Abstract
Two-component systems (TCS) are major signal transduction pathways that allow bacteria to detect and respond to environmental and intracellular changes. A group of TCS has been shown to be involved in the response against antimicrobial peptides (AMPs). These TCS are characterized by the possession of intramembrane-sensing histidine kinases, and they are usually associated with ABC transporters of the peptide-7 exporter family (Pep7E). Lactobacillus casei BL23 encodes two TCS belonging to this group (TCS09 and TCS12) that are located next to two ABC transporters (ABC09 and ABC12), as well as a third Pep7E ABC transporter not genetically associated with any TCS (orphan ABC). This study addressed the involvement of modules TCS09/ABC09 and TCS12/ABC12 in AMP resistance. Results showed that both systems contribute to L. casei resistance to AMPs, and that each TCS constitutes a functional unit with its corresponding ABC transporter. Analysis of transcriptional levels showed that module 09 is required for the induction of ABC09 expression in response to nisin. In contrast, module 12 controls a wider regulon that encompasses the orphan ABC, the dlt operon (d-alanylation of teichoid acids), and the mprF gene (l-lysinylation of phospholipids), thereby controlling properties of the cell envelope. Furthermore, the characterization of a dltA mutant showed that Dlt plays a major role in AMP resistance in L. casei. This is the first report on the regulation of the response of L. casei to AMPs, giving insight into its ability to adapt to the challenging environments that it encounters as a probiotic microorganism.
INTRODUCTION
Lactobacillus casei is a facultatively heterofermentative lactic acid bacterium commonly found in foodstuffs, plant material, and the oral cavity, gastrointestinal tract, and genital tract of humans and animals. It is also used as a starter culture in the food industry, and some strains are considered probiotics (1, 2). It is generally agreed that probiotic microorganisms must be able to reach their place of action in a viable and active state in order to exert their beneficial effects on the host. They face a great variety of physicochemical stresses during industrial production, processing, and storage, as well as during their passage through the gastrointestinal tract (2, 3). Furthermore, the intestine is a challenging habitat for probiotics due to competition with the resident microbiota and the action of the host defenses. Among other factors, antimicrobial peptides (AMPs) produced both by the innate immune system of the host (4–8) and by other microorganisms present in the gastrointestinal tract (9, 10) constitute a major challenge for survival in the intestine.
AMPs usually consist of 12 to 50 amino acids with a net positive charge of +2 to +7; thus, they are called cationic antimicrobial peptides. They are amphipathic molecules, which makes them selective toward interaction with and insertion into the negatively charged bacterial membranes (11). The production of cationic AMPs has been reported in virtually all groups of organisms, including bacteria, fungi, plants, and animals (12). Epithelial and immune system cells in the intestine segregate cationic AMPs to the intestinal lumen (6). Together with other immune system effectors, they constitute the first line of innate host defenses in the mucosal surfaces (4–7). In addition to host-produced cationic AMPs, members of the resident microbiota also produce AMPs known as bacteriocins. Bacteriocins are ribosomally synthesized, heat-stable antimicrobial peptides produced by bacteria (9, 13). They can have a broad or a narrow spectrum of action, and the producer strains usually express specific self-protective mechanisms against them (9, 13). Most AMPs target essential cell envelope structures (6, 13). For example, nisin, a lantibiotic produced by strains of Lactococcus lactis, presents a dual mode of antimicrobial activity: it binds with high affinity to the sugar-pyrophosphate moiety of the bacterial cell wall precursor lipid II and uses it as a docking molecule to cause inhibition of cell wall biosynthesis and pore formation in the bacterial membrane (14–16).
On the other hand, bacteria have evolved mechanisms to detect and elicit a resistance response against AMPs. Among them, two-component signal transduction systems (TCS) play a major role in these processes (17–21). TCS are typically constituted by a membrane-bound histidine kinase that acts as a signal sensor/transducer and a response regulator that usually acts as a transcription activator/repressor (22–25). Histidine kinases monitor environmental signals and, in response to a stimulus, autophosphorylate at a highly conserved histidine residue (H-box). The high-energy phosphate group is subsequently transferred to an aspartyl residue on the response regulator receiver domain. Phosphorylation of the response regulator in turn modulates the activity of the response regulator effector domain (22).
A particular group of TCS found in some Firmicutes has been shown to be involved in the response to cell envelope stress exerted mainly by peptide antibiotics (17, 26–28). The histidine kinases of these TCS belong to the intramembrane-sensing histidine kinase subfamily (29) and are characterized by the possession of two transmembrane helices with a short extracellular linker and no cytoplasmic domains besides the H-box and the kinase domain. These systems are typically associated with ATP-binding cassette (ABC) transporters of the peptide-7 exporter (Pep7E) family (17, 26). These ABC transporters consist of an ATPase subunit and a permease with 10 transmembrane helices and a large (202 amino acids) extracellular domain between helices 7 and 8. Interestingly, the characterization of some of the TCS and cognate ABC transporters, such as BceRS/BceAB of Bacillus subtilis (21, 30–32) and BceABRS of Streptococcus mutans (33), has shown that the histidine kinase alone is unable to detect the presence of the antimicrobial peptide, and that it requires the ABC transporter for signaling. Therefore, TCS and ABC transporters work as functional units, here termed AMP resistance modules.
A previous survey of the TCS encoded by L. casei BL23 identified two gene clusters encoding TCS homologous to BceRS of B. subtilis: TCS09 (LCABL_16420/16430) and TCS12 (LCABL_19600/19610). Both of these were associated with genes encoding putative ABC transporters (ABC09 [LCABL_16400/16410] and ABC12 [LCABL_19580/19590]) (Fig. 1) (34). In this previous study, inactivation of the corresponding response regulators led to increased sensitivity to antimicrobial peptides, such as bacitracin and nisin (34). Furthermore, it was observed that a strain defective in RR12 displayed a pleiotropic phenotype of greater sensitivity to environmental stresses, like presence of bile, acidic pH, and high temperatures (34). Together, these data suggested that these systems are involved in the cell envelope stress response of L. casei BL23. This prompted us to investigate in detail the functional role of these TCS and their cognate ABC transporters in L. casei and the possible regulatory links between them. The results presented here show that TCS09/ABC09 is a detoxification module involved specifically in resistance to AMPs, whereas TCS12/ABC12 controls a larger regulon involved in the maintenance of the physicochemical properties of the cell envelope.
Fig 1.
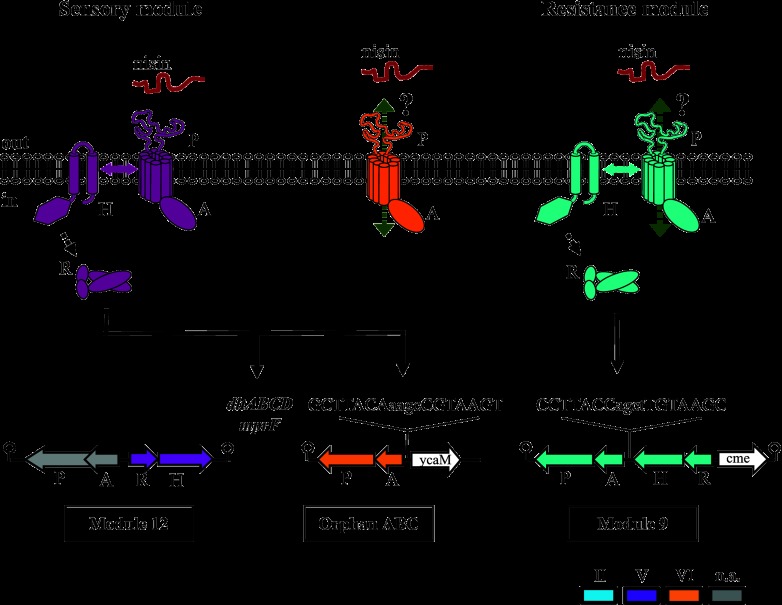
Schematic representation of the signaling network of BceRS/BceAB-like sensory and resistance modules of L. casei BL23. TCS and ABC transporters are colored according to their phylogenetic group assignment under the classification system of Dintner et al. (17) (n.a., no group assigned). Unrelated genes present in the operons are shown as white arrows. Sequences of putative response regulator binding sites upstream of OrATP and ATP09 are shown; the 7-bp inverted repeats are highlighted in capital letters. Nisin is shown as a substrate interacting with the permeases. Putative transport of nisin by the permeases is shown by green double-headed dotted arrows. Signal transfer between TCS09 and ABC09 and between TCS12 and ABC12 is indicated in the membrane bilayer. Putative phosphotransfer between the histidine kinases and response regulators is shown by black dotted arrows. Transcriptional activation is shown by black arrows; additional target genes for module 12 are listed. P, A, R, and H stand for permease, ATPase, response regulator, and histidine kinase, respectively.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. Escherichia coli DH10B was used as an intermediate host for cloning purposes. E. coli strains were grown in LB medium at 37°C with aeration. L. casei strains were grown in MRS broth (Oxoid) at 37°C under static conditions. The corresponding solid media were prepared by adding 1.5% (wt/vol) agar. Strains were stored at −80°C in their corresponding growth media containing 20% (vol/vol) glycerol. Antibiotics used were 100 μg ml−1 ampicillin for E. coli and 5 μg ml−1 erythromycin for L. casei.
Table 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Source or reference |
|---|---|---|
| Strains | ||
| E. coli DH10B | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80dlacZΔM15 ΔlacX74 endA1 recA1 deoR Δ(ara, leu)7697 araD139 galU galK nupG rpsL λ− | Stratagene |
| L. casei BL23 | Wild type | B. Chassy, University of Illinois |
| L. casei ΔRR09 | BL23 ΔLCABL_16430 | This study |
| L. casei ΔRR12 | BL23 Δrrp1 (LCABL_19600) | 34 |
| L. casei DLT | LCABL_08550 (dltA) mutant; pRV08550; Eryr | This study |
| L. casei P09 | LCABL_16400 mutant; pRV16400; Eryr | This study |
| L. casei P12 | LCABL_19580 mutant; pRV19580; Eryr | This study |
| L. casei MPRF | LCABL_24490 mutant; pRV24490; Eryr | This study |
| Plasmids | ||
| pRV300 | Insertional vector for Lactobacillus; Ampr, Eryr | 35 |
| pRVRR09del | pRV300 containing fused flanking fragments upstream and downstream of LCABL_16430 | This study |
| pRV08550 | pRV300 containing a 679-bp internal fragment of LCABL_08550 (dltA) | This study |
| pRV16400 | pRV300 containing a 975-bp internal fragment of LCABL_16400 | This study |
| pRV19580 | pRV300 containing a 773-bp internal fragment of LCABL_19580 | This study |
| pRV24490 | pRV300 containing a 1,333-bp internal fragment of LCABL_24490 | This study |
Ampr, ampicillin resistance; Eryr, erythromycin resistance.
Comparative genomics and motif-based searches for putative Bce-like response regulator binding sites.
The phylogenetic classification of BceRS-like TCS and BceAB-like ABC transporters by Dintner and coworkers (17) included the permeases and histidine kinases of L. casei ATCC 334 but not those of L. casei BL23. Since these two strains are highly homologous (36, 37), we performed a nucleotide BLAST (http://blast.ncbi.nlm.nih.gov/) of the L. casei BL23 genome using the previously identified genes of L. casei ATCC 334 as queries. Genes with 98 to 99% identity were identified in all cases (LCABL_16400 to LSEI_1417, LCABL_16420 to LSEI_1419, LCABL_19580 to LSEI_1738, LCABL_19610 to LSEI_1741, and LCABL_21670 to LSEI_1993). Therefore, all L. casei BL23 proteins were assigned to the same phylogenetic group as those of L. casei ATCC 334.
Putative binding sites for the response regulators were identified by a manual search of candidate promoter regions for sequences with similarity to the described binding consensus TNACA-N4-TGTAA (17).
Construction of mutants.
Standard methods were used for cloning in E. coli (38). Oligonucleotides used in this study are listed in Table S1 in the supplemental material. E. coli strains were transformed by electroporation with a Gene Pulser apparatus (Bio-Rad), as recommended by the manufacturer, and L. casei strains were transformed as described previously (39).
L. casei mutant strains DLT, MPRF, P09, and P12 were obtained by insertional inactivation of the corresponding genes (Table 1). Primers were designed to amplify regions of the L. casei BL23 genome corresponding to an internal region of each gene. In order to generate knockout mutants in dltA and mprF genes, the corresponding PCR products were digested with HindIII/SpeI and ligated to the integrative vector pRV300 (35) digested with the same enzymes. For mutants L. casei P09 and L. casei P12, the PCR products were cloned into pRV300 vector linearized with KpnI using a CloneEZ PCR cloning kit (GenScript) by following the manufacturer's instructions. The resulting constructs (Table 1) were transformed into E. coli DH10B, verified by restriction analysis, and used to transform L. casei BL23. Single-crossover integrants were selected by resistance to erythromycin, and insertional inactivation of the target gene was confirmed by PCR with a primer hybridizing to an external region of the cloned fragment in the genome and a primer hybridizing to the vector.
In order to obtain a BL23-derived strain harboring a complete deletion of gene LCABL_16430 (ΔRR09), flanking fragments of the region to be deleted were amplified using primer sets RG058/RG059 and RG060/RG061. The resulting PCR products were fused by PCR; the fusion product was digested with EcoRI/XhoI and ligated into pRV300 digested with the same enzymes. The resulting construct (Table 1) was transformed into E. coli DH10B, verified by restriction analysis and by DNA sequencing, and subsequently introduced into L. casei BL23 by electroporation. Single-crossover integrations were checked as described before. One single-crossover integrant was grown in MRS without erythromycin for approximately 200 generations in order to obtain a second crossover recombination, leading to the loss of the gene of interest. Cells were plated on MRS and replica plated on MRS plus erythromycin. Antibiotic-sensitive clones were isolated, and excision of the pRV300 derivatives leading to deletion of the chromosomal region of interest was checked by PCR and confirmed by sequencing of PCR-amplified fragments spanning the deleted regions.
Cytochrome c binding assay.
Comparison of the whole-cell surface charges of the wild-type strain and mutants ΔRR12, P12, DLT, MPRF, and ΔRR09 was performed by a cytochrome c binding assay as described elsewhere (40–42) and modified as follows. Bacterial cells at stationary phase were harvested by centrifugation and washed twice with one volume of 20 mM MOPS (morpholinepropanesulfonic acid), pH 7. The cells were resuspended in the same buffer at a final concentration of 1010 CFU ml−1 and incubated with 150 μg ml−1 cytochrome c (Sigma) for 10 min at room temperature. The mixture was centrifuged twice, and the absorbance of the supernatant (containing unbound cytochrome c) was determined at 530 nm. The binding ratio was calculated by comparing the absorbance of each supernatant after incubation with the cells relative to the absorbance of the cytochrome c solution without bacterial cells. Results shown are the means and standard deviations from three independent experiments.
Phenotypic characterization of L. casei BL23 and mutants.
The growth of L. casei BL23 and mutants ΔRR12, P12, DLT, and MPRF under different stress conditions was determined as previously described (34) and monitored as the optical density at 595 nm (OD595). Briefly, cells from frozen stocks were inoculated onto MRS agar plates and incubated at 37°C. Single colonies were grown in MRS until stationary phase. Cells were harvested by centrifugation and washed twice with two volumes of 0.1% (wt/vol) peptone-water. Cells were inoculated to a final OD595 of 0.05 in 250 μl of the corresponding medium and dispensed in 96-well microtiter plates. Growth was monitored for 20 h by changes in the OD595. The conditions tested were reference conditions (MRS at 37°C without shaking), MRS adjusted to pH 4 with HCl, MRS buffered with 0.1 M phosphate buffer (pH 6.8), MRS supplemented with 0.5% (wt/vol) bile, and growth in MRS at 42°C. No antibiotics were used in the growth assays. No revertants to the wild-type genotype were detected under these experimental conditions. At least three independent replicates of each growth curve were obtained.
The MIC of the cell wall-acting AMPs bacitracin, mersacidin, nisin, plectasin, vancomycin, and subtilin for all mutants was determined in MRS using different concentrations of the antimicrobial agents. The assays were performed in 96-well microtiter plates inoculated and incubated as indicated above. The MIC was defined as the lowest concentration of antimicrobial agent needed to completely inhibit the growth of the bacterial strain at 15 h. All experiments were performed in duplicate.
Reverse transcription and qRT-PCR.
Samples for RNA isolation were collected as follows. Overnight cultures from single colonies of L. casei BL23 and defective mutants were used to inoculate 100 ml MRS medium to a final OD595 of 0.06. Cells were grown at 37°C to an OD595 of 0.5. Each culture was split into two halves, and nisin, at a sublethal concentration, was added to one half, leaving the other half untreated (control). Incubation was continued for 10 min, and three samples of 10 ml were taken from each culture. The cells were harvested by centrifugation (5,000 × g, 10 min, 4°C) and washed with 1 volume of cold 50 mM EDTA (pH 8.0), and the bacterial pellets were frozen at −80°C until use. The nisin concentrations used in the induction assays were 22.5 and 750 ng ml−1 for L. casei BL23 and 22.5 ng ml−1 for the defective mutants. Isolation of total RNA from L. casei strains, synthesis of cDNA, primer design, and quantitative real-time PCR (qRT-PCR) were carried out as described previously (43). Primers used are listed in Table S1 in the supplemental material. The lepA, ileS, pyrG, and pcrA sequences were selected from a set of 10 reference genes (43) by using the geNorm application (44). The relative expression based on the expression ratio between the target genes and reference genes was calculated using the software tool REST (45). Linearity and amplification efficiency were determined for each primer pair. Every real-time PCR determination was performed at least six times.
RESULTS
L. casei BL23 encodes two BceRS-like TCS and three BceAB-like ABC transporters in its genome.
Two out of the 17 TCS encoded by L. casei BL23 were shown to belong to the Bce-like TCS group: TCS09 and TCS12 (34). They possess intramembrane-sensing histidine kinases with 4- and 9-amino-acid extracellular loops (HK09 and HK12, respectively) between the transmembrane helices. Comparative genomics analyses showed that L. casei BL23 possesses three BceAB-like ABC transporters in its genome: ABC09 and ABC12, located adjacent to TCS09 and TCS12, respectively, and a third one not genetically associated with any TCS, termed orphan ABC (OrABC) (Fig. 1; also see Table S2 in the supplemental material). According to the phylogenetic classification of Dintner et al. (17) of BceRS-like TCS and BceAB-like ABC transporters, TCS09 and ABC09 belong to group II, TCS12 belongs to group V, ABC12 could not be assigned to any group, and OrABC belongs to group VI. This phylogenetic group assignment raised several questions. On one hand, the fact that both components of module 09 (TCS09 and ABC09) belonged to the same phylogenetic group led us to hypothesize that they could work as a functional unit, as has been previously described for homologous modules of B. subtilis and Staphylococcus aureus (27). On the other hand, we wondered if module 12 could also be a functional unit, since the signal transfer would occur between a TCS and an ABC transporter from different groups. Finally, OrABC belongs to group VI, which is unique because members of that group are never associated with a TCS. Most of these orphan ABCs contain a putative response regulator binding site in their promoter regions, but so far it has not been determined how their expression is regulated. This study aims to provide answers to these questions.
Inactivation of Bce-like TCS systems or their cognate ABC transporters results in increased sensitivity to antimicrobial compounds.
The homology of modules 09 and 12 to proteins involved in AMP resistance, together with experimental evidence previously obtained, strongly suggested that these systems are involved in AMP resistance in L. casei as well (34). Furthermore, the genetic organization also suggested that TCS09/ABC09 and TCS12/ABC12 work as functional units, as previously described for other BceAB/BceRS homologous systems. In order to ascertain these points, a collection of mutant strains defective in the response regulators of the TCS and the permease subunits of the cognate ABC transporters were obtained, and the sensitivity of the wild type and all of the mutants to AMPs (bacitracin, nisin, mersacidin, plectasin, subtilin, and vancomycin) was tested.
Mutant L. casei ΔRR12 had been previously obtained (34). The RR09-encoding gene is located immediately upstream of the HK09 and ABC09 genes (Fig. 1). In order to avoid polar effects of the mutation of RR09 on the expression of the genes located downstream, a deletion mutant (L. casei ΔRR09) was obtained. Mutants L. casei P09 and L. casei P12 were constructed by insertional inactivation, since the genes encoding permease 09 and permease 12 are located at the end of their corresponding gene clusters (Fig. 1), thus polar effects of the insertion on the expression of downstream genes were unlikely.
The sensitivity of the mutants to AMPs was estimated by determining the MICs of bacitracin, mersacidin, nisin, plectasin, subtilin, and vancomycin (Table 2). Strain P09 was more sensitive to bacitracin, nisin, plectasin, and subtilin (MICs were approximately 2-fold lower than those for the wild-type strain; Table 2). Strain ΔRR09 displayed the same phenotype as P09. These results suggest that TCS09 and ABC09 work together as a functional unit in mediating AMP resistance.
Table 2.
MICs of AMPs against L. casei BL23 and derivative strains
| Strain | MIC of: |
|||||
|---|---|---|---|---|---|---|
| Bacitracin (μg ml−1) | Nisin (μg ml−1) | Vancomycin (mg ml−1) | Plectasin (μg ml−1) | Mersacidin (μg ml−1) | Subtilin (%) | |
| BL23 | 10 | 0.5 | 1.7 | 40 | 10 | 25 |
| P09 | 5 | 0.3 | 1.7 | 20 | 10 | 10 |
| ΔRR09 | 5 | 0.3 | 1.7 | 20 | 10 | 10 |
| P12 | 5 | 0.08 | 0.4 | 2.5 | 2.5 | 10 |
| ΔRR12 | 5 | 0.08 | 0.4 | 2.5 | 2.5 | 10 |
| DLT | 10 | 0.04 | 0.4 | 2.5 | 2.5 | 10 |
| MPRF | 5 | 0.5 | 1.7 | >40 | 5 | 20 |
On the other hand, strain P12 was more sensitive than the wild-type strain to all of the AMPs tested (MICs were 2-fold lower for bacitracin and subtilin, 4-fold lower for vancomycin and mersacidin, 6-fold lower for nisin, and 16-fold lower for plectasin; Table 2). Interestingly, mutant ΔRR12 displayed the same phenotype as P12 (Table 2), suggesting that they also constitute a functional unit that mediates AMP resistance, even though TCS12 and ABC12 do not belong to the same phylogenetic group.
Transcriptional response of L. casei BL23 and derived strains to nisin.
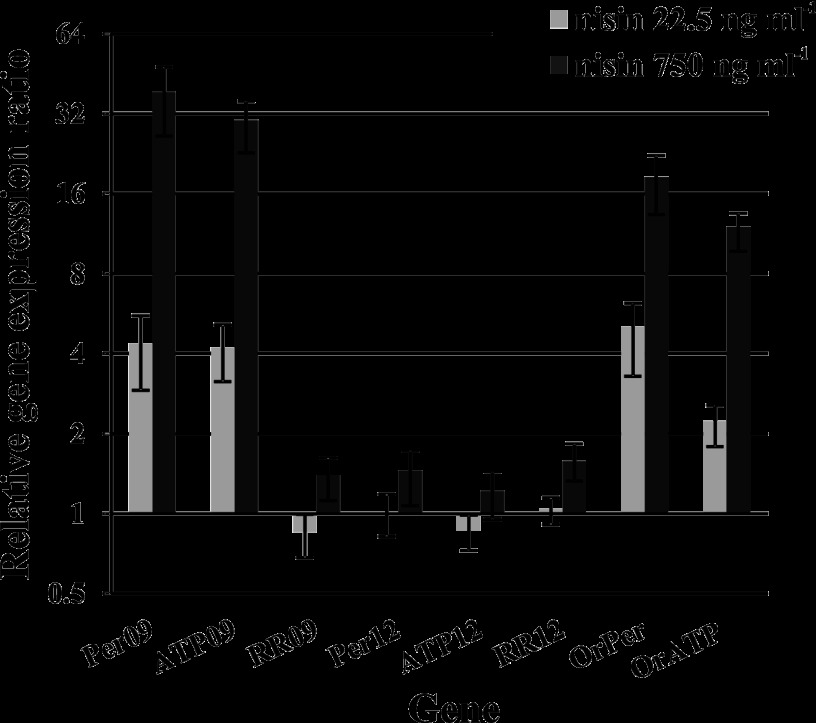
To further investigate the role of module 09, module 12, and OrABC in AMP resistance, the transcriptional response of the corresponding genes to AMPs was determined by qRT-PCR using nisin as a model AMP. The nisin concentrations used for these assays were chosen so that they had a significant effect on the growth rate, but they did not completely inhibit the growth of the strains tested (data not shown). Accordingly, a concentration of 22.5 ng ml−1 of nisin was chosen for the mutant strains, whereas two concentrations of nisin were used for the wild-type strain, 22.5 and 750 ng ml−1, due to its higher resistance to nisin. Exposure of exponentially growing cultures of L. casei BL23 to nisin resulted in a concentration-dependent induction of the expression of genes encoding ABC09 and OrABC, whereas very small changes in the expression of TCS09, TCS12, and ABC12 were detected (Fig. 2).
Fig 2.
Relative transcript levels of L. casei BL23 bceRS and bceAB homologous genes after nisin induction compared to their expression in the reference condition. Transcript levels were quantified by real-time RT-PCR of RNA samples taken 10 min after nisin addition and compared to samples from untreated cultures. The applied nisin concentrations are indicated. Data are shown as means ± standard deviations from six independent experiments.
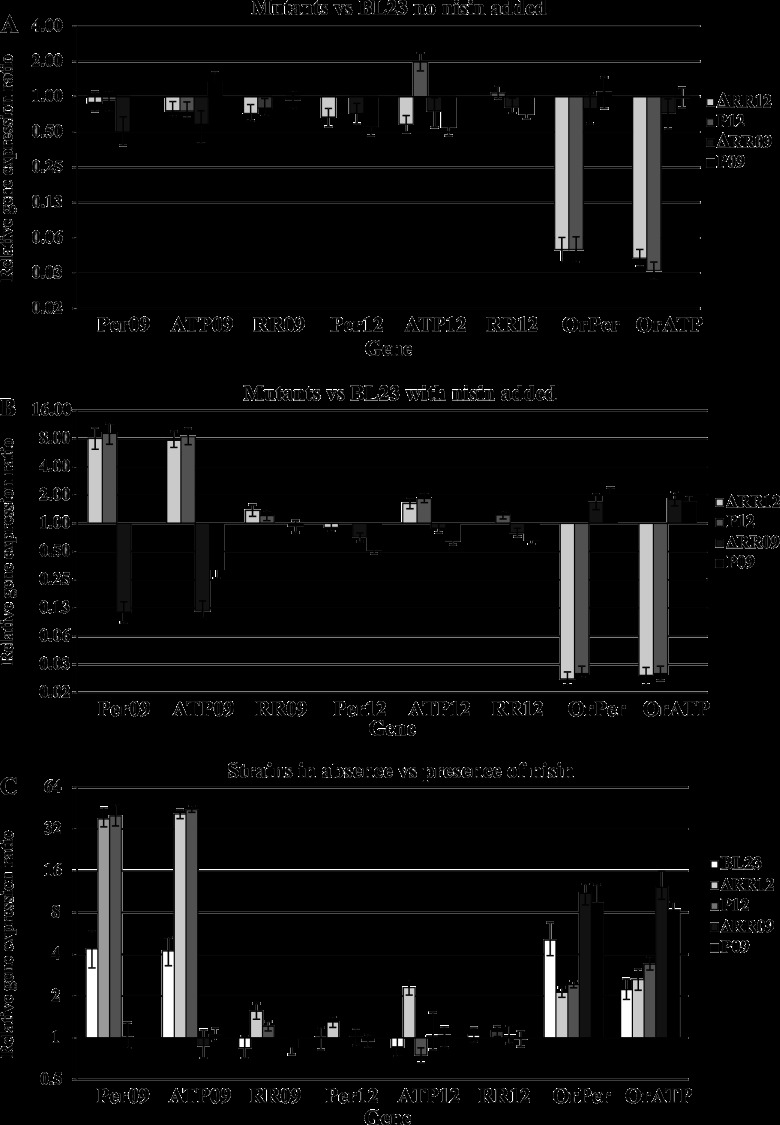
Derivative strains ΔRR09 and P09 displayed the same transcriptional profile (Fig. 3). In the reference condition, only small differences in transcript levels between both mutant strains and the wild-type strain were observed (Fig. 3A). In contrast, inactivation of either RR09 or permease 09 led to a loss of induction of the ABC09 genes in response to nisin (Fig. 3B and C), whereas the genes encoding OrABC still were induced by nisin. These results indicate that TCS09 is required for the induction of ABC09 observed in the wild-type strain in response to nisin. Furthermore, they also support the hypothesis of ABC09 being part of the signaling pathway, possibly acting as the sensory partner of TCS09.
Fig 3.
Expression of bce-like genes in mutant backgrounds. Relative transcript levels of bceRS and bceAB homologous genes in L. casei BL23-derived strains are shown compared to the parental strain in the absence of nisin (A) and 10 min after nisin addition (22.5 ng ml−1) (B). (C) Relative transcript levels of the same genes in L. casei BL23 and derivative strains 10 min after nisin addition (22.5 ng ml−1) compared to the levels of the same strains in the absence of nisin.
Comparison of strains ΔRR12 and P12 to the wild-type strain showed that both mutations had a similar effect on the gene expression profiles (Fig. 3). In reference conditions (Fig. 3A), a strong decrease in expression of the OrABC-encoding genes was observed. This effect was also observed after addition of nisin (Fig. 3B). Furthermore, an 8-fold higher induction of genes encoding ABC09 in response to nisin compared to the parental strain (Fig. 3B and C) was observed in these mutants. The possible reasons for this increased expression are discussed below. In contrast, small differences in the transcript levels of genes encoding RR09, RR12, permease 12, and ATPase 12 were observed compared to the parental strain (Fig. 3A, B, and C). These results show that module 12 is required for the expression of OrABC, and that expression of ABC12 is not under the transcriptional control of TCS12.
The identical transcriptional profiles of strains ΔRR09 and P09 on one hand and strains ΔRR12 and P12 on the other are in agreement with the hypothesis that each TCS and ABC couple works together as a functional unit.
Identification of putative target promoters of RR09 and RR12 and verification of the predictions by qRT-PCR.
The determination of the transcriptional levels in cells exposed to nisin indicated that module 09 regulated the expression of its cognate transporter ABC09, whereas module 12 regulated the expression of OrABC but not the expression of its cognate ABC12. Furthermore, previous studies had shown that mutant ΔRR12 was sensitive to environmental stresses, such as the presence of bile, acidic pH, and high temperature (34). These findings prompted the question of whether TCS12 could also regulate the expression of genes controlling other cellular functions.
To identify putative promoters under the control of TCS09 or TCS12, a motif-based search of the L. casei BL23 genome based on the consensus binding sequence for BceR-like response regulators described in Dintner et al. (17) was carried out (Fig. 4A). Four putative BceR-like target promoters upstream of genes encoding ATP09, OrATPase, LCABL_08540, and LCABL_24490 were found in L. casei BL23 (Fig. 4B). Gene LCABL_08540 is a small gene upstream of the dlt operon of L. casei BL23 (37) which encodes the molecular machinery for the synthesis of d-alanyl lipoteichoic acids (46). LCABL_08540 encodes a putative uncharacterized protein that appears conserved between different Lactobacillus strains, where it is annotated as a d-Ala-teichoic acid biosynthesis-related protein. However, it is not known if it contributes to the functionality of the dlt operon in L. casei BL23 (46); thus, we focused our attention on the remaining genes of the dlt operon (starting at dltA). Gene LCABL_24490 encodes a putative protein significantly similar to characterized lysyl-phosphatidylglycerol (Lys-PG) synthetases (47, 48) encoded by Listeria monocytogenes (47% identical residues, 68% conserved residues) and Staphylococcus aureus (29% identical residues, 52% conserved residues). Therefore, we have renamed this gene mprF.
Fig 4.
(A) Consensus binding sequence for BceR-like regulators, adapted from Dintner et al. (17). (B) Sequence alignment of putative BceR-like target sequences identified in L. casei BL23 complete genome by motif-based search using the consensus sequence shown in panel A. See the text for details. Asterisks indicate conserved positions.
The transcriptional levels of dltA and mprF in L. casei BL23 and all derived mutant strains were determined by qRT-PCR in reference conditions and after nisin induction to test whether a regulatory link exists between the Bce-like modules and the dlt and mprF genes of L. casei BL23. The expression of dltA and mprF in reference conditions, relative to the wild type, was severely decreased in mutants P12 and ΔRR12 (Fig. 5A), whereas no significant differences were found in mutants P09 and ΔRR09. After addition of nisin, expression of dltA and mprF remained very low compared to the parental strain in strains P12 and ΔRR12 (Fig. 5B). Taken together with the predicted promoter binding sites, these data strongly suggest that module 12 regulates the basal expression of the dlt operon and mprF gene in L. casei BL23. Interestingly, while the overall expression levels were much lower in mutants P12 and RR12 compared to that of the wild type, nisin-dependent induction of both the dlt operon (approximately 4.5-fold) and mprF gene (approximately 2-fold) relative to reference conditions was still observed (Fig. 5C). These data suggest that L. casei must harbor an additional regulatory system to control their expression.
Fig 5.
Expression of dlt operon and mprF in mutant backgrounds. Relative transcript levels of genes dltA and mprF in L. casei BL23 derivative strains compared to the parental strain in the absence of nisin (A) and 10 min after nisin addition (22.5 ng ml−1) (B). (C) Relative transcript levels of L. casei BL23 and derivative strains after nisin addition (*, 22.5 ng ml−1; **, 750 ng ml−1) compared to the same strains in the absence of nisin.
The pleiotropic phenotype of module 12-defective strains is due to a low expression of the dlt operon.
We then addressed the question of whether the pleiotropic phenotype previously described for the ΔRR12 mutant (34) was due to reduced expression of MrpF or Dlt. To this end, two new mutant strains were obtained by insertional inactivation of mprF and dltA: L. casei MPRF and L. casei DLT, respectively (Table 1). Growth of these two strains under different conditions was compared to that of the wild-type strain and mutants ΔRR12 and P12 as previously described (34). Mutant MPRF showed the same phenotype as the wild type under all conditions tested (Fig. 6). However, mutants DLT, P12, and ΔRR12 did not reach the same final OD595 as the wild type in reference conditions (Fig. 6A), were more sensitive to 0.5% bile and high temperature (Fig. 6B and C) than the wild type, and were not able to grow at pH 4 (Fig. 6D). To ascertain if the lower final OD under reference conditions was due to a higher sensitivity to acid, the bacteria were grown in MRS supplemented with 0.1 M phosphate buffer, thus preventing acidification of the medium during growth. Under these conditions, mutants DLT, P12, and ΔRR12 were able to reach final OD595 values similar to those of the wild type (Fig. 6E). This result demonstrates that the different levels of growth in reference conditions are mainly due to higher acid sensitivities.
Fig 6.
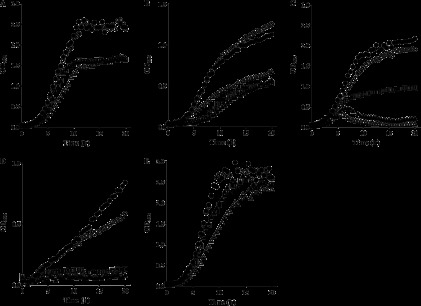
Growth of L. casei BL23 (●) and strains ΔRR12 (□), Per12 (▲), DLT (Δ), and MPRF (○) under different conditions. (A) Reference condition. (B) Growth in the presence of 0.5% bile. (C) Growth at 42°C. (D) Growth in medium adjusted to pH 4. (E) Growth in buffered media (MRS supplemented with 0.1 M phosphate). Data are shown as representative results from three independent experiments.
The MICs of the previously assayed AMPs against the strains DLT and MPRF were also determined (Table 2). The MICs of nisin, vancomycin, plectasin, mersacidin, and subtilin obtained for the DLT mutant were similar to the MICs for mutants P12 and ΔRR12 (Table 2). In contrast, strain MPRF showed MICs similar to those of the wild type with only a slight increase in sensitivity to bacitracin and mersacidin (Table 2).
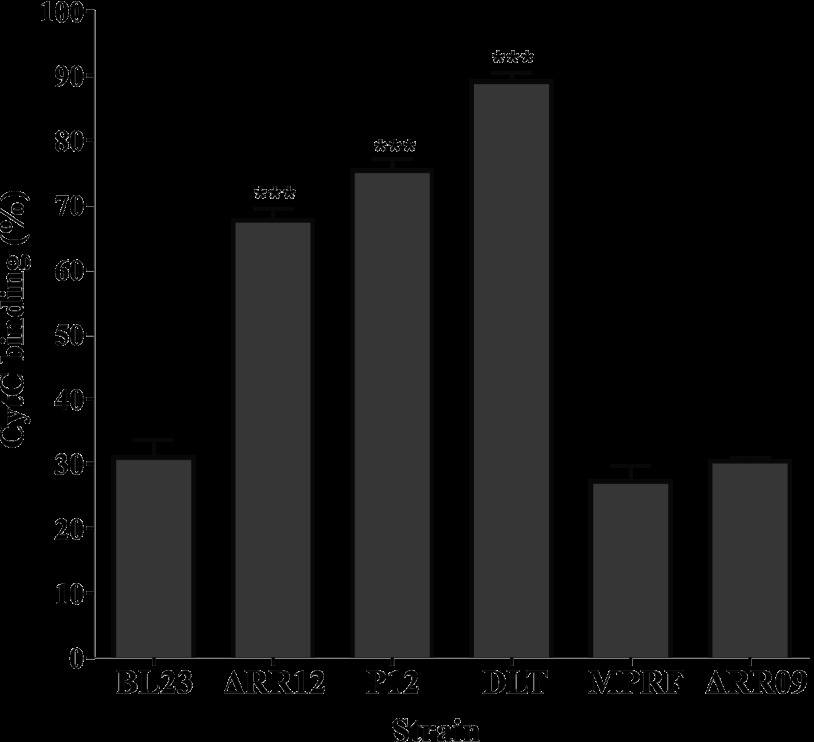
Since Dlt and MprF activities modulate the bacterial cell surface charge and thus affect the susceptibility to AMPs (40, 42), a cytochrome c binding affinity assay was performed to determine whether strains DLT and MPRF actually displayed a difference in cell surface charge compared to the wild-type strain. Cytochrome c is a highly positively charged protein, and its binding to the bacterial surface depends on the net negative charge. L. casei BL23 and strain ΔRR09 bound ∼30% of the cytochrome c present in the solution (Fig. 7). Strains ΔRR12, P12, and DLT bound 67.5, 75, and 89%, respectively, of the cytochrome c present in the solution (Fig. 7), indicating that they have a more negative surface charge than the wild-type strain. However, the mprF mutant showed the same phenotype as the wild-type strain (Fig. 7).
Fig 7.
Cytochrome c (CytC) binding assay. Binding is expressed as the percentage of cytochrome c bound to stationary-phase cells of strain BL23 and derivatives after incubation with 150 μg ml−1 cytochrome c for 10 min at room temperature. Data represent the means and standard deviations from three independent experiments. Asterisks indicate significant differences (P < 0.05 by the unpaired Student t test) between the indicated strain and the control strain (BL23).
Considering these results together, we conclude that the pleiotropic phenotype of module 12 mutants is due to the low expression of the dlt operon that leads to an increase in the net negative charge of the bacterial surface.
DISCUSSION
AMPs are key components of the innate immune defense system (49), but bacteria also produce AMPs in order to gain an advantage in complex microbial communities, such as the gut microbiome (50). Conversely, bacteria have evolved sophisticated systems to counteract the action of AMPs. BceRS/BceAB modules have been shown to be involved in AMP resistance in different Firmicutes bacteria (17, 19, 21, 32, 51–53). In this study, we showed that the BceRS/BceAB-like modules of L. casei BL23, TCS09/ABC09 and TCS12/ABC12, are also involved in AMP resistance in this probiotic microorganism. Inactivation of either RR09 or permease 09 led to higher sensitivity to bacitracin, nisin, plectasin, and subtilin than that observed in the wild-type strain, whereas inactivation of either RR12 or permease 12 resulted in higher sensitivity to bacitracin, nisin, mersacidin, plectasin, subtilin and vancomycin (Table 2). In this study, we have used nisin as a model AMP. Nisin belongs to the group of lantibiotics, ribosomally synthesized peptides which are characterized by lanthionine or methyllanthionine rings, among other posttranslational modifications (54–56). Mature nisin is an elongated, amphipathic, and positively charged molecule, and it is active against Gram-positive bacteria, including lactobacilli. Nisin-producing strains possess immunity conferred to them by the lipoprotein NisI and the ABC transporter NisFEG (57). NisFEG does not belong to the peptide-7 exporter family; thus, it is not related to BceAB-like ABC transporters. Strong synergy between NisI and NisFEG has been observed, indicating that both proteins are required for immunity (54).
An important feature of Bce-like modules characterized to date is that the transporters are required for stimulus perception and signal transduction within the modules (17, 26, 30, 52). The results reported in this study showed that the mutant strains defective in the response regulator and its associated ABC permease from the same module displayed identical phenotypes and changes in transcript levels (Table 2 and Fig. 3 and 5). These results strongly suggest that both TCS09 and TCS12 require their respective ABC transporters; therefore, these TCS and ABC transporters work as functional modules, as has been previously described for other systems of this group (30, 32, 33, 52).
Determination of changes in transcript levels in response to nisin showed that the genes coding for ABC09, but not RR09, are induced in L. casei BL23 in response to nisin in a concentration-dependent manner (Fig. 2). This induction was lost in the mutant strains ΔRR09 and P09 (Fig. 3B and C). These results show that TCS09 regulates the nisin-dependent expression of ABC09 and strongly suggest that both the TCS and the ABC transporter are required for nisin induction of ABC09 expression. Our findings also suggest that ABC09 confers resistance to the compound that induces its expression, similar to what has been described for the ABC transporters BceAB and PsdAB of B. subtilis (21). Therefore, taking into consideration the evidence provided by other homologous systems and the results obtained in this study, we propose that the target AMP (nisin) is sensed by ABC09 and activates HK09, which transfers the signal to RR09, resulting in the induction of the expression of ABC09, which confers the resistance (Fig. 1).
On the other hand, small changes in response to nisin were detected in the transcript levels of the genes coding for ABC transporter 12 as well as the RR12 gene. Since a transporter involved in substrate sensing but not in transport does not necessarily need to increase its expression in response to the presence of the substrate (17), this result suggests that module 12 acts as a sensory system without detoxification function. Furthermore, the transcript levels of the genes encoding the orphan ABC transporter, the dlt operon, and the mprF gene in the Per12 mutant was lowered to the same basal levels as those in the ΔRR12 mutant, both in reference conditions (Fig. 3A and 5A) and in response to nisin (Fig. 3B and 5B), relative to the wild type. These results further support that ABC12 is as essential as TCS12 for controlling the constitutive expression of the dlt operon, gene mprF, and the orphan ABC-encoding genes but apparently not for a specific response to nisin. This idea is further substantiated by the fact that the expression of genes controlled by module 12 is still induced in module 12-defective mutants (Fig. 3C and 5C), although their transcript levels are much lower than those in the parental strain. These data suggest that other regulatory systems besides module 12 control the expression of the dlt operon and mprF in L. casei BL23.
The involvement of BceRS/BceAB modules in the control of the cell envelope charge has been previously described in other organisms. For example, the GraRS TCS of S. aureus and the homologous system ApsRS of Staphylococcus epidermidis control the expression of the dlt operon and mprF, as well as vraFG, which encodes a BceAB-like transporter that also mediates the AMP resistance (19, 20, 51, 58). The TCS VirRS of Listeria monocytogenes also controls expression of the dlt operon and mprF, among other genes (59). Interestingly, VirRS apparently does not regulate the expression of its adjacent ABC transporter Lmo1747-1746 (59), but it regulates the expression of the nongenetically associated ABC transporter AnrAB, which has also been shown to be involved in AMP resistance (60).
Initial binding of cationic AMPs to bacterial cell envelopes is mediated by electrostatic attraction between the positively charged peptides and the negatively charged bacterial surfaces (12). Bacteria can utilize a number of mechanisms to modulate their cell envelope charge. Many Gram-positive bacteria can neutralize polyanionic teichoic acid polymers of the cell wall by esterification with d-alanine (61). The machinery required for this activity is encoded by the dlt operon. The MprF protein is found in both Gram-positive and Gram-negative bacteria. It catalyzes the modification of the anionic phospholipids of the membrane with l-lysine or l-alanine (62). Thus, MprF and the DltABCD system function as bacterial immune evasion systems that confer resistance to cationic AMPs by reducing the negative net charge of the bacterial cell surface (19, 20, 40, 42, 48, 61–63). Besides the sensitivity to antimicrobial peptides, other phenotypes have been associated with a nonfunctional Dlt system. For example, inactivation of dltC in Streptococcus mutans resulted in the loss of the acid tolerance response and lower growth rate than that of the parental strain (64).
The cytochrome c binding assay showed that low expression of the dlt operon in L. casei effectively led to an increase in the net negative charge of the bacterial surface in mutants ΔRR12, P12, and DLT (Fig. 7). These strains also displayed lower AMP resistance and growth defects (Table 2 and Fig. 6). On the contrary, low expression of MprF did not significantly change the cell surface charge (Fig. 7) and only resulted in a 2-fold decrease in MIC for bacitracin and mersacidin (Table 2), whereas its growth was similar to that of the wild-type strain under all other conditions tested (Fig. 6). Ernst and colleagues (63) reported that the level of lysyl-phosphatidylglycerol (Lys-PG) did not correlate with the level of cationic AMP resistance, since a basal amount of Lys-PG is enough to confer full cationic AMP resistance. Together, these results indicate that the growth defects observed in module 12-defective mutants were mainly due to low expression of the dlt operon. In this sense, it is worth noting the higher expression of ABC09 in response to nisin in the module 12-defective mutants compared to the wild-type strain (Fig. 3B). A possible explanation for this observation is that the higher cell surface negative charge of these mutants would increase their affinity for nisin and, thus, increase the signal for induction of ABC09, although additional evidence is needed to test this hypothesis.
In conclusion, results reported in this study show that the BceRS/BceAB-like modules of L. casei BL23, TCS09/ABC09 and TCS12/ABC12, are involved in AMP resistance in this probiotic microorganism (Fig. 1). It was also shown that BceRS-like TCS of L. casei BL23 constitute functional modules with their cognate ABC transporters. Module 09 regulates the expression of its cognate ABC09, which may have dual functions of sensing and resistance to AMPs. Module 12 is an AMP sensory system where ABC12 does not have a direct role in AMP detoxification. The sensitivity of module 12 mutants to AMPs and their pleiotropic phenotype is caused by reduced expression of the RR12-regulated genes, in particular the dlt operon. The function of OrABC, the third target operon of module 12, remains unclear. To our knowledge, this is the first report on the characterization of a complete set of AMP resistance mechanisms in the probiotic species L. casei.
Supplementary Material
ACKNOWLEDGMENTS
This work was financed by funds from the former Spanish Ministry of Science and Innovation (AGL2007-60975, AGL2010-15679, and Consolider Fun-C-Food CSD2007-00063) and Generalitat Valenciana (ACOMP2012/137). A.R.-G. was the recipient of an FPI grant (BES-2008-004527) from the former Spanish Ministry of Science and Innovation. Work in the laboratory of T.M. was financed by a grant from the Deutsche Forschungsgesellschaft (MA2837/1-3). S.G. was funded by grants from the Deutsche Forschungsgesellschaft (GE2164/3-1) and the Fonds der Chemischen Industrie.
We thank Amalia Blasco and Tina Wecke for technical assistance.
Footnotes
Published ahead of print 1 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00178-13.
REFERENCES
- 1.de Vrese M, Schrezenmeir J. 2008. Probiotics, prebiotics, and synbiotics. Adv. Biochem. Eng. Biotechnol. 111:1–66 [DOI] [PubMed] [Google Scholar]
- 2.Kleerebezem M, Vaughan EE. 2009. Probiotic and gut lactobacilli and bifidobacteria: molecular approaches to study diversity and activity. Annu. Rev. Microbiol. 63:269–290 [DOI] [PubMed] [Google Scholar]
- 3.Corcoran BM, Stanton C, Fitzgerald G, Ross RP. 2008. Life under stress: the probiotic stress response and how it may be manipulated. Curr. Pharm. Des. 14:1382–1399 [DOI] [PubMed] [Google Scholar]
- 4.Cunliffe RN. 2003. Alpha-defensins in the gastrointestinal tract. Mol. Immunol. 40:463–467 [DOI] [PubMed] [Google Scholar]
- 5.Dommett R, Zilbauer M, George JT, Bajaj-Elliott M. 2005. Innate immune defence in the human gastrointestinal tract. Mol. Immunol. 42:903–912 [DOI] [PubMed] [Google Scholar]
- 6.Gallo RL, Hooper LV. 2012. Epithelial antimicrobial defence of the skin and intestine. Nat. Rev. Immunol. 12:503–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hancock RE, Diamond G. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402–410 [DOI] [PubMed] [Google Scholar]
- 8.Peschel A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179–186 [DOI] [PubMed] [Google Scholar]
- 9.Cotter PD, Hill C, Ross RP. 2005. Bacteriocins: developing innate immunity for food. Nat. Rev. Microbiol. 3:777–788 [DOI] [PubMed] [Google Scholar]
- 10.Gálvez A, Abriouel H, López RL, Ben Omar N. 2007. Bacteriocin-based strategies for food biopreservation. Int. J. Food Microbiol. 120:51–70 [DOI] [PubMed] [Google Scholar]
- 11.Hancock REW. 1997. Peptide antibiotics. Lancet 349:418–422 [DOI] [PubMed] [Google Scholar]
- 12.Peschel A, Sahl HG. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529–536 [DOI] [PubMed] [Google Scholar]
- 13.Nishie M, Nagao J, Sonomoto K. 2012. Antibacterial peptides “bacteriocins”: an overview of their diverse characteristics and applications. Biocontrol Sci. 17:1–16 [DOI] [PubMed] [Google Scholar]
- 14.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B. 1999. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364 [DOI] [PubMed] [Google Scholar]
- 15.Brötz H, Josten M, Wiedemann I, Schneider U, Götz F, Bierbaum G, Sahl HG. 1998. Role of lipid-bound peptidoglycan precursors in the formation of pores by nisin, epidermin and other lantibiotics. Mol. Microbiol. 30:317–327 [DOI] [PubMed] [Google Scholar]
- 16.Wiedemann I, Breukink E, van Kraaij C, Kuipers OP, Bierbaum G, de Kruijff B, Sahl HG. 2001. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 276:1772–1779 [DOI] [PubMed] [Google Scholar]
- 17.Dintner S, Staroń A, Berchtold E, Petri T, Mascher T, Gebhard S. 2011. Coevolution of ABC transporters and two-component regulatory systems as resistance modules against antimicrobial peptides in Firmicutes bacteria. J. Bacteriol. 193:3851–3862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guo L, Lim KB, Gunn JS, Bainbridge B, Darveau RP, Hackett M, Miller SI. 1997. Regulation of lipid A modifications by Salmonella typhimurium virulence genes phoP-phoQ. Science 276:250–253 [DOI] [PubMed] [Google Scholar]
- 19.Li M, Cha DJ, Lai Y, Villaruz AE, Sturdevant DE, Otto M. 2007. The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol. Microbiol. 66:1136–1147 [DOI] [PubMed] [Google Scholar]
- 20.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. 2007. Gram-positive three-component antimicrobial peptide-sensing system. Proc. Natl. Acad. Sci. U. S. A. 104:9469–9474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Staroń A, Finkeisen DE, Mascher T. 2011. Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob. Agents Chemother. 55:515–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao R, Stock AM. 2009. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 63:133–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ninfa AJ, Magasanik B. 1986. Covalent modification of the glnG product, NRI, by the glnL product, NRII, regulates the transcription of the glnALG operon in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 83:5909–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nixon BT, Ronson CW, Ausubel FM. 1986. Two-component regulatory systems responsive to environmental stimuli share strongly conserved domains with the nitrogen assimilation regulatory genes ntrB and ntrC. Proc. Natl. Acad. Sci. U. S. A. 83:7850–7854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stock AM, Robinson VL, Goudreau PN. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183–215 [DOI] [PubMed] [Google Scholar]
- 26.Coumes-Florens S, Brochier-Armanet C, Guiseppi A, Denizot F, Foglino M. 2011. A new highly conserved antibiotic sensing/resistance pathway in Firmicutes involves an ABC transporter interplaying with a signal transduction system. PLoS One 6:e15951 doi:10.1371/journal.pone.0015951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gebhard S, Mascher T. 2011. Antimicrobial peptide sensing and detoxification modules: unravelling the regulatory circuitry of Staphylococcus aureus. Mol. Microbiol. 81:581–587 [DOI] [PubMed] [Google Scholar]
- 28.Joseph P, Fichant G, Quentin Y, Denizot F. 2002. Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J. Mol. Microbiol. Biotechnol. 4:503–513 [PubMed] [Google Scholar]
- 29.Mascher T. 2006. Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol. Lett. 264:133–144 [DOI] [PubMed] [Google Scholar]
- 30.Bernard R, Guiseppi A, Chippaux M, Foglino M, Denizot F. 2007. Resistance to bacitracin in Bacillus subtilis: unexpected requirement of the BceAB ABC transporter in the control of expression of its own structural genes. J. Bacteriol. 189:8636–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ohki R, Giyanto Tateno K, Masuyama W, Moriya S, Kobayashi K, Ogasawara N. 2003. The BceRS two-component regulatory system induces expression of the bacitracin transporter, BceAB, in Bacillus subtilis. Mol. Microbiol. 49:1135–1144 [DOI] [PubMed] [Google Scholar]
- 32.Rietkötter E, Hoyer D, Mascher T. 2008. Bacitracin sensing in Bacillus subtilis. Mol. Microbiol. 68:768–785 [DOI] [PubMed] [Google Scholar]
- 33.Ouyang J, Tian XL, Versey J, Wishart A, Li YH. 2010. The BceABRS four-component system regulates the bacitracin-induced cell envelope stress response in Streptococcus mutans. Antimicrob. Agents Chemother. 54:3895–3906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alcántara C, Revilla-Guarinos A, Zúñiga M. 2011. Influence of two-component signal transduction systems of Lactobacillus casei BL23 on tolerance to stress conditions. Appl. Environ. Microbiol. 77:1516–1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leloup L, Ehrlich SD, Zagorec M, Morel-Deville F. 1997. Single-crossover integration in the Lactobacillus sake chromosome and insertional inactivation of the ptsI and lacL genes. Appl. Environ. Microbiol. 63:2117–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Broadbent JR, Neeno-Eckwall EC, Stahl B, Tandee K, Cai H, Morovic W, Horvath P, Heidenreich J, Perna NT, Barrangou R, Steele JL. 2012. Analysis of the Lactobacillus casei supragenome and its influence in species evolution and lifestyle adaptation. BMC Genomics 13:533 doi:10.1186/1471-2164-13-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mazé A, Böel G, Zúñiga M, Bourand A, Loux V, Yebra MJ, Monedero V, Correia K, Jacques N, Beaufils S, Poncet S, Joyet P, Milohanic E, Casaregola S, Auffray Y, Pérez-Martínez G, Gibrat JF, Zagorec M, Francke C, Hartke A, Deutscher J. 2010. Complete genome sequence of the probiotic Lactobacillus casei strain BL23. J. Bacteriol. 192:2647–2648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sambrook J, Fritsch EF, Maniatis T. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 39.Posno M, Leer RJ, van Luijk N, van Giezen MJ, Heuvelmans PT, Lokman BC, Pouwels PH. 1991. Incompatibility of Lactobacillus vectors with replicons derived from small cryptic Lactobacillus plasmids and segregational instability of the introduced vectors. Appl. Environ. Microbiol. 57:1822–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Matsuo M, Oogai Y, Kato F, Sugai M, Komatsuzawa H. 2011. Growth-phase dependence of susceptibility to antimicrobial peptides in Staphylococcus aureus. Microbiology 157:1786–1797 [DOI] [PubMed] [Google Scholar]
- 41.Meehl M, Herbert S, Gotz F, Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2679–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peschel A, Otto M, Jack RW, Kalbacher H, Jung G, Götz F. 1999. Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274:8405–8410 [DOI] [PubMed] [Google Scholar]
- 43.Landete JM, García-Haro L, Blasco A, Manzanares P, Berbegal C, Monedero V, Zúñiga M. 2010. Requirement of the Lactobacillus casei MaeKR two-component system for L-malic acid utilization via a malic enzyme pathway. Appl. Environ. Microbiol. 76:84–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. 2002. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3:RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfaffl MW, Horgan GW, Dempfle L. 2002. Relative expression software tool (REST) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 30:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Neuhaus FC, Heaton MP, Debabov DV, Zhang Q. 1996. The dlt operon in the biosynthesis of D-alanyl-lipoteichoic acid in Lactobacillus casei. Microb. Drug Resist. 2:77–84 [DOI] [PubMed] [Google Scholar]
- 47.Peschel A, Jack RW, Otto M, Collins LV, Staubitz P, Nicholson G, Kalbacher H, Nieuwenhuizen WF, Jung G, Tarkowski A, van Kessel KP, van Strijp JA. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with L-lysine. J. Exp. Med. 193:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thedieck K, Hain T, Mohamed W, Tindall BJ, Nimtz M, Chakraborty T, Wehland J, Jansch L. 2006. The MprF protein is required for lysinylation of phospholipids in listerial membranes and confers resistance to cationic antimicrobial peptides (CAMPs) on Listeria monocytogenes. Mol. Microbiol. 62:1325–1339 [DOI] [PubMed] [Google Scholar]
- 49.Jenssen H, Hamill P, Hancock RE. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dobson A, Cotter PD, Ross RP, Hill C. 2012. Bacteriocin production: a probiotic trait? Appl. Environ. Microbiol. 78:1–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falord M, Karimova G, Hiron A, Msadek T. 2012. GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56:1047–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hiron A, Falord M, Valle J, Debarbouille M, Msadek T. 2011. Bacitracin and nisin resistance in Staphylococcus aureus: a novel pathway involving the BraS/BraR two-component system (SA2417/SA2418) and both the BraD/BraE and VraD/VraE ABC transporters. Mol. Microbiol. 81:602–622 [DOI] [PubMed] [Google Scholar]
- 53.Kawada-Matsuo M, Yoshida Y, Nakamura N, Komatsuzawa H. 2011. Role of two-component systems in the resistance of Staphylococcus aureus to antibacterial agents. Virulence 2:427–430 [DOI] [PubMed] [Google Scholar]
- 54.Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP. 2008. Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell. Mol. Life Sci. 65:455–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sahl HG, Bierbaum G. 1998. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu. Rev. Microbiol. 52:41–79 [DOI] [PubMed] [Google Scholar]
- 56.Schnell N, Entian KD, Schneider U, Götz F, Zähner H, Kellner R, Jung G. 1988. Prepeptide sequence of epidermin, a ribosomally synthesized antibiotic with four sulphide-rings. Nature 333:276–278 [DOI] [PubMed] [Google Scholar]
- 57.Siegers K, Entian KD. 1995. Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl. Environ. Microbiol. 61:1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herbert S, Bera A, Nerz C, Kraus D, Peschel A, Goerke C, Meehl M, Cheung A, Götz F. 2007. Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog. 3:e102 doi:10.1371/journal.ppat.0030102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mandin P, Fsihi H, Dussurget O, Vergassola M, Milohanic E, Toledo-Arana A, Lasa I, Johansson J, Cossart P. 2005. VirR, a response regulator critical for Listeria monocytogenes virulence. Mol. Microbiol. 57:1367–1380 [DOI] [PubMed] [Google Scholar]
- 60.Collins B, Curtis N, Cotter PD, Hill C, Ross RP. 2010. The ABC transporter AnrAB contributes to the innate resistance of Listeria monocytogenes to nisin, bacitracin, and various beta-lactam antibiotics. Antimicrob. Agents Chemother. 54:4416–4423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Neuhaus FC, Baddiley J. 2003. A continuum of anionic charge: structures and functions of D-alanyl-teichoic acids in gram-positive bacteria. Microbiol. Mol. Biol. Rev. 67:686–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ernst CM, Peschel A. 2011. Broad-spectrum antimicrobial peptide resistance by MprF-mediated aminoacylation and flipping of phospholipids. Mol. Microbiol. 80:290–299 [DOI] [PubMed] [Google Scholar]
- 63.Ernst CM, Staubitz P, Mishra NN, Yang SJ, Hornig G, Kalbacher H, Bayer AS, Kraus D, Peschel A. 2009. The bacterial defensin resistance protein MprF consists of separable domains for lipid lysinylation and antimicrobial peptide repulsion. PLoS Pathog. 5:e1000660 doi:10.1371/journal.ppat.1000660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyd DA, Cvitkovitch DG, Bleiweis AS, Kiriukhin MY, Debabov DV, Neuhaus FC, Hamilton IR. 2000. Defects in D-alanyl-lipoteichoic acid synthesis in Streptococcus mutans results in acid sensitivity. J. Bacteriol. 182:6055–6065 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.