Abstract
The integral membrane-bound Nrf1 transcription factor fulfils important functions in maintaining cellular homeostasis and organ integrity, but how it is controlled vectorially is unknown. Herein, creative use of Gal4-based reporter assays with protease protection assays (GRAPPA), and double fluorescence protease protection (dFPP), reveals that the membrane-topogenic vectorial behaviour of Nrf1 dictates its post-translational modification and transactivation activity. Nrf1 is integrated within endoplasmic reticulum (ER) membranes through its NHB1-associated TM1 in cooperation with other semihydrophobic amphipathic regions. The transactivation domains (TADs) of Nrf1, including its Asn/Ser/Thr-rich (NST) glycodomain, are transiently translocated into the ER lumen, where it is glycosylated in the presence of glucose to become a 120-kDa isoform. Thereafter, the NST-adjoining TADs are partially repartitioned out of membranes into the cyto/nucleoplasmic side, where Nrf1 is subject to deglycosylation and/or proteolysis to generate 95-kDa and 85-kDa isoforms. Therefore, the vectorial process of Nrf1 controls its target gene expression.
All organisms living in an oxygenated environment have evolved efficient strategies enabling their constituent cells to express a series of robust homeostatic genes in order to defend against cellular oxidative stress and adapt to the changing surroundings. The expression of antioxidant cytoprotective genes is controlled primarily by the cap‘n’collar (CNC) family of transcription factors1,2. This family comprises the Drosophila Cnc protein as the founding member3,4, the Caenorhabditis elegans protein skinhead-1 (Skn-1)5,6, and four vertebrate activators nuclear factor-erythroid 2 (NF-E2) p45 subunit7,8,9, NF-E2 p45-related factor 1 [Nrf1 (ref. 10), including its long form, called transcription factor 11 (TCF11)11,12 and its short isoform, designated as locus control region-factor 1 (LCR-F1)13,14, see Fig. 1], Nrf2 (ref. 15), and Nrf3 (refs. 16,17), as well as two distantly related repressors BTB and CNC homolog 1 (Bach1)18,19 and Bach2 (refs. 20,21). In addition to cytoprotection against various oxidants and electrophiles, the CNC family members also regulate expression of a wide range of genes that are involved in development, growth, longevity, and cellular defence, metabolism, and repair response under either normal homeostatic or pathophysiological conditions. Most of those genes are direct targets of CNC family transcription factors, which bind to the antioxidant/electrophile response elements (AREs/EpREs)1,2.
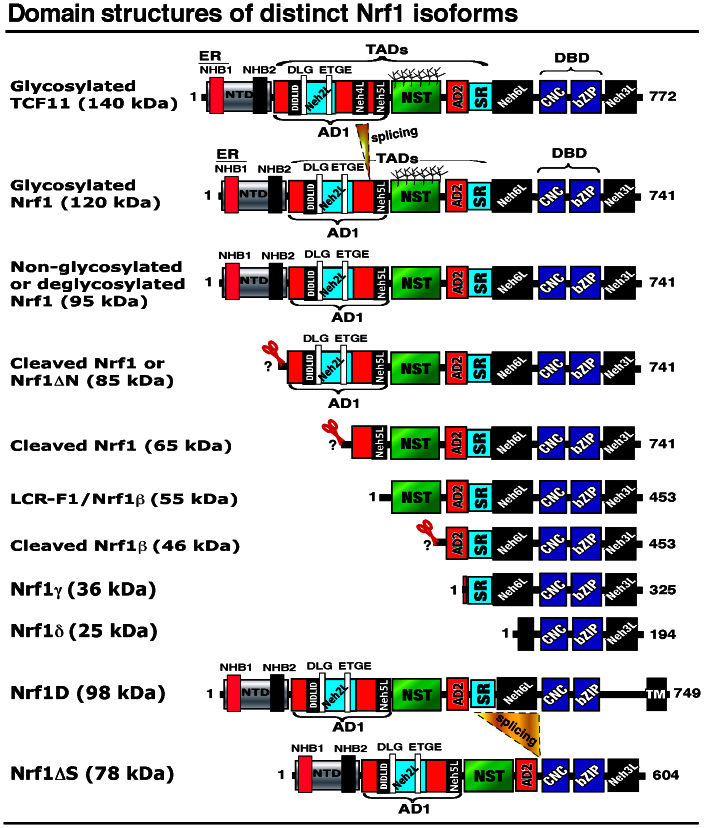
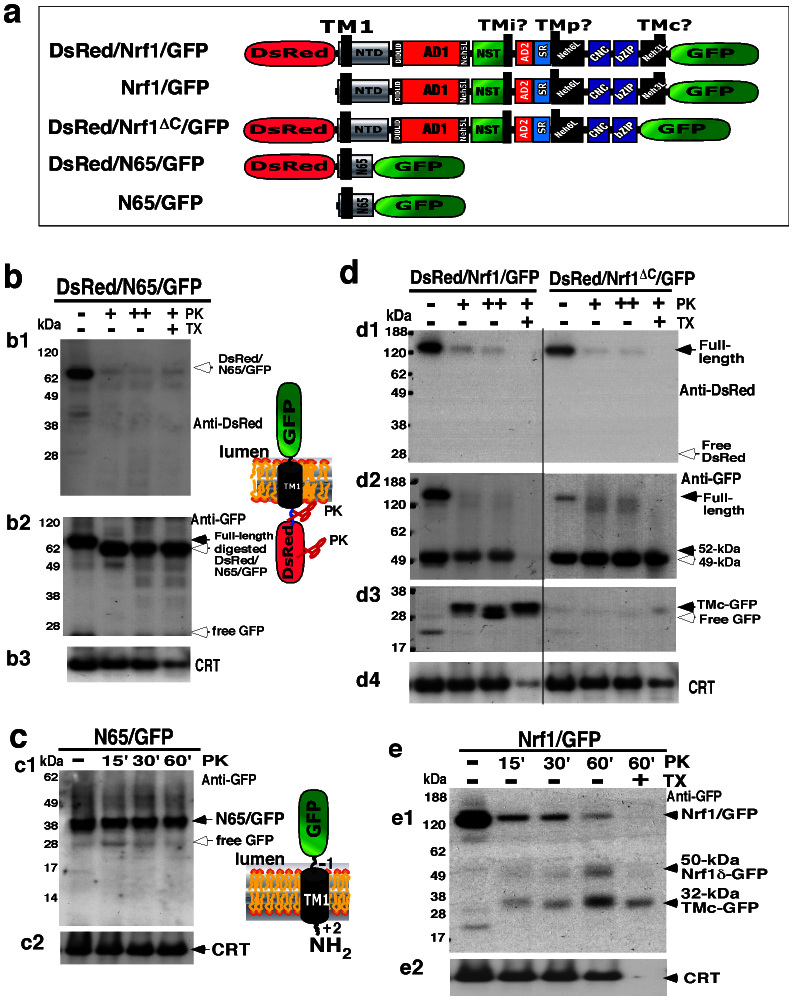
Figure 1. Structural domains of distinct Nrf1 isoforms.
The N-terminal domain (NTD) contains a TM1-associated NHB1 sequence that dictates Nrf1 to target to the endoplasmic reticulum (ER) and enables it to anchor within the membrane. Both CNC and bZIP regions were combined into the Neh1L (Nrf2-ECH homology 1-like) domain that enables heterodimerization with a small Maf or other bZIP proteins before binding to antioxidant response element (ARE) sequences in the promoter regions of target genes. The transactivation domains (TADs) of Nrf1 comprise AD1 (acidic domain 1), NST (Asn/Ser/Thr-rich), AD2 and SR (serine-repeat) regions. On the C-terminal border of NST glycodomain immediately to the acidic-hydrophobic amphipathic AD2, the TMi segment may act as a luminal-anchoring switch to control repartitioning of its flanking AD1 and AD2/SR domains. Within AD1, Neh2L contains the DIDLID/DLG element and ETGE motif; both are conserved with equivalents of Nrf2 and CncC. In addition to Neh2L, the Neh5L, but not Neh4L, subdomain is included within AD1 of Nrf1, when compared with its longer TCF11. The N-linked glycosylation of the NST domain confers Nrf1 and TCF11 to migrate electrophoretically at estimated masses of ∼120-kDa and ∼140-kDa, respectively, whilst their non-glycosylated/deglycosylated proteins exhibit fast electrophoretic mobilities at ∼95-kDa and ∼110-kDa. They may be further processed through selective proteolysis to give rise to multiple cleaved forms of between 85-kDa and 25-kDa. Of which the putative activated 85-kDa protein can be also translated through an alternative initiation signal (in Nrf1 Δ767 clones designated originally51,52) so that it lacks an ER-anchoring NTD (thus called Nrf1ΔN), and may be further processed into an unstable polypeptide of 65-kDa or 55-kDa. The 55-kDa form, that was originally designated LCR-F1 (called Nrf1β herein), is produced by in-frame translation and/or selective proteolysis, and may be rapidly degraded to yield short dominant-negative isoforms of 46-kDa, 36-kDa (called Nrf1γ) and 25-kDa (called Nrf1δ). For convenience, Nrf1 variant ΔD and Δ10 clones51,52 are renamed as Nrf1D or Nrf1ΔS, respectively. The triangle represents a loss of the corresponding region by alternative splicing, whereas the scissors with a question mark (?) indicate unidentified cleavage sites.
In mammals, Nrf1 and Nrf2 represent two principal orthologs of the Cnc and Skn-1 factors, that regulate the basal and inducible expression of antioxidant, detoxification and 26S proteosomal component genes1,22,23,24,25. To date, there has been a disproportionate focus on Nrf2 and relatively less is known about the function of Nrf1. In fact, global knockout of Nrf1 in the mouse leads to embryonic lethality and severe oxidative stress13,26,27, indicating that loss of Nrf1 function cannot be compensated by the presence of Nrf2 in these animals. Specifically, conditional knockout of Nrf1 in the liver, bone and brain results in non-alcoholic steatohepatitis and hepatoma28,29, reduced bone size30 and neurodegenerative disorders31,32, respectively. By contrast, Nrf2 knockout mice are viable with no obvious phenotypes33, suggesting it seems to be dispensable. These facts convincingly demonstrate that Nrf1, but not Nrf2, is essential for maintaining both organ integrity and cellular (redox, lipid and protein) homeostasis. This indicates that Nrf1 fulfils a unique and indispensable function, that is distinct from Nrf2, in controlling a subset of ARE-battery genes that are responsible for cytoprotection against stress, apoptosis, degeneration, ageing, inflammation and carcinogenesis.
Amongst the CNC family members, Nrf1 is an integral membrane-bound glycoprotein with several distinct isoforms (Fig. 1). The full-length Nrf1 is first synthesized as a 95-kDa non-glycosylated protein [i.e. the term non-glycosylation is defined that the primordial portion of Nrf1 residing on the cytoplasmic side of membranes has actually never been glycosylated at the consensus asparagines (Asn-X-Ser/Thr, in which X represents any of other amino acids rather than proline) in its Asn/Ser/Thr-rich (NST) domain, because it is clear that the N-linked glycosylation reaction catalyzed by oligosaccharyltransferases (OST) occurs in the lumen of the endoplasmic reticulum (ER)34,35]. Subsequently, the non-glycosylated Nrf1 protein is co-translationally targeted to the ER through its N-terminal homology box 1 (NHB1) signal sequence36,37,38. The NHB1-associated transmembrane region (called TM1, aa 7–26) determines the membrane-topology of Nrf1 (refs. 38,39,40), which is highly conserved with those of TCF11 (ref. 22), Nrf3 (ref. 41), CncC24 and Skn-1 (ref. 42); they are comprised of an NHB1-CNC subfamily of membrane-bound transcription factors, with similar topologies being predicted (Figs. S1 and S2). Once Nrf1 is anchored within the ER membrane through its TM1 region, its NST glycodomain (aa 299–400) is translocated into the lumen of this organelle, where the NST domain is asparagine-glycosylated to yield an inactive 120-kDa glycoprotein. Thereafter, Nrf1 is sorted out of the ER through the outer nuclear envelope membrane and into the inner nuclear membrane39. When required, the luminal regions of Nrf1 is partially repartitioned out of membranes and retrotranslocated into the cyto/nucleoplasmic subcellular compartments, whereupon the protein is allowed for N-linked deglycosylation by a peptide:N-glycosidase (PNGase)43,44,45,46 (i.e. the term deglycosylation), in order to generate an active 95-kDa transcription factor. In the vectorial process, Nrf1 may also be selectively subjected to its proteolytic processing to give rise to either a putatively cleaved 85-kDa activator or other short dominant-negative isoforms, so that they are released from the ER to translocate the nucleus39,47,48,49. However, it is notable that the positively-staining signal for Nrf1 is neither observed in the mitochondria nor the Golgi apparatus under confocal and immuno-electron microscopes36,38,39.
Once the 95-kDa and/or 85-kDa Nrf1 reach the nucleoplasmic compartments, they can gain access to the general transcriptional machinery (GTM) primarily via its Nrf2-ECH homology 5-like subdomain (Neh5L, aa 280–298)39,50, allowing transactivation of target genes. By contrast, LCR-F1 (also called Nrf1β, functioning as a weak activator14,39,51), along with other dominant-negative isoforms Nrf1γ and Nrf1δ14,39,51,52 (see Fig. 1), are located predominantly in the nucleus39,47. Therein, the DNA-binding of all Nrf1 isoforms to ARE sequences in the target gene promoters is exerted through its CNC-bZIP domain heterodimerizing with a small Maf protein or other bZIP factors2,53,54. The ability of Nrf1 to activate ARE-driven genes is differentially regulated by several distinct transactivation domains (TADs), that comprise its acidic domain 1 (AD1, that comprises aa 125–298 covering the PEST1 sequence, and the Neh2L and Neh5L subdomains), NST glycodomain (aa 299–400), AD2 (aa 403–453), and serine-repeat domain (SR, aa 454–489)14,39. Intriguingly, the TMi glycopeptide (i.e. a putative intermediate transmembrane-associated helix formed by aa 374–393, see Fig. S1) is located in the NST C-terminal border immediately to the acidic-hydrophobic amphipathic region within AD2 (refs. 22,39), whereas the TMp hinge region (i.e. a Pro-kinked transmembrane helix that is predicted to be formed by aa 507–525, see Fig. S1) is located in the N-terminal border of the Neh6L domain (aa 490–580), which is flanked N-terminally by the acidic TADs (aa 125–489) and C-terminally by the basic CNC-bZIP domain (aa 581–687). However, the molecular mechanism by which the transactivation activity of Nrf1 is tempo-spatially controlled in its membrane-topogenic processes remains elusive. Herein, our novel evidence reveals distinct topogenic-vectorial processes whereby some of Nrf1 isoforms are translocated into the ER and/or retrotranslocated across the membranes into the cyto/nucleoplasmic side, allowing transactivation of its target genes.
It is notable that our previous work showed that inhibition of N-linked glycosylation by tunicamycin (TU) results in the wild-type Nrf1 being expressed as a non-glycosylated 95-kDa protein, with a transactivation activity that is 25% lower than that of wild-type Nrf1 treated with vehicle control39. By contrast, the PNGase-based deglycosylation inhibitor Z-VAD-fmk [carbobenzoxy-valyl-alanyl-aspartyl-(O-methyl)-fluoromethyl ketone]55 causes an increase in the abundance of the inactive 120-kDa Nrf1 glycoprotein, as accompanied by a partial decrease in its transactivation activity (unpublished data). Further, the Nrf1(1–7)xN/D mutant (in which all seven asparagine glycosylation consensus sites in the NST domain were mutated into aspartate residues in order to create a potent activator, representing deglycosylated 95-kDa Nrf1 protein) mediates an increased transactivation activity of ARE-driven gene expression, whereas the Nrf1(1–7)xN/Q mutant (in which all seven asparagine consensus sites in the NST domain were mutated into glutamine residues so as to create a non-glycosylated 95-kDa protein) exhibits a modestly decreased activity39, when compared with that of the wild-type Nrf1 factor. These findings demonstrate that the transactivation activity of Nrf1 is, at least in part, regulated by the changing glycosylation status of its NST domain situated within the centre of acidic TADs. Such asparagines-linked glycosylation of Nrf1, as other membrane proteins characterized by Goder and Spiess56, could influence (or be influenced by) its dynamic topological orientation within and around membranes. These have led us to envisage that glucose levels might alter the glycosylation/deglycosylation status of Nrf1, and in turn, these modifications could be controlled by membrane-topogenic vectorial behaviours of this NHB1-CNC protein that dynamically moves in and out of ER membranes during its topological folding or post-translational processing. As expected, our present study demonstrates that distinct vectorial processes of Nrf1 are dictated by glucose consumption and deprivation. Importantly, the membrane-topogenic vectorial behaviour of Nrf1 controls its post-translational modification and its ability to transactivate its target gene expression.
Results
Distinct Nrf1 isoforms are co-expressed in different mammalian species
Accumulating evidence reveals that over eleven Nrf1 isoforms (Fig. 1) are produced from the single nfe2l1 gene, though differentially expressed, in different mammalian species1,2,51,52,57. These isoforms are synthesized by translation through distinct initiation signals (i.e. the first or internal start ATG codons) embedded in different lengths of open reading frames, portions of which can be alternatively spliced from the cognate mRNAs10,14,51,52,53,58. The prototypic full-length Nrf1 protein arises by alternative splicing of the mRNA enabling translation of the long TCF11 form12,58, which is expressed in the human and rat, but not in the mouse10,12,14. Although removal of the Neh4L-encoding nucleotides results in a loss of the putative TAD element in Nrf1, this factor was shown to have a similar ability to transactivate ARE-driven genes as TCF11 (with a molecular mass of approximately 140-kDa estimated on Laemmli SDS-PAGE gels)59. Both the full-length Nrf1 and TCF11 forms can also be processed by selective proteolysis to give rise to the putative activated forms of between about 85-kDa and 65-kDa [which lack the ER-anchoring N-terminal domain (NTD, aa 1–124, that negatively regulates Nrf1), but retain either the entire or the essential portions of TADs]. They may also be further processed to generate various TADs-deficient isoforms such as those of approximately 55-kDa, 36-kDa and 25-kDa (designated Nrf1β/LCR-F1, Nrf1γ and Nrf1δ, respectively) (Fig. 1; see refs. 11,39,60). These truncated variants are neither targeted to the ER nor recovered in membrane fractions38,39 (also see Figs. 2e, and S5).
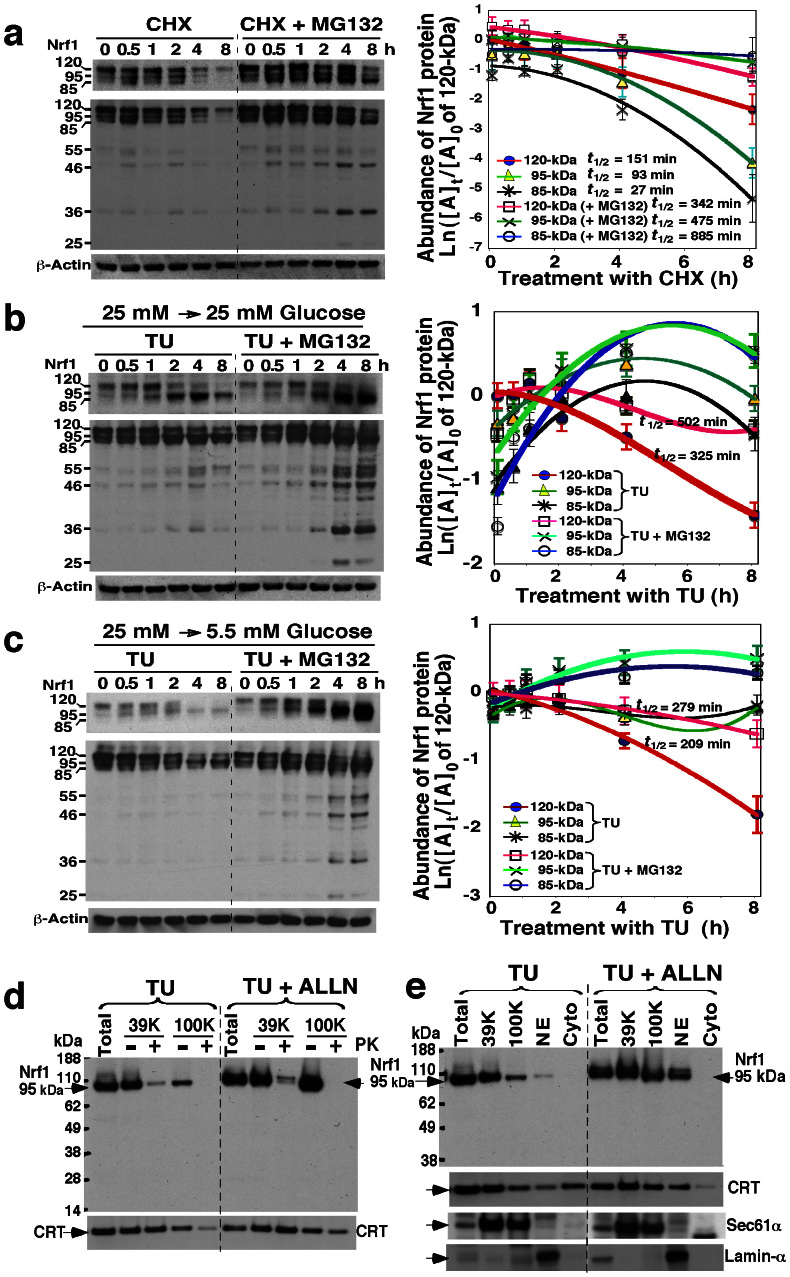
Figure 2. The stability of longer Nrf1 isoforms in response to changes in glucose concentration.
(a) Examination of the stability of Nrf1 isoforms. COS-1 cells were transfected with an Nrf1 expression construct for 6 h, and were allowed to recover from transfection for 16 h in the fresh 25 mM-glucose medium. The cells were treated with cycloheximide (CHX, 50 μg/ml) and/or MG132 (5 μM) for the indicated times before being disrupted. The abundance of Nrf1 proteins was determined by immunoblotting. The left upper shows a cropped image from the film that was exposed to the immunoblot for a shorter time to eliminate intense bands. The intensity of blots was calculated by dividing the values for Nrf1 with that for β-actin. The relative amount of the 120-, 95- and 85-kDa proteins at the indicated times ([A]t) was normalized to [A]0 (i.e. the value of 120-kDa at t0 after CHX), and is shown graphically (right, mean ± S.D, n = 4), along with their half-lives (t1/2). (b,c) Stability of Nrf1 isoforms in cells grown in 25 mM or 5.5 mM glucose medium upon blockage of glycosylation by tunicamycin (TU). Nrf1-expressing cells were transferred to 25 mM (b) or 5.5 mM (c) glucose medium, to which was added TU (1 μg/ml) and/or MG132 (5 μM) for the indicated times. Following western blotting, the relative abundance of the 120-, 95- and 85-kDa Nrf1 proteins was calculated after normalization to [A]0 of 120-kDa at t0 after TU and shown graphically (right, mean ± S.D, n = 4). (d,e) The non-glycosylated 95-kDa Nrf1 isoform is not protected by membranes. Nrf1-expressing cells were treated with TU (1 μg/ml) and/or ALLN (5 μg/ml) for 18 h in the fresh 25 mM-glucose medium, and then subjected to subcellular fractionation (e), followed immediately by membrane proteinase protection assays (d). The resulting products were identified by immunoblotting with V5 antibody. Calreticulin (CRT) was an ER-luminal marker, that existed in ER-enriched subcellular fractions purified by centrifuging at 39, 000 × g (39 K) and 100, 000 × g (100 K). Sec61α and Lamin-α were used to verify ER and nuclear envelope (NE) membranes.
The LCR-F1/Nrf1β polypeptide can also arise by internal translation starting between Met289 and Met297 codons within the longer mRNA transcripts14,53. It migrates at a mass of 55-kDa in the pH 7.0 LDS/NuPAGE gel (Fig. 2; see refs. 38,39), but the pH 8.9 Laemmli SDS-PAGE allows its mobility to be exhibited at an estimated size of 65-kDa59,61 (which was called p65Nrf1 in ref. 61). By comparison with the full-length Nrf1, the 55-kDa LCR-F1/Nrf1β form lacks both the NTD and the essential transctivation domain AD1, but still contains AD2, NST and SR domains that are also required for full activity. It is thus postulated that LCR-F1/Nrf1β is a soluble nuclear activator with a poor transactivation activity; this has been confirmed by us and others14,39,51,52. An exception was made by additional observations that LCR-F1/Nrf1β was thought to be a significant dominant-negative inhibitor of ARE-driven gene transcription mediated by the full-length Nrf1 and/or Nrf2 factors61. The disparity suggests a possibility that LCR-F1/Nrf1β is unstable so that it is rapidly degraded to yield those lower molecular weight isoforms of between 46-kDa and 25-kDa (Fig. 2; see refs. 11,39,60). In fact, the 36-kDa Nrf1γ and the 25-kDa Nrf1δ (both arise from in-frame translation and selective proteolysis) were shown to be a bona fide dominant-negative form that lacks all the potential transactivation domains AD1, AD2, NST and/or SR regions11,39,51 (also see Fig. S5C). This is based on the fact that when they are over-expressed, these small isoforms can competitively interfere with the assembly and function of the active transcription factor complex (i.e. Nrf1 or Nrf2), thereby inhibiting induction of target genes. As expected, production of both the 55-kDa Nrf1β activator and the dominant-negative 36-kDa Nrf1γ form (Fig. S5D), which are localized in the nucleus (Fig. S5B), was blocked by the Nrf1M289–297L mutant (in which the internal translation start codons at Met289, Met292, Met294 and Met297 were mutated into leucine residues), resulting in an increase in the transcativation activity of Psv40nqo1-ARE-luc reporter gene by approximately 50% more than the activity mediated by the wild-type Nrf1 factor (Fig. S5C). In addition, generation of the dominant-negative 25-kDa Nrf1δ form was prevented by the Nrf1βM548L mutant (in which another potential in-frame translation start codon at Met548 of Nrf1 was mutated into leucine, Fig. S5D), which exhibited an increased activity when compared with the wild-type Nrf1β factor (Fig. S5C).
Additional two splice variants were originally designated as Nrf1 clones Δ767 and ΔD51,52, which contain a deletion of the functional translational start and stop codons, respectively. Removal of the first translation start codon results in the generation of a small form (called Nrf1ΔN in Fig. 1), in which the first N-terminal 181-aa region, covering the entire NTD and an one-third portion of the AD1, is substituted with a dodecapeptide MGWESRLTAASA (GenBank accession No. NM_001130453.1). For this reason, we postulate that Nrf1ΔN represents a constitutively activated form of Nrf1 (ref. 2). By contrast, the Nrf1 splice variant ΔD has a deletion of the translation stop codon, but gains a change in the second half of the leucine zipper motif (GenBank accession No. AF071084.1)52. Within this variant, the C-terminal 72 aa of the wild-type Nrf1, that are highly positively charged, are exchanged for an alternative 80-aa stretch that is enriched with negatively charged residues and contains a putative integral transmembrane (TM) segment situated at the C-terminal end. Moreover, the third Nrf1 variant (originally called clone Δ10) possesses a 137-aa deletion covering the SR and Neh6L regions51. For convenience, Nrf1 variant clones ΔD and Δ10 are renamed as Nrf1D or Nrf1ΔS, respectively (Fig. 1), but whether they have a capability similar to that for the wild-type Nrf1 to be anchored within the ER membrane is not identified.
Interestingly, the three long ectopic 120-, 95- and 85-kDa Nrf1 isoforms are forced to express at an approximate ratio of 1.0∶0.6∶0.3 in COS-1 cells that had been transfected with an expression plasmid for wild-type Nrf1 (Fig. 2a). The major ER-resident form of Nrf1 is the 120-kDa glycoprotein with a dynamic membrane behaviour, which is markedly distinct from the minor 95- and 85-kDa nonglycosylated and/or deglycosylated isoforms. Both of the 95- and 85-kDa Nrf1 are rapidly digested by proteinase K (PK) in membrane protection reactions39,62, indicating that they are positioned on the cyto/nucleoplasmic sides of membranes. In fact, the 95-kDa, 85-kDa and other short isoforms are recovered predominantly in the soluble nuclear and/or cytosolic subcellular fractions39. These observations led us to investigate the order of events whereby these three Nrf1 isoforms are translocated into the ER lumen and/or retrotranslocated across the membranes, and to consider how the process is dictated by glucose consumption and deprivation.
The stability of Nrf1 isoforms arising from its post-translational modifications in vectorial processes
To elucidate the order of events by which Nrf1 isoforms arise from distinct post-translational modifications (i.e. glycosylation, deglycosylation and/or proteolysis) in response to changes in the concentration of glucose, we have performed a series of experiments in the presence or deficiency of glucose in the media, to which added are inhibitors of N-linked glycosylation, along with proteasome inhibitors. This facilitates to assess what impact glucose levels had on the appearance of specific Nrf1 isoforms.
Firstly, to determine the stability of 120-, 95- and 85-kDa Nrf1 isoforms, cells expressing wild-type Nrf1 were grown in medium containing 25 mM glucose (control), and treated with cycloheximide (CHX, that inhibits protein biosynthesis) alone or together with proteasome inhibitors MG132. Western blotting showed that the abundance of all three longer Nrf1 proteins was significantly decreased following 2 h-treatment with CHX, whilst co-treatment of cells with MG132 prolonged the turnover of 120-kDa Nrf1 glycoprotein over 4 h, but it markedly increased the abundance of the 95-kDa and 85-kDa protein until 8 h after treatment (Fig. 2a, left). The stoichiometric graph (right) showed a tendency to reveal a marked difference in the turnover of the 120-, 95- and 85-kDa proteins that was determined with their half-lives estimated to be 151, 93 and 27 min, respectively, after treatment of cells with CHX. Upon addition of MG132 the stabilities of 120-, 95- and 85-kDa Nrf1 were substantially increased (left), giving estimated half-lives of 342, 475 and 885 min (right graph), respectively. The discrepancy between the upward trends in their stabilities suggest that the extra-luminal 85-kDa Nrf1 protein is rapidly degraded primarily through the 26S proteasome-mediated proteolysis pathway, whilst most of the 120-kDa glycoprotein is protected by the intact membranes against proteolytic degradation by 26S proteasomes, and a portion of the 95-kDa protein may also be partially protected by membranes during its topogenesis. In addition, treatment with the calpain inhibitor-I [CI, also called ALLN (N-Acetyl-Leucyl-Leucyl-Norleucine)] showed that non-proteasome-mediated pathways may also contribute to the proteolytic processing of 120-kDa and 95-kDa Nrf1 (ref. 39).
Secondly, to determine the effect that inhibition of Nrf1 glycosylation has on both its protein stability and its post-translational modification, cells expressing wild-type Nrf1 were allowed to be grown in medium containing either 25 mM glucose (control) or medium containing 5.5 mM glucose (restriction), and treated with TU (an inhibitor of oligosaccharyltransferases) to block N-linked glycosylation of newly-synthesized proteins, or plus MG132 or ALLN to block proteolysis. As shown in Fig. 2b, treatment with TU caused a time-dependent block in the appearance of the 120-kDa Nrf1 glycoprotein (left), which was reduced to 50% by 325 min estimated (right graph), and thereafter it gradually disappeared. Treatment of cells exposed to TU with MG132 (or ALLN in Fig. 2d) did not apparently increase the amount of the 120-kDa Nrf1 glycoprotein (Fig. 2b, left), though these chemicals modestly slowed its turnover so that its half-life time was prolonged from about 325 min to 502 min (right graph). After 2 h treatment of cells with TU, the disappearance of the 120-kDa Nrf1 glycoprotein was accompanied by an obvious increase in the abundance of the major 95-kDa Nrf1 protein (Fig. 2b, left), which is thus assumed to comprise both deglycosylated and non-glycosylated 95-kDa proteins. Thereafter, expression of the major 95-kDa Nrf1 (along with the minor 85-kDa Nrf1, left middle) proteins returned to low levels at 8 h after TU treatment, but could be augmented by treatment with MG132. These data demonstrate that the full-length Nrf1 protein is glycosylated to become the 120-kDa glycoprotein before it is deglycosylated to yield the 95-kDa isoform (i.e. this process is termed deglycosylation). Under certain circumstances, the 95-kDa isoform can also arise from the failure to glycosylate Nrf1 after biosynthesis (i.e. termed non-glycosylated 95-kDa Nrf1 protein), as was apparent during the prolonged exposure to TU, particularly in the presence of MG132 (Fig. 2b, left). These observations suggest that the rapid degradation of the deglycosylated/non-glycosylated 95-kDa Nrf1 protein occurs predominantly through 26S proteasome-mediated proteolysis pathway, whilst most of the luminal-resident 120-kDa glycoprotein may be subject to a non-proteasome-mediated degradation pathway (e.g. Calpain). Furthermore, a small fraction of the non-glycosylated 95-kDa Nrf1 protein, but neither of the 120-kDa glycoprotein or the cleaved 85-kDa isoform, was expressed in COS-1 cells that had been incubated with TU for 18 h (Fig. 2e, left 5 lanes). By contrast, co-treatment of cells with TU and ALLN caused a marked increase in the abundance of the non-glycosylated 95-kDa Nrf1 protein (Fig. 2e, right 5 lanes). Although the non-glycosylated 95-kDa Nrf1 protein was recovered in either ER or NE membranes, rather than the cytosolic fraction, purified from TU-treated cells (Fig. 2e), it was neither protected by membranes against PK digestion (Fig. 2d), nor did it exhibit potent transcriptional activity (see ref. 39). These results suggest that the non-glycosylated 95-kDa Nrf1 protein may be mislocated and/or misfolded before it is then degraded by proteasomes.
Thirdly, changes in the glycosylation status of Nrf1 were examined upon restriction of glucose (i.e. cells allowed to be grown in medium containing 5.5 mM glucose following transfer from medium containing 25 mM glucose). After synthesis of Nrf1 glycoprotein was blocked by addition of TU, glucose restriction resulted in a significant loss of the 120-kDa glycoprotein band (Fig. 2c, left), with an estimated half-life of 209 min in the absence of MG132, or 279 min in the presence of MG132, respectively (right graph). The expression of the 95- and 85-kDa Nrf1 proteins was maintained at low levels, but their abundances were markedly increased upon addition of MG132. These data suggest that production of the 120-kDa Nrf1 glycoprotein is dependent on the presence of glucose, whereas a portion of the 95- and/or 85-kDa proteins may be derived from the 120-kDa glycoprotein.
Vectorial processes by which Nrf1 isoforms arise from distinct post-translational modifications in the intracellular response to changes in glucose supply
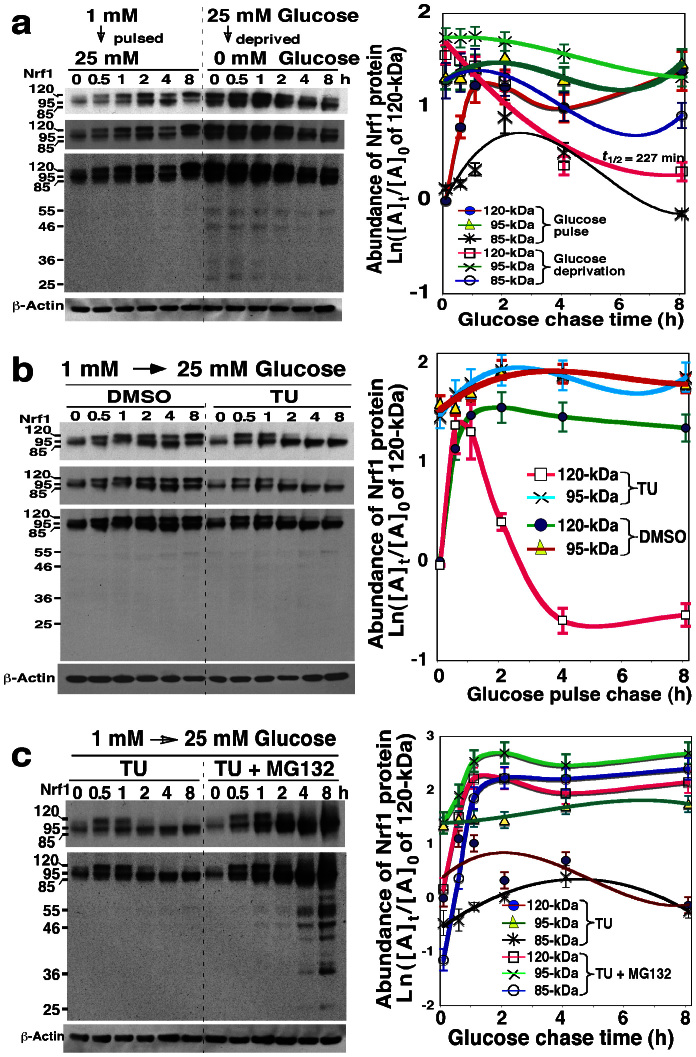
To elucidate the membrane-topogenic vectorial process of Nrf1 in which the protein is glycosylated and/or deglycosylated in response to changes in glucose supply, cells expressing wild-type Nrf1 were allowed to be starved for 16 h in medium containing 1 mM glucose, and then transferred to fresh control medium containing 25 mM glucose to which was added DMSO vehicle, TU alone, or TU plus MG132 for various periods of time. As shown in Figure 3, the result reveals a time-dependent process in which Nrf1 is glycosylated in the ER lumen and thereafter is deglycosylated in the extra-luminal compartments in response to acute glucose addition (i.e. cells expressing wild-type Nrf1 were allowed to recover overnight in medium containing 1 mM glucose before they were placed in medium containing 25 mM glucose). In this glucose addition experiment, the 95-kDa Nrf1 protein was the most abundant isoform expressed in glucose-starved cells that had been grown in medium containing 1 mM glucose. However, upon exposure to 25 mM glucose a gradual increase in the 120- and 95-kDa proteins was observed giving similar levels 2 to 4 h after exposure to 25 mM glucose as was seen in control cells, and thereafter, the 120-kDa glycoprotein became dominant (Fig. 3a, left 6 lanes). After 2 h treatment with TU, the 120-kDa Nrf1 band disappeared and then was replaced by the enhanced 95-kDa protein (Fig. 3b, right 6 lines). The rapid turnover of newly-synthesized 120-kDa Nrf1 after 60 min was not prevented by treatment with MG132 because the proteasome inhibitor did not obviously elevated amount of the nascent 120-kDa protein (Fig. 3c right 6 lines). These data are consistent with the hypothesis that Nrf1 is first synthesized as a 95-kDa non-glycosylated form, targeted to the ER, and then translocated into the lumen of this organelle, where it is glycosylated to become the 120-kDa isoform. By contrast, amount of the major 95-kDa Nrf1 proteins was significantly augmented by MG132, as accompanied by a certain increase in expression of the 85-kDa Nrf1 and other short isoforms (Fig. 3c, right 6 lines). This result suggests that when Nrf1 failed to be glycosylated, its 95-kDa non-glycosylated protein is rapidly processed by proteasome-mediated proteolysis to generate multiple polypeptides of between 85-kDa and 25-kDa.
Figure 3. Post-translational processing of Nrf1 into longer isoforms in response to changes in glucose concentration.
(a) Glycosylation and deglycosylation of Nrf1 in response to glucose supply or glucose starvation. Following transfection with the wild-type Nrf1 expression construct, a group of COS-1 cells (left 6 lanes) was allowed to starve in 1 mM-glucose medium for 16 h, before they were transferred to fresh 25 mM-glucose medium (i.e. glucose supply). Another group of the cells (right 6 lanes) was left to recover for 16 h in 25 mM-glucose medium, before transfer to glucose-free medium (i.e. glucose deprivation). Thereafter, these two groups of cells were disrupted at the indicated times after glucose addition or glucose deprivation, and then examined by western blotting. The left upper panels show two cropped images from the film that was exposed to the immunoblot for different shorter times to eliminate intense bands. The right graph shows the relative abundance of the 120-, 95- and 85-kDa proteins (mean ± S.D, n = 4) that were calculated after normalization to [A]0 of 120-kDa at t0 after transfer 25 mM-glucose medium to the starved cells that had been grown in glucose-free medium. (b,c) The vectorial process by which Nrf1 is post-translationally modified was determined by addition to the 1 mM-glucose starved cells of 25 mM-glucose in the presence of DMSO vehicle, TU alone, or TU plus MG132. After transfection, Nrf1-expressing cells were starved in 1 mM- glucose medium for 16 h before transfer to 25 mM-glucose media containing DMSO, TU (1 μg/ml), or plus MG132 (5 μM) for the indicated times. The relative abundance of the 120-, 95- and 85-kDa Nrf1 proteins was determined by western blotting and calculated as described above (right, mean ± S.D, n = 4).
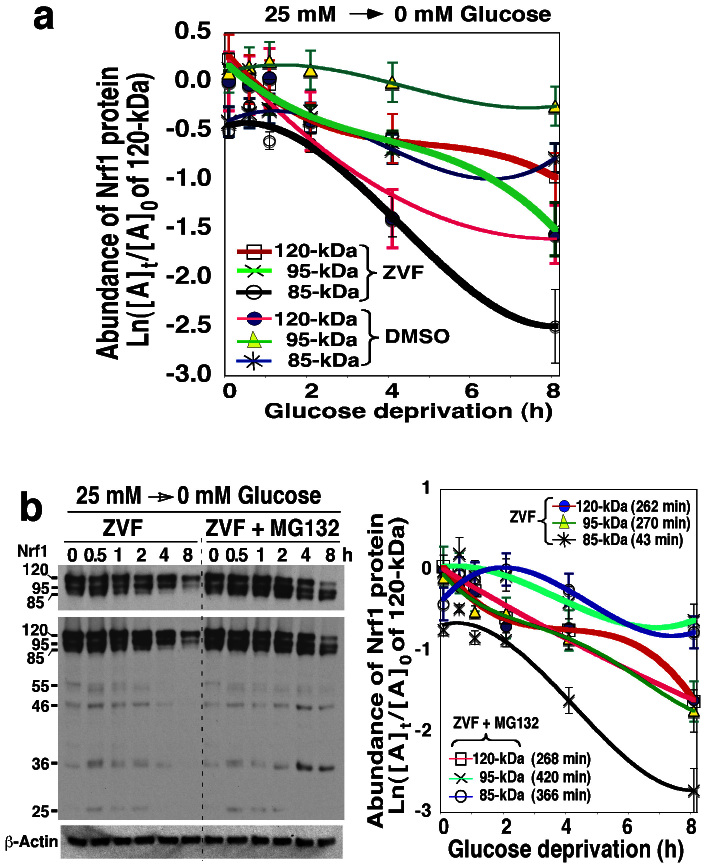
Next, to elucidate the topogenic vectorial processes whereby Nrf1 is glycosylated and/or deglycosylated in response to glucose-free conditions, cells expressing wild-type Nrf1 were allowed to be grown for 16 h in medium containing 25 mM glucose and then starved for the indicated times in glucose-free medium that contained DMSO vehicle, Z-VAD-fmk (which is abbreviated to ZVF, an inhibitor of N-linked deglycosylation) alone or plus MG132 (to block the proteasomal degradation). As shown in Figure 3a (right 6 lanes), in cells that had been grown in medium containing 25 mm glucose, complete glucose deprivation triggered a time-dependent loss of the 120-kDa Nrf1 glycoprotein. Its disappearance was also accompanied by a modest decrease in the abundance of 95- and/or 85-kDa proteins by 4 h after glucose deprivation, and their decreased levels remained until 8 h after glucose deprivation (c.f., Fig. 3a, right 6 lanes with Fig. 4b, left 6 lanes). Inhibition of N-linked deglycosylation by Z-VAD-fmk had little impact on the turnover of the 120-kDa Nrf1 glycoprotein (Fig. 4b), changing its half-life from 227 min to 262 min (Fig. 4a), or 268 min in the presence of MG132) (Fig. 4b, right 6 lanes). The data suggest that the 120-kDa Nrf1 is protected by membranes against proteasome-mediated degradation and this glycoprotein turnover is thus assumed to occur through a proteasome-independent mechanism, particularly before it is allowed to be deglycosylated. Interestingly, the inhibition of glucose deprivation-stimulated deglycosylation of 120-kDa Nrf1 by Z-VAD-fmk also caused a marked decrease in the levels of 95- and 85-kDa proteins (Fig. 4b, left 6 lanes) to 50% of their original levels by 270 min and 43 min estimated respectively (Fig. 4a), suggesting that deglycosylation of 120-kDa Nrf1 glycoprotein may proceed with generation of the 95- and 85-kDa isoforms. Inhibition of the proteasome by MG132 rendered a time-dependent increase in expression of the 95- and 85-kDa Nrf1 proteins (Fig. 4b, right 6 lanes), so that their half-lives were prolonged to 420 min and 365 min, respectively (right graph). Collectively, these results suggest that a major fraction of the 95-kDa isoform arises from deglycosylation of the 120-kDa Nrf1 glycoprotein after it has been transferred from the ER lumen to extra-luminal cyto/nucleoplasmic compartments, whereupon the 95-kDa Nrf1 protein is proteolytically processed by 26S proteasomes to yield the 85-kDa isoform. Thereafter, the 95-kDa and/or 85-kDa Nrf1 proteins are subject to further proteolytic degradation to yield several short isoforms of between 55 kDa and 25 kDa, but detailed mechanisms require further study.
Figure 4. Post-translational processing of Nrf1 into longer isoforms in response to glucose deprivation.
The vectorial process by which Nrf1 is post-translationally modified was determined by the treatment of cells that is subject to glucose deprivation along with Z-VAD-fmk (which blocks N-linked deglycosylation of glycoprotein). Following transfection, Nrf1-expressing cells were allowed to recover from transfection in the fresh 25 mM-glucose medium for 16 h, before transfer to fresh glucose-free medium containing DMSO vehicle, Z-VAD-fmk (ZVF, 20 μM) alone, or ZVF plus MG132 (5 μM) for the indicated times. The abundance of the 120-, 95- and 85-kDa Nrf1 proteins was determined by western blotting (b, left), as described above in Fig. 3a (right 6 lanes). The relative intensity of immunoblots for these three longer isoforms was calculated (mean ± S.D, n = 4), and is shown graphically (a and b right), with distinct trends in their half-lives (t1/2) estimated.
The changing topology of Nrf1 within and around membranes is determined by its TM1 in cooperation with other semihydrophobic amphipathic regions
Our previous work has shown that the NHB1-associated TM1 sequence (aa 7–26) anchors Nrf1 within ER membranes and is itself orientated in an Ncyt/Clum fashion within the membrane2,38,39,40. To gain an insight into the membrane-topological folding of Nrf1 within the ER, we created several double fluorescence chimaeric proteins, in which Nrf1 and its truncation mutants are either sandwiched in between DsRed and GFP or fused C-terminally with GFP alone (Fig. 5a). These sandwiched fusion proteins were subjected to classic proteinase protection assay; this procedure, called double fluorescence protease protection (dFPP), was established on the base of the fluorescence protease protection (FPP) assay to identify the membrane-topology of GFP chimeras63,64. As shown in Figure 5b, membrane protease protection reactions of intact ER-enriched fractions from cells expressing the expression construct for DsRed/N65/GFP (in which N65 represents the N-terminal 65 aa of Nrf1, containing the TM1-embedded NHB1 sequence), followed by immunoblotting with antibodies against either DsRed or GFP, revealed that the full-length DsRed/N65/GFP sandwiched protein was integrated into the membranes (right cartoon). The DsRed epitope was almost completely degraded by PK (Fig. 5b1). The resulting loss of DsRed from DsRed/N65/GFP gave rise to a major PK-digested fragment of approximately 55-kDa with the GFP epitope facing the lumen; but this fragment appeared to be evidently protected by membranes against further digestion by PK, even in the presence of 1% Triton X-100 (TX) (Fig. 5b2). The data demonstrate that the TM1 region of N65 is orientated in an Ncyt/Clum fashion spanning ER membranes, while the water-soluble DsRed and GFP portions are positioned on the cytoplasmic and luminal sides of membranes, respectively (Fig. 5b, right cartoon). Further membrane PK protection assays of the N65/GFP fusion protein revealed that its membrane-topology appeared similar to that of the sandwiched DsRed/N65/GFP protein (Fig. 5c, right cartoon), indicating that the Ncyt/Clum orientation of TM1 in the N65 peptide appears to be unaffected by attachment of the DsRed epitope immediately to the front of the TM1-associated NHB1 sequence. It is important to note that, although the local membrane-topology of the NHB1 sequence in Nrf1 appears to resemble those of ATF6 and SREBP-1 (Fig. S2), it has neither Site-1 nor Site-2 protease cleavage sites (it is notable that both proteases are exclusively located in the Golgi apparatus rather than the ER65,66,67). It is therefore assumed that the NHB1 sequence of Nrf1 is not processed in the ER38 through a mechanism accounting for the successively proteolytic processing of ATF6 and SREBP-1 (refs. 65,66,67).
Figure 5. The membrane-topological folding of Nrf1 requires TM1 to cooperate with other semihydrophobic regions.
(a) Structural diagrams of five chimeras indicated. Nrf1ΔC lacks TMc-adjoining 56 amino acids of Nrf1, whereas, its N65 contains the TM1-associated NHB1 sequence. The locations of TM1 and the other three possible membrane-associated regions TMi, TMp and TMc are indicated. It is to note that attachment of DsRed immediately to the N-terminal end of NHB1 does not alter the orientation of TM1 spanning membranes. (b,c) COS-1 cells expressing DsRed/N65/GFP (b) and N65/GFP (c) were subjected to subcellular fractionation in order to purify the intact ER fraction. Subsequently, equal amounts (50 μg of protein in 40 μl of 1× isotonic buffer) of ER were examined by membrane proteinase protection reactions that were carried out by incubating (on ice) each of the subcellular fractions with 50 μg/ml (+) or 100 μg/ml (++) of proteinase K (PK) in the presence (+) or absence (−) of 1% Triton X-100 (TX) for 30 min (b) or for the indicated times (c). The reaction products were resolved in a 4-12% LDS/NuPAGE Tris-Bis gel and identified by western blotting with antibodies against DsRed (b, upper), GFP (b, middle; c, upper) and CRT. The results reveal that the TM1-associated NHB1 sequence enables N65 to anchor in an Ncyt/Clum oriention spanning membranes (left two cartoons), with DsRed and/or GFP portions being positioned in the cytoplasmic and luminal sides, respectively. (d,e) Following classic membrane proteinase protection assays of ER-enriched fractions expressing DsRed/Nrf1/GFP (d), DsRed/Nrf1ΔC/GFP (d) or Nrf1/GFP (e) were carried out as described above. The reaction products were visualized by western blotting with antibodies against DsRed (d, upper), GFP (d, middle; e, upper) and CRT. Quantitative analysis of Nrf1/GFP and its fragments (Fig. S5A) revealed that ∼78% of Nrf1/GFP protein was digested by PK for 15 min, whereas the remaining 22% of this protein was further digested to yield two polypeptides of 52-kDa and 32-kDa with their C-terminus being fused with GFP. To explain the results obtained from PK digestion of DsRed/Nrf1/GFP and DsRed/Nrf1ΔC/GFP, two different models were proposed in Fig. S4.
To learn more about the overall membrane-topological folding of Nrf1 with the ER, the full-length Nrf1 protein was sandwiched in between DsRed and GFP to yield DsRed/Nrf1/GFP. The membrane PK protection assays showed that the N-terminal DsRed portion is retained on the cytoplasmic side of membranes where it is not protected so that it was almost completely digested within 30 min by PK (Fig. 5d1). By contrast, a luminal 32-kDa peptide, with its C-terminal end being tagged by GFP, was protected by ER membranes from digestion of DsRed/Nrf1/GFP by PK (50 μg/ml), even in the presence of TX (Fig. 5d3, left). Further processing of the 32-kDa GFP-containing peptide by 100 μg/ml of PK gave rise to two smaller fragments of 28-kDa (free GFP) and 31-kDa (containing most of TMc fused with GFP). Interestingly, none of these three GFP-containing peptides of between 28 kDa and 32 kDa with its C-terminus facing the lumen were observed in the membrane PK protection assay of DsRed/Nrf1ΔC/GFP [in which the Nrf1ΔC mutant lacks the C-terminal 56 amino acids of Nrf1 including a net negatively charged amphipathic TMc peptide (Fig. S1), that contains a potential membrane-spanning helix-helix interaction motif AxxxAGxxxS, of the type described previously60] (Fig. 5d3, right). These observations had led us to postulate that during topogenesis of Nrf1, if its TMc peptide is integrated through association with membranes in an Ncyt/Clum orientation similar to that of TM1, a flexible hinge region located between the TM1 and TMc segments should be allowed to orientate the intervening polypeptide in an Nlum/Ccyt fashion (i.e. its N- and C-termini facing the luminal and cytoplasmic sides of membranes, respectively). Such a sequence (e.g. a proline-kinked amphipathic semihydrophobic TMp hinge region within the Neh6L domain) was deduced from bioinformatic analysis (Fig. S1), and those data obtained from glycosylation mapping, and mutagenesis mapping linked to membrane proteinase protection assays (unpublished).
Protease digestion of DsRed/Nrf1/GFP and DsRed/Nrf1ΔC/GFP also generated a GFP-fused polypeptide of either 52-kDa (which includes a large C-terminal portion of Nrf1 situated between the putative TMp and TMc peptides, Fig. 5d2, left) or 49-kDa (which contains the C-terminal portion of Nrf1 but lacks the TMc peptide, Fig. 5d2, right). Intriguingly, the 52-kDa fragment was more sensitive to PK and further processed to yield a TX-resistant 32-kDa TMc-GFP facing the lumen. Similarly, two polypeptides of 50-kDa and 32-kDa were also observed in PK digests of the Nrf1/GFP chimeras (in which the wild-type Nrf1 is fused C-terminally with GFP, Fig. 5e1). By contrast, the 49-kDa fragment lacking the TMc segment appeared more resistant to digestion by PK, even in the presence of TX (Fig. 5d2, right). Moreover, our previous work showed that proteinase digestion of endogenous 120-kDa Nrf1 for 30 min using ER membranes yielded peptides of 55, 46, 36 and 32 kDa39, though most of these disappeared upon incubation with PK for 60 min2,40. Taken together, these results suggest a possibility that a substantial C-terminal portion of Nrf1 (with an estimated mass of 52 ∼ 55 kDa) situated between the TMp and TMc peptides is initially translocated into the ER lumen, and subsequently some basic-rich regions adjoining TMp may be partially repartitioned out of membrane into the cyto/nucloplasmic side where they are proteolytically processed into several short peptides (for the proposed models, see Fig. S3). By contrast, the TMc-adjoining region in Nrf1 appears to be integrated in a potent Ncyt/Clum orientation spanning the TX-resistant membranes with its C-terminus facing the ER lumen (Fig. S3), but it is dynamically moved out of membranes into the cytoplasmic side, so that it within the major fraction of Nrf1/GFP is digested to disappear in the membrane protection reactions (Figs. 5e1 and S5A).
The membrane-topogenic vectorial behaviour of Nrf1 controls the ability of its TADs to transactivate Gal4-UAS reporter gene expression
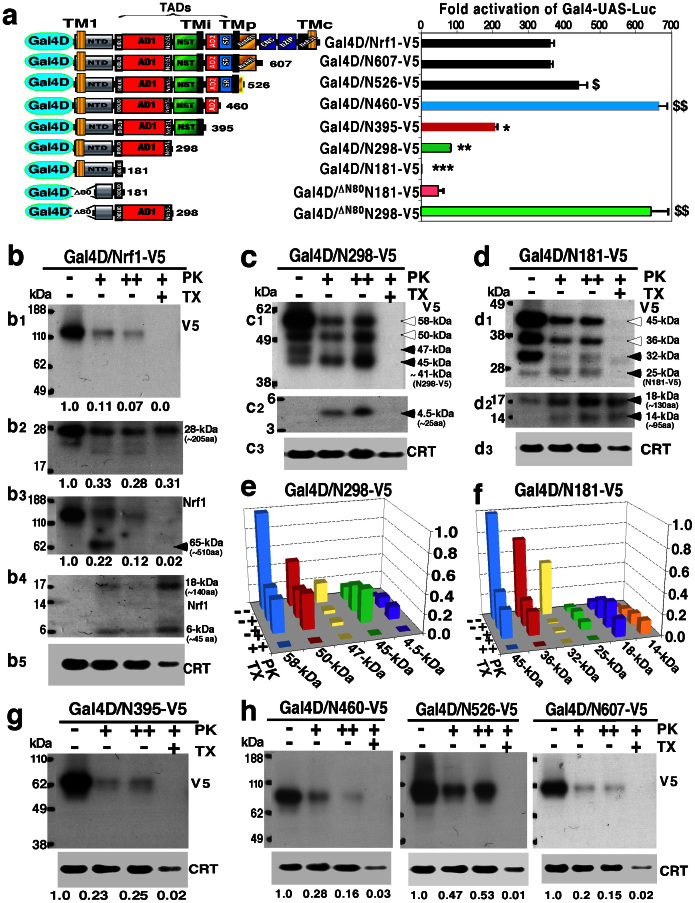
The above data appear consistent with the notion that some regions adjacent to the TM1, TMp and/or TMc peptides may control the topological folding of Nrf1 and its partial repartitioning out of membranes to regulate target gene expression. To test this possibility, we combined Gal4-based reporter assays linked with protease protection assays (called GRAPPA) to examine whether membrane-related vectorial behaviour of Nrf1 has an effect on its activity to transactivate target gene expression. In these experiments, the Gal4 DNA-binding domain (Gal4D) was attached to the N-terminus of either wild-type Nrf1 or those C-terminally truncated Nrf1 mutants (Fig. 6a). According to the positive-inside rule (i.e. net positively-charged residues adjacent to transmembrane regions are positioned on the cyto/nucleoplasmic side of membranes)68,69,70, the basic Gal4D portion of 16.8 kDa (which is much smaller than the more basic N-terminal 50-kDa portion of ATF6 or SREBP-1, Fig. S2B) should not alter the Ncyt/Clum orientation of TM1 within Nrf1, so that it may be positioned on the cyto/nucleoplasmic side of membranes (Fig. S4A). The resulting ability of various Gal4D/Nrf1 fusion proteins to transactivate a Gal4D-UAS reporter gene was used as a measure of the membrane-topogenic vectorial behaviours of Nrf1 that may dictate the positioning of its TADs and/or subsequent repartitioning out of membranes to mediate target gene expression (Fig. S4B). In this assay, we found that Gal4D/Nrf1-V5 produced a 360-fold increase in reporter gene activity (Fig. 6a, right), even though it was transiently located in the ER36. Membrane proteinase protection assays on the intact ER fractions showed that the full-length Gal4D/Nrf1-V5 and its 28-kDa fragment (which comprises about 205 aa possibly located between residues 535 and 741) were sensitive to PK (Fig. 6, b1 & b2), with an exception that a residual amount of the 28-kDa fragment with the TMc peptide facing the lumen was partially protected by membranes against PK digestion even in the presence of 1% TX (Fig. 6b2). This suggests that the V5-tagged C-terminus of Nrf1 might be dynamically partitioned on either the cytoplasmic or luminal sides of membranes to extents varying with different folding fractions (Fig. S4A). This assumption is supported by the evidence obtained from the Nrf1/GFP and DsRed/Nrf1/GFP chimaeric proteins (Fig. 5, d & e). In addition, modest digestion of Gal4D/Nrf1-V5 by 50 μg/ml of PK gave rise to three non-tagged polypeptides of 65, 18 and 6 kDa (which comprise approximately 510, 140 and 45 aa, respectively) (Fig. 6, b3 & b4). They were visualized by cross-reacting with antibodies against Nrf1β (this antigen comprises residues 291–741 of Nrf1), but largely disappeared following enhanced digestion by 100 μg/ml PK. These suggest that all three polypeptides are not sufficiently protected by membranes from protease attack, as they may be dynamically repositioned into the cyto/nucleoplamic side out of membranes.
Figure 6. Membrane-topogenic vectorial behaviour of Nrf1 controls its ability to transactivate Gal4-UAS reporter gene.
(a) Schematic structure of Gal4D fusion proteins with Nrf1 or its truncated mutants used in reporter assays. The locations of TM1, TMi, TMp and TMc are indicated. The AD1, NST, AD2 and/or SR regions in Nrf1 serve as TADs. The transcriptional activity of Gal4D/Nrf1 chimeric factors is shown (right). Luciferase activity was measured in lysates of COS-1 cells expressing each Gal4D/Nrf1 factor, PTKUAS/Gal4-Luc and β-gal plasmid. The data were calculated as a fold change (mean ± S.D, n = 9) over the Gal4D control after normalization to the β-gal value. Statistical significance was determined by the Student t-test or MANOVA. When compared with those obtained from Gal4D/Nrf1-V5, significant increases are shown ($, p < 0.05; $$, p < 0.001; n = 9), as are significant decreases (*, p < 0.05; **, p < 0.001; n = 9). (b) Cells expressing Gal4D/Nrf1-V5 were subjected to subcellular fractionation and membrane proteinase protection reactions for 30 min with either 50 μg/ml (+) or 100 μg/ml (++) of proteinase K (PK) in the presence (+) or absence (−) of 1% (v/v) Triton X-100 (TX). The reaction products were examined by immunoblotting with antibodies against the V5 tag (b1, b2) or Nrf1β (b3, b4). Calreticulin (CRT) served as an ER luminal-resident protein marker. The intensity of cross-reacting bands in some blots was calculated by the ImageJ programme. (c to f) The intact ER membrane PK protection assays of Gal4D/N298-V5 (c, e) and Gal4D/N181-V5(d, f) from 30 min digestion by PK. The electrophoretic mobilities of these two full-length protein and their PK-digested polypeptides were analyzed by 4–12% LDS/NuPAGE gels. Subsequently, the intensity of immunoblots for multiple V5-tagged polypeptides was calculated after normalization to the corresponding value of CRT in each well. Their mean values obtained from three independent experiments are shown graphically (e and f, n = 3). (g,h) The intact ER membrane proteinase protection reactions of Gal4D/N395-V5, Gal4D/N460-V5, Gal4D/N526-V5 or Gal4D/N607-V5 over 30 min were performed as above. The intensity of immunoblots with the V5 tag was calculated after normalization with the value for CRT.
We next used the Gal4-based assay to determine whether some of the N-terminal regions (e.g. TM1) of Nrf1 are integrated within membranes. The results revealed that residues 1–181 of Nrf1 were embedded in the Gal4D/N181-V5 chimaeric factor that lacked transactivation activity, suggesting that its NTD (aa 1–124) completely repressed the transactivation potential of the N-terminal one-third portion (aa 125–181) of AD1 (Fig. 6a). Similarly, although the N298 polypeptide contains the entire AD1 region (aa 125–298), the Gal4D/N298-V5 chimaeric factor exhibited only a quarter of transactivation activity exhibited by wild-type Gal4D/Nrf1-V5 (Fig. 6a, right). However, deletion of the N-terminal 80 residues (including the TM1-associated NHB1 sequence) from either N181 or N298 (to yield Gal4D/ΔN80N181-V5 and Gal4D/ΔN80N298-V5, respectively), allowed their transactivation activities to be rescued from inhibition by the NTD of Nrf1. This appears to be consistent with those results obtained from mutagenesis mapping of the NTD38 and membrane protease protection assays of both DsRed/N65/GFP and N65/GFP fusion proteins (Fig. 5, b & c). Together, these results revealed that the TM1-associated sequence (in particular aa 15–24 of Nrf1, see ref. 38) is essential for integration within membranes.
Further proteinase protection assays were used to examine whether the remaining region of NTD that connects with AD1 is either partitioned into the lumen or repartitioned out of membranes. Exposure of intact ER-enriched membrane fractions expressing Gal4D/N298-V5 or Gal4D/N181-V5 to PK for 30 min yielded a fraction of similar V5-tagged polypeptide ladders in the ER lumen (Fig. 6, c to f). Digestion of Gal4D/N298-V5 by PK yielded a 4.5-kDa peptide that was estimated to contain approximately 25 aa derived from Nrf1, suggesting that the Neh5L region resides transiently in the lumen (Fig. 6c2). By contrast, PK digestion of Gal4D/N181-V5 yielded peptides of 14-kDa and 18-kDa that were estimated to contain roughly 95 and 130 aa derived from Nrf1, respectively (Fig. 6d2), but similar PK-digested peptides were not observed in the cases of DsRed/N65/GFP and N65/GFP (Fig. 5, b & c). Together with other observations by Steffen et al22 and Tsuchiya et al48, these results suggest that an N-terminal region possibly located between residues 55 and 85 may be dynamically repositioned on the cytoplasmic side of ER, where it enables Nrf1 to be targeted for a proteolytic degradation pathway. As shown in Figure 6 (c to f), a residual amount of other longer polypeptides of between 25-kDa and 58-kDa, which are predicted to contain the entire N181 or N298 fused with various lengths of Gal4D (only the full-length Gal4D can elicit its DNA-binding activity but short degraded peptides lack the activity) were observed in 30-min PK digests, suggesting that during initial topogenesis, approximately a quarter of the full-length Gal4D/N181-V5 or Gal4D/N298-V5 is not tightly anchored by TM1 within membranes and thus may be released into the lumen, whilst other three-fourths of the fusion proteins containing V5-tagged AD1 may have been partially repartitioned out of membranes into the cyto/nucleoplasmic side, where they are proteolytically digested to yield multiple peptide species. The residual N181- and/or N298-containing fragments were also further digested following incubation with PK for 60 min (Fig. S4C), suggesting that the luminal-resident AD1 is capable of being repartitioned across membranes into the cyto/nucleoplasmic side, enabling transactivation of UAS-Gal4 reporter gene (Fig. 6a). The vectorial behaviour of AD1 was also examined by both engineered glycosylation mapping of this domain and membrane protease protection assays of DsRed/N275/GFP (data not shown herein).
During de novo synthesis, the orientation of TM1 within the NTD of Nrf1 causes subsequently translated portions to be directed to the lumen of the ER. We therefore sought to establish whether the TMi- and/or TMp-containing regions (Fig. S1) in Nrf1 alter the topological distribution of this protein by comparing Gal4D/N181-V5 and Gal4D/N298-V5 with Gal4D/N395-V5 (which contains the TMi peptide on the C-terminal side of the NST glycodomain), Gal4D/N460-V5 (which contains TMi and AD2), Gal4D/N526-V5 (which contains TMi, AD2, and the TMp peptide on the C-terminal border of the SR/PEST2 sequence), or Gal4D/N607-V5 (which contains TMi, AD2, TMp, SR/PEST2 and Neh6L). As shown in Figure 6 (g & h), the results revealed that the N395-, N460-, N526- and N607-containing fusion proteins were substantially more sensitive to PK in the membrane proteinase protection assays than the N181- and N298-containing proteins. When compared with Gal4D/N395-V5 and Gal4D/N460-V5, the Gal4D/N526-V5 fusion protein was relatively resistant to digestion, whereas Gal4D/N607-V5 was more or at least as sensitive to digestion than others tested. These data suggest that the membrane-topological folding of Nrf1 needs TM1 to cooperate with other regions such as TMi and TMp, and in turn the positioning of these regions is monitored by their flanking domains, though the detailed molecular basis requires for further studies. Further comparison of their abilities to transactivate the Gal4-UAS reporter gene (Figs. 6a, and S4B) revealed that the TMi-adjoining AD2 (and SR) positively regulates Nrf1, whilst the TMp-adjoining Neh6L domain negatively regulates this factor. By comparison with the wild-type Nrf1 glycoprotein of 120-kDa, relatively small amounts of the various NST-containing Gal4D fusion proteins were glycosylated (Fig. S4D), suggesting that the NST glycodomain in the basic Gal4 fusion protein may be repartitioned out of membranes where it is deglycosylated.
Discussion
For reasons that are largely historical or involve difficulty of experimentation, the vast majority of researchers in the field have been almost entirely fixated on the water-soluble Nrf2 (to the tune of hundreds of publications per year) so far, whereas the membrane-bound Nrf1 appears to be comparatively ignored1,2, even though the latter is actually essential for regulating critical homeostatic and developmental genes. Now, Nrf1 seems to be coming more into its own lately, particularly with the demonstration that it is a critical regulator of 26S proteasomal subunit expression under conditions of proteasomal challenge (i.e. with chemotherapeutic drugs)22,23,24,25. Our past work showed that much of the cellular Nrf1 (but not Nrf2), with several domains being more conserved with its ancestral proteins CNC and Skn-1, is located in the ER but neither in the Golgi nor mitochondria36,38,39, making it an intriguing question how Nrf1 might be regulated and what its main functions are. Our present study reveals a hitherto unknown intrinsic mechanism by which the membrane-topogenic behaviour of Nrf1 controls both its post-translational modification and functional activity. This addresses a significant topic on how Nrf1 is allowed to move in and out of ER membranes, and/or release away from the ER in a regulated fashion into the nucleus, enabling transactivation of target genes; thus it is a membrane-spanning protein that is dynamically moving with changing topology. The order of membrane-topogenic events is defined, whereby the newly-translated Nrf1 is translocated into the ER lumen where it is glycosylated through its NST domain, and thereafter the glycoprotein is repartitioned and retrotranslocated across membranes into the cyto/nucloplamic compartments where it is deglycosylated, enabling transactivation of Nrf1-target genes before this deglycoprotein is proteolytically degraded by the proteasome. Herein, we have also provided evidence that normal glucose supply and glucose deprivation alter post-translational modification of Nrf1 (i.e. N-linked glycosylation and deglycosylation, as well as possibly proteolysis mediated by proteasome and/or calpain).
Importantly, the distinct biological functions of the membrane-bound Nrf1 and the water-soluble Nrf2 are determined by differences in their primary structures. Since Nrf1 is essential for survival, it must exclusively regulate certain genes that are critical for cellular homeostasis. As such, accumulating evidence indicates that Nrf1 is a dynamic membrane-spanning protein that exists with multiple topological isoforms (called topoforms), each with a distinct function. According to current knowledge of the folding of proteins within membranes68,69,70, the membrane-topology of Nrf1 is likely to be determined by hydrophobic, semihydrophobic, amphipathic, and other topogenic signals (collectively designated topogon) within the protein. The topogon is co-translationally decoded by molecular machines (e.g. translocon) into an initial topological structure partitioned within and around the specific membrane lipid microdomain. During subsequent folding of Nrf1, potential intramolecular interactions between its own helical regions and intermolecular interactions of its helical regions with those in other membrane proteins (e.g. retrotranslocon or flippase) and/or with membrane lipids dictate the formation of the final topological structure and position of the NHB1-CNC factor within and around membranes.
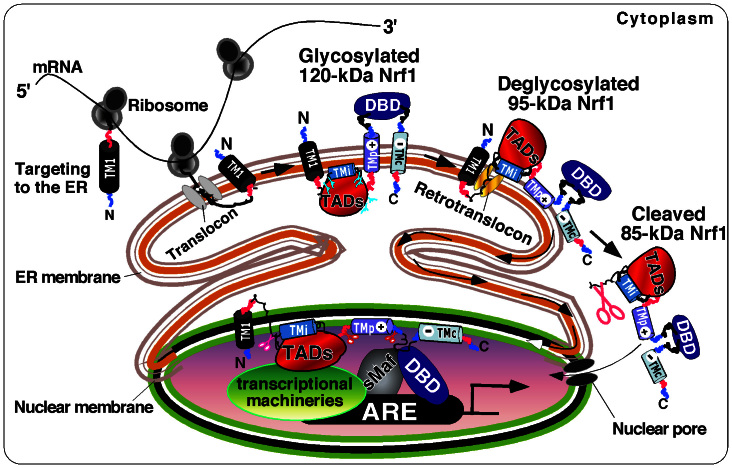
To be explicit, we propose a model for the membrane-topogenic vectorial process of Nrf1 (Figs. 7 and S4A), in which Nrf1 can adopt distinct dynamic membrane-topologies that are determined by its N-terminal TM1 sequence (aa 7–26), which itself exists in an Ncyt/Clum orientation within membranes. Our previous mutagenesis mapping data38 revealed that, within the TM1-associated NHB1 sequence, the core hydrophobic residues 15–24 are essential for anchoring Nrf1 within ER membranes so that it adopts an Ncyt/Clum orientation, whereas its C-terminal flanking aa 38–44 contain a potential REIIL motif, but do not serve as a cleavage site by Site-1 protease. Otherwise, loss of the TM1 peptide resulted in production of an unstable peripheral membrane-associated or free nuclear mutant proteins38,39,40. Nevertheless, the TM1 peptide is still unlikely to fold tightly within ER membranes because it is not highly hydrophobic (Fig. S1), and this characteristic will allow a small fraction of Nrf1 to be released into the extra-membranous cytoplamic compartments (in which it is digested in membrane proteases protection reactions, Fig. 6) or luminal compartments (in which it is protected by membranes against protease digestion). In reality, the luminal portion of Nrf1 may also be refolded and thus become reintegrated into the ER membrane, in which it would seem to be susceptible to a protease attack.
Figure 7. A proposed model to explain the membrane-topogenic behaviour of Nrf1 in vectorial processes.
After ER targeting, Nrf1 is anchored in the membrane through TM1. AD1 and NST-adjoining domains are transiently translocated into the ER lumen, where Nrf1 is glycosylated through the NST domain to become a 120-kDa glycoprotein. During initial topogenesis of Nrf1, the TMi glycopeptide and its adjacent AD2 N-terminal amphipathic portion are probably tethered to the luminal leaflet of the membrane lipid bilayer, whilst the TMp peptide may be dynamically associated with membranes. Once TMp is orientated in an Nlum/Ccyt fashion within membranes, its flanking PEST2 and Neh6L regions are partitioned into the luminal and cyto/nucleoplasmic sides, respectively. During this stage, the basic CNC-bZIP domain is retained in the cyto/nucloplasmic sides of membranes, and its connecting TMc is likely to be either left in the cytoplasm or integrated into membranes with its C-terminus facing the lumen. If TMp is not integrated within membranes, its C-terminally flanking regions might also release into the lumen, until they are repartitioned out of membranes. During topogenesis of Nrf1, once TMi is liberated from the restraint of its flanking glycopeptide, it may be reintegrated with membranes. This process would enable the repartitioning of AD2 and SR out of membranes, allowing them to act as a bona fide TAD. When appropriate, the luminal AD1 and NST glycodomain are partially repartitioned across membranes into the cyto/nucleoplasm, enabling Nrf1 to be deglycosylated to produce a 95-kDa activator. Although the NHB1-associated sequence is not processed via regulated intramembrane proteolysis accounting for ATF6 and SREBP-1 (ref. 38 and herein), other regions within NTD and/or AD1 might be allowed for proteolytic processing to move the NTD possibly through the Hrd1- and VCP/p97-dependent ERAD22,23,48 and/or PEST1-signalling pathways57, in order to release an 85-kDa cleaved isoform of Nrf1 from the ER/nuclear membranes into the nucleus to transactivate ARE-driven gene expression. Lastly, several potential degrons can trigger proteolytic degradation of Nrf1 to yield 55-kDa Nrf1β/LCR-F1 (a weak activator), and the dominant-negative 36-kDa Nrf1γ and 25-kDa Nrf1δ isoforms. In addition, they can also arise from internal in-frame translation.
During biosynthesis of Nrf1, it is anchored by TM1 within the ER membrane and the remaining part of the factor is continuously translocated into the lumen, where its NST domain is glycosylated to yield a polypeptide of 120 kDa. Within the NST domain of Nrf1, the glycosylated asparagines that flank the TMi peptide allow the protein to become tethered to the luminal leaflet of the membrane lipid bilayer, though the TMi sequence may fold into a glycine-kinked amphipathic semihydrophobic helix structure spanning membranes. This assumption is consistent with bioinformatics analysis (Fig. S1B) and experimental evidence provided herein. When the TMi sequence is liberated from confinement caused by glycosylation, it is likely to be reintegrated within membranes and therefore its topological orientation seems to be determined by its flanking deglycosylated regions. This vectorial process, though it has been not sufficiently explored, may allow repartitioning of the AD1 and NST domains such that they can adopt a transient membrane-spanning topology. It is also possible that the liberated TMi sequence promotes repartitioning of the AD2 and SR domains out of membranes, particularly in response to glucose deprivation. This notion is also supported by data showing that mutation of TMi-adjoining asparagines to prevent glycosylation increases the activity of Nrf1. It is therefore postulated that the TMi peptide could act as a flexible hinge switch to control the repartitioning of AD1 and AD2 out of membranes, enabling them to engage with the transcriptional machinery and induce Nrf1-target gene expression. In turn, topological regulation of gene transactivation may also be differentially modulated by TMi-flanking AD1 and AD2. This is supported by the findings that loss of both TMi-containing NST domain and AD2 (in Gal4D/N298-V5 retaining AD1) or AD2 (in Gal4D/N395-V5 retaining AD1 and NST) caused the reporter gene activity to be reduced to approximately 25% or 55% of that of the full-length Gal4D/Nrf1-V5 factor, respectively (Fig. 6).
The membrane lipid electrochemical potential of the ER, that gives a relatively positive charge on the outside (i.e. the luminal side of membranes), promotes translocation of the acidic TADs with net negative charges into the ER lumen and also allows retention of the net positively charged basic Neh6L and CNC-bZIP domains on the cytoplamic side of the ER. This is supported by the data obtained from membrane proteinase protection assay, revealing that loss of some portions of the Neh6L and bZIP domains causes the resulting mutant proteins to be trapped in the ER lumen. Intriguingly, we found that it is possible for the TMc sequence to adopt the same Ncyt/Clum orientation as TM1 (Ncyt/Clum); this is consistent with the positive-inside, charge difference rules, and the hydrophobic gradient along membranes68,69. Our findings also suggest that the TMc peptide may be dynamically moving in and out of membrane, even though it is likely to be integrated in the TX-resistant membrane microdomains. Once TMc is integrated in an Ncyt/Clum orientation spanning membranes, on such occasions the topological organization of Nrf1 may force the semihydrophobic amphipathic TMp hinge region to span the membrane by folding into an Nlum/Ccyt orientation (Figs. 7, S1 and S4). Only in the presence of TM1, both TMc and TMp can contribute to negative regulation of Nrf1 through association with membranes under normal homeostatic conditions2,39. In addition, the negative regulation of Nrf1 by TMp is also supported by the finding that the TMp-truncated Gal4D/N460-V5 fusion factor exhibited approximately 1.5-fold greater transactivation activity than that of the TMp-containing Gal4D/N526-V5 factor (Fig. 6). These observations raise the possibility that, after Nrf1 is anchored by TM1 within membranes, either TMc or TMp may adopt a co-spanning topology and move dynamically in and out of membranes (Figs. S2 to S4). This topogenic-vectorial process is likely to depend on their intramolecular interactions with TM1 through small, polar and charged residues (refs. 11,60, and this study). The topogenic process may also rely on intermolecular interactions of their adjoining CRAC regions with membrane lipids, such as cholesterol and sphingolipids that are enriched in the detergent-resident membrane microdomain71,72, but this possibility warrants further study. Overall, it appears that the topological organization of Nrf1 within membranes is highly dynamic (in particular TMp and TMc) and is also influenced by changes in the membrane lipids and the environment.
When required, acidic-hydrophobic amphipathic TADs and/or other regions of Nrf1 are repositioned from the ER lumen into the cyto/nucleoplasmic side, where it is allowed for deglycosylation to yield a 95-kDa activator and/or for selective proteolysis to generate multiple short species with masses of 85-kDa (as a cleaved activator), 55-kDa Nrf1β (as a weak activator), 36 kDa Nrf1γ and 25 kDa Nrf1δ (the latter two act as dominant-negative isoforms) (Figs. 1a & S5C); they will adopt distinct membrane-topological conformations, located in different subcellular compartments (Fig. S5B). Some of these short isoforms may be generated through internal translation from the in-frame ATG codons and/or selective proteolysis of longer Nrf1 proteins by proteosome and/or calpain (this study, see refs. 11,39,60). Those newly-translated short Nrf1 species lacking the TM1 determinant, though they contain other semihydrophobic and amphipathic sequences (e.g. TMp and/or TMc), cannot be targeted to the ER so that they are thus positioned in the cyto/nucleoplasmic compartments (Fig. S5B). Alternatively, other short species that arise from post-translational proteolysis of the ER-bound full-length Nrf1 protein may be positioned in close proximity to membranes; this is attributable to a potential cooperation of the semihydrophobic amphipathic regions with TM1 for the overall membrane-topological folding of Nrf1 before it is proteolytically degraded. Once these amphipathic regions would disassociate from TM1, the corresponding Nrf1 species will be eventually released from membranes, so that they could gain a distinct membrane-topology. Therefore, the use of microscopy following in vivo proteinase protection reactions is hard to determine dynamic membrane-topologies of distinct Nrf1 isoforms by distinguishing their superimposed images.
In summary, an intrinsic molecular cell mechanism by which the membrane-topogenic behaviour of Nrf1 controls its post-translational modification and transcriptional regulation of target genes has been discovered in the present study together with our recent work. During translation, Nrf1 is targeted to the ER through its NHB1 signal peptide (Fig. 7). The TM1 sequence that is embedded within the uncleavable NHB1 enables Nrf1 to anchor in membranes and dictates its initial membrane topology. However, the final topological folding of Nrf1 requires TM1 to cooperate with semihydrophobic amphipathic TMi, TMp and TMc regions situated in distinct domains of the transcription factor. After Nrf1 is orientated within the ER membrane, its DNA-binding domain (DBD) is partitioned to reside on the cytoplasmic side of the membrane, whilst its acidic TADs, including its AD1, AD2 and NST glycodomain, are transiently translocated into the ER lumen, enabling glycosylation of Nrf1 to create an inactive 120-kDa glycoprotein in the presence of glucose. When required, some of the TADs are dynamically repartitioned out of membranes and/or retrotranslocated into the cyto/nucleoplasm, whereupon Nrf1 is deglycosylated to become an active 95-kDa transcription factor, particularly upon glucose deprivation. The TMi-adjoining regions (e.g. AD1 and AD2) may modulate the partial repartitioning of TADs and their coupled degrons across membranes into the cyto/nucleoplasmic side, where Nrf1 is likely allowed for selective proteolysis to yield an 85-kDa cleaved isoform. By contrast, the TMp-flanking regions between TADs and DBD can monitor further proteolytic processing of Nrf1 to create dominant-negative forms in order to shut down target gene expression. Thus, the membrane-related vectorial behaviour of Nrf1 dictates its glycosylation, deglycosylation, selective proteolysis, and possibly its transport into the nucleus, which together control its TAD-mediated transcriptional activity through binding of the DBD/sMaf heterodimer to ARE sequences in target genes. Overall, our present work provides a platform for moving forward to tease out how Nrf1 is regulated downstream of and possibly during the post-translational processing that is selectively controlled by the membrane-topogenic vectorial behaviour of this factor.
Methods
Chemicals and antibodies
All chemicals were of the highest quality commercially available. The ER extraction kit was purchased from Sigma-Aldrich. The enzymes Endo H and PK were obtained from New England Biolabs. Rabbit polyclonal antibodies against calreticulin (CRT) and green fluorescent protein (GFP) were bought from Calbiochem and Abcam PLC, respectively. A mouse monoclonal antibody against the V5 epitope and rabbit polyclonal antibodies against DsRed (a Discosoma sp. red fluorescent protein) were from Invitrogen Ltd. Antisera against Nrf1 that were produced in rabbits using a polypeptide covering amino acids 292–741.
Expression constructs
Expression constructs for full-length mouse Nrf1 and its mutants, along with other expression constructs for N65/GFP and Nrf1/GFP fusion proteins, have been created as described previously36,39. Three sandwich fusion proteins DsRed/Nrf1/GFP, DsRed/Nrf1ΔC/GFP and DsRed/N65/GFP, were engineered by inserting nucleotide sequences encoding Nrf1, Nrf1ΔC (lacking the C-terminal 56 amino acids of Nrf1), and N65 (containing the first N-terminal 65 aa of Nrf1), respectively, between the cDNAs for DsRed2 and GFP within the pDsRed2-GFP vector through the SalI/KpnI multiple cloning site39. A series of Gal4D/Nrf1 expression constructs were generated by ligating cDNA sequences that encode full-length Nrf1, or its N-terminal 181, 298, 395, 460, 526 or 607 amino acids (called N181, N298, N395, N460, N526 and N607, respectively), to the 3′-end of cDNA encoding the Gal4 DNA-binding domain (called Gal4D), within pcDNA3.1Gal4D-V5, through the BamHI/EcoRI cloning site36. All oligonucleotide primers were synthesized by MWG Biotech, and the fidelity of all cDNA products was confirmed by sequencing.
Cell culture, transfection, and reporter gene assays
Unless otherwise indicated, monkey kidney COS-1 cells (3 × 105) were seeded in 6-well plates and grown for 24 h in Dulbecco's modified Eagle's medium (DMEM) contain 25 mM glucose and 10% FBS. After reaching 70% confluence, the cells were transfected with a Lipofectamine 2000 (Invitrogen) mixture that contained an expression construct for wild-type Nrf1 or a mutant protein, together with Psv40nqo1-ARE-luc, PSV40GSTA2-6×ARE-Luc (which contains six copies of the core ARE consensus sequence from rat GSTA2, see refs. 36,39,73), or PTKUAS/Gal4-Luc39, along with pcDNA4/HisMax/lacZ encoding β-galactosidase (β-gal) that was used as a control for transfection efficiency. In addition, mutant versions of these reporter genes that lacked the ARE sequence were used as negative controls. Luciferase or CAT activity was measured approximately 36 h after transfection. The basal and stimulated ARE-driven reporter gene activity obtained following transfection with an expression vector for Nrf1 (or its mutants) was calculated as a ratio of its value against the background activity (i.e. the luciferase acitivity obtained following co-transfection of an empty pcDNA3.1/V5 His B vector and an ARE-driven reporter after subtraction of the non-specific value from cotransfecting an empty pcDNA3.1/V5 His B vector and a non-ARE-containing Luc plasmid). Subsequently, the basal activity of full-length wild-type Nrf1 was given the value of 1.0, and other data were calculated as a ratio of this value and are shown as a fold change (mean ± S.D). The data presented each represent at least 3 independent experiments undertaken on separate occasions that were each performed in triplicate. Differences in their transcriptional activity were subjected to statistical analyses.
Subcellular fractionation and membrane PK protection reactions as combined in FPP, dFPP or GRAPPA
To investigate the topological folding of Nrf1 within membranes and its repartitioning out of membranes, the intact ER-rich membrane fractions were prepared from COS-1 cells expressing Nrf1 fusion protein in classic membrane PK protection assays, as described previously40,74,75,76,77. The intact ER-rich fraction was resuspended in 100 μl of 1 × isotonic buffer [10 mM Hepes, pH 7.8, containing 250 mM sucrose, 1 mM EGTA, 1 mM EDTA and 25 mM KCl]. Subsequently, membrane proteinase protection reactions were performed for 15, 30 or 60 min on ice in an aliquot (50 μg of protein) of the membrane-containing preparation with PK added to a final concentration of 50 or 100 μg protein/ml in either the presence or absence of 1% (v/v) Triton X-100 (TX). The reactions were stopped by incubation at 90°C for 10 min following the addition of 1 mM PMSF. Subsequently, the reactants were analysed by western blotting with antibodies against Nrf1β, or the V5 ectope. The amount of Nrf1 protein remaining after PK digestion was calculated as described below. Importantly, fluorescence protease protection (FPP) assay63,64 was described to determine the topology of membrane proteins (which terminus is facing either the luminal or the cytoplasmic sides of membranes). Double fluorescence sandwiched proteins linked to proteinase protection reaction was called dFPP herein. Gal4-based reporter assays linked to protease protection assays (called GRAPPA) were used as a measure of whether the membrane-topogenic vectorial behaviour of Nrf1 has an impact on reporter gene expression.
Effects of cellular glucose concentrations on Nrf1 activity and analysis following treatment with CHX, TU, Z-VAD-fmk or MG132
COS-1 cells (3 × 105) were cultured for 24 h in DMEM containing 25 mM glucose and 10% FBS. After reaching 80% confluence, the cells were transfected for 6 h with 5 μl of a Lipofectamine 2000 mixture containing 1.5 μg of the expression construct for wild-type Nrf1 and 0.2 μg of pcDNA4/HisMax/lacZ. The transfected cells were allowed to recover for 16 h in DMEM containing 25 mM glucose (control) or 1.0 mM glucose (starvation). Thereafter, the 1.0 mM glucose-starved cells were transferred to medium containing 25 mM glucose supplemented with vehicle DMSO, TU (1 μg/ml) alone or plus MG132 (5 μM). The transfected cells that had been left to recover in medium containing 25 mM glucose were subjected to: (i) glucose deprivation, using medium that lacked glucose to which was added 0.1% (v/v) DMSO vehicle control, Z-VAD-fmk (20 μM) alone or plus MG132 (5 μM); (ii) hypoglucose restriction, using medium containing 5.5 mM glucose to which was added TU (1 μg/ml) alone or plus MG132 (5 μM); (iii) normal glucose supply using medium containing 25 mM glucose to which was added CHX (50 μg/ml), TU (1 μg/ml) alone or plus MG132 (5 μM). The cells were subsequently harvested at the indicated time points after manipulation of glucose levels. The expression of Nrf1 in the cells treated with glucose-free medium or medium containing 5.5 mM glucose or 25 mm glucose was examined by western blotting with antibodies against the V5 tag. The intensity of blots was quantified using the ImageJ programme developed by NIH, and was then used to calculate changes in the abundance of various Nrf1 isoforms.
Deglycosylation reactions and western blotting
The in vitro deglycosylation reactions of each sample with 500 units of Endo H were carried out, followed by western blotting. On some occasions, nitrocellulose membranes that had already been blotted with an antibody were washed for 30 min with stripping buffer [7M guanidine hydrochloride, 50 mM glycine, 0.05 mM EDTA, 0.1 M KCl and 20 mM 2-mercaptoethanol at pH 10.8], before being re-probed with an additional primary antibody against CRT or β-Actin; both served as internal controls to verify equal loading of protein into each electrophoretic well78. The intensity of blots was calculated using the Quantity One® software developed at Bio-Rad Laboratories.
Bioinformatic analyses
The membrane-topology of Nrf1 was initially predicted using several bioinformatic algorithms, including the TopPred (http://mobyle.pasteur.fr/cgi-bin/portal.py?form=toppred), HeliQuest (http://heliquest.ipmc.cnrs.fr/) and AmphipaSeek (http://npsa-pbil.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_amphipaseek.html) programmes. The predicted topologies were evaluated using molecular and biochemical experiments.
Statistical analysis
The statistical significance of changes in Nrf1 activity and in the intensity of immunoblots, was determined using the Student's t test or Multiple Analysis of Variations (MANOVA). The data presented herein are shown as a fold change (mean ± S.D), each of which represents at least 3 independent experiments undertaken on separate occasions that were each performed in triplicate
Author Contributions
Y.Z. designed this study, performed the experiments, analyzed the data, prepared figures and wrote the paper. J.D.H. helped analysis of the data and wrote this paper.
Supplementary Material
Supplementary information
Acknowledgments
This work was supported by the National Natural Science Foundation of China (NSFC, key program 91129703 and project 31270879), and the Fundamental Research Funds for the Central Universities (CDJRC11230003) awarded to Prof. Yiguo Zhang (University of Chongqing, China), and by the Association for International Cancer Research (grant 09-0254) awarded to Prof. John D. Hayes and Prof. Ronald Hay (University of Dundee, UK).
References
- Sykiotis G. P. & Bohmann D. Stress-activated cap'n'collar transcription factors in aging and human disease. Sci. Signal. 3, re3 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. Molecular and cellular control of the Nrf1transcription factor: An integral membrane glycoprotein. Vdm Verlag Dr. Müller Publishing House Germany (May 2009). The first edition, pp1–264 (2009). [Google Scholar]
- Mohler J., Mahaffey J. W., Deutsch E. & Vani K. Control of Drosophila head segment identity by the bZIP homeotic gene cnc. Development 121, 237–47 (1995). [DOI] [PubMed] [Google Scholar]
- Mohler J., Vani K., Leung S. & Epstein A. Segmentally restricted, cephalic expression of a leucine zipper gene during Drosophila embryogenesis. Mech. Dev. 34, 3–9 (1991). [DOI] [PubMed] [Google Scholar]
- Bowerman B., Draper B. W., Mello C. C. & Priess J. R. The maternal gene skn-1 encodes a protein that is distributed unequally in early C. elegans embryos. Cell 74, 443–52 (1993). [DOI] [PubMed] [Google Scholar]
- Bowerman B., Eaton B. A. & Priess J. R. Skn-1, a maternally expressed gene required to specify the fate of ventral blastomeres in the early C. elegans embryo. Cell 68, 1061–75 (1992). [DOI] [PubMed] [Google Scholar]
- Andrews N. C., Erdjument-Bromage H., Davidson M. B., Tempst P. & Orkin S. H. Erythroid transcription factor NF-E2 is a haematopoietic-specific basic-leucine zipper protein. Nature 362, 722–8 (1993). [DOI] [PubMed] [Google Scholar]
- Chan J. Y., Han X. L. & Kan Y. W. Isolation of cDNA encoding the human NF-E2 protein. Proc. Natl. Acad. Sci. USA 90, 11366–70 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi K. et al. Regulation of transcription by dimerization of erythroid factor NF-E2 p45 with small Maf proteins. Nature 367, 568–72 (1994). [DOI] [PubMed] [Google Scholar]
- Chan J. Y., Han X. L. & Kan Y. W. Cloning of Nrf1, an NF-E2-related transcription factor, by genetic selection in yeast. Proc. Natl. Acad. Sci. USA 90, 11371–5 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen O. et al. Small Maf proteins interact with the human transcription factor TCF11/Nrf1/LCR-F1. Nucleic Acids Res. 24, 4289–97 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna L. et al. Structural organization and mapping of the human TCF11 gene. Genomics 27, 237–44 (1995). [DOI] [PubMed] [Google Scholar]
- Farmer S. C., Sun C. W., Winnier G. E., Hogan B. L. & Townes T. M. The bZIP transcription factor LCR-F1 is essential for mesoderm formation in mouse development. Genes Dev. 11, 786–98 (1997). [DOI] [PubMed] [Google Scholar]
- Caterina J. J., Donze D., Sun C. W., Ciavatta D. J. & Townes T. M. Cloning and functional characterization of LCR-F1: a bZIP transcription factor that activates erythroid-specific, human globin gene expression. Nucleic Acids Res. 22, 2383–91 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moi P., Chan K., Asunis I., Cao A. & Kan Y. W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 91, 9926–30 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A. et al. Molecular cloning and functional characterization of a new Cap‘n’ collar family transcription factor Nrf3. J. Biol. Chem. 274, 6443–52 (1999). [DOI] [PubMed] [Google Scholar]
- Chenais B. et al. Functional and placental expression analysis of the human NRF3 transcription factor. Mol. Endocrinol. 19, 125–37 (2005). [DOI] [PubMed] [Google Scholar]
- Blouin J. L. et al. Isolation of the human BACH1 transcription regulator gene, which maps to chromosome 21q22.1. Hum. Genet. 102, 282–8 (1998). [DOI] [PubMed] [Google Scholar]
- Ohira M. et al. Characterization of a human homolog (BACH1) of the mouse Bach1 gene encoding a BTB-basic leucine zipper transcription factor and its mapping to chromosome 21q22.1. Genomics 47, 300–6 (1998). [DOI] [PubMed] [Google Scholar]
- Muto A. et al. Identification of Bach2 as a B-cell-specific partner for small maf proteins that negatively regulate the immunoglobulin heavy chain gene 3′ enhancer. EMBO J. 17, 5734–43 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki S. et al. Cloning and expression of human B cell-specific transcription factor BACH2 mapped to chromosome 6q15. Oncogene 19, 3739–49 (2000). [DOI] [PubMed] [Google Scholar]
- Steffen J., Seeger M., Koch A. & Kruger E. Proteasomal degradation is transcriptionally controlled by TCF11 via an ERAD-dependent feedback loop. Mol. Cell 40, 147–58 (2010). [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S. K. et al. Transcription factor Nrf1 mediates the proteasome recovery pathway after proteasome inhibition in mammalian cells. Mol. Cell 38, 17–28 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimberg K. B., Beskow A., Lundin D., Davis M. M. & Young P. Basic leucine zipper protein Cnc-C is a substrate and transcriptional regulator of the Drosophila 26S proteasome. Mol. Cell Biol. 31, 897–909 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. et al. Specific SKN-1/Nrf stress responses to perturbations in translation elongation and proteasome activity. PLoS Genet. 7, e1002119 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J. Y. et al. Targeted disruption of the ubiquitous CNC-bZIP transcription factor, Nrf-1, results in anemia and embryonic lethality in mice. EMBO J. 17, 1779–87 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L. et al. Nrf1 is critical for redox balance and survival of liver cells during development. Mol. Cell Biol. 23, 4673–86 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z. et al. Liver-specific inactivation of the Nrf1 gene in adult mouse leads to nonalcoholic steatohepatitis and hepatic neoplasia. Proc. Natl. Acad. Sci. USA 102, 4120–5 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtsuji M. et al. Nrf1 and Nrf2 play distinct roles in activation of antioxidant response element-dependent genes. J. Biol. Chem. 283, 33554–62 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Xing W., Wergedal J., Chan J. Y. & Mohan S. Targeted disruption of nuclear factor erythroid-derived 2-like 1 in osteoblasts reduces bone size and bone formation in mice. Physiol. Genomics 40, 100–10 (2010). [DOI] [PubMed] [Google Scholar]
- Kobayashi A. et al. Central nervous system-specific deletion of transcription factor Nrf1 causes progressive motor neuronal dysfunction. Genes Cells 16, 692–703 (2011). [DOI] [PubMed] [Google Scholar]
- Lee C. S. et al. Loss of nuclear factor E2-related factor 1 in the brain leads to dysregulation of proteasome gene expression and neurodegeneration. Proc. Natl. Acad. Sci. USA 108, 8408–13 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan K., Lu R., Chang J. C. & Kan Y. W. NRF2, a member of the NFE2 family of transcription factors, is not essential for murine erythropoiesis, growth, and development. Proc. Natl. Acad. Sci. USA 93, 13943–8 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Q. & Lennarz W. J. Oligosaccharyltransferase: a complex multisubunit enzyme of the endoplasmic reticulum. Biochem. Biophys. Res. Commun. 266, 684–9 (1999). [DOI] [PubMed] [Google Scholar]
- Shibatani T., David L. L., McCormack A. L., Frueh K. & Skach W. R. Proteomic analysis of mammalian oligosaccharyltransferase reveals multiple subcomplexes that contain Sec61, TRAP, and two potential new subunits. Biochemistry 44, 5982–92 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Crouch D. H., Yamamoto M. & Hayes J. D. Negative regulation of the Nrf1 transcription factor by its N-terminal domain is independent of Keap1: Nrf1, but not Nrf2, is targeted to the endoplasmic reticulum. Biochem. J. 399, 373–85 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W. & Chan J. Y. Nrf1 is targeted to the endoplasmic reticulum membrane by an N-terminal transmembrane domain. Inhibition of nuclear translocation and transacting function. J. Biol. Chem. 281, 19676–87 (2006). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lucocq J. M., Yamamoto M. & Hayes J. D. The NHB1 (N-terminal homology box 1) sequence in transcription factor Nrf1 is required to anchor it to the endoplasmic reticulum and also to enable its asparagine-glycosylation. Biochem. J. 408, 161–72 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Lucocq J. M. & Hayes J. D. The Nrf1 CNC/bZIP protein is a nuclear envelope-bound transcription factor that is activated by t-butyl hydroquinone but not by endoplasmic reticulum stressors. Biochem. J. 418, 293–310 (2009). [DOI] [PubMed] [Google Scholar]
- Zhang Y. & Hayes J. D. Identification of topological determinants in the N-terminal domain of transcription factor Nrf1 that control its orientation in the endoplasmic reticulum membrane. Biochem. J. 430, 497–510 (2010). [DOI] [PubMed] [Google Scholar]
- Zhang Y., Kobayashi A., Yamamoto M. & Hayes J. D. The Nrf3 transcription factor is a membrane-bound glycoprotein targeted to the endoplasmic reticulum through its N-terminal homology box 1 sequence. J. Biol. Chem. 284, 3195–210 (2009). [DOI] [PubMed] [Google Scholar]
- Kim H. M., Do C. H. & Lee D. H. Taurine reduces ER stress in C. elegans. J. Biomed. Sci. 17 Suppl 1, S26 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill R. A. Enzymatic release of oligosaccharides from glycoproteins for chromatographic and electrophoretic analysis. J. Chromatogr. A 720, 201–15 (1996). [DOI] [PubMed] [Google Scholar]
- van Geest M. & Lolkema J. S. Membrane topology and insertion of membrane proteins: search for topogenic signals. Microbiol. Mol. Biol. Rev. 64, 13–33 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M. K., Sharma R. S. & Mishra V. Induction of apoptosis by ribosome inactivating proteins: importance of N-glycosidase activity. Appl. Biochem. Biotechnol. 166, 1552–61 (2012). [DOI] [PubMed] [Google Scholar]
- Robertus J. The structure and action of ricin, a cytotoxic N-glycosidase. Semin. Cell Biol. 2, 23–30 (1991). [PubMed] [Google Scholar]
- Husberg C., Murphy P., Bjorgo E., Kalland K. H. & Kolsto A. B. Cellular localisation and nuclear export of the human bZIP transcription factor TCF11. Biochim. Biophys. Acta 1640, 143–51 (2003). [DOI] [PubMed] [Google Scholar]
- Tsuchiya Y. et al. Dual Regulation of the Transcriptional Activity of Nrf1 by β-TrCP- and Hrd1-Dependent Degradation Mechanisms. Mol. Cell Biol. 31, 4500–12 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas M., Phan D., Watanabe M. & Chan J. Y. The Fbw7 tumor suppressor regulates nuclear factor E2 related factor 1 (Nrf1) transcription factor turnover through proteasome-mediated proteolysis. J. Biol. Chem. 286, 39282–39289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J. et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem J. 404, 459–66 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novotny V., Prieschl E. E., Csonga R., Fabjani G. & Baumruker T. Nrf1 in a complex with fosB, c-jun, junD and ATF2 forms the AP1 component at the TNFα promoter in stimulated mast cells. Nucleic Acids Res. 26, 5480–5 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prieschl E. E. et al. A novel splice variant of the transcription factor Nrf1 interacts with the TNFα promoter and stimulates transcription. Nucleic Acids Res. 26, 2291–7 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnsen O., Murphy P., Prydz H. & Kolsto A. B. Interaction of the CNC-bZIP factor TCF11/LCR-F1/Nrf1 with MafG: binding-site selection and regulation of transcription. Nucleic Acids Res. 26, 512–20 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupert P. B., Daughdrill G. W., Bowerman B. & Matthews B. W. A new DNA-binding motif in the Skn-1 binding domain-DNA complex. Nat. Struct. Biol. 5, 484–91 (1998). [DOI] [PubMed] [Google Scholar]
- Misaghi S., Pacold M. E., Blom D., Ploegh H. L. & Korbel G. A. Using a small molecule inhibitor of peptide: N-glycanase to probe its role in glycoprotein turnover. Chem. Biol. 11, 1677–87 (2004). [DOI] [PubMed] [Google Scholar]
- Goder V., Bieri C. & Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J. Cell Biol. 147, 257–66 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev N. L., Bennitz J. D., Huang T., McBride S. & Willmore W. G. The Nrf1 CNC-bZIP protein is regulated by the proteasome and activated by hypoxia. PLoS One 6, e29167 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna L. et al. Molecular cloning of a putative novel human bZIP transcription factor on chromosome 17q22. Genomics 22, 553–62 (1994). [DOI] [PubMed] [Google Scholar]
- Husberg C., Murphy P., Martin E. & Kolsto A. B. Two domains of the human bZIP transcription factor TCF11 are necessary for transactivation. J. Biol. Chem. 276, 17641–52 (2001). [DOI] [PubMed] [Google Scholar]
- Langosch D. & Arkin I. T. Interaction and conformational dynamics of membrane-spanning protein helices. Protein Sci. 18, 1343–58 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Kwok A. M. & Chan J. Y. The p65 isoform of Nrf1 is a dominant negative inhibitor of ARE-mediated transcription. J. Biol. Chem. 282, 24670–8 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang Y. K., Yeager R. L., Tanaka Y. & Klaassen C. D. Enhanced expression of Nrf2 in mice attenuates the fatty liver produced by a methionine- and choline-deficient diet. Toxicol. Appl. Pharmacol. 245, 326–34 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H., Hailey D. W. & Lippincott-Schwartz J. Fluorescence protease protection of GFP chimeras to reveal protein topology and subcellular localization. Nat. Methods 3, 205–210 (2006). [DOI] [PubMed] [Google Scholar]
- Lorenz H., Hailey D. W., Wunder C. & Lippincott-Schwartz J. The fluorescence protease protection (FPP) assay to determine protein localization and membrane topology. Nat. Protoc. 1, 276–279 (2006). [DOI] [PubMed] [Google Scholar]
- Ye J. et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–64 (2000). [DOI] [PubMed] [Google Scholar]
- Brown M. S., Ye J., Rawson R. B. & Goldstein J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–8 (2000). [DOI] [PubMed] [Google Scholar]
- Wolfe M. S. & Kopan R. Intramembrane proteolysis: theme and variations. Science 305, 1119–23 (2004). [DOI] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 7, 909–18 (2006). [DOI] [PubMed] [Google Scholar]
- Dowhan W. & Bogdanov M. Lipid-dependent membrane protein topogenesis. Annu. Rev. Biochem. 78, 515–40 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skach W. R. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 16, 606–12 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holthuis J. C. & Levine T. P. Lipid traffic: floppy drives and a superhighway. Nat. Rev. Mol. Cell Biol. 6, 209–20 (2005). [DOI] [PubMed] [Google Scholar]
- Lingwood D. & Simons K. Lipid rafts as a membrane-organizing principle. Science 327, 46–50 (2010). [DOI] [PubMed] [Google Scholar]
- Wang X. J., Hayes J. D. & Wolf C. R. Generation of a stable antioxidant response element-driven reporter gene cell line and its use to show redox-dependent activation of Nrf2 by cancer chemotherapeutic agents. Cancer Res. 66, 10983–94 (2006). [DOI] [PubMed] [Google Scholar]
- Bailey D., Barreca C. & O'Hare P. Trafficking of the bZIP transmembrane transcription factor CREB-H into alternate pathways of ERAD and stress-regulated intramembrane proteolysis. Traffic 8, 1796–814 (2007). [DOI] [PubMed] [Google Scholar]
- Afshar N., Black B. E. & Paschal B. M. Retrotranslocation of the chaperone calreticulin from the endoplasmic reticulum lumen to the cytosol. Mol. Cell Biol. 25, 8844–53 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. W. et al. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell 127, 999–1013 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafvelin G. & von Heijne G. Topological "frustration" in multispanning E. coli inner membrane proteins. Cell 77, 401–12 (1994). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. Involvement of the acid sphingomyelinase pathway in UVA-induced apoptosis. J. Biol. Chem. 276, 11775–82 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information