SUMMARY
The immunoglobulin heavy chain (IgH) gene locus undergoes radial re-positioning within the nucleus and locus contraction in preparation for gene recombination. We demonstrate that IgH locus conformation involves two levels of chromosomal compaction. At the first level the locus folds into several multi-looped domains. One such domain at the 3′ end of the locus requires an enhancer, Eμ; two other domains at the 5′ end are Eμ-independent. At the second level, these domains are brought into spatial proximity by Eμ-dependent interactions with specific sites within the VH region. Eμ is also required for radial re-positioning of IgH alleles indicating its essential role in large scale chromosomal movements in developing lymphocytes. Our observations provide a comprehensive view of the conformation of IgH alleles in pro-B cells and the mechanisms by which it is established.
INTRODUCTION
Radial positioning of loci within the nucleus and chromosome conformation have recently gained prominence as mechanisms for developmentally regulated gene expression (Kadauke and Blobel, 2009; Takizawa et al., 2008). This interest rides on the foundation of pioneering studies that examined global chromosome structure and folding within the nucleus (Gasser and Laemmli, 1987; Paulson and Laemmli, 1977). Of particular note, the concept of chromosomal loops arose from a combination of biochemical and direct visualization studies (Cook et al., 1976). Based on the observation that loops were tethered at their base to the nuclear scaffold, Laemmli and colleagues proposed a rosette-like configuration for chromosomes (Marsden and Laemmli, 1979). Chromosomal loops are also the central feature of computational models of chromosome structure that account for chromosome conformation by varying the size and numbers of loops associated with chromosomal domains (Knoch et al., 2000; Sachs et al., 1995). The extent to which these features apply to developmentally regulated loci, and the mechanisms by which these structures are generated, are critical for understanding gene regulatory mechanisms.
Antigen receptor genes of B and T lymphocytes are assembled from gene segments that are spread over several megabases of the genome (Krangel, 2009; Perlot and Alt, 2008). The immunoglobulin heavy chain (IgH) locus in the mouse consists of 150 variable (VH) gene segments, 8-12 diversity (DH) gene segments and 4 joining (JH) gene segments (Johnston et al., 2006; Retter et al., 2007). Two rearrangement steps assemble functional IgH genes during B cell development. First, a DH gene segment recombines with a JH gene segment to form a DJH junction; this is followed by VH recombination to the DJH junction to generate V(D)J recombined alleles.
Prior to initiation of DNA rearrangements, the IgH locus undergoes two forms of chromosome movements. First, radial repositioning moves the locus away from the nuclear periphery to a more central location (Kosak et al., 2002). This step does not occur in progenitors that lack the transcription factor E2A (Sayegh et al., 2005) where B cell differentiation is blocked at a very early stage. Second, locus contraction brings the two ends of the IgH locus into physical proximity (Kosak et al., 2002; Sayegh et al., 2005). These movements are independently regulated because locus contraction, but not radial re-positioning, is abolished in B cell progenitors that lack the transcription factors Pax5 (Fuxa et al., 2004) or YY1 (Liu et al., 2007). Recently, Busslinger and colleagues proposed that Pax5 mediates locus contraction via a conserved sequence element that they named Pax5-activated intergenic repeat (PAIR) (Ebert et al., 2011). 14 PAIRs, of which 7 bind Pax5 in pro-B cells, are spread over approximately 750 kb of the distalmost part of the VH locus. It is not clear whether YY1 is mechanistically connected to the Pax5/PAIR pathway.
Jhunjhunwala et al. (Jhunjhunwala et al., 2008) developed a model for IgH locus conformation in its germline (pre-rearrangement) state. They measured spatial distances between different points throughout the IgH locus using 3D-FISH and trilateration. The data was used to mathematically compute the conformation of the genomic region. They found that in transcription factor E2A-deficient pre-pro-B cells IgH locus conformation fit best within the framework of the computational Major Loop Subcompartment (MLS) model. In further differentiated pro-B cells, however, the conformation is more compact and deviates significantly from the MLS model. A central feature of the state in pro-B cells is that the distal VH genes (labeled J558 and 3609 in Figure 1A) and proximal VH genes (labeled 7183 in Figure 1A) are positioned at comparable spatial distance from the DH-JH part of the IgH locus. The molecular mechanisms by which these changes are brought about are not clear.
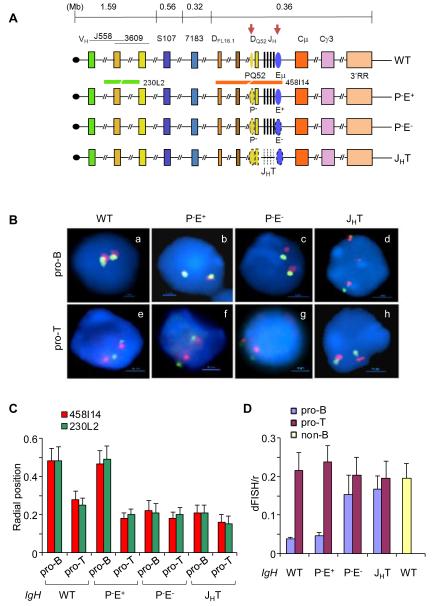
Figure 1. Nuclear positioning and locus contraction of IgH alleles with cis-regulatory sequence deletions.
(A) Top line is a schematic representation of the murine IgH locus. Approximate distances between regions of interest are derived from the sequence of the locus in C57BL6 mouse strain (Johnston et al., 2006). The telomere (black circle)-proximal variable region (VH) spans approximately 2.5Mb and contains 150 VH segments. Gene segments corresponding to J558 and 3609 families are largely interspersed at the 5′ end. The 7183 family lies at the 3′ end of the cluster. The 5′-most and 3′-most diversity (DH) gene segments, DFL16.1 and DQ52, are indicated; between them lie variable numbers of DSP gene segments depending on the mouse strain. JH gene segments are depicted as black vertical lines. Two cis-regulatory elements discussed here, PQ52 and Eμ, are indicated as ovals and are marked by tissue-specific DNase I hypersensitive sites (red arrows). The region containing exons of various IgH isotypes span another 200 kb and is followed by a cluster of DNase I hypersensitive sites that comprise the 3′ regulatory region (3′RR). Next three lines show IgH alleles that carry deletions of specific regulatory sequences (shown by dotted lines) as indicated; IgH genotype notations used in the text are noted on the right. Red and green lines below the WT allele show the position of BAC probes used in FISH analyses.
(B) Two-color FISH using bone marrow pro-B cells (a-d) and thymocytes (e-h) that carry wild-type (WT) or mutated IgH alleles as indicated. BAC probes are indicated in (A) and blue color marks nuclear DNA with DAPI. A representative nucleus from each genotype is shown.
(C) Radial positioning of WT and mutated IgH alleles was determined by measuring the distance of red and green FISH signals from the nuclear boundary in approximately 200 nuclei from pro-B cells and thymocytes of the indicated IgH genotypes. Y axis shows the distance between FISH signals and the nuclear periphery divided by the nuclear radius. Error bars represent the standard deviation between nuclei. The percentage of IgH alleles close to the nuclear periphery in each genotype is shown in Figure S1. (D) Locus contraction of WT and mutated IgH alleles was estimated by measuring the distance between red and green FISH signals in approximately 200 nuclei. Y axis shows the distance between FISH signals divided by the nuclear radius. B lineage-depleted bone marrow cells from RAG2-deficient mice were used as non-B controls. Error bars represent the standard deviation between nuclei. Measurements were made with three independent cell preparations each obtained from 5-6 mice of the indicated genotypes.
The tissue-specific enhancer Eμ (Figure 1A) regulates both steps of IgH locus recombination (Afshar et al., 2006; Perlot et al., 2005). We previously showed that deletion of the 220 nucleotide Eμ core results in a partially active locus in precursor B cells (Chakraborty et al., 2009). Eμ-deleted alleles lack acetylated histones H3 and 4, but other activation-specific epigenetic marks, such as H3K4me2 or tissue-specific loss of H3K9me2, are clearly evident. Based on these observations we proposed that full activation of the IgH locus requires Eμ-independent and Eμ-dependent steps. Here we demonstrate that the conformation of the IgH locus is generated by Eμ-dependent and Eμ-independent chromatin loops. One set of Eμ-dependent interactions defines a domain that encompasses the 3′ 262 kb of the locus. A second set of Eμ-dependent interactions brings parts of the VH locus close to the DH gene segments. All Eμ-interacting sequences bind the transcription factor YY1, indicating a role for this factor in establishing Eμ- dependent loops. We also found evidence for Eμ-independent looping between CTCF-bound sites in the IgH locus. Furthermore, Eμ-deleted alleles did not undergo radial repositioning indicating that Eμ-independent forms of locus activation can occur at the nuclear periphery. Our observations provide a comprehensive view of the conformational state of the IgH locus in pro-B cells and the mechanisms by which it is established.
RESULTS
Eμ regulates radial positioning and IgH locus contraction
To understand the relationship between cis-regulatory sequences and epigenetic changes at the IgH locus we determined radial positioning of IgH alleles with defined deletions (Figure 1A). P−E+ alleles (that delete only a promoter, PQ52, associated with DQ52) and P−E− alleles (that delete both Eμ and PQ52) have been previously described (Afshar et al., 2006). Both these alleles were analyzed in a recombinase-deficient context to maintain the locus in unrearranged state. JHT alleles lack a 3.5 kb region starting at the 5′ end of the P− deletion and extending to the 3′ end of the E− deletion (Gu et al., 1993). These alleles were assayed in recombinase sufficient cells since the absence of all JH gene segments precludes any rearrangement of these alleles. We isolated primary pro-B cells from the bone marrow by positive selection using anti-CD19-coupled magnetic beads and used the cells without further expansion ex vivo for fluorescent in situ hybridization (FISH) studies.
We used bacterial artificial chromosome (BAC) probes that mark the 5′ and 3′ ends of the IgH locus to study IgH radial positioning and locus contraction. WT IgH alleles were located away from periphery in pro-B cells but not in pro-T cells (Figure 1B and averaged in 1C). Loss of PQ52 (P−E+ alleles) did not affect radial positioning in either pro-B or pro-T cells. However, P− E− alleles were located closer to the nuclear periphery in pro-B cells compared to WT or P−E+ alleles (Figure 1B, C and S1). Indeed, the location of P− E−alleles in pro-B cells was similar to that of WT or P−E+ alleles in primary pro-T cells (Figure 1B, lower panel). These observations indicate that Eμ is necessary for radial repositioning of IgH alleles in primary pro-B cells.
We assayed IgH locus contraction by determining the distance between the two BAC probes in pro-B and pro-T cells of different genotypes. We found that P−E− and JHT alleles did not undergo locus contraction in pro-B cells as visualized by the lack of overlap of FISH signals (Figure 1B, and averaged in 1D). Instead, the average distance between the two probes in P−E− and JHT pro-B cells was similar to that seen in pro-T cells of each genotype, or in non-B lineage cells from the bone marrow of WT mice (Figure 1D). This effect was specific to loss of Eμ since P−E+ alleles underwent normal locus contraction. Additionally, IgH alleles deleted only for Eμ also did not contract (Figure 3D and E). Thus, Eμ is essential for locus contraction; in contrast, PQ52 does not contribute to either radial positioning or locus contraction of IgH alleles.
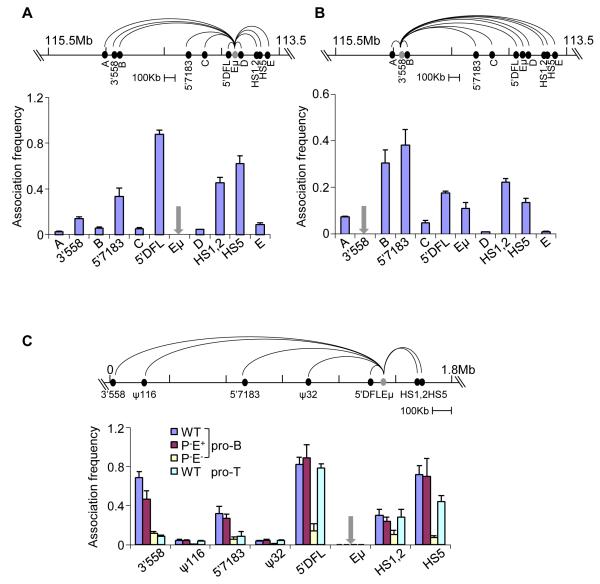
Figure 3. 3C analyses of Eμ-interacting regions.
Quantitative 3C analyses in D345 pro-B cells using different anchors (grey) as indicated. Taqman probes for detection of amplicons were located close to the anchor primers. Data with Eμ (A) and 3′558 (B) anchor primers are shown (additional 3C studies with 5′7183 and HS5 anchors are in Figure S3A and B). Association frequency (Y axis) between two primers was normalized to long-range 3′HS1 interaction in the ß-globin locus; grey arrows mark the site of anchor primers in the bar graphs. Data shown is the average of three independent 3C experiments, with error bars representing the standard error of mean between experiments.
(C) Quantitative 3C analyses using primary bone marrow pro-B cells and thymocytes that carry IgH alleles of the indicated genotypes (all cells were obtained from RAG2-deficient background). Coordinates of the IgH locus in the 129 strain (Simpson et al., 1997) are shown on the top line with positions of the relevant sequences identified by 4C. 3C assays were carried out using anchor primer was used in combination with primers located near HS1,2, HS5, 5′DFL, 5′7183 and 3′558; the ψ116 and ψ32 sequences served as negative controls. Amplification products were detected using a Taqman probe located close to the Eμ anchor primer. The association frequency (Y axis) between two primers was normalized to long-range 3′HS1 interaction in the ß-globin locus (Figure S3C). Data shown is the average of three independent 3C experiments in each genotype; error bars represent the standard error of mean between experiments.
Eμ-dependent locus contraction
To understand the basis for Eμ-dependent locus contraction we used 4C assays (Gondor et al., 2008) to identify regions of the IgH locus that were in close proximity to Eμ. For this, cross-linked chromatin from a RAG-deficient pro-B cell line, D345, was digested with Nla III or Mse I, ligated, and then amplified using anchor primers from the test region (Figure 2A). Sequences ligated between the anchor primers were identified by hybridization to mouse genomic tiling 2.0R E arrays that contained mouse chromosome 12; as a control we used sonicated genomic DNA from the same cells. Array data was analyzed using CisGenome (Ji et al., 2008).
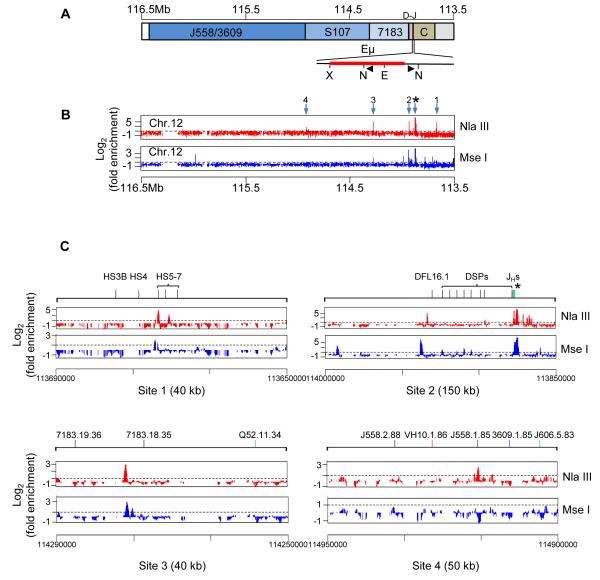
Figure 2. Eμ-mediated long-range chromatin interactions in the IgH locus.
(A) Schematic representation of the unrearranged IgH locus oriented with the DH-Cμ region on the right and VH region on the left. The region surrounding Eμ is expanded below to show the positions of restriction enzyme sites (N=Nla III; X= Xba I; E=EcoR I) and bait primers (black triangles) used in 4C assays.
(B) 4C was carried out as described in the Experimental Procedures section using D345, a recombinase-deficient Abelson virus transformed pro-B cell line of C57BL6 origin. Position of bait primers close to Eμ is marked by the asterisk. Genomic sequences that were ligated between the Nla III sites (top panel) or Mse I sites (bottom panel) were amplified by PCR using the bait primers and hybridized to Affymetrix Genomic tiling 2.0R E array. Hybridization signal intensity was compared to input DNA in CisGenome and fold enrichment (Y axis) calculated as described in Experimental Procedures. Numbered arrows mark enriched regions. 1 corresponds to the HS5 region of the 3′RR, 2 corresponds to sequences 5′ of DFL16.1 (5′DFL in text), 3 corresponds to sequence referred to as 5′7183 in the text and 4 corresponds to sequence referred to as 3′558 in the text. One of two independent experiments with each restriction enzyme is shown; high resolution 4C data is shown in Figure S2A, B. 4C analysis with 5′7183 anchor is shown in Figure S2C.
(C) Expanded views of 4C results of the regions near each of the numbered Eμ- interacting sites in part B. The extent of each expanded region is noted in parentheses; further high resolution 4C data is shown in Figure S2A. 4C analysis with 5′7183 anchor is shown in Figure S2B.
We found that the 3′ regulatory region (3′RR) of the IgH locus was prominently represented in sequences amplified with Eμ anchor primers (Figure 2B, arrow 1; Figure S2A). The 3′ RR comprises a cluster of 8 DHSs distributed over 30 kb; five of these (HS3a, b, 1, 2, 4) are found only in activated mature B cells whereas HS5-7 are present in pro-B cell lines (Garrett et al., 2005). The HS1,2 region has been previously shown to be close to Eμ in mature splenic B cells and a myeloma cell line (Ju et al., 2007; Wuerffel et al., 2007). Sequences that we amplified within Eμ anchors corresponded to the HS5 region (Figure 2C). Thus, Eμ-3′RR association occurs in the earliest B cell progenitors prior to initiation of V(D)J recombination. We also identified sequences just 5′ of DFL16.1 (5′DFL) (Figure 2B and C, arrow 2; Figure S2A) and a region close to the 5′ end of the proximal VH7183 gene family (Figure 2B and C, arrow 3; Figure S2A) in Eμ- anchored 4C assays. HS5, 5′DFL and 5′7183 are located approximately 206 kb, 57 kb and 400 kb from Eμ, respectively, suggesting that these regions are brought into proximity of Eμ by chromosome looping.
We also identified a sequence located towards the 3′ end of the VHJ558 genes approximately 1Mb from Eμ (Figure 2B, arrow 4; Figure S2A), but the signal intensity was much lower. Eμ association with the 3′RR, 5′DFL and 5′7183 was also detected in 4C assays using a different restriction enzyme, Mse I (Figure 2B, lower line). The inability to detect 3′558 sequence using Mse I may be because the sites for this restriction enzyme are not appropriately juxtaposed in crosslinked chromatin. Finally, we carried out 4C with anchor primers located within the newly detected 5′7183 region. We detected prominent interactions with Eμ and the 3′RR, thereby strengthening the idea that these regions were in spatial proximity in pro-B cells (Figure S2B). We conclude that Eμ interactions form a domain that contains all DH and JH gene segments as well as exons that encode all Ig isotypes. In addition, Eμ interacts with two sites within the VH genes, at 5′7183 and 3′558; these interactions are possible sources of Eμ-dependent locus contraction. Neither of the VH-associated Eμ interacting sequences correspond to PAIR elements.
To substantiate the interactions detected by 4C we carried out quantitative 3C analyses. Using Eμ as anchor (Figure 3A) we detected prominent interactions with 5′DFL, the 3′RR (labeled HS1,2 and HS5) and 5′7183. The interaction with 3′558 was weaker, but significantly higher than regions A-D that served as negative controls. Conversely, using 3′558 as the anchor (Figure 3B) we detected interactions with 5′7183, 5′DFL, Eμ and the 3′RR, thereby confirming spatial proximity of these widely-separated parts of the IgH locus. Fragment B, located 34 kb from 3′558 scored strongly with the 3′558 anchor, but not with Eμ anchor. Proximity between the 3′558 anchor and fragment B could be one possible explanation for this; alternatively, 3′558 could be involved in more than one kind of loop. We also carried out 3C studies with anchors located at 5′7183 and the 3′RR (Figure S3A and B) and confirmed reciprocal interactions between all five interacting sequences identified by 4C analyses.
While it is difficult to directly compare 3C results using different anchor primers (and associated Taqman probes), we noticed that the relative association frequency between different parts of the locus varied with the anchor used. For example, the Eμ anchor detected interaction with 5′DFL and HS5 more effectively than with 5′7183 or 3′558. Conversely, the 3′558 anchor detected 5′7183 more effectively than 5′DFL, Eμ or HS5. Our working hypothesis is that these selectivities represent preferential associations in pro-B cells. One set of prominent interactions involve Eμ, 5′DFL and HS5 that leads to a 206 kb domain at the 3′ end of the IgH locus. Another set of interactions, exemplified by 3′558 to 5′7183, occur within the VH part of the locus. Inter-domain interactions represented by Eμ to 3′558 or 5′7183, or by HS5 to 3′558 or 5′7183, are relatively less efficient and may occur, for example, in a smaller proportion of cells. Such contacts may get “fixed” during cross-linking to be revealed in the 3C or 4C assays.
To determine the role of Eμ in establishing locus conformation we carried out quantitative 3C analyses using chromatin prepared from primary bone marrow pro-B cells carrying WT, P−E+ and P−E− IgH alleles. An Eμ anchor readily amplified sequences 5′ of DFL16.1 and within the 3′RR on WT alleles (Figure 3C, blue bars labeled 5′DFL and HS5). The signal to HS1,2 likely represents the lower part of a peak centered around HS5. Additionally, we detected Eμ interaction with 5′7183 and 3′558 regions within the VH locus on WT alleles. Two regions, labeled ψ116 and ψ32 that flank 5′7183 served as negative controls. All Eμ associations were substantially reduced in P−E− pro-B cells, but not in P−E+ pro-B cells, demonstrating that they were Eμ-dependent (Figure 3C, compare red and yellow bars). Loops within the β-globin locus were comparable in all 3 cell preparations (Figure S3C). We only detected Eμ interaction with 5′DFL and the 3′RR in CD4−CD8− thymocytes (Figure 3C, light blue bars) indicating that Eμ associations with the VH locus were B lineage-specific. We conclude that Eμ is essential to establish chromatin loops to 5′DFL, 3′RR and specific sites in the VH locus. The absence of Eμ- dependent loops to these VH sites (5′7183 and 3′558) in pro-B cells may be the basis for the lack of locus contraction of P−E− alleles noted in FISH analyses (Figure 1).
To further confirm the presence of Eμ-dependent loops we carried out high resolution 3D-FISH analyses on primary pro-B cells from RAG2−/− and P−E− (RAG2−/−) mice using 10 kb probes. Previously described probes h4 and h11 (Jhunjhunwala et al., 2008) were used to validate our results; these probes mark sequences close to Eμ and towards the 5′ end of the VHJ558 gene family, respectively (Figure 4A). We also generated new probes corresponding to looping sites identified by our 4C assays (Figure 4A). To visualize Eμ-dependent interactions we used probe h4 with probes h11, 5′7183 and 3′558. Each probe combination gave closely juxtaposed signals in WT and P−E+ pro-B cells, but not in P−E− pro-B cells (Figure 4B). After quantitation of inter-probe distance we found that compaction of P−E− alleles was reduced by 1.4-1.8-fold for h4-h11, as well as for each new pair-wise probe combination (Table S1). We also determined the proportion of IgH alleles in which the two FISH signals were separated by different distances. For all probe combinations the separation between FISH signals was skewed towards greater separation on P−E− alleles (Figure 4C).
Figure 4. Visualization of Eμ-dependent locus contraction.
(A) The unrearranged IgH locus as represented in Figure 2A showing the location of six 10 kb probes used for FISH.
(B) Three color 3D-FISH were carried out with bone marrow pro-B cells of the indicated IgH genotypes cells in a RAG2-deficient background. Short probes labeled with Alexa Fluor 594 (red) and 488 (green), and BAC RP23-201H14 labeled with Alexa Fluor 697 (blue) were hybridized to fixed pro-B cells. Signals were visualized by epifluorescence microscopy and distances between probes were determined as described (Jhunjhunwala et al., 2008) and shown in Table S1. Probe combinations were: a, d, g h4-red, h11-green; b, e, h h4-red, 5′7183-green; c, f, i h4-green, 3′558-red. Red line represents 1 μm.
(C) Quantitation of FISH data shown in part B. Distances between red and green 3D FISH signals in part A were divided into 5 categories (<0.2, 0.2-0.5, 0.5-0.8, 0.8-1.0, and >1.0 μm) for 60-90 nuclei. The percentage of IgH alleles in each category was determined (Y axis) for each IgH genotype (X axis) and is represented in different colors. Probe combinations are shown above the bars. Pro-B cells were purified from 5-6 mice of each genotype.
(D) Three color 3D-FISH with RAG2-deficient pro-B cell lines carrying WT and Eμ- deleted IgH alleles. Probe combinations were: a, c h4-red, h11-green; b, d h4-green, 3′558-red.
(E) Quantitation of the FISH data shown in part D as described in C.
(F) Three color 3D-FISH with bone marrow pro-B cells of the indicated IgH genotypes cells obtained from RAG2-deficient background. Probe combinations were: a, c h1-red, h11-green; b, d DFL-red, 3′RR-green. Pro-B cells were purified from 5-6 mice of each genotype.
(G) Quantitation of FISH data shown in part F as described in C.
Finally, we used two probe sets for FISH analyses in pro-B cell lines with WT or Eμ-deficient (Eμ−) IgH alleles. As noted in primary cells, the separation distance between probes was increased in approximately 80% of Eμ− alleles (Figure 4D and E, Table S1). Thus, physical proximity of the VH region to the DH-Cμ region requires Eμ. Interestingly, FISH analysis also showed de-contraction of the interaction between DFL16.1 and the 3′RR on P−E− alleles (Figure 4F, G, Table S1). Taken together, these observations demonstrate the presence of several Eμ-dependent loops in the IgH locus.
Eμ-associated looping sites bind YY1
The basis for lack of IgH locus contraction in YY1-deficient pro-B cells (Liu et al., 2007) is not known. We reasoned that Eμ-bound YY1 (Park and Atchison, 1991) may interact with other YY1-bound sequences to induce locus contraction. We therefore analyzed YY1 binding to key Eμ-associated looping sites by chromatin immunoprecipitation. We detected YY1 binding to sequences 3kb 5′ of DFL16.1 and in HS5-7 of the 3′RR that are involved in Eμ-5′DFL and Eμ-3′RR loops (Figure 5A, blue bars in the part marked D-J-Cμ). These sites also bound CTCF (Featherstone et al., 2010; Garrett et al., 2005), though Eμ itself did not (Figure 5A, red bars). Immunofluorescence studies demonstrated that YY1 and CTCF also co-localized at a subset of nuclear sites (Figure S4). These observations are consistent with Eμ-5′DFL and Eμ-3′RR loops being mediated by homotypic YY1 interactions or by Eμ-bound YY1 interacting with CTCF-bound 5′DFL or 3′RR.
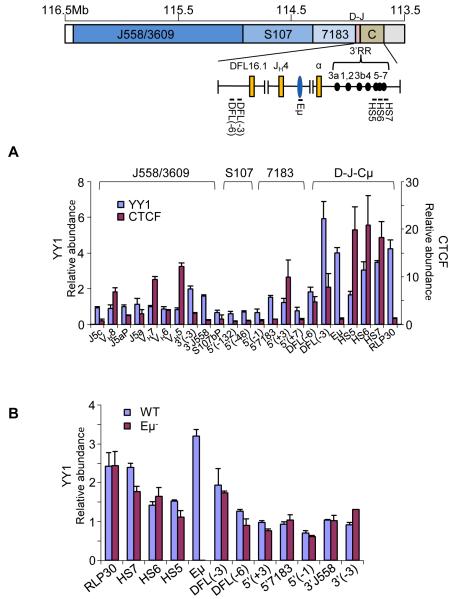
Figure 5. Interaction of YY1 with Eμ-interacting sequences.
(A) Chromatin immunoprecipitations were carried out with anti-YY1 and anti-CTCF antibodies using D345 pro-B cells. ChIP primers from the 3′ region of the IgH locus used are shown on the top line. VH primers assay gene families noted above the bar graph. The relative abundance of specific amplicons in the immunoprecipitate compared to input DNA is shown on the Y axis. Y axis scales differ for YY1 ChIP (left) and CTCF ChIP (right). RPL30 is a positive control for YY1 binding (Liu et al., 2007). HS5-7 correspond to amplicons located within these DNase I hypersensitive sites in the 3′RR, DFL(-3) and DFL(-6) amplicons are located 3 and 6 kb 5′ of DFL16.1, amplicons near 5′7183 and 3′558 are indicated. Data shown is the average of three independent ChIP experiments with each antibody; error bars represent the standard deviation between experiments. See also Figure S4.
(B) Chromatin immunoprecipitation was carried out with anti-YY1 antibody using RAG2-deficient pro-B cell lines carrying WT and Eμ− IgH alleles. Amplicons are as noted in part A. Error bars represent the standard deviation between three independent experiments.
We also detected YY1 binding near 5′7183 and 3′558, but not at several other sites in the VH locus (Figure 5A). However, YY1 binding to the 5′7183 and 3′558 was lower than that at the 3′RR or 5′DFL. The sites that did not bind YY1 were located within different VH gene families at the 5′, middle and 3′ ends of the VH locus. Thus, 5′7183 and 3′558 are distinctly different from other parts of the VH locus with regard to YY1 binding, which may be why they are preferred sites of interaction with Eμ. We cannot rule out the presence of other YY1 binding sites in regions that were not queried by our primer sets. To determine whether Eμ regulates YY1 binding to looping sites, we assayed YY1 binding on Eμ-deficient alleles. We found that YY1 bound normally to all sites, other than Eμ, on Eμ-deficient alleles (Figure 5B). We conclude that Eμ does not regulate YY1 binding to other parts of the locus; rather it provides a YY1 binding site that other YY1- bound parts of the locus can interact with.
IgH locus loops that involve CTCF
The transcription factor CTCF has been implicated in chromosome looping at several loci (Gerasimova et al., 2007; Phillips and Corces, 2009; Wallace and Felsenfeld, 2007). Over sixty CTCF binding sites have been identified in the germline IgH locus (Degner et al., 2009), of which the majority are located within the VH domain. To determine if CTCF is involved in looping of the VH region, we carried out 4C assays using chromatin immunoprecipitated with anti-CTCF antibodies (ChIP-loop). The anchor locations VH3 and VH10 were chosen as representative sites within the proximal and distal VH genes, respectively (Figure 6A).
Figure 6. CTCF-containing loops in the IgH locus.
(A) A schematic of the IgH locus as described in Figure 2A. J558/3609, S107 and 7183 refer to VH gene families. Triangles show positions of oppositely-oriented primers labeled VH3 and VH10 used in ChIP-loop and 4C assays. The nearest VH gene segment to these primers is indicated.
(B) ChIP-loop 4C assays were performed using D345 pro-B cells. Cross-linked chromatin was immunoprecipitated with anti-CTCF antibodies, followed by digestion of the associated chromatin with Nla III. After re-ligation the DNA was amplified with VH3 or VH10 primers. Sequences amplified within VH3 primers (red trace) or VH10 primers (blue trace) were hybridized to Affymetrix chromosome 12 tiling arrays and quantitated as described in Experimental Procedures. Asterisk indicates position of anchor primers; labeled arrows indicate positions of sequences identified in the assay. Data shown is representative of two independent experiments with each anchor primer.
(C) Conventional 4C assay using Nla III restriction enzyme was carried out using VH3 (red trace) or VH10 (blue trace) anchor primers. Asterisks mark the position of anchor primers; arrows indicate interacting regions shared between ChIP-loop and 4C arrays.
(D) CTCF binding to sites identified by anti-CTCF ChIP-loop assays. Cross-linked chromatin from D345 cells was immunoprecipitated with anti-CTCF or anti-Rad21 antibodies. Co-precipitated genomic DNA was amplified with primers close to regions identified in ChIP-loop and 4C assays. VH1, 2, 3, VH3-1 to VH3-5 lie in the cluster of interacting sequences identified with VH3 primers. VH10-1 to VH10-4 correspond to interacting sequences identified with VH10 primers. VH3 and VH10 amplicons are close to the corresponding anchor primers used in 4C assays. CTCF binding to the 3′ DNase I hypersensitive site (3′HS1) in the β globin locus was used as the positive control. CTCF binding within the IgH locus is Eμ-independent (Figure S5A). Error bars represent the standard deviation between three independent experiments.
(E) Top line shows the location of five short probes used in 3D FISH (see also Figure S5B). Three color 3D-FISH was carried out using bone marrow pro-B cells with wild type and P−E− IgH alleles on a RAG2-deficient background. Probes were labeled as follows: a and c, DFL-red, V3-green; b and d, V10-3-red, V10-green. Distances between probes were determined as described in Figure 4C and shown in Table S1. Right panels show quantitation of FISH data as described in Figure 4C, obtained from the analyses of 60-90 nuclei from 2 independent cell preparations. 3C analysis of V3-DFL loops in WT and Eμ− pro-B cells is shown in Figure S5C, D.
VH3 anchor primers identified several regions within 140 kb, as well as sequences 5′ of DFL16.1 located approximately 250 kb away (Figure 6B, red trace arrow DFL (-3)). Because CTCF binds 5′DFL16.1 sequences, we infer that a CTCF-containing loop brings DFL16.1 into the proximity of the VH7183 gene family. VH10 anchor primers identified 4 major interacting sites spanning approximately 500 kb (Figure 6B, blue trace, arrows 1-4). One of these sites (VH10-3) corresponded to a previously identified CTCF binding site (VH8 in (Degner et al., 2009)). To obtain independent evidence that these sequences were involved in chromosome looping we carried out regular 4C using VH3 and VH10 anchor primers. In each case we noted interaction with sites identified in the ChIP-loop assay (Figure 6C, indicated by arrows) as well as sites that were not detected in the ChIP-loop assay. The latter could be due to looping factors other than CTCF, or sites bound weakly by CTCF.
We further tested whether the newly-identified VH3 and VH10-interacting regions bound CTCF. We found that most VH3 interacting sequences bound CTCF at high levels (Figure 6D, VH1-3 and VH3-(1, 3, 4). In contrast, only VH10 and VH10-3 bound CTCF efficiently (Figure 6D), suggesting that loops to VH10-1, 2 and VH10-4 may contain other factors. CTCF binding to these sites did not require Eμ (Figure S5A). Our biochemical data are consistent with separate loops from VH10 to each of VH10-(1-4) leading to 500 kb, 450kb, 287 kb, and 113 kb loops, respectively. Alternatively, the four interacting sites may coalesce to form a ‘hub’ from which 4 loops of 113 kb, 170 kb, 180 kb and 20 kb radiate out. Additional studies are needed to distinguish between these possibilities.
To independently verify proximity between these sequences and to determine whether they required Eμ, we used 3D-FISH to measure spatial distances between DFL16.1 and VH3 and between VH10-3 and VH10 by 3D-FISH. New probes corresponding to looping sites identified by our ChIP-loop 4C assays were prepared by amplification of appropriate BAC templates (Figure 6E top line, labeled DFL, V3, V10-3 and V10). DFL/V3 and V10/V10-3 probe pairs resulted in virtually superimposed FISH signals in primary pro-B cells with WT or P−E− IgH alleles (Figure 6E, Table S1). In contrast, V10-3/h4 probe proximity was disrupted on P−E− alleles (Figure S5B). Quantitation of the distribution of inter-probe distances on WT and P−E− alleles (Figure 6E) or locus compaction (Table S1), revealed no difference between WT and P−E− alleles. Because the comparably sized DFL-3′RR loop undergoes easily discernible locus decontraction on P−E− alleles, we conclude that CTCF-involving loops, such as those between DFL and V3, or between V10 and V10-3, are Eμ-independent. We further confirmed Eμ-independence of the DFL-V3 loop by 3C assays in cells containing WT or Eμ− IgH alleles (Figure S5C, D).
DISCUSSION
The mechanisms by which coordinated chromosomal movements govern gene expression are central to understanding transcriptional regulation. Such movements remove genes from the “repressive” environment of the nuclear periphery, or bring together clusters of genes in transcription factories. Beyond nuclear location, conformational changes within a locus permit interactions between regulatory sequences or help demark independently-regulated chromatin domains. The IgH locus undergoes several forms of chromosomal movements that ensure developmental stage- and lineage-specific DNA recombination and transcription. Here we demonstrate that IgH locus conformation is generated in two steps. The first step generates multi-looped domains whose sizes range from 200-400 kb. The second step brings these domains together to spatially juxtapose the 5′ and 3′ ends of the locus, and thereby generate a fully compacted state. A cis-regulatory element, Eμ, participates in both steps of locus compaction. The functional implications of each chromatin domain are discussed below.
Eμ-dependent 3′ domain
Based on a combination of 3C, 4C and FISH studies we propose that Eμ nucleates a domain that extends from a few kb 5′ of DFL16.1 till the 3′RR. This domain contains all the DH and JH gene segments, and constant region exons. We propose a three-loop configuration for this domain. The smallest 2.8 kb loop between Eμ and PQ52 contains the JH gene segments and DQ52 (Figure 7, middle panel C, colored green). Eμ-PQ52 interaction is indicated by the observed Eμ-dependence of PQ52 transcription and DNase I hypersensitivity. This mini-domain is marked by extremely high levels of H3/H4ac and H3K4me3, high DNase I sensitivity (Chakraborty et al., 2007; Chakraborty et al., 2009; Maes et al., 2001) and RAG1/2 binding (Ji et al., 2010). We suggest that the likely function of this domain is to recruit RAG proteins to the IgH locus to initiate recombination.
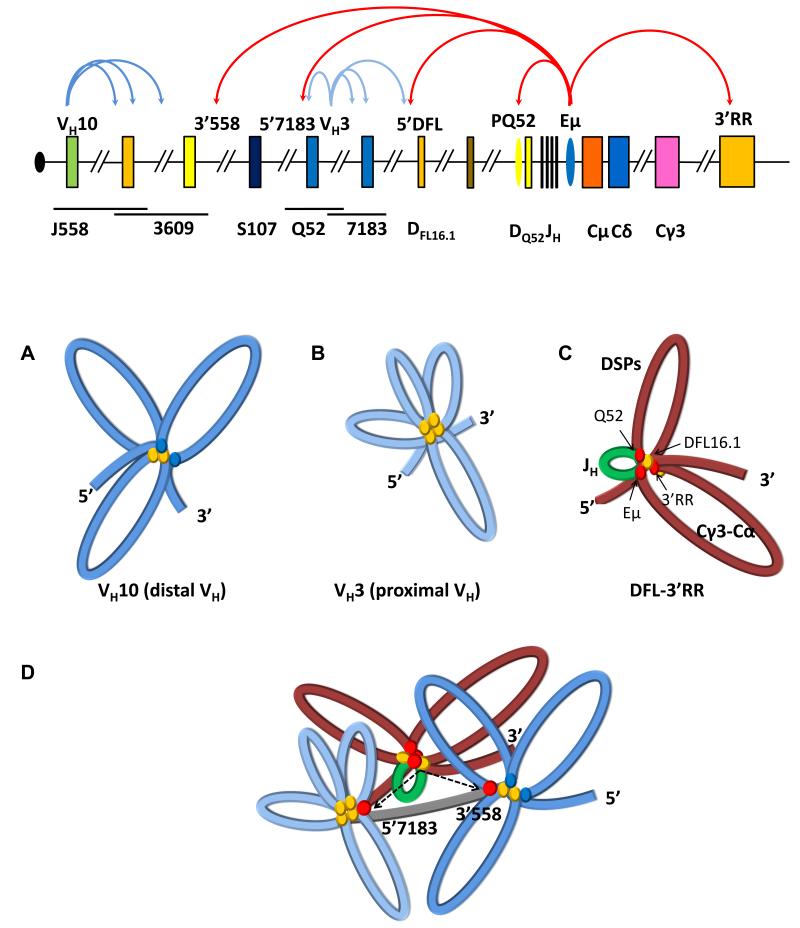
Figure 7. Two levels of chromatin compaction at IgH alleles.
Top line shows a schematic of the unrearranged IgH locus. For description, see Figure 1 legend. Regulatory sequences PQ52 and Eμ are indicated as ovals. Eμ-dependent and Eμ-independent chromatin loops identified in this study are shown as red and blue colored curved arrows, respectively. The first level of chromatin compaction involves the formation of multi-looped domains (middle panel). Three such domains were identified. Those in 5′ VH part of the locus near VH10 and VH3 (A and B, respectively), are Eμ- independent. The number of loops in each domain is inferred from the number of interaction sites as discussed in the text. CTCF binding is indicated by yellow ovals; possible role for factors other than CTCF at VH10 is indicated by blue ovals. At the 3′ end an Eμ-dependent domain (middle panel, part C) extends from sequences 5′ of DFL16.1 (5′DFL) to the 3′RR. A proposed three-loop configuration for this domain is discussed in the text. The smallest loop that contains the 4 JH gene segments and DQ52 is indicated in green because it has the highest levels of activating histone modifications, and binds RAG1 and RAG2 to form a recombination center (Ji et al., 2010). The two other loops that contain DSP gene segments and constant region exons (Cγ3-Cα) (shown in red) are marked with H3K9me2. Red ovals at the base of these loops represent the possible role of YY1 protein in establishing this domain. The second level of chromatin compaction involves the interaction of Eμ with specific sites in the VH region (panel D). We identified two such sites, 5′7183 and 3′558 (top line); both these interaction sites lie outside VH10- and VH3-associated domains, suggesting that the multi-looped structure within each domain does not change with these interactions. Rather Eμ-3′558 and Eμ- 5′7183 interactions bring these domains into the vicinity of the DFL-3′RR domain and, thereby, in physical proximity to the RAG-rich recombination center. Linker regions between identified domains are shown in grey.
A somewhat larger loop, of about 57 kb, is generated by 5′DFL/Eμ interaction. The majority of DH gene segments are sequestered within this mini-domain (Figure 7, middle panel C, smaller red loop labeled DSPs) which is marked by heterochromatic H3K9me2, except near DFL16.1. We propose that DH rearrangements are initiated within this chromatin domain by RAG proteins bound to the JH-associated recombination center. The most readily available DH gene segments in the proposed chromatin configuration are DFL16.1 and DQ52, which are localized at the base of loops tethered to Eμ. Thus, these gene segments recombine preferentially, thereby providing a mechanistic basis for the over-representation of DFL16.1 and DQ52 in V(D)J recombined alleles of B lymphocytes (Subrahmanyam and Sen, 2010). Activation of DSP gene segments for recombination might involve transient association of Eμ with a DSP-associated promoter (Chakraborty et al., 2007) and consequent recruitment of that gene segment to the JH domain.
The largest (206 kb) loop in this region is created by Eμ/3′RR interactions (Figure 7, middle panel C labeled Cγ3-Cα). Its epigenetic features are similar to the intermediate loop in that active histone modifications only occur at the base of the loop at Eμ and 3′RR. The function of this domain, particularly for IgH gene assembly by recombination, is not clear. It is not our intention to imply that a stable 3 loop structure is present in all pro-B cells. Rather, we envisage a dynamic structure where loops between these interaction sites form and break continuously.
Eμ-independent looping within the VH region
Using anti-CTCF ChIP-loop assays we present evidence for multiple cis- interactions in the 5′ region of the IgH locus that contains VH gene segments. Importantly, these loops are Eμ-independent. Three interesting conclusions follow from these observations. First, both sets of interactions identified by anti-CTCF ChIP-loop extend over a few hundred kb. For example, the interacting sites in the VH3 region are spread over approximately 250kb, and in the VH10 region they are spread over approximately 500kb. The striking difference between the two regions is the high density of “peaks” near VH3 and the relatively few “peaks” near VH10. One possibility is that the proximal VH region (near VH3) may be folded into multiple (>6) 30-40 kb loops, whereas the distal VH region (near VH10) may be folded into three 100-150 kb loops (Figure 7, middle panel A). Alternatively, even the VH3 region may be folded into two-three 100-120 kb loops, with the exact configuration of loops being different from cell-to-cell (Figure 7, middle panel B).
Second, CTCF-bound sites near VH10 do not interact with CTCF-bound sites near VH3, or vice-versa. We suggest that this may be because VH3 and VH10 fall in different chromatin domains that do not interact significantly with each other. Though inter-domain interaction is not evident by the assays we have used, it is important to note that both domains are brought close to the DH/JH part of the locus by interacting with Eμ. We note that the VH10-associated domain lies completely within the 5′ part of the IgH locus that contains the newly-identified PAIR elements (Ebert et al., 2011). However, neither VH10 itself nor its associated interaction sites correspond to CTCF binding sites within PAIR elements.
Third, the presence of VH3-associated loops to DFL16.1 implies that at least a subset of proximal VH7183 family members are brought into the vicinity of DFL16.1 in the absence of Eμ-dependent large-scale locus contraction. We surmise that it is these Eμ-independent loops that allow proximal VH recombination to continue in the absence of locus contraction, for example in Pax5- or YY1-deficient pro-B cells (Hesslein et al., 2003; Liu et al., 2007). Furthermore, close examination of the residual VH recombination on Eμ-deleted alleles reveals preferential utilization of proximal VH gene segments (Perlot et al., 2005). These rearrangements are readily explained by Eμ-independent VH3 to DFL16.1 loops identified in this study.
Eμ mediated IgH locus contraction
The location and sizes of three domains described in the preceding sections do not account for IgH locus contraction as defined by FISH studies. We provide evidence for a second level of compaction that occurs via Eμ interaction with specific parts of the VH region. We propose that these latter interactions bring together the two ends of the IgH locus and account for the phenomenon of locus contraction (Figure 7D). The role of Eμ in juxtaposing the 5′ and 3′ parts of the IgH locus provides a reasonable mechanism for the absence of VH recombination on Eμ-deleted alleles (Afshar et al., 2006; Klein et al., 1984; Perlot et al., 2005; Sakai et al., 1999). Notably, Eμ-independent clustering of VH gene segments in domains such as the one near VH10 ensures that each Eμ-dependent interaction with the VH region brings multiple VH gene segments close to the DH-Cμ part of the locus.
The Eμ-interacting regions, 5′7183 and 3′558, are located approximately 400 kb and 1.0 Mb away from Eμ. Because both 5′7183 and 3′558 bind YY1, we propose that locus contraction results from interactions between Eμ-bound YY1 and YY1 bound to these distal sites. Eμ-bound YY1 may also make heterotypic interactions with CTCF bound close to 5′7183. Additionally, interaction of Eμ with 5′7183 or 3′558 may be increased by Eμ-independent compaction of the VH domain by CTCF, or by inter-PAIR interactions in the distal VH part of the IgH locus. The interactions that we identified also provide an explanation for the observed proximity of distal VH gene segments to the very 3′ end of the IgH locus. We suggest that Eμ interaction with 3′558 and the 3′RR draws together the 5′ and 3′ ends of the IgH locus (Figure 7D).
Finally, our studies provide insight into the multi-step process of locus activation, particularly the distinction between Eμ-dependent and Eμ-independent steps. For example, CTCF and YY1 binding to the IgH locus is Eμ-independent. Since Eμ-deficient alleles are located at the nuclear periphery, we infer that lymphoid-restricted binding of these factors can occur at the nuclear periphery. The resulting structure, comprising of VH region loops, may be the basis for the conclusion from trilateration studies that IgH alleles adapt an MLS-compatible conformation in E2A-deficient pre-pro-B cells (Jhunjhunwala et al., 2008). Eμ activation via E2A and Eμ-dependent looping to 5′7183 and 3′558 would reconfigure the locus to deviate away from the MLS model in pro-B cells. Finally, Eμ-dependent generation of the 5′DFL-3′RR domain, and associated RAG-rich recombination center, leads to the fully active state of the locus that is ready to initiate recombination.
EXPERIMENTAL PROCEDURES
Mice and cell lines
JHT (Gu et al., 1993), P−E+ and P−E− mice have been previously described (Afshar et al., 2006). RAG2-deficient mice on 129 or C57BL6 background were purchased from Taconic or maintained at the NIA animal facility. Abelson virus transformed cell lines Eμ− contains a 220 bp deletion of Eμ and lacks recombination activating gene (RAG) 2 (Chakraborty et al., 2009); D345 pro-B cell line contains an inactive RAG1 allele in a C57BL6 background (Ji et al., 2010) and was kindly provided by Dr. David Schatz (Yale University).
Cell purification
Pro-B cells were purified from the bone marrow by positive selection using anti-CD19- coupled magnetic beads (Stem Cell Technologies, BC, Canada) according to the manufacture’s protocol. Thymocytes were prepared by making single cell suspensions of the thymus and filtration through nylon mesh. All procedures were carried out at 4°C. Cell purity and viability was assessed by flow cytometry.
ChIP and transcripts analysis
ChIPs, RT-PCR, Real-Time PCR and data analysis were performed as described (Chakraborty et al., 2009). The following antibodies were used for ChIP: anti-Rad21 (Abcam ab992), anti-CTCF (Millipore 17-10044), and anti-YY1 (Santa Cruz H414). Previously described primers for ChIP and RT-PCR assays were from (Chakraborty et al., 2007) and (Liu et al., 2007). New primers used for ChIP assays are noted in Table S1.
Chromosome conformation capture assay (3C) and circular chromosome conformation capture assay (4C)
(3C) assays were performed as described (Wuerffel et al., 2007) using Hind III to digest cross-linked chromatin. 3C ligation products were measured by the Taqman quantitative PCR technology (Hagege et al., 2007). We normalized 3C results between experiments using the IgH-unrelated β-globin locus long range interaction of 3′HS1 (Splinter et al., 2006). PCR control fragments for determination of primer efficiency of each primer combination (Table S2) were generated using genomic DNA from the regions of interest as described (Nativio et al., 2009) or BAC clones covering the genomic segments under study.
For 4C assays, crosslinked chromatin was digested with Mse I, or Nla III, followed by religation for 3 days. After reversal of crosslinking by incubation at 65°C overnight, genomic DNA was purified and nested PCR carried with different anchor primer pairs (Table S2) as described (Gondor et al., 2008). PCR products were fragmented, labeled with biotin and hybridized to the Affymetrix mouse GenChip Mouse tiling 2.0 R E array according to the manufacturer’s specifications. Data normalization and enriched region detection was performed using CisGenome (Ji et al., 2008) with default parameters. Significantly enriched regions were determined with one-tail t-test statistics. Moving averages of normalized log2 ratio between sample and input were calculated using the msProcess Package of bioconductor (www.bioconductor.org) and plotted along chromosomal coordinates (mm8) for visualization.
ChIP-Loop 4C
Partially sonicated formaldehyde-crosslinked chromatin was incubated overnight at 4°C with anti-CTCF antibody complexes to Dynabeads (Invitrogen, CA) at 4°C. DNA-Dynabead complexes were washed extensively with restriction enzyme buffer followed by incubaton with Mse I or Nla III. Further work-up was carried out as described for 4C.
Fluorescent in Situ Hybridization (FISH) and Immunolocalization
Locus specific DNA probes for FISH were prepared from BACs RP23-230L2 and RP23- 458I14 (Invitrogen, CA). BAC probes were labeled by nick translation using ChromaTide Alexa Fluor 568-4 dUTP (red) and ChromaTide Alexa Fluor 488-5 dUTP (green) (Invitrogen, CA) (Sayegh et al., 2005).
Position-specific 10 kb probes were generated by PCR using BAC templates with primers listed in Table S1. Probes h4, h11 as well as BAC RP23-201H14 were kindly provided by Dr. Cornelis Murre (UCSD). FISH with 10 kb probes were performed as described (Jhunjhunwala et al., 2008) using a Nikon T2000 microscope equipped with a 100x lens and motorized 100 μm Piezo Z-stage (Applied Scientific Instrumentation, OR).
Depending on the size of the nucleus 30-40 serial optical sections spaced by 0.2μm were acquired. The data sets were deconvolved using NIS-Elements software (Nikon, NY) and optical sections merged to produce 3D images. The spatial distance between probes was measured as described (Jhunjhunwala et al., 2008).
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Cornelis Murre and Suchit Jhunjhunwala for sharing and troubleshooting the procedures for small probe FISH. We thank Drs. Dinah Singer, Amy Kenter, Cornelis Murre, Fred Alt and Sebastian Fugmann for discussions throughout the work and critical appraisal of the manuscript. This work was supported by the Intramural Research Program of the National Institute on Aging (Baltimore, MD) and by NIH grant (P01 HL68744 and CA100905) to EMO.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Afshar R, Pierce S, Bolland DJ, Corcoran A, Oltz EM. Regulation of IgH gene assembly: role of the intronic enhancer and 5′DQ52 region in targeting DHJH recombination. J Immunol. 2006;176:2439–2447. doi: 10.4049/jimmunol.176.4.2439. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Chowdhury D, Keyes A, Jani A, Subrahmanyam R, Ivanova I, Sen R. Repeat organization and epigenetic regulation of the DH-Cmu domain of the immunoglobulin heavy-chain gene locus. Mol Cell. 2007;27:842–850. doi: 10.1016/j.molcel.2007.07.010. [DOI] [PubMed] [Google Scholar]
- Chakraborty T, Perlot T, Subrahmanyam R, Jani A, Goff PH, Zhang Y, Ivanova I, Alt FW, Sen R. A 220-nucleotide deletion of the intronic enhancer reveals an epigenetic hierarchy in immunoglobulin heavy chain locus activation. J Exp Med. 2009;206:1019–1027. doi: 10.1084/jem.20081621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook PR, Brazell IA, Jost E. Characterization of nuclear structures containing superhelical DNA. J Cell Sci. 1976;22:303–324. doi: 10.1242/jcs.22.2.303. [DOI] [PubMed] [Google Scholar]
- Degner SC, Wong TP, Jankevicius G, Feeney AJ. Cutting edge: developmental stage-specific recruitment of cohesin to CTCF sites throughout immunoglobulin loci during B lymphocyte development. J Immunol. 2009;182:44–48. doi: 10.4049/jimmunol.182.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert A, McManus S, Tagoh H, Medvedovic J, Salvagiotto G, Novatchkova M, Tamir I, Sommer A, Jaritz M, Busslinger M. The distal V(H) gene cluster of the Igh locus contains distinct regulatory elements with Pax5 transcription factor-dependent activity in pro-B cells. Immunity. 2011;34:175–187. doi: 10.1016/j.immuni.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Featherstone K, Wood AL, Bowen AJ, Corcoran AE. The mouse immunoglobulin heavy chain V-D intergenic sequence contains insulators that may regulate ordered V(D)J recombination. J Biol Chem. 2010;285:9327–9338. doi: 10.1074/jbc.M109.098251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M. Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev. 2004;18:411–422. doi: 10.1101/gad.291504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrett FE, Emelyanov AV, Sepulveda MA, Flanagan P, Volpi S, Li F, Loukinov D, Eckhardt LA, Lobanenkov VV, Birshtein BK. Chromatin architecture near a potential 3′ end of the igh locus involves modular regulation of histone modifications during B-Cell development and in vivo occupancy at CTCF sites. Mol Cell Biol. 2005;25:1511–1525. doi: 10.1128/MCB.25.4.1511-1525.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser SM, Laemmli UK. A glimpse at chromosomal order. Trends in Genetics. 1987;3:16–22. [Google Scholar]
- Gerasimova TI, Lei EP, Bushey AM, Corces VG. Coordinated control of dCTCF and gypsy chromatin insulators in Drosophila. Mol Cell. 2007;28:761–772. doi: 10.1016/j.molcel.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gondor A, Rougier C, Ohlsson R. High-resolution circular chromosome conformation capture assay. Nat Protoc. 2008;3:303–313. doi: 10.1038/nprot.2007.540. [DOI] [PubMed] [Google Scholar]
- Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Hagege H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forne T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- Hesslein DG, Pflugh DL, Chowdhury D, Bothwell AL, Sen R, Schatz DG. Pax5 is required for recombination of transcribed, acetylated, 5′ IgH V gene segments. Genes Dev. 2003;17:37–42. doi: 10.1101/gad.1031403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH. An integrated software system for analyzing ChIP-chip and ChIP-seq data. Nat Biotechnol. 2008;26:1293–1300. doi: 10.1038/nbt.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y, Resch W, Corbett E, Yamane A, Casellas R, Schatz DG. The in vivo pattern of binding of RAG1 and RAG2 to antigen receptor loci. Cell. 2010;141:419–431. doi: 10.1016/j.cell.2010.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221–4234. doi: 10.4049/jimmunol.176.7.4221. [DOI] [PubMed] [Google Scholar]
- Ju Z, Volpi SA, Hassan R, Martinez N, Giannini SL, Gold T, Birshtein BK. Evidence for physical interaction between the immunoglobulin heavy chain variable region and the 3′ regulatory region. J Biol Chem. 2007;282:35169–35178. doi: 10.1074/jbc.M705719200. [DOI] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA. Chromatin loops in gene regulation. Biochim Biophys Acta. 2009;1789:17–25. doi: 10.1016/j.bbagrm.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Sablitzky F, Radbruch A. Deletion of the IgH enhancer does not reduce immunoglobulin heavy chain production of a hybridoma IgD class switch variant. EMBO J. 1984;3:2473–2476. doi: 10.1002/j.1460-2075.1984.tb02158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch TA, Munkel C, Langowski J, editors. Three-dimensional of chromosome territories in human interphase nucleus. Spriner; Heidelberg, Germany: 2000. [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H. Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science. 2002;296:158–162. doi: 10.1126/science.1068768. [DOI] [PubMed] [Google Scholar]
- Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Schmidt-Supprian M, Shi Y, Hobeika E, Barteneva N, Jumaa H, Pelanda R, Reth M, Skok J, Rajewsky K, Shi Y. Yin Yang 1 is a critical regulator of B-cell development. Genes Dev. 2007;21:1179–1189. doi: 10.1101/gad.1529307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes J, O’Neill LP, Cavelier P, Turner BM, Rougeon F, Goodhardt M. Chromatin remodeling at the Ig loci prior to V(D)J recombination. J Immunol. 2001;167:866–874. doi: 10.4049/jimmunol.167.2.866. [DOI] [PubMed] [Google Scholar]
- Marsden MP, Laemmli UK. Metaphase chromosome structure: evidence for a radial loop model. Cell. 1979;17:849–858. doi: 10.1016/0092-8674(79)90325-8. [DOI] [PubMed] [Google Scholar]
- Nativio R, Wendt KS, Ito Y, Huddleston JE, Uribe-Lewis S, Woodfine K, Krueger C, Reik W, Peters JM, Murrell A. Cohesin is required for higher-order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 2009;5:e1000739. doi: 10.1371/journal.pgen.1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park K, Atchison ML. Isolation of a candidate repressor/activator, NF-E1 (YY-1, delta), that binds to the immunoglobulin kappa 3′ enhancer and the immunoglobulin heavy-chain mu E1 site. Proc Natl Acad Sci U S A. 1991;88:9804–9808. doi: 10.1073/pnas.88.21.9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulson JR, Laemmli UK. The structure of histone-depleted metaphase chromosomes. Cell. 1977;12:817–828. doi: 10.1016/0092-8674(77)90280-x. [DOI] [PubMed] [Google Scholar]
- Perlot T, Alt FW. Cis-regulatory elements and epigenetic changes control genomic rearrangements of the IgH locus. Adv Immunol. 2008;99:1–32. doi: 10.1016/S0065-2776(08)00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlot T, Alt FW, Bassing CH, Suh H, Pinaud E. Elucidation of IgH intronic enhancer functions via germ-line deletion. Proc Natl Acad Sci U S A. 2005;102:14362–14367. doi: 10.1073/pnas.0507090102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JE, Corces VG. CTCF: master weaver of the genome. Cell. 2009;137:1194–1211. doi: 10.1016/j.cell.2009.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Retter I, Chevillard C, Scharfe M, Conrad A, Hafner M, Im TH, Ludewig M, Nordsiek G, Severitt S, Thies S, et al. Sequence and characterization of the Ig heavy chain constant and partial variable region of the mouse strain 129S1. J Immunol. 2007;179:2419–2427. doi: 10.4049/jimmunol.179.4.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachs RK, van den Engh G, Trask B, Yokota H, Hearst JE. A random-walk/giant-loop model for interphase chromosomes. Proc Natl Acad Sci U S A. 1995;92:2710–2714. doi: 10.1073/pnas.92.7.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai E, Bottaro A, Davidson L, Sleckman BP, Alt FW. Recombination and transcription of the endogenous Ig heavy chain locus is effected by the Ig heavy chain intronic enhancer core region in the absence of the matrix attachment regions. Proc Natl Acad Sci U S A. 1999;96:1526–1531. doi: 10.1073/pnas.96.4.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayegh CE, Jhunjhunwala S, Riblet R, Murre C. Visualization of looping involving the immunoglobulin heavy-chain locus in developing B cells. Genes Dev. 2005;19:322–327. doi: 10.1101/gad.1254305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson EM, Linder CC, Sargent EE, Davisson MT, Mobraaten LE, Sharp JJ. Genetic variation among 129 substrains and its importance for targeted mutagenesis in mice. Nat Genet. 1997;16:19–27. doi: 10.1038/ng0597-19. [DOI] [PubMed] [Google Scholar]
- Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subrahmanyam R, Sen R. RAGs’ eye view of the immunoglobulin heavy chain gene locus. Semin Immunol. 2010;22:337–345. doi: 10.1016/j.smim.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Takizawa T, Meaburn KJ, Misteli T. The meaning of gene positioning. Cell. 2008;135:9–13. doi: 10.1016/j.cell.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace JA, Felsenfeld G. We gather together: insulators and genome organization. Curr Opin Genet Dev. 2007;17:400–407. doi: 10.1016/j.gde.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuerffel R, Wang L, Grigera F, Manis J, Selsing E, Perlot T, Alt FW, Cogne M, Pinaud E, Kenter AL. S-S synapsis during class switch recombination is promoted by distantly located transcriptional elements and activation-induced deaminase. Immunity. 2007;27:711–722. doi: 10.1016/j.immuni.2007.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.