, growth-regulating factor; HbsAg, hepatitis B surface antigen; hfat-1, humanized (codon optimized ) fat-1; HHT CAG, Huntington disease gene with CAG repeats; hITG b1, α2, integrins b1, a2; HT, H-transferase; hv-HA-ras, Harvey rat sarcoma viral oncogene; hα1 + hβA, hemoglobin α1 and βA; IGF, insulin-like growth factor; IgH, immu-noglobulin heavy chain; IGHM, immunoglobulin-μ; Igλ, Imunnoglobulin light chain; JH, immu-noglobulin heavy chain joining region; BLG-hAAT, COL1A1 knock-in vector containing bovine β-lactoglobulin promoter driving human α1-antitrypsin; LDLR, low-density lipoprotein receptor. LF, lactoferrin; LP2, two LoxP sites; Lz, lysozyme; mAb, mouse monoclonal antibody; MCP, membrane cofactor protein; MX, interferon-induced GTP-binding protein Mx1; PPARγ, peroxisome proliferator-activated receptor γ; PrP = PRNP, major prion protein; RHO-h23H, human Rhodopsin with Pro23His mutation; Rho-Pro374Leu, rhodopsin gene with Pro374Leu mutation; SV40, Simian vacuolating virus 40; TK, thymidine kinase; TM, thrombomodulin; TPA, tissue plasminogen activator; Visna-env, Visna Virus envelope; vWF, Von Willebrand factor; Marker transgenes: BFP, blue fluorescent protein; CAT, chloramphenicol acetyl transferase; DsRed2/RFP, red fluorescent protein; E, enhanced; -GEO, -galactosidase-GFP fusion gene; GFP, green fluorescent protein; huKO, humanized Kusabira-Orange; neo, neomycin phosphotransferase II; SEAP, secreted alkaline phosphatase; TFP, tomato fluorescent protein; VenusFP, Venus fluorescent protein; YFP, yellow fluorescent protein; SB, Sleeping Beauty Transposon system. Viruses used for transduction: AAV, adeno-associated virus; EIAV, equine infectious anemia virus; LV, lentivirus; RV, retrovirus.
Methods of transgene delivery: CPI, cytoplasmic injection; ICSI, intracytoplasmic sperm injection; MI, microinjection; PNI, pronuclear injection; SCNT, somatic cell nuclear transfer; SMGT, sperm-mediated gene transfer; WCI, whole-cell injection cloning.
Transgene expression detected in F0 or F1 animals with numbers where available. NA, not available; ND, not done.
Table 4.
Transgenic animals for human or animal diseases
| Cassette* | Delivery† | F0 Exp‡ | F1 Exp‡ | Reference |
|---|---|---|---|---|
| Human disease models | ||||
|
| ||||
| Pigs | ||||
|
| ||||
| MMTV/hv-Ha-ras | PNI | 1/1 | Yes | (Yamakawa et al., 1999) |
| pRho/pRho-Pro347Leu | PNI | 3/3 | ND | (Petters et al., 1997) |
| rNSE/pHTT CAGs | PNI | NA | ND | (Uchida et al., 2001) |
| mTie2/peNOS | SCNT | 4/4 | ND | (Hao et al., 2006) |
| pMX/Cre | SCNT | 1/10 | ND | (Chen et al., 2010) |
| CAG/hHTT CAGs | SCNT | Yes | ND | (Yang et al., 2010) |
| Rho4.4/hELOVL4-5bpdel,-Y270ter | PNI, SCNT | NA | Yes | (Sommer et al., 2011) |
| hRHO/hRHO-hP23H | SCNT | 6/10 | Yes | (Ross et al., 2012) |
| CMV, INV/hITG b1, a2 | SB-HMC | 6/6 | ND | (Staunstrup et al., 2012) |
| hCOL-BAC, hALB-BAC | SMGT, ICSI | 6/8 | ND | (Watanabe et al., 2012) |
| PGK/YFP-Cre | SB-SCNT | 6/6 | ND | (Carlson, Geurts, et al., 2011) |
|
| ||||
| Animal disease resistance | ||||
|
| ||||
| Pigs | ||||
|
| ||||
| mAb/mAb | PNI | 1/1 | ND | (Weidle et al., 1991) |
| mIgA/mIgA | PNI | 2/2 | Yes | (Lo et al., 1991) |
| hMT, SV40, mMX/mMX | PNI | 2/9 | Yes | (Brem, 1993; Muller et al., 1992) |
| mMX-SV40 | PNI | 1/6 | NA | (Pinkert et al., 2001) |
|
| ||||
| Cattle | ||||
|
| ||||
| RSV/HbsAg | RV, PNI | 1/1 | ND | (Chan et al., 1998) |
| oBLG/lysostaphin | SCNT | 3/3 | Yes | (Wall et al., 2005) |
|
| ||||
| Goats | ||||
|
| ||||
| mIgA | PNI | 0 | ND | (Lo et al., 1991) |
| hH1/anti-PrP shRNA | LV-SCNT | 0 | No | (Golding et al., 2006) |
|
| ||||
| Sheep | ||||
|
| ||||
| oVisna-LTR/ oVisna-env | MI | 3/3 | ND | (Clements et al., 1994) |
See Table 1 for standard abbreviations.
BAC, bacterial artificial chromosome.
Figure 4.
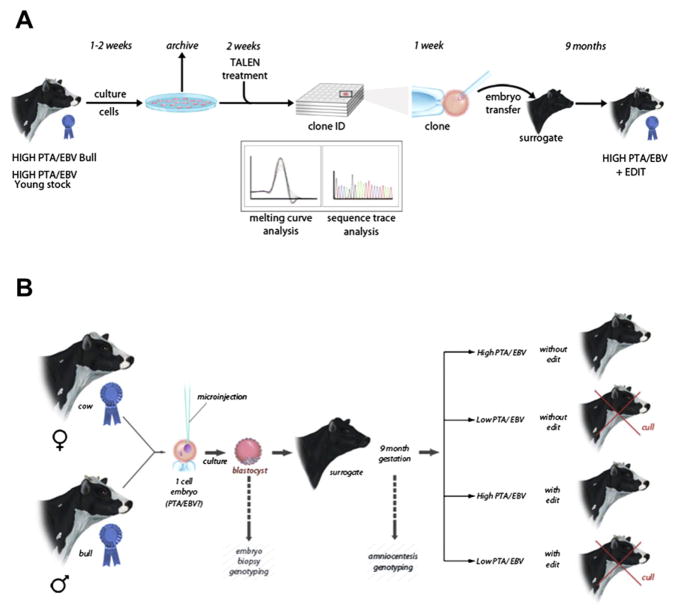
Methods for genetic modification in livestock. (A) A flow diagram of the primary steps involved with the production of transgenic livestock by SCNT (cloning) and embryo microinjection. For simplicity, the illustrations show pigs only, but the general procedure applies to each of the major livestock species. Each procedure requires either surgical or in vitro production of oocytes or embryos. Donor cells used for SCNT (left) can be genetically modified in culture by a number of methods described in this review. Modified donor cells are injected into enucleated oocytes, which are then fused and activated prior to embryo transfer into a recipient. Embryo microinjection (right) is performed on zygotes 18–24 h after fertilization. The injection site can vary, but typically, DNA is delivered directly to the pronucleus by pronuclear injection, SB trans-posons plus transposase mRNA, ZFN, or TALEN mRNA can be injected into the cytoplasm, and viral particles are typically injected into the perivitelline space. Embryos manipulated in each case are implanted into a synchronized recipient female to establish pregnancy. Resulting offspring can be screened for the desired modifications and expression patterns. (B) Spermatogonial stem cells offer a second method for genetic modification of livestock. Genetic modification or gene edits can be executed in vitro in spermatogonial stem cells isolated from donor testes. Modified cells are transplanted into germ cell-depleted testes of a recipient. Implanted spermatogonial stem cells produce sperm that carry the genetic modification(s) that can be used for breeding via artificial insemination or in vitro fertilization (IVF) to derive founder animals. For color version of this figure, the reader is referred to the online version of this book.
From a human gene therapy perspective, it would appear that the safety issues for gene delivery to humans are more relaxed than they are to animals! Between 1989 and mid-2012, 1786 gene delivery clinical trials in humans have been approved (http://www.wiley.com/legacy/wileychi/genmed/clinical/) of which about two-thirds employed viral vectors and the rest plasmid or other forms of “naked” DNA. There are two important differences in the design of gene therapy vectors. First, selectable marker genes are permitted in vectors introduced into human cells, with some restrictions (e.g., the kanamycin-resistance gene is preferred over genes encoding resistance to other antibiotics). Second, safeguards must be taken to ensure that only somatic cells take up transgenes; germline transmission of transgenic material is strictly forbidden. For genetic engineering of large animals, the important lessons from human gene therapy trials derive from comprehensive evaluations of insertional mutagenesis by a plethora of vectors. These vectors have a variety of integration preferences that include actively transcribed genes (lentiviruses), promoters and other transcriptional motifs (some retroviruses and adeno-associated viruses), and more random patterns (Sleeping Beauty transposons) (Berry, Hannenhalli, Leipzig, & Bushman, 2006; Mitchell et al., 2004). The issue of transgenes abnormally affecting resident genes has led to some adverse effects and to intense scrutiny of every patient for insertional mutagenesis. The results of these studies suggest that single gene activities do not cause adverse events, rather it appears that multiple events are responsible for adverse effects (Baum, 2011; Kustikova et al., 2009). This conclusion is not surprising given that there are hundreds of active endogenous transposable elements in human genomes that do not cause problems at a significant rate (Iskow et al., 2010); clearly, animal genomes have defenses against most random integrations. The totality of data from gene therapy studies, in which genetic material has been inserted into millions of human genomes strongly suggests that germline transgenesis will cause few significant effects on the recipient animal besides those designed by the genetic engineers.
The acceptance of the introduction of transgenic DNA into humans should serve as a model for evaluating gene transfer in farm animals. Yet, by mid-2012 only two types of transgenic animals have been approved for commerce. The first type includes transgenic goats that produce a human protein product in their milk (ATryn, sold by GTC Biotherapeutics). These animals are not sold to the public; only their transgenic product is sold for medical purposes. Ironically, ATryn was approved for human therapy in an arguably more stringently regulated European market 3 years prior to approval in the USA. The second type comprises genetically modified freshwater aquarium fish, called Glofish® (Knight, 2003), which have been cleared for retail sale by pet stores in most states. In the meantime, transgenic salmon, containing an extra copy of a salmon growth hormone gene, have languished in a regulatory morass for more than a decade (Van Eenennaam & Muir, 2011). The legacies of transgenic chicken and fish are clear—there is widespread suspicion by the public, which is reflected by governmental regulatory agencies, involving the safety of transgenic animal products. Most of these concerns over health and safety issues, environmental containment, etc. were also expressed for transgenic crops where the regulatory history has been far different.
1.2.3. Genetically Engineered Animals Preceded Genetically Modified Plants
The first genetic engineering of plants came a couple of years after transgenic animals were made (Lamppa, Nagy, & Chua, 1985). The far more rapid progress in the genetic engineering of animals in comparison to plants was the result of several causes, including (1) strong financial support by National Institutes of Health (NIH) for developing human gene therapy that required a detailed understanding of molecular genetic processes in mammals and (2) the relative ease in introducing transgenic DNA into animal cells through the plasma membrane compared to the far more difficult procedures required to traverse plant cell walls. Yet, despite the increased scientific challenges involved with genetic engineering of plants and the far greater propensity of transgenic pollen and seed to spread, thereby increasing environmental concerns, by 2011, there were 67 million hectares of transgenic crops in the USA and 89 million hectacres worldwide, accounting for more than 85% of the maize, cotton, soybean, and sugar beet crops and worth billions of dollars (Peng, 2011). Containment and other environmental concerns (Hutchison et al., 2010; Sears et al., 2001) have been overcome in transgenic crop species that are far harder to contain physically and genetically (Tabeshnik, 2010) than in animals. Transgenic crops are commonly thought to contribute to more than 80% of the items on supermarket shelves (http://www.womenshealthmag.com/health/frankenfish).
1.2.4. Lessons from the Early Genetic Engineering of Commercially Important Species
Since the birth of the first genetically engineered large farm animal in 1985 (Hammer 1985), more than 180 successful trials of transgenic large livestock production have been reported in the subsequent 27 years (Tables 1–5). In the 1980s, the focus was on enhancing animal growth performances by ectopically expressing heterogenic or extra copies of growth factor genes. Common transgenes included growth hormone genes from a variety of sources, insulin-like growth factor, growth hormone-releasing factor, and others (Table 1). These early studies demonstrated the feasibility in expression of exogenous transgenes in livestock but failed to produce any animals with value worthy of translating to agriculture. Many transgenic animals either did not transmit their transgenes and/or the transgenes failed to remain active due to epigenetic silencing (Kues et al., 2006) or the animals failed to thrive (Table 1). In retrospect, these experiments likely failed for a variety of reasons including either the use of an inappropriate transgene promoter and instability of transgenes due to repeated structure, epigenetic silencing, or position effects. During the 1990s, the attention shifted to large animals as bioreactors for the production of a variety of proteins in milk, including many hematopoietic human proteins such as Factors VIII and IX, von Willebrand factor (vWF), and alpha-1 antitrypsin (AAT) in blood clotting pathways (Table 2). For this, the casein and whey acidic protein transcriptional regulators were employed as they provided high levels of expression of the transgenic proteins in milk (Clark & Whitelaw, 2003). These systems largely restricted expression of the transgene to mammary glands; thus, expressed proteins were less likely to interfere with the welfare of transgenic animals. Despite a higher success rate in terms of producing animals with economically viable levels of protein production, the framework for their regulatory approval lagged behind scientific developments by almost two decades. Indeed, only a single product from transgenic biore-actors has reached the U.S. market, ATryn, sold by GTC Biotherapeutics. A second product, recombinant human C1 esterase inhibitor produced in the milk of transgenic rabbits, has been approved for use in Europe but not yet in the USA (van Doorn et al., 2005).
Table 5.
Gene targeting in livestock through homologous recombination (HR) and NHEJ
| Gene(s)* | Success† | Agent | Efficiency = genotyping+/ total colonies (%) | F1‡ | Reference |
|---|---|---|---|---|---|
| HR | |||||
|
| |||||
| Xenotransplantation transgenics | |||||
|
| |||||
| Pigs | |||||
|
| |||||
| α-1,3GT | +/− | Naked DNA | 1.54 | Yes | (Dai et al., 2002) |
| α-1,3GT | +/− | Naked DNA | 13.84 | Yes | (Lai et al., 2002a) |
| α-1,3GT | +/− | Naked DNA | 1.19 | Yes | (Ramsoondar et al., 2003) |
| α-1,3GT | −/T to G | Spontaneous mutation | NA | Yes | (Phelps et al., 2003) |
| α-1,3GT | +/− | Naked DNA | 0.32 | ND | (Takahagi et al., 2005) |
|
| |||||
| Cattle | |||||
|
| |||||
| α-1,3GT | −/− | Naked DNA | 0.52, 1.57 | Noa | (Sendai et al., 2006) |
|
| |||||
| Sheep | |||||
|
| |||||
| α-1,3GT | +/− | Naked DNA | 1.1 | NA | (Denning et al., 2001) |
|
| |||||
| Bioreactor transgenics | |||||
|
| |||||
| Pigs | |||||
|
| |||||
| Cκ | +/− | Naked DNA | 0.75 | −/−b | (Ramsoondar et al., 2011) |
| JH | +/− | Naked DNA | 0.64 | −/−b | (Mendicino et al., 2011) |
|
| |||||
| Disease transgenics | |||||
|
| |||||
| Pigs | |||||
|
| |||||
| CFTR | +/−, +/Δ | F508 AAV | 0.053–8.20 | Yes | (Rogers et al., 2008) |
|
| |||||
| Cattle | |||||
|
| |||||
| IGHM, PrPc | −/−, −/− | Naked DNA | 0.45–6.4 | Yes | (Kuroiwa et al., 2004) |
| PrP | −/− | Naked DNA | 3.30 | ND | (Richt et al., 2007) |
| Goat | |||||
|
| |||||
| PrP | +/− | Naked DNA | 1.53 | −/−b | (Yu et al., 2009; Yu et al., 2006) |
|
| |||||
| Sheep | |||||
|
| |||||
| COL1A1 | +/−, +/oBLG-hAATd | Naked DNA | 34.0 | ND, ND | (McCreath et al., 2000) |
| PrP | +/− | Naked DNA | 10.3 | ND | (Denning et al., 2001) |
|
| |||||
| NHEJ | |||||
|
| |||||
| Pigs | |||||
|
| |||||
| EGFP | +/− | ZFN | ~2% | ND | (Whyte et al., 2011) |
| PPARγ | +/− | ZFN | ~4.2% | ND | (Yang et al., 2011) |
| α-1,3GT | −/− | ZFN | ~2% | ND | (Hauschild et al., 2011) |
| LDLR | +/− | TALEN | 22%; 18/18 pigs | ND | (Carlson, Tan, et al., in press) |
|
| |||||
| Cattle | |||||
|
| |||||
| BLG | +/− | ZFN | 19.4% | ND | (Yu et al., 2011) |
Refer to Table 1 for standard abbreviations.
Genes are defined in the legend to Table 1; in some cases, more than one gene was inactivated.
Heterozygote knockout; −/−, homozygote knockout; −/ T to G, heterozygote knockout with a T to G mutation in the other allele; +/Δ F508, the human mutation Δ F508 knocked into one of the two alleles; +/oBLG-hAAT, oBLG-hAAT expression cassette knocked into one of the two alleles.
In some cases, where there is update information on transgenic offspring, the results are labeled (Y or N):
One piglet resulted from sequential targeting but died shortly after birth;
Homozygous KO F1 obtained by breeding of heterozygous KO F0;
Sequential targeting to KO both alleles for both genes in the same cells;
Knocked in oBLG-hAAT construct into one of the alleles and detected hAAT expression right after the lamb perished.
Table 2.
Transgenic animals as bioreactors and sources of bioproducts
| Cassette* | Delivery† | F0 Exp‡ | F1 Exp‡ | Reference |
|---|---|---|---|---|
| Pigs | ||||
|
| ||||
| mWAP/mWAP | PNI | 3/3 | Yes | (Shamay et al., 1991; Wall et al., 1991) |
| mWAP/hFVIII | PNI | 1/1 | 4/4 | (Paleyanda et al., 1997) |
| mWAP/hFibrinogen | PNI | 3/4 | ND | (Butler et al., 1997) |
| mWAP/hFIX | PNI | 2/3 | Yes | (Van Cott et al., 1999) |
| ba-LA/hFIX | PNI | NA | Yes | (Wu et al., 1999) |
| mWAP/hProtein C | PNI | 6/8 | Yes | (Van Cott et al., 2001) |
| CAG/hAlb | ICSI | 1/1 | ND | (Naruse et al., 2005) |
| mWAP/hEPO | PNI | NA | Yes | (Park et al., 2006) |
| bCsn/hvWF | PNI | 2/2 | Yes | (Lee et al., 2009) |
| gCsn/hEPO | SCNT | ND | Yes | (Cho et al., 2009) |
|
| ||||
| Cattle | ||||
|
| ||||
| bCsn/hEPO | PNI | NA | ND | (Hyttinen et al., 1994) |
| hIgH and Igλ | HAC, SCNT | 6/6 | Yes | (Kuroiwa et al., 2002) |
| oBLG/hBSSL | SCNT | ND | ND | (Chen et al., 2002) |
| mκP/bi-scFV r28M | SCNT | 9/9 | ND | (Grosse-Hovest et al., 2004) |
|
| ||||
| Goats | ||||
|
| ||||
| mWAP/hTPA | PNI | ND | Yes | (Ebert et al., 1991) |
| oCsn/hAT | SCNT | 1/1 | ND | (Baguisi et al., 1999) |
| oCsn/hG-CSF | PNI | 1/2, 2/2 | No, Yes | (Freitas et al., 2012) |
| mWAP/spider silk | PNI | ND | Yes | (Baldassarre et al., 2003) |
| oCsn/hBChE | PNI | NA | Yes | (Baldassarre et al., 2004) |
|
| ||||
| Sheep | ||||
|
| ||||
| oBLG-hα1AT | MI, PNI | 3/5, 2/3 | Yes | (McClenaghan et al., 1991) |
| oBLG/hFIX | PNI, SCNT | 2/2, ND | Yes, ND | (Schnieke et al., 1997) |
| oBLG/hFibrinogen | PNI | 3/3 | ND | (Butler et al., 1997) |
| mWAP/hFVIII | PNI | ND | ND | (Halter et al., 1993) |
| mWAP/mWAP | MI | 2/2 | Yes | (Wall et al., 1996) |
| oβ-Lac/hFVIII | PNI | ND | ND, Yes | (Niemann et al., 1999) |
See Table 1 for standard abbreviations
HAC, human artificial chromosome
Pigs due to similar size and physiology also became the leading candidate for production of tissues and organs for xenotransplantation to humans (Bucher, Morel, & Buhler, 2005). As our knowledge in the molecules and reactions involved in xenograft rejection following tissue and organ transplantation grew, another wave of modifications arose to humanize the cell surface proteins of animals to suppress animal-specific antigens that initiated strong immunological rejections by the immune systems of human recipients (Klymiuk, Aigner, Brem, & Wolf, 2010; Sachs & Galli, 2009). A primary goal was to neutralize α1,3-galactose, the primary antigen responsible for hyper-acute rejection (Cooper, 2003) from the cell surface of pigs by inactivating the α1,3-galactose transferase gene (GGTA1). Several other transgenic approaches were developed to combat immune rejection, including either introducing or knocking out cell surface determinant proteins such as CD55, CD46, and CD59, followed by homologous recombination and SCNT to create GGTA1 knockout animals (Tables 3 and 5). Additional transgenic animals have been created to neutralize incompatibilities between blood coagulation systems and to limit T-cell responses (Table 3). Another key target for inactivation was the porcine endogenous retrovirus (PERV) locus that might allow recombinant retroviruses to emerge from transplanted porcine chromosomes, though transmission of PERV from swine to humans has never been observed in vivo (Fishman & Patience, 2004).
Table 3.
Transgenic pigs for xenotransplantation
| Cassette* | Delivery† | F0 Exp‡ | F1 Exp‡ | Reference |
|---|---|---|---|---|
| hβ-globin/hα1 and βA | PNI | 3/3 | ND | (Swanson et al., 1992) |
| mH-2Kb/hCD59 | PNI | 1/3 | ND | (Fodor et al., 1994) |
| pMCP/hCD55 | PNI | 1/5 | Yes | (Murakami et al., 2000) |
| hICAM2/hHT | PNI | 8/185 | ND | (Nottle et al., 2001) |
| mH-2Kb/hCD55 + hHT | PNI | 4/20 | ND | (Nottle, 2001) |
| mH-2Kb/hCD55 + hCD59 + hHT | PNI | 11/16 | ND | (Nottle et al., 2001) |
| hICAM2/hCD46 + hCD55 + hCD59 | PNI | 2/94 | ND | (Nottle, 2001) |
| CAG/hGnT-III | PNI | NA | Yes | (Miyagawa et al., 2001) |
| RSV/hCD55 | SMGT | 34/53 | Yes | (Lavitrano et al., 2002) |
| pAlb/TK | SCNT | 1/3 | ND | (Beschorner, 2003) |
| ba-LA/pLF, ba-LA/ hFIX | WCI | 4/4 | ND | (Lee et al., 2003) |
| hCD59/hCD59 + hMCP/hMCP + hCD59 | PNI | 1/1 | ND | (Zhou, 2004) |
| rNSE/hCTLA4-Ig | PNI | 2/8 | Yes | (Martin et al., 2005) |
| NTA-RCA/hCD55, NTA-RCA/hCD59 | PNI | 9/10 | Yes | (Kues et al., 2006) |
| hH1/anti-PERV shRNA | LV, SCNT | 2/2, 12/12 | ND, Yes | (Dieckhoff et al., 2008; Ramsoondar et al., 2009) |
| CAG/pCTLA4-Ig | SCNT | 15/15 | ND | (Phelps et al., 2009) |
| CMV/hTM | SCNT | 7/7 | ND | (Petersen et al., 2009) |
| CAG/hA20 | SCNT | 2/2 | ND | (Oropeza et al., 2009) |
| PGK/hApoBEC3G | SB-SCNT | 10/10 | ND | (Carlson, Geurts, et al., 2011) |
| PGK,Ub,CAG/LP2-hApoBEC3G | SB-SCNT | 3/3, 4/4, 0/1 | ND | (Carlson, Geurts, et al., 2011) |
See Table 1 for standard abbreviations.
The physiological similarities that make pigs good candidates for xeno-transplantation also made them ideal candidates for modeling of human diseases (Table 4). Some human diseases cannot be accurately modeled in rodents due to differences in size and physiology. The first such example was created nearly 15 years ago by transgenic expression of a dominant–mutant rhodopsin gene (Pro347Leu) (Petters et al., 1997) as a model of retinitis pigmentosa. The phenotype of this model has remained stable through more than nine generations of outcrossing (Sommer et al., 2011) and is used yet today. The ability to perform homologous recombination in livestock fibroblasts and creation of animals by SCNT enabled modeling human disease caused by of loss-of-function (LOF) mutations (Table 5). The cystic fibrosis pig was the first porcine model of human disease to take advantage of targeted gene knockout. In contrast to mice, pigs either knocked out or containing a common mutation of the Cftr gene (Δ508) accurately recapitulate many of the pathologies observed in humans (Rogers et al., 2008). The similar size and physiology of pigs and humans suggests that introducing disease-associated alleles into pig genomes will result in relevant platforms for development of human therapeutics and devices.
All of the studies in Tables 1–4 led to substantial understanding of the limitations of transgenic technology using randomly integrating expression cassettes or recombinant sequences to inactivate selective genes. But, in addition to practical modifications that were based on direct benefits to humans, there were also innovative studies designed to generate transgenic animals that would enhance sustainability, e.g. the Enviropig (Golovan et al., 2001) was created to reduce manure phosphorous emissions, and fortuitously enhanced bone strength. Improved animal welfare is a clear area for animal genomics to flourish using precision genetics.
Yet, in contrast to transgenic plants and despite U.S. government (NIH, U.S. Department of Agriculture, National Science Foundation, Financial Services Authority, Environmental Protection Agency) investments of around $100 million dollars in funding research and risk analysis on large transgenic animals, not even one line of transgenic animal has been cleared for human consumption. The stated principle concerns have been either potential harm to consumers or potential harm to the environment, yet these concerns are not supported by scientific findings (Fedoroff, Haselkorn, & Chassy, 2011). These are exactly the same issues faced by transgenic plants that have far greater abilities to spread and where far less is known about their genetics (Schurman & Munro, 2010). The advent of precision genetic techniques promises to satisfy scientifically based concerns regarding the development of transgenic farm animals.
There are five principle concerns with current transgenic organisms wherein expression cassettes were introduced randomly into recipient genomes: (1) insertional mutagenesis—the incoming genetic regulatory motifs affect the activity of a resident gene by either inappropriately activating or suppressing its expression; (2) inability to precisely control the expression of the transgene—resident genetic regulatory motifs in the vicinity of the integrated transgene influence its expression; (3) unstable expression of the transgene due to epigenetic effects that occur over time; (4) presence of unwanted DNA sequences that are required by the vector— plasmid or viral; and (5) unknown effects on expression of the transgene in various tissues—the transgene may be designed for expression in one tissue, but its expression in other organs and cells may vary considerably.
Over the past decade, newly developed methods allow specific replacement, addition, and/or deletion of genetic sequences in animal genomes. The application of precision genetics will avoid nearly all of the substantive issues of genetically engineered organisms that have been raised in the past.
2. PRECISION GENETIC ENGINEERING
As noted above, there are two issues critical to genetic modification of food animals. The first, only defined changes are made at specific genetic loci. This is important to ensure that only the expected phenotype will occur in the animal without collateral changes that could lead to unintended effects on consumers’ health (e.g. production of an allergen as a result of random insertion leading to gene fusion or activation of genes in unexpected ways). The second is the efficiency and precision with which such defined genetic changes can be introduced into genomes of large animals. Over the past decade there has been enormous progress in both areas, as predicted by Clark & Whitelaw (2003).
There are three types of modifications to genomes that will enable efficient transgenesis in animals without unanticipated consequences: (1) adding precisely defined genetic sequence that will confer a new trait to an animal; in this case, the actual location of the gene is not important. (2) Editing a gene so that it either is inactivated or is converted to a desirable allele. (3) Adding a gene to a specific site in the genome, e.g., to express a protein under the direction of a native gene or placement of a gene in a location previously defined to permit effective gene expression (e.g., a safe harbor).
2.1. Precision Introduction of Expression Cassettes Using Transposons
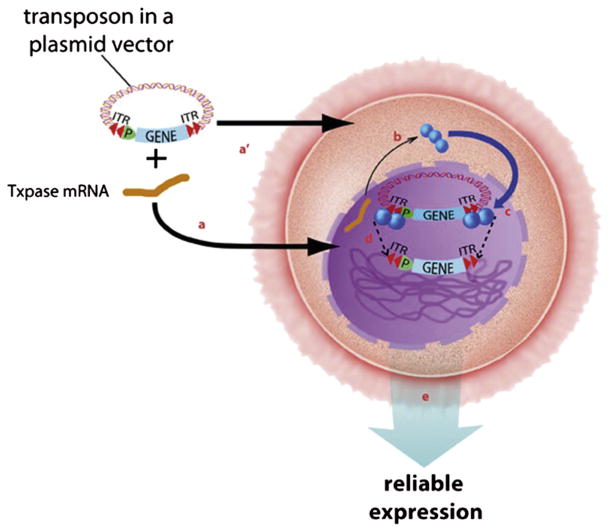
Transposons are used to accomplish the first category of precision genetic engineering. Transposons are natural mobile elements that move either by a copy-and-paste mechanism via an RNA intermediate (class I transposon; by far the most numerous in animal and plant genomes) or a cut-and-paste mechanism (class II transposons) in which a precise DNA sequence is excised from one source of DNA and inserted into another DNA. Class II transposon systems consist of two components: (1) the transposon vector that contains a transgenic expression cassette flanked by inverted terminal repeats and (2) a source for the transposase enzyme (Fig. 5). Generally, class II transposons, cloned in plasmids, are used for genetic engineering because they can direct the integration of a defined expression cassette harboring a transgene and its regulators while leaving behind the rest of the plasmid with its selection markers (Dupuy et al., 2002; Hackett, Ekker, Largaespada, & McIvor, 2005). Nearly all of the class II DNA transposons identified in vertebrate genomes appear to be inactive (Plasterk, Izsvák, & Ivics, 1999; Venter et al., 2001; Waterston et al., 2002). Hence, the first transposon used in animal cells, called Sleeping Beauty because it was awakened from a ca. 14-million year sleep (Ivics et al., 1997), was synthetic. One consequence of the synthetic engineering of Sleeping Beauty from hundreds of extinct and active transposase genes is that it has considerably higher activity than natural transposons (Grabundzija et al., 2010). A number of other transposon systems have been developed for use in vertebrate cells, mainly for gene therapy in order to avoid viruses (Ivics et al., 2009). The advantages of transposons for human gene therapy, where transposons have been used for more than a decade in animal models (Aronovich, McIvor, & Hackett, 2011), extend to genetic engineering of large animals as well (Carlson, Garbe, et al., 2011; Clark et al., 2007).
Figure 5.
DNA transposition consists of an enzymatic cut-and-paste reaction in which a transposon containing a gene of interest [shown in blue, with its promoter (P)] is cut out of a plasmid and inserted into a chromosome. The cleavage reaction occurs at the ends of the ITRs (inverted set of red double arrowheads) of the transposon. The transposons integrate only into TA-dinucleotide basepairs (about 200 million in a mammalian genome). The ITRs are the only DNA sequences required by the transposase enzyme for transposition. The transposase enzyme (Txpase, blue balls) drives the cut-and-paste reaction. Transposase activity is obtained by co-injecting transposon and an mRNA encoding the Txpase (blue squiggle) into either the nucleus (a) or cytoplasm (a′). The plasmid carrying the transposon and transposase-encoding mRNA enter a cell (large back oval) and proceed through the nuclear membrane (dashed line) (b). The transposase mRNA is translated in the cytoplasm to give an appropriate level of enzyme (c). The transposase molecules enter the nucleus and bind to the transposon, two at each end (c). Four transposase enzymes work in concert to cleave the plasmid at the termini of the transposon and paste it (dotted lines) into chromosomal DNA (green tangled lines) (e). Monomeric integration into a chromosome can confer reliable expression of the gene of interest that is contained within the transposon through multiple generations. For color version of this figure, the reader is referred to the online version of this book.
2.2. Precision Editing of Genomic Sequences Using Meganucleases and Zinc Finger Nucleases
The studies listed in Tables 1–4 depended on random introduction of new DNA sequences into animal genomes. Random integration can produce unpredictable genetic effects that are bilateral between chromosomal genes and transgenes (Voigt, Izsvák, & Ivics, 2008). Position-effect variegation wherein transgenic sequences are silenced when introduced into chromatin and transactivation by the transgene on endogenous genes that are switched off can occur. One potential method to target transposons to specific sites would use E. coli RecA fusion proteins to induce genomic modifications. The bacterial recombinase RecA forms a nucleic acid-protein filament on single-stranded DNA during the repair of DNA double-strand breaks that efficiently undergoes a homology search and engages in pairing with the complementary DNA sequence. The pairing activity of RecA–DNA filaments that leads to site-specific breakage of DNA strands has been explored in zebra fish but awaits extension to large animal genomes (Cui, Yang, Kaufman, Agalliu, & Hackett, 2003; Liao & Essner, in press).
Rare-cutting DNases such as the yeast meganuclease I-SceI (Jasin, 1996; Rouet, Smih, & Jasin, 1994; Smih, Rouet, Romanienko, & Jasin, 1995) show great promise for the alteration of chromosomal sequences at a few specific sites (Choulika, Perrin, Dujon, & Nicolas, 1995). Meganucleases are precise and effective at cleaving their cognate recognition site in the genome, but the overlap of DNA recognition domains and the enzymatic centers of these compact proteins has made reprogramming them to recognize different sites in the genome difficult, although some progress has been made (Arnould et al., 2011; Chames et al., 2005). Efforts to use these reagents have been confounded by the rarity of sites present in livestock genomes that correspond to the addresses represented in current enzyme libraries (Fahrenkrug unpublished).
A major step toward the goal of developing site-specific genetic engineering was construction of chimeric nucleases composed of a nuclease domain and a separate, designer DNA recognition domain. The first such enzymes employed zinc finger (ZF) DNA recognition domains tethered to the endonuclease domain of FokI (Kim, Cha, & Chandrasegaran, 1996). Because Cys2His2 ZFs can be designed to bind to specific sites (Desjarlais & Berg, 1993; Jamieson, Wang, & Kim, 1966), artificial zinc finger nucleases (ZFNs) became a tool to cleave specific genetic loci (Bibikova, Beumer, Trautman, & Carroll, 2003; Bibikova et al., 2001; Kim et al., 1996; Park et al., 2003; Porteus & Carroll, 2005). The human gene therapy community quickly recognized the potential of site-specific integration of therapeutic transgenes and developed the use of ZFNs in human cells (Carroll, 2011; Hockemeyer et al., 2009; Porteus & Baltimore, 2003; Urnov et al., 2005; Urnov, Rebar, Holmes, Zhang, & Gregory, 2010). Table 5 lists studies in large animals that have employed ZFNs for targeted mutagenesis.
ZFNs were revolutionary, but although their assembly appeared easy theoretically (Klug, 2010), in practice, it was not. Generally, specific ZF-binding domains recognize a three-base sequence. Unexpectedly, it turned out that the various finger domains influenced each other such that when assembled into arrays, the fingers did not bind to targeted sequences with high efficiency (Lam, van Bakel, Cote, van der Ven, & Hughes, 2011). This problem necessitated the testing and selection of multiple combinations of fingers to determine those with the highest ZFN specificity and efficiency. The Oligomerized Pool Engineering strategy permits manufacture of ZFNs that recognize sites about every 200 basepairs of random genomic sequence (Maeder et al., 2008; Sander et al., 2010). Alternatively, context-dependent assembly (CoDA) (Sander, Dahlborg, et al., 2011) uses an archive of validated two-finger units derived from selection that have been validated to function when positioned adjacent to each other. CoDA-based ZFNs can be constructed that recognize approximately one site in every 500 basepairs of random genomic sequence. Other options that claim to have a targeting range of 1 in 125 basepairs of random genomic sequence are available (Kim, Lee, Kim, Cho, & Kim, 2009; Ramirez et al., 2008).
2.3. Precision Editing of Genomic Sequences Using TALENs
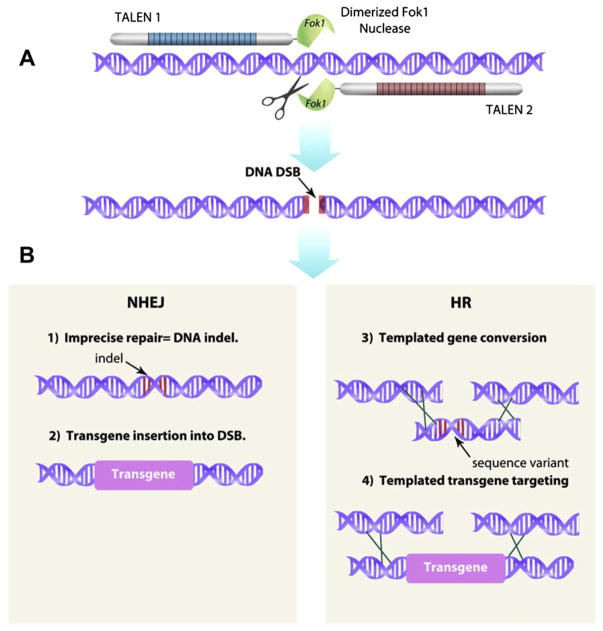
Recently a new type of chimeric nucleases has exploded onto the genetic engineering scene due to their ease in design and greater range of sites that can be targeted (Bogdanove & Voytas, 2011; Carlson, Fahrenkrug, & Hackett, in press). Transcription activator-like (TAL) effector nucleases (TALENs), like ZFNs, consist of assembled DNA-binding motifs coupled to a FokI endonuclease domain (Boch & Bonas, 2010; Boch et al., 2009; Christian et al., 2010; Li, Huang, Jiang, et al., 2011; Mahfouz et al., 2011; Moscou & Bogdanove, 2009). TAL-effector DNA-binding motifs are found in proteins secreted by plant pathogens in the bacterial genus Xanthomonas. Typically, TAL-effectors consist of tandem repeated 34 amino acid blocks. Residues 12 and 13 of the 34 amino acid repeats are referred to as repeat variable diresidues (RVDs). The RVDs define the binding to a specific base. Unlike ZFs that bind to three basepairs, each TAL-effector repeat binds to a single basepair (Boch et al., 2009; Moscou & Bogdanove, 2009) (Fig. 6). A simple cipher greatly simplifies the design of TALENs and makes their modular assembly far easier than is possible with ZFNs (Cermak et al., 2011; Li, Huang, Zhao, et al., 2011; Miller et al., 2011; Morbitzer, Elsaesser, Hausner, & Lahaye, 2011; Reyon et al., 2012; Weber, Engler, Gruetzner, Werner, & Marillonnet, 2011; Zhang et al., 2011).
Figure 6.
Site-specific targeting of genetic changes using hybrid DNases. (A) A pair of TALEN nucleases is shown as an example of hybrid DNases designed to cleave at a unique sequence in a genome. The pair of TALENs executes a double-strand DNA break (DSB) at the targeted locus. (B) If no other DNA sequences are added, the DSB will be repaired by the process of NHEJ that will generally result in a minor insertion or deletion of a few basepairs (indels; example 1). Alternatively, because the NHEJ DNA repair enzymes that assemble at the DSB can facilitate the integration of a foreign DNA sequence, a transgene can be introduced into the site with higher than random efficiency (example 2). Alternatively, if a DNA sequence that has a high identity with the region surrounding the DSB is introduced, homologous recombination (HR) can occur (examples 3 and 4). The introduced DNA sequence may vary by only a single (or a few) basepair, which results in a defined mutation that is equivalent to a natural allele (example 3). However, if an entire expression cassette with a foreign transgene is flanked by homologous sequences at the DSB, then the transgene will have a high probability of being copied precisely into the DSB (example 4). For color version of this figure, the reader is referred to the online version of this book.
Since the demonstration by Boch et al. (2009) that artificial TAL effectors could be targeted to specific DNA sites to activate transcription, sequence-specific DNA-binding proteins with predicted binding specificities have been generated economically in a matter of days using standard methods of molecular biology (Cermak et al., 2011; Li, Huang, Zhao, et al., 2011; Morbitzer et al., 2011). TALENs introduced into human cells can direct site-specific mutagenesis at rates of up to 45% of chromosomes (Hockemeyer et al., 2011; Mahfouz et al., 2011; Miller et al., 2011; Mussolino et al., 2011; Orlando et al., 2010). TALENs have been used to create site-specific modifications in zebrafish (Huang et al., 2011; Sander, Yeh, Peterson, & Joung, 2011) and rats (Tesson et al., 2011) at levels equivalent to those achieved with ZFNs. In addition to their ease of assembly, TALENs have another advantage over ZFNs—studies of native TAL-effector sequence preferences suggest a good TALEN sites occur in every 35 bp (Cermak et al., 2011). However, a recent study stretched the rules proposed by Cermak et al. (2011) and found that the true targeting range may be even better than 1 site per 35 basepairs in the genome (Reyon et al., 2012). In addition, the recent elucidation of the molecular structures of TAL-effector binding to DNA (Deng et al., 2012; Mak, Bradley, Cernadas, Bogdanove, & Stoddard, 2012) may further improve the design process and specificity.
2.4. Off-Target Cleavage Activity by ZFNs and TALENs in the Context of Natural Variation
A potential concern in the use of ZFN and TALEN site-specific nucleases is cleavage at unintended sites, referred to as off-target activity. This issue has been addressed over the past decade. While some potential off-target sites can be predicted, unbiased studies of ZFN off-target cleavage reveal shortcomings of in silico off-target predictions (Gabriel et al., 2011; Pattanayak, Ramirez, Joung, & Liu, 2011). Both Gabriel et al. (2011) and Pattanayak et al. (2011) chose to evaluate off-target cleavage of the highly characterized CCR5-224 ZFN pair, currently in clinical trials for gene therapy in humans. A total of 13 off-target sites were identified that occurred at an appreciable frequency (1:7–1:10,000 cells). In all cases, cleavage at the desired site was greater than five-fold more frequent than at other sites. The most important conclusion from these studies is that while off-target activity was present in a minority of cells, it was highly restricted to a small subset of loci, which implies that selective screening of potential off-target sites can be conducted following use of other ZFNs and TALENs.
As with ZFNs, early studies reveal that TALENs can bind degenerate sequences and have demonstrated activity at related off-target sites (Mussolino & Cathomen, 2011; Tesson et al., 2011). The specificity of TALENs has yet to be characterized in detail. Preliminary studies in cells and zebra fish reveal that cytotoxic effects of TALENs are either lower or similar to those with comparable ZFNs (Mussolino et al., 2011). Notably, TALEN pairs in these studies utilized the wild-type homodimeric Fok1 domain, which are more prone to cleaving erroneous sites, while ZFNs used one of the three obligate heterodimer domains that increase specificity and reduce cytotoxicity (Doyon et al., 2011; Miller et al., 2007; Szczepek et al., 2007).
Regardless of the platform (ZFNs or TALENs) and Fok1 domain (homodimer or heterodimer) used, there will be the potential of generating off-target genetic lesions. To address the implications of off-target lesions in genetically modified animals, we compared the worst-case estimate of off-target frequency with natural variation and germline mutation rate. As an example, consider a theoretical ZFN (or TALEN) with a poor on/off-target activity ratio of 1:1 that directs targeted cleavage and mutagenesis at a 25% efficiency, then one in four cells with an on-target event also would be expected to have an accompanying single off-target lesion. As a result, one in four animals derived from cloning of these cells would have a de novo change to its genome outside of the intended locus. In comparison, deep sequencing of two parent–child trios in the 1000 genomes project (a total of six people) revealed that each individual has 30–50 de novo germline mutations (Durbin et al., 2010; Marth et al., 2011). Assuming the data for humans is applicable to other large mammals, the risk of a random change to the genome by reproduction is more than 100-fold greater than any unintended mutations resulting from a site-specific nuclease employed for directed genome modification.
There is a further consideration. Most de novo germline mutations in humans are single-base substitutions in contrast to an indel that would result from non-homologous end-joining (NHEJ) activity during repair of an off-target site (Fig. 6B). Two-thirds of exonic indels would be expected to cause a frameshift leading to premature termination of translation, whereas only a small portion of naturally occurring single nucleotide polymorphisms (SNPs) would result in a protein truncation. Deep sequencing has found that indels are about 10-fold less frequent in the human genome than SNPs (22,000 vs. 1800 per genome compared to reference) with up to 50% of the indels being novel in any given individual (Alkan, Coe, & Eichler, 2011; Marth et al., 2011). Thus, introducing this aspect into the calculation for the worst possible scenarios, off-target NHEJ activity would occur more than 10-fold less frequently than the background indel mutation rate. Moreover, because only about 2% of the genome encodes proteins, about 98% of off-target events would be unlikely to affect protein sequences.
Deep sequencing of hundreds of human genomes has revealed that the average human genome has approximately 250–300 LOF mutations, with 50–100 in human disease genes (Durbin et al., 2010; Pelak et al., 2010) and about 20 completely inactivated genes (MacArthur et al., 2012) as classified by the Human Gene Mutation Database (http://www.hgmd.org). Thus, the human genome is highly variable (Kidd et al., 2010) and recent next-generation sequencing of the cattle genome suggests similar, high degrees of variation (Bickhart et al., 2012). Indeed, sequence survey of around 100 cattle (Fahrenkrug, unpublished) and high-density genotyping (J. Taylor, personal communication) have revealed similar frequencies of both heterozygous and homozygous LOF alleles.
2.5. Precision Alterations in Livestock Genomes
2.5.1. Transposon-Modified Animal Genomes
Transposon systems have been mainly and extensively used in mice for identifying oncogenes and for developing methods for human gene therapy. Transposons have been used less frequently in large, genetically modified animals. As shown in Tables 1–4, many of these animals were accomplished through random insertion of naked linear DNA introduced by early embryo injections, SMGT, or transfection of harvested animal cells accompanied by SCNT. As noted earlier, epigenetic effects, position-effect variegation, and variations in the numbers of integrated expression cassettes hampered the efficiencies of generating modified animals with predictable levels of transgene expression. Alternatives were broadly sought to optimize such situations; recombinant viruses or the Sleeping Beauty (SB) transposon system (Ivics et al., 1997) bearing desired transgenes have been shown to mediate insertions more efficiently via embryo injections, transfections, and SCNT (Tables 1–4). Moreover, they are less prone to integrate in the form of concatemers, and through intricate ways, one is able to control the copy number insertions. Transposons may be preferable to viruses given public concern about even functionally impaired viral relics in the modified genomes.
2.5.2. ZFN-Modified Animal Genomes
Gestation length and maturation to reproduction age for pigs and cattle is significant. For example, generation of a homozygous knockout from heterozygous mutant cells (both sexes) by cloning and breeding requires 16 and 30 months for pigs and cattle, respectively. It is possible to reduce this burden with sequential cycles of genetic modification and SCNT (Kuroiwa et al., 2004); however, this is both technically challenging and cost prohibitive. Taking advantage of the proclivity of ZFNs to modify both alleles, Hauschild et al. (2011) recently generated bi-allelic GGTA1 knockout pigs using commercial ZFN reagents and cloning. In this example, bi-allelic null cells could be enriched by fluorescence-activated cell sorting for the absence of the α1,3-galactose surface epitope. Unfortunately, biological enrichment for null cells using flow sorting will not be available for the majority of genes. Others have generated heterozygous knockout animals by ZFN-induced NHEJ in flbroblasts from pigs and cattle (Table 5). These studies demonstrate proof-of-principle; in about half of the examples engineered, ZFNs were relatively inefficient (i.e. only 2–4% of transfected cells were modified), which in terms of colony screening is not a significant improvement over standard homologous recombination. However, in contrast to traditional methods of homologous recombination, gene knockouts can be accomplished by introducing frame-shifts in coding regions from NHEJ without the use of selection markers.
2.5.3. TALEN-Modified Animal Genomes
At first glance, TALENs appear as somewhat of a redundant tool to ZFNs; they support the same types of precision genetic alterations (Fig. 6). However, there are two key features of TALENs that set them apart from ZFNs for widespread adaptation by livestock biotechnologists. First, and most importantly, simple design and assembly strategies for TALENs have been developed that can be implemented in any molecular biology laboratory (Cermak et al., 2011). A second advantage of TALENs is their targeting range that is far superior to that of ZFNs. For instance, we were able to rapidly assemble 36 TALEN pairs using the Cermak assembly procedure, 64% of which were active in livestock fibroblasts with an average chromosome modification frequency of 25% (Carlson, Tan, et al., in press). We recently reported the births of 18 low-density lipoprotein receptor ± Ossabaw piglets from TALEN-induced NHEJ and SCNT (Table 5). Carlson et al. also demonstrated that several TALEN pairs were efficient at inducing indels by direct injection of mRNA encoding them into the cytoplasm of both swine (about 30%) and bovine (about 75%) embryos.
Application of TALENs to cultured cells has also shown great promise for the creation of livestock with precise modifications. For example, we developed strategies for derivation of fibroblast clones with bi-allelic modifications (up to 10%) without biological enrichment (Carlson, Tan, et al., in press). TALENS are also capable of more complex changes in livestock fibroblasts. Cotransfection of two pairs of TALENs targeting the same chromosome was capable of creating large chromosomal deletions or inversions (Carlson, Tan, et al., in press). Perhaps most compelling, cotransfection of TALENs with a donor template has allowed directed homologous recombination for efficient insertion of either a transgene or for copying small, defined change to the genome without the aid of selection markers (authors, unpublished).
3. FUTURE DIRECTIONS—APPLICATIONS OF PRECISION GENETICS IN ANIMALS
3.1. Rapid Allele Introgression for Improvement of Food Animals
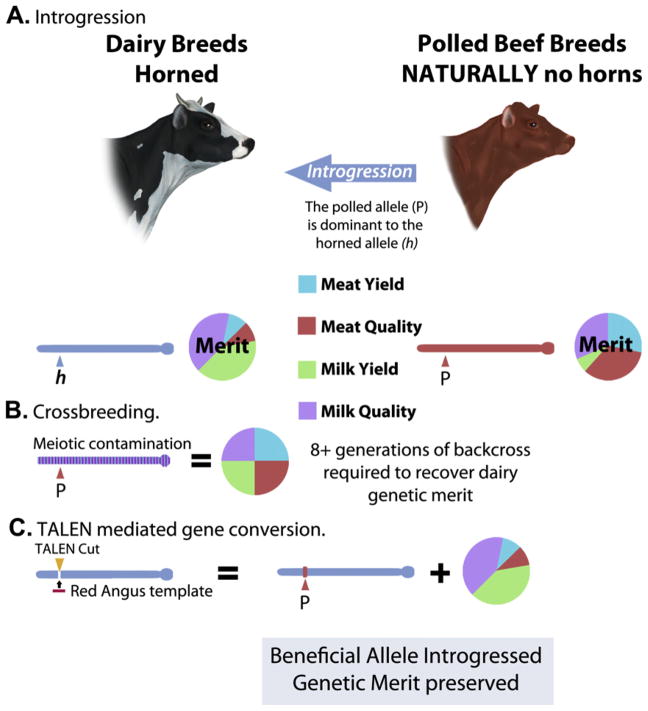
There are numerous livestock breeds that have been extensively selected for a specialized set of traits, i.e. milk yield and composition, meat yield and composition, growth rate, thermotolerance, disease and parasite resistance, etc. Frequently, alleles that would benefit a particular breed are present within the species but exist only in undeveloped breeds or breeds that have historically been selected for traits that differ to those that are of priority in the target breed (e.g. meat vs. milk production). TALEN-based gene conversion may provide an opportunity for transferring beneficial alleles between animals/breeds without disrupting the improved genetic architectures achieved by long-term selection within these breeds. However, traits for which only a few loci account for a large proportion of the observed genetic variance are clearly more attractive targets for this technology (Casas et al., 1999; Grisart et al., 2002) than traits for which a large number of loci contribute only minor magnitudes of effect (Cole et al., 2009; Kemper, Visscher, & Goddard, 2012), such as those that appear to predominate for complex traits.
The example presented in Figure 7 is of particular interest. Holstein cattles have been extensively selected for high milk yield and milk quality. Unfortunately, the great majority of both male and female Holsteins develop horns. To protect the welfare of both dairy farm operators and the cattle themselves, horns are routinely manually removed from the majority of Holstein cattle. Mechanical de-horning is painful, elicits a temporary elevation in animal stress, and adds expense to animal production (Graf & Senn, 1999), and despite the intent of protecting animals from subsequent injury, the practice is viewed by some as inhumane. In contrast, several breeds (e.g., Red Angus, specialized for high quality/yield meat) are naturally horn free, a trait referred to as polled (Fig. 7). The polled trait follows a dominant inheritance pattern (Long & Gregory, 1978) and multiple groups are making progress on identifying the causative mutation (Seichter et al., 2012; J. Taylor, personal communication).
Figure 7. Rapid allele introgression in livestock.
A) The diagram contrasts introgression of desired alleles (polled allele to horned animals) by crossbreeding (panel B) versus TALEN-mediated gene conversion (panel C). Beef and dairy breeds are selected for divergent classes of traits resulting in genetic merit selected for production of meat or milk, respectively. The accumulation of these traits is referred to as the genetic merit of each animal. Crossbreeding mixes these traits, which would result in animals that would not be ideal for either milk or meat production. The trait-selected genome architecture of these animals is conflicted by meiotic contamination, which would require about eight generations of selection to recover the original genetic merit. Panel C shows how TALEN-mediated gene conversion is able to transfer just a desired trait from beef cattle into dairy breeds. In this example, TALENs generate a double-strand DNA break at the horned-polled locus that can be repaired by a homologous template carrying the polled allele from a polled beef breed, e.g., Red Angus. The resulting animal will be both free of horns and maintain the original genetic architecture and merit for milk production. For color version of this figure, the reader is referred to the online version of this book.
Introgression of the polled allele into horned breeds could easily be accomplished by crossbreeding (Fig. 7B); however, the total genetic merit for milk production in the crossbred animals would dramatically suffer. Furthermore, meiotic recombination would mix alleles influencing beef and milk production traits in each crossbred animal that would require numerous generations of backcrossing and intensive genome-wide, marker-assisted selection to recover the original level of quality milk production. During the same period, continued selection for milk production alone within the purebred Holstein population would have created genetic improvement that could never be recovered in the graded-up polled Holstein population. Thus, the inability to transfer a distinct allele from one breed to another translates to significant temporal and economic losses due to the long generation intervals in livestock. However, our results demonstrate that TALEN-mediated homologous recombination can be used to direct efficient allelic introgression in livestock without contamination of untargeted sequences and/or introduction of undesirable traits (authors, unpublished). In the specific case of the polled trait, once the responsible locus is identified, TALEN-mediated homologous recombination could in theory be used to introduce just the polled allele without meiotic contamination (or allelic diffusion) (Fig. 7C). The resulting animals would both lack horns and retain their high genetic merit for milk production.
There are numerous additional examples where TALEN-mediated allelic introgression could benefit animal agriculture. As previously mentioned for humans, each genome harbors 200–300 defective/broken genes in both heterozygous (the majority) and homozygous states. The fact that putative LOF alleles are observed in homozygous states indicates that many of these loci are not lethals, possibly due to functional redundancy with other genes. However, within each individual about seven of these loci are early developmental lethal and many of the others are likely to have deleterious effects on animal productivity and these loci are excellent targets for repair using TALEN-mediated allelic correction. Often, while desired alleles are being accumulated through selection, closely linked defective alleles are perpetuated and even enriched within a population. Causative mutations for at least 62 disease loci have now been determined in cattle and are cataloged at OMIA (http://omia.angis.org.au/home/) (Table 6). Recently, several haplotypes were discovered that affect the fertility in common dairy breeds of cattle including Holstein, Brown Swiss, and Jersey (VanRaden, Olson, Null, & Hutchison, 2011). These haplotypes were identified due to their lack of occurrence in the homozygous state, despite their significant frequency in the population (4.5–25% carriers), which suggests that the homozygous haplotype results in lethality. Given the frequency of predicted LOF alleles from sequence surveys, more examples like this will emerge.
Table 6.
Identified mutations causing disease in cattle
| OMIA entry | Phenotype | Gene | Mutation type | Deviation |
|---|---|---|---|---|
| OMIA 000001 - 9913 | Abortion | APAF1 | SNP | Nonsense |
| OMIA 001565 - 9913 | Abortion and stillbirth | MIMT1 | ~110 kB deletion | |
| OMIA 000593 - 9913 | Acrodermatitis enteropathica | SLC39A4 | SNP | Splice site |
| OMIA 000543 - 9913 | Anhidrotic ectodermal dysplasia | EDA | SNP | Nonsense |
| OMIA 001541 - 9913 | Arachnomelia BTA23 | MOCS1 | 2 nt deletion | Frameshift |
| OMIA 000059 - 9913 | Arachnomelia BTA5 | SUOX | 1 nt INS | Frameshift |
| OMIA 001465 - 9913 | Arthrogryposis multiplex congenita | ISG15 | ~233 kB deletion | |
| OMIA 001106 - 9913 | Axonopathy | MFN2 | SNP | Splice site |
| OMIA 001437 - 9913 | Beta-lactoglobulin aberrant low expression | PAEP | SNP | Enhancer |
| OMIA 000151 - 9913 | Brachyspina | FANCI | 3.3 kB Deletion | |
| OMIA 000161 - 9913 | Cardiomyopathy and woolly haircoat syndrome | PPP1R13L | 7 bp duplication | Frameshift |
| OMIA 000162 - 9913 | Cardiomyopathy dilated | OPA3 | SNP | Nonsense |
| OMIA 000185 - 9913 | Chediak–Higashi syndrome | LYST | SNP | Nonsense |
| OMIA 000187 - 9913 | Chondrodysplasia | EVC2 | SNP and 1 bp deletion | Splice site and frameshift |
| OMIA 000194 - 9913 | Citrullinaemia | ASS1 | SNP | Nonsense |
| OMIA 001340 - 9913 | Complex vertebral malformation | SLC35A3 | SNP | Missense |
| OMIA 001450 - 9913 | Congenital muscular dystonia 1 | ATP2A1 | SNP | Missense |
| OMIA 001451 - 9913 | Congenital muscular dystonia 2 | SLC6A5 | SNP | Missense |
| OMIA 000262 - 9913 | Deficiency of uridine monophosphate synthase | UMPS | SNP | Nonsense |
| OMIA 001680 - 9913 | Dominant white with bilateral deafness | MITF | SNP | Missense |
| OMIA 001485 - 9913 | Dwarfism Angus | PRKG2 | SNP | Nonsense |
| OMIA 001271 - 9913 | Dwarfism Dexter | ACAN | 4 bp INS or SNP | Frameshift |
| OMIA 001473 - 9913 | Dwarfism growth hormone deficiency | GH1 | SNP | Missense |
| OMIA 001686 - 9913 | Dwarfism proportionate with inflammatory lesions | RNF11 | SNP | Splice site |
| OMIA 000327 - 9913 | Ehlers–Danlos syndrome | EPYC | SNP | Missense |
| OMIA 000328 - 9913 | Ehlers–Danlos syndrome type VII (dermatosparaxis) | ADAMTS2 | 17 bp deletion | |
| OMIA 000340 - 9913 | Epidermolysis bullosa | KRT5 | SNP | Missense |
| OMIA 000363 - 9913 | Factor XI deficiency | F11 | 76 bp insertion | |
| OMIA 000419 - 9913 | Glycogen storage disease II | GAA | SNPs | Nonsense and missense |
| OMIA 001139 - 9913 | Glycogen storage disease V | PYGM | SNP | Missense |
| OMIA 000424 - 9913 | Goitre familial | TG | SNP | Nonsense |
| OMIA 000437 - 9913 | Haemophilia A | F8 | SNP | Missense |
| OMIA 000540 - 9913 | Hypotrichosis | HEPHL1 | SNP | Nonsense |
| OMIA 001544 - 9913 | Hypotrichosis with coat-color dilution | PMEL | 3 bp deletion | |
| OMIA 000547 - 9913 | Ichthyosis congenita | ABCA12 | SNP | Missense |
| OMIA 000595 - 9913 | Leukocyte adhesion deficiency type I | ITGB2 | SNP | Missense |
| OMIA 000625 - 9913 | Mannosidosis alpha | MAN2B1 | SNPs | Missense |
| OMIA 000626 - 9913 | Mannosidosis beta | MANBA | SNP | Nonsense |
| OMIA 000627 - 9913 | Maple syrup urine disease | BCKDHA | SNPs | Nonsense |
| OMIA 000628 - 9913 | Marfan syndrome | FBN1 | SNPs | Missense and splice site |
| OMIA 001342 - 9913 | Mucopolysaccharidosis IIIB | NAGLU | SNP | Missense |
| OMIA 000733 - 9913 | Multiple ocular defects | WFDC1 | 1 bp INS | Frameshift |
| OMIA 000683 - 9913 | Muscular hypertrophy (double muscling) | MSTN | Numerous SNPs, 11 bp deletion, 10 bp INS | |
| OMIA 000685 - 9913 | Myasthenic syndrome congenital | CHRNE | 20 bp deletion | |
| OMIA 000689 - 9913 | Myoclonus | GLRA1 | SNP | Nonsense |
| OMIA 001319 - 9913 | Myopathy of the diaphragmatic muscles | HSPA1A | 11 kb deletion | |
| OMIA 001482 - 9913 | Neuronal ceroid lipofuscinosis 5 | CLN5 | 1 bp duplication | Frameshift |
| OMIA 000755 - 9913 | Osteopetrosis | SLC4A2 | 2.8 kb deletion | |
| OMIA 000836 - 9913 | Protoporphyria | FECH | SNP | Stoploss |
| OMIA 001464 - 9913 | Pseudomyotonia congenital | ATP2A1 | SNP | Missense |
| OMIA 001135 - 9913 | Renal dysplasia | CLDN16 | 37 kb or 56 kb deletion | |
| OMIA 001593 - 9913 | Scurs type 2 | TWIST1 | 10 bp duplication | |
| OMIA 001230 - 9913 | Sex reversal: XY female | SRY | Large Deletion | |
| OMIA 001228 - 9913 | Spherocytosis | SLC4A1 | SNP | Nonsense |
| OMIA 001247 - 9913 | Spinal dysmyelination | SPAST | SNP | Missense |
| OMIA 000939 - 9913 | Spinal muscular atrophy | KDSR | SNP | Missense |
| OMIA 000963 - 9913 | Syndactyly (mule foot) | LRP4 | SNP or 2 bp replacement | Splice site or missense |
| OMIA 001452 - 9913 | Tail crooked | MRC2 | 2 bp deletion or SNP | Nonsense or missense |
| OMIA 001003 - 9913 | Thrombopathia | RASGRP2 | SNP | Missense |
| OMIA 001009 - 9913 | Tibial hemimelia | ALX4 | 45.7 kb deletion | |
| OMIA 001360 - 9913 | Trimethylaminuria | FMO3 | SNP | Nonsense |
| OMIA 001079 - 9913 | Yellow fat | BCO2 | SNP | Nonsense |
Management of known disease alleles has traditionally relied on the culling of carriers via marker-assisted elimination from genetic improvement programs. However, given the frequency of such alleles within the population, it seems likely that selection programs will be confounded by linkage disequilibrium between LOF and beneficial alleles. We propose that under these circumstances, the confounding genetic defects may be candidates for correction by TALEN-mediated gene conversion. Indeed, of the 75 mutations for the 62 cattle disease loci described in Online Mendelian Inheritance in Animals website (http://omia.angis.org.au/home/), 87% are either SNPs or small indels of less than 20 bp (Table 6), which are highly likely to be amenable to homology directed allelic correction. Such targetable loci will likely predominate as suggested by deep sequence surveys of numerous species.
Correction either of genetic lesions or the introgression of desirable alleles into livestock must be consistent with the objectives of ongoing genetic improvement programs. This could be achieved by either (1) editing the genomes of animals previously determined to be of significant genetic value or (2) editing the genomes of animals prior to determining their implicit genetic value (Fig. 8). In the case of cloning (Fig 8A), gene-editing would need to be implemented sufficiently quickly to keep pace with ongoing genetic improvement programs. The application of genomic selection is already accelerating genetic improvement by allowing the estimation of genetic merit without the requirement of performance testing. In theory, genetically superior newborn animals could immediately be identified and subjected to gene editing for the correction of an LOF allele or the introgression of desirable alleles that are not already present. This approach provides for a controlled and characterized outcome at every step of the process. Theoretically, there are no limitations in the types and numbers of edits that can be made. Alternatively, since embryo transfer is already part of the genetic improvement paradigm for some livestock (e.g., cattle), editing could be applied by the direct treatment of embryos (Fig 8B). The efficiency of such modifications would need to be sufficiently high to offset any losses in reproductive rate engendered by embryo treatment. In the case of simple gene inactivation, the frequency of success is already very high (75%), with even homozygous modification in 10–20% of embryos (Carlson, Tan, et al., in press). More sophisticated edits have yet to be tested in livestock embryos, but results with ZFNs in mice, rats, and rabbits (Carbery et al., 2010; Flisikowska et al., 2011; Meyer et al., 2010) and with TALENs in zebra fish (Huang et al., 2011; Sander, Cade, et al., 2011) and rodents (Tesson et al., 2011) suggest that even template repair can reach signifcant frequencies in treated embryos. Furthermore, the use of repair templates in association with RecA-mediated sequence searching, alignment, and strand-invasion functions may further increase the number and frequency of gene-editing events in injected embryos. Moreover, precision genome editing can also be used to introduce alleles that do not currently exist within a species by homology-driven allelic substitution. Geneticists working with non-livestock species, e.g., humans, have identified candidate alleles with potential utility in farm animals. There are now the possibilities to create livestock that can be used for disease models as well as enhance agricultural sustainability, food safety, and security. At the current rate of improvement in efficiency, gene editing will be limited only by our imagination.
Figure 8.
Strategies for implementation of allelic introgression. The introgression of desirable alleles into livestock could follow either a vertical (panel A) or a horizontal (panel B) paradigm. (A) In the vertical paradigm, allelic introgression would be performed in cells derived from a donor individual(s) with a high predictability of transmitting ability/estimated breeding value (PTA/EBV, denoted by a blue ribbon). One or several genetic heterozygous or homozygous allele conversions (genetic edits) could be made and verified (e.g., by sequence analysis) prior to cloning of an individual. The resulting animal would not only carry the edits but would also maintain the original PTA/EBV of the donor animal. This animal would be entered back into the genetic improvement program and edits would be selected in subsequent generations. (B) Horizontal implementation takes advantage of the fact that embryo transfer is routine in genetic improvement programs of some livestock species, e.g., cattle. Zygotes produced from animals with high PTA/EBV could be injected with TALENs plus repair templates corresponding to the desired alleles and implanted into a surrogate for establishment or pregnancy. Resulting offspring could be scored for high PTA/EBV and either the presence or the absence of the targeted edits. Animals with high PTA/EBV would be maintained in the genetic improvement program regardless of the edit status, while animals with low PTA/EBV would be culled. Two potential improvements of this process can be envisioned. (1) An embryo biopsy at the blastocysts stage could be collected to evaluate the edit status or PTA/EBV so that only edited and/or high PTA/EBV embryos would be implanted into surrogates. (2) Fetal cells could be collected early in pregnancy by amniocentesis for evaluation of the edit status or PTA/ EBV. Low PTA/EBV or non-edited animals could be culled prior to parturition. Development of these technologies could further accelerate the rate of livestock improvement. In contrast to the vertical paradigm, allelic introgression and genetic improvement will continue to occur in the horizontal paradigm, thereby producing animals that would be one generation ahead in terms of genetic improvement. This method could be easily applied to generate numerous animals from multiple lines such that dissemination of converted alleles (genetic edits) would be accomplished rapidly within a population with minimal risk of inbreeding. For color version of this figure, the reader is referred to the online version of this book.
3.2. Regulatory Issues
Safety to consumers is the primary concern of regulatory as well as agricultural workers and geneticists. Precision genetics clearly will reduce unexpected alterations in genomes compared to those that occurred in the first waves of transgenic animals as well as crops and in human gene therapy. However, no technology is completely free of risk. As previously mentioned, ZFNs have already advanced to human clinical trials (Cannon & June, 2011). Effective gene therapy of humans requires treatment of several million cells and re-implantation into a host. This amplifies the chance of accumulating a deleterious mutation several million fold compared to single genetically modified embryonic cells with genetically edited genomes. The current paradigm for generation and approval of genetically engineered animals either for human consumption or for biological products that will be used in humans or for treatment of human disorders emanates from a single modified cell/embryo. All subsequent animals would be generated from one or a few founder(s). This paradigm offers several opportunities to eliminate mutations that might compromise animal welfare. First, generation of animals by either SCNT or microinjection allows biological selection in culture against compromised genomes prior to delivery to an embryonic environment. Second, animal genomes can be sequenced for less than $5,000 and this cost is rapidly declining (http://www.genome.gov/sequencingcosts/). Since off-target lesions in founder animals would be clonal, their identification by sequencing will become a standard step before the animals are proposed for commercialization. Breeding will allow segregation of any off-target lesions from the desired genetic alteration. In severe cases, afflicted animals would be culled. Fortunately, since the majority of off-target lesions occur at a very limited number of sites that do not have to be in genes, screening for off-target events will be relatively easy to apply to the paradigm described in Fig. 8B.
What are the real risks of consuming GE animals? The first question to answer is what are the feared, not necessarily legitimate, effects of off-target lesions in food animal genomes to human or animal welfare. First, an on- or off-target change could result in a LOF mutation affecting the animal’s welfare (Jackson et al., 2010). In this case, the animal would be culled and not proposed for commercial sale. Second, an on- or off-target lesion could alter a protein’s sequence such that a novel peptide could elicit an immunological response. Actually, nature already runs this experiment. Agricultural animals have genomes similar in size to that in humans and thus should accumulate de novo mutations at a similar rate as humans, i.e., about 40 mutations/ individual/generation. In the case of pigs, about 1.3 billion animals are consumed per year. The accumulated number of consumed mutations per year would then be about 50 billion, corresponding to about 10 changes at every position in the porcine genome per year. Third, an interaction between an untargeted alteration and other factors could produce an unspecified deleterious effect. As mentioned above, each individual genome harbors many thousands of unique SNPs, indels, and copy number variants. There is no way to quantify an unspecified interaction between genetic elements of a sort that have not been seen before. However, whatever the chances might be of a heretofore-unknown genetic interaction having an adverse effect, they are certainly less than known genetic interactions that occur by crossbreeding, which has never been considered to have a negative impact on food safety.
Although a recombinant DNA construct may be considered a drug (FFDCA, 21 U.S.C. 321 et. seq.), the question is whether animals derived through the application of precision genetics also meet the definition. While the process used in precision genetics is different from natural processes by virtue of being man caused, the outcomes obtained through precision genetics, e.g., substituting one naturally occurring allelic form of a gene for another of the same gene or inducing a mutation in an existing gene that is similar to one obtained through classical animal breeding, are the same as those that occur in nature. All scientific evidence suggests that precision genetics should be a method that has far fewer risks than conventional breeding and therefore should be generally regarded as safe (Waltz, 2012).
Acknowledgments
We thank our colleagues in the Center for Genome Engineering for continuing advice and insights into methods for developing precision genetic technologies using chimeric nucleases and transposons. The authors were supported by NIH grants 1R01DK082516 and P01-HD32652 to PBH, and NIH grants 1R43 RR033149, 1R41HL108440, and USDA-NIFA BRAG grant 2012-01628 to SCF. We thank Dr. Jerry Taylor for reading portions of this review. We apologize for the selections we have made that inadvertently left out references to work published by our colleagues throughout the world.
References
- Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nature Reviews Genetics. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnould S, Delenda C, Grizot S, Desseaux C, Pâques F, Silva GH, et al. The I-CreI meganuclease and its engineered derivatives: applications from cell modification to gene therapy. Protein Engineering Design and Selection. 2011;24:27–31. doi: 10.1093/protein/gzq083. [DOI] [PubMed] [Google Scholar]
- Aronovich EL, McIvor RS, Hackett PB. The Sleeping Beauty transposon system: a non-viral vector for gene therapy. Human Molecular Genetics. 2011;20(R1):14–20. doi: 10.1093/hmg/ddr140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguisi A, Behboodi E, Melican DT, Pollock JS, Destrempes MM, Cammuso C, et al. Production of goats by somatic cell nuclear transfer. Nature Biotechnology. 1999;17:456–461. doi: 10.1038/8632. [DOI] [PubMed] [Google Scholar]
- Baldassarre H, Wang B, Kafidi N, Gauthier M, Neveu N, Lapointe J, et al. Production of transgenic goats by pronuclear microinjection of in vitro produced zygotes derived from oocytes recovered by laparoscopy. Theriogenology. 2003;59:831–839. doi: 10.1016/s0093-691x(02)01128-7. [DOI] [PubMed] [Google Scholar]
- Baldassarre H, Wang B, Keefer CL, Lazaris A, Karatzas CN. State of the art in the production of transgenic goats. Reproduction Fertility and Development. 2004;16:465–470. doi: 10.10371/RD04028. [DOI] [PubMed] [Google Scholar]
- Baum C. Parachuting in the epigenome: the biology of gene vector insertion profiles in the context of clinical trials. EMBO Molecular Medicine. 2011;3:75–77. doi: 10.1002/emmm.201000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauman DE, McCutcheon SN, Steinhour WD, Eppard PJ, Sechen SJ. Sources of variation and prospects for improvement of productive efficiency in the dairy cow: a review. Journal of Animal Science. 1985;60:583–592. doi: 10.2527/jas1985.602583x. [DOI] [PubMed] [Google Scholar]
- Beard JW, Sharp DG, Eckert EA, Beard D, Mommaerts EB. Properties of the virus of the fowl erythromyeloblastic disease. Proceedings National Cancer Conference. 1952;2:1396–1411. [Google Scholar]
- Berg P, Baltimore D, Boyer HW, Cohen SN, Davis RW, Hogness DS, et al. Potential biohazards of recombinant DNA molecules. Science. 1974;185:303. [PubMed] [Google Scholar]
- Berg P, Singer MF. The recombinant DNA controversy: twenty years later. Proceeding of the Nationl Academy Sciences of the United States of America. 1995;92:9011–9013. doi: 10.1073/pnas.92.20.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry C, Hannenhalli S, Leipzig J, Bushman FD. Selection of target sites for mobile DNA integration in the human genome. PLoS Computational Biology. 2006;2:e157. doi: 10.1371/journal.pcbi.0020157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beschorner W, Prather R, Sosa C, Thompson SC, Schieber T, Yang T. Transgenic pigs expressing the suicide gene thymidine kinase in the liver. Xeno-transplantation. 2003;10:530. [Google Scholar]
- Bibikova M, Beumer K, Trautman JK, Carroll D. Enhancing gene targeting with designed zinc finger nucleases. Science. 2003;300:764. doi: 10.1126/science.1079512. [DOI] [PubMed] [Google Scholar]
- Bibikova M, Carroll D, Segal DJ, Trautman JK, Smith J, Kim YG, et al. Stimulation of homologous recombination through targeted cleavage by chimeric nucleases. Molecular Cell Biology. 2001;21:289–297. doi: 10.1128/MCB.21.1.289-297.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickhart DM, Hou Y, Schroeder SG, Alkan C, Cardone MF, Matukumalli LK, et al. Copy number variation of individual cattle genomes using next-generation sequencing. Genome Research. 2012;22:778–790. doi: 10.1101/gr.133967.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleck GT, White BR, Miller DJ, Wheeler MB. Production of bovine alpha-lactalbumin in the milk of transgenic pigs. Journal of Animal Science. 1998;76:3072–3078. doi: 10.2527/1998.76123072x. [DOI] [PubMed] [Google Scholar]
- Boch J, Bonas U. Xanthomonas AvrBs3 Family-Type III effectors: discovery and function. Annual Review of Phytopathology. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- Boch J, Scholze H, Schornack S, Landgraf A, Hahn S, Kay S, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- Bogdanove AJ, Voytas DF. TAL efffectors: customizable proteins for DNA targeting. Science. 2011;333:1843–1844. doi: 10.1126/science.1204094. [DOI] [PubMed] [Google Scholar]
- Borlaug NE. Ending world hunger. The promise of biotechnology and the threat of antiscience zealotry. Plant Physiology. 2000;124:487–490. doi: 10.1104/pp.124.2.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosselman RA, Hsu RY, Boggs T, Hu S, Bruszewski J, Ou S, et al. Germline transmission of exogenous genes in the chicken. Science. 1989;243:533–535. doi: 10.1126/science.2536194. [DOI] [PubMed] [Google Scholar]
- Bowen RA, Reed ML, Schnieke A, Seidel GE, Jr, Stacey A, Thomas WK, et al. Transgenic cattle resulting from biopsied embryos: expression of c-ski in a transgenic calf. Biology of Reproduction. 1994;50:664–668. doi: 10.1095/biolreprod50.3.664. [DOI] [PubMed] [Google Scholar]
- Brem G. Inheritance and tissue-specific expression of transgenes in rabbits and pigs. Molecular Reproduction and Devlopment. 1993;36:242–244. doi: 10.1002/mrd.1080360220. [DOI] [PubMed] [Google Scholar]
- Brem G, Brenig B, Muller M, Kranblich H, Winnacker EL. Production of transgenic pigs and possible application to pig breeding. Occasional Publication of the British Society for Animal Production. 1988;12:15. [Google Scholar]
- Brem G, Brenig B, Goodman HM, Selden RC, Graf F, Kruff B, et al. Production of transgenic mice, rabbits and pigs by microinjection into pronuclei. Zuchthygiene. 1985;20:251–252. [Google Scholar]
- Brinster RL, Chen HY, Trumbauer M, Senear AW, Warren R, Palmiter RD. Somatic expression of herpes thymidine kinase in mice following injection of a fusion gene into eggs. Cell. 1981;27:223–231. doi: 10.1016/0092-8674(81)90376-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brophy B, Smolenski G, Wheeler T, Wells D, L’Huillier P, Laible G. Cloned transgenic cattle produce milk with higher levels of beta-casein and kappa-casein. Nature Biotechnology. 2003;21:157–162. doi: 10.1038/nbt783. [DOI] [PubMed] [Google Scholar]
- Bucher P, Morel P, Buhler LH. Xenotransplantation: an update on recent progress and future perspectives. Transplant International. 2005;18:894–901. doi: 10.1111/j.1432-2277.2005.00124.x. [DOI] [PubMed] [Google Scholar]
- Butler SP, van Cott K, Subrumanian A, Gwazduaskas FC, Velander WH. Current progress in the production of recombinant human fibrinogen in the milk of transgenic animals. Thrombosis and Haemostasis. 1997;78:537–542. [PubMed] [Google Scholar]
- Cabot RA, Kuhholzer B, Chan AW, Lai L, Park KW, Chong KY, et al. Transgenic pigs produced using in vitro matured oocytes infected with a retroviral vector. Animal Biotechnology. 2001;12:205–214. doi: 10.1081/ABIO-100108347. [DOI] [PubMed] [Google Scholar]
- Cannon P, June C. Current Opinion in HIV and AIDS. 2011;6:74–79. doi: 10.1097/COH.0b013e32834122d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbery ID, Ji D, Harrington A, Brown V, Weinstein EJ, Liaw L, et al. Targeted genome modification in mice using zinc-finger nucleases. Genetics. 2010;186:451–459. doi: 10.1534/genetics.110.117002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Fahrenkrug SC, Hackett PB. Targeting DNA with fingers and TALENs. Molecular Therory Nucleic Acids. 2012;1:e3. doi: 10.1038/mtna.2011.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Garbe JR, Tan W, Martin MJ, Dobrinsky JR, Muir WM, et al. Strategies for selection marker-free swine transgenesis using the Sleeping Beauty transposon system. Transgenic Research. 2011;20:1125–1137. doi: 10.1007/s11248-010-9481-7. [DOI] [PubMed] [Google Scholar]
- Carlson DF, Geurts AM, Garbe JR, Park CW, Rangle-Filho A, O’Grady SM, et al. Efficient mammalian germline transgenesis by cis-enhanced Sleeping Beauty transposition. Transgenic Research. 2011;20:29–45. doi: 10.1007/s11248-010-9386-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson DF, Tan W, Lillico SG, Stverakova D, Proudfoot C, Christian M, et al. Efficient TALEN-mediated Gene Knockout Livestock; Towards a Porcine Model of Atherosclerosis. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll D. Zinc-finger nucleases: a panoramic view. Current Gene Therapy. 2011;11:2–10. doi: 10.2174/156652311794520076. [DOI] [PubMed] [Google Scholar]
- Casas E, Keele JW, Fahrenkrug SC, Smith TP, Cundiff LV, Stone RT. Quantitative analysis of birth, weaning, and yearling weights and calving difficulty in Piedmontese crossbreds segregating an inactive myostatin allele. Journal of Animal Science. 1999;77:1686–1689. doi: 10.2527/1999.7771686x. [DOI] [PubMed] [Google Scholar]
- Cermak T, Doyle EL, Christian M, Wang L, Zhang Y, Schmidt C, et al. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Research. 2011;39:e82. doi: 10.1093/nar/gkr218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chames P, Epinat JC, Guillier S, Patin A, Lacroix E, Paques F. In vivo selection of engineered homing endonuclease using double-strand break induced homologous recombination. Nucleic Acids Research. 2005;33:e178. doi: 10.1093/nar/gni175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan AW, Homan EJ, Ballou LU, Burns JC, Bremel RD. Transgenic cattle produced by reverse-transcribed gene transfer in oocytes. Proceeding of the Nationl Academy Sciences of the United States of America. 1998;95:14028–14033. doi: 10.1073/pnas.95.24.14028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang K, Qian J, Jiang M, Liu YH, Wu MC, Chen CD, et al. Effective generation of transgenic pigs and mice by linker based sperm-mediated gene transfer. BMC Biotechnology. 2002;2:5. doi: 10.1186/1472-6750-2-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Li L, Pang D, Li Z, Wang T, Zhang M, et al. Construction of transgenic swine with induced expression of Cre recombinase. Animal. 2010;4:767–771. doi: 10.1017/S1751731109991571. [DOI] [PubMed] [Google Scholar]
- Chen SH, Vaught TD, Monahan JA, Boone J, Emslie E, Jobst PM, et al. Efficient production of transgenic cloned calves using preimplantation screening. Biology of Reproduction. 2002;67:1488–1492. doi: 10.1095/biolreprod.102.006981. [DOI] [PubMed] [Google Scholar]
- Cho SK, Hwang KC, Choi YJ, Bui HT, Nguyen VT, Park C, et al. Production of transgenic pigs harboring the human erythropoietin (hEPO) gene using somatic cell nuclear transfer. The Journal of Reproduction and Development. 2009;55:128–136. doi: 10.1262/jrd.20102. [DOI] [PubMed] [Google Scholar]
- Choulika A, Perrin A, Dujon B, Nicolas JF. Induction of homologous recombination in mammalian chromosomes by using the I-SceI system of Saccharomyces cerevisiae. Molecular Cell Biology. 1995;15:1968–1973. doi: 10.1128/mcb.15.4.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M, Cermak T, Doyle EL, Schmidt C, Zhang F, Hummel A, et al. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibelli JB, Stice SL, Golueke PJ, Kane JJ, Jerry J, Blackwell C, et al. Cloned transgenic calves produced from nonquiescent fetal fibroblasts. Science. 1998;280:1256–1258. doi: 10.1126/science.280.5367.1256. [DOI] [PubMed] [Google Scholar]
- Clark J, Whitelaw B. A future for transgenic livestock. Nature Reviews Genetics. 2003;4:825–833. doi: 10.1038/nrg1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KJ, Carlson DF, Foster LK, Kong BW, Foster DN, Fahrenkrug SC. Enzymatic engineering of the porcine genome with transposons and recombinases. BMC Biotechnology. 2007;7:42. doi: 10.1186/1472-6750-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clements JE, Wall RJ, Narayan O, Hauer D, Schoborg R, Sheffer D, et al. Development of transgenic sheep that express the visna virus envelope gene. Virology. 1994;200:370–380. doi: 10.1006/viro.1994.1201. [DOI] [PubMed] [Google Scholar]
- Cline MJ, Stang H, Mercola K, Morse L, Ruprecht R, Brown J, et al. Gene transfer in intact animals. Nature. 1980;284:422–425. doi: 10.1038/284422a0. [DOI] [PubMed] [Google Scholar]
- Cohen SN, Chang AC, Boyer HW, Helling RB. Construction of biologically functional bacterial plasmids in vitro. Proceeding of the Nationl Academy Sciences of the United States of America. 1973;70:3240–3244. doi: 10.1073/pnas.70.11.3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JB, VanRaden PM, O’Connell JR, Van Tassell CP, Sonstegard TS, Schnabel RD, et al. Distribution and location of genetic effects for dairy traits. Journal of Dairy Science. 2009;92:2931–2946. doi: 10.3168/jds.2008-1762. [DOI] [PubMed] [Google Scholar]
- Cooper DK. Clinical xenotransplantion–how close are we? Lancet. 2003;362:557–559. doi: 10.1016/S0140-6736(03)14118-9. [DOI] [PubMed] [Google Scholar]
- Crittenden LB, Salter DW. Genetic engineering to improve resistance to viral diseaes of poultry: a model for application to livestock improvement. Canadian Journal of Animal Science. 1985;65:553–562. [Google Scholar]
- Crittenden LB, Salter DW. Gene insertion: current progress and long-term goals. Avian Disease. 1986;30:43–46. [PubMed] [Google Scholar]
- Cui Z, Yang Y, Kaufman CD, Agalliu A, Hackett PB. RecA-mediated, targeted mutagenesis in zebrafish. Marine Biotechnology. 2003;5:174–184. doi: 10.1007/s10126-002-0059-0. [DOI] [PubMed] [Google Scholar]
- Dai Y, Vaught TD, Boone J, Chen SH, Phelps CJ, Ball S, et al. Targeted disruption of the alpha1,3-galactosyltransferase gene in cloned pigs. Nature Biotechnology. 2002;20:251–255. doi: 10.1038/nbt0302-251. [DOI] [PubMed] [Google Scholar]
- Damak S, Jay NP, Barrell GK, Bullock DW. Targeting gene expression to the wool follicle in transgenic sheep. Biotechnology (Nature Publishing Company) 1996a;14:181–184. doi: 10.1038/nbt0296-181. [DOI] [PubMed] [Google Scholar]
- Damak S, Su H, Jay NP, Bullock DW. Improved wool production in transgenic sheep expressing insulin-like growth factor 1. Biotechnology (Nature Publishing Company) 1996b;14:185–188. doi: 10.1038/nbt0296-185. [DOI] [PubMed] [Google Scholar]
- Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu JK, et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335(6069):720–723. doi: 10.1126/science.1215670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Yang D, Zhao B, Ouyang Z, Song J, Fan N, et al. Use of the 2A peptide for generation of multi-transgenic pigs through a single round of nuclear transfer. PLoS One. 2011;6:e19986. doi: 10.1371/journal.pone.0019986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denning C, Burl S, Ainslie A, Bracken J, Dinnyes A, Fletcher J, et al. Deletion of the alpha(1,3)galactosyl transferase (GGTA1) gene and the prion protein (PrP) gene in sheep. Nature Biotechnology. 2001;19:559–562. doi: 10.1038/89313. [DOI] [PubMed] [Google Scholar]
- Desjarlais JR, Berg JM. Use of a zinc-finger consensus sequence framework and specificity rules to design specific DNA binding proteins. Proceeding of the Nationl Academy Sciences of the United States of America. 1993;90:2256–2260. doi: 10.1073/pnas.90.6.2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin RH, Sakhrani D, Tymchuk WE, Rise ML, Goh B. Domestication and growth hormone transgenesis cause similar changes in gene expression in coho salmon (Oncorhynchus kisutch) Proceeding of the Nationl Academy Sciences of the United States of America. 2009;106:3047–3052. doi: 10.1073/pnas.0809798106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckhoff B, Petersen B, Kues WA, Kurth R, Niemann H, Denner J. Knockdown of porcine endogenous retrovirus (PERV) expression by PERV-specific shRNA in transgenic pigs. Xenotransplantation. 2008;15:36–45. doi: 10.1111/j.1399-3089.2008.00442.x. [DOI] [PubMed] [Google Scholar]
- Doyon Y, Vo TD, Mendel MC, Greenberg SG, Wang J, Xia DF, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. PLoS One. 2011;8:74–79. doi: 10.1038/nmeth.1539. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Clark KJ, Carlson CM, Fritz S, Davidson AE, Markley KM, et al. Mammalian germ-line transgenesis by transposition. Proceeding of the Nationl Academy Sciences of the United States of America. 2002;99:4495–4499. doi: 10.1073/pnas.062630599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert KM, Low MJ, Overstrom EW, Buonomo FC, Baile CA, Roberts TM, et al. A Moloney MLV-rat somatotropin fusion gene produces biologically active somatotropin in a transgenic pig. Molecular Endocrinology. 1988;2:277–283. doi: 10.1210/mend-2-3-277. [DOI] [PubMed] [Google Scholar]
- Ebert KM, Selgrath JP, DiTullio P, Denman J, Smith TE, Memon MA, et al. Transgenic production of a variant of human tissue-type plasminogen activator in goat milk Generation of transgenic goats and analyses of expression. Biotechnology. 1991;9:835–838. doi: 10.1038/nbt0991-835. [DOI] [PubMed] [Google Scholar]
- Ebert KM, Smith TE, Buonoma FC, Overstrom EW, Low EJ. Porcine growth hormone gene expression from viral promoters in transgenic swine. Animal Biotechnology. 1990;1:145–159. [Google Scholar]
- Ellinwood NM, Vite CH, Haskins ME. Gene therapy for lysosomal storage diseases: the lessons and promise of animal models. The Journal of Gene Medicine. 2004;6:481–506. doi: 10.1002/jgm.581. [DOI] [PubMed] [Google Scholar]
- Erickson RP. Mouse models of human genetic disease: which mouse is more like man? BioEssays. 1996;18:993–998. doi: 10.1002/bies.950181209. [DOI] [PubMed] [Google Scholar]
- Fahrenkrug SC, Blake A, Carlson DF, Doran T, Van Eenennaam A, Faber D, et al. Precision genetics for complex objectives in animal agriculture. Journal of Animal Science. 2010;88:2530–2539. doi: 10.2527/jas.2010-2847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff N, Haselkorn R, Chassy BM. EPA’s proposed biotech policy turns a deaf ear to science. FASEB Journal. 2011;25:2855–2857. doi: 10.1096/fj.11-0901ufm. [DOI] [PubMed] [Google Scholar]
- Fishman JA, Patience C. Xenotransplantation: infectious risk revisited. American Journal of Transplantation. 2004;4:1383–1390. doi: 10.1111/j.1600-6143.2004.00542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flisikowska T, Thorey IS, Ofner S, Ros F, LIdkw V, Zeitier B, et al. Efficient immunoglobulin gene disruption and targeted replacement in rabbit using zinc finger nucleases. PLoS One. 2011;6:e2105. doi: 10.1371/journal.pone.0021045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fodor WL, Williams BL, Matis LA, Madri JA, Rollins SA, Knight JW, et al. Expression of a functional human complement inhibitor in a transgenic pig as a model for the prevention of xenogeneic hyperacute organ rejection. Proceeding of the Nationl Academy Sciences of the United States of America. 1994;91:11153–11157. doi: 10.1073/pnas.91.23.11153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley JA, Ramankutty N, Brauman KA, Cassidy ES, Gerber JS, Johnston M, et al. Solutions for a cultivated planet. Nature. 2011;478:337–342. doi: 10.1038/nature10452. [DOI] [PubMed] [Google Scholar]
- Freitas VJF, Serova IA, Moura RR, Andreeva LE, Melo LM, Teixeira DIA, et al. The establishment of two transgenic goat lines for mammary gland hG-CSF expression. Small Ruminant Research. 2012;105:105–113. [Google Scholar]
- Gabriel R, Lombardo A, Arens A, Miller JC, Genovese P, Kaeppel C, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nature Biotechnology. 2011;29:816–823. doi: 10.1038/nbt.1948. [DOI] [PubMed] [Google Scholar]
- Garcia-Vazquez FA, Ruiz S, Matas C, Izquierdo-Rico MJ, Grullon LA, De Ondiz A, et al. Production of transgenic piglets using ICSI-sperm-mediated gene transfer in combination with recombinase RecA. Reproduction. 2010;140:259–272. doi: 10.1530/REP-10-0129. [DOI] [PubMed] [Google Scholar]
- Garrels W, Mátés L, Holler S, Dalda A, Taylor U, Petersen B, et al. Germline transgenic pigs by Sleeping Beauty transposition in porcine zygotes and targeted integration in the pig genome. PLoS One. 2011;6:e23573. doi: 10.1371/journal.pone.0023573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfray HC, Beddington JR, Crute IR, Haddad L, Lawrence D, Muir JF, et al. Food security: the challenge of feeding 9 billion people. Science. 2010;327:812–818. doi: 10.1126/science.1185383. [DOI] [PubMed] [Google Scholar]
- Golding MC, Long CR, Carmell MA, Hannon GJ, Westhusin ME. Suppression of prion protein in livestock by RNA interference. Proceeding of the Nationl Academy Sciences of the United States of America. 2006;103:5285–52890. doi: 10.1073/pnas.0600813103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovan SP, Meidinger RG, Ajakaiye A, Cottrill M, Wiederkehr MZ, Barney DJ, et al. Pigs expressing salivary phytase produce low-phosphorus manure. Nature Biotechnology. 2001;19:741–745. doi: 10.1038/90788. [DOI] [PubMed] [Google Scholar]
- Gordon JW, Ruddle FH. Integration and stable germ line transmission of genes injected into mouse pronuclei. Science. 1981;214:1244–1246. doi: 10.1126/science.6272397. [DOI] [PubMed] [Google Scholar]
- Gordon JW, Ruddle FH. Germ line transmission in transgenic mice. Progress in Clinical and Biological Research. 1982;85(Pt. B):111–124. [PubMed] [Google Scholar]
- Gordon JW, Scangos GA, Plotkin DJ, Barbosa JA, Ruddle FH. Genetic transformation of mouse embryos by microinjection of purified DNA. Proceeding of the Nationl Academy Sciences of the United States of America. 1980;77:7380–7384. doi: 10.1073/pnas.77.12.7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabundzija I, Irgang M, Mátés L, Belay E, Matrai J, Gogol-Doring A, et al. Comparative analysis of transposable element vector systems in human cells. Molecular Therapy. 2010;18:1200–1209. doi: 10.1038/mt.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf B, Senn M. Behavioural and physiological responses of calves to dehorning by heat cauterisation with or without local anaesthesia. Applied Animal Behaviour Science. 1999;62:153–171. [Google Scholar]
- Grisart B, Coppieters W, Farnir F, Karim L, Ford C, Berzi P, et al. Positional candidate cloning of a QTL in dairy cattle: identification of a missense mutation in the bovine DGAT1 gene with major effect on milk yield and composition. Genome Research. 2002;12:222–231. doi: 10.1101/gr.224202. [DOI] [PubMed] [Google Scholar]
- Grosse-Hovest L, Muller S, Minoia R, Wolf E, Zakhartchenko V, Wenigerkind H, et al. Cloned transgenic farm animals produce a bispecific antibody for T cell-mediated tumor cell killing. Proceeding of the Nationl Academy Sciences of the United States of America. 2004;101:6858–6863. doi: 10.1073/pnas.0308487101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett PB. The molecular biology of transgenic fish. In: Hochachka P, Mommsen T, editors. Biochemistry and Molecular Biology of Fishes. Amsterdam: Elsevier Science Publishers B.V; 1993. pp. 207–240. [Google Scholar]
- Hackett PB, Alvarez MC. The molecular genetics of transgenic fish. Recent Advances Marine Biotechnology. 2000;4:77–145. [Google Scholar]
- Hackett PB, Ekker SC, Largaespada DA, McIvor RS. Sleeping Beauty transposon-mediated gene therapy for prolonged expression. Advances in Genetics. 2005;54:187–229. doi: 10.1016/S0065-2660(05)54009-4. [DOI] [PubMed] [Google Scholar]
- Hackett PB, Largaespada DA, Cooper LJN. A transposon and transposase system for human application. Molecular Therapy. 2010;18:674–683. doi: 10.1038/mt.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halter R, Carnwath J, Espanion G, Herrmann D, Lemme E, Niemann H, et al. Strategies to express factor VIII gene constructs in the ovine mammary gland. Theriogenology. 1993;39:137–149. [Google Scholar]
- Hammer RE, Pursel VG, Rexroad CE, Jr, Wall RJ, Bolt DJ, Ebert KM, et al. Production of transgenic rabbits, sheep and pigs by microinjection. Nature. 1985;315:680–683. doi: 10.1038/315680a0. [DOI] [PubMed] [Google Scholar]
- Hao YH, Yong HY, Murphy CN, Wax D, Samuel M, Rieke A, et al. Production of endothelial nitric oxide synthase (eNOS) over-expressing piglets. Transgenic Research. 2006;15:739–750. doi: 10.1007/s11248-006-9020-8. [DOI] [PubMed] [Google Scholar]
- Haskell RE, Bowen RA. Efficient production of transgenic cattle by retroviral infection of early embryos. Molecular Reproduction and Development. 1995;40:386–390. doi: 10.1002/mrd.1080400316. [DOI] [PubMed] [Google Scholar]
- Haskins ME, Desnick RJ, DiFerrante N, Jezyk PF, Patterson DF. Beta-glucuronidase deficiency in a dog: a model of human mucopolysaccharidosis VII. Pediatric Research. 1984;18:980–984. doi: 10.1203/00006450-198410000-00014. [DOI] [PubMed] [Google Scholar]
- Hauschild J, Petersen B, Santiago Y, Queisser AL, Carnwath JW, Lucas-Hahn A, et al. Efficient generation of a biallelic knockout in pigs using zinc-finger nucleases. Proceeding of the Nationl Academy Sciences of the United States of America. 2011;108:12013–12017. doi: 10.1073/pnas.1106422108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill KG, Curry J, DeMayo FJ, Jones-Diller K, Slapak JR, Bondioli KR. Production of transgenic cattle by pronuclear injection. Theriogenology. 1992;37:222. (Abstr.) [Google Scholar]
- Hirabayashi M, Takahashi R, Ito K, Kashiwazaki N, Hirao M, Hirasawa K, et al. A comparative study on the integration of exogenous DNA into mouse, rat, rabbit, and pig genomes. Experimental Animal. 2001;50:125–131. doi: 10.1538/expanim.50.125. [DOI] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nature Biotechnology. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, Cassady JP, et al. Genetic engineering of human pluripotent cells using TALE nucleases. Nature Biotechnology. 2011;29:731–734. doi: 10.1038/nbt.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Kabermann A, Brem G, Wolf E, et al. Epigenetic regulation of lentiviral transgene vectors in a large animal model. Molecular Therapy. 2006;13:59–66. doi: 10.1016/j.ymthe.2005.07.685. [DOI] [PubMed] [Google Scholar]
- Hofmann A, Kessler B, Ewerling S, Weppert M, Vogg B, Ludwig H, et al. Efficient transgenesis in farm animals by lentiviral vectors. EMBO Reports. 2003;4:1054–1060. doi: 10.1038/sj.embor.7400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann A, Zakhartchenko V, Weppert M, Sebald H, Wenigerkind H, Brem G, et al. Generation of transgenic cattle by lentiviral gene transfer into oocytes. Biology of Reproduction. 2004;71:405–409. doi: 10.1095/biolreprod.104.028472. [DOI] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. Heritable gene targeting in zebrafish using customized TALENs. Nature Biotechnology. 2011;29:699–700. doi: 10.1038/nbt.1939. [DOI] [PubMed] [Google Scholar]
- Hutchison WD, Burkness EC, Mitchell PD, Moon RD, Leslie TW, Fleischer SJ, et al. Areawide suppression of European corn borer with Bt maize reaps savings to non-Bt maize growers. Science. 2010;330:222–225. doi: 10.1126/science.1190242. [DOI] [PubMed] [Google Scholar]
- Hyttinen JM, Peura T, Tolvanen M, Aalto J, Alhonen L, Sinervirta R, et al. Generation of transgenic dairy cattle from transgene-analyzed and sexed embryos produced in vitro. Biotechnology (Nature Publishing Company) 1994;12:606–608. doi: 10.1038/nbt0694-606. [DOI] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, et al. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell. 2010;141:1253–1261. doi: 10.1016/j.cell.2010.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivarie R. Avian transgenesis: progress towards the promise. Trends in Biotechnology. 2003;21:14–19. doi: 10.1016/s0167-7799(02)00009-4. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Hackett PB, Plasterk RH, Izsvak Z. Molecular reconstruction of Sleeping Beauty, a Tc1-like transposon from fish, and its transposition in human cells. Cell. 1997;91:501–510. doi: 10.1016/s0092-8674(00)80436-5. [DOI] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mátés L, Boeke JD, Nagy A, Bradley A, et al. Transposon-mediated genome manipulation in vertebrates. Nature Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson KA, Berg JM, Murray JD, Maga EA. Evaluating the fitness of human lysozyme transgenic dairy goats: growth and reproductive traits. Transgenic Research. 2010;19:977–986. doi: 10.1007/s11248-010-9371-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenisch R. Transgenic animals. Science. 1988;240:1468–1474. doi: 10.1126/science.3287623. [DOI] [PubMed] [Google Scholar]
- Jakobsen JE, Li J, Kragh PM, Moldt B, Lin L, Liu Y, et al. Pig transgenesis by Sleeping Beauty DNA transposition. Transgenic Research. 2011;20:533–545. doi: 10.1007/s11248-010-9438-x. [DOI] [PubMed] [Google Scholar]
- Jamieson AC, Wang H, Kim SH. A zinc finger directory for high-affinity DNA recognition. Proceeding of the Nationl Academy Sciences of the United States of America. 1966;93:12834–12839. doi: 10.1073/pnas.93.23.12834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasin M. Genetic manipulation of genomes with rare-cutting endonucleases. Trends in Genetics. 1996;12:224–228. doi: 10.1016/0168-9525(96)10019-6. [DOI] [PubMed] [Google Scholar]
- Kemper KE, Visscher PM, Goddard ME. Genetic architecture of body size in mammals. Genome Biology. 2012;13:244. doi: 10.1186/gb-2012-13-4-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd JM, Graves T, Newman TL, Fulton R, Hayden HS, Malig M, et al. A human genome structural variation sequencing resource reveals insights into mutational mechanisms. Cell. 2010;143:837–847. doi: 10.1016/j.cell.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Research. 2009;19:1279–1288. doi: 10.1101/gr.089417.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proceeding of the Nationl Academy Sciences of the United States of America. 1996;93:1156–1160. doi: 10.1073/pnas.93.3.1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A. The discovery of zinc fingers and their applications in gene regulation and genome manipulation. Annual Review of Biochemistry. 2010;79:213–231. doi: 10.1146/annurev-biochem-010909-095056. [DOI] [PubMed] [Google Scholar]
- Klymiuk N, Aigner B, Brem G, Wolf E. Genetic modification of pigs as organ donors for xenotransplantation. Molecular Reproduction and Development. 2010;77:209–221. doi: 10.1002/mrd.21127. [DOI] [PubMed] [Google Scholar]
- Knight J. Glofish casts light on murky policing of transgenic animals. Nature. 2003;426:372. doi: 10.1038/426372b. [DOI] [PubMed] [Google Scholar]
- Koeberl DD, Pinto C, Brown T, Chen YT. Gene therapy for inhereted metabolic disorders in companion animals. ILAR Journal. 2009;50:122–127. doi: 10.1093/ilar.50.2.122. [DOI] [PubMed] [Google Scholar]
- Krimpenfort P, Rademakers A, Eyestone W, van der Schans A, van den Broek S, Kooiman P, et al. Generation of transgenic dairy cattle using ‘in vitro’ embryo production. Biotechnology (Nature Publishing Company) 1991;9:844–847. doi: 10.1038/nbt0991-844. [DOI] [PubMed] [Google Scholar]
- Kues WA, Niemann H. The contribution of farm animals to human health. Trends in Biotechnology. 2004;22:286–294. doi: 10.1016/j.tibtech.2004.04.003. [DOI] [PubMed] [Google Scholar]
- Kues WA, Schwinzer R, Wirth D, Verhoeyen E, Lemme E, Herrmann D, et al. Epigenetic silencing and tissue independent expression of a novel tetracycline inducible system in double-transgenic pigs. FASEB Journal. 2006;20:1200–1202. doi: 10.1096/fj.05-5415fje. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kasinathan P, Choi YJ, Naeem R, Tomizuka K, Sullivan EJ, et al. Cloned transchromosomic calves producing human immunoglobulin. Nature Biotechnology. 2002;20:889–894. doi: 10.1038/nbt727. [DOI] [PubMed] [Google Scholar]
- Kuroiwa Y, Kasinathan P, Matsushita H, Sathiyaselan J, Sullivan EJ, Kakitani M, et al. Sequential targeting of the genes encoding immunoglobulin-mu and prion protein in cattle. Nature Genetics. 2004;36:775–780. doi: 10.1038/ng1373. [DOI] [PubMed] [Google Scholar]
- Kurome M, Ishikawa T, Tomii R, Ueno S, Shimada A, Yazawa H, et al. Production of transgenic and non-transgenic clones in miniature pigs by somatic cell nuclear transfer. The Journal of Reproduction and Development. 2008;54:156–163. doi: 10.1262/jrd.19165. [DOI] [PubMed] [Google Scholar]
- Kustikova OS, Schiedlmeier B, Brugman MH, Stahlhut M, Bartels S, Li Z, et al. Cell-intrinsic and vector-related properties cooperate to determine the incidence and consequences of insertional mutagenesis. Molecular Therapy. 2009;17:1537–1547. doi: 10.1038/mt.2009.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmuk KN, Schook LB. Pigs as a model for biomedical sciences. In: Rothschild MF, Ruvinsky A, editors. The Genetics of the Pig. CABI; 2011. pp. 426–444. [Google Scholar]
- Lacy E, Roberts S, Evans EP, Burtenshaw MD, Costantini FD. A foreign beta-globin gene in transgenic mice: integration at abnormal chromosomal positions and expression in inappropriate tissues. Cell. 1983;34:343–358. doi: 10.1016/0092-8674(83)90369-0. [DOI] [PubMed] [Google Scholar]
- Lai L, Kang JX, Li R, Wang J, Witt WT, Yong HY, et al. Generation of cloned transgenic pigs rich in omega-3 fatty acids. Nature Biotechnology. 2006;24:435–436. doi: 10.1038/nbt1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Kolber-Simonds D, Park KW, Cheong HT, Greenstein JL, Im GS, et al. Production of alpha-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science. 2002a;295:1089–1092. doi: 10.1126/science.1068228. [DOI] [PubMed] [Google Scholar]
- Lai L, Park KW, Cheong HT, Kuhholzer B, Samuel M, Bonk A, et al. Transgenic pig expressing the enhanced green fluorescent protein produced by nuclear transfer using colchicine-treated fibroblasts as donor cells. Molecular Reproduction and Development. 2002b;62:300–306. doi: 10.1002/mrd.10146. [DOI] [PubMed] [Google Scholar]
- Lam KN, van Bakel H, Cote AG, van der Ven A, Hughes TR. Sequence specificity is obtained from the majority of modular C2H2 zinc-finger arrays. Nucleic Acids Research. 2011;39:4680–4690. doi: 10.1093/nar/gkq1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamppa G, Nagy F, Chua NH. Light-regulated and organ-specific expression of a wheat Cab gene in transgenic tobacco. Nature. 1985;316:750–752. doi: 10.1038/316750a0. [DOI] [PubMed] [Google Scholar]
- Lavitrano M, Bacci ML, Forni M, Lazzereschi D, Di Stefano C, Fioretti D, et al. Efficient production by sperm-mediated gene transfer of human decay accelerating factor (hDAF) transgenic pigs for xenotransplantation. Proceeding of the Nationl Academy Sciences of the United States of America. 2002;99:14230–14235. doi: 10.1073/pnas.222550299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CS, Lee DS, Fang NZ, Oh KB, Shin ST, Lee KK. Integration and expression of goat β-casein/hGH hybrid gene in a transgenic goat. Reproductive Developmental Biology. 2006;30:293–299. [Google Scholar]
- Lee HG, Lee HC, Kim SW, Lee P, Chung HJ, Lee YK, et al. Production of recombinant human von Willebrand factor in the milk of transgenic pigs. The Journal of Reproduction and Development. 2009;55:484–490. doi: 10.1262/jrd.20212. [DOI] [PubMed] [Google Scholar]
- Lee JW, Wu SC, Tian XC, Barber M, Hoagland T, Riesen J, et al. Production of cloned pigs by whole-cell intracytoplasmic microinjection. Biology of Reproduction. 2003;69:995–1001. doi: 10.1095/biolreprod.103.015917. [DOI] [PubMed] [Google Scholar]
- Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Research. 2011;39:359–372. doi: 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, et al. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Research. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao HK, Essner JJ. Use of RecA fusion proteins to induce genomic modifications in zebrafish. Nucleic Acids Research. 2011;39:4166–4179. doi: 10.1093/nar/gkq1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu ZH, Song J, Wang ZK, Tian JT, Kong QR, Zheng Z, et al. Green fluorescent protein (GFP) transgenic pig produced by somatic cell nuclear transfer. Chinese Science Bulletin. 2008;53:1035–1039. [Google Scholar]
- Lo D, Pursel V, Linton PJ, Sandgren E, Behringer R, Rexroad C, et al. Expression of mouse IgA by transgenic mice, pigs and sheep. European Journal of Immunology. 1991;21:1001–1006. doi: 10.1002/eji.1830210421. [DOI] [PubMed] [Google Scholar]
- Long CR, Gregory KE. Inheritance of the horned, scurred, and polled condition in cattle. Journal of Heredity. 1978;69:395–400. [Google Scholar]
- Lunney JK. Advances in swine biomedical model genomics. International Journal of Biological Science. 2007;3:179–184. doi: 10.7150/ijbs.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald J, Taylor L, Sherman A, Kawakami K, Takahashi Y, Sang HM, et al. Efficient genetic modification and germ-line transmission of primordial germ cells using piggyBac and Tol2 transposons. Proceeding of the Nationl Academy Sciences of the United States of America. 2012;109:E1466–E1477. doi: 10.1073/pnas.1118715109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Molecular Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maga EA, Sargent RG, Zeng H, Pati S, Zarling DA, Oppenheim SM, et al. Increased efficiency of transgenic livestock production. Transgenic Research. 2003;12:485–496. doi: 10.1023/a:1024257906647. [DOI] [PubMed] [Google Scholar]
- Mahfouz MM, Li L, Shamimuzzaman M, Wibowo A, Fang X, Zhu JK. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proceeding of the Nationl Academy Sciences of the United States of America. 2011;108:2623–2628. doi: 10.1073/pnas.1019533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mak AN, Bradley P, Cernadas RA, Bogdanove AJ, Stoddard BL. The crystal structure of TAL effector PthXo1 bound to its DNA target. Science. 2012;335:716–719. doi: 10.1126/science.1216211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansour SL, Thomas KR, Capecchi MR. Disruption of the proto-oncogene int-2 in mouse embryo-derived stem cells: a general strategy for targeting mutations to non-selectable genes. Nature. 1988;336:348–352. doi: 10.1038/336348a0. [DOI] [PubMed] [Google Scholar]
- Marth GT, Yu F, Indap AR, Garimella K, Gravel S, Leong WF, et al. The functional spectrum of low-frequency coding variation. Genome Biology. 2011;12:R84. doi: 10.1186/gb-2011-12-9-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B, Ji S, Maudsley S, Mattson MP. “Control” laboratory rodents are metabolically morbid: why it matters. Proceeding of the Nationl Academy Sciences of the United States of America. 2010;107:6127–6133. doi: 10.1073/pnas.0912955107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin C, Plat M, Nerriere-Daguin V, Coulon F, Uzbekova S, Venturi E, et al. Transgenic expression of CTLA4-Ig by fetal pig neurons for xeno-transplantation. Transgenic Research. 2005;14:373–384. doi: 10.1007/s11248-004-7268-4. [DOI] [PubMed] [Google Scholar]
- Massey JM. Animal production industry in the year 2000 A.D. Journal of Reproduction Fertility Supplement. 1990;41:199–208. [PubMed] [Google Scholar]
- Matsunari H, Onodera M, Tada N, Mochizuki H, Karasawa S, Haruyama E, et al. Transgenic-cloned pigs systemically expressing red fluorescent protein, Kusabira-Orange. Cloning Stem Cells. 2008;10:313–323. doi: 10.1089/clo.2008.0024. [DOI] [PubMed] [Google Scholar]
- McClenaghan M, Archibald AL, Harris S, Simons JP, Whitelaw CBA, Wilmut I, et al. Production of human 1-antitrypsin in the milk of transgenic sheep and mice targeting expression of cDNA sequences to the mammary gland. Animal Biotechnology. 1991;2:161–176. [Google Scholar]
- McCreath KJ, Howcroft J, Campbell KH, Colman A, Schnieke AE, Kind AJ. Production of gene-targeted sheep by nuclear transfer from cultured somatic cells. Nature. 2000;405:1066–1069. doi: 10.1038/35016604. [DOI] [PubMed] [Google Scholar]
- McMichael AJ. Insights from past millennia into climatic impacts on human health and survival. Proceeding of the Nationl Academy Sciences of the United States of America. 2012;109:44730–44737. doi: 10.1073/pnas.1120177109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendicino M, Ramsoondar J, Phelps C, Vaught T, Ball S, LeRoith T, et al. Generation of antibody- and B cell-deficient pigs by targeted disruption of the J-region gene segment of the heavy chain locus. Transgenic Research. 2011;20:625–641. doi: 10.1007/s11248-010-9444-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Hrabe de Angelis M, Wurst W, Kuhn R. Gene targeting by homolougous recombination in mouse zygotes mediated by zinc-finger nucleases. Proceeding of the Nationl Academy Sciences of the United States of America. 2010;107:15022–15026. doi: 10.1073/pnas.1009424107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nature Biotechnology. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nature Biotechnology. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- Miller KF, Bolt DJ, Pursel VG, Hammer RE, Pinkert CA, Palmiter RD, et al. Expression of human or bovine growth hormone gene with a mouse metallothionein-1 promoter in transgenic swine alters the secretion of porcine growth hormone and insulin-like growth factor-I. Journal of Endrocrinology. 1989;120:481–488. doi: 10.1677/joe.0.1200481. [DOI] [PubMed] [Google Scholar]
- Mitchell RS, Beitzel BF, Schroder AR, Shinn P, Chen H, Berry CC, et al. Retroviral DNA integration: ASLV, HIV, and MLV show distinct target site preferences. PLoS Biology. 2004;2:1127–1136. doi: 10.1371/journal.pbio.0020234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa S, Murakami H, Murase A, Nakai R, Koma M, Koyota S, et al. Transgenic pigs with human N-acetylglucosaminyltransferase III. Transplantation Proceedings. 2001;33:742–743. doi: 10.1016/s0041-1345(00)02232-6. [DOI] [PubMed] [Google Scholar]
- Mizuarai S, Ono K, Yamaguchi K, Nishijima K, Kamihira M, Iijima S. Production of transgenic quails with high frequency of germ-line transmission using VSV-G pseudotyped retroviral vector. Biochemical and Biophysical Research Communications. 2001;286:456–463. doi: 10.1006/bbrc.2001.5422. [DOI] [PubMed] [Google Scholar]
- Monaco MH, Gronlund DE, Bleck GT, Hurley WL, Wheeler MB, Donovan SM. Mammary specific transgenic over-expression of insulin-like growth factor-I (IGF-I) increases pig milk IGF-I and IGF binding proteins, with no effect on milk composition or yield. Transgenic Research. 2005;14:761–773. doi: 10.1007/s11248-005-7219-8. [DOI] [PubMed] [Google Scholar]
- Morbitzer R, Elsaesser J, Hausner J, Lahaye T. Assembly of custom TALE-type DNA binding domains by modular cloning. Nucleic Acids Research. 2011;39:5790–5799. doi: 10.1093/nar/gkr151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- Muller M, Brenig B, Winnacker EL, Brem G. Transgenic pigs carrying cDNA copies encoding the murine Mxl protein which confers resistance to influenza virus. Gene. 1992;121:263–270. doi: 10.1016/0378-1119(92)90130-h. [DOI] [PubMed] [Google Scholar]
- Murakami H, Nagashima H, Takahagi Y, Fujimura T, Miyagawa S, Okabe M, et al. Production of transgenic pigs expressing human DAF (CD55) regulated by the porcine MCP gene promoter. Transplantation Proceedings. 2000;32:2505–2506. doi: 10.1016/s0041-1345(00)01769-3. [DOI] [PubMed] [Google Scholar]
- Murray JD, Nancarrow CD, Marshall JT, Hazelton IG, Ward KA. Production of transgenic merino sheep by microinjection of ovine metallothionein-ovine growth hormone fusion genes. Reproduction Fertility Development. 1989;1:147–155. doi: 10.1071/rd9890147. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Cathomen T. On target? Tracing zinc-finer-nuclease specificity. Nature Methods. 2011;8:725–726. doi: 10.1038/nmeth.1680. [DOI] [PubMed] [Google Scholar]
- Mussolino C, Morbitzer R, Lutge F, Dannemann N, Lahaye T, Cathomen T. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Research. 2011;39:9283–9293. doi: 10.1093/nar/gkr597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruse K, Ishikawa H, Kawano HO, Ueda H, Kurome M, Miyazaki K, et al. Production of a transgenic pig expressing human albumin and enhanced green fluorescent protein. The Journal of Reproduction and Development. 2005;51:539–546. doi: 10.1262/jrd.16073. [DOI] [PubMed] [Google Scholar]
- Niemann H, Halter R, Carnwath JW, Herrmann D, Lemme E, Paul D. Expression of human blood clotting factor VIII in the mammary gland of transgenic sheep. Transgenic Research. 1999;8:237–247. doi: 10.1023/a:1008999622117. [DOI] [PubMed] [Google Scholar]
- Nottle MB, Haskard KA, Verma PJ, Du ZT, Grupen CG, McIlfatrick SM, et al. Effect of DNA concentration on transgenesis rates in mice and pigs. Transgenic Research. 2001;10:523–531. doi: 10.1023/a:1013007329936. [DOI] [PubMed] [Google Scholar]
- Nottle MB, Nagashima H, Verma PJ, Du ZT, Grupen CG, et al. Production and analysis of transgenic pigs containing a metallothionein porcine growth hormon gene construct. In: Murray JD, Anderson GB, Oberbauer AM, McGloughlin MM, editors. Transgenic Animals in Agriculture. New York: CABI Publishing; 1999. pp. 145–156. [Google Scholar]
- Nowak-Imialek M, Kues WA, Petersen B, Lucas-Hahn A, Herrmann D, Haridoss S, et al. Oct4-enhanced green fluorescent protein transgenic pigs: a new large animal model for reprogramming studies. Stem Cells Development. 2011;20:1563–1575. doi: 10.1089/scd.2010.0399. [DOI] [PubMed] [Google Scholar]
- Orlando SJ, Santiago Y, Dekelver RC, Freyvert Y, Boydston EA, Moehle EA, et al. Zinc-finger nuclease-driven targeted integration into mammalian genomes using donors with limited chromosomal homology. Nucleic Acids Research. 2010;38:e152. doi: 10.1093/nar/gkq512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oropeza M, Petersen B, Carnwath JW, Lucas-Hahn A, Lemme E, Hassel P, et al. Transgenic expression of the human A20 gene in cloned pigs provides protection against apoptotic and inflammatory stimuli. Xenotransplantation. 2009;16:522–534. doi: 10.1111/j.1399-3089.2009.00556.x. [DOI] [PubMed] [Google Scholar]
- Paleyanda RK, Velander WH, Lee TK, Scandella DH, Gwazdauskas FC, Knight JW, et al. Transgenic pigs produce functional human factor VIII in milk. Nature Biotechnology. 1997;15:971–975. doi: 10.1038/nbt1097-971. [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Brinster RL, Hammer RE, Trumbauer ME, Rosenfeld MG, Birnberg NC, et al. Dramatic growth of mice that develop from eggs micro-injected with metallothionein-growth hormone fusion genes. Nature. 1982;300:611–615. doi: 10.1038/300611a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter RD, Norstedt G, Gelinas RE, Hammer RE, Brinster RL. Metallothionein-human GH fusion genes stimulate growth of mice. Science. 1983;222:809–814. doi: 10.1126/science.6356363. [DOI] [PubMed] [Google Scholar]
- Pan D, Zhang L, Zhou Y, Feng C, Long C, Liu X, et al. Efficient production of omega-3 fatty acid desaturase (sFat-1)-transgenic pigs by somatic cell nuclear transfer. Science China Life Sciences. 2010;53:517–523. doi: 10.1007/s11427-010-0080-x. [DOI] [PubMed] [Google Scholar]
- Park JK, Lee YK, Lee P, Chung HJ, Kim S, Lee HG, et al. Recombinant human erythropoietin produced in milk of transgenic pigs. Journal of Biotechnology. 2006;122:362–371. doi: 10.1016/j.jbiotec.2005.11.021. [DOI] [PubMed] [Google Scholar]
- Park KS, Lee DK, Lee H, Lee Y, Jang YS, Kim YH, et al. Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nature Biotechnology. 2003;21:1208–1214. doi: 10.1038/nbt868. [DOI] [PubMed] [Google Scholar]
- Pattanayak V, Ramirez CL, Joung JK, Liu DR. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nature Methods. 2011;8:765–770. doi: 10.1038/nmeth.1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelak K, Shianna KV, Ge D, Maia JM, Zhu M, Smith JP, et al. The characterization of twenty sequenced human genomes. PLoS Genetics. 2010;6:e1001111. doi: 10.1371/journal.pgen.1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W. GM crop cultivation surges, but novel traits languish. Nature Biotechnology. 2011;29:302. doi: 10.1038/nbt.1842. [DOI] [PubMed] [Google Scholar]
- Petersen B, Ramackers W, Tiede A, Lucas-Hahn A, Herrmann D, Barg-Kues B, et al. Pigs transgenic for human thrombomodulin have elevated production of activated protein C. Xenotransplantation. 2009;16:486–495. doi: 10.1111/j.1399-3089.2009.00537.x. [DOI] [PubMed] [Google Scholar]
- Petters RM, Alexander CA, Wells KD, Collins EB, Sommer JR, Blanton MR, et al. Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nature Biotechnology. 1997;15:965–970. doi: 10.1038/nbt1097-965. [DOI] [PubMed] [Google Scholar]
- Phelps CJ, Ball SF, Vaught TD, Vance AM, Mendicino M, Monahan JA, et al. Production and characterization of transgenic pigs expressing porcine CTLA4-Ig. Xenotransplantation. 2009;16:477–485. doi: 10.1111/j.1399-3089.2009.00533.x. [DOI] [PubMed] [Google Scholar]
- Phelps CJ, Koike C, Vaught TD, Boone J, Wells KD, Chen SH, et al. Production of alpha-1,3-galactosyltransferase-deficient pigs. Science. 2003;299:411–414. doi: 10.1126/science.1078942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkert CA, Johnson LW, Irwin MH, Wong S, Baetge EE, Wolfe DF, et al. Optimization of superovulation and fertilization protocols in the production of transgenic swine. Advances in Reprodutive. 2001;5:45–53. [Google Scholar]
- Pinkert CA, Pursel VG, Miller KF, Palmiter RD, Brinster RL. Production of transgenic pigs harboring growth hormone (MTbGH) or growth hormone releasing factor (MThGRF) genes. Journal of Animal Science. 1987;65(Suppl 1):260. (Abstr.) [Google Scholar]
- Plasterk RHA, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends in Genetics. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- Polge EJC, Barton SC, Surani MAH, Miller JR, Wagner T, Rottman R, et al. Induced expression of a bovine growth hormone construct in transgenic pigs. London: Biotechnology of Growth Regulation, Butterworths; 1989. pp. 279–289. [Google Scholar]
- Ponder KP, Wang B, Wang P, Ma X, Herati R, Wang B, et al. Mucopolysaccharidosis I cats mount a cytotoxic T lymphocyte response after neonatal gene therapy that can be blocked with CTLA4-Ig. Molecular Therapy. 2006;14:5–13. doi: 10.1016/j.ymthe.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Baltimore D. Chimeric nucleases stimulate gene targeting in human cells. Science. 2003;300:763. doi: 10.1126/science.1078395. [DOI] [PubMed] [Google Scholar]
- Porteus MH, Carroll D. Gene targeting using zinc finger nucleases. Nature Biotechnology. 2005;8:967–973. doi: 10.1038/nbt1125. [DOI] [PubMed] [Google Scholar]
- Pursel V, Wall RJ, Mitchell AD, Elsasser TH, Solomon MB, Coleman ME, et al. Expression of insulin-like growth factor I in skeletal muscle of transgenic swine. Wallingford, UK: CAB International; 1999. [Google Scholar]
- Pursel VG, Mitchell AD, Bee G, Elsasser TH, McMurtry JP, Wall RJ, et al. Growth and tissue accretion rates of swine expressing an insulin-like growth factor I transgene. Animal Biotechnology. 2004;15:33–45. doi: 10.1081/ABIO-120029812. [DOI] [PubMed] [Google Scholar]
- Pursel VG, Pinkert CA, Miller KF, Bolt DJ, Campbell RG, Palmiter RD, et al. Genetic engineering of livestock. Science. 1989;244:1281–1288. doi: 10.1126/science.2499927. [DOI] [PubMed] [Google Scholar]
- Pursel VG, Rexroad CE, Jr, Bolt DJ, Miller KF, Wall RJ, Hammer RE, et al. Progress on gene transfer in farm animals. Veterinary Immunology and Immunopathology. 1987;17:303–312. doi: 10.1016/0165-2427(87)90149-8. [DOI] [PubMed] [Google Scholar]
- Pursel VG, Sutrave P, Wall RJ, Kelly AM, Hughes SH. Transfer of cSKI gene into swine to enhance muscle development. Theriogenology. 1992;37:278. [Google Scholar]
- Pursel VG, Wall RJ, Solomon MB, Bolt DJ, Murray JE, Ward KA. Transfer of an ovine metallothionein-ovine growth hormone fusion gene into swine. Journal of Animal Science. 1997;75:2208–2214. doi: 10.2527/1997.7582208x. [DOI] [PubMed] [Google Scholar]
- Ramirez CL, Foley JE, Wright DA, Muller-Lerch F, Rahman SH, Cornu TI, et al. Unexpected failure rates for modular assembly of engineered zinc fingers. Nature Methods. 2008;5:374–375. doi: 10.1038/nmeth0508-374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsoondar J, Mendicino M, Phelps C, Vaught T, Ball S, Monahan J, et al. Targeted disruption of the porcine immunoglobulin kappa light chain locus. Transgenic Research. 2011;20:643–653. doi: 10.1007/s11248-010-9445-y. [DOI] [PubMed] [Google Scholar]
- Ramsoondar J, Vaught T, Ball S, Mendicino M, Monahan J, Jobst P, et al. Production of transgenic pigs that express porcine endogenous retrovirus small interfering RNAs. Xenotransplantation. 2009;16:164–180. doi: 10.1111/j.1399-3089.2009.00525.x. [DOI] [PubMed] [Google Scholar]
- Ramsoondar JJ, Machaty Z, Costa C, Williams BL, Fodor WL, Bondioli KR. Production of alpha 1,3-galactosyltransferase-knockout cloned pigs expressing human alpha 1,2-fucosylosyltransferase. Biology of Reproduction. 2003;69:437–445. doi: 10.1095/biolreprod.102.014647. [DOI] [PubMed] [Google Scholar]
- Rexroad CE, Jr, Hammer RE, Bolt DJ, Mayo KE, Frohman LA, Palmiter RD, et al. Production of transgenic sheep with growth-regulating genes. Molecular Reproduction and Development. 1989;1:164–169. doi: 10.1002/mrd.1080010304. [DOI] [PubMed] [Google Scholar]
- Rexroad CE, Jr, Mayo K, Bolt DJ, Elsasser TH, Miller KF, Behringer RR, et al. Transferrin- and albumin-directed expression of growth-related peptides in transgenic sheep. Journal of Animal Science. 1991;69:2995–3004. doi: 10.2527/1991.6972995x. [DOI] [PubMed] [Google Scholar]
- Reyon D, Tsai SQ, Khayter C, Foden JA, Sander JD, Joung JK. FLASH assembly of TALENs for high-throughput genome editing. Nature Biotechnology. 2012;30:460–465. doi: 10.1038/nbt.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richt JA, Kasinathan P, Hamir AN, Castilla J, Sathiyaseelan T, Vargas F, et al. Production of cattle lacking prion protein. Nature Biotechnology. 2007;25:132–138. doi: 10.1038/nbt1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchie WA, King T, Neil C, Carlisle AJ, Lillico S, McLachlan G, et al. Transgenic sheep designed for transplantation studies. Molecular Reproduction and Development. 2009;76:61–64. doi: 10.1002/mrd.20930. [DOI] [PubMed] [Google Scholar]
- Rogers CS, Stoltz DA, Meyerholz DK, Ostedgaard LS, Rokhlina T, Taft PJ, et al. Disruption of the CFTR gene produces a model of cystic fibrosis in newborn pigs. Science. 2008;321:1837–1841. doi: 10.1126/science.1163600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GE. Improvement of wool production through genetic engineering. Trends in Biotechnology. 1990;8:6. doi: 10.1016/0167-7799(90)90123-f. [DOI] [PubMed] [Google Scholar]
- Rollin BE. “The Frankenstein Thing”: the moral impact of genetic engineering of agricultural animals on society and future science. In: Evans JW, Hollaender A, editors. Genetic Engineering of Animals: An agricultural perspective. New York: Plenum Press; 1985. pp. 285–297. [DOI] [PubMed] [Google Scholar]
- Roshlau K, Rommel P, Andreewa L, Zackel M, Roschlau D, Zackel B, et al. Gene transfer experiments in cattle. Journal of Reproduction and Fertility. 1989;38(Supp1):153. [PubMed] [Google Scholar]
- Ross JW, Fernandez de Castro JP, Zhao J, Samuel M, Walters E, Rios C, et al. Generation of an inbred miniature pig model of retinitis pigmentosa. Investigative Ophthalmology and Visual Science. 2012;53:501–507. doi: 10.1167/iovs.11-8784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouet P, Smih F, Jasin M. Expression of a site-specific endonuclease stimulates homologous recombination in mammalian cells. Proceeding of the Nationl Academy Sciences of the United States of America. 1994;91:6064–6068. doi: 10.1073/pnas.91.13.6064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rous P. A sarcoma of the fowl transmissable by an agent separable from the tumor cells. Journal of Experimental Medicine. 1910;13:416–427. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs DH, Galli C. Genetic manipulation in pigs. Current Opinion in Organ Transplantation. 2009;14:148–153. doi: 10.1097/mot.0b013e3283292549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeki K, Matsumoto K, Kinoshita M, Suzuki I, Tasaka Y, Kano K, et al. Functional expression of a Delta12 fatty acid desaturase gene from spinach in transgenic pigs. Proceeding of the Nationl Academy Sciences of the United States of America. 2004;101:6361–6366. doi: 10.1073/pnas.0308111101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamone D, Baranao L, Santos C, Bussmann L, Artuso J, Werning C, et al. High level expression of bioactive recombinant human growth hormone in the milk of a cloned transgenic cow. Journal of Biotechnology. 2006;124:469–472. doi: 10.1016/j.jbiotec.2006.01.005. [DOI] [PubMed] [Google Scholar]
- Salter DW, Smith EJ, Hughes SH, Wright SE, Crittenden LB. Transgenic chickens: insertion of retroviral genes into the chicken germ line. Virology. 1987;157:236–240. doi: 10.1016/0042-6822(87)90334-5. [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Petersen RT, Joung JK, et al. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nature Biotechnology. 2011;29:697–698. doi: 10.1038/nbt.1934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, et al. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nature Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Reyon D, Maeder ML, Foley JE, Thibodeau-Beganny S, Li X, et al. Predicting success of oligomerized pool engineering (OPEN) for zinc finger target site sequences. BMC Bioinformatics. 2010;11:543. doi: 10.1186/1471-2105-11-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Yeh JR, Peterson RT, Joung JK. Engineering zinc finger nucleases for targeted mutagenesis of zebrafish. Methods Cell Biology. 2011;104:51–58. doi: 10.1016/B978-0-12-374814-0.00003-3. [DOI] [PubMed] [Google Scholar]
- Sang H. Prospects for transgenesis in the chick. Mechanisms of Development. 2004;121:1179–1186. doi: 10.1016/j.mod.2004.05.012. [DOI] [PubMed] [Google Scholar]
- Schmidhuber J, Tubiello FN. Global food security under climate change. Proceeding of the Nationl Academy Sciences of the United States of America. 2007;104:19703–19708. doi: 10.1073/pnas.0701976104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnieke AE, Kind AJ, Ritchie WA, Mycock K, Scott AR, Ritchie M, et al. Human factor IX transgenic sheep produced by transfer of nuclei from transfected fetal fibroblasts. Science. 1997;278:2130–2133. doi: 10.1126/science.278.5346.2130. [DOI] [PubMed] [Google Scholar]
- Schurman R, Munro WA. Fighting for the future of food. Vol. 35. Minneapolis: University of Minnesota Press; 2010. p. 262. [Google Scholar]
- Sears MK, Hellmich RL, Stanley-Horn DE, Oberhauser KS, Pleasants JM, Mattila HR, et al. Impact of Bt corn pollen on monarch butterfly populations: A risk assessment. Proceeding of the Nationl Academy Sciences of the United States of America. 2001;98:111937–111942. doi: 10.1073/pnas.211329998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seichter D, Russ I, Rothammer S, Eder J, Forster M, Medugorac I. Animal Genetics. 2012 doi: 10.1111/j.1365-2052.2011.02302.x. (epub ahead of print) [DOI] [PubMed] [Google Scholar]
- Seidel GE., Jr . Characteristics of future agricultural animals. In: Evans JW, Hollaender A, editors. Genetic Engineering of Animals: An Agricultural Perspective. New York: Plenum Press; 1985. pp. 299–310. [Google Scholar]
- Sendai Y, Sawada T, Urakawa M, Shinkai Y, Kubota K, Hoshi H, et al. alpha1,3-Galactosyltransferase-gene knockout in cattle using a single targeting vector with loxP sequences and cre-expressing adenovirus. Transplantation. 2006;81:760–766. doi: 10.1097/01.tp.0000190422.66657.f1. [DOI] [PubMed] [Google Scholar]
- Shamay A, Solinas S, Pursel VG, McKnight RA, Alexander L, Beattie C, et al. Production of the mouse whey acidic protein in transgenic pigs during lactation. Journal of Animal Science. 1991;69:4552–4562. doi: 10.2527/1991.69114552x. [DOI] [PubMed] [Google Scholar]
- Smih F, Rouet P, Romanienko PJ, Jasin M. Double-strand breaks at the target locus stimulate gene targeting in embryonic stem cells. Nucleic Acids Research. 1995;23:5012–5019. doi: 10.1093/nar/23.24.5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith MD, Asche F, Guttormsen AG, Wiener JB. Food safety. Genetically modified salmon and full impact assessment. Science. 2010;330:1052–1053. doi: 10.1126/science.1197769. [DOI] [PubMed] [Google Scholar]
- Smithies O, Gregg RG, Boggs SS, Koralewski MA, Kucherlapati RS. Insertion of DNA sequences into the human chromosomal beta-globin locus by homologous recombination. Nature. 1985;317:230–234. doi: 10.1038/317230a0. [DOI] [PubMed] [Google Scholar]
- Snaith MR, Tornell J. The use of transgenic systems in pharmaceutical research. Briefings in Functional Genomics and Proteomics. 2002;1:119–130. doi: 10.1093/bfgp/1.2.119. [DOI] [PubMed] [Google Scholar]
- Snouwaert JN, Brigman KK, Latour AM, Malouf NN, Boucher RC, Smithies O, et al. An animal model for cystic fibrosis made by gene targeting. Science. 1992;257:1083–1088. doi: 10.1126/science.257.5073.1083. [DOI] [PubMed] [Google Scholar]
- Sommer JR, Estrada JL, Collins EB, Bedell M, Alexander CA, Yang Z, et al. Production of ELOVL4 transgenic pigs: a large animal model for Stargardt-like macular degeneration. The British Journal of Ophthalmology. 2011;95:1749–1754. doi: 10.1136/bjophthalmol-2011-300417. [DOI] [PubMed] [Google Scholar]
- Sommer JR, Jackson LR, Simpson SG, Collins EB, Piedrahita JA, Petters RM. Transgenic Stra8-EYFP pigs: a model for developing male germ cell technologies. Transgenic Research. 2012;21:383–392. doi: 10.1007/s11248-011-9542-6. [DOI] [PubMed] [Google Scholar]
- Staunstrup NH, Madsen J, Primo MN, Li J, Liu Y, Kragh PM, et al. Development of transgenic cloned pig models of skin inflammation by DNA transposon-directed ectopic expression of human beta1 and alpha2 integrin. PLoS One. 2012;7:e36658. doi: 10.1371/journal.pone.0036658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson ME, Martin MJ, O’Donnell JK, Hoover K, Lago W, Huntress V, et al. Production of functional human hemoglobin in transgenic swine. Biotechnology (Nature Publishing Company) 1992;10:557–559. doi: 10.1038/nbt0592-557. [DOI] [PubMed] [Google Scholar]
- Szczepek M, Brondani V, Buchel J, Serrano L, Segal DJ, Cathomen T. Structure-based redesign of the dimerization interface reduces the toxicity of zinc-finger nucleases. Nature Biotechnology. 2007;25:786–793. doi: 10.1038/nbt1317. [DOI] [PubMed] [Google Scholar]
- Tabeshnik BE. Communal benefits of transgenic corn. Science. 2010;330:189–190. doi: 10.1126/science.1196864. [DOI] [PubMed] [Google Scholar]
- Takahagi Y, Fujimura T, Miyagawa S, Nagashima H, Shigehisa T, Shirakura R, et al. Production of alpha 1,3-galactosyltransferase gene knockout pigs expressing both human decay-accelerating factor and N-acetylglucosaminyltransferase III. Molecular Reproduction and Development. 2005;71:331–338. doi: 10.1002/mrd.20305. [DOI] [PubMed] [Google Scholar]
- Tessanne K, Golding MC, Long CR, Peoples MD, Hannon G, Westhusin ME. Production of transgenic calves expressing an shRNA targeting myostatin. Molecular Reproduction and Development. 2012;79:176–185. doi: 10.1002/mrd.22007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesson L, Usal C, Ménoret S, Leung E, Niles BJ, Remy S, et al. Knockout rats generated by embryo microinjection of TALENs. Nature Biotechnology. 2011;29:695–696. doi: 10.1038/nbt.1940. [DOI] [PubMed] [Google Scholar]
- Thoraval P, Afanassieff M, Cosset FL, Lasserre F, Verdier G, Coudert F, et al. Germline transmission of exogenous genes in chickens using helper-free ecotropic avian leucosis virus-based vectors. Transgenic Research. 1995;4:369–377. doi: 10.1007/BF01973755. [DOI] [PubMed] [Google Scholar]
- Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proceeding of the Nationl Academy Sciences of the United States of America. 2011;108:20260–20264. doi: 10.1073/pnas.1116437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong J, Wei H, Liu X, Hu W, Bi M, Wang Y, et al. Production of recombinant human lysozyme in the milk of transgenic pigs. Transgenic Research. 2011;20:417–419. doi: 10.1007/s11248-010-9409-2. [DOI] [PubMed] [Google Scholar]
- Uchida M, Shimatsu Y, Onoe K, Matsuyama N, Niki R, Ikeda JE, et al. Production of transgenic miniature pigs by pronuclear microinjection. Transgenic Research. 2001;10:577–582. doi: 10.1023/a:1013059917280. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nature Reviews Genetics. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- Van Cott KE, Butler SP, Russell CG, Subramanian A, Lubon H, Gwazdauskas FC, et al. Transgenic pigs as bio-reactors: a comparison of gamma-carboxylation of glutamic acid in recombinant human protein C and factor IX by the mammary gland. Genetics Analysis Biomolecular Engineering. 1999;15:155–160. doi: 10.1016/s1050-3862(99)00020-0. [DOI] [PubMed] [Google Scholar]
- Van Cott KE, Lubon H, Gwazdauskas FC, Knight J, Drohan WN, Velander WH. Recombinant human protein C expression in the milk of transgenic pigs and the effect on endogenous milk immunoglobulin and transferrin levels. Transgenic Research. 2001;10:43–51. doi: 10.1023/a:1008963817646. [DOI] [PubMed] [Google Scholar]
- van Doorn MB, Burggraaf J, van Dam T, Eerenberg A, Levi M, Hack CE, et al. A phase I study of recombinant human C1 inhibitor in asymptomatic patients with hereditary angioedema. The Journal of Allergy and Clinical Immunology. 2005;116:876–883. doi: 10.1016/j.jaci.2005.05.019. [DOI] [PubMed] [Google Scholar]
- Van Eenennaam AL, Muir WM. Transgenic salmon: a final leap to the grocery shelf? Nature Biotechnology. 2011;29:706–710. doi: 10.1038/nbt.1938. [DOI] [PubMed] [Google Scholar]
- Van Raden PM, Olson KM, Null DJ, Hutchison JL. Harmful recessive effects on fertility detected by absence of homozygous haplotypes. Journal of Dairy Science. 2011;94:6153–6161. doi: 10.3168/jds.2011-4624. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Vize PD, Michalska AE, Ashman R, Lloyd B, Stone BA, Quinn P, et al. Introduction of a porcine growth hormone fusion gene into transgenic pigs promotes growth. Journal of Cell Science. 1988;90(Pt. 2):295–300. doi: 10.1242/jcs.90.2.295. [DOI] [PubMed] [Google Scholar]
- Voigt K, Izsvák Z, Ivics Z. Targeted gene insertion for molecular medicine. Journal of Molecular Medicine. 2008;86:1205–1219. doi: 10.1007/s00109-008-0381-8. [DOI] [PubMed] [Google Scholar]
- Wagner EF, Covarrubias L, Stewart TA, Mintz B. Prenatal lethalities in mice homozygous for human growth hormone gene sequences integrated in the germ line. Cell. 1983;35:647–655. doi: 10.1016/0092-8674(83)90097-1. [DOI] [PubMed] [Google Scholar]
- Wagner TE, Murray FA. Genetic engineering of laboratory and livestock mammals. Journal of Animal Science. 1985;61(Suppl 3):25–37. doi: 10.1093/ansci/61.supplement_3.25. [DOI] [PubMed] [Google Scholar]
- Wall RJ, Powell AM, Paape MJ, Kerr DE, Bannerman DD, Pursel VG, et al. Genetically enhanced cows resist intramammary Staphylococcus aureus infection. Nature Biotechnology. 2005;23:445–451. doi: 10.1038/nbt1078. [DOI] [PubMed] [Google Scholar]
- Wall RJ, Pursel VG, Shamay A, McKnight RA, Pittius CW, Hennighausen L. High-level synthesis of a heterologous milk protein in the mammary glands of transgenic swine. Proceeding of the Nationl Academy Sciences of the United States of America. 1991;88:1696–1700. doi: 10.1073/pnas.88.5.1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall RJ, Rexroad CE, Jr, Powell A, Shamay A, McKnight R, Hennighausen L. Synthesis and secretion of the mouse whey acidic protein in transgenic sheep. Transgenic Research. 1996;5:67–72. doi: 10.1007/BF01979923. [DOI] [PubMed] [Google Scholar]
- Waltz E. Tiptoeing around transgenics. Nature Biotechnology. 2012;30:215–217. doi: 10.1038/nbt.2143. [DOI] [PubMed] [Google Scholar]
- Wang J, Yang P, Tang B, Sun X, Zhang R, Guo C, et al. Expression and characterization of bioactive recombinant human alpha-lactalbumin in the milk of transgenic cloned cows. Journal of Dairy Science. 2008;91:4466–4476. doi: 10.3168/jds.2008-1189. [DOI] [PubMed] [Google Scholar]
- Ward KA, Nancarrow CD. The genetic engineering of production traits in domestic animals. Experientia. 1991;47:913. doi: 10.1007/BF01929882. [DOI] [PubMed] [Google Scholar]
- Watanabe M, Kurome M, Matsunari H, Nakano K, Umeyema K, Shiota A, et al. The creation of transgenic pigs expressing human proteins using BAC-derived, full-length genes and intracytoplasmic sperm injection-mediated gene transfer. Transgenic Research. 2012;21:605–618. doi: 10.1007/s11248-011-9561-3. [DOI] [PubMed] [Google Scholar]
- Waterston RH, et al. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Weber E, Engler C, Gruetzner R, Werner S, Marillonnet S. A modular cloning system for standardized assembly of multigene constructs. PLoS One. 2011;6:e16765. doi: 10.1371/journal.pone.0016765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster NL, Forni M, Bacci ML, Giovannoni R, Razzini R, Fantinati P, et al. Multi-transgenic pigs expressing three fluorescent proteins produced with high efficiency by sperm mediated gene transfer. Molecular Reproduction and Development. 2005;72:68–76. doi: 10.1002/mrd.20316. [DOI] [PubMed] [Google Scholar]
- Weidle UH, Lenz H, Brem G. Genes encoding a mouse monoclonal antibody are expressed in transgenic mice, rabbits and pigs. Gene. 1991;98:185–191. doi: 10.1016/0378-1119(91)90172-8. [DOI] [PubMed] [Google Scholar]
- Whitelaw CB, Radcliffe PA, Ritchie WA, Carlisle A, Ellard FM, Pena RN, et al. Efficient generation of transgenic pigs using equine infectious anaemia virus (EIAV) derived vector. FEBS Letters. 2004;571:233–236. doi: 10.1016/j.febslet.2004.06.076. [DOI] [PubMed] [Google Scholar]
- Whitworth KM, Li R, Spate LD, Wax DM, Rieke A, Whyte JJ, et al. Method of oocyte activation affects cloning efficiency in pigs. Molecular Reproduction and Development. 2009;76:490–500. doi: 10.1002/mrd.20987. [DOI] [PubMed] [Google Scholar]
- Whyte JJ, Zhao J, Wells KD, Samuel MS, Whitworth KM, Walters EM, et al. Gene targeting with zinc finger nucleases to produce cloned eGFP knockout pigs. Molecular Reproduction and Development. 2011;78:2. doi: 10.1002/mrd.21271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieghart M, Hoover JL, McGrane MM, Hanson RW, Rottman FM, Holtzman SH, et al. Production of transgenic pigs harbouring a rat phosphoenolpyruvate carboxykinase-bovine growth hormone fusion gene. Journal of Reproduction Fertility Supplement. 1990;41:89–96. [PubMed] [Google Scholar]
- Wolfe JH. Gene therapy in large animal models of human genetic diseases. ILAR Journal. 2009;50:107–111. doi: 10.1093/ilar.50.2.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkovich EM, Cook BI, Allen JM, Crimmins TM, Betancourt JL, Travers SE, et al. Warming experiments underpredict plant phenological responses to climate change. Nature. 2012;485:494–497. doi: 10.1038/nature11014. [DOI] [PubMed] [Google Scholar]
- Worm B, Hilborn R, Baum JK, Branch TA, Collie JS, Costello C, et al. Rebuilding global fisheries. Science. 2009;325:578–585. doi: 10.1126/science.1173146. [DOI] [PubMed] [Google Scholar]
- Wu S, Lin S, Chen C, Choo K, Cheng WT. Production of transgenic pig haboring cDNA of human Factor IX gene (Abstract) Journal of Genetics and Molecular Biology. 1999;10:36. [Google Scholar]
- Yamakawa H, Nagai T, Harasawa R, Yamagami T, Takahashi J, Ishikawa K, et al. Production of transgenic pig carrying MMTV/v-Ha-ras. The Journal of Reproduction and Development. 1999;45:111–118. [Google Scholar]
- Yan S. Cloning in fish – nucleocytoplasmic hybrids. Hong Kong: IUBS Educational and Cultural Press, Ltd; 1998. [Google Scholar]
- Yang D, Wang CE, Zhao B, Li W, Ouyang Z, Liu Z, et al. Expression of Huntington’s disease protein results in apoptotic neurons in the brains of cloned transgenic pigs. Human Molecular Genetics. 2010;19:3983–3994. doi: 10.1093/hmg/ddq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Yang H, Li W, Zhao B, Ouyang Z, Liu Z, et al. Generation of PPARγ mono-allelic knockout pigs via zinc-finger nucleases and nuclear transfer cloning. Cell Research. 2011;21:979–982. doi: 10.1038/cr.2011.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Wang J, Gong G, Sun X, Zhang R, Du Z, et al. Cattle mammary bioreactor generated by a novel procedure of transgenic cloning for large-scale production of functional human lactoferrin. PLoS One. 2008;3:e3453. doi: 10.1371/journal.pone.0003453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G, Chen J, Xu Y, Zhu C, Yu H, Liu S, et al. Generation of goats lacking prion protein. Molecular Reproduction and Development. 2009;76:3. doi: 10.1002/mrd.20960. [DOI] [PubMed] [Google Scholar]
- Yu G, Chen J, Yu H, Liu S, Xu X, Sha H, et al. Functional disruption of the prion protein gene in cloned goats. The Journal of General Virology. 2006;87:1019–1027. doi: 10.1099/vir.0.81384-0. [DOI] [PubMed] [Google Scholar]
- Yu S, Luo J, Song Z, Ding F, Dai Y, Li N. Highly efficient modification of beta-lactoglobulin (BLG) gene via zinc-finger nucleases in cattle. Cell Research. 2011;21:1638–1640. doi: 10.1038/cr.2011.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nature Biotechnology. 2011;29:149–153. doi: 10.1038/nbt.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li L, Cai Y, Xu X, Chen J, Wu Y, et al. Expression of active recombinant human lactoferrin in the milk of transgenic goats. Protein Expression and Purification. 2008;57:127–135. doi: 10.1016/j.pep.2007.10.015. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Xi Q, Ding J, Cai W, Meng F, Zhou J, et al. Production of transgenic pigs mediated by pseudotyped lentivirus and sperm. PLoS One. 2012;7:e35335. doi: 10.1371/journal.pone.0035335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou CY, McInnes E, Copeman L, Langford G, Parsons N, Lancaster R, et al. Production of triple transgenic pigs expressing human Cd59, Mcp and Daf. Transplantation. 2004;78:576. [Google Scholar]