Abstract
A new genetic technique for constructing mutants of Methanosarcina acetivorans C2A by using hpt as a counterselectable marker was developed. Mutants with lesions in the hpt gene, encoding hypoxanthine phosphoribosyltransferase, were shown to be >35-fold more resistant to the toxic base analog 8-aza-2,6-diaminopurine (8ADP) than was the wild type. Reintroduction of the hpt gene into a Δhpt host restored 8ADP sensitivity and provided the basis for a two-step strategy involving plasmid integration and excision for recombination of mutant alleles onto the M. acetivorans chromosome. We have designated this method markerless exchange because, although selectable markers are used during the process, they are removed in the final mutants. Thus, the method can be repeated many times in the same cell line. The method was validated by construction of ΔproC Δhpt mutants, which were recovered at a frequency of 22%. Additionally, a Methanosarcina-Escherichia shuttle vector, encoding the Escherichia coli proC gene as a new selectable marker, was constructed for use in proC hosts. Finally, the markerless exchange method was used to recombine a series of uidA reporter gene fusions into the M. acetivorans proC locus. In vitro assay of β-glucuronidase activity in extracts of these recombinants demonstrated, for the first time, the utility of uidA as a reporter gene in Methanosarcina. A >5,000-fold range of promoter activities could be measured by using uidA: the methyl-coenzyme M reductase operon fusion displayed ∼300-fold-higher activity than did the serC gene fusion, which in turn had 16-fold-higher activity than did a fusion to the unknown orf2 gene.
The genetic techniques available for use in Methanosarcina species are among the most advanced available for archaea but fall far short of those available for use with bacteria such as Escherichia coli and Bacillus subtilis. Nevertheless, methods continue to improve. Clonal populations of Methanosarcina, which are essential for genetic studies, can now easily be acquired via streaking on solid media; various other surface manipulations are also routine (e.g., patching, replica plating, etc.) (30). Moreover, an efficient transformation system for the introduction of DNA into Methanosarcina has been developed (21) and the recent developments of in vivo transposon mutagenesis (37) and directed gene replacement (38) allow efficient isolation and construction of mutants in Methanosarcina.
Two antibiotic resistance cassettes, the pac gene of Streptomyces alboniger for puromycin resistance (15, 21) and the ileS12 gene from Methanosarcina barkeri Fusaro for pseudomonic acid resistance (4), are useful as selectable markers for genetic manipulation in Methanosarcina. Unfortunately, pseudomonic acid is not commercially available, which limits the use of ileS12 and leaves the pac cassette as the single readily used marker. This dearth of selectable markers has limited the application of genetic analysis to certain problems in Methanosarcina. In particular, genome sequencing has shown that Methanosarcina species possess multiple copies of genes involved in the same step of numerous metabolic pathways (9, 13). In many cases, three or more duplicate genes are present. Thus, even if both pac and ileS12 are used, it is impossible to inactivate all copies of a given gene.
A potential solution to this problem lies in the use of a two-step integration-segregation strategy. In this method, a nonreplicating plasmid bearing the mutation of interest is inserted into the chromosome via homologous recombination with the wild-type allele. These merodiploid recombinants are selected by using a single selectable marker encoded on the plasmid backbone. Upon removal of selection, the unstable merodiploid allele can resolve via a second homologous recombination event between mutant and wild-type sequences. These recombinants can be either wild type or mutant, depending on the location of the recombination event, but, importantly, lose the plasmid backbone. Thus, this method allows reuse of the same antibiotic resistance marker multiple times because the marker is removed in the process of mutant construction. To improve the efficiency of this process, a counterselectable marker is often incorporated into the plasmid backbone. This allows selection, rather than screening, to be used to isolate recombinants that have resolved the merodiploid state.
Numerous counterselectable markers have been developed for use in bacteria (8, 11, 14, 28, 34) and eukaryotes (12, 18, 31, 33). Commonly used examples include the sacB-mediated sucrose-sensitive (14) and tetRA-mediated tetracycline-sensitive (5) selections used in bacteria and ura3-mediated 5-fluoroorotic acid (5-FOA)-sensitive selection used in various fungi. The latter counterselection differs from the others in that its use requires the utilization of ura3 mutant host strains. Thus, introduction of ura3+ confers 5-FOA sensitivity upon ura3 mutants, providing the basis for subsequent selection against the introduced wild-type copy. The ura3 counterselection has also proven useful in archaea, where it was successfully used in Halobacterium salinarum during the construction of bop deletion mutants (27). The 5-FOA-sensitive selection has also been used in conjunction with pyrE2 mutants in the haloarchaeon Halofreax volcanii, where it was used as a counterselectable marker to delete the cmi gene (3).
Genes involved in purine and pyrimidine salvage pathways, in particular those encoding various phosphoribosyl transferases (PRTases), have also been used as counterselectable markers (3, 10, 12, 18, 27, 31, 33). These enzymes provide the cell alternative means for de novo nucleotide biosynthesis, by incorporating exogenous bases into their nucleotide pools, and for recycling nonstandard bases. Thus, purine PRTases are responsible for converting purines (i.e., adenine, guanine, and the nonstandard xanthine and hypoxanthine) into the corresponding nucleotide monophosphates. Purine analogs (e.g., 8-aza-2,6-diaminopurine [8ADP], 8-aza-guanine, 8-aza-hypoxanthine, etc.) are also substrates for these PRTases; however, incorporation of these compounds can be lethal to the cells. Accordingly, PRTase-defective mutants are resistant to toxic base analogs and, in such hosts, reintroduction of the wild-type copy provides the basis for a counterselection strategy. For example, the B. subtilis uracil PRTase gene upp has been used as a counterselectable marker with 5-fluorouracil to create a variety of mutants in B. subtilis Δupp hosts (10).
The possibility of developing such a system in archaea is supported by several studies of purine and pyrimidine salvage pathways (6, 7, 17, 32, 36). In addition to demonstrating the presence of base salvage pathways, it was observed that spontaneous resistance to purine analogs is associated with decreased activity of certain purine salvage enzymes. In a previous study (37), we isolated an M. acetivorans mutant with a transposon insertion into a gene for a putative hypoxanthine PRTase, which we hoped could be used for development of a counterselection strategy. In this paper, we describe the development of this system for use in M. acetivorans and its use in construction of additional genetic tools, including new selectable markers and reporter gene fusions.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Standard conditions were used for growth of E. coli strains (35). DH5α/λ-pir (24) was used as the host for maintenance of all pir-dependent plasmids, while DH10B (Stratagene, La Jolla, Calif.) was used for all other replicons. M. acetivorans strains, described in Table 1, were grown in single-cell morphology (30) at 35°C in high-salt (HS) broth medium containing either 125 mM methanol-40 mM acetate (HS-MA), 125 mM methanol (HS-M), 50 mM trimethylamine (HS-T), 75 mM dimethylamine (HS-D), or 80 mM acetate (HS-A). Growth of M. acetivorans on media solidified with 1.5% agar was as described earlier (4). All plating manipulations were carried out under strictly anaerobic conditions in an anaerobic glove box. Solid medium plates were incubated in an intrachamber anaerobic incubator as described earlier (23). For selection of Methanosarcina strains carrying the puromycin transacetylase gene (pac), puromycin (Pur; CalBiochem, San Diego, Calif.) was added from sterile, anaerobic stocks at a final concentration of 2 μg/ml. Methanosarcina extract prepared as described below was added at a final concentration of 0.05% (vol/vol) for supplementation of potential auxotrophic mutants incapable of synthesizing methanogen-specific factors. The purine analog 8ADP (Sigma, St. Louis, Mo.) was added from sterile, anaerobic stocks at a final concentration of 20 μg/ml for selection against the hypoxanthine PRTase (hpt) gene. The purine analogs 2-aminopurine, 2-methylpurine (2MP), 2-mercaptopurine, 6-mercaptopurine, 6-methyl-aminopurine, 2,6-diaminopurine, and 2-amino-6-methylmercaptopurine (Sigma) were added from sterile, anaerobic stocks at concentrations ranging from 462 to 0.302 μg/ml. Proline (Pro; Sigma) was added from sterile, anaerobic stocks at a final concentration of 50 mM for construction and maintenance of all M. acetivorans pro strains (38).
TABLE 1.
M. acetivorans strains
| Strain | Genotype | Reference or construction |
|---|---|---|
| C2A | Wild type | 31 |
| WWM1 | Δhpt | Markerless exchange with pMP42 |
| WWM24 | ΔproC Δhpt | Markerless exchange with pMP69 |
| WWM20 | proC::mcrBp-uidA Δhpt | Markerless exchange with pJK85 |
| WWM23 | proC::serCp-uidA Δhpt | Markerless exchange with pJK86 |
| WWM25 | proC::orf2p-uidA Δhpt | Markerless exchange with pJK87 |
| WWM26 | proC5(BglII frameshift) Δhpt | Markerless exchange with pJK84 |
Methanosarcina extract preparation.
M. acetivorans cells from a 1-liter stationary-phase culture grown in HS-MA medium were collected by centrifugation (6,000 × g for 15 min). Cells were lysed by resuspension in ca. 100-ml 0.1 M NaHCO3, pH 6.8, containing 0.1 mg of DNase I/ml (spontaneous lysis occurs due to osmotic shock). The lysate was clarified by centrifugation (13,000 × g for 1 h), and high-molecular-weight components were removed by ultrafiltration with a 3,000-molecular-weight cutoff membrane (YM3; Amicon, Beverly, Mass.). Cell-free filtrate was filter sterilized by using a 0.25-μm-pore-size filter and was stored anaerobically at 4°C in the dark. All manipulations were conducted under strictly anaerobic conditions in an anaerobic chamber with an atmosphere of N2/CO2/H2 (75:20:5).
DNA methods.
Standard methods were used throughout for isolation and manipulation of plasmid DNA from E. coli (2). Plasmid DNA was isolated from M. acetivorans as described earlier (21), while genomic DNA was isolated with a modified cetyltrimethylammonium bromide method (22). DNA hybridizations were performed by using the DIG System (Roche, Mannheim, Germany) according to manufacturer guidelines, with the following modifications. Hybridizations were performed at 65°C in a high-sodium dodecyl sulfate (SDS) buffer (25 mM Na2HPO4, 7% SDS, pH 7.2). Posthybridization washes were performed at 65°C for 30 min with gentle agitation twice each in 20 mM Na2HPO4, 5% SDS, pH 7.2, and 20 mM Na2HPO4, 1% SDS, pH 7.2. MagnaGraph Nylon transfer membranes were from Osmonics (Westborough, Mass.). DNA sequences were determined from double-stranded templates by using the BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems, Foster City, Calif.) following manufacturers' guidelines at the W. M. Keck Center for Comparative and Functional Genomics, University of Illinois.
Transformation.
E. coli strains were transformed by electroporation by using an E. coli Gene Pulser (Bio-Rad) as recommended. Liposome-mediated transformation was used for Methanosarcina species as described earlier (21), modified in reference 4.
Plasmid constructions.
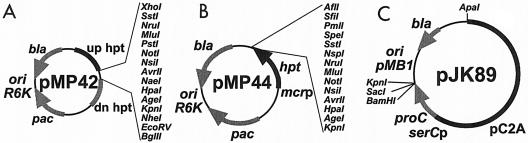
Plasmid constructions and oligonucleotide primers used are presented in Table 2. The physical map of selected plasmids is presented in Fig. 1. pJK41 and all derivatives are pir dependent in E. coli and are nonreplicating in Methanosarcina (20).
TABLE 2.
Plasmids and primers used in this study
| Plasmid or primer | Description and/or construction or primer sequence | Reference or source or added site |
|---|---|---|
| Plasmids | ||
| pJK41 | AmpR, PurR cloning vector, oriR6K replicon | 19 |
| pWM241 | pC2A replicon cloned into pBluescript SK | 20 |
| pWM368 | M. barkeri Fusaro mcrB promoter source | 39 |
| pWM369 | M. barkeri Fusaro serC promoter source | 39 |
| pWM370 | M. barkeri Fusaro orf2 promoter source | 39 |
| pCK12 | M. acetivorans C2A proC plasmid | 40 |
| pMP29 | NdeI/KpnI-cut hpt PCR product (using primers NdeI-apt and ApaIKpnIap) cloned into NdeI/KpnI-cut pWM368 | This study |
| pMP33 | AflII/XhoI-cut up-hpt PCR fragment (using primers AflIIaphpt and SacI-aphpt) cloned into AflII/XhoI-cut pJK41 | This study |
| pMP42 | BglII/BamHI-cut dn-hpt PCR fragment (using primers BglIIdnapt and BamHIdnapt) cloned into BglII/BamHI-cut pMP33 | This study |
| pMP44 | 762-bp SphI/ApaI fragment of pMP29 cloned in SphI/ApaI-cut pJK41 | This study |
| pMP45 | NdeI/BglII-cut uidA PCR product (using primers uidA (NdeI) and uidA(Bgl2)) cloned into NdeI/BglII-cut pWM368 | This study |
| pJK81 | NdeI/BglII uidA fragment from pMP45 cloned into NdeI-BamHI-cut pWM368 | This study |
| pJK82 | NdeI/BglII uidA fragment from pMP45 cloned into NdeI-BamHI-cut pWM369 | This study |
| pJK83 | NdeI/BglII uidA fragment from pMP45 cloned into NdeI-BamHI-cut pWM370 | This study |
| pMP62 | BamHI-cut/T4 DNA polymerase-treated proC fragment from pCK12 cloned into AflII-cut/T4 DNA polymerase-treated pMP44 | This study |
| pMP69 | SpeI/MluI-cut up-proC PCR product (using primers SpeI-proB and MluI-proB) and MluI/KpnI-cut dn-proC PCR product (using primers MluI-proC and KpnI-proC) cloned into SpeI/KpnI-cut pMP44 | This study |
| pJK84 | BglII-cut pMP62 treated with T4 DNA pol and dNTPs then religated | This study |
| pJK85 | SphI-Ecl136II mcrBp-uidA fragment of pJK81 cloned into BglII-cut/T4 DNA Pol-treated/SphI-cut pMP62 | This study |
| pJK86 | SphI-Ecl136II serCp-uidA fragment of pJK82 cloned into BglII-cut/T4 DNA Pol-treated/SphI-cut pMP62 | This study |
| pJK87 | SphI-Ecl136II orf2p-uidA fragment of pJK83 cloned into BglII-cut/T4 DNA Pol-treated/SphI-cut pMP62 | This study |
| pJK88 | NdeI/BamHI-cut E. coli proC PCR fragment (using primers procF and procR) cloned into NdeI/BamHI-cut pWM369 | This study |
| pJK89 | SpeI fragment of pWM241 with pC2A replicon cloned into XbaI-cut pJK88 | This study |
| Primers | ||
| NdeI-apt | CCGCCGCATATGCTTGAAAGACTGAAAGACTACATGG | NdeI |
| ApaIKpnIap | CCGCCGGGTACCGGGCCCTCACTGATCCCCAAAGACATCC | KpnI, ApaI |
| AflIIupapt | CCGCCGCTTAAGCCACTCCCGGAACAGAGG | AflII |
| SacI-upapt | CCGCCGCTCGAGTTTGGCGGTTTCAATATTCC | XhoI |
| BglIIdnapt | GGGGGGAGATCTCATATGTCTTTGGGGATCAGTGAAGC | BglII |
| BamHIdnapt | GGGGGGGGATCCCACCAGAGACTTTGCAATTACC | BamHI |
| uidA(NdeI) | GGGGGGCATATGTTACGTCCTGTAGAAACCC | NdeI |
| uidA(Bgl2) | GGGGGGAGATCTGATCATTAACGGCGCAGTACCG | BglII |
| SpeI-proB | GCCGCCACTAGTTTAGTTCGGGAGCAATAGGC | SpeI |
| MluI-proB | GCCGCCACGGCTTTAGTTTATCCCTTCCAGTAT | MluI |
| MluI-proC | GCCGCCACGGCTCGATTCCTTTTTAC | MluI |
| KpnI-proC | GCCGCCGGTACCACTGGTTCCGCCTGAAA | KpnI |
| procF | GCGCGCCATATGGAAAAGAAAATCGGTTTTATTGG | NdeI |
| procR | GCGCGCGGATCCGACGTAACCGCACCGAAGTG | BamHI |
FIG. 1.
Plasmids used in this study. (A) pMP42 is an M. acetivorans C2A hpt deletion plasmid. The regions upstream (up hpt; dark grey box) and downstream (dn hpt; light grey box) of hpt were PCR amplified and were cloned into pJK41 (thin black line [20]), creating the Δhpt allele. pJK41 and its derivatives are pir dependent in E. coli (oriR6K) and are nonreplicating in Methanosarcina species. pac and bla (light grey arrows) encode puromycin resistance in Methanosarcina species and ampicillin resistance in E. coli. Multiple cloning sites are available for integration of genes into the hpt locus. (B) pMP44 is the base plasmid for markerless exchange in M. acetivorans and is also derived from pJK41. The M. acetivorans hpt (medium grey arrow) is expressed from the M. barkeri Fusaro methyl-coenzyme M reductase promoter (mcrBp; light grey box). Multiple cloning sites are available for insertion of mutated alleles to be recombined onto the M. acetivorans chromosome. (C) pJK89 is a Methanosarcina-E. coli shuttle vector that carries selectable markers for proC complementation and ampicillin resistance. The E. coli proC gene (proC; dark grey arrow) is expressed from the M. barkeri Fusaro phosphoserine aminotransferase promoter (serCp; light grey box) and is functional in both E. coli and M. acetivorans. The pC2A replicon (21) allows autonomous replication in Methanosarcina species. pJK89 replicates via the high-copy-number pMB1 replicon in E. coli.
Determination of generation time.
Cells from mid-log-phase M. acetivorans cultures (optical density at 600 nm [OD600] ∼ 0.5) were harvested by centrifugation (5,000 × g for 10 min), washed once, and then resuspended in an equal volume of plain HS broth (without a carbon and energy source). Washed cells (0.3 ml) were inoculated into 10-ml broth cultures in triplicate and were incubated at 37°C on a tube roller. Growth was monitored at OD600 by using a Milton Roy Company Spectronic 21.
Determination of β-glucuronidase activity.
5-Bromo-4-chloro-3-indolyl-β-d-glucuronide (X-Gluc; Sigma) was used to qualitatively determine the presence of the uidA gene product (β-glucuronidase) in M. acetivorans. M. acetivorans colonies were blotted onto Whatman filter paper, flooded with 2 mg of X-Gluc/ml in 150 mM Tris-HCl, pH 8.0, and incubated aerobically at room temperature for 5 to 30 min. Clones expressing uidA turned blue (β-Gluc+). p-Nitro-phenol-glucuronide (pNP-Gluc) was used in quantitative assays of β-glucuronidase expressed from translational fusions of uidA to various M. barkeri genes. Cell extracts were prepared aerobically by pelleting mid-log-phase cultures (OD600 ∼ 0.5) by centrifugation (5,000 × g for 10 min), osmotic lysis of the cell pellet by resuspension in ice-cold 50 mM Tris-HCl, pH 8.0, and removal of cell debris by centrifugation at 4°C (14,000 × g for 15 min). Extracts were held on ice until assayed. Assays (1 ml) were performed at room temperature in 50 mM Tris-HCl, pH 8.0, with 4 mM pNP-Gluc. Assays were started by addition of pNP-Gluc and were continuously monitored at 415 nm by using a Hewlett-Packard 8453 UV-Visible Spectrophotometer. The specific activity (micromoles · minute−1 · milligram of protein) was calculated by using the molar extinction coefficient for pNP-Gluc (ɛ = 12,402 liters · mol−1 · cm−1), which was experimentally determined in the buffer conditions used in this assay. Protein concentration was determined by the Bradford assay kit from Pierce (Rockford, Ill.).
RESULTS
Isolation and characterization of hpt1::mini-mar mutant.
In a previous study we identified a transposon-induced mutant of M. acetivorans C2A as an auxotroph requiring supplementation with a cell-free Methanosarcina extract (37). However, the initial auxotrophic phenotype of this mutant was apparently an artifact of our growth conditions because the mutant grew without supplementation after a prolonged adaptation period. The transposon insertion site of this mutant was cloned and sequenced, revealing an insertion into an open reading frame with strong homology (53% identical with Fasta) (26) to the product of the Methanothermobacter thermautotrophicus hpt gene, namely, hypoxanthine PRTase (29). The transposon-induced mutant was therefore designated hpt1::mini-mar.
The M. acetivorans hpt gene (designated MA0717 in the M. acetivorans genome sequence; GenBank accession no. NC 003552) is 570 bp in length and encodes a protein of 189 amino acids. The downstream gene, MA0718, overlaps 20 bp at the 3′ end of hpt and, thus, may comprise an operon with hpt. MA0718 is a 1,008-bp gene encoding a 335-amino-acid protein with significant homology to diphthamide synthase and C-5 cytosine-specific DNA methylase.
Hypoxanthine PRTases convert hypoxanthine to inosine-5′-monophosphate and are involved in the purine salvage pathways of a number of organisms (16, 32, 36). This is consistent with our observation that the hpt1::mini-mar mutant originally behaved as an auxotroph. The medium used in the isolation of this strain contains exogenous nucleotide bases. We suspect that de novo synthesis is repressed in this medium and that the adaptation period required for prototrophic growth involves switching from the salvage pathway back to the de novo pathway. In some methanoarchaea, purine salvage mutants have been isolated by selecting resistance to purine analogs (6, 7, 17, 32, 36). Therefore, we tested wild-type M. acetivorans and the hpt1::mini-mar strain for sensitivity to a number of purine analogs (Table 3). For wild-type M. acetivorans, the most toxic purine analogs tested were 8ADP and 2MP, both of which inhibited growth at ca. 2 μg/ml. The hpt1::mini-mar mutant was significantly more resistant to both compounds, consistent with the proposed role of the hpt gene in purine salvage. The MIC of 8ADP for the hpt1::mini-mar mutant is >74.5 μg/ml, at least 35-fold higher than that for the wild type. 2MP, however, retained some toxicity for the mutant, causing very poor growth at 10 μg/ml and complete growth inhibition at 40 μg/ml. Therefore, the toxicity of 2MP is independent of hpt at this higher concentration, suggesting that the cell possesses another means to activate 2MP. Spontaneous 8ADPr mutants of M. acetivorans arise at a frequency of 2.9 × 10−5 (determined by plating dilutions of M. acetivorans C2A onto medium with and without 20 μg of 8ADP/ml).
TABLE 3.
MIC of purine analogs on M. acetivorans C2A and hpt1::mini-mar
| Purine analog | MIC (μg/ml)a
|
|
|---|---|---|
| C2A | hpt1::mini-MAR | |
| 2-MP | 1.31 | 10 |
| 8ADP | 1.74 | >74.5 |
| 6-Mercaptopurine | 5.67 | NTb |
| 2,6-Diaminopurine | 24.6 | NT |
| 6-Methylaminopurine | 107 | NT |
| 2-Aminopurine | >462 | NT |
| 2-Mercaptopurine | >462 | NT |
| 2-Amino-6-methylmercaptopurine | >462 | NT |
ca. 107 cells of the indicated strain were inoculated into HS-MA medium containing serial dilutions of a purine analog. Cultures were incubated at 37°C, and growth was scored after 14 days.
NT, not tested.
Construction of a Δhpt host strain for use in 8ADP counterselection strategies.
The hpt gene has been used as a counterselectable marker in other organisms (12); thus, the fortuitous isolation of this hpt1::mini-mar mutant afforded an opportunity to expand the genetic techniques available for use in M. acetivorans. To make use of this finding, an unmarked hpt mutant was constructed to serve as the host for the “markerless” exchange system described below.
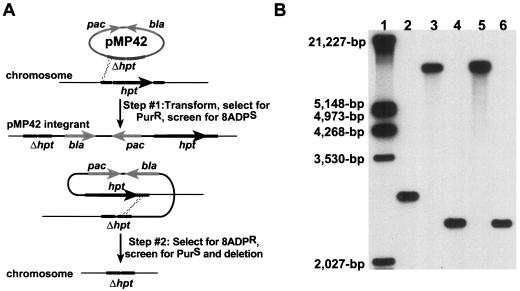
The hpt gene was deleted from the M. acetivorans C2A chromosome by using the two-step procedure shown in Fig. 2A. Approximately 800 bp of DNA upstream and downstream of hpt was cloned, creating the Δhpt allele carried on plasmid pMP42, which also carries the pac cassette encoding resistance to puromycin (Fig. 1A). (This deletion leaves the hpt promoter and downstream gene intact; however, we have made no attempt to verify expression of this gene.) Due to the plasmid's inability to replicate in Methanosarcina, selection for Purr pMP42 transformants results in hpt/Δhpt merodiploid strains with the plasmid integrated into the chromosome via homologous recombination (Fig. 2A, step no. 1). These merodiploids remain sensitive to 8ADP due to the presence of an intact copy of the hpt gene. Growth under nonselective conditions (i.e., without puromycin or 8ADP) allows resolution of the merodiploid by homologous recombination between the repeated sequences upstream and downstream of hpt. Approximately half of these recombinants leave the Δhpt allele on the chromosome, and these can easily be selected from the mixed population by selection for 8ADPr (Fig. 2A, step no. 2).
FIG. 2.
Construction of Δhpt host for markerless exchange. (A) The M. acetivorans hpt gene (black arrow) was deleted in a two-step process with pMP42. M. acetivorans was transformed to puromycin resistance (PurR) with pMP42 as described elsewhere. Because pMP42 is incapable of replication in M. acetivorans PurR transformants can only arise via homologous recombination (dotted lines) into the region flanking hpt (step no. 1). Integration through the upstream region is shown; however, integration into the region downstream of hpt is also possible. Integration of pMP42 into the chromosome creates an unstable merodiploid, which can segregate via a second homologous recombination event under nonselective conditions, resulting in plasmid excision from the chromosome (step no. 2). Some of these resolved merodiploids leave the Δhpt allele on the chromosome, and these recombinants are resistant to 8ADP (8ADPR). (B) The genome structure of two pMP42 integrants and their 8ADPR recombinants was verified by DNA hybridization of AluNI-digested genomic DNA to DIG-labeled pMP42 as described elsewhere. Lane 1, DIG-labeled EcoRI/HindIII lambda DNA (Roche); lane 2, M. acetivorans C2A; lane 3, pMP42 upstream integrant; lane 4, 8ADPr recombinant of the upstream integrant; lane 5, pMP42 downstream integrant; and lane 6, 8ADPR recombinant of the downstream integrant. The predicted sizes of the hybridizing bands for hpt, Δhpt, and both pMP42 integrants are 2,807, 2,437, and 7,371 bp, respectively, based on the known DNA sequence of the region.
To validate this strategy, we characterized 32 Purr pMP42 transformants of M. acetivorans. As expected, all 32 were Purr and 8ADPs. Ten of these pMP42 integrants were analyzed by DNA hybridization to determine the site of plasmid integration. Five integrants gave the predicted hybridization pattern for simple insertions into the chromosome, while the remaining five appeared to have multiple plasmids integrated into the chromosome in tandem (data not shown). Two simple integrants, one with pMP42 integrated into the region upstream of hpt, the other with pMP42 integrated through the downstream region, were grown nonselectively for 4 generations and were then plated onto medium with 8ADP to select recombinants that had resolved the merodiploid state in favor of the Δhpt allele (Fig. 2A, step no. 2). Eight 8ADPr recombinant clones (one from the upstream insertion and seven from downstream insertion) were characterized in detail. Two of these were both Purr and 8ADPr, suggesting that they had acquired point mutations in the intact hpt gene, and were not analyzed further. The remaining six recombinants were Purs and 8ADPr, indicating that they had resolved the merodiploid state by homologous recombination, resulting in the loss of the plasmid backbone.
These recombinants, along with their parental pMP42 integrants and wild-type M. acetivorans, were analyzed by DNA hybridization. As shown in Fig. 2B, M. acetivorans C2A (lane 2) shows the wild-type hpt hybridizing band. Both integrants (lanes 3 and 5) show a single hybridizing band with an increased size corresponding to insertion of pMP42 into the chromosome. The 8ADPr recombinants from each integrant (lanes 4 and 6) show a single, smaller hybridizing band consistent with the loss of the hpt gene and excision of the plasmid DNA. Similar results confirming the integration of pMP42 and subsequent deletion of hpt were obtained in hybridization experiments involving digests with three different restriction endonucleases (data not shown). One verified Δhpt mutant, designated WWM1, was used for all subsequent studies.
The generation times of M. acetivorans C2A and WWM1 were determined on methanol, trimethylamine, and dimethylamine (Table 4). The growth rate differed by less than 10% for M. acetivorans C2A and WWM1 for all substrates tested, indicating that this mutation does not grossly affect the physiology of M. acetivorans under laboratory conditions.
TABLE 4.
Generation time of M. acetivorans C2A and Δhpt strainsa
| Substrate | Generation time (h) for:
|
|
|---|---|---|
| C2A | Δhptb | |
| Methanol | 7.1 ± 0.2 | 7.8 ± 0.1 |
| Trimethylamine | 9.8 ± 0.4 | 10.4 ± 0.7 |
| Dimethylamine | 24.8 ± 1.2 | 27.4 ± 0.8 |
Determined as described in Materials and Methods.
WWM1 was used as the Δhpt strain.
Construction of ΔproC Δhpt by using “markerless” genetic exchange.
The Δhpt strain WWM1 allows efficient recombination of unmarked mutations onto the chromosome via a method that we designated markerless exchange because, although it uses selectable markers, it does not leave them on the chromosome at the end of the process (Fig. 3A). The method uses plasmid pMP44, which carries the selectable maker for Purr, pac, and the counterselectable marker hpt (Fig. 1B).
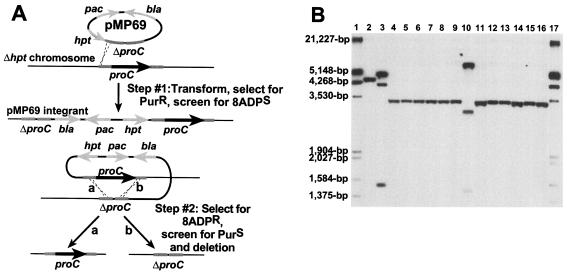
FIG. 3.
(A) Deletion of proC using markerless exchange. Markerless exchange utilizes the hpt gene as a counterselectable marker to construct gene deletions on the M. acetivorans chromosome. The proC gene was deleted from M. acetivorans Δhpt by using pMP69 with the ΔproC allele shown in light grey. M. acetivorans Δhpt was transformed to puromycin resistance (PurR) with pMP69 as described. Because pMP69 is incapable of replication in M. acetivorans, PurR transformants can only arise via homologous recombination (dotted lines) into the region flanking proC (step no. 1). (Insertion into the upstream region is shown. Integration is also possible via the downstream homology.) Integration of the plasmid creates an unstable merodiploid that is PurR and sensitive to the toxic purine analog ADP (8ADPS). Nonselective growth allows plasmid excision and resolution of the merodiploid allele via a second recombination event (dotted lines), resulting in either the wild type or the deletion allele, depending on which homologous region is used for the second event. (B) Verification of proC deletions. Genomic DNA from two pMP69 merodiploids and 12 8ADPR, Pro− recombinants was examined by hybridization of HindIII-digested genomic DNA to DIG-labeled pMP69 as described earlier. Lane 1, DIG-labeled EcoRI/HindIII lambda DNA (Roche); lane 2, WWM1; lane 3, pMP69 upstream integrant; lanes 4 to 9, 8ADPR, Pro− recombinants of the upstream integrant; lane 10, pMP69 downstream integrant; and lanes 11 to 16, 8ADPR, Pro− recombinants of the downstream integrant. Based on the known DNA sequence of the region, the hybridizing bands for proC and ΔproC are expected to be 4,463 and 3,388 bp, respectively. The pMP69 upstream integrant is expected to produce hybridizing bands of 4,818, 4,142, and 1,505 bp, while pMP69 downstream integrant is expected to give bands of 5,898, 3,062, and 1,505 bp.
To perform markerless exchange, the desired mutation is first cloned into pMP44. The resulting plasmid is then used to transform WWM1 to Purr. Because this plasmid is incapable of replication in Methanosarcina, these transformants arise by integration into the host chromosome via recombination between the mutant allele on pMP44 and the chromosomal wild-type allele. (Integration cannot occur at the hpt locus because the Δhpt mutation removes all homology to the hpt gene in pMP44.) The resulting merodiploids are 8ADPs due to the presence of hpt on pMP44; thus, recombinants that have resolved the merodiploid state (and therefore lost the integrated pMP44 backbone) can be selected as 8ADPr clones. Such recombinants are easily differentiated from spontaneous mutations within hpt because they also become Purs. Depending on where the resolving recombination event occurs, these Purs/8ADPr recombinants will retain either the wild-type or mutant allele. Assuming that these recombination events occur at equal frequency and that the mutant does not have a substantial growth disadvantage, the frequencies of the mutant and wild type should be roughly equal. Therefore, screening a relatively small number of recombinants should give the desired mutant.
To test this strategy, we constructed a mutant with an unmarked deletion of proC. This gene was previously shown to be dispensable in M. acetivorans and provides an easily testable phenotype when mutated (i.e., proline auxotrophy) (38). To construct the proC deletion, ca. 1 kb of PCR amplified DNA upstream and downstream of proC was cloned into pMP44, resulting in the ΔproC allele carried on plasmid pMP69 (Fig. 3A). M. acetivorans WWM1 (Δhpt) was grown with proline supplementation and was transformed to Purr with circular pMP69 (Fig. 3A, step no. 1). Analysis of 18 Purr transformants by DNA hybridization showed that seven clones had pMP69 integrated upstream of proC, while four clones had pMP69 integrated downstream of proC. The remaining seven Purr integrants gave an aberrant banding pattern that could not easily be explained by simple integration events and were not analyzed further (data not shown). One integrant of each type was grown nonselectively for 8 generations and was then subjected to 8ADP selection (Fig. 3A, step no. 2). To examine the efficiency of this method, we examined 100 8ADPr recombinants from the upstream integrant and 106 recombinants from the downstream integrant. Of the 100 8ADPr clones obtained from the upstream insertion strain, three were Purr, suggesting that they carried point mutations in the plasmid-encoded hpt gene, and 32 were proline auxotrophs. Similarly, two 8ADPr clones from the downstream insertion strain appeared to have point mutations in hpt, while 13 out of the remaining 104 exhibited proline auxotrophy. Twelve 8ADPr Pro− recombinants (six derived from each integrant) were analyzed by DNA hybridization to confirm loss of proC (Fig. 3B). All twelve recombinants gave a smaller hybridizing band of exactly the predicted size. One correct ΔproC recombinant, designated WWM24, was used for further studies.
The generation times of WWM1 and WWM24 were determined with and without proline supplementation. The generation times of WWM1 are 7.3 ± 0.3 and 7.2 ± 0.4 h with and without proline supplementation, respectively. The generation time of WWM24 with proline supplementation is 9.7 ± 0.8 h. Without proline, WWM24 did not grow. As will be discussed below, the difference in generation time between WWM1 and WWM24 with proline supplementation probably accounts for the low percentage of ΔproC recombinants obtained.
Integration of reporter gene fusions into proC.
In addition to its use for construction of defined deletions by using the markerless exchange method, the hpt counterselection can also be used to insert foreign genes into the M. acetivorans chromosome. To demonstrate this method, we constructed a series of translational fusions of various M. barkeri Fusaro promoters to the commonly used E. coli reporter gene uidA, which encodes β-glucuronidase, and recombined them into proC on the M. acetivorans WWM1 chromosome.
The promoter regions for the M. barkeri methyl-coenzyme M reductase (mcrBp) operon, the M. barkeri phosphoserine aminotransferase (serCp) gene, and the M. barkeri unknown open reading frame (orf2p) (37) were fused to uidA, resulting in plasmids pJK85, pJK86, and pJK87, respectively. These three promoters were expected to exhibit a wide range of activities based on previous studies (1, 37). pJK84 contains a frameshift in proC and was used to construct an isogenic strain lacking β-glucuronidase for use as a control. The mcrBp-uidA, serCp-uidA, and orf2p-uidA reporter gene fusions were integrated into the chromosome of WWM1 at the proC locus by using the markerless exchange. In this case, recombinants were screened for proline auxotrophy, puromycin sensitivity, and the presence of β-glucuronidase. Two independently isolated Pro−, Purs, β-Gluc+ clones for each fusion were identified, and β-glucuronidase activity was determined for each strain. As expected the fusions showed widely divergent β-glucuronidase levels, with the activity levels of the proC::mcrBp-uidA, proC::serCp-uidA, and proC::orf2p-uidA fusions being 6.7 ± 0.7 U, 2.1 × 10−2 ± 2.4 × 10−2 U, and 1.3 × 10−2 ± 0.5 × 10−3 U, respectively.
Use of E. coli proC as a selectable marker in M. acetivorans ΔproC strains.
The proline auxotrophy of the ΔproC strains was exploited to create an additional selectable marker for use in M. acetivorans. It was previously shown that proC mutations in M. acetivorans could be complemented by plasmid-encoded proC genes (38), thus allowing prototrophy to be used as a selection in these strains. To avoid unwanted recombination between the selectable pro+ marker and host chromosome, we constructed a gene cassette in which the E. coli proC gene was driven from the serC promoter of M. barkeri (22). This hybrid proC gene was then used as the selectable marker in the shuttle plasmid pJK89, which replicates via the high-copy-number pMB1 origin in E. coli and the medium-copy-number pC2A replicon in M. acetivorans (Fig. 1C).
Transformation of M. acetivorans WWM24 (ΔproC Δhpt) to prototrophy with pJK89 resulted in 4.4 × 107 total transformants per 2 μg of DNA, comparable to other shuttle vectors containing the pac cassette that routinely give ca. 107 transformants per 2 μg of DNA (21). Plasmid DNA isolated from Methanosarcina transformants could be used to retransform E. coli, and restriction endonuclease digestion patterns of plasmids obtained after passage through M. acetivorans were unchanged, indicating that the plasmid is structurally stable in M. acetivorans (data not shown).
DISCUSSION
The markerless exchange method using hpt as a counterselectable marker significantly expands the genetic capabilities available for study of M. acetivorans. Similar genetic tools have been reported for only a few archaea. A counterselection strategy has been described for use in two haloarchaea (3, 27); however, the method reported here is the first to be described for use in methanoarchaea. The hpt-based method was used here to construct insertions and deletions at the proC locus but subsequently has proven to be generally useful. In studies to be reported elsewhere, the method has been used to construct numerous deletions of genes involved in the utilization of methanol as a growth substrate (M. A. Pritchett and W. W. Metcalf, unpublished). A key feature of the method is that it can be used repeatedly to create strains with multiple mutations. To date we have constructed strains with as many as five unmarked deletions. Moreover, in cases where multiple genes are cotranscribed in an operon, the method allows construction of nonpolar mutations of individual genes within the transcriptional unit.
Two potential concerns, one more significant than the other, should be considered when employing this method. As mentioned above, resolution of the merodiploid state should result in ca. 50% wild-type and 50% mutant recombinants. This is based on the assumption that recombination between the duplicated upstream and downstream regions occurs at equal frequency and that the mutant and wild-type strains grow at the same rate following segregation. Although the first assumption has not been tested, it is very likely to be true, so long as the upstream and downstream regions of homology are of similar size. In contrast, the second assumption is expected to be true only for a subset of mutations. In this study, only 22% (45 of 201) of recombinants selected from proC+/ΔproC merodiploids were Pro−. Comparison of the generation times of proC+ and ΔproC mutant strains showed a significant difference (7.2 ± 0.4 h versus 9.7 ± 0.8 h, respectively). Thus, the wild-type cells would have outgrown the proC mutants during the nonselective growth and, as a result, would have constituted a larger percentage of the population at the time of plating onto selective medium. More pronounced differences in generation time would result in even larger skews in population sizes. Thus, use of this method for isolation of mutants with profound growth defects is likely to be problematic and the inability to isolate a mutation with this method should not be construed as proof that the gene in question is essential.
A more minor concern stems from the relatively high frequency of spontaneous 8ADP resistance (2.9 × 10−5). Accordingly, selection for resolution of merodiploids by 8ADPr can result in clones that have not lost the plasmid backbone but rather have acquired mutations in the hpt gene. These clones are easily distinguished from merodiploid-resolving recombinants because they retain the pac gene in the plasmid backbone and, hence, are Purr. In this study, only five of the 206 8ADPr clones selected from the proC+/ΔproC merodiploid were Purr, suggesting that spontaneous hpt mutations are rare compared to loss of plasmid by homologous recombination. Thus, spontaneous 8ADP resistance is not a significant problem for markerless exchange; however, the high spontaneous resistance rate does prevent direct selection of insertions and deletions at the hpt locus. In previous studies, we attempted to disrupt the hpt locus by direct selection for 8ADP resistance after transformation with linear DNA carrying a mutated hpt allele, but these attempts always failed to produce the desired recombinants. In all cases, the clones obtained appeared to be spontaneous 8ADPr mutants (data not shown). For this situation, it is likely that the combined losses in efficiency due to both transformation and a double recombination event (as opposed to the single event used in the strategy presented above) lowered the frequency of the desired events to well below that of spontaneous 8ADPr. These difficulties led us to develop the two-step integration-segregation strategy.
Despite the ability of markerless exchange to allow construction of multiple mutations in a single strain, it is often easier, and faster, to construct mutations with simple insertions of selectable markers. For this reason, we constructed a selectable marker based on the E. coli proC gene that confers prototrophy on ΔproC hosts. The lack of homology between the E. coli and M. acetivorans proC genes and its relative smallness are particularly useful features of this marker. The proC plasmids developed here can be used in strains containing mutations marked with the pac cassette, thus allowing facile complementation analysis. Although not explicitly tested, it is probable that the proC cassette of pJK89 will also function in single copy. Thus, this marker may also prove useful in transposon mutagenesis or gene replacement. However, pJK89 can only be used in proC strains like WWM24, which restricts its use somewhat. To our knowledge, this is the first instance of using a bacterial gene to complement an archaeal mutant.
Finally, the demonstration of reporter genes in Methanosarcina is an important advance. Although reporter gene fusions have been used in a wide variety of organisms, including methanoarchaea like Methanococcus species, such fusions have not yet been well described for use in Methanosarcina (although unpublished data cited in reference 19 suggest that lacZ can be used). In this study, β-glucuronidase activity was easily quantified over a 5,000-fold range in extracts of strains with uidA fusions to three different M. barkeri Fusaro promoters. Activity was undetectable in M. acetivorans strains lacking uidA fusions (data not shown). Further, β-glucuronidase activity could be qualitatively visualized by exposure of cells harboring the uidA gene to an X-Gluc solution. Unfortunately, visualization of β-glucuronidase activity with X-Gluc cannot be performed anaerobically because pigment formation does not occur under the strongly reducing conditions used to cultivate methanogens (25). Therefore, clones had to be replica plated or patched for aerobic testing with X-Gluc, while the original plates were maintained under anaerobic conditions to preserve cell viability.
The promoters used here were carefully chosen and probably represent nearly the full span of promoter activity in Methanosarcina: mcrBp drives expression of the genes for the enzyme methylreductase (1), which comprises ca. 5% of total cell protein, whereas transcripts from orf2p could barely be detected with a sensitive primer extension assay (21). Each fusion was constructed so that the ATG start codon of uidA was superimposed upon the start codon of the gene in question. Thus, the β-glucuronidase activity of each fusion represents contributions from both the transcriptional and translational control signals for each gene. The β-glucuronidase activity from the mcrB promoter was ∼300-fold higher than that from the serC promoter, while the serC promoter activity was ∼16-fold higher than that of the orf2 promoter. These promoter fusions provide a valuable baseline for comparison of future reporter gene fusions. Moreover, the availability of promoters of known strengths allows controlling expression of foreign genes by swapping promoters. Thus, we chose to express the E. coli proC gene in the selectable marker described above from the moderately expressed serC promoter (i.e., it seemed appropriate to drive expression of an amino acid biosynthetic gene from a known amino acid biosynthetic gene promoter). In contrast, expression of the mariner transposase gene from the mcrB promoter was much more effective than from serCp and orf2p in promoting transposition of the mini-MAR transposon in M. acetivorans (37).
The methods presented here, in combination with those previously published, comprise a basic toolkit that will now allow greatly expanded genetic studies of Methanosarcina species (4, 21, 22, 37, 38). Coupled with the metabolic diversity of members of this genus and the availability of complete, or nearly complete, genome sequences for three Methanosarcina species (9, 13), the prospects for future study of these complex and interesting microorganisms are very exciting.
Acknowledgments
This research was supported by grant MCB-987459 from the National Science Foundation to W.W.M.
REFERENCES
- 1.Allmansberger, R., S. Knaub, and A. Klein. 1988. Conserved elements in the transcription initiation regions preceding highly expressed structural genes of methanogenic archaebacteria. Nucleic Acids Res. 16:7419-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1992. Current protocols in molecular biology, vol. 1 and 2. John Wiley & Sons, New York, N.Y.
- 3.Bitan-Banin, G., R. Ortenberg, and M. Mevarech. 2003. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 185:772-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boccazzi, P., J. K. Zhang, and W. W. Metcalf. 2000. Generation of dominant selectable markers for resistance to pseudomonic acid by cloning and mutagenesis of the ileS gene from the archaeon Methanosarcina barkeri Fusaro. J. Bacteriol. 182:2611-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bochner, B. R., H. C. Huang, G. L. Chieven, and B. N. Ames. 1980. Positive selection for loss of tetracycline resistance. J. Bacteriol. 143:926-933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bowen, T. L., W. C. Lin, and W. B. Whitman. 1996. Characterization of guanine and hypoxanthine phosphoribosyltransferases in Methanococcus voltae. J. Bacteriol. 178:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bowen, T. L., and W. B. Whitman. 1987. Incorporation of exogenous purines and pyrimidines by Methanococcus voltae and isolation of analog-resistant mutants. Appl. Environ. Microbiol. 53:1822-1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bozue, J. A., L. Tarantino, and R. S. Munson, Jr. 1998. Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol. Lett. 164:269-273. [DOI] [PubMed] [Google Scholar]
- 9.Deppenmeier, U., A. Johann, T. Hartsch, R. Merkl, R. A. Schmitz, R. Martinez-Arias, A. Henne, A. Wiezer, S. Baumer, C. Jacobi, H. Bruggemann, T. Lienard, A. Christmann, M. Bomeke, S. Steckel, A. Bhattacharyya, A. Lykidis, R. Overbeek, H. P. Klenk, R. P. Gunsalus, H. J. Fritz, and G. Gottschalk. 2002. The genome of Methanosarcina mazei: evidence for lateral gene transfer between bacteria and archaea. J. Mol. Microbiol. Biotechnol. 4:453-461. [PubMed] [Google Scholar]
- 10.Fabret, C., S. D. Ehrlich, and P. Noirot. 2002. A new mutation delivery system for genome-scale approaches in Bacillus subtilis. Mol. Microbiol. 46:25-36. [DOI] [PubMed] [Google Scholar]
- 11.Fu, R., and G. Voordouw. 1997. Targeted gene-replacement mutagenesis of dcrA, encoding an oxygen sensor of the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough. Microbiology 143:1815-1826. [DOI] [PubMed] [Google Scholar]
- 12.Fukagawa, T., N. Hayward, J. Yang, C. Azzalin, D. Griffin, A. F. Stewart, and W. Brown. 1999. The chicken HPRT gene: a counter selectable marker for the DT40 cell line. Nucleic Acids Res. 27:1966-1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gay, P., D. L. Coq, M. Steinmetz, E. Ferrari, and J. A. Hoch. 1983. Cloning structural gene sacB, which codes for exoenzyme levansucrase of Bacillus subtilis: expression of the gene in Escherichia coli. J. Bacteriol. 153:1424-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gernhardt, P., O. Possot, M. Foglino, L. Sibold, and A. Klein. 1990. Construction of an integration vector for use in the archaebacterium Methanococcus voltae and expression of a eubacterial resistance gene. Mol. Gen. Genet. 221:273-279. [DOI] [PubMed] [Google Scholar]
- 16.Jochimsen, B., P. Nygaard, and T. Vestergaard. 1975. Location on the chromosome of Escherichia coli of genes governing purine metabolism. Adenosine deaminase (add), guanosine kinase (gsk) and hypoxanthine phosphoribosyltransferase (hpt). Mol. Gen. Genet. 143:85-91. [DOI] [PubMed] [Google Scholar]
- 17.Knox, M. R., and J. E. Harris. 1988. Isolation and characterization of mutants of mesophilic methanogenic bacteria resistant to analogies of DNA bases and nucleosides. Arch. Microbiol. 149:557-560. [Google Scholar]
- 18.Koprek, T., D. McElroy, J. Louwerse, R. Williams-Carrier, and P. G. Lemaux. 1999. Negative selection systems for transgenic barley (Hordeum vulgare L.): comparison of bacterial codA- and cytochrome P450 gene-mediated selection. Plant J. 19:719-726. [DOI] [PubMed] [Google Scholar]
- 19.Lange, M., and B. K. Ahring. 2001. A comprehensive study into the molecular methodology and molecular biology of methanogenic Archaea. FEMS Microbiol. Rev. 25:553-571. [DOI] [PubMed] [Google Scholar]
- 20.Metcalf, W. W. 1999. Genetic analysis in members of the domain Archaea, p. 278-326. In M. Smith and L. Sockett (ed.), Methods in microbiology: genetic methods for diverse prokaryotes. Academic Press, London, United Kingdom.
- 21.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metcalf, W. W., J. K. Zhang, X. Shi, and R. S. Wolfe. 1996. Molecular, genetic, and biochemical characterization of the serC gene of Methanosarcina barkeri Fusaro. J. Bacteriol. 178:5797-5802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Metcalf, W. W., J. K. Zhang, and R. S. Wolfe. 1998. An anaerobic, intrachamber incubator for growth of Methanosarcina spp. on methanol-containing solid media. Appl. Environ. Microbiol. 64:768-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller, V. L., and J. J. Mekalanos. 1988. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J. Bacteriol. 170:2575-2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noll, K. M., and M. Vargas. 1997. Recent advances in genetic analysis of hyperthermophilic Archaea and Bacteria. Arch. Microbiol. 168:73-80. [DOI] [PubMed] [Google Scholar]
- 26.Pearson, W. R. 1990. Rapid and sensitive sequence comparison with FASTP and FASTA. Methods Enzymol. 183:63-98. [DOI] [PubMed] [Google Scholar]
- 27.Peck, R. F., S. Dassarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 28.Pelicic, V., J. M. Reyrat, and B. Gicquel. 1996. Generation of unmarked directed mutations in mycobacteria, using sucrose counter-selectable suicide vectors. Mol. Microbiol. 20:919-925. [DOI] [PubMed] [Google Scholar]
- 29.Sauer, J., and P. Nygaard. 1999. Expression of the Methanobacterium thermoautotrophicum hpt gene, encoding hypoxanthine (guanine) phosphoribosyltransferase, in Escherichia coli. J. Bacteriol. 181:1958-1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sowers, K. R., J. E. Boone, and R. P. Gunsalus. 1993. Disaggregation of Methanosarcina spp. and growth as single cells at elevated osmolarity. Appl. Environ. Microbiol. 59:3832-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spring, K. J., J. S. Mattick, and R. H. Don. 1994. Escherichia coli gpt as a positive and negative selectable marker in embryonal stem cells. Biochim. Biophys. Acta 1218:158-162. [DOI] [PubMed] [Google Scholar]
- 32.Stuer-Lauridsen, B., and P. Nygaard. 1998. Purine salvage in two halophilic archaea: characterization of salvage pathways and isolation of mutants resistant to purine analogs. J. Bacteriol. 180:457-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tanuri, A., L. Jesus da Costa, R. Brindeiro, C. A. Ramos, C. P. Pau, and M. A. Rayfield. 2000. Construction of a selectable nef-defective live-attenuated human immunodeficiency virus expressing Escherichia coli gpt gene. Virology 268:79-86. [DOI] [PubMed] [Google Scholar]
- 34.van der Geize, R., G. I. Hessels, R. van Gerwen, P. van der Meijden, and L. Dijkhuizen. 2001. Unmarked gene deletion mutagenesis of kstD, encoding 3-ketosteroid Δ1-dehydrogenase, in Rhodococcus erythropolis SQ1 using sacB as counter-selectable marker. FEMS Microbiol. Lett. 205:197-202. [DOI] [PubMed] [Google Scholar]
- 35.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 36.Worrell, V. E., and D. P. Nagle, Jr. 1990. Genetic and physiological characterization of the purine salvage pathway in the archaebacterium Methanobacterium thermoautotrophicum Marburg. J. Bacteriol. 172:3328-3334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang, J. K., M. A. Pritchett, D. J. Lampe, H. M. Robertson, and W. W. Metcalf. 2000. In vivo transposon mutagenesis of the methanogenic archaeon Methanosarcina acetivorans C2A using a modified version of the insect mariner-family transposable element Himar1. Proc. Natl. Acad. Sci. USA 97:9665-9670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang, J. K., A. K. White, H. C. Kuettner, P. Boccazzi, and W. W. Metcalf. 2002. Directed mutagenesis and plasmid-based complementation in the methanogenic archaeon Methanosarcina acetivorans C2A demonstrated by genetic analysis of proline biosynthesis. J. Bacteriol. 184:1449-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]