Abstract
Helicobacter pylori inhabits the stomach mucosa and is a causative agent of stomach ulcer and cancer. In general, bacteriophages (phages) are strongly associated with bacterial evolution, including the development of pathogenicity. Several tailed phages have so far been reported in H. pylori. We have isolated an H. pylori phage, KHP30, and reported its genomic sequence. In this study, we examined the biological characteristics of phage KHP30. Phage KHP30 was found to be a spherical lipid-containing phage with a diameter of ca. 69 nm. Interestingly, it was stable from pH 2.5 to pH 10, suggesting that it is adapted to the highly acidic environment of the human stomach. Phage KHP30 multiplied on 63.6% of clinical H. pylori isolates. The latent period was ca. 140 min, shorter than the doubling time of H. pylori (ca. 180 min). The burst size was ca. 13, which was smaller than the burst sizes of other known tailed or spherical phages. Phage KHP30 seemed to be maintained as an episome in H. pylori strain NY43 cells, despite a predicted integrase gene in the KHP30 genomic sequence. Seven possible virion proteins of phage KHP30 were analyzed using N-terminal protein sequencing and mass spectrometry, and their genes were found to be located on its genomic DNA. The genomic organization of phage KHP30 differed from the genomic organizations in the known spherical phage families Corticoviridae and Tectiviridae. This evidence suggests that phage KHP30 is a new type of spherical phage that cannot be classified in any existing virus category.
INTRODUCTION
Helicobacter pylori, a Gram-negative spiral bacterium, colonizes the human stomach mucosa (1). It causes chronic inflammation, which may progress to peptic ulceration, atrophic gastritis, and gastric cancer (2). Recent studies of H. pylori have revealed that the pathogenicity of H. pylori strains is related to the specific geographic region from which they derive (3, 4). An East Asian type of H. pylori strain most likely causes gastric cancer and highly likely contains cytotoxin-associated gene A (cagA), a carcinogenic genetic element (1, 3). However, how the pathogenicity of the H. pylori strains has evolved is unknown, although H. pylori inhabits a very restricted niche in the stomach mucosa. Therefore, the factors involved in H. pylori evolution must be investigated.
Bacteriophages (phages) are the most diverse and abundant life form on Earth. Phages practice lateral gene transfer and are involved in a coevolutionary arms race with bacteria (5–8), so they may well be factors involved in bacterial evolution. Several H. pylori phages have been reported (9–13), and some of these have been morphologically studied and have been found to belong to the order Caudovirales (i.e., the tailed phages) (9–12). Recent advances in sequencing technologies have allowed intensive genomic analyses of Helicobacter spp. and have also suggested the presence of phages in the bacterial genome (10, 11, 14, 15). However, the biological characteristics of these H. pylori phages are not well-known.
We have recently isolated phage KHP30 from the culture supernatant of an East Asian H. pylori strain, NY43, isolated from a patient at Yamaguchi University Hospital in Japan, and have reported its genomic sequence (16). To extend our understanding of the contribution of phages to H. pylori evolution, the biological characteristics of the H. pylori phages must be examined. To our knowledge, the biology of the H. pylori phages has not yet been investigated. In this study, we analyzed the biological characteristics of H. pylori phage KHP30.
MATERIALS AND METHODS
Media and reagents used in this study.
All reagents were obtained from Nacalai Tesque (Kyoto, Japan) or Wako Pure Chemical (Osaka, Japan), unless otherwise stated. Equine serum (HyClone Laboratories, South Logan, UT) was heated (56°C, 30 min) before use. Brucella broth was purchased from Becton, Dickinson and Co. (Sparks, MD). BE medium (pH 7.2) is brucella broth (pH 6.8) containing 10% equine serum (HyClone Laboratories). BEV medium (pH 7.2) is BE medium containing 10 μg/ml vancomycin. BEV medium was used to culture H. pylori and phage, unless otherwise stated. The double-layered agar method was used for phage plaque formation, with BEV-based medium containing 1.5% agar and brucella broth-based medium containing 0.5% agar used for the lower and upper layers, respectively.
Bacterial strains and culture conditions.
H. pylori strain 3401 was used as the standard host bacterium (17). H. pylori strain NY43, which was isolated from a 54-year-old male patient with gastric ulcer, spontaneously releases phage KHP30 and was also used in this study. The other H. pylori strains used in this study, which were isolated from different patients, are shown in Table S1 in the supplemental material (18–22). All H. pylori strains were incubated in an appropriate medium in air containing 10% CO2 at 37°C.
Bacterial genotyping by RFLP.
Restriction fragment length polymorphism (RFLP) was conducted as described elsewhere with a slight modification (23). Briefly, the genomic DNAs of H. pylori were extracted using NucleoSpin tissue columns (Macherey-Nagel, Düren, Germany). The DNAs were amplified by PCR using two sets of primers: 5′-AGGAGAATGAGATGA and 5′-ACTTTATTGGCTGGT for UreAB fragments and 5′-ATGGCTTTTCAGGTCAATAC and 5′-GCTTAAGATATTTTGTTGAACG for FlaA fragments. After the PCR amplification, the UreAB and FlaA fragments were digested by restriction enzymes HaeIII and HhaI (TaKaRa Bio, Shiga, Japan), respectively. The restricted DNAs were electrophoresed in 1.5% agarose gels. After staining with ethidium bromide, the gel was visualized under UV transillumination. H. pylori types were manually differentiated according to their restriction band patterns (see Fig. S1 in the supplemental material). The RELP types of H. pylori used in this study are shown in Table S1 in the supplemental material. According to the RFLP, the H. pylori strains used in this study were considered to have the genetic variation.
Phage KHP30.
Phage KHP30 was isolated previously (16). Briefly, phage KHP30 was isolated from the culture supernatant of H. pylori strain NY43 with three rounds of single-plaque isolation in double-layered agar containing H. pylori strain 3401. Phage KHP30 formed plaques and showed lytic activity in liquid medium (see Fig. S2 in the supplemental material). It was amplified in H. pylori strain 3401 in double-layered agar and used as a phage stock, unless otherwise stated.
Measurements of bacterial and phage concentrations.
Bacterial concentrations were measured as turbidities with a Klett-Summerson photoelectric colorimeter (filter 54; Klett Manufacturing Co., New York, NY). One Klett unit was assumed to be equivalent to 3.6 × 106 cells/ml, based on a standardized correlation between turbidity and bacterial cell numbers counted directly with a Petroff-Hausser counting chamber (Hausser Scientific, Horsham, PA). Phage concentrations (PFU/ml) were measured with the double-layered agar method using H. pylori strain 3401 as the host, unless otherwise stated.
Large-scale culture and purification of phage.
One liter of brucella broth containing 0.5% β-cyclodextrin and 10 μg/ml vancomycin (pH 7.0) was inoculated with H. pylori strain 3401 to be ca. 8.2 × 106 cells/ml and simultaneously supplemented with phage KHP30 stock at a multiplicity of infection (MOI) of 0.5 to 2. The culture mixture was incubated with shaking for 1.5 to 2 days until cell lysis. The culture lysate (ca. 2.4 × 108 PFU/ml) was centrifuged at 10,000 × g for 10 min to remove the cell debris. Polyethylene glycol 6000 (10%), 3 M NaCl, and 1% Tween 20 were added to the culture supernatant. After centrifugation (10,000 × g, 20 min, 4°C), the pellet was treated with 100 μg/ml DNase I (Sigma-Aldrich Co., St. Louis, MO) and 100 μg/ml RNase A (Sigma-Aldrich) in 2 ml of TM solution (10 mM Tris-HCl, 5 mM MgCl2, pH 7.2) for 60 min at 37°C.
The phages were purified using CsCl density gradient ultracentrifugation. The phage suspension (1 ml) was layered above a discontinuous CsCl density gradient of layered 2.5, 1.5, and 0.5 ml of CsCl solutions (ρ = 1.3, 1.5, and 1.7, respectively) and was centrifuged (100,000 × g, 1 h, 4°C). The phage band was collected, and the CsCl density gradient ultracentrifugation was repeatedly conducted to increase the purity of the phage particles. The phage band (0.5 to 1 ml) was then placed on top of a discontinuous CsCl density gradient, of which 1 to 1.2, 0.6 to 0.9, and 0.3 ml of CsCl was layered (ρ = 1.3, 1.5, and 1.7, respectively), and was centrifuged (100,000 × g, 1 h, 4°C). The first and second ultracentrifugations were conducted using the S80AT3 and S100AT4 rotors of a Himac CS100GX microultracentrifuge (Hitachi, Tokyo, Japan). Polycarbonate centrifuge tubes were used for ultracentrifugation.
Electron microscopic observation of phage particles.
After dialysis against AAS (0.1 M ammonium acetate, 10 mM NaCl, 1 mM CaCl2, 1 mM MgCl2, pH 7.2) for 1 h at 4°C, the purified phage sample (ca. 1.0 × 1010 to 1011 PFU/ml) was loaded onto a Formvar-coated copper mesh and stained with 2% uranyl acetate. The sample was observed with an H-7100 transmission electron microscope (Hitachi) at 100 kV. The size of the phage was measured using the standard bar automatically displayed by the instrument at magnifications of ×100,000 to ×200,000. The mean phage particle sizes and their standard deviations were calculated.
Electron microscopic observation of phage-infected H. pylori sections.
Phage KHP30 was added to H. pylori strain 3401 (ca. 3.6 × 108 cells/ml) at an MOI of 10 to 20. The phage-bacterium suspension was incubated at 37°C with shaking for 140 and 280 min. H. pylori alone was used as the control. The H. pylori cells were fixed by suspension in 200 μl of AAS containing 2% glutaraldehyde (TAAB Laboratories Equipment, Berkshire, United Kingdom). Ultrathin sections of the bacteria were prepared essentially as described elsewhere (24). The sections were observed with an H-7100 transmission electron microscope (Hitachi) at 100 kV.
Trend analysis of the phage head/particle volume and genomic size.
By a review of published papers and the genome database at the National Center for Biotechnology Information (NCBI), the head/particle size and genome sequence lengths of the following phages were obtained: phages SAP-2, ϕMR11, ϕMR25, KPP12, KPP10, ϕEF24C, SPP1, λ, A511, P1, N4, ϕC31, PRD1, SSIP-1, PM2, P23-77, Bam35c, GIL16, and AP50 (25–45). The head/particle volume, which was estimated using the formula 4/3 × π × r3 (where r is the radius, or one-half of the diameter), and the sequence lengths of the tailed and spherical phages were examined by linear regression analysis using the statistical software GraphPad Prism (v4; GraphPad Software, La Jolla, CA).
Sensitivity of phage to organic solvents.
The sensitivity of phage KHP30 to organic solvents was examined as described elsewhere (46). Briefly, after the purified phage was dialyzed against AAS for 1 h at 4°C, 1 volume of either AAS (control), chloroform, or diethyl ether was added to 3 volumes of the purified phage suspension (ca. 4.0 × 107 PFU/ml). After the components were mixed, the samples were incubated at 25°C for 1 h. The phage concentrations were then measured.
Phage lipid extraction and TLC analysis.
After the ultracentrifugal purification, six fractions (0.5 ml) were collected from the top of the centrifugal tube (total volume, 3 ml). The phage concentration in each fraction was measured. The fractions were then dialyzed against AAS solution. The fractions were dried with a vacuum concentrator (CC-105; Tomy Digital Biology, Tokyo, Japan). Chloroform-methanol (2:1, vol/vol; 1 ml) was added, and the sample was incubated at 25°C for 2 h with shaking. The suspension was centrifuged (1,000 × g, 10 min), and the organic solvent was removed with the vacuum concentrator (CC-105; Tomy Digital Biology). Chloroform (0.1 ml) was added, and the sample was stored at −20°C until use. The extract from each fraction was spotted onto a silica gel 60 F254 thin-layer chromatography (TLC) plate (Merck, Darmstadt, Germany), and a one-dimensional chromatograph was developed with chloroform-methanol-water (14:6:1, vol/vol). The lipids could be separated using the differences in their ionic charge in TLC. After the silica gel plates were dried, the lipids were stained with iodine vapor.
Phage adsorption, latent period, and burst size.
H. pylori strain 3401 (ca. 3.6 × 108 cells/ml) was prepared. For phage adsorption measurement, phage KHP30 was mixed with the H. pylori culture at an MOI of ca. 10−6 to 10−5. After incubation with shaking for specific periods, 1 ml of the culture was collected and centrifuged at 9,200 × g for 1 min, and the phage concentration in the supernatant was measured. Moreover, for the measurements of the phage latent period and burst size, phage KHP30 was added to the H. pylori culture at an MOI of ca. 10−5 and the culture was incubated for 10 to 15 min. The H. pylori sample was washed with BEV medium and resuspended in fresh BEV medium. An aliquot of the H. pylori culture was collected at specific points in time, and the phage concentration was measured. The doubling time of H. pylori 3401 was also calculated from the turbidity change (in Klett units).
Host spectrum of the phage.
The phage was amplified in H. pylori strain 3401, KMT84, or KMT86. The plaque-forming activities of the phage were examined by streaking a loop of the phage lysates onto double-layered agar containing the appropriate H. pylori strain. The plaques that formed on the double-layered plate were examined after incubation.
Stability of the phage at various pHs.
Brucella broth was prepared with a pH of 1.5, 2.0, 2.5, 3.0, 3.5, 4.0, 6.0, 6.8 (standard Brucella broth), 8.0, 10.0, or 12.0, adjusted with HCl or NaOH, as appropriate. The pH of the medium was measured using a pH meter (D-51AC; Horiba, Kyoto, Japan). After 104-fold dilution of the phage suspension with brucella broth at various pHs to ca. 6.0 × 105 PFU/ml, the suspensions were incubated at 25°C for 1 h (46). After incubation, the pHs of the phage suspensions were rechecked with pH indicator paper (Whatman, Kent, United Kingdom). The phage concentrations were measured.
Bioinformatic analysis of the phage KHP30 genome.
The possible integration of a KHP30-like phage into the host genome was examined with the BLASTp program. The similarity of phage KHP30 to other H. pylori phages was examined with the BLASTp and BLASTn programs. The analyses were performed with the software GenomeMatcher (http://www.ige.tohoku.ac.jp/joho/gmProject/gmhomeJP.html) (47). The phage and bacterial data were obtained from GenBank, and the data used in the analyses are listed in Table S2 in the supplemental material.
PFGE of H. pylori DNA.
Samples for pulsed-field gel electrophoresis (PFGE) were prepared from H. pylori strains 3401 and NY43 (ca. 1.7 × 109 cells/ml), as described elsewhere (48). A bacteriophage lambda ladder PFG marker (New England BioLabs, Beverly, MA) was included in this analysis as a molecular weight standard. Electrophoresis (1% gel) was performed using a CHEF-DR II system (Bio-Rad Laboratories) at 6.0 V/cm with pulse ramps from 1 to 45 s for 20 h at 14°C in 0.5× Tris-borate-EDTA buffer. After staining with ethidium bromide, the gel was visualized under UV transillumination.
Southern blot analysis of phage DNAs.
After PFGE of H. pylori DNA, a Southern blot analysis was conducted, as described elsewhere (49). Open reading frame 14 (orf14) of phage KHP30, which encodes the major structural protein of this phage (see below), was used as a probe to detect phage KHP30 DNA for the following reason. When the orf14 sequence of phage KHP30 was compared with sequences in the NCBI database using BLASTn, orf14-like sequences were not found on the DNA sequences intrinsic to H. pylori, although they were found on putative phage DNA sequences integrated into the genomes in some H. pylori strains. A primer set (primers ORF14F [ GGTATAGAAGTTGGTAGAGAGATCC ] and ORF14R [ CCATATCAGACACTAAACCGATCACG ] ) was designed on the basis of the orf14 sequence in the phage KHP30 genome (16), and the orf14 fragment was amplified by PCR using the phage DNA as a template. The phage DNA was obtained, as described below in “Restriction digestion analysis of phage DNAs.” The PCR product and bacteriophage λ DNA (TaKaRa Bio) were labeled with horseradish peroxidase, and the signals were detected using an Amersham ECL direct nucleic acid labeling and detection system (GE Healthcare, Little Chalfont, United Kingdom) according to the manufacturer's instructions.
Restriction digestion analysis of phage DNAs.
In silico restriction digestion of the phage KHP30 genome was conducted using the NEBcutter (v2.0) program (New England BioLabs) (50). The restriction enzyme EcoRI was selected for this analysis because the digestion pattern was appropriate.
Genomic DNA was extracted from the phage particles essentially as described elsewhere (43, 44). Moreover, H. pylori strain NY43, which releases phage KHP30, was cultured in BEV medium and then pelleted. The phage DNAs inside the H. pylori cells were extracted using a FastGene plasmid minikit (Nippon Genetics Co., Tokyo, Japan). The phage DNAs (1 μg) were digested with EcoRI (TaKaRa Bio) and then resolved electrophoretically in a 0.8% agarose gel. After staining with ethidium bromide, the gels were visualized under UV transillumination.
Phage protein analysis.
Phage proteins were prepared from the phage purified by CsCl density gradient ultracentrifugation for SDS-PAGE, as described previously (44). The phage proteins were separated electrophoretically on a 12.5% SDS-polyacrylamide gel with an (unstained) XL-Ladder Broad marker (APRO Life Science Institute, Tokushima, Japan), basically followed by the protocol described elsewhere (49). Phage proteins were subjected to N-terminal protein sequencing and mass spectrometry, as described elsewhere (51).
Accession numbers.
The protein sequences determined by N-terminal sequencing were deposited in the UniProt database (accession numbers I7H893 for ORF13 and I7H0H9 for ORF14). Moreover, for the analysis of mass spectrometric data, the database of protein data for phage KHP30 combined with H. pylori strain 2018 was constructed locally. H. pylori strain 2018 has no phage protein sequence similar to that of phage KHP30 and was used to increase the accuracy of protein identification. The protein data for phage KHP30 and H. pylori strain 2018 were obtained from GenBank (accession numbers AB647160 and CP002572, respectively). The data were analyzed with the Paragon method using ProteinPilot (v3.0) software (AB Sciex) (52). The protein identification threshold was the detection of more than one peptide with 99% confidence.
RESULTS AND DISCUSSION
Morphological features of phage KHP30.
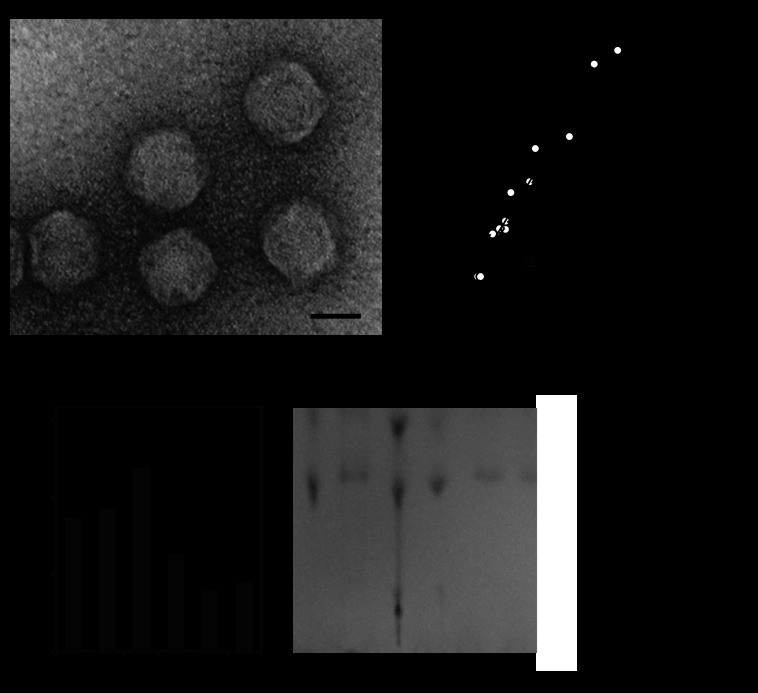
The purified phage KHP30 with plaque-forming ability was successfully isolated. Electron microscopic analysis of the phage morphology showed that phage KHP30 has a spherical shape with no tail (Fig. 1A). The diameter of the phage KHP30 particle was 68.8 ± 2.3 nm (mean ± standard deviation, n = 11). In addition, when observing the thin sections of phage-infected H. pylori, no tail production was seen, nor were any tails attached to the phages observed (see Fig. S3 in the supplemental material). All these observations suggest that phage KHP30 is a spherical phage.
Fig 1.
Morphological features of phage KHP30. (A) Electron microscopic observation of intact phage KHP30 particles. Bar, 50 nm. (B) Analysis of the head/particle size and genome size of phage KHP30 compared with those of other phages. White and black circles, tailed and spherical phages, respectively; black triangle, phage KHP30. Linear regression lines for the tailed and spherical phages are drawn in black (r2 = 0.9664 and 0.9712, respectively), and the 95% confidence intervals for the mean predictions are plotted as dotted lines. (C) Lipid content of phage KHP30 using the fractions obtained from CsCl density gradient ultracentrifugation. (Left) Phage concentration in each fraction; (right) TLC of the lipids extracted from the indicated fractions.
Phage KHP30 has a double-stranded DNA (dsDNA) genome (16). Spherical phages with dsDNA often contain lipids inside the phage particles (53), and so they seemed to have a smaller genomic DNA size relative to the particle volume than the tailed phages. Therefore, trends in the phage head/particle volumes and the genome sizes in the tailed and spherical phages were then analyzed, and the trend for phage KHP30 was examined (Fig. 1B). Both the tailed and spherical phages have their own trends for the ratio of the head/particle volume to the genome size. The trend for phage KHP30 differed from that for the tailed phages, and it seemed to be closer to that for the spherical phages containing lipids.
In general, the authentic tailed phages seem to be resistant to treatment with organic solvents, such as chloroform and ether, whereas the spherical phages containing lipids are sensitive to them (54–56). Thus, treatment of phage KHP30 with organic solvents can be used to speculate indirectly over its lipid content. The sensitivity of phage KHP30 to organic solvents such as chloroform and diethyl ether was examined. Chloroform depleted the phage concentration by almost 100%, and diethyl ether depleted it by ca. 80%. Thus, phage KHP30 was assumed to contain lipids. Subsequently, the presence of lipids in the phage KHP30 particles was examined by TLC lipid analysis of fractions obtained from the ultracentrifugal phage purification (Fig. 1C) (25, 31, 32, 34). Fraction 3, containing a phage band, showed the highest phage concentration, and the lipids in that fraction were most clearly separated by TLC, although the lipid class has not yet been specified. Thus, phage KHP30 was considered to have lipids.
Unlike the other enveloped viruses, some spherical phages with a dsDNA genome, which are classified into tectiviruses or corticoviruses, contain lipids inside the viral particles (53–56). Electron microscopic observation of the purified phage particles treated with chloroform or diethyl ether showed that phage KHP30 also did not have lipids outside the particles (data not shown).
Adsorption, latent period, and burst size of phage KHP30.
In general, phage infection is initiated by the adsorption of the phage to the bacterial surface, and the infecting phage produces progeny phage inside the cells, which then spreads outside the cells after cell lysis. These infection processes can be measured as the adsorption efficiency, latent period (i.e., the time from adsorption to the release of progeny phage), and burst size (i.e., the ratio of the released phage to the infecting phage) (57, 58). These infection parameters are important in evaluating the biological activity of the phage.
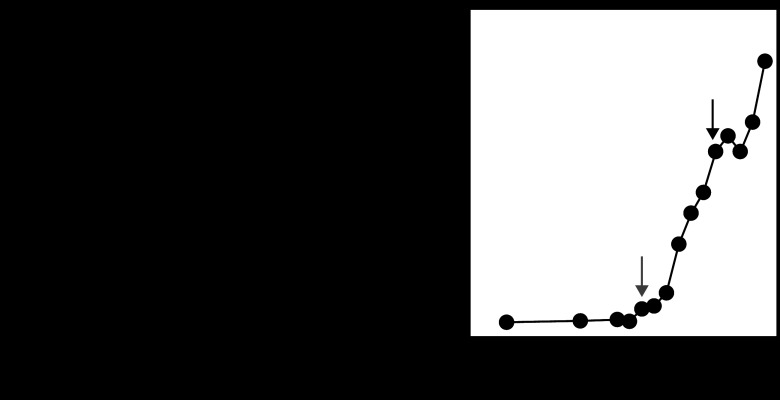
First, the adsorption efficiency was examined using H. pylori strain 3401. Approximately 30% of the phage population adsorbed to this strain within 15 min, after which most phage adsorption seemed to be reduced (Fig. 2A). Although CaCl2 and/or MgCl2 is essential for the adsorption of some phages (46), the adsorption efficiency of phage KHP30 was not improved by these divalent cations (data not shown). Second, the latent period of phage KHP30 was ca. 140 min (Fig. 2B) and is therefore longer than the latent periods of other spherical phages with dsDNA genomes, which generally range from 35 to 45 min (46). The latent period of phage KHP30 is thus less than the H. pylori doubling time, which is usually ca. 3 h, as in H. pylori strain 3401 (59). Thus, the phage life cycle is completed within the period of bacterial cell division.
Fig 2.
Life cycle of phage KHP30 in H. pylori strain 3401. (A) Adsorption rate. (B) Single-step phage growth analysis. Gray arrow, initiation of phage production; black arrow, time when the phage production curve leveled off. The period before phage production (until the gray arrow) is the latent period. The burst size was determined as the relative phage number at the plateau stage (at the black arrow).
Finally, the phage burst size generally ranges from 50 to 100 in the tailed phages and from 40 to 700 in the spherical phages with dsDNA genomes (46). However, the burst size of phage KHP30 was ca. 13 in H. pylori strain 3401 and is therefore quite small (Fig. 2B).
Host range and phage infectivity of different H. pylori strains.
The plaque-forming capacity of phage KHP30 was examined in 44 H. pylori strains. Phage KHP30 formed plaques on 28 of these H. pylori strains (63.6%; see Table S1 in the supplemental material). Thus, phage KHP30 seems to have a significantly broader host range than other reported H. pylori phages (9, 11).
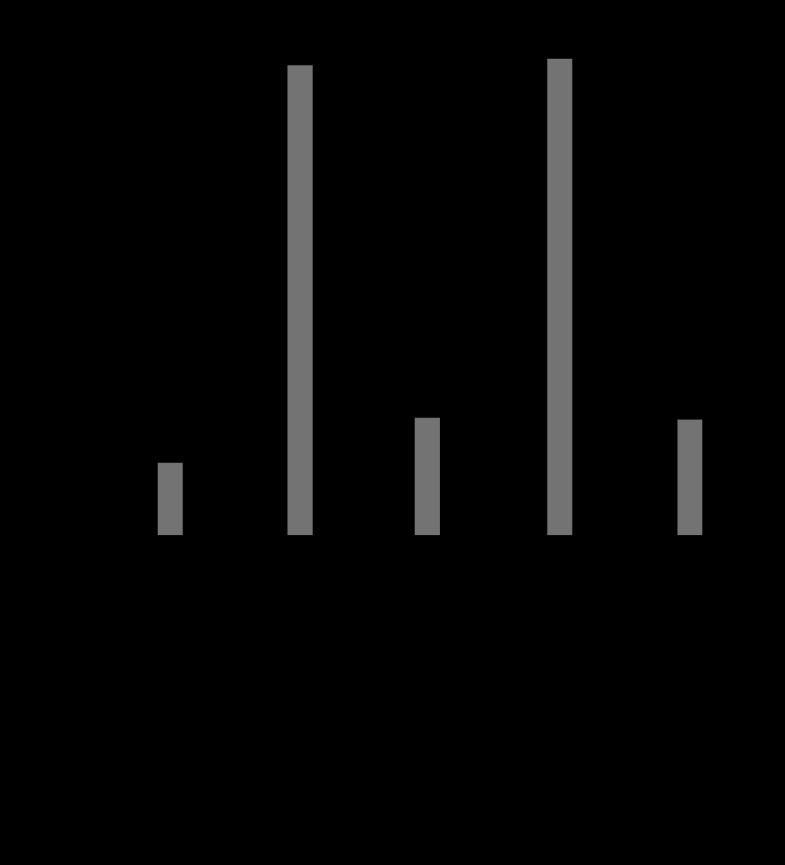
In our host range analysis, the phage plaque-forming capacity seemed to differ among the individual H. pylori strains, and its infectivity seemed to be influenced by the host immunity. We examined the infectivity of phage KHP30 for H. pylori strains by comparing the efficiency of plating of phage KHP30 in H. pylori strains 3401, KMT84, and KMT86. The relative efficiency of plating was calculated as the ratio of the number of PFU when the phage was plated on a particular bacterial strain to the number of PFU when it was plated on standard strain 3401. Phage KHP30 was initially adapted to strain 3401 (KHP30/3401) and did not show strong infectivity for H. pylori strains KMT84 and KMT86 compared with its infectivity for strain 3401 (Fig. 3). Phage KHP30 was then adapted to H. pylori strains KMT84 and KMT86, and the adapted phages were designated KHP30/KMT84 and KHP30/KMT86, respectively. These phages showed highly increased infectivity for their adaptive hosts, KMT84 and KMT86, respectively (Fig. 3). The infectivity of phages KHP30/KMT84 and KHP30/KMT86 for H. pylori 3401 was also maintained. Phages KHP30/KMT84 and KHP30/KMT86 were then readapted to H. pylori strain 3401 and designated phages KHP30/KMT84/3401 and KHP30/KMT86/3401, respectively. The infectivity of phages KHP30/KMT84/3401 and KHP30/KMT86/3401 seemed to revert to that of phage KHP30/3401 (Fig. 3). These data suggest that phage KHP30 can adapt to H. pylori immunity, which may be an adaptation to the bacterial restriction-modification system.
Fig 3.
Phage infectivity in different H. pylori strains. Phage KHP30 was passaged through H. pylori strain 3401, KMT84, or KMT86. The efficiency of plating was measured and used as the index of phage infectivity. Gray and black bars, efficiencies of plating for the phage in H. pylori strains KMT84 and KMT86, respectively.
Stability at various pHs.
H. pylori protects itself from the highly acidic environment of the stomach by neutralizing the gastric juices with urease and by moving by chemotaxis through the mucus to the epithelium, which is considered to be near pH 7.0 (1, 60). Although H. pylori has developed these tactics as adaptive strategies, the phage cannot move by itself and cannot produce enzymes to protect itself. If a phage lyses H. pylori in the mucus of the stomach, the progeny phage will be exposed to the harsh gastric environment, in which the pH ranges from neutral to acidic. Therefore, phage stability at acidic pHs is relevant to its survival. To examine the phage's stability at various pHs, we incubated phage KHP30 in culture media of different pHs, where it showed stability over a pH range of 2.5 to 10 (Fig. 4). The pHs of the phage solutions remained the same after incubation. Thus, the phage itself is considered to be stable over a wide range of pHs.
Fig 4.
Stability of phage KHP30 at different pHs. The activity of phage KHP30 was measured after incubation for 1 h at different pHs. The phage concentration measured in standard brucella broth (pH 6.8) was set equal to 100%.
Some spherical phages containing lipids have been isolated from harsh environments (with high temperatures or high salt concentrations) that are also inhabited by the host bacterium (25, 31, 32). Considering the unique morphological features of the lipid-containing spherical phages and their niches, phage KHP30 is considered to have adapted to the extreme gastric environment, and phage lipids may function to confer stability and protect the phage from this harsh environment.
Genomic DNA conformation in H. pylori cells and phage particles.
Integrated phage sequences have been discovered in H. pylori strains isolated worldwide (11, 14, 15) and were briefly confirmed by in silico analysis in this study (see Fig. S4 in the supplemental material). Integrated phage sequences have also been found in Helicobacter spp. infecting cats, mice, and ferrets. H. pylori strain NY43, from which phage KHP30 was originally isolated, spontaneously and continuously produces phage KHP30, even when it is repeatedly subcultured. The whole genome of phage KHP30 (ca. 26 kbp) contains a putative integrase gene, orf2 (16). Therefore, phage KHP30 is inferred to be a lysogenic phage.
To determine the state of the phage KHP30 genomic DNA in H. pylori cells, the phage inside H. pylori strain NY43 was analyzed by PFGE and Southern blotting (Fig. 5A). These analyses revealed that phage KHP30 DNA was present independently of the bacterial genome in the host bacterial cell. Thus, the phage KHP30 genome did not seem to integrate into the bacterial genome of its host, H. pylori strain NY43. Moreover, the phage DNAs were extracted from the phage particles and from H. pylori strain NY43 and digested with EcoRI, and the DNA band pattern was examined (Fig. 5B). The extrachromosomal DNA was considered to be predominantly phage KHP30 because the DNA digestion pattern was very similar to that of the DNAs extracted from the phage particles. When we compared the in vitro digestion pattern with the in silico digestion pattern, the phage DNA inside the bacterial cells was a mixture of the linear and circular forms and/or the concatemeric form. On the basis of this evidence, the phage KHP30 DNA seemed to be present as episomes in the host cell.
Fig 5.
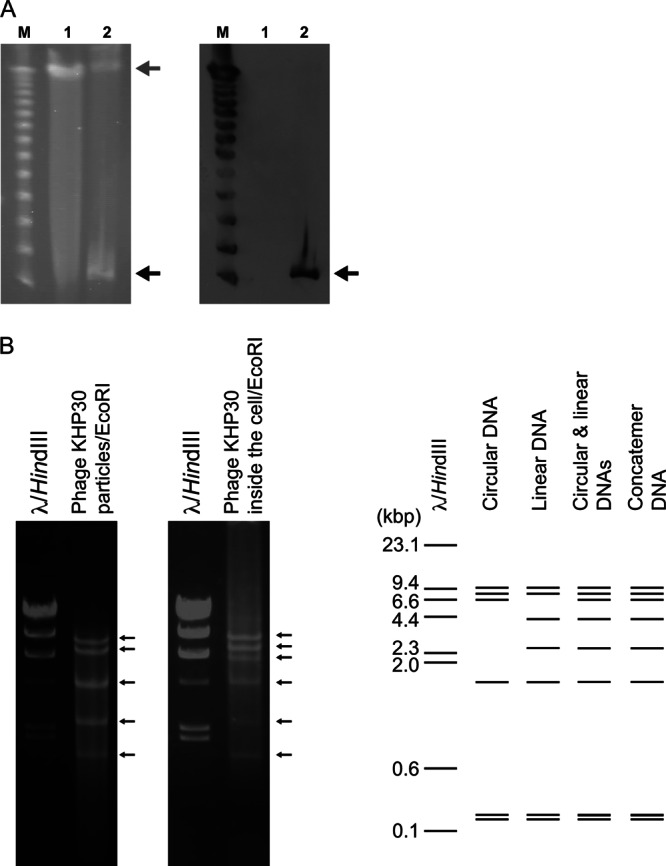
(A) Location of the intracellular phage genome in phage-producing H. pylori strain NY43. The DNAs of H. pylori strains 3401 and NY43 were separated by PFGE (left) and subjected to Southern blot analysis with a phage-specific probe (right). H. pylori strain 3401 was used as the non-phage-harboring control. Black and gray arrows, phage and H. pylori DNAs, respectively. Lanes M, 1, and 2 in both panels, bacteriophage lambda ladder, strain 3401, and strain NY43, respectively. (B) Restriction digestion analysis of phage DNAs. (Left) Phage DNAs derived from the purified particles or the phage DNAs derived from H. pylori strain NY43 were digested with EcoRI and separated electrophoretically in a 0.8% agarose gel. Bacteriophage lambda DNA digested with HindIII was also separated electrophoretically as a marker. The DNA bands are indicated by arrows. (Right) In silico EcoRI digestion patterns of phage KHP30 DNA in the linear form, the circular form, a mixture of the linear and circular forms, and the concatemeric form, which were resolved in a 0.7% agarose gel.
In a pseudolysogenic life cycle, phage is present as a plasmid or episome when the cells do not grow efficiently (61, 62). Both lytic and lysogenic phages can transmit their life cycle to pseudolysogeny (61, 62). Considering the biological activity of phage KHP30 under this experimental condition, phage KHP30 seemed to conduct a pseudolysogenic life cycle preferentially, at least in H. pylori strain NY43.
Phage KHP30 protein analysis.
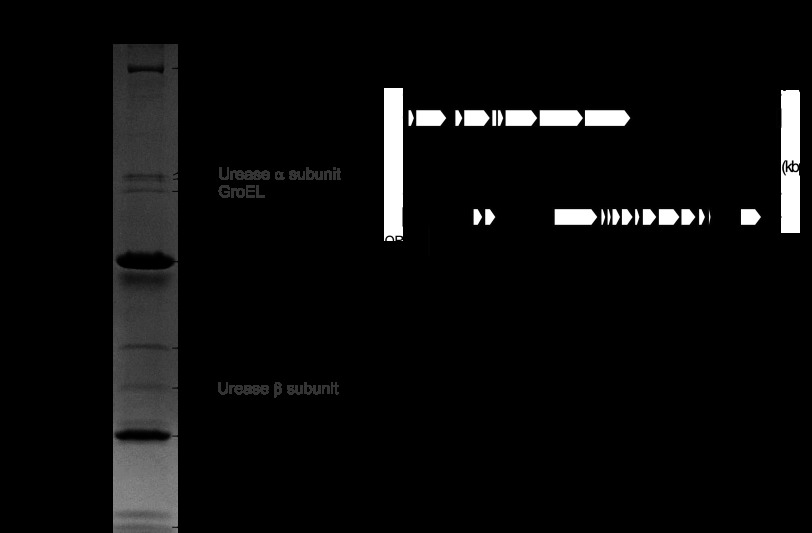
The proteins derived from the purified phage KHP30 particles were analyzed with N-terminal protein sequencing and mass spectrometry (Fig. 6), which identified ORFs 13 and 14 and ORFs 3, 11, 12, 13, 14, 17, and 29, respectively. According to the pattern of protein bands separated by SDS-PAGE, ORF14 was found to have the greatest quantity among the separated proteins, so ORF14 was considered to be a major structural protein. ORFs 13 and 11 had the second and third greatest quantities, so they were also considered structural proteins. N-terminal processing of the major structural protein, as seen in the major structural protein of the other tailed phage, has not been observed in ORF14 of phage KHP30 (43, 63–65), although the first 3 amino acid residues of ORF13 seemed to be removed. In contrast, ORFs 3, 12, 17, and 29 were present in minor amounts. Host proteins, such as the α and β subunits of urease and GroEL, were detected in small amounts. It is unclear whether these minor host and phage proteins are components of phage KHP30.
Fig 6.
Analysis of phage KHP30 proteins. (Left) The proteins of purified phage KHP30 were separated by SDS-PAGE and were analyzed by N-terminal protein sequencing and mass spectrometry. N-terminal protein sequencing showed ORF13 to have the sequence GIKEKEIELETLKREIAQAEASLEQDFIKH and ORF14 to have the sequence MLEKLNNINFNNISNNLNLGIEVGREIQNA. The ORFs are indicated on the right of the gel and were determined by mass spectrometry. The ORF with an asterisk is a protein identified with N-terminal protein sequencing. (Right) Genome map. Black arrows, ORFs identified by protein analysis.
The ORFs encoding the identified proteins were mapped on the phage KHP30 genome and seemed to be clustered as a module (i.e., ORFs 11, 12, 13, and 14), as seen in other tailed phage genomes (66, 67). Moreover, the genome of phage KHP30 was compared with those of other H. pylori phages, including phages KHP40, ϕHP33, and 1961P. These phages showed a high level of similarity not only in their DNA sequence but also in their ORFs and ORF arrangement (see Fig. S5 in the supplemental material). In particular, structural proteins (i.e., ORFs 11, 12, 13, and 14) were relatively conserved.
Possible novel classification of H. pylori phage KHP30.
Lipid-containing spherical phages such as phage KHP30 are not frequently isolated in phage research. According to the current viral taxonomic classification, the members of the families Corticoviridae and Tectiviridae are lipid-containing spherical phages with dsDNA genomes (55, 56). However, phage KHP30 differs from the spherical phages of these two phage families. First, the genome size of phage KHP30 (ca. 26 kbp) is larger than the genome sizes of the phages of the families Corticoviridae and Tectiviridae, which are ca. 10 kbp and ca. 15 kbp, respectively, although the particle diameters of phage KHP30 and the other spherical phages (corticoviruses and tectiviruses) are relatively similar (69 nm, 57 nm, and 66 nm, respectively) (16, 55, 56). Second, phage KHP30 may be pseudolysogenic or lysogenic, whereas the phages of these two families are lytic (55, 56). Third, the ORFs and the ORF arrangement of phage KHP30 are dissimilar to those of the other lipid-containing spherical phages. Finally, the burst size of phage KHP30 (ca. 13) is considerably smaller than the burst sizes of the phages belonging to these two phage families (ca. 40 to 700) (46). On the basis of the evidence reported in this study, H. pylori phage KHP30 is very different from the phages of the families Corticoviridae and Tectiviridae.
Because of the genetic and genomic similarities among H. pylori phages, H. pylori phages KHP30, KHP40, ϕHP33, and 1961P were considered to belong to the same group. However, the morphology of phage KHP30 differs from those of H. pylori phages ϕHP33 and 1961P, which have been classified in the order Caudovirales (i.e., tailed phage), whereas phage KHP30 has been identified in this study as a lipid-containing spherical phage. The other H. pylori phage, KHP40, has also been isolated by us (16). Phage KHP40, which was amplified in H. pylori strain 3401 and also observed with electron microscopy in this study, was identified as a spherical phage almost identical in size to phage KHP30 (see Fig. S6 in the supplemental material). Although the reason for the morphological differences of the H. pylori phages is unknown, it may be a consequence of the host strains used for phage amplification or because of other unknown factors. We believe that H. pylori phages KHP30 and KHP40 constitute a novel type of lipid-containing spherical phage which may be classified within a new viral family.
Conclusion.
In this study, we revealed that H. pylori phage KHP30 is a novel spherical phage containing lipids with double-stranded DNA. It was stable over a wide pH range, and it could multiply by over 60% in its host. Phage KHP30 has a long latent period and a relatively small burst size. The phage was shown to be maintained as an episome, at least in H. pylori NY43 cells (i.e., the cells had undergone pseudolysogeny). Phages generally contribute to host bacterial evolution by horizontal gene transfer and the host-parasite coevolutionary arms race. We hope that further study of the molecular biology and epidemiology of H. pylori phages will clarify their pivotal role in H. pylori evolution.
Supplementary Material
Footnotes
Published ahead of print 8 March 2013
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.03530-12.
REFERENCES
- 1.Konturek PC, Konturek SJ, Brzozowski T. 2009. Helicobacter pylori infection in gastric cancerogenesis. J. Physiol. Pharmacol. 60:3–21 [PubMed] [Google Scholar]
- 2.Peek RM, Blaser MJ. 2002. Helicobacter pylori and gastrointestinal tract adenocarcinomas. Nat. Rev. Cancer 2:28–37 [DOI] [PubMed] [Google Scholar]
- 3.Atherton JC, Blaser MJ. 2009. Coadaptation of Helicobacter pylori and humans: ancient history, modern implications. J. Clin. Invest. 119:2475–2487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suzuki R, Shiota S, Yamaoka Y. 2012. Molecular epidemiology, population genetics, and pathogenic role of Helicobacter pylori. Infect. Genet. Evol. 12:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canchaya C, Fournous G, Chibani-Chennoufi S, Dillmann ML, Brüssow H. 2003. Phage as agents of lateral gene transfer. Curr. Opin. Microbiol. 6:417–424 [DOI] [PubMed] [Google Scholar]
- 6.Kashiwagi A, Yomo T. 2011. Ongoing phenotypic and genomic changes in experimental coevolution of RNA bacteriophage Q.β and Escherichia coli. PLoS Genet. 7:e1002188 doi:10.1371/journal.pgen.1002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Krylov VN. 2003. Role of horizontal gene transfer by bacteriophages in the origin of pathogenic bacteria. Genetika 39:595–620 (In Russian.) [PubMed] [Google Scholar]
- 8.Paterson S, Vogwill T, Buckling A, Benmayor R, Spiers AJ, Thomson NR, Quail M, Smith F, Walker D, Libberton B, Fenton A, Hall N, Brockhurst MA. 2010. Antagonistic coevolution accelerates molecular evolution. Nature 464:275–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heintschel von Heinegg E, Nalik HP, Schmid EN. 1993. Characterization of a Helicobacter pylori phage (HP1). J. Med. Microbiol. 38:245–249 [DOI] [PubMed] [Google Scholar]
- 10.Lehours P, Vale FF, Bjursell MK, Melefors O, Advani R, Glavas S, Guegueniat J, Gontier E, Lacomme S, Alves Matos A, Menard A, Mégraud F, Engstrand L, Andersson AF. 2011. Genome sequencing reveals a phage in Helicobacter pylori. mBio 2(6):e00239–11 doi:10.1128/mBio.00239-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luo CH, Chiou PY, Yang CY, Lin NT. 2012. Genome, integration, and transduction of a novel temperate phage of Helicobacter pylori. J. Virol. 86:8781–8792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schmid EN, von Recklinghausen G, Ansorg R. 1990. Bacteriophages in Helicobacter (Campylobacter) pylori. J. Med. Microbiol. 32:101–104 [DOI] [PubMed] [Google Scholar]
- 13.Wan XQ, Tang DS, Liu AP, Tan SY, Li WK, Kuang J, Li HM. 2011. Isolation of a wild-type virulent phage of Helicobacter pylori and its simulated treatments of gastrointestinal Hp in vitro. Nan Fang Yi Ke Da Xue Xue Bao 31:304–307 (In Chinese.) [PubMed] [Google Scholar]
- 14.Arnold IC, Zigova Z, Holden M, Lawley TD, Rad R, Dougan G, Falkow S, Bentley SD, Müller A. 2011. Comparative whole genome sequence analysis of the carcinogenic bacterial model pathogen Helicobacter felis. Genome Biol. Evol. 3:302–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eppinger M, Baar C, Linz B, Raddatz G, Lanz C, Keller H, Morelli G, Gressmann H, Achtman M, Schuster SC. 2006. Who ate whom? Adaptive Helicobacter genomic changes that accompanied a host jump from early humans to large felines. PLoS Genet. 2:e120 doi:10.1371/journal.pgen.0020120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchiyama J, Takeuchi H, Kato S, Takemura-Uchiyama I, Ujihara T, Daibata M, Matsuzaki S. 2012. Complete genome sequences of two Helicobacter pylori bacteriophages isolated from Japanese patients. J. Virol. 86:11400–11401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Karita M, Kouchiyama T, Okita K, Nakazawa T. 1991. New small animal model for human gastric Helicobacter pylori infection: success in both nude and euthymic mice. Am. J. Gastroenterol. 86:1596–1603 [PubMed] [Google Scholar]
- 18.Akopyants NS, Eaton KA, Berg DE. 1995. Adaptive mutation and cocolonization during Helicobacter pylori infection of gnotobiotic piglets. Infect. Immun. 63:116–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee A, O'Rourke J, De Ungria MC, Robertson B, Daskalopoulos G, Dixon MF. 1997. A standardized mouse model of Helicobacter pylori infection: introducing the Sydney strain. Gastroenterology 112:1386–1397 [DOI] [PubMed] [Google Scholar]
- 20.Marshall BJ, Royce H, Annear DI, Goodwin CS, Pearman JW, Warren JR, Armstrong JA. 1984. Original isolation of Campylobacter pyloridis from human gastric mucosa. Microbios Lett. 25:83–88 [Google Scholar]
- 21.Trang VT, Takeuchi H, Kudo H, Aoki A, Katsuno S, Shimamura T, Sugiura T, Ukeda H. 2009. Antimicrobial activity of aminoreductone against Helicobacter pylori. J. Agric. Food Chem. 57:11343–11348 [DOI] [PubMed] [Google Scholar]
- 22.Yamaguchi H, Osaki T, Taguchi H, Sato N, Toyoda A, Takahashi M, Kai M, Nakata N, Komatsu A, Atomi Y, Kamiya S. 2003. Effect of bacterial flora on postimmunization gastritis following oral vaccination of mice with Helicobacter pylori heat shock protein 60. Clin. Diagn. Lab. Immunol. 10:808–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bair MJ, Chen CL, Chiang CK, Huang MF, Hu CC, Chang HT. 2008. Capillary electropherograms for restriction fragment length polymorphism of Helicobacter pylori. Electrophoresis 29:3964–3970 [DOI] [PubMed] [Google Scholar]
- 24.Uchiyama J, Takemura I, Hayashi I, Matsuzaki S, Satoh M, Ujihara T, Murakami M, Imajoh M, Sugai M, Daibata M. 2011. Characterization of lytic enzyme open reading frame 9 (ORF9) derived from Enterococcus faecalis bacteriophage ϕEF24C. Appl. Environ. Microbiol. 77:580–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aalto AP, Bitto D, Ravantti JJ, Bamford DH, Huiskonen JT, Oksanen HM. 2012. Snapshot of virus evolution in hypersaline environments from the characterization of a membrane-containing Salisaeta icosahedral phage 1. Proc. Natl. Acad. Sci. U. S. A. 109:7079–7084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bamford DH, Bamford JKH. 2005. Lipid-containing bacteriophage PM2, the type organism of Corticoviridae, p 171–174 Calendar R, Abedon ST. (ed), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 27.Dröge A, Santos MA, Stiege AC, Alonso JC, Lurz R, Trautner TA, Tavares P. 2000. Shape and DNA packaging activity of bacteriophage SPP1 procapsid: protein components and interactions during assembly. J. Mol. Biol. 296:117–132 [DOI] [PubMed] [Google Scholar]
- 28.Fukuda K, Ishida W, Uchiyama J, Rashel M, Kato S, Morita T, Muraoka A, Sumi T, Matsuzaki S, Daibata M, Fukushima A. 2012. Pseudomonas aeruginosa keratitis in mice: effects of topical bacteriophage KPP12 administration. PLoS One 7:e47742 doi:10.1371/journal.pone.0047742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grahn AM, Butcher SJ, Bamford JKH, Bamford DH. 2005. PRD1: dissecting the genome, structure, and entry, p 161–170 Calendar R, Abedon ST. (ed), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 30.Hoshiba H, Uchiyama J, Kato S, Ujihara T, Muraoka A, Daibata M, Wakiguchi H, Matsuzaki S. 2010. Isolation and characterization of a novel Staphylococcus aureus bacteriophage, ϕMR25, and its therapeutic potential. Arch. Virol. 155:545–552 [DOI] [PubMed] [Google Scholar]
- 31.Jaatinen ST, Happonen LJ, Laurinmäki P, Butcher SJ, Bamford DH. 2008. Biochemical and structural characterisation of membrane-containing icosahedral dsDNA bacteriophages infecting thermophilic Thermus thermophilus. Virology 379:10–19 [DOI] [PubMed] [Google Scholar]
- 32.Jalasvuori M, Jaatinen ST, Laurinavicius S, Ahola-Iivarinen E, Kalkkinen N, Bamford DH, Bamford JK. 2009. The closest relatives of icosahedral viruses of thermophilic bacteria are among viruses and plasmids of the halophilic archaea. J. Virol. 83:9388–9397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kazmierczak KM, Rothman-Denes LC. 2005. Bacteriophage N4, p 302–314 Calendar R, Abedon ST. (ed), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 34.Kivelä HM, Kalkkinen N, Bamford DH. 2002. Bacteriophage PM2 has a protein capsid surrounding a spherical proteinaceous lipid core. J. Virol. 76:8169–8178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klumpp J, Dorscht J, Lurz R, Bielmann R, Wieland M, Zimmer M, Calendar R, Loessner MJ. 2008. The terminally redundant, nonpermuted genome of Listeria bacteriophage A511: a model for the SPO1-like myoviruses of gram-positive bacteria. J. Bacteriol. 190:5753–5765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lehnherr H. 2005. Bacteriophage P1, p 350–364 Calendar R, Abedon ST. (ed), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 37.Lickfeld KG, Menge B. 1976. Morphogenesis of bacteriophage lambda: electron microscopy of thin sections. J. Mol. Biol. 103:299–318 [DOI] [PubMed] [Google Scholar]
- 38.Matsuzaki S, Yasuda M, Nishikawa H, Kuroda M, Ujihara T, Shuin T, Shen Y, Jin Z, Fujimoto S, Nasimuzzaman MD, Wakiguchi H, Sugihara S, Sugiura T, Koda S, Muraoka A, Imai S. 2003. Experimental protection of mice against lethal Staphylococcus aureus infection by novel bacteriophage ϕMR11. J. Infect. Dis. 187:613–624 [DOI] [PubMed] [Google Scholar]
- 39.Nagy E, Prágai B, Ivánovics G. 1976. Characteristics of phage AP50, an RNA phage containing phospholipids. J. Gen. Virol. 32:129–132 [DOI] [PubMed] [Google Scholar]
- 40.Ravantti JJ, Gaidelyte A, Bamford DH, Bamford JK. 2003. Comparative analysis of bacterial viruses Bam35, infecting a gram-positive host, and PRD1, infecting gram-negative hosts, demonstrates a viral lineage. Virology 313:401–414 [DOI] [PubMed] [Google Scholar]
- 41.Smith MCM. 2005. Molecular genetics of Streptococcus phages, p 621–635 Calendar R, Abedon ST. (ed), The bacteriophages, 2nd ed Oxford University Press, New York, NY [Google Scholar]
- 42.Son JS, Lee SJ, Jun SY, Yoon SJ, Kang SH, Paik HR, Kang JO, Choi YJ. 2010. Antibacterial and biofilm removal activity of a podoviridae Staphylococcus aureus bacteriophage SAP-2 and a derived recombinant cell-wall-degrading enzyme. Appl. Microbiol. Biotechnol. 86:1439–1449 [DOI] [PubMed] [Google Scholar]
- 43.Uchiyama J, Rashel M, Maeda Y, Takemura I, Sugihara S, Akechi K, Muraoka A, Wakiguchi H, Matsuzaki S. 2008. Isolation and characterization of a novel Enterococcus faecalis bacteriophage ϕEF24C as a therapeutic candidate. FEMS Microbiol. Lett. 278:200–206 [DOI] [PubMed] [Google Scholar]
- 44.Uchiyama J, Rashel M, Takemura I, Kato S, Ujihara T, Muraoka A, Matsuzaki S, Daibata M. 2012. Genetic characterization of Pseudomonas aeruginosa bacteriophage KPP10. Arch. Virol. 157:733–738 [DOI] [PubMed] [Google Scholar]
- 45.Verheust C, Fornelos N, Mahillon J. 2005. GIL16, a new gram-positive tectiviral phage related to the Bacillus thuringiensis GIL01 and the Bacillus cereus pBClin15 elements. J. Bacteriol. 187:1966–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ackermann HW, DuBow MS. 1987. Viruses of prokaryotes, vol I General properties of bacteriophages.CRC Press, Boca Raton, FL [Google Scholar]
- 47.Ohtsubo Y, Ikeda-Ohtsubo W, Nagata Y, Tsuda M. 2008. GenomeMatcher: a graphical user interface for DNA sequence comparison. BMC Bioinformatics 9:376 doi:10.1186/1471-2105-9-376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hosaka Y, Irinoda K, Nakano R, Tanabe S, Koizumi W, Saigenji K, Inoue M. 2005. Use of the restriction enzyme EcoRI for pulsed-field gel electrophoretic analysis of Helicobacter pylori. J. Clin. Microbiol. 43:931–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Green MR, Sambrook J. 2012. Molecular cloning: a laboratory manual, 4th ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 50.Vincze T, Posfai J, Roberts RJ. 2003. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 31:3688–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uchiyama J, Takemura I, Satoh M, Kato S, Ujihara T, Akechi K, Matsuzaki S, Daibata M. 2011. Improved adsorption of an Enterococcus faecalis bacteriophage ϕEF24C with a spontaneous point mutation. PLoS One 6:e26648 doi:10.1371/journal.pone.0026648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. 2007. The Paragon algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol. Cell. Proteomics 6:1638–1655 [DOI] [PubMed] [Google Scholar]
- 53.Ackermann HW. 2009. Phage classification and characterization. Methods Mol. Biol. 501:127–140 [DOI] [PubMed] [Google Scholar]
- 54.Lavigne R, Molineux IJ, Kropinski AM. 2012. Order Caudovirales, p 39–45 King AMQ, Adams MJ, Carstens EB, Lefkowitz EJ. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, Waltham, MA [Google Scholar]
- 55.Oksanen HM, Bamford DH. 2012. Tectiviridae, p 317–321 King AMQ, Lefkowitz E, Michael J, Adams MJ, Carstens EB. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, Waltham, MA [Google Scholar]
- 56.Prangishvili D. 2012. Corticoviridae, p 179–186 King AMQ, Lefkowitz E, Michael J, Adams MJ, Carstens EB. (ed), Virus taxonomy: ninth report of the International Committee on Taxonomy of Viruses. Academic Press, Waltham, MA [Google Scholar]
- 57.Carlson K. 2005. Working with bacteriophages: common techniques and methodological approaches, p 437–487 Kutter E, Sulakvelidze A. (ed), Bacteriophages biology and applications. CRC Press, Boca Raton, FL [Google Scholar]
- 58.Guttman B, Raya R, Kutter E. 2005. Basic phage biology, p 29–66 Kutter E, Sulakvelidze A. (ed), Bacteriophages biology and applications. CRC Press, Boca Raton, FL [Google Scholar]
- 59.Comtois SL, Gidley MD, Kelly DJ. 2003. Role of the thioredoxin system and the thiol-peroxidases Tpx and Bcp in mediating resistance to oxidative and nitrosative stress in Helicobacter pylori. Microbiology 149:121–129 [DOI] [PubMed] [Google Scholar]
- 60.Sachs G, Scott DR, Wen Y. 2011. Gastric infection by Helicobacter pylori. Curr. Gastroenterol. Rep. 13:540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Golais F, Hollý J, Vítkovská J. 2012. Coevolution of bacteria and their viruses. Folia Microbiol. (Praha) doi:10.1007/s12223-012-0195-5 [DOI] [PubMed] [Google Scholar]
- 62.Los M, Wegrzyn G. 2012. Pseudolysogeny. Adv. Virus Res. 82:333–343 [DOI] [PubMed] [Google Scholar]
- 63.Black LW, Showe MK, Steven AC. 1994. Morphogenesis of the T4 head, p 218–258 Karam JD, Drake JW, Kreuzer KN, Mosig G, Hall DH, Eiserling FA, Black LW, Spicer EK, Kutter E, Carlson K, Miller ES. (ed), Molecular biology of bacteriophage T4. ASM Press, Washington, DC [Google Scholar]
- 64.Uchiyama J, Rashel M, Matsumoto T, Sumiyama Y, Wakiguchi H, Matsuzaki S. 2009. Characteristics of a novel Pseudomonas aeruginosa bacteriophage, PAJU2, which is genetically related to bacteriophage D3. Virus Res. 139:131–134 [DOI] [PubMed] [Google Scholar]
- 65.Uchiyama J, Rashel M, Takemura I, Wakiguchi H, Matsuzaki S. 2008. In silico and in vivo evaluation of bacteriophage ϕEF24C, a candidate for treatment of Enterococcus faecalis infections. Appl. Environ. Microbiol. 74:4149–4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Botstein D. 1980. A theory of modular evolution for bacteriophages. Ann. N. Y. Acad. Sci. 354:484–490 [DOI] [PubMed] [Google Scholar]
- 67.Comeau AM, Bertrand C, Letarov A, Tetart F, Krisch HM. 2007. Modular architecture of the T4 phage superfamily: a conserved core genome and a plastic periphery. Virology 362:384–396 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.