Abstract
Inhibitory interneurons of the spinal dorsal horn play critical roles in the processing of noxious and innocuous sensory information. They form a family of morphologically and functionally diverse neurons that likely fall into distinct subtypes. Traditional classifications rely mainly on differences in dendritic tree morphology and firing patterns. Although useful, these markers are not comprehensive and cannot be used to drive specific genetic manipulations targeted at defined subsets of neurons. Here, we have used genome-wide expression profiling of spinal dorsal horns of wild-type mice and of two strains of transcription factor-deficient mice (Ptf1a−/− and Ascl1/Mash1−/− mice) to identify new genetic markers for specific subsets of dorsal horn inhibitory interneurons. Ptf1a−/− mice lack all inhibitory interneurons in the dorsal horn, whereas only the late-born inhibitory interneurons are missing in Ascl1−/− mice. We found 30 genes that were significantly downregulated in the dorsal horn of Ptf1a−/− mice. Twenty-one of those also showed reduced expression in Ascl1−/− mice. In situ hybridization analyses of all 30 genes identified four genes with primarily non-overlapping expression patterns in the dorsal horn. Three genes, pDyn coding the neuropeptide dynorphin, Kcnip2 encoding a potassium channel associated protein, and the nuclear receptor encoding gene Rorb, were expressed in Ascl1-dependent subpopulations of the superficial dorsal horn. The fourth gene, Tfap2b, encoding a transcription factor, is expressed mainly in a Ascl1-independent subpopulation of the deep dorsal horn. Functional experiments in isolated spinal cords showed that the Ascl1-dependent inhibitory interneurons are key players of nociceptive reflex plasticity.
Introduction
The spinal dorsal horn serves as the first relay station for incoming somatosensory and nociceptive information. Inhibitory interneurons at this site exert pivotal functions in the processing of sensory signals. They attenuate responses to nociceptive stimuli, silence nociceptive neurons during innocuous stimulation, and confine neuronal excitation to somatotopically defined areas of the dorsal horn (Sandkühler, 2009).
Traditionally, dorsal horn interneurons are classified primarily according to dendritic tree morphology and neuronal firing patterns (for review, see Zeilhofer et al., 2012). A number of studies have provided conclusive evidence that these markers do, at least to some extent, predict roles in sensory processing (Grudt and Perl, 2002; Heinke et al., 2004; Yasaka et al., 2010; for review, see Graham et al., 2007; Zeilhofer et al., 2012). However, these markers are not fully satisfactory. They reflect the diversity of dorsal horn interneuron populations only partially and are poor criteria for the preselection of neurons for electrophysiological recordings, and they cannot be used for specific genetic manipulations of subsets of neurons.
More recently, neuropeptides, Ca2+ binding proteins, and enzymes have been used as molecular markers (Laing et al., 1994; Bröhl et al., 2008; Sardella et al., 2011; Tiong et al., 2011; for review, see Todd, 2010). These genetically encoded markers overcome many of the limitations of traditional criteria and have turned out to be instrumental in the identification of neuronal subtypes. It is likely that the potential of this approach has not yet been fully realized and that a significant number of genetic markers remain to be discovered.
The use of genome-wide expression profiling techniques together with the advent of Ptf1a and Ascl1 (also known as Mash1) deficient mice (Ptf1a−/− and Ascl1−/− mice) offers an ideal opportunity for the identification of new markers for dorsal horn inhibitory interneurons. Ptf1a and Ascl1 are two key regulators in the development of inhibitory interneurons. These interneurons are born in two sequential phases of neurogenesis. dI4 inhibitory interneurons arise between E10 and E11.5 (early phase of neurogenesis), whereas inhibitory dILA interneurons are generated between E12 and E14.5 (late phase of neurogenesis) (Gross et al., 2002; Müller et al., 2002). The differentiation of all inhibitory dorsal spinal interneurons depends on a trimeric transcription factor complex containing Ptf1a, Rbpj, and an E-protein. In the absence of Ptf1a, all dorsal horn neurons acquire an excitatory phenotype (Glasgow et al., 2005; Hori et al., 2008). In contrast, the bHLH transcription factor Ascl1 is only required for the generation of inhibitory interneurons born during the late phase (Mizuguchi et al., 2006; Wildner et al., 2006).
Here, an unbiased genome-wide comparison of gene expression in the spinal dorsal horns of Ascl1−/− and Ptf1a−/− mice and subsequent in situ hybridization experiments identified eight genes with locally restricted expression patterns in the spinal dorsal horn of which four showed a primarily non-overlapping distribution. Functional experiments in isolated spinal cord indicate that the Ascl1-dependent subpopulation of dorsal horn interneurons is particularly relevant for spinal nociceptive reflex plasticity.
Materials and Methods
Mouse strains.
The generation of the mutant Ptf1aCre and Ascl1GFP alleles has been described previously (Kawaguchi et al., 2002; Wildner et al., 2006). Throughout text, Ascl1GFP/GFP mice are labeled as Ascl1−/− mice and Ptf1aCre/Cre mice are labeled as Ptf1a−/− mice. Mice labeled as control encompass heterozygous and wild-type mice of the respective genotype, whereas mice labeled as wild-type control only have wild-type alleles.
RNA isolation and microarray analyses.
The dorsal spinal cord was dissected from wild-type, Ptf1a−/−, and Ascl1−/− mice at E18.5. Tissue was collected in RNAlater (Ambion) and homogenized in Trizol (Invitrogen), and total RNA was isolated according to the protocol of the manufacturer. Tissue from at least five embryos was pooled for one sample. RNA was further purified using RNeasy MinElute Cleanup kit (Qiagen). cDNA synthesis and in vitro generation and labeling of cRNA were performed according to the recommendations provided by Affymetrix. Amplified cRNA was purified using the GeneChip Sample Cleanup Module (Affymetrix), and the quality of the amplification was verified by gel electrophoresis. cRNAs were fragmented and then hybridized to either Affymetrix Mouse Genome 430 2.0 arrays (Ascl1−/− vs wild type) or Affymetrix Mouse Exon 1.0 arrays (Ptf1a−/− vs wild type), using three samples per embryonic stage and genotype. Additional data processing and identification of differentially expressed genes was performed in the R environment for statistical computing (R Development Core Team 2005) using the Bioconductor base installation (Gentleman et al., 2004) and packages affyPLM, gcrma, and limma. Briefly, array quality was assessed with affyPLM, and data were normalized with gcrma. Probe sets with low variance of expression across all arrays were filtered out, and differentially expressed genes were identified using the empirical Bayes-moderated t test implemented in the limma package. p values associated with the t statistics were adjusted using a false discovery rate approach to compensate for multiple testing.
Immunohistochemistry and in situ hybridization.
Immunohistochemistry was performed on 20 μm cryosections of mouse embryos as described previously (Müller et al., 2005). The following antibodies were used: rabbit anti-Pax2 (Zymed), rabbit and guinea pig anti-Tlx3 (Müller et al., 2005), rabbit anti-MafA (Wende et al., 2012), rabbit anti-calcitonin gene-related peptide (CGRP), and rabbit anti-peripherin and rabbit anti-TrkA. Cyanine 2 (Cy2)- or Cy3-conjugated donkey secondary antibodies (Dianova) were used for the detection of primary antibodies. Fluorescence was imaged on a Carl Zeiss LSM 5 or LSM710 Pascal confocal microscope, and images were processed in Adobe Photoshop (Adobe Systems).
For in situ hybridization, embryonic tissues were embedded into OCT compound (Sakura Finetek) and cryosectioned as described previously (Wildner et al., 2008). Hybridizations were performed with DIG-labeled riboprobes. DNA for dynorphin, glutamate decarboxylase 67 (GAD67) (Gad1), vesicluar glutamate transporter 2 (vGluT2) (slc17a6), glycine transporter 2 (GlyT2) (slc6a5), Kcnip2, Rorb, Tfap2b, Npy, Prkcq, and Klhl14 were amplified with gene-specific sets of PCR primers and cDNA templates prepared from E18.5 mouse spinal cords. The PCR fragments were cloned into Teasy vector (Promega), and sequence was verified before use for riboprobe generation. For double staining of a cytoplasmic mRNA and a nuclear protein, in situ hybridization was performed first, followed by immunostaining with anti-Pax2 (Zymed) and Cy3-conjugated secondary donkey antibodies (Dianova). The in situ signals were photographed under transillumination and converted into a green or red pseudo-color (Cheng et al., 2004). For quantification of neuronal subtypes that express the respective analyzed gene, sections (20 μm thickness) of the E18.5 thoracic spinal cord were hybridized with specific probes. Numbers of neurons that express the various genes were counted on bright-field images, using four or more sections each of two or three distinct animals for each genotype. Only cells of the dorsal spinal cord were counted. SD was calculated and depicted as error bars in the quantification graphs.
Electrophysiology.
An in vitro hemisected spinal cord preparation was used to record spinal cord reflexes as described by Heppenstall and Lewin (2001). Spinal cords of wild-type and Ascl1GFP/GFP mice were removed from E20 mice that were dissected from females treated with progesterone to delay parturition. Direct current recordings were made after a 2 h recovery period with a close-fitting glass suction electrode attached to the ventral root (T10–T12). The dorsal root was stimulated via a glass suction electrode at a current sufficient to activate C fibers (500 μA, 500 μs). Traces were acquired with the Powerlab 4.0 system using Superscope software.
Results
Loss of late-born inhibitory interneurons in Ascl1−/− mice
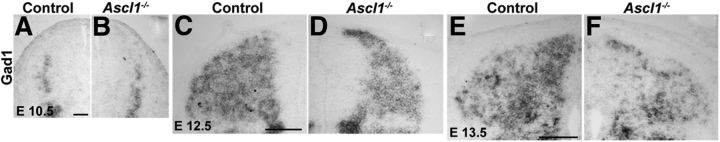
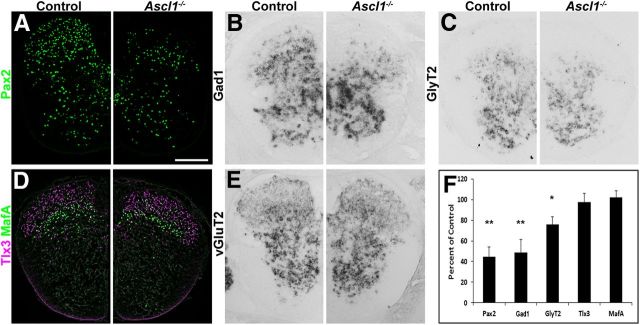
It is well established that Ascl1−/− mice lack late-born inhibitory interneurons in the spinal dorsal horn. However, the quantitative contribution of the Ascl1-depedent subset to all dorsal horn inhibitory interneurons is not known. Similarly, it is unknown to what extent these Ascl1-dependent interneurons differ from other inhibitory dorsal horn neurons. To address these questions, we first compared the distribution of inhibitory interneurons during dorsal horn neurogenesis and analyzed the expression of GAD67 (Gad1), a widely used marker for GABAergic interneurons, in wild-type and Ascl1−/− mice. During the first phase of neurogenesis (E10.5), no reduction in GAD67 expression was observed in the Ascl1−/− mice (Fig. 1A,B), whereas on E12.5 and E13.5 (during the second phase of neurogenesis), a clear reduction in numbers of GAD67-positive neurons was apparent (Fig. 1C–F). On E18, when spinal neurogenesis was complete and when neurons had settled in their final positions, a 50% reduction in the number of GABAergic cells in the dorsal horns was observed compared with wild-type controls (Fig. 2B, quantified in F). These results were further confirmed by the use of other markers for inhibitory interneurons. Staining for Pax2 (Fig. 2A,F) and vesicular inhibitory amino acid transporter (data not shown) revealed similar reductions in expression in the mutant mice. The reduction of these markers was most pronounced in the superficial dorsal horn. Approximately one-third of dorsal horn inhibitory neurons use glycine in addition to GABA as a second fast inhibitory neurotransmitter and are characterized by the expression of the plasma membrane glycine transporter GlyT2. Their somata are mainly located in the deep dorsal horn. In Ascl1−/− mice, the loss of GlyT2-positive neurons was less pronounced than that of GAD67- and Pax2-positive neurons (Fig. 2C,F). No significant change in the number of excitatory cells (identified through the presence of the transcription factors Tlx3 and MafA, and vGluT2) was observed in the spinal cord of Ascl1−/− compared with control littermates (Fig. 2D–F). We also observed no obvious differences in the ingrowth of sensory fibers into the dorsal horn, as assessed by a comparison of the distribution of axons expressing TrkA (Fig. 3A–C), CGRP (Fig 3D), and peripherin (Fig. 3E) in wild-type and Ascl1−/− mice.
Figure 1.
Decreased Gad1 expression in the dorsal spinal cord of Ascl1−/− mice at E12.5 and E13.5. In situ hybridization on transverse sections of control and Ascl1−/− spinal cords at E10.5 (A, B), E12.5 (C, D), and E13.5 (E, F) using an antisense probe specific for Gad1. Scale bars: A, 50 μm; C, E, 100 μm.
Figure 2.
Loss of inhibitory interneurons in the dorsal spinal horn of Ascl1−/− mice. A significant reduction in the number of neurons expressing marker genes for inhibitory interneurons can be seen in the dorsal spinal cord of Ascl1−/− mice compared with control littermates (A–C). Loss of GABAergic interneurons (B) is much more pronounced than that of glycinergic neurons (C). No change in the number of excitatory dorsal spinal neurons was detected (D, E). A, D, Immunohistochemistry on transverse sections of control and Ascl1−/− spinal cords at E18 using an antibody against Pax2 (A) or antibodies against MafA and Tlx3 (D). B, C, E, In situ hybridization on control and Ascl1−/− spinal cord sections at E18.5 using antisense probes directed against Gad1 (B), GlyT2 (C), or vGluT2 (E). F, Quantification of the number of cells expressing the indicated gene relative to control (percentage). Error bar represents the SD. *p < 0.05, **p < 0.005 (unpaired Student's t test). Scale bar, 200 μm.
Figure 3.
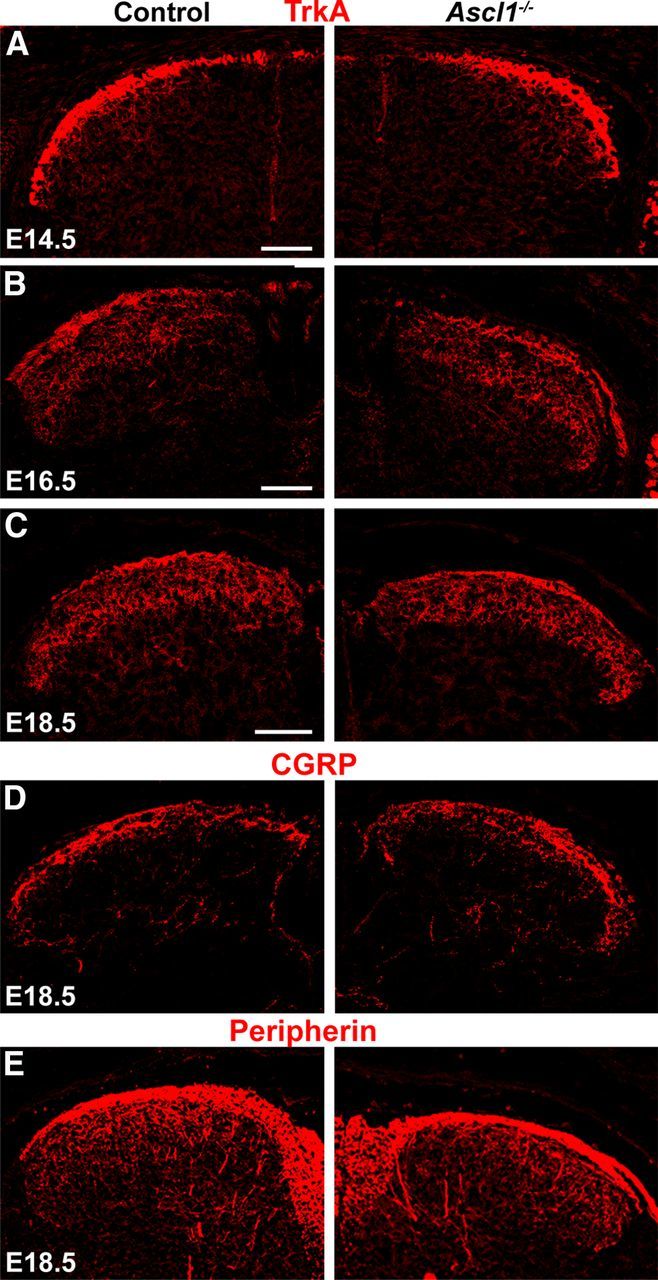
No gross changes in the innervation of the dorsal spinal horn of Ascl1−/− mice were detected. Ingrowth of afferent sensory fibers into the dorsal spinal cord was analyzed by immunohistochemistry using antibodies against TrkA, CGRP, and peripherin. A–C, Immunohistochemistry on transverse sections of control and Ascl1−/− spinal cords at E14.5, E16.5, and E18.5 using an antibody against TrkA. Immunohistochemistry on control and Ascl1−/− spinal cords at E18.5 using antibodies against CGRP (D) or peripherin (E). Scale bars, 100 μm.
Expression profiling of inhibitory spinal interneurons
As indicated above, Ascl1-dependent and Ascl1-independent inhibitory interneurons differ in their spatial distribution within the spinal dorsal horn. We went on to investigate whether both types also differ in their gene expression profiles and extracted total RNA from dorsal spinal cords of Ascl1−/− or Ptf1a−/− mice and from littermates of both mutant mice. Genes downregulated in Ascl1−/− and Ptf1a−/− mice should constitute possible marker genes of Ascl1-dependent subpopulations of inhibitory interneurons, whereas genes downregulated in Ptf1a−/− only would conversely identify Ascl1-independent subsets. Microarray analysis of Ptf1a−/− dorsal horns resulted in the identification of 30 genes that were downregulated in a highly significant manner (corrected p ≤ 0.05 after Holm–Bonferroni post hoc correction) (Table 1). Among these genes, we found known markers of inhibitory interneurons, such as Gad1, Gad2, Slc6a5, and Pax2, but also several genes that had not been attributed previously to spinal inhibitory interneurons (e.g., Klhl14, Rorb, and Tfap2b). We also found genes that were significantly upregulated in the Ptf1a−/− dorsal horns. Among these are genes known to be expressed in excitatory neurons (e.g., Slc17a6, Cck, and Grp) (Cheng et al., 2004; Xu et al., 2008). Approximately half of the genes significantly downregulated in the Ptf1a−/− screen (16 of 30) were found to be significantly downregulated also in the dorsal horns of Ascl1−/− mice (e.g., Npy and Prkcq) (Table 2). Unexpectedly, the screen of the Ascl1−/− dorsal horns also revealed six genes that were significantly downregulated only in the Ascl1−/− dorsal horns (Table 2). Because four of these six genes (Pdyn, Neurod1, Neurod2, and Pou4f2) have been shown previously to be expressed specifically in inhibitory neurons (Laing et al., 1994; Bröhl et al., 2008; Huang et al., 2008; Sardella et al., 2011; Tiong et al., 2011), the absence of the significant downregulation in the Ptf1a screen probably points at technical limitations.
Table 1.
List of downregulated genes
| Symbol | Gene name | Ptf1a | FC | Ascl1 | FC |
|---|---|---|---|---|---|
| Tfap2b | Transcription factor AP-2β | ++ | 12 | n.s. | |
| Gad1 | Glutamic acid decarboxylase 1 | ++ | 3.1 | ++ | 2.1 |
| Pax8 | Paired box 8 | ++ | 3.9 | ++ | 2.2 |
| Cacna2d3 | Calcium channel, voltage-dependent, α2/δ3 | ++ | 2.5 | ++ | 2.1 |
| Pnoc | Prepronociceptin | ++ | 3.7 | ++ | 3.8 |
| Prkcq | Protein kinase Cθ | ++ | 3.9 | ++ | 2.4 |
| Npy | Neuropeptide Y | ++ | 5.6 | + | 1.5 |
| Neurod6 | Neurogenic differentiation 6 | ++ | 3.7 | + | 1.3 |
| Gad2 | Glutamic acid decarboxylase 2 | ++ | 3.6 | n.s. | |
| Nrxn3 | Neurexin III | ++ | 2.8 | + | 1.4 |
| Slc6a5 | Solute carrier family 6 member 5 | ++ | 3.4 | + | 1.4 |
| Grik2 | Glutamate receptor, ionotropic, kainate 2 (β2) | ++ | 2.3 | + | 1.6 |
| Klhl14 | Kelch-like 14 (Drosophila) | ++ | 2.6 | n.s. | |
| Rnf152 | Ring finger protein 152 | ++ | 1.8 | n.s. | |
| 6330527O06Rik | LAMP family protein C20orf103 homolog precur | ++ | 5.6 | + | 1.7 |
| Sall3 | Sal-like 3 (Drosophila) | ++ | 2.1 | + | 1.6 |
| Sez6 | Seizure-related gene 6 | ++ | 1.9 | + | 1.5 |
| 9230112E08Rik | RIKEN cDNA 9230112E08 gene | ++ | 1.5 | n.s. | |
| Xlr3a | X-linked lymphocyte-regulated 3A | ++ | 1.4 | n.s. | |
| Lhx1 | LIM homeobox protein 1 | ++ | 3.3 | n.s. | |
| Cadps2 | Ca2+-dependent activator protein for secretion 2 | ++ | 2.2 | n.s. | |
| Pax2 | Paired box gene 2 | ++ | 3.1 | n.s. | |
| Grik3 | Glutamate receptor, ionotropic, kainate 3 | ++ | 1.6 | n.s. | |
| Crabp1 | Cellular retinoic acid binding protein I | ++ | 1.5 | n.s. | |
| Mpped2 | Metallophosphoesterase domain containing 2 | ++ | 1.6 | n.s. | |
| Rorb | RAR-related orphan receptor β | ++ | 1.8 | + | 1.4 |
| Ipcef1 | Interaction protein for cytohesion exchange factors 1 | ++ | 1.5 | n.s. | |
| Tmem86b | Transmembrane protein 86B | ++ | 1.7 | n.s. | |
| Grm5 | Glutamate receptor, metabotropic 5 | ++ | 1.7 | + | 1.4 |
| Zmat4 | Zinc finger, matrin type 4 | ++ | 1.7 | + | 1.8 |
The transcriptome of dorsal horn cells of Ptf1a−/− and Ascl1−/− mice was compared with littermate controls using Affymetrix expression arrays. The gene symbol and name of the respective genes are depicted. In columns 3 and 5, it is indicated whether the downregulation of the respective gene was called highly significant (++, corrected p ≤ 0.005), significant (+, corrected p ≤ 0.05), or not significant (n.s.). In columns 4 (Ptf1a vs control) and 6 (Ascl1 vs control), the fold change (FC) the respective gene was downregulated according to the Affymetrix results is indicated.
Table 2.
List of downregulated genes
| Symbol | Gene name | Ascl1 | FC | Ptf1a | FC |
|---|---|---|---|---|---|
| Pdyn | Prodynorphin | ++ | 2.7 | n.s. | |
| Gad1 | Glutamic acid decarboxylase 1 | ++ | 2.1 | ++ | 3.1 |
| Cacna2d3 | Calcium channel, voltage-dependent, α2/δ3 | ++ | 2.1 | ++ | 2.5 |
| Pax8 | Paired box gene 8 | ++ | 2.2 | ++ | 3.9 |
| Kcnip2 | Kv channel-interacting protein 2 | ++ | 3.7 | n.s. | |
| Pnoc | Prepronociceptin | ++ | 3.8 | ++ | 3.7 |
| Prkcq | Protein kinase Cθ | ++ | 2.4 | ++ | 3.9 |
| Neurod1 | Neurogenic differentiation 1 | ++ | 2.9 | n.s. | |
| Slc35d3 | Solute carrier family 35, member D3 | ++ | 4.3 | n.s. | |
| Neurod2 | Neurogenic differentiation 2 | ++ | 3.1 | n.s. | |
| Pou4f2 | POU domain, class 4, transcription factor 2 | ++ | 2.9 | n.s. |
The transcriptome of dorsal horn cells of Ascl1−/− mice was compared with littermate controls using Affymetrix expression arrays. The gene symbol and name of the respective genes are depicted. In columns 3 and 5, it is indicated whether the downregulation of the respective gene was called highly significant (++, corrected p ≤ 0.005), significant (+, corrected p ≤ 0.05), or not significant (n.s.). In columns 4 (Ptf1a vs control) and 6 (Ascl1 vs control), the fold change (FC) the respective gene was downregulated according to the Affymetrix results is indicated.
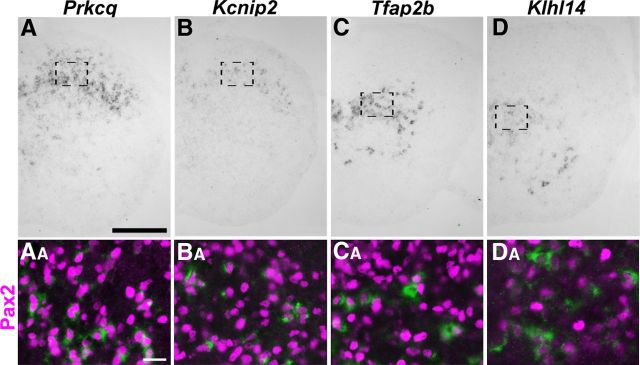
We next aimed to verify the hits of the DNA microarray screen and to identify genes specifically expressed in subsets of inhibitory interneurons. To preselect promising candidate genes from the 30 genes found downregulated in the Ptf1a−/− mice, plus the additional six genes from the Ascl1−/− mice, we used the Allen Brain Atlas (http://mousespinal.brain-map.org/) to survey the expression patterns in P4 spinal cords. We selected 16 genes that appeared to be expressed in distinct spatial domains of the spinal dorsal horn for additional in situ hybridization analysis in sections of E18.5 spinal cords of wild-type, Ascl1−/−, and Ptf1a−/− mice. For seven of these (Kcnip2, Klhl14, Npy, Pdyn, Prkcq, Rorb, and Tfap2b), we obtained robust and reproducible in situ hybridization signals (Fig. 4A–G) that were lost or significantly attenuated in the Ptf1a−/− spinal cords (Fig. 4AA–GA,AC–GC). All genes showed greatly reduced or undetectable expression in dorsal horns of Ptf1a−/− mice, indicating that they were indeed specific to inhibitory interneurons (Fig. 4). Rorb revealed residual expression in dorsal horn neurons of Ptf1a−/− mice (Fig. 4F,FA,FC), indicating that other neural cell types, probably subsets of excitatory interneurons, also express this gene.
Figure 4.
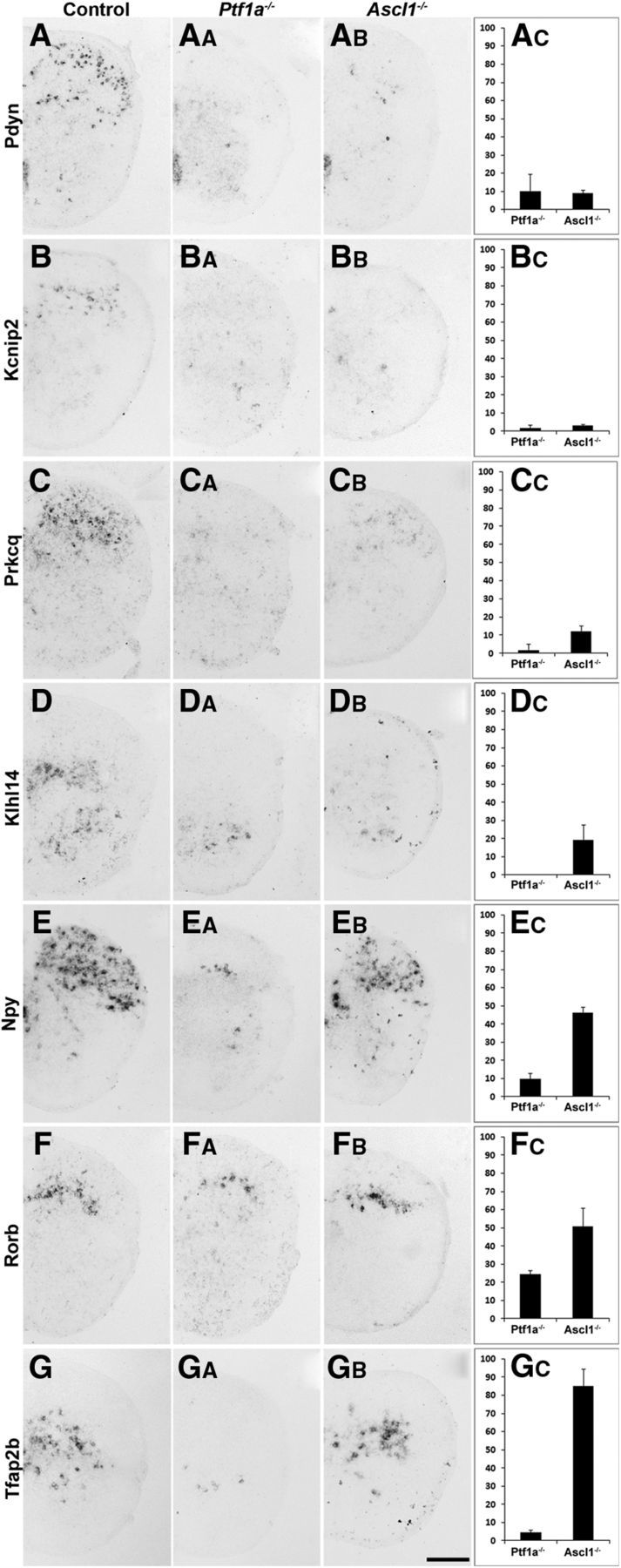
Identification of expression domains marked by genes downregulated in Ptf1a−/− and/or Ascl1−/− mice. A–GB, In situ hybridization on spinal cords of control, Ptf1a−/−, and Ascl1−/− mice at E18.5 using probes specific for pDyn, Kcnip2, Rorb, Tfap2b, Prkcq, Klhl14, and Npy. AC–GC, Quantification of the number of neurons expressing the respective gene in Ptf1a−/− or Ascl1−/− mice relative to control. Expression of the more superficially expressed genes pDyn (A–AC), Kcnip2 (B–BC), and Prkcq (C, CC) is reduced to <10% of control levels in Ptf1a−/− and Ascl1−/− mice. Expression of NPY (E–EC) and Tfap2b (G–GC) is almost completely lost in Ptf1a−/− but only reduced to 50 or 85% in Mash−/−, respectively. Error bars represent the SD. Scale bar, 200 μm.
To confirm the inhibitory character of the neurons expressing the genes studied, we performed colabeling using in situ hybridization together with immunohistochemistry with an antibody directed against Pax2. We and others have reported previously that Npy and Pdyn are mainly expressed in inhibitory interneurons (Rowan et al., 1993; Bröhl et al., 2008; Huang et al., 2008). In addition, we identified Kcnip2, Khlh14, Prkcq, and Tfap2b as displaying a high degree of overlap with Pax2 (Fig. 5).
Figure 5.
Coexpression of Pax2 and Kcnip2, Tfap2b, Prkcq, and Klhl14 in dorsal horn neurons. A–C, In situ hybridization of Prkcq, Kcnip2, Tfap2b, and Klhl14 on spinal cords at E18.5. Areas of magnification are indicated by dotted boxes. AB–CB, Double staining of nuclear Pax2 (red) protein and cytoplasmic mRNA (green) encoding the indicated genes in dorsal horn neurons at E18.5. Scale bars: A–D, 200 μm; AA–AD, 20 μm.
Identification of genes expressed in spatially restricted subsets of inhibitory interneurons
We then investigated whether the Ascl1-dependent and Ascl1-independent populations of neurons can be further subdivided on the basis of the expression of these seven genes. To this end, we quantified the loss of neurons expressing a certain marker in Ascl1−/− mice with the intention to identify markers that define Ascl1-dependent and Ascl1-independent subsets of neurons, respectively. Pdyn-positive and Kcnip2-positive neurons showed the strongest reduction in numbers in Ascl1−/− mice. The degrees of reduction were similar to those observed in Ptf1a−/− mice (Fig. 4A–BC), indicating that Pdyn and Kcnip2 are expressed almost exclusively by the Ascl1-dependent population of inhibitory dorsal horn neurons. As expected, both types were mainly restricted to the superficial dorsal horn. The few (∼10%) Pdyn-positive neurons remaining in the Ptf1a−/− and Ascl1−/− mice are likely to represent a small subpopulation of excitatory dynorphin-expressing interneurons (Baseer et al., 2012). Klhl14 and Prkcq were completely lost in the Ptf1a−/− mice, but a small percentage of neurons remained in the deep dorsal horn of Ascl1−/− mice (Fig. 4C–DC), indicating that these two markers were mainly expressed in Ascl1-dependent inhibitory interneurons. Npy and Rorb were found in late-born superficial dorsal horn neurons and deep dorsal horn neurons (Fig. 4E,F). However, unlike Pdyn and Kcnip2, reduction of Npy- and Rorb-expressing neurons was more pronounced in Ptf1a−/− mice than in Ascl1−/− mice (Fig. 4E–FC), suggesting that both markers are also expressed in Ascl1-independent inhibitory interneurons. Furthermore, a few neurons expressing Npy and Rorb were still detected in the deep dorsal horn of Ptf1a−/− mice. These remaining neurons may again be excitatory interneurons. We identified one marker, Tfap2b, that was completely absent in Ptf1a−/− mice but almost unaffected in Ascl1−/− (Fig. 4G–GC), indicating that it was expressed specifically in the Ascl1-independent population of inhibitory interneurons. These findings indicate that Ascl1-dependent interneurons differentiate further into different inhibitory subsets of neurons populating predominantly the upper layers as well as to some extent the ventromedial region of the dorsal horn.
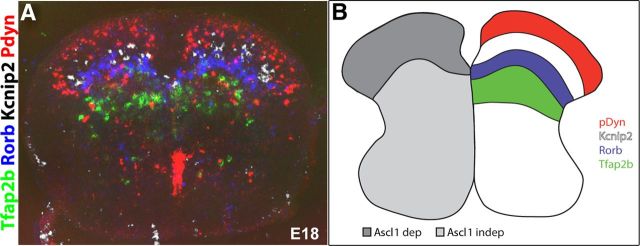
Finally, we searched the seven genes identified above for a selection that displays non-overlapping expression in the dorsal horn. To this end, we systematically created graphical overlays of three to four in situ hybridizations made in adjacent sections of wild-type spinal cords. These overlays revealed that pDyn, Kcnip2, Rorb, and Tfap2b were expressed in spatially distinct, mostly non-overlapping domains arranged from dorsal to ventral, respectively (Fig. 6A,B). Consequently, these genes were best suited to identify distinct subpopulations of dorsal horn inhibitory neurons, whereas Klhl14, Npy, and Prkcq were expressed in patterns partially overlapping with the aforementioned genes (data not shown). The layer-like expression pattern of the marker genes analyzed here is reminiscent of the laminar organization of the adult spinal cord. The specific innervation of these laminae by different sensory nerve fibers with peptidergic and non-peptidergic nociceptors innervating the dorsal and middle parts of lamina II and with non-nociceptive somatosensory fibers terminating in laminae III and deeper further supports a specific function in sensory processing of each of these interneuron types.
Figure 6.
Inhibitory interneuron populations of the dorsal horn marked by different genes. A, Computer overlay of adjacent cryosections processed for mRNA in situ hybridization with pDyn (red), Kcnip2 (white), Rorb (blue), and Tfab2b (green). Colors were artificially assigned. B, Schematic representation of layer-specific distribution of inhibitory subpopulations and their dependents on Ascl1.
The inhibitory control of spinal nociceptive reflexes is impaired in Ascl1−/− mice
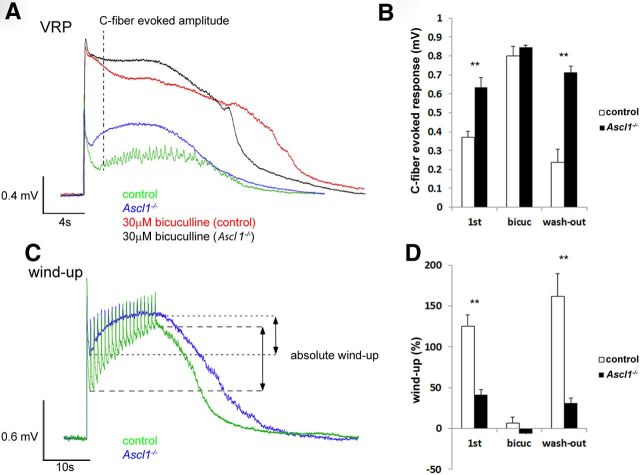
To provide first evidence for a differential role in sensory processing of the Ascl1-dependent subpopulation of inhibitory interneurons, we decided to analyze Ascl1−/− mice in tests of spinal nociception. Because these mice die during the first day after birth, behavioral characterization in most standard pain models is not feasible (Guillemot et al., 1993). Therefore, we analyzed spinal nociceptive reflex responses during C-fiber stimulation using an in vitro spinal cord preparation (Heppenstall and Lewin, 2001). In these experiments, we recorded electrical potentials in the ventral root evoked by electrical stimulation of sensory axons in the dorsal root (Jahr and Yoshioka, 1986; Heppenstall and Lewin, 2001). A fast potential of large amplitude and a longer-lasting potential of smaller amplitude could be distinguished. These potentials reflect the activation of motoneurons through direct monosynaptic connections and through indirect activation occurring via dorsal horn interneurons, respectively (Heppenstall and Lewin, 2001). The fast potential was similar in control and Ascl1−/− mice (1.32 ± 0.12 and 1.05 ± 0.09 mV, respectively; p = 0.15 Student's t test), whereas the amplitude of the long-lasting potential was significantly increased in the Ascl1−/− mice (Fig. 7A,B), suggesting a critical role of inhibitory interneurons and inhibitory neurotransmission in the control of nociceptive reflexes. In line with this concept, bicuculline, a blocker of GABAA receptors, increased ventral root potentials in Ascl1−/− animals to a lesser extent than in control animals (224 ± 40 vs 138 ± 28% increase in control and Ascl1−/− mice, respectively). In fact, the difference in the amplitude of nociceptive reflexes between the two genotypes disappeared when inhibitory GABAA-receptor-mediated neurotransmission was blocked with bicuculline (Fig. 7B).
Figure 7.
Changes in the ventral root potentials (VRP) and a reduced wind-up in Ascl1−/− mice. A, Representative example traces of ventral root potentials recorded from Ascl1−/− or wild-type control mice either with or without bicuculline superfusion. B, Quantification of the C-fiber-evoked amplitude shows a significant increase in Ascl1−/− mice as well as after bicuculline superfusion. C, Example traces of wind-up in Ascl1−/− and wild-type control mice. D, Wind-up is significantly reduced in Ascl1−/− mice and after bicuculline superfusion [1st, measurement without bicuculine; bicuc, measurement after 10 min bicuculline superfusion (30 μm); wash-out, measurement after 40 min of bicuculline washout]. The error bars represent the SEM. **p < 0.005 (unpaired Student's t test).
We then investigated possible changes in activity-dependent plasticity of the spinal nociceptive reflex. Repetitive (1 Hz) stimulation of sensory axons evokes a progressive increase in the amplitude of the ventral root potentials, called wind-up, which is thought to reflect central sensitization (Herrero et al., 2000). Compared with control animals, wind-up was significantly reduced in Ascl1−/− mice (Fig. 7C,D), and application of bicuculline abolished wind-up in both genotypes (Fig. 7D). These results indicate that Ascl1-dependent inhibitory interneurons exert a tonic control on spinal nociceptive reflexes and suggest that their short-term, activity-dependent facilitation occurs through a progressive loss of their activity.
Discussion
In the present study, we identified several genes highly enriched in Ascl1-dependent or Ascl1-independent populations of dorsal horn inhibitory interneurons and characterized their expression patterns within the spinal dorsal horn. Our findings have implications for the understanding of the development and differentiation of inhibitory dorsal horn neurons and provide a basis for functional studies addressing the role in sensory processing of different subpopulations defined by genetically encoded markers.
Inhibitory interneurons of the spinal dorsal horn are born in two sequential phases of neurogenesis and subsequently differentiate into an unknown number of subtypes. This differentiation is controlled at least partially through the orchestrated sequential expression of transcription factors, which are so far only partially identified (Gross et al., 2002; Cheng et al., 2004, 2005; Wildner et al., 2006; Bröhl et al., 2008; Huang et al., 2008). Our present analysis focused on the Ascl1-dependent (late-born) population of inhibitory dorsal horn interneurons (Mizuguchi et al., 2006; Wildner et al., 2006). Our results indicate that Ascl1-dependent interneurons constitute approximately half of the dorsal horn inhibitory interneurons and differentiate into at least four different subtypes (Kcnip2, Npy, Pdyn, and Rorb) mainly populating the superficial dorsal horn. This CNS area is densely innervated by sensory fibers carrying primarily nociceptive information. The preferential localization of Ascl1-dependent interneurons in the spinal dorsal horn is thus in good agreement with the facilitation of nociceptive reflexes observed in Ascl1−/− mice.
Transcriptional profiling of Ascl1−/−, Ptf1a−/−, and wild-type dorsal horn tissue and in situ hybridization identified seven genes (Kcnip2, Klhl14, Npy, Pdyn, Prkcq, Rorb, and Tfap2b) with expression in inhibitory interneurons of spatially restricted subdomains of the spinal dorsal horn. All except one (Tfap2b) were significantly downregulated in both mutant mice and expressed primarily in the superficial dorsal horn. For two of these genes (Npy and Pdyn) or their respective gene products, expression in inhibitory interneurons of the adult superficial spinal cord has been demonstrated previously (Laing et al., 1994; Sardella et al., 2011). NPY-positive neurons make up ∼18% of all lamina I/II inhibitory interneurons and ∼9% of inhibitory interneurons in lamina III (Polgár et al., 2011). It is likely that NPY-positive neurons exert antinociceptive actions through both GABAergic and NPY-mediated mechanisms. They inhibit spinal projection neurons, which transmit somatosensory and nociceptive signals to the brain, and local protein kinase Cγ (PKCγ)-positive excitatory interneurons (Polgár et al., 1999, 2011), which critically contribute to neuropathic pain (Malmberg et al., 1997; Neumann et al., 2008). Injection of NPY into the spinal canal of rodents exerts analgesia in different acute pain tests (Hua et al., 1991), whereas mice lacking NPY Y1 receptors are hypersensitive to acute nociceptive stimuli and develop stronger neuropathic pain than wild-type mice (Naveilhan et al., 2001).
Pdyn is a second gene identified in our screen that has been discovered previously as a marker of inhibitory dorsal horn interneurons. Pdyn (or dynorphin)-expressing neurons and NPY-positive neurons are found in primarily overlapping regions of the dorsal horn but are rarely coexpressed in the same neurons (Laing et al., 1994; Sardella et al., 2011). Dynorphin, an agonist at κ-opioid receptors, is strongly upregulated in different chronic pain states and possibly makes a major contribution to chronic neuropathic pain. However, neither their synaptic inputs nor their synaptic targets have been characterized in detail so far.
The other five genes encode different classes of gene products. Prkcq encodes PKCθ, Rorb and Tfap2b are transcription factor genes, Kcnip2 encodes a potassium channel associated protein, and Klhl14 is the kelch-like 14 protein whose function is unknown. Specific functions in spinal circuit formation or sensory processing have not been attributed to any of them so far. Nevertheless, their specific expression in dorsal spinal inhibitory subsets should make them suitable markers for inhibitory subpopulations.
Systematic comparisons of the expression patterns of the genes studied here suggest a layer- and domain-specific distribution at least for pDyn-, Kcnip2-, Rorb-, and Tfap2b-expressing interneurons. The other three genes, Npy, Prkcq, and Klhl14, showed slightly broader expression patterns overlapping with the aforementioned genes. For example, Npy expression overlaps with that of pDyn, Kcnip2, and Rorb. However, as exemplified above, an overlapping or even a congruent region of expression does not exclude that both markers label distinct populations of neurons. Thus, it is possible or even likely that more than the four genes with non-overlapping distribution can be used as markers of distinct populations of neurons.
Previous attempts to define neurochemical markers of distinct populations of dorsal horn inhibitory interneurons relied on the analysis of gene products with known expression in the dorsal horn (for review, see Todd, 2010). These studies identified galanin, neuronal nitric oxide synthase (nNOS), NPY, and parvalbumin as markers of four primarily non-overlapping populations of dorsal horn inhibitory neurons (Laing et al., 1994; Sardella et al., 2011; Tiong et al., 2011). NPY (Npy) also appeared as downregulated in our genome-wide screens, and the widespread coexpression of galanin and dynorphin in dorsal horn neurons (Sardella et al., 2011) suggests that the Pdyn-positive population is closely related to the subset of galanin-positive interneurons. At present, it is unknown whether the expression of any of the remaining five genes identified in the present study correlate with the presence of nNOS or parvalbumin.
Increased nociceptive reflex activity in spinal cords from Ascl1−/− mice suggests the presence of a strong inhibitory tone of Ascl1-dependent inhibitory interneurons onto the underlying reflex circuit. This is in good agreement with in vivo findings demonstrating that block of spinal GABAA and/or glycine receptors induces extensive pain behavior in rodents (Beyer et al., 1985; Roberts et al., 1986). In the present study, increased nociceptive reflex activity was accompanied by diminished activity-dependent facilitation of this reflex. Our observation that the loss of dorsal horn inhibitory neurons causes an increased reflex activity but reduces the degree of possible potentiation suggests that facilitation occurs at least partly through reduced inhibition. This is in good agreement with the total loss of wind-up after bicuculline superfusion and is supported by a previous report showing that C-fiber-induced spinal hyperalgesia originates from an endocannabinoid-mediated reduction of dorsal horn inhibitory neurotransmission (Pernía-Andrade et al., 2009). Work from several independent laboratories indicates that a reduction in neuronal inhibition can also occur endogenously after peripheral inflammation or neuropathy and that diminished inhibition contributes to symptoms of chronic pain (Moore et al., 2002; Harvey et al., 2004; Reinold et al., 2005; Scholz et al., 2005; Zhang et al., 2011).
Inhibitory interneurons of the spinal dorsal horn control somatosensation and pain through different and only partially understood mechanisms. They are activated by excitatory input from certain sensory nerve fibers arriving from the periphery and from antinociceptive fiber tracts descending from supraspinal CNS areas. They target different types of dorsal horn neurons, including projection neurons and local interneurons, and axon terminals to mediate presynaptic and postsynaptic inhibition. It is possible, or even likely, that the different physiological functions outlined above correlate with the expression of genetic markers. The recent report by Ross et al. (2010) provides an example for a successful correlation of a genetic marker of inhibitory interneurons with a defined physiological function. Mice deficient in the transcription factor BHLHb5 show a loss of certain inhibitory interneurons that selectively affects the assembly of the sensory circuits involved in the sensation of itch. Another example is the recently described selective innervation of low-threshold myelinated afferents by parvalbumin-positive inhibitory interneurons through axo-axonic synapses (Hughes et al., 2012).
Unlike traditional markers, such as dendritic tree morphology and firing patterns, genetically encoded markers are extremely useful for specific genetic manipulations of subtypes of neurons. Such manipulations include the generation of fluorescent reporter mice enabling targeted recordings from identified cells and of cre diver lines that are extremely versatile tools for the specific ablation of neurons through targeted expression of cytotoxins (Buch et al., 2005; Abrahamsen et al., 2008) or for the silencing or excitation of neurons by optogenetic (Boyden et al., 2005; Zhang et al., 2007) or chemogenetic techniques (Ray et al., 2011).
Although the markers identified here define distinct populations of dorsal horn inhibitory interneurons, they are also expressed in cells different from inhibitory interneurons and in other tissues. Dynorphin is also expressed in some excitatory interneurons and in dorsal root ganglion (DRG) neurons (Sweetnam et al., 1982; Marvizón et al., 2009; Baseer et al., 2012), and Kcnip2, for example, is also expressed in the heart (Decher et al., 2001). Potential caveats arising from these additional expression sites can be overcome by cross-sectional gene targeting (for review, see Dymecki et al., 2010). This approach involves the use of two different recombinases (e.g., cre and flpe) driven by promoters of different but overlapping specificity. In the case of dorsal horn inhibitory neurons, the use of a Gad1-cre transgenic line as a second recombinase line would, for example, avoid recombination in DRG neurons or other excitatory cells, whereas the use of Hoxb8-cre mice (Witschi et al., 2010) would avoid recombination in supraspinal areas.
The finding of this study should help to resolve the highly complex architecture of the neuronal circuit in the spinal dorsal horn and may thus foster our understanding of spinal sensory processing under physiological conditions and its malfunctioning in chronic pain conditions.
Footnotes
This work was supported partly by German Research Foundation Collaborative Research Centre Grant SFB 665 and the Helmholtz Program “Function and Dysfunction of the Nervous System” (C.B.), European Research Council Advanced Investigator Grant 250128 (Dorsal Horn Interneurons in Sensory Processing), and Swiss National Science Foundation Grant 131093 (H.U.Z.). We thank Isabelle Camenisch, Cornelia Leuzinger, Claudia Päseler, and Petra Stallerow for excellent technical assistance and maintenance of mice. The Ptf1a microarray screen was performed and analyzed at the Functional Genomics Center Zurich.
The authors declare no competing financial interests.
References
- Abrahamsen B, Zhao J, Asante CO, Cendan CM, Marsh S, Martinez-Barbera JP, Nassar MA, Dickenson AH, Wood JN. The cell and molecular basis of mechanical, cold, and inflammatory pain. Science. 2008;321:702–705. doi: 10.1126/science.1156916. [DOI] [PubMed] [Google Scholar]
- Baseer N, Polgár E, Watanabe M, Furuta T, Kaneko T, Todd AJ. Projection neurons in lamina III of the rat spinal cord are selectively innervated by local dynorphin-containing excitatory neurons. J Neurosci. 2012;32:11854–11863. doi: 10.1523/JNEUROSCI.2707-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer C, Roberts LA, Komisaruk BR. Hyperalgesia induced by altered glycinergic activity at the spinal cord. Life Sci. 1985;37:875–882. doi: 10.1016/0024-3205(85)90523-5. [DOI] [PubMed] [Google Scholar]
- Boyden ES, Zhang F, Bamberg E, Nagel G, Deisseroth K. Millisecond-timescale, genetically targeted optical control of neural activity. Nat Neurosci. 2005;8:1263–1268. doi: 10.1038/nn1525. [DOI] [PubMed] [Google Scholar]
- Bröhl D, Strehle M, Wende H, Hori K, Bormuth I, Nave KA, Müller T, Birchmeier C. A transcriptional network coordinately determines transmitter and peptidergic fate in the dorsal spinal cord. Dev Biol. 2008;322:381–393. doi: 10.1016/j.ydbio.2008.08.002. [DOI] [PubMed] [Google Scholar]
- Buch T, Heppner FL, Tertilt C, Heinen TJ, Kremer M, Wunderlich FT, Jung S, Waisman A. A Cre-inducible diphtheria toxin receptor mediates cell lineage ablation after toxin administration. Nat Methods. 2005;2:419–426. doi: 10.1038/nmeth762. [DOI] [PubMed] [Google Scholar]
- Cheng L, Arata A, Mizuguchi R, Qian Y, Karunaratne A, Gray PA, Arata S, Shirasawa S, Bouchard M, Luo P, Chen CL, Busslinger M, Goulding M, Onimaru H, Ma Q. Tlx3 and Tlx1 are post-mitotic selector genes determining glutamatergic over GABAergic cell fates. Nat Neurosci. 2004;7:510–517. doi: 10.1038/nn1221. [DOI] [PubMed] [Google Scholar]
- Cheng L, Samad OA, Xu Y, Mizuguchi R, Luo P, Shirasawa S, Goulding M, Ma Q. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8:1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- Decher N, Uyguner O, Scherer CR, Karaman B, Yüksel-Apak M, Busch AE, Steinmeyer K, Wollnik B. hKChIP2 is a functional modifier of hKv4.3 potassium channels: cloning and expression of a short hKChIP2 splice variant. Cardiovasc Res. 2001;52:255–264. doi: 10.1016/S0008-6363(01)00374-1. [DOI] [PubMed] [Google Scholar]
- Dymecki SM, Ray RS, Kim JC. Mapping cell fate and function using recombinase-based intersectional strategies. Methods Enzymol. 2010;477:183–213. doi: 10.1016/S0076-6879(10)77011-7. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, Hornik K, Hothorn T, Huber W, Iacus S, Irizarry R, Leisch F, Li C, Maechler M, Rossini AJ, Sawitzki G, Smith C, Smyth G, Tierney L, Yang JY, Zhang J. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasgow SM, Henke RM, Macdonald RJ, Wright CV, Johnson JE. Ptf1a determines GABAergic over glutamatergic neuronal cell fate in the spinal cord dorsal horn. Development. 2005;132:5461–5469. doi: 10.1242/dev.02167. [DOI] [PubMed] [Google Scholar]
- Graham BA, Brichta AM, Callister RJ. Moving from an averaged to specific view of spinal cord pain processing circuits. J Neurophysiol. 2007;98:1057–1063. doi: 10.1152/jn.00581.2007. [DOI] [PubMed] [Google Scholar]
- Gross MK, Dottori M, Goulding M. Lbx1 specifies somatosensory association interneurons in the dorsal spinal cord. Neuron. 2002;34:535–549. doi: 10.1016/S0896-6273(02)00690-6. [DOI] [PubMed] [Google Scholar]
- Grudt TJ, Perl ER. Correlations between neuronal morphology and electrophysiological features in the rodent superficial dorsal horn. J Physiol. 2002;540:189–207. doi: 10.1113/jphysiol.2001.012890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guillemot F, Lo LC, Johnson JE, Auerbach A, Anderson DJ, Joyner AL. Mammalian achaete-scute homolog 1 is required for the early development of olfactory and autonomic neurons. Cell. 1993;75:463–476. doi: 10.1016/0092-8674(93)90381-Y. [DOI] [PubMed] [Google Scholar]
- Harvey RJ, Depner UB, Wässle H, Ahmadi S, Heindl C, Reinold H, Smart TG, Harvey K, Schütz B, Abo-Salem OM, Zimmer A, Poisbeau P, Welzl H, Wolfer DP, Betz H, Zeilhofer HU, Müller U. GlyR alpha3: an essential target for spinal PGE2-mediated inflammatory pain sensitization. Science. 2004;304:884–887. doi: 10.1126/science.1094925. [DOI] [PubMed] [Google Scholar]
- Heinke B, Ruscheweyh R, Forsthuber L, Wunderbaldinger G, Sandkühler J. Physiological, neurochemical and morphological properties of a subgroup of GABAergic spinal lamina II neurones identified by expression of green fluorescent protein in mice. J Physiol. 2004;560:249–266. doi: 10.1113/jphysiol.2004.070540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heppenstall PA, Lewin GR. BDNF but not NT-4 is required for normal flexion reflex plasticity and function. Proc Natl Acad Sci U S A. 2001;98:8107–8112. doi: 10.1073/pnas.141015098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero JF, Laird JM, López-García JA. Wind-up of spinal cord neurones and pain sensation: much ado about something? Prog Neurobiol. 2000;61:169–203. doi: 10.1016/S0301-0082(99)00051-9. [DOI] [PubMed] [Google Scholar]
- Hori K, Cholewa-Waclaw J, Nakada Y, Glasgow SM, Masui T, Henke RM, Wildner H, Martarelli B, Beres TM, Epstein JA, Magnuson MA, Macdonald RJ, Birchmeier C, Johnson JE. A nonclassical bHLH Rbpj transcription factor complex is required for specification of GABAergic neurons independent of Notch signaling. Genes Dev. 2008;22:166–178. doi: 10.1101/gad.1628008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua XY, Boublik JH, Spicer MA, Rivier JE, Brown MR, Yaksh TL. The antinociceptive effects of spinally administered neuropeptide Y in the rat: systematic studies on structure-activity relationship. J Pharmacol Exp Ther. 1991;258:243–248. [PubMed] [Google Scholar]
- Huang M, Huang T, Xiang Y, Xie Z, Chen Y, Yan R, Xu J, Cheng L. Ptf1a, Lbx1 and Pax2 coordinate glycinergic and peptidergic transmitter phenotypes in dorsal spinal inhibitory neurons. Dev Biol. 2008;322:394–405. doi: 10.1016/j.ydbio.2008.06.031. [DOI] [PubMed] [Google Scholar]
- Hughes DI, Sikander S, Kinnon CM, Boyle KA, Watanabe M, Callister RJ, Graham BA. Morphological, neurochemical and electrophysiological features of parvalbumin-expressing cells: a likely source of axo-axonic inputs in the mouse spinal dorsal horn. J Physiol. 2012;590:3927–3951. doi: 10.1113/jphysiol.2012.235655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahr CE, Yoshioka K. Ia afferent excitation of motoneurones in the in vitro new-born rat spinal cord is selectively antagonized by kynurenate. J Physiol. 1986;370:515–530. doi: 10.1113/jphysiol.1986.sp015948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi Y, Cooper B, Gannon M, Ray M, MacDonald RJ, Wright CV. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat Genet. 2002;32:128–134. doi: 10.1038/ng959. [DOI] [PubMed] [Google Scholar]
- Laing I, Todd AJ, Heizmann CW, Schmidt HH. Subpopulations of GABAergic neurons in laminae I-III of rat spinal dorsal horn defined by coexistence with classical transmitters, peptides, nitric oxide synthase or parvalbumin. Neuroscience. 1994;61:123–132. doi: 10.1016/0306-4522(94)90065-5. [DOI] [PubMed] [Google Scholar]
- Malmberg AB, Chen C, Tonegawa S, Basbaum AI. Preserved acute pain and reduced neuropathic pain in mice lacking PKCgamma. Science. 1997;278:279–283. doi: 10.1126/science.278.5336.279. [DOI] [PubMed] [Google Scholar]
- Marvizón JC, Chen W, Murphy N. Enkephalins, dynorphins, and beta-endorphin in the rat dorsal horn: an immunofluorescence colocalization study. J Comp Neurol. 2009;517:51–68. doi: 10.1002/cne.22130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–778. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Moore KA, Kohno T, Karchewski LA, Scholz J, Baba H, Woolf CJ. Partial peripheral nerve injury promotes a selective loss of GABAergic inhibition in the superficial dorsal horn of the spinal cord. J Neurosci. 2002;22:6724–6731. doi: 10.1523/JNEUROSCI.22-15-06724.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller T, Brohmann H, Pierani A, Heppenstall PA, Lewin GR, Jessell TM, Birchmeier C. The homeodomain factor lbx1 distinguishes two major programs of neuronal differentiation in the dorsal spinal cord. Neuron. 2002;34:551–562. doi: 10.1016/S0896-6273(02)00689-X. [DOI] [PubMed] [Google Scholar]
- Müller T, Anlag K, Wildner H, Britsch S, Treier M, Birchmeier C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005;19:733–743. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveilhan P, Hassani H, Lucas G, Blakeman KH, Hao JX, Xu XJ, Wiesenfeld-Hallin Z, Thorén P, Ernfors P. Reduced antinociception and plasma extravasation in mice lacking a neuropeptide Y receptor. Nature. 2001;409:513–517. doi: 10.1038/35054063. [DOI] [PubMed] [Google Scholar]
- Neumann S, Braz JM, Skinner K, Llewellyn-Smith IJ, Basbaum AI. Innocuous, not noxious, input activates PKCgamma interneurons of the spinal dorsal horn via myelinated afferent fibers. J Neurosci. 2008;28:7936–7944. doi: 10.1523/JNEUROSCI.1259-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernía-Andrade AJ, Kato A, Witschi R, Nyilas R, Katona I, Freund TF, Watanabe M, Filitz J, Koppert W, Schüttler J, Ji G, Neugebauer V, Marsicano G, Lutz B, Vanegas H, Zeilhofer HU. Spinal endocannabinoids and CB1 receptors mediate C-fiber-induced heterosynaptic pain sensitization. Science. 2009;325:760–764. doi: 10.1126/science.1171870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár E, Shehab SA, Watt C, Todd AJ. GABAergic neurons that contain neuropeptide Y selectively target cells with the neurokinin 1 receptor in laminae III and IV of the rat spinal cord. J Neurosci. 1999;19:2637–2646. doi: 10.1523/JNEUROSCI.19-07-02637.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polgár E, Sardella TC, Watanabe M, Todd AJ. Quantitative study of NPY-expressing GABAergic neurons and axons in rat spinal dorsal horn. J Comp Neurol. 2011;519:1007–1023. doi: 10.1002/cne.22570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray RS, Corcoran AE, Brust RD, Kim JC, Richerson GB, Nattie E, Dymecki SM. Impaired respiratory and body temperature control upon acute serotonergic neuron inhibition. Science. 2011;333:637–642. doi: 10.1126/science.1205295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinold H, Ahmadi S, Depner UB, Layh B, Heindl C, Hamza M, Pahl A, Brune K, Narumiya S, Müller U, Zeilhofer HU. Spinal inflammatory hyperalgesia is mediated by prostaglandin E receptors of the EP2 subtype. J Clin Invest. 2005;115:673–679. doi: 10.1172/JCI200523618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts LA, Beyer C, Komisaruk BR. Nociceptive responses to altered GABAergic activity at the spinal cord. Life Sci. 1986;39:1667–1674. doi: 10.1016/0024-3205(86)90164-5. [DOI] [PubMed] [Google Scholar]
- Ross SE, Mardinly AR, McCord AE, Zurawski J, Cohen S, Jung C, Hu L, Mok SI, Shah A, Savner EM, Tolias C, Corfas R, Chen S, Inquimbert P, Xu Y, McInnes RR, Rice FL, Corfas G, Ma Q, Woolf CJ, Greenberg ME. Loss of inhibitory interneurons in the dorsal spinal cord and elevated itch in Bhlhb5 mutant mice. Neuron. 2010;65:886–898. doi: 10.1016/j.neuron.2010.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S, Todd AJ, Spike RC. Evidence that neuropeptide Y is present in GABAergic neurons in the superficial dorsal horn of the rat spinal cord. Neuroscience. 1993;53:537–545. doi: 10.1016/0306-4522(93)90218-5. [DOI] [PubMed] [Google Scholar]
- Sandkühler J. Models and mechanisms of hyperalgesia and allodynia. Physiol Rev. 2009;89:707–758. doi: 10.1152/physrev.00025.2008. [DOI] [PubMed] [Google Scholar]
- Sardella TC, Polgár E, Garzillo F, Furuta T, Kaneko T, Watanabe M, Todd AJ. Dynorphin is expressed primarily by GABAergic neurons that contain galanin in the rat dorsal horn. Mol Pain. 2011;7:76. doi: 10.1186/1744-8069-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholz J, Broom DC, Youn DH, Mills CD, Kohno T, Suter MR, Moore KA, Decosterd I, Coggeshall RE, Woolf CJ. Blocking caspase activity prevents transsynaptic neuronal apoptosis and the loss of inhibition in lamina II of the dorsal horn after peripheral nerve injury. J Neurosci. 2005;25:7317–7323. doi: 10.1523/JNEUROSCI.1526-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweetnam PM, Neale JH, Barker JL, Goldstein A. Localization of immunoreactive dynorphin in neurons cultured from spinal cord and dorsal root ganglia. Proc Natl Acad Sci U S A. 1982;79:6742–6746. doi: 10.1073/pnas.79.21.6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiong SY, Polgár E, van Kralingen JC, Watanabe M, Todd AJ. Galanin-immunoreactivity identifies a distinct population of inhibitory interneurons in laminae I-III of the rat spinal cord. Mol Pain. 2011;7:36. doi: 10.1186/1744-8069-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd AJ. Neuronal circuitry for pain processing in the dorsal horn. Nat Rev Neurosci. 2010;11:823–836. doi: 10.1038/nrn2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wende H, Lechner SG, Cheret C, Bourane S, Kolanczyk ME, Pattyn A, Reuter K, Munier FL, Carroll P, Lewin GR, Birchmeier C. The transcription factor c-Maf controls touch receptor development and function. Science. 2012;335:1373–1376. doi: 10.1126/science.1214314. [DOI] [PubMed] [Google Scholar]
- Wildner H, Müller T, Cho SH, Bröhl D, Cepko CL, Guillemot F, Birchmeier C. dILA neurons in the dorsal spinal cord are the product of terminal and non-terminal asymmetric progenitor cell divisions, and require Mash1 for their development. Development. 2006;133:2105–2113. doi: 10.1242/dev.02345. [DOI] [PubMed] [Google Scholar]
- Wildner H, Gierl MS, Strehle M, Pla P, Birchmeier C. Insm1 (IA-1) is a crucial component of the transcriptional network that controls differentiation of the sympatho-adrenal lineage. Development. 2008;135:473–481. doi: 10.1242/dev.011783. [DOI] [PubMed] [Google Scholar]
- Witschi R, Johansson T, Morscher G, Scheurer L, Deschamps J, Zeilhofer HU. Hoxb8-Cre mice: a tool for brain-sparing conditional gene deletion. Genesis. 2010;48:596–602. doi: 10.1002/dvg.20656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Lopes C, Qian Y, Liu Y, Cheng L, Goulding M, Turner EE, Lima D, Ma Q. Tlx1 and Tlx3 coordinate specification of dorsal horn pain-modulatory peptidergic neurons. J Neurosci. 2008;28:4037–4046. doi: 10.1523/JNEUROSCI.4126-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasaka T, Tiong SY, Hughes DI, Riddell JS, Todd AJ. Populations of inhibitory and excitatory interneurons in lamina II of the adult rat spinal dorsal horn revealed by a combined electrophysiological and anatomical approach. Pain. 2010;151:475–488. doi: 10.1016/j.pain.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeilhofer HU, Wildner H, Yévenes GE. Fast synaptic inhibition in spinal sensory processing and pain control. Physiol Rev. 2012;92:193–235. doi: 10.1152/physrev.00043.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Wang LP, Brauner M, Liewald JF, Kay K, Watzke N, Wood PG, Bamberg E, Nagel G, Gottschalk A, Deisseroth K. Multimodal fast optical interrogation of neural circuitry. Nature. 2007;446:633–639. doi: 10.1038/nature05744. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Cai YQ, Zou F, Bie B, Pan ZZ. Epigenetic suppression of GAD65 expression mediates persistent pain. Nat Med. 2011;17:1448–1455. doi: 10.1038/nm.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]