Abstract
Human apolipoprotein A-I (apoA-I) is a 28 kDa protein and a major component of high-density lipoproteins, mediating several essential metabolic functions related to heart disease. In the present study the potential protective role against bacterial pathogens was explored. ApoA-I suppressed bacterial growth of Escherichia coli and Klebsiella pneumoniae. The protein was able to bind lipopolysaccharides and showed a strong preference for bilayer vesicles made of phosphatidylglycerol over phosphatidylcholine. Lysine side chains of apoA-I were acetylated to evaluate the importance of electrostatic forces in the binding interaction with both membrane components. Electrophoresis properties, dot blot analysis, circular dichroism, and fluorescence spectroscopy to probe for changes in protein structure indicated that the acetylated protein displayed a strongly reduced LPS and PG binding. A mutant containing only the N-terminal domain of apoA-I also showed a reduced ability to interact with the membrane components, although to a lesser extent. These results indicate the potential for apoA-I to function as an antimicrobial protein and exerts this function through lysine residues.
Keywords: apolipoprotein, apoA-I, lipopolysaccharides, phosphatidylglycerol, antimicrobial
1. Introduction
High-density lipoproteins (HDL) play a central role in reverse cholesterol transport, mediating the efflux of cholesterol from the peripheral tissues to the liver for metabolism and excretion [1]. In addition to the anti-atherogenic properties, HDL is recognized for playing a role in the innate immune response against pathogens. Evidence suggests that HDL is directly involved in several steps of the primary immune response pathway, serving a critical role in defending the host organism against viral, bacterial, and parasitic infections [2-4]. Specifically, apolipoprotein A-I (apoA-I), the main protein component of HDL, inhibits cell fusion mediated by HIV glycoproteins to host cells and HDL, suppresses bacterial growth and protects against bacterial infections in vivo[2, 3, 5]. HDL also possesses anti-inflammatory properties in response to lipopolysaccharides (LPS), the endotoxic component found in the outer membrane of gram-negative bacteria. Both in vivo and in vitro studies have indicated a positive correlation between HDL levels and protection against bacterial endotoxemia. Intravenous administration of reconstituted HDL protected rats and mice against lethal doses of LPS [6, 7]. Furthermore, in human clinical trials, treatment with exogenous reconstituted HDL results in diminished endotoxic effects upon exposure to LPS [8]. It is suggested that HDL particles neutralize LPS activity directly through partitioning of the toxic lipid A region into the lipid monolayer of the lipoprotein or indirectly by associating with lipopolysaccharide binding protein [6, 9].
Among the different lipoprotein classes, HDL displays the greatest anti-microbial activity and LPS binding affinity [3, 10]. Variations in bactericidal activity and anti-inflammatory responses between HDL, low-density and very low-density lipoproteins have led to the investigation into the role of lipoprotein-associated apolipoproteins. Several studies have reported that apoA-I is directly involved in the protection against bacterial infections and LPS toxicity. Treatment of gram-negative bacteria cultures with apoA-I suppresses cell growth, resulting in a reduced colony count [11]. In vitro studies have shown that apoA-I can directly associate with LPS, thereby rendering it biologically inactive [12]. Furthermore, animals injected with LPS pre-incubated with apoA-I have significantly decreased mortality rates compared to their counterparts treated solely with LPS [13].
Although the ability of apoA-I to exert anti-microbial activity and neutralize endotoxins has been documented, the specific molecular interactions involved are still poorly understood. In the present study, we have focused on the binding interaction of apoA-I with LPS and phosphatidylglycerol (PG). These negatively charged membrane components of the outer and inner bacterial membrane may serve as recognition sites for apoA-I through ionic interactions. To test this hypothesis, we modified the apoA-I lysine residues through acetylation and investigated its binding affinity to LPS and PG. In addition, to locate potential binding sites on apoA-I, binding studies were carried out with a deletion mutant comprised of the N-terminal domain of apoA-I.
Materials and Methods
2.1 Site-directed mutagenesis, protein expression, purification and sample preparation
The apoA-I C-terminal deletion mutant, apoA-IΔ190-243, was constructed by introducing a stop codon at amino acid position Ala 190 using the following primers: 5′-GGGGCCCGCCTGTAGGAGTACCACGCC-3′ and 5′- GGCGTGGTACTCCTACAGGCG GGCCCC -3′. The mutant construct (in the pET-20b(+) expression vector) was generated by polymerase chain reaction using a QuikChange-II site-directed mutagenesis kit (Agilent Technologies). The deletion of the C-terminal residues (Δ190-243) was verified by DNA sequencing (Genewiz). Recombinant apoA-I bearing a 6xHis-tag was over-expressed in E. coli BL21 (DE3) pLysS cells (Agilent Technologies), and induced with 0.5 mM IPTG at 37 °C for 3.5 h. Cells were collected by centrifugation at 8,000 g for 15 min (Sorvall RC5C Plus). Cells were resuspended in phosphate buffered saline (PBS; 150 mM NaCl, 10 mM NaH2PO4, 10 mM Na2HPO4, pH 7.4) and lysed by sonication using a digital sonifier (Branson) in five 30 s increments at 30 % amplitude. Sonicated samples were subjected to two rounds of centrifugation at 20,000 g for 30 min at 4 °C to remove cell components. Supernatant was mixed with equal parts (v/v) of loading buffer (2× PBS, 6 M guanidine-HCl, pH 7.4) and purified using 5 mL capacity Hi-trap chelating columns (GE Healthcare). Proteins were eluted using elution buffer (500 mM imidazole in PBS) pH 7.4 and dialyzed against 4 L of 10 mM ammonium bicarbonate, 1 mM EDTA with 3 additional buffer changes within 48 h. Proteins were further purified by size-exclusion chromatography using Superdex 200 resin in a XK-26/70 column (GE Healthcare). Prior to experimentation, proteins were dissolved in 6 M guanidine-HCl and dialyzed against 2 L of PBS at 4 °C with three additional buffer changes within 48 h.
2.2 Modification of apoA-I lysine residues
Recombinant wild-type (WT) apoA-I was dissolved in PBS to a final concentration of 0.5 mg/mL and mixed with an equal volume of saturated sodium acetate. Excess acetic anhydride was added in four equal parts every 15 min and mixed on ice for 1 h, followed by an additional mixing for 20 min. Modified proteins were dialyzed against PBS. ApoA-I samples were extensively dialyzed against 10 mM ammonium bicarbonate, 1 mM EDTA and subsequently lyophilized. The mass of the apoA-I samples was determined using MALDI-TOF (4800 MALDI TOF/TOF Analyzer, AB SCIEX) at the IIRMES facility at CSU Long Beach. Proteins were dissolved in sinapinic acid in 0.1% TFA and 50% acetonitrile.
2.3 Electrophoresis
For each apoA-I variant, 10 μg of protein was incubated at 70 °C for 10 min with lithium dodoceyl sulfate sample loading buffer (Invitrogen). Samples were run on a NuPAGE 10% Bis-Tris gel (Invitrogen) at a constant 200 V for 35 min using MES SDS running buffer. For non-denaturing electrophoresis experiments, 20 μg of protein was incubated at 37 °C with varying amounts of LPS serotype O55:B5 (Sigma Aldrich) for 1 h and subsequently cooled to ambient temperature prior to loading onto the gel. Samples were run on 4-20 % gradient Tris-glycine gels (Invitrogen) at a constant 125 V for 120 min. Gels were stained using 0.5 % (m/v) naphthol blue-black in 10 % glacial acetic acid, 45 % methanol (v/v) for 30 min, and subsequently destained overnight in 10 % glacial acetic acid and 45 % methanol.
2.4 Dot blot analysis
Immuno-dot blotting analysis was performed using a Bio-Dot microfiltration dot-blot apparatus (Bio-Rad). Nitrocellulose membranes were pre-soaked in Tris buffered saline (TBS, 20 mM Tris-HCl, 150 mM NaCl, pH 7.5). Unused wells were sealed off with tape to ensure proper vacuum. Sample wells were filled with 100 μL TBS and drained. LPS (50 μL of 10 ng/μL in PBS) was absorbed onto the nitrocellulose membrane. Unoccupied sites were blocked using 300 μL of 1% bovine serum albumin (BSA) in Tween-TBS (0.05% Tween-20 in TBS) and subsequently washed twice with 200 μL Tween-TBS. ApoA-I in a range of 0.1 to 1 μg was added to each sample well in triplicate. Unbound proteins were washed three times with 200 μL of Tween-TBS. Goat anti-human apoA-I HRP conjugated antibody (100 μL diluted 2,000-fold in 1% BSA, Tween-TBS; Abcam, Cambride, MA) was added to each sample well. Wells were subsequently washed three times with 200 μL Tween-TBS, and 3× 200 μL TBS to remove excess Tween. To each sample well 40 μL of Amersham ECL Plus Western Blotting Detection Reagent (GE Healthcare) was added, followed by 40 μL of deionized water. Bound apoA-I was visualized using Kodak BioMax XAR-2 autoradiography film with a 5 min exposure time. Films were imaged using Gel Doc XR and Quantity One software (Bio-Rad). Spot intensity on the image was determined using Image Lab software (Bio-Rad) by creating masks of equal size for each spot and the intensity of each spot was quantified. The average background intensity of control spots (no LPS or no apoA-I) was subtracted from the reported spot intensity values for treatments.
2.5 Circular dichroism
ApoA-I samples (0.25 mg/mL in 20 mM sodium phosphate buffer, pH 7.3) were incubated in the absence or presence of LPS at a 1:2 or 1:1 LPS-protein mass ratio. The ellipticity was measured between 185 and 260 nm in a Jasco 810 polarimeter using a cuvette with a 1 mm path length. Plots are the average of four scans, measured at a scanning rate of 50 nm/min. Ellipticity values were converted to molar ellipticity by the following equation:
θλ is the observed ellipticity (in degrees), d is the pathlength (cm), c is the protein concentration (g/mL), and MRW is the mean residue weight. The a-helical content was determined by using the following equation in which [θ]222 nm is the molar ellipticity at 222 nm [14]:
For guanidine-HCl denaturation analysis, apoA-I samples in a concentration of 0.2 mg/mL were incubated overnight at a given denaturant concentration in order to attain equilibrium, and ellipticity was measured at 222 nm.
2.6 Fluorescence spectroscopy
To measure exposed hydrophobic surfaces, apoA-I was incubated in the presence of 8-anilinonaphthalene-1-sulfonate (ANS; Sigma-Aldrich). Fifty μg apoA-I (dissolved in PBS) was pre-incubated with LPS (E. coli O55:B5, Sigma-Aldrich) in a 1:2 or 1:1 LPS-protein mass ratio for 15 min at 37 °C. ANS was prepared as a stock solution of 250 mM, and added to the mixture (2.5 mM final concentration) and incubated for another 15 min at 37 °C. ANS fluorescence was measured using a Perkin-Elmer LS-55 Fluorescence Spectrometer with a quartz semi-micro cuvette (Starna Cells). Samples were excited at a wavelength of 395 nm and the emission spectrum was measured from 400 to 600 nm (4 nm excitation and emission slit width) at 37 °C. All measurements were done in triplicate. For tryptophan fluorescence analysis, 50 μg apoA-I was preincubated with LPS (1:2 and 1:1 LPS-protein mass ratio) for 30 min at 37 °C in PBS. Tryptophan was excited at 295 nm to minimize the contribution of tyrosine residues present in apoA-I. The emission spectrum was measured from 300 to 450 nm using an excitation and emission slit width of 3.5 nm at 37 °C, and all measurements were done in triplicate.
2.7 Calcein release
Five mg of egg phosphatidylglycerol (PG) or egg phosphatidylcholine (PC) (Avanti Polar Lipids Inc.) was dissolved in 0.5 mL 3:1 chloroform/methanol (v/v) in phosphate-free glass tubes washed with Contrad 70 (Decon Labs). The phospholipids were dried with a stream of N2 gas and placed in a vacuum freeze dryer for 24 h. The lipid film was then suspended in Tris-HCl buffer (20 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, pH 7.4) containing 100 mM calcein (Sigma-Aldrich). After rigorous vortexing for 1 min, the lipid suspension was extruded with a Mini-Extruder (Avanti Polar Lipids Inc) at 45 °C through a membrane with a pore size of 100 nm to form large unilamellar vesicles (LUV) with encapsulated calcein. LUVs were separated from free calcein using a PD-10 desalting column (GE Healthcare). Phospholipid concentrations were determined using the Ames phosphate assay [15]. Changes in the fluorescence emission intensity, resulting from membrane leakage of entrapped calcein, was measured using a LS 55 Fluorescence Spectrometer (PerkinElmer). Excitation and emission wavelength were set at 490 and 520 nm, respectively (5 nm slit width). LUVs (10 μM) were equilibrated for 2 min in a 1 mL cuvet containing Tris-HCl buffer at 37 °C, after which the protein was added at varying lipid-protein molar ratios (10:1 - 10,000:1), which corresponded to 1 μM - 1 nM protein. The fluorescence emission intensity was monitored for another 8 min, after which 10 μl of a 1 % Triton X-100 solution was added to determine the maximum calcein release. The percentage of protein-mediated calcein release from the vesicles was determined using the following equation:
Ip is the maximum fluorescence intensity caused by the addition of protein, Ib is the background fluorescence intensity before addition of protein (just prior to the 2 min mark), Imax is the fluorescence intensity after detergent was added (10 min). Each experiment was conducted at least three times to generate the mean value ± standard deviation.
2.8 Antimicrobial assay
Escherichia coli (ATCC Number: 9637) or Klebsiella pneumoniae (ATCC Number: 13883) were cultured in liquid luria broth (LB; Fisher Scientific) for 12 h (37 °C, 225 rpm). While in the log-phase of growth, cells were diluted to an OD600 of 0.02 in LB, the diluted cells transferred to individual wells of a 96-well plate (Falcon) and further diluted 9:10 with apoA-I protein or BSA (0.4 mg/ml). Cells were cultured in the presence of test proteins for 1 h at 37 °C in a humidified chamber. Subsequently, cells were placed at 4 °C, serially diluted in PBS (Fisher Scientific) and 50 μl of each dilution plated on LB agar plates in duplicate or triplicate. The plates were subsequently incubated (37 °C for 16-20 h) and the colonies counted. The relative colony forming units (CFU) for each treatment was calculated using the following formula: Relative CFU = ((number of CFU in a treatment) / (average number of CFU in the BSA treatment) * 100). Error values are standard error of the mean (SEM). A minimum of duplicate samples were prepared for each treatment and the results show the combined data of at least three experiments conducted on different days.
3. Results
3.1 Modification of apoA-I
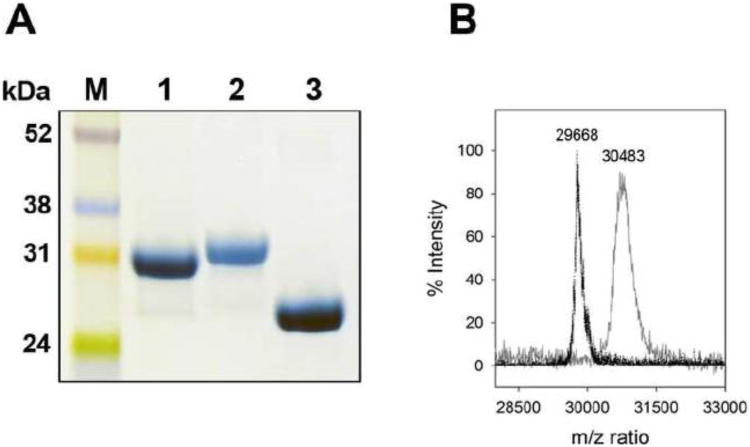
Lysine residues may play a critical role in the interaction of apoA-I with bacterial membrane components thereby exerting antibacterial activity. Lipopolysaccharides are anchored in the outer leaflet of the outer membrane, while the inner membrane bilayer is rich in anionic phospholipids such as PG [16]. To study this in detail, apoA-I was modified: the 21 positively charged ε-amino side chains of lysine were acetylated by acetic anhydride. It has been suggested that apoA-I adopts a two-domain structure [17]. The C-terminal domain (amino acid residues 190 to 243) contains 6 lysine side-chains and can be removed without disrupting the helix bundle stability [18]. This offers an opportunity to investigate if the N-terminal domain of the protein exerts similar LPS and PG binding properties compared to the full length protein, or that the C-terminal harbors potential binding sites for LPS and PG. Acetylation of apoA-I was analyzed by SDS-PAGE, showing a small decrease in electrophoretic mobility, resulting from an increase in mass from 30.3 kDa (WT apoA-I) to 32.1 kDa of the acetylated protein (Figure 1A). In addition, the acetylated protein was stained with decreased intensity, as amido black may preferentially bind to positively charged amino acid side chains. This result indicated that apoA-I was acetylated (referred to as ac-apoA-I), after which the degree of modification was calculated based on MALDI-TOF analysis. The mass shifted from 29,668 Da for WT apoA-I to 30,483 Da for ac-apoA-I, an increase of 815 Da which can account for 19.4 acetylations (42 Da increase per acetylation) (Figure 1B), indicating that most of the 21 lysine residues in apoA-I were acetylated (92.3 %). The C-terminal deletion mutant (apoA-IΔ190-243) was obtained by site-directed mutagenesis, and after expression and purification the deletion was confirmed by SDS-PAGE (mass of 22.4 kDa, Figure 1A) and MALDI-TOF (23,780 Da, expected mass 23,611 Da).
Fig. 1.
Analysis of apoA-I variants. Panel A shows the SDS-PAGE analysis of WT apoA-I (lane 1), ac-apoA-I (lane 2), and apoA-IΔ190-243 (lane 3). Panel B: MALDI-TOF analysis of WT apoA-I (m/z 29,668) and ac-apoA-I (m/z 30,483).
3.2 LPS binding interaction
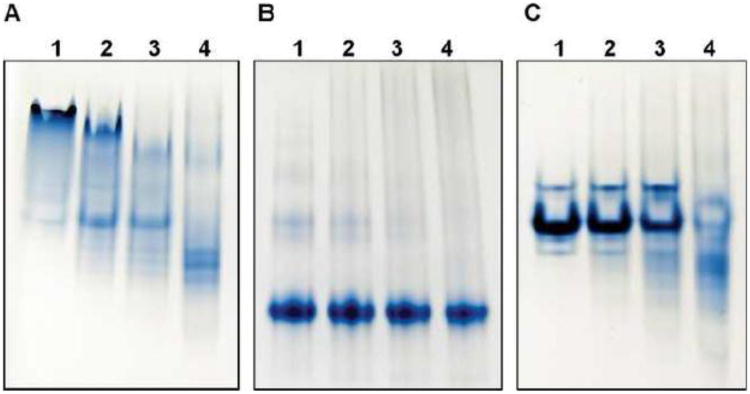
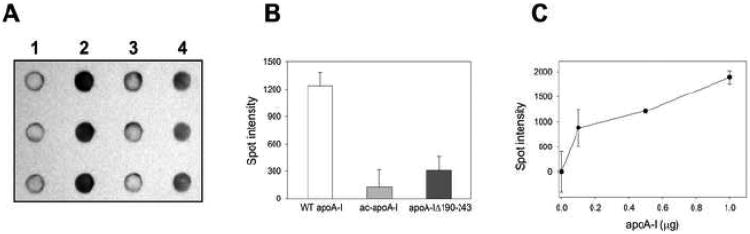
Binding of apoA-I to LPS was initially determined by changes in the electrophoretic mobility of apoA-I when subjected to non-denaturing PAGE. ApoA-I appears as a heterogeneous band at the top portion of the gel, which indicates the presence of oligomers. Addition of LPS to apoA-I led to a gradual disappearance of the original apoA-I bands, while new bands appeared at the lower portion of the gel. The increased mobility on PAGE indicates that apoA-I formed a complex with LPS (Figure 2A). The complex may have an increased net negative charge because of the phosphate moieties on LPS. The increased mobility of ac-apoA-I compared to WT apoA-I in the absence of LPS is likely caused by the neutralization of lysine side chains which decreased the theoretical pI from 5.27 to 4.24 (Figure 2B). In addition, the single band of ac-apoA-I indicates it may favor a monomeric state. In contrast to WT apoA-I, the position and staining intensity of ac-apoA-I did not change upon LPS addition, indicative of decreased LPS binding. Mobility and staining changes upon addition of LPS to apoA-I Δ190-243 were observed, but compared to the wild-type protein more LPS was required (Figure 2C). This indicates that the deletion mutant was still able to bind to LPS, but less efficiently than WT apoA-I. To further investigate this difference in LPS binding, dot blot analysis was performed. Membranes with LPS absorbed were incubated with the three protein variants. This showed that the ac-apoA-I dot was significantly less intense compared WT apoA-I (Figure 3A and B). This result indicates a decrease in LPS binding activity of ac-apoA-I and is in agreement with the electrophoresis analysis. The intensity of apoA-I A190-243 was also reduced, but not as much as the acetylated protein. In addition, control experiments showed that the spot intensity was proportional to the amount of apoA-I (Figure 3C).
Fig. 2.
Non-denaturing PAGE of apoA-I variants in the absence or presence of LPS. Panel A: WT apoA-I; panel B: ac-apoA-I; panel C: apoA-IΔ190-243. Proteins (20 μg) in the absence of LPS are shown in lane 1. Lanes 2-4: 20 μg apoA-I pre-incubated with 4 μg (lane 2), 10 μg (lane 3), and 20 μg LPS (lane 4).
Fig. 3.
Dot blot analysis of apoA-I binding to LP S. A and B: 0.5 μg LPS was loaded onto the nitrocellulose membrane followed by addition of 1 μg of apoA-I. Sample analysis was performed in triplicate for each of the apoA-I variants and LPS control. 1) LPS in the absence of apoA-I; 2) WT apoA-I; 3) ac-apoA-I; 4) apoA-IΔ190-243. The spot intensity is shown in panel B (arbitrary units). C: Spot intensity as a function of apoA-I concentration (0.5 μg LPS).
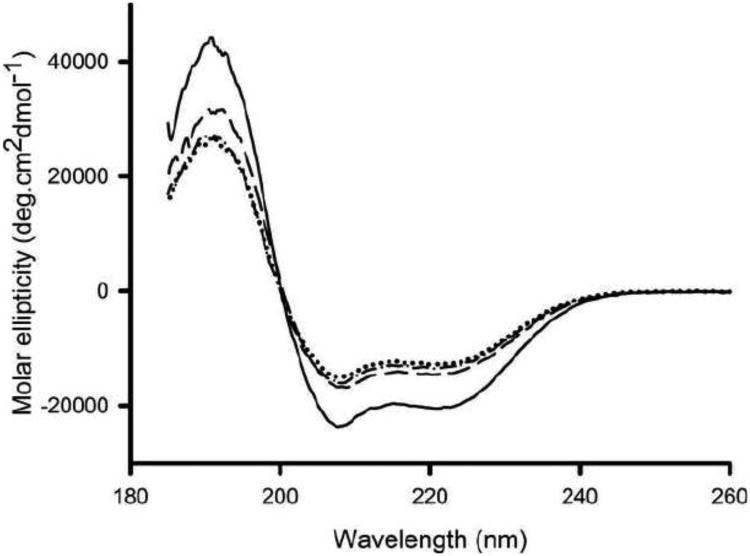
The Far-UV CD spectrum of WT apoA-I is characteristic for an α-helical protein with troughs at 208 and 222 nm (Figure 4). The lysine modification of apoA-I caused a modest decrease in the α-helical content, from 59 to 43 % (Table 1). The midpoint of guanidine induced denaturation was 0.92 ± 0.04 M for WT and 0.91 ± 0.05 M for ac-apoA-I, indicating that acetylation did not affect the stability of the protein. ApoA-IΔ190-243 also displayed a reduced α-helical content. Thus, while the proteins lost some of their helical content, they retained their overall α-helical character and stability. Addition of LPS to apoA-I caused a significant reduction in molar ellipticity (Figure 4), resulting in a decrease in α-helical content from 59 to 42% (2:1 protein/LPS mass ratio) or 40% (1:1 protein/LPS mass ratio). This result indicated that LPS binding resulted in a change in the structural organization of apoA-I. On the other hand, the molar ellipticity of ac-apoA-I remained essentially unchanged upon LPS addition. Similar to the WT protein, LPS addition to apoA-IΔ190-243 caused a reduction in the helical content (Table 1).
Fig. 4.
Effect of LPS on the secondary structure of WT apoA-I. Far UV circular dichroism scan of apoA-I in the absence and presence of LPS. Shown are WT apoA-I (——), WT apoA-I + LPS (……), ac-apoA-I (— —) and ac-apoA-I + LPS (—· —·).
Table 1.
ANS and tryptophan fluorescence, and circular dichroism analysis of apoA-I and variants in the presence or absence of LPS.
| ANS fluorescence | Tryptophan fluorescence | Circular dichroism | ||||
|---|---|---|---|---|---|---|
|
| ||||||
| protein | protein:LPS ratio | λmax | intensity | λmax | intensity | α-helical content |
| WT apoA-I | 1:0 | 478 nm | 143 ± 4 | 348 nm | 499 ± 38 | 59.2 ± 0.4 % |
| 2:1 | 469 nm | 188 ± 6 | 349 nm | 613 ± 90 | 42.3 ± 0.1 % | |
| 1:1 | 462 nm | 202 ± 18 | 350 nm | 623 ± 67 | 40.1 ± 0.3 % | |
| ac-apoA-I | 1:0 | 482 nm | 48 ± 4 | 348 nm | 203 ± 26 | 43.3 ± 1.6 % |
| 2:1 | 482 nm | 57 ± 4 | 348 nm | 205 ± 32 | 43.6 ± 1.9 % | |
| 1:1 | 482 nm | 55 ± 2 | 348 nm | 204 ± 24 | 41.4 ± 0.5% | |
| apoA-I Δ 190-243 | 1:0 | 482 nm | 63 ± 2 | 350 nm | 469 ± 65 | 41.9 ± 1.8 % |
| 2:1 | 477 nm | 102 ± 2 | 350 nm | 553 ± 59 | 34.4 ± 3.1 % | |
| 1:1 | 475 nm | 134 ± 5 | 350 nm | 540 ± 78 | 32.7 ± 4.7 % | |
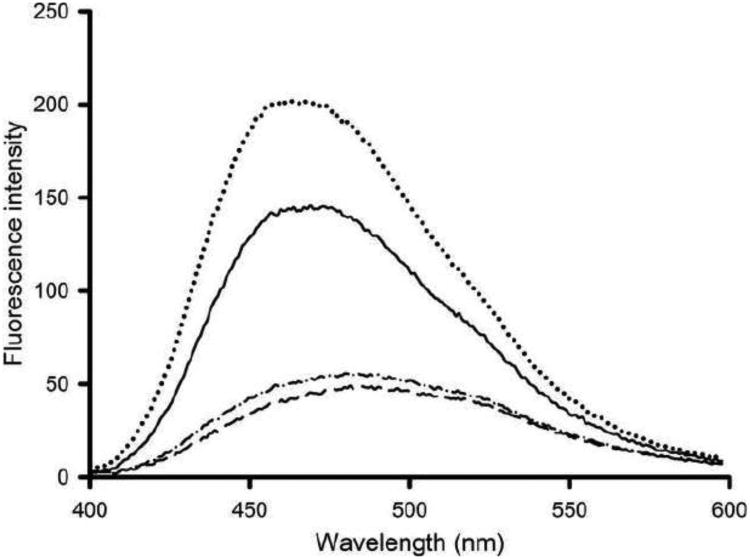
ANS typically binds to exposed hydrophobic pockets on proteins, evident by a large increase in fluorescence intensity and a substantial blue-shift in the wavelength of maximum emission (λ max) [19, 20]. Since changes in secondary structure were observed upon LPS binding, this may also lead to changes in apoA-I ANS binding sites. Control incubations of LPS with ANS showed that ANS fluorescence intensity did not change, indicating that ANS did not bind to LPS (data not shown). However, ANS fluorescence was strongly increased upon incubation with apoA-I, concomitant with a blue-shift in λ max from 511 to 480 nm. The ANS fluorescence intensity in the presence of apoA-I was further increased upon incubation with LPS (Figure 5). This indicates an increased number of ANS binding sites on apoA-I induced by the presence of LPS. ANS fluorescence intensity was less intense for ac-apoA-I (Table 1). In contrast to the unmodified protein, ANS fluorescence intensity remained virtually unchanged when LPS was incubated with ac-apoA-I. LPS addition to apoA-IΔ190-243 resulted in an increase in the ANS fluorescence intensity, indicating that the deletion mutant retained the ability to bind to LPS.
Fig. 5.
Effect of LPS on the ANS binding to LPS. Shown are the ANS fluorescence emission intensity (arbitrary units) between 400 and 600 nm in the presence of WT apoA-I (——) and a 1:1 (mass) mixture of ……), ac-apoA-I (— —), and ac-apoA-I + LPS (—· —·).
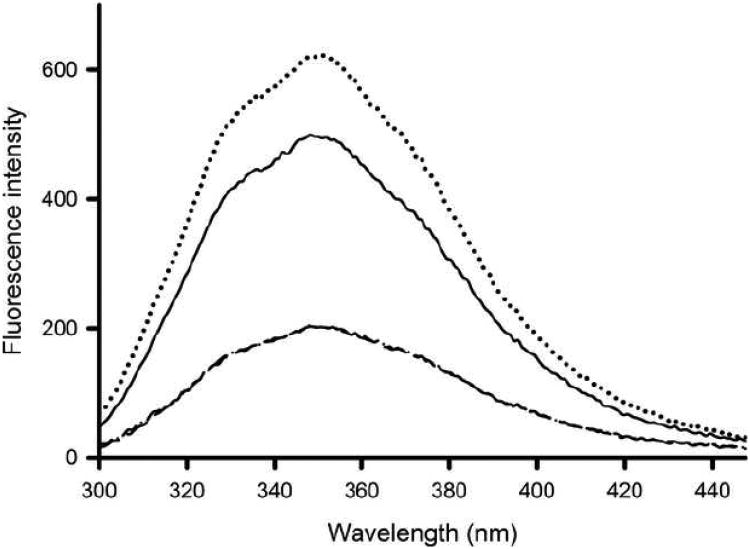
ApoA-I bears four tryptophan residues at positions 8, 50, 72 and 108, all located in the N-terminal domain of the protein. The tryptophan fluorescence intensity of apoA-I was ∼500 (arbitrary units) with a λ max of 348 nm, indicating that the average environment of the tryptophan residues is polar. Incubation with LPS resulted in a 2 nm red-shift, with an increased fluorescence intensity to 623 (Figure 6). The tryptophan fluorescence intensity of ac-apoA-I was substantially lower, and no change was observed in either λ max or emission intensity upon incubation with LPS. ApoA-IΔ190-243 incubations in the absence and presence of LPS showed a modest increase in the fluorescence intensity (Table 1).
Fig. 6.
Tryptophan fluorescence emission scan. ApoA-I samples were excited at 295 nm and the emission monitored between 300 and 450 nm (arbitrary units). Shown are WT apoA-I (— —), WT apoA-I in the presence of LPS (……), ac-apoA-I (— —) and ac-apoA-I + LPS (—· —·).
3.2 ApoA-I binding to PC and PG vesicles
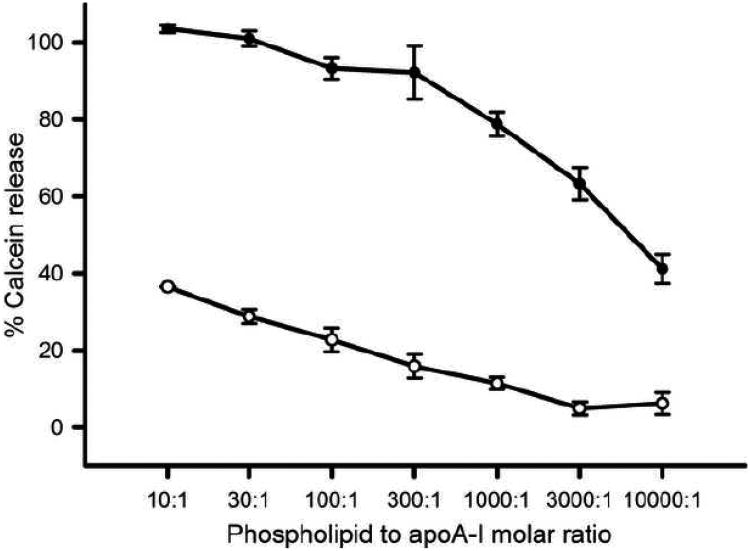
Negatively charged phospholipids such as PG are abundantly present in membranes of gram-negative bacteria. To investigate the ability to interact with negatively charged membranes, calcein was incorporated in PG vesicles at 100 mM concentration at which the fluorophore is highly quenched. Protein binding to the LUV surface may then result in packing defects in the phospholipid bilayer, and calcein is released in the surrounding buffer. The diluted concentration of calcein in buffer causes it to fluoresce strongly at 520 nm. The ability of apoA-I to release calcein from PG vesicles was then compared to PC vesicles. The release of calcein from PG vesicles (10 μM) by apoA-I was >90 % at protein concentrations between 1 and 0.033 μM. Further dilution of apoA-I reduced the activity but even dilution to a concentration of 1 nM apoA-I was still able to trigger a ∼40 % release (Figure 7). In contrast, when calcein was incorporated into LUVs made of PC, an abundant phospholipid in eukaryotic cell membranes, the activity was strongly reduced: 1 μM apoA-I released 38% of calcein, thus 1000 times more protein was required to release a similar amount of calcein from PC membranes compared to PG membranes. This result indicated that apoA-I has a strong preference for PG membranes compared to PC membranes.
Fig. 7.
ApoA-I-induced calcein release from PG and PC vesicles. A 10 μM suspension of PG vesicles (●) or PC vesicles (○) was incubated with various concentrations of WT apoA-I (1 μM -1 nM, molar lipid to protein ratio of 10:1-10,000:1) after which the % of calcein was determined by the change in fluorescence intensity at 520 nm.
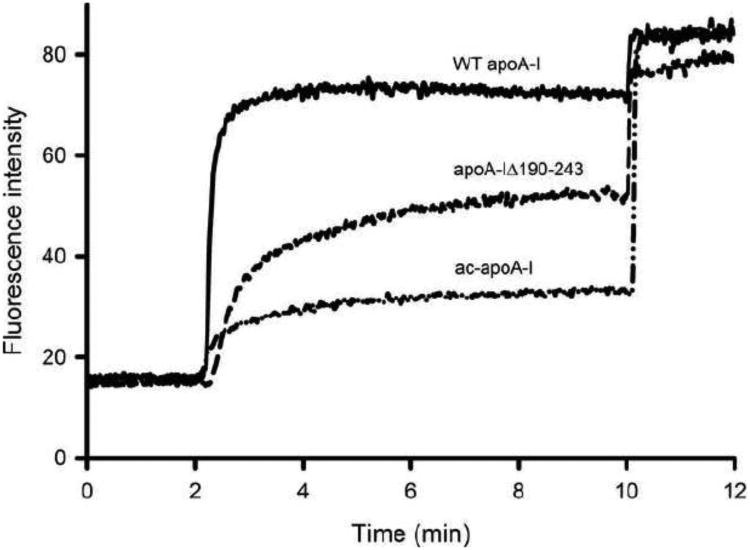
To assess the ability of the apoA-I variants to release calcein from PG membranes, 3.3 ng protein was used (lipid to protein molar ratio of 3000:1), a concentration that allows for the measurement of changes in calcein release. After equilibrating a 10 μM suspension of PG vesicles containing calcein for 2 min, apoA-I was added resulting in a rapid increase in fluorescent intensity and thus release of calcein (Figure 8). Addition of detergent at 8 min released calcein from the remaining non-lysed vesicles. ApoA-I was able to release 80.1 ± 2.4 % of encapsulated calcein, but calcein release was reduced to 25.9 ± 0.70 for ac-apoA-I and 57.3 ± 3.6% for apoA-IΔ190-243.
Fig. 8.
Calcein release from PG vesicles by apoA-I variants. Vesicles (10 μM) were equilibrated for 2 min. ApoA-I (3.3 nM, 3000:1 PG-protein molar ratio) was added at 2 min to calcein-filled PG vesicles and the fluorescence intensity monitored until 10 min at which the remaining vesicles were lysed by addition of Triton X-100.
3.3 Antibacterial activity of apoA-I
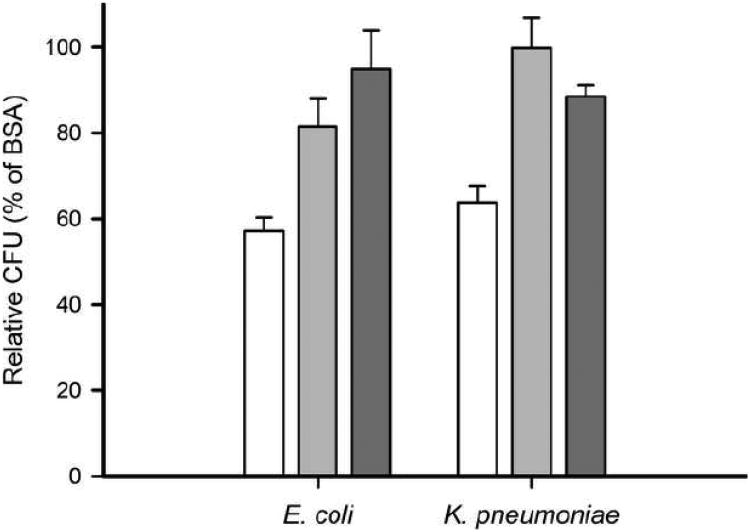
Antibacterial activity was analyzed by assessing bacterial growth of the gram-negative bacteria E. coli and K. pneumoniae (Figure 9). The relative number of CFU for E. coli and K. pneumoniae incubated in the presence of apoA-I protein was reduced relative to cells treated with BSA (57.2 ± 3.1 % and 63.8 ± 3.9 %, respectively), demonstrating that apoA-I is an antibacterial protein. E. coli was moderately, but not significantly, more sensitive than K. pneumoniae to apoA-I. The antibacterial activity of apoA-I was attenuated when the protein was acetylated as measured by the increased relative number of CFU for E. coli and K. pneumoniae incubated in the presence of ac-apoA-I (81.6 ± 6.5 % and 99.8 ± 7.1 %, respectively), suggesting that positively charged lysine residues of apoA-I play a critical role in the antibacterial activity. Of note, acetylation of apoA-I decreased the antibacterial activity in tests of K. pneumoniae. The C-terminal truncation mutant apoA-IΔ190-243 did not show high levels of antibacterial activity against E. coli or K. pneumoniae (94.9 ± 9.0 % and 88.5 ± 2.6 % of controls, respectively).
Fig. 9.
Antibacterial activity of apoA-I of E. coli or K. pneumonia. Anti-microbial activity was analyzed by assessing bacterial growth of log-phase gram-negative bacteria in the presence of apoA-I (white bars), ac-apoA-I (light gray bars) and apoA-I Δ190-243 (dark gray bars). Colony counts were relative to BSA (100 %) and were significantly reduced in the presence of apoA-I (57 % and 64 % CFU, respectively). The potency of apoA-I to reduce bacterial growth was significantly decreased after acetylation (82 % and 100 % CFU) or removal of the C-terminal end (95 % and 89 %). E. coli unpaired t-test: WT vs. ac-apoA-I (P = 0.0020, t = 3.849, df = 13); WT vs. apoA-I Δ190-243 (P = 0.0003, t = 4.951, df = 13), ac-apoA-I vs. Δ190-243 (P = 0.2614, t = 1.208, df = 8). K. pneumonia unpaired t-test: WT vs. ac-apoA-I (P = 0.0011, t = 4.285, df = 12), WT vs Δ190-243 (P = 0.0083, t = 3.155, df = 12), ac vs. Δ190-243 (P = 0.2091, t = 1.496, df = 4).
Discussion
Human apoA-I is a 28 kDa single polypeptide composed of 243 amino acid residues. It is composed of amphipathic α-helices, and circular dichroism analysis estimated an overall helical content of 50-57% [21]. ApoA-I is a highly flexible protein, allowing it to exist in lipid-free, lipid-poor (pre β HDL) and lipid-rich states (spherical HDL) [22]. Extensive structural analysis suggests that apoA-I is composed of two independently folded domains, consisting of an N-terminal helix bundle (residues 1-189) and a loosely structured C-terminal domain (residues 190-243) [17, 23]. The C-terminal domain displays high lipid binding affinity, and possibly initiates lipid binding to attach apoA-I to the lipid surface [24]. However, other studies suggested that the N-terminal helix can also act as a lipid anchor [25]. The C-terminal domain of apoA-I has been identified as the site for the self-associating properties of the protein [24]. This structural and functional organization bears similarities with that of human apoE [26].
Gram-negative bacterial membranes are composed of an outer membrane with LPS protruding from the outer leaflet, and an inner membrane rich in PG [27, 28]. Both membrane components are potential targets for antimicrobial proteins, and in the current study we investigated the role of lysine residues and the contribution of the C-terminal domain on the ability of apoA-I to interact with these bacterial membrane components. Based on extensive analysis of antimicrobial proteins and peptides, it is generally recognized that the following properties are attributed with effective protection against gram-negative bacterial pathogens: 1) high cationic residue content and 2) large hydrophobic regions, typically oriented in an amphipathic motif [29, 30]. Thus exchangeable apolipoproteins with the amphipathic α-helix as a signature feature for mediating lipid binding have the potential to function as antimicrobial proteins. For example, fish apoA-I has been recognized as an antimicrobial protein, human apoE is effective in neutralizing a potentially lethal dose of LPS, and insect apolipoprotein, apolipophorin III (apoLp-III), is able to associate with LPS and bacterial membranes [31-35].
In the present study it was shown that incubation of two gram-negative bacteria strains in the presence of WT apoA-I significantly suppressed bacterial growth, resulting in reduced colony counts. On the other hand, ac-apoA-I and apoA-I Δ190-243 displayed attenuated bacterial growth suppression. Although minor, differences in bacteriostatic effects between E. coli and K. pneumoniae may be due, in part, to differences in recognition of the expressed O-antigen present on the bacterial surface. Previous studies have reported that apoA-I anti-microbial activity is partially mediated by its ability to discern the O-antigen component of bacterial pathogens [36]. In addition, studies with apoLp-III have shown that the apolipoproteins can recognize the carbohydrate portion of LPS, although this interaction is weaker compared to complete LPS [34].
Upon incubation with LPS, apoA-I displayed significant changes in electrophoretic mobility under non-denaturing conditions, indicating binding interaction between the protein and LPS. The altered electrophoresis properties may have resulted from changes in size and overall net charge of newly formed protein-LPS complexes. Such complexes have been characterized for apoLp-III, and consist of 3 to 4 apolipoproteins and 9 to 24 LPS molecules, depending on the LPS source [34]. The LPS-induced change in PAGE mobility was reduced for apoA-IΔ190-243, while addition of LPS to ac-apoA-I did not alter the electrophoretic mobility of the protein. Dot blot analysis showed a decrease in LPS binding for ac-apoA-I and, to a lesser extent, for apoA-IA190-243. The interaction of apoA-I with LPS was further analyzed by measuring structural changes of apoA-I and variants due to LPS binding interaction. It was shown previously that apoA-I undergoes partial unfolding in certain a-helical regions in the presence of LPS [37]. In the current study we also observed a decreased α-helical content of WT apoA-I upon LPS binding. This implies a structural reorganization of the apolipoprotein induced by LPS, although we have no information about the exact nature of this change. Ac-apoA-I displayed virtually no change in a-helical content when incubated in the presence of LPS, whereas apoA-I Δ190-243 showed changes in secondary structure but to a lesser degree compared to WT apoA-I. Likewise, assessment of changes in the exposure of hydrophobic regions by ANS fluorescence revealed similar trends among WT and apoA-I variants. The increased ANS binding to apoA-I in the presence of LPS again indicate structural changes in apoA-I induced by LPS. Analysis of the tryptophan emission spectra revealed a similar pattern. The observed trends in LPS binding among the apoA-I variants reveal that electrostatic interactions contribute significantly to the ability of apoA-I to bind with LPS, and that such interactions may occur between the lysine e-amino groups and the negatively charged moieties of the lipid A region. Similar binding interactions have also been reported for HDL, which has been shown to interact with phosphate groups and the diglucosamine backbone of LPS [38]. The C-terminal domain may be at least partly responsible for LPS binding, although LPS binding was not completely lost upon removal of the C-terminal domain. This suggests that also a part of the N-terminal core domain is capable of LPS binding, presumably because of the amphipathic character of the α-helices. It was recently shown that a fragment comprising amino acid residues 148-243 may harbor the site that is responsible for the neutralization of LPS toxicity [37].
A comparison of protein interactions with PC or PG LUVs showed that apoA-I displayed a strong preference for PG. Binding activity, as measured by the ability to release calcein from bilayer vesicles, was approximately 1,000 times higher for when the LUVs were made from PG compared to PC. This result may point out that apoA-I is able to discriminate between PC-rich mammalian membranes and bacterial membranes containing significant amounts of PG. Thus it seems apparent that apoA-I utilizes electrostatic forces to differentiate between host and pathogenic cells. However, gangliosides present in the mammalian membrane also provides a net negative charge. Therefore, charge distribution on the apolipoprotein surface may be required for membrane specificity. The apoA-I variants displayed similar trends in activity among interactions with PG vesicles compared to LPS binding. Upon acetylation, the ability of apoA-I to release calcein from PG vesicles was significantly reduced. It is interesting to note that, although apoA-I contains roughly an equal amount of lysine and arginine residues (21 and 16, respectively), neutralization of lysine residues alone resulted in a major decline in activity. Removal of the C-terminal domain reduced the ability to release calcein, although the deletion mutant remained active. It seems reasonable to conclude that charge-charge interactions are the main driving force for apoA-I to bind to PG membranes, which is similar to that observed for apoLp-III [39]. Similar to LPS binding, the N-terminal domain of apoA-I contributes to the PG binding activity. In contrast, previous studies showed that the conversion of PC vesicles by apoA-IΔ190-243 into discoidal lipid-protein complexes was greatly reduced [23, 40]. However, the present study showed that while reduction in activity was observed, the protein remained active for PG vesicles.
It cannot be excluded that unforeseen structural changes in the protein variants tested here affected the LPS and PG binding. Acetylation decreased the α-helical content, although the extent was less than previous reported [41]. Lipid binding, measured by the conversion of PC vesicles into discoidal complexes, was hardly affected [41]. In addition, the present study showed that the protein stability was retained upon acetylation. Removal of ∼ 50 C-terminal amino acid residues could potentially affect the structure in the remaining protein. However, the C-terminal domain may be unstructured in the lipid-free form of the protein and thus the deletion will probably not affect the core structure. This explains why WT and apoA-I A190-243 display a similar protein stability [40, 42]. In any event, structure-guided site-directed mutagenesis will be a necessary tool to pinpoint the exact regions in the protein responsible for the membrane interaction.
The results of the present study, taken together, suggest that ionic forces predominately contribute to the binding interaction with LPS and PG bilayer vesicles. Electrostatic interactions between the ε-amino groups of lysine side chains may initiate binding with negatively charged phosphate moieties on LPS, after which the amphipathic character of apoA-I facilitates binding to the hydrophobic portion of LPS. PG, which is abundantly present in the inner membrane of the bacteria, may then recruit apoA-I to the surface leading to membrane destabilization, further contributing to the bacteriostatic properties of apoA-I.
Gram-negative bacterial infections present a major concern in the healthcare field, accounting for approximately 70% of hospital-acquired infections in intensive care units and 30% of total hospital acquired infections [43]. In addition, the increasing resistance of these organisms to traditional antibiotics and the decline in the development of novel therapeutics has created a pressing need to explore alternative strategies. In recent years, apoA-I has garnered recognition for its ability to exert anti-microbial properties and inhibit bacterial sepsis by neutralizing LPS. In addition, apoA-I exhibits virtually none of the toxic effects, which are typically associated with other anti-microbial peptides, making it an advantageous alternative to current methods of treatment against gram-negative bacterial infections.
Highlights.
apoA-I binding to LPS was strongly reduced upon acetylation of lysine residues
apoA-I shows a much stronger binding interaction with PG compared to PC
acetylated protein lost most of the PG binding activity
antimicrobial activity was reduced for acetylated apoA-I
electrostatic forces are critical for binding of apoA-I to LPS and PG
Acknowledgments
This work was funded by grants from the National Institutes of Health (NIH) to P.M.M.W (SC3GM089564) and E.J.H-S (SC3GM092298) and the NIH Research Initiative for Scientific Enhancement (RISE) Program (5R25GM071638). The authors thank Dr. Yuan Yu Lee, Center for Education in Proteomics Analysis at CSU Long Beach, for mass spectrometric analysis.
Abbreviations
- ANS
8-anilinonaphthalene-1-sulfonate
- apoA-I
apolipoprotein A-I
- BSA
bovine serum albumin
- CD
circular dichroism
- CFU
colony forming units
- HDL
high-density lipoproteins
- LPS
lipopolysaccharides
- PC
phosphatidylcholine
- PG
phosphatidylglycerol
- PBS
phosphate buffered saline
- LUV
large unilamellar vesicles
- TBS
Tris buffered saline
- WT
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tall AR. Cholesterol efflux pathways and other potential mechanisms involved in the athero-protective effect of high density lipoproteins. J Intern Med. 2008;263:256–273. doi: 10.1111/j.1365-2796.2007.01898.x. [DOI] [PubMed] [Google Scholar]
- 2.Martin I, Dubois MC, Saermark T, Ruysschaert JM. Apolipoprotein A-1 interacts with the N-terminal fusogenic domains of SIV (simian immunodeficiency virus) GP32 and HIV (human immunodeficiency virus) GP41: implications in viral entry. Biochem Biophys Res Commun. 1992;186:95–101. doi: 10.1016/s0006-291x(05)80780-6. [DOI] [PubMed] [Google Scholar]
- 3.Tada N, Sakamoto T, Kagami A, Mochizuki K, Kurosaka K. Antimicrobial activity of lipoprotein particles containing apolipoprotein Al. Mol Cell Biochem. 1993;119:171–178. doi: 10.1007/BF00926868. [DOI] [PubMed] [Google Scholar]
- 4.Shiflett AM, Bishop JR, Pahwa A, Hajduk SL. Human high density lipoproteins are platforms for the assembly of multi-component innate immune complexes. J Biol Chem. 2005;280:32578–32585. doi: 10.1074/jbc.M503510200. [DOI] [PubMed] [Google Scholar]
- 5.Hubsch AP, Casas AT, Doran JE. Protective effects of reconstituted high-density lipoprotein in rabbit gram-negative bacteremia models. J Lab Clin Med. 1995;126:548–558. [PubMed] [Google Scholar]
- 6.Levine DM, Parker TS, Donnelly TM, Walsh A, Rubin AL. In vivo protection against endotoxin by plasma high density lipoprotein. Proc Natl Acad Sci U S A. 1993;90:12040–12044. doi: 10.1073/pnas.90.24.12040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Imai T, Fujita T, Yamazaki Y. Beneficial effects of apolipoprotein A-I on endotoxemia. Surg Today. 2003;33:684–687. doi: 10.1007/s00595-003-2585-4. [DOI] [PubMed] [Google Scholar]
- 8.Pajkrt D, Doran JE, Koster F, Lerch PG, Arnet B, van der Poll T, ten Cate JW, van Deventer SJ. Antiinflammatory effects of reconstituted high-density lipoprotein during human endotoxemia. J Exp Med. 1996;184:1601–1608. doi: 10.1084/jem.184.5.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Massamiri T, Tobias PS, Curtiss LK. Structural determinants for the interaction of lipopolysaccharide binding protein with purified high density lipoproteins: role of apolipoprotein A-I. J Lipid Res. 1997;38:516–525. [PubMed] [Google Scholar]
- 10.Levels JH, Abraham PR, van den Ende A, van Deventer SJ. Distribution and kinetics of lipoprotein-bound endotoxin. Infect Immun. 2001;69:2821–2828. doi: 10.1128/IAI.69.5.2821-2828.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thaveeratitham P, Plengpanich W, Naen-Udorn W, Patumraj S, Khovidhunkit W. Effects of human apolipoprotein A-I on endotoxin-induced leukocyte adhesion on endothelial cells in vivo and on the growth of Escherichia coli in vitro. J Endotoxin Res. 2007;13:58–64. doi: 10.1177/0968051907078611. [DOI] [PubMed] [Google Scholar]
- 12.Emancipator K, Csako G, Elin RJ. In vitro inactivation of bacterial endotoxin by human lipoproteins and apolipoproteins. Infect Immun. 1992;60:596–601. doi: 10.1128/iai.60.2.596-601.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma J, Liao XL, Lou B, Wu MP. Role of apolipoprotein A-I in protecting against endotoxin toxicity. Acta Biochim Biophys Sin (Shanghai) 2004;36:419–424. doi: 10.1093/abbs/36.6.419. [DOI] [PubMed] [Google Scholar]
- 14.Morrow JA, Segall ML, Lund-Katz S, Phillips MC, Knapp M, Rupp B, Weisgraber KH. Differences in stability among the human apolipoprotein E isoforms determined by the amino-terminal domain. Biochemistry. 2000;39:11657–11666. doi: 10.1021/bi000099m. [DOI] [PubMed] [Google Scholar]
- 15.Ames BN. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 16.Raetz CR, Whitfield C. Lipopolysaccharide endotoxins. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito H, Lund-Katz S, Phillips MC. Contributions of domain structure and lipid interaction to the functionality of exchangeable human apolipoproteins. Prog Lipid Res. 2004;43:350–380. doi: 10.1016/j.plipres.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Saito H, Dhanasekaran P, Nguyen D, Holvoet P, Lund-Katz S, Phillips MC. Domain structure and lipid interaction in human apolipoproteins A-I and E, a general model. J Biol Chem. 2003;278:23227–23232. doi: 10.1074/jbc.M303365200. [DOI] [PubMed] [Google Scholar]
- 19.Rogers DP, Roberts LM, Lebowitz J, Engler JA, Brouillette CG. Structural analysis of apolipoprotein A-I: effects of amino- and carboxy-terminal deletions on the lipid-free structure. Biochemistry. 1998;37:945–955. doi: 10.1021/bi9713512. [DOI] [PubMed] [Google Scholar]
- 20.Stryer L. The interaction of a naphthalene dye with apomyoglobin and apohemoglobin. A fluorescent probe of non-polar binding sites. J Mol Biol. 1965;13:482–495. doi: 10.1016/s0022-2836(65)80111-5. [DOI] [PubMed] [Google Scholar]
- 21.Leroy A, Jonas A. Native-like structure and self-association behavior of apolipoprotein A-I in a water/n-propanol solution. Biochim Biophys Acta. 1994;1212:285–294. doi: 10.1016/0005-2760(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 22.Brouillette CG, Anantharamaiah GM, Engler JA, Borhani DW. Structural models of human apolipoprotein A-I: a critical analysis and review. Biochim Biophys Acta. 2001;1531:4–46. doi: 10.1016/s1388-1981(01)00081-6. [DOI] [PubMed] [Google Scholar]
- 23.Davidson WS, Hazlett T, Mantulin WW, Jonas A. The role of apolipoprotein AI domains in lipid binding. Proc Natl Acad Sci U S A. 1996;93:13605–13610. doi: 10.1073/pnas.93.24.13605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu HL, Atkinson D. Conformation and lipid binding of a C-terminal (198-243) peptide of human apolipoprotein A-I. Biochemistry. 2007;46:1624–1634. doi: 10.1021/bi061721z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka M, Dhanasekaran P, Nguyen D, Ohta S, Lund-Katz S, Phillips MC, Saito H. Contributions of the N- and C-terminal helical segments to the lipid-free structure and lipid interaction of apolipoprotein A-I. Biochemistry. 2006;45:10351–10358. doi: 10.1021/bi060726t. [DOI] [PubMed] [Google Scholar]
- 26.Narayanaswami V, Kiss RS, Weers PMM. The helix bundle: a reversible lipid binding motif. Comp Biochem Physiol A: Mol Integr Physiol. 2010;155:123–133. doi: 10.1016/j.cbpa.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raetz CR, Dowhan W. Biosynthesis and function of phospholipids in Escherichia coli. J Biol Chem. 1990;265:1235–1238. [PubMed] [Google Scholar]
- 28.Teixeira V, Feio MJ, Bastos M. Role of lipids in the interaction of antimicrobial peptides with membranes. Prog Lipid Res. 2012;51:149–177. doi: 10.1016/j.plipres.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 29.Kaconis Y, Kowalski I, Howe J, Brauser A, Richter W, Razquin-Olazaran I, Inigo-Pestana M, Garidel P, Rossle M, Martinez de Tejada G, Gutsmann T, Brandenburg K. Biophysical mechanisms of endotoxin neutralization by cationic amphiphilic peptides. Biophys J. 2011;100:2652–2661. doi: 10.1016/j.bpj.2011.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jenssen H, Hamill P, Hancock RE. Peptide antimicrobial agents. Clin Microbiol Rev. 2006;19:491–511. doi: 10.1128/CMR.00056-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnston LD, Brown G, Gauthier D, Reece K, Kator H, Van Veld P. Apolipoprotein A-I from striped bass (Morone saxatilis) demonstrates antibacterial activity in vitro. Comp Biochem Physiol B: Biochem Mol Biol. 2008;151:167–175. doi: 10.1016/j.cbpb.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 32.Gordon SM, Hofmann S, Askew DS, Davidson WS. High density lipoprotein: it's not just about lipid transport anymore. Trends Endocrinol Metab. 2011;22:9–15. doi: 10.1016/j.tem.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Oosten M, Rensen PC, Van Amersfoort ES, Van Eck M, Van Dam AM, Breve JJ, Vogel T, Panet A, Van Berkel TJ, Kuiper J. Apolipoprotein E protects against bacterial lipopolysaccharide-induced lethality. A new therapeutic approach to treat gram-negative sepsis. J Biol Chem. 2001;276:8820–8824. doi: 10.1074/jbc.M009915200. [DOI] [PubMed] [Google Scholar]
- 34.Oztug M, Martinon D, Weers PMM. Characterization of the apoLp-III/LPS complex: insight into the mode of binding interaction. Biochemistry. 2012;51:6220–6227. doi: 10.1021/bi300619a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zdybicka-Barabas A, Cytrynska M. Involvement of apolipophorin III in antibacterial defense of Galleria mellonella larvae. Comp Biochem Physiol B: Biochem Mol Biol. 2011;158:90–98. doi: 10.1016/j.cbpb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 36.Biedzka-Sarek M, Metso J, Kateifides A, Meri T, Jokiranta TS, Muszynski A, Radziejewska-Lebrecht J, Zannis V, Skurnik M, Jauhiainen M. Apolipoprotein A-I exerts bactericidal activity against Yersinia enterocolitica serotype O:3. J Biol Chem. 2011;286:38211–38219. doi: 10.1074/jbc.M111.249482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Henning MF, Herlax V, Bakas L. Contribution of the C-terminal end of apolipoprotein AI to neutralization of lipopolysaccharide endotoxic effect. Innate Immun. 2011;17:327–337. doi: 10.1177/1753425910370709. [DOI] [PubMed] [Google Scholar]
- 38.Brandenburg K, Jurgens G, Andra J, Lindner B, Koch MH, Blume A, Garidel P. Biophysical characterization of the interaction of high-density lipoprotein (HDL) with endotoxins. Eur J Biochem. 2002;269:5972–5981. doi: 10.1046/j.1432-1033.2002.03333.x. [DOI] [PubMed] [Google Scholar]
- 39.Vasquez LJ, Abdullahi GE, Wan CP, Weers PMM. Apolipophorin III lysine modification: effect on structure and lipid binding. Biochim Biophys Acta. 2009;1788:1901–1906. doi: 10.1016/j.bbamem.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tanaka M, Koyama M, Dhanasekaran P, Nguyen D, Nickel M, Lund-Katz S, Saito H, Phillips MC. Influence of tertiary structure domain properties on the functionality of apolipoprotein A-I. Biochemistry. 2008;47:2172–2180. doi: 10.1021/bi702332b. [DOI] [PubMed] [Google Scholar]
- 41.Brubaker G, Peng DQ, Somerlot B, Abdollahian DJ, Smith JD. Apolipoprotein A-I lysine modification: effects on helical content, lipid binding and cholesterol acceptor activity. Biochim Biophys Acta. 2006;1761:64–72. doi: 10.1016/j.bbalip.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 42.Mei X, Atkinson D. Crystal structure of C-terminal truncated apolipoprotein A-I reveals the assembly of high density lipoprotein (HDL) by dimerization. Biol Chem. 2011;286:38570–38582. doi: 10.1074/jbc.M111.260422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peleg AY, Hooper DC. Hospital-acquired infections due to gram-negative bacteria. N Engl J Med. 2010;362:1804–1813. doi: 10.1056/NEJMra0904124. [DOI] [PMC free article] [PubMed] [Google Scholar]