Abstract
Background
The primary role of natriuretic peptide receptor-3 (NPR3) or NPR-C is in the clearance of natriuretic peptides that play an important role in modulating intravascular volume and vascular tone. Genetic variation in NPR3 has been associated with variation in blood pressure and obesity. Despite the importance of NPR3, sequence variation in the gene has not been addressed using DNA from different ethnic populations. We set out to identify and functionally characterize genetic variation in NPR3 in 3 ethnic groups.
Methods and Results
DNA samples from 96 European American, 96 African American, and 96 Han Chinese American healthy subjects were used to resequence NPR3 exons, splice junctions, and flanking regions. We identified 105 polymorphisms, 50 of which were novel, including 8 nonsynonymous single-nucleotide polymorphisms, 7 were novel. Expression constructs were created for the nonsynonymous single-nucleotide polymorphisms. HEK293 cells were transfected with constructs for wild type and variant allozymes; and recombinant proteins were measured by quantitative Western blot analysis. The most significant change in NPR3 protein was observed for the Arg146 variant allozyme, with 20% of wild-type protein, primarily because of autophagy-dependent degradation. NPR3 structural modeling confirmed that the Arg146 variant protein was not compatible with wild-type conformation and could result in protein misfolding or instability.
Conclusions
Multiple novel NPR3 genetic polymorphisms were identified in 3 ethnic groups. The Arg146 allozyme displayed a significant decrease in protein quantity because of degradation mediated predominantly by autophagy. This genetic variation could have a significant effect on the metabolism of natriuretic peptides with potential clinical implications.
Keywords: natriuretic peptide receptor-3, natriuretic peptides, pharmacogenetics, polymorphism
The natriuretic peptides play a key role in modulating cardiac structure and function by affecting the renin–angiotensin–aldosterone system, sodium and water excretion, and vasomotor tone.1 These peptides are degraded approximately equally by 2 mechanisms, enzymatic degradation catalyzed by membrane metallo-endopeptidase and clearance by natriuretic peptide receptor-3 (NPR3).2 All 3 natriuretic peptides, atrial natriuretic peptide, brain natriuretic peptide, and C-type natriuretic peptide, seem to have similar binding affinity for NPR3. NPR3 is a single pass type 1 membrane protein with a 450-amino acid extracellular ligand-binding domain and a transmembrane region. The cytoplasmic tail is composed of 37 amino acid residues, and NPR3 is the only natriuretic peptide receptor that does not have a cytoplasmic guanylate cyclase catalytic domain.3 Its primary function seems to be clearance of natriuretic peptides by a process of internalization and degradation.4 However, the receptor may also have a signaling function, and it is reportedly linked to inhibitory G protein and may regulate adenyl cyclase activity.5
Genetic variation in NPR3 has been associated with variation in blood pressure regulation, abdominal fat distribution, and human body height.6–10 A possible role for NPR3 in hypertension risk has been suggested by 2 large genome-wide association studies (GWAS). A recent GWAS identified 16 novel loci, 1 of which was NPR3-C5orf23 that were associated with blood pressure in 200 000 individuals of European descent.10 This locus was also found to be significant in a GWAS for systolic and diastolic blood pressure involving East Asians, highlighting the possible importance of this gene in blood pressure control.7 Genetic variation in NPR3 determined by the application of a tag single-nucleotide polymorphism (SNP) approach has also been associated with hypertension in patients with diabetes mellitus, and the hypertension was found to be associated with salt responsiveness.9 This observation involved a nonsynonymous (ns) NPR3 SNP, rs2270915 (1561A>G, Asn[521]Asp), and was replicated in 2 separate populations with diabetes mellitus.9 However, despite its importance, there have been no comprehensive NPR3 resequencing studies that included the functional characterization of nsSNPs in NPR3, genetic variation that could have significant clinical implications. Therefore, the purpose of our study was to resequence all 8 exons, all exon–intron splice junctions, and ≈2000 bp of the 5′-flanking region (5′-FR) of NPR3 and then to perform functional genomic studies with nsSNPs, SNPs that alter the encoded amino acid sequence of the protein. Resequencing was performed in a multi-ethnic population considered healthy to enable identification of common and rare genetic variants to provide fundamental information that could be expanded to study genetic variation in disease states and drug response phenotypes.
Materials and Methods
DNA Samples
DNA from 96 African American (AA), 96 European American, and 96 Han Chinese American (HCA) subjects (sample sets HD100AA, HD100CAU, and HD100CHI) was obtained from the Coriell Cell Repository (Camden, NJ). The DNA had been collected and anonymized from healthy individuals by the National Institute of General Medical Sciences with no other phenotypic information collected to serve as a high-quality resource to study genetic variation. Written informed consent was obtained from all subjects for the use of their DNA for research purposes. The present study was reviewed and approved by the Mayo Clinic Institutional Review Board.
NPR3 Resequencing and Variant Discovery
The 8 exons, exon–intron splice junctions, and ≈2 kb of the 5′-FR of NPR3 were amplified using the polymerase chain reaction (PCR). PCR primer sequences used to perform the amplifications are listed in Table I in the online-only Data Supplement. The PCR amplifications were performed with FastStart Taq DNA polymerase (Roche Diagnostics Corporation, Indianapolis, IN) in a GeneAmp PCR System 9700 thermal cycler (Applied Biosystems, Foster City, CA). Because of the high guanylate cyclase content of exon 1, the guanylate cyclase-Rich PCR System (Roche Diagnostics Corporation, Indianapolis, IN) was used for that amplification. Amplicons were sequenced on both strands in the Mayo Molecular Biology Core Facility using dye terminator sequencing chemistry. To exclude the possibility of PCR-induced artifacts, independent amplifications, followed by sequencing, were performed for any SNP or insertion/deletion (indel) observed in only a single DNA sample or for any sample displaying an ambiguous chromatogram (eg, a HCA sample showing a triallelic rs3792761 genotype). The sequencing chromatograms were analyzed using Mutation Surveyor v2.2 default parameters (SoftGenetics, LLC, State College, PA). The software calls were manually inspected to eliminate false positives. Reference genomic sequences were obtained from the NCBI Reference Sequence (RefSeq) collection (contig and cDNA accession numbers NT_006576.15 and NM_000908.2, respectively). The same 96 AA, 96 European American, and 96 HCA DNA samples had been genotyped with Illumina HumanHap 550K and Illumina HumanHap 510S BeadChips (San Diego, CA), as well as the Affymetrix 6.0 SNP Chip (Santa Clara, CA). The Illumina genotyping was performed in the genotype shared resource at the Mayo Clinic. The Affymetrix genotyping was performed by the Coriell Cell Repository.
NPR3 Expression in HEK293 Cells
The human NPR3 cDNA clone (NM_000908) was obtained from OriGene Technologies, Inc (Rockville, MD) and was cloned into the eukaryotic expression vector pCMV6-XL4. The insert was sequenced in both directions. Site-directed mutagenesis was then performed using the QuikChange II kit (Stratagene, La Jolla, CA) to create expression constructs for each of the variant receptors. The sequences of primers used to perform site-directed mutagenesis are listed in Table II in the online-only Data Supplement. The sequences of all variant receptor constructs were also confirmed by sequencing in both directions.
HEK293 cells are human embryonic kidney cells that do not express the natriuretic peptide receptors and, as a result, were selected for use in our expression studies. Specifically, these cells were grown in Dulbecco's modified Eagle's medium (DMEM; ATCC, Catalog No. 30–2002) supplemented with 10% fetal bovine serum. For each well in a 6-well-plate, 3×105 cells were plated. The following day, triplicate transfections with wild type (WT), empty vector, and constructs for the 9 variant allozymes for NPR3 that we had observed were performed using the FuGENE 6 transfection reagent (Roche Applied Science, Indianapolis, IN) at a charge ratio of 3:1. The cells were also cotransfected with pSV-β-galactosidase DNA (Promega) to make it possible to correct for possible variation in transfection efficiency (construct DNA: pSV-β-galactosidase DNA=3:1). After 48 hours, the cells were washed with PBS and were harvested for the preparation of whole cell lysates.
Western Blot Analysis
Levels of immunoreactive protein were determined for each NPR3 variant allozyme construct by performing quantitative Western blot analysis. Specifically, whole cell lysates for cells transfected with allozyme constructs were subjected to electrophoresis on 7.5% Tris-Glycine extended gels loaded on the basis of assay of the cotransfected β-galactosidase (β-galactosidase Enzyme Assay System with Reporter Lysis Buffer, Promega. Madison, WI). After electrophoresis, proteins were transferred to PVDF membranes (BioRad, Hercules, CA), and the membranes were incubated with purified mouse anti-NPR3 antibody (Origene, Cat. No. TA500956) diluted 1:1000, followed by incubation with a secondary antibody (1:5000). The immunogen used to generate the anti-NPR3 antibody was the full-length human recombinant protein of human NPR3 (NP_000899) produced in HEK293T cells. Hence, the binding of the antibody is not limited to any specific protein sequence of NPR3. Immunoreactive proteins were detected using the ECL Western Blotting System (Amersham Pharmacia, Piscataway, NJ). The IPLab Gel H (Biosystemetica, Plymouth, UK) system and the National Institutes of Health image program (http//rsb.info.nih.gov/nih-image) were used to quantify immunoreactive proteins, and the data were expressed as a percentage of the intensity of a standard of WT human NPR3 protein on the same gel. Three samples each for WT, empty vector, and each variant receptor were assayed for each allozyme studied.
Protein Degradation Studies
The NPR3 variant allozymes with the least protein expression were selected to perform protein degradation studies. Six hours after transfection with WT and the selected variant allozyme NPR3 constructs (Arg146, Cys3, Val499, Ser477+521Asp, and Ser477), HEK293 cells were transferred from 100-mm plates into 6-well plates. The following day, to test for proteasome-mediated degradation, the cells were treated for 16 hours with either dimethyl sulfoxide or 20 μmol/L of MG132, a proteasome inhibitor, dissolved in dimethyl sulfoxide. To determine whether autophagy might participate in the clearance of receptor protein, transfected cells were also treated for 48 hours with the autophagy inhibitor 3-methyladenosine (3MA), 2 mmol/L. The cells were then washed with PBS and harvested for the preparation of whole cell lysates. Immunoreactive NPR3 protein was then assayed by Western blot analysis after correction for the cotransfected β-galactosidase. Actin was used as an internal standard for these assays. All experiments were performed in triplicate, and values reported are mean±SEM for 3 determinations.
Structural Analysis
The ligand-binding extracellular domain (ECD) of human NPR3 has been crystallized in free form and as 2:1 complexes with its natriuretic peptide ligands atrial natriuretic peptide, brain natriuretic peptide, and C-type natriuretic peptide.3,11 The altered amino acid residues that we observed in variant NPR3 allozymes were mapped onto all 4 crystal structures, with similar observations in each case. The 2.4 Å resolution structure of atrial natriuretic peptide-bound NPR3 (PDB accession code 1YK0) was chosen as the template for more detailed computational modeling of the variant allozymes. Molecular visualization and modeling of the variant structures were performed using the computer program Coot.12 Structural figures were prepared with Molscript and Raster3D.13,14
Statistical Analysis
Values for π, θ, and Tajima's D were calculated as described by Tajima,15 followed by correction for length. Linkage disequilibrium (LD) analysis was performed by calculating D′ and r2 values.16,17 The LD data were displayed graphically using Haploview.18 Because of the presence of a triallelic marker in the HCA samples, Haploview could not be used to display those data. Haplotype analysis was performed as described by Schaid et al19 using the E-M (expectation-maximization) algorithm. Mean protein values were compared using Student t test and ANOVA.
Results
Human NPR3 Resequencing and Genotype Data
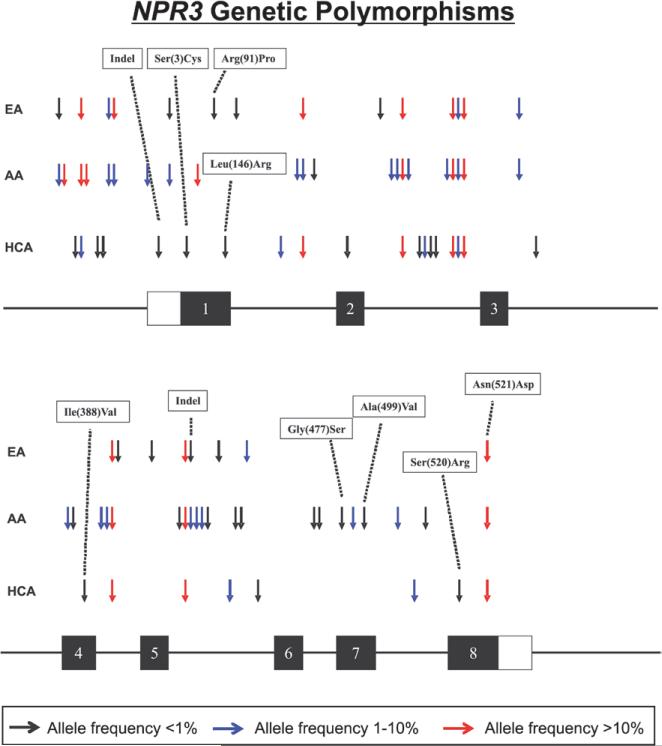
NPR3 was resequenced using 288 DNA samples representing 3 ethnic groups, 96 DNA samples each from AA, European American, and HCA subjects. A total of 65 SNPs and 2 indels were observed, with samples from AA subjects displaying, as anticipated, the greatest genetic variation, with 41 SNPs and 1 indel. European American and HCA samples displayed 22 (21 SNPs, 1 indel) and 27 (26 SNPs, 1 indel) variants, respectively (Figure 1; Table). Nine of the SNPs were common to all 3 ethnic groups with minor allele frequencies ≥1% in all 3 sample sets. There were 15 SNPs within the open reading frame, including 8 nsSNPs: 8C>G, 272G>C, 437T>G, 1162A>G, 1429G>A, 1496C>T, 1560T>G, and 1561A>G (rs2270915) that resulted in the following respective changes in encoded amino acids: Ser(3)Cys, Arg(91)Pro, Leu(146) Arg, Ile(388)Val, Gly(477)Ser, Ala(499)Val, Ser(520)Arg, and Asn(521)Asp. The remaining 7 coding SNPs were synonymous changes in sequence. Five of the ns cSNPs were found in the ECD of the mature NPR3 receptor protein, 1 in the transmembrane domain and 2 in the cytoplasmic domain (Figure I in the online-only Data Supplement). The 2 nsSNPs in the portion of the gene encoding the 37–amino acid cytoplasmic domain, amino acids 520 and 521, were in the inhibitory guanine nucleotide regulatory protein (Gi) activating sequence of the receptor.20 Among the 65 SNPs observed in NPR3, 16 were present in dbSNP Build 130 (www.ncbi.nlm.nih.gov/SNP; Table) and 10 were present in the 1000 Genomes pilot data.21 Therefore, our resequencing identified 49 SNPs and 2 indels that were novel. Among the 8 nsSNPs observed, 7 were novel. Addition of the Illumina/Affymetrix genotyping data enriched our set of mainly exonic variants with 39 SNPs within introns (Figure II in the online-only Data Supplement). All NPR3 SNPs and indels were in Hardy–Weinberg equilibrium (P>0.05), with the exception of nucleotide (–1397; rs72740637) in the 5′-FR of the gene (P=0.005) of HCA subjects (minor allele frequency 1%).
Figure 1.
Human natriuretic peptide receptor-3 (NPR3) genetic polymorphisms. The figure shows a schematic representation of the structure of NPR3, with arrows indicating the locations of polymorphisms identified during resequencing. Exons encoding the open reading frame are represented as dark rectangles and portions of exons that encode UTR sequences are indicated by open rectangles. Colors of arrows indicate minor allele frequencies. AA indicates African American; EA, European American; and HCA, Han Chinese American.
Table.
NPR3 Genetic Polymorphisms
| NPR3 Genetic Polymorphism Frequency of Variant Allele | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Location | Nucleotide | Sequence Change | Amino Acid Change | African American | European American | Han Chinese American | RefSNP ID dbSNP130 | 1000 Genomes | NCBI36/hg18 Chromosomal Location |
| 5′FR | –1525 | A→G | 0.047 | 0.005 | 0.000 | 32746115 | |||
| 5′FR | –1431 | G→T | 0.156 | 0.000 | 0.000 | rs6868454 | X | 32746209 | |
| 5′FR | –1426 | G→A | 0.000 | 0.000 | 0.005 | 32746214 | |||
| 5′FR | –1397 | G→A | 0.286 | 0.292 | 0.010 | rs72740637 | X | 32746243 | |
| 5′FR | –1122 | G→A | 0.115 | 0.000 | 0.000 | rs28513089 | X | 32746518 | |
| 5′FR | –1079 | C→T | 0.000 | 0.000 | 0.005 | rs16890133 | 32746561 | ||
| 5′FR | –1029 | G→T | 0.000 | 0.000 | 0.005 | 32746611 | |||
| 5′FR | –987 | G→A | 0.010 | 0.063 | 0.000 | 32746653 | |||
| 5′FR | –250 | C→A | 0.042 | 0.182 | 0.000 | rs9716700 | X | 32747390 | |
| 5′UTR | –201 | T→A | 0.089 | 0.000 | 0.000 | 32747439 | |||
| 5′UTR | –47 | insertion of G | 0.000 | 0.000 | 0.005 | 32747593 | |||
| 5′UTR | –6 | G→C | 0.036 | 0.005 | 0.000 | 32747634 | |||
| Exon 1 | 8 | C→G | Ser(3)Cys | 0.000 | 0.000 | 0.005 | 32747647 | ||
| Exon 1 | 237 | T→C | 0.120 | 0.000 | 0.000 | rs6859964 | X | 32747876 | |
| Exon 1 | 272 | G→C | Arg(91)Pro | 0.000 | 0.005 | 0.000 | 32747911 | ||
| Exon 1 | 437 | T→G | Leu(146)Arg | 0.000 | 0.000 | 0.005 | 32748076 | ||
| Exon 1 | 693 | G→A | 0.000 | 0.005 | 0.000 | 32748332 | |||
| Intron 1 | 23 | C→T | 0.000 | 0.000 | 0.042 | 32748431 | |||
| Intron 1 | 144 | C→T | 0.036 | 0.000 | 0.000 | 32748552 | |||
| Intron 1 | 180 | C→T | 0.063 | 0.104 | 0.158 | rs4867473 | X | 32748588 | |
| Intron 1 | –66 | G→T | 0.005 | 0.000 | 0.000 | 32760495 | |||
| Exon 2 | 822 | G→A | 0.000 | 0.000 | 0.005 | 32760613 | |||
| Intron 2 | 16 | C→A | 0.000 | 0.005 | 0.000 | 32760699 | |||
| Intron 2 | 104 | C→G | 0.016 | 0.000 | 0.000 | 32760787 | |||
| Intron 2 | 145 | C→T | 0.010 | 0.000 | 0.000 | 32760828 | |||
| Intron 2 | 146 | C→T | 0.151 | 0.255 | 0.708 | rs1147224 | X | 32760829 | |
| Intron 2 | 171 | G→T | 0.010 | 0.000 | 0.000 | 32760854 | |||
| Intron 2 | 233 | A→G | 0.000 | 0.000 | 0.005 | 32760916 | |||
| Intron 2 | 262 | C→T | 0.000 | 0.000 | 0.010 | 32760945 | |||
| Intron 2 | 301 | A→G | 0.000 | 0.000 | 0.005 | 32760984 | |||
| Intron 2 | 312 | G→A | 0.000 | 0.000 | 0.005 | 32760995 | |||
| Intron 2 | 333 | T→C | 0.010 | 0.000 | 0.000 | 32761016 | |||
| Intron 2 | 426 | G→C or A | 0.432 | 0.281 | 0.156 | rs3792761 | 32761109 | ||
| Intron 2 | –74 | G→A | 0.026 | 0.036 | 0.016 | rs2292025 | 32774643 | ||
| Intron 2 | –69 | C→T | 0.146 | 0.115 | 0.224 | rs2292026 | 32774648 | ||
| Intron 3 | 78 | C→T | 0.010 | 0.031 | 0.000 | 32774971 | |||
| Intron 3 | –77 | C→T | 0.000 | 0.000 | 0.005 | 32810494 | |||
| Exon 4 | 1086 | C→T | 0.010 | 0.000 | 0.000 | 32810597 | |||
| Exon 4 | 1104 | C→T | 0.005 | 0.000 | 0.000 | 32810615 | |||
| Exon 4 | 1162 | A→G | Ile(388)Val | 0.000 | 0.000 | 0.005 | 32810673 | ||
| Intron 4 | 46 | G→A | 0.021 | 0.000 | 0.000 | rs13358498 | 32810752 | ||
| Intron 4 | 65 | A→T | 0.010 | 0.000 | 0.000 | 32810771 | |||
| Intron 4 | 98 | T→G | 0.255 | 0.438 | 0.333 | rs1173743 | X | 32810804 | |
| Intron 4 | –37 | G→A | 0.000 | 0.005 | 0.000 | 32816548 | |||
| Exon 5 | 1203 | C→T | 0.000 | 0.005 | 0.000 | 32816592 | |||
| Intron 5 | 82 | A→G | 0.005 | 0.000 | 0.000 | 32816761 | |||
| Intron 5 | 274 | T→G | 0.245 | 0.234 | 0.172 | rs751432 | X | 32816953 | |
| Intron 5 | 449 | deletion of CT | 0.048 | 0.005 | 0.000 | 32817128 | |||
| Intron 5 | –320 | C→T | 0.010 | 0.000 | 0.000 | 32818436 | |||
| Intron 5 | –317 | C→T | 0.052 | 0.000 | 0.000 | 32818439 | |||
| Intron 5 | –288 | G→T | 0.005 | 0.000 | 0.000 | 32818468 | |||
| Intron 5 | –281 | A→T | 0.000 | 0.005 | 0.000 | 32818475 | |||
| Intron 5 | –206 | T→A | 0.000 | 0.000 | 0.021 | 32818550 | |||
| Intron 5 | –198 | A→G | 0.005 | 0.000 | 0.000 | 32818558 | |||
| Intron 5 | –110 | C→T | 0.005 | 0.000 | 0.000 | rs73755322 | 32818646 | ||
| Intron 5 | –59 | C→T | 0.000 | 0.010 | 0.000 | 32818697 | |||
| Intron 5 | –23 | A→G | 0.000 | 0.000 | 0.005 | 32818733 | |||
| Intron 6 | –62 | C→T | 0.005 | 0.000 | 0.000 | 32820600 | |||
| Intron 6 | –60 | A→C | 0.005 | 0.000 | 0.000 | 32820602 | |||
| Exon 7 | 1429 | G→A | Gly(477)Ser | 0.005 | 0.000 | 0.000 | 32820664 | ||
| Exon 7 | 1461 | C→T | 0.021 | 0.000 | 0.000 | 32820696 | |||
| Exon 7 | 1496 | C→T | Ala(499)Val | 0.005 | 0.000 | 0.000 | 32820731 | ||
| Intron 7 | 210 | C→T | 0.010 | 0.000 | 0.000 | 32820956 | |||
| Intron 7 | 218 | A→G | 0.000 | 0.000 | 0.016 | 32820964 | |||
| Intron 7 | –27 | T→C | 0.005 | 0.000 | 0.000 | 32822070 | |||
| Exon 8 | 1560 | T→G | Ser(520)Arg | 0.000 | 0.000 | 0.005 | 32822145 | ||
| Exon 8 | 1561 | A→G | Asn(521)Asp | 0.125 | 0.224 | 0.172 | rs2270915 | X | 32822146 |
Alterations in nucleotide and amino acid sequences, as well as minor allele frequencies for all 3 ethnic groups studied are listed. SNPs in exons and 5′-FRs have been numbered on the basis of their locations in the cDNA, with the ‘A’ in the translation initiation codon assigned (+1). Negative numbers are located 5′, and positive numbers 3′ to this position. SNPs in introns have been numbered on the basis of their distance to the nearest 5′ and 3′ exon–intron splice junctions, using positive and negative number, respectively. The final 3 columns indicate whether the polymorphism was already present in dbSNP, 1000 Genomes Project, and the NCBI chromosomal location of the polymorphism, respectively. 5′-FR indicates 5′-flanking region; NPR3, natriuretic peptide receptor-3; and SNP, single-nucleotide polymorphism.
We also calculated nucleotide diversity, a measure of genetic variation, adjusted for the number of alleles studied. Using 2 standard measures—π, average heterozygosity per site and θ, a population mutation measure that is theoretically equal to the neutral mutation parameter—we found π to be similar across the 3 ethnic groups, whereas values for θ were largest for the AA samples. Tajima's D, a test of the neutral mutation hypothesis, did not differ significantly from 0 for any of the 3 ethnic groups studied (Table III in the online-only Data Supplement).
LD and Haplotype Analysis
We also performed pairwise studies of LD using both our gene resequencing and Illumina/Affymetrix genotyping data for the 288 DNA samples. For the LD analysis, pairwise calculations of D′ and r2 values were performed for all variants with minor allele frequency values ≥5%. Haplotype blocks were judged to be present if 95% of the pairwise comparisons were in strong LD. D′ values of 1.0 indicate that 2 variants are maximally associated, whereas values of 0 indicate they are randomly associated, implying greater historical recombination. Graphical representations of the haplotype structures for NPR3 based on these data are shown in Figure III in the online-only Data Supplement, which includes the Illumina/Affymetrix genotype data for these DNA samples.
Because haplotypes have proven to be of value for use in genetic association studies,22,23 we also defined NPR3 haplotypes using the gene resequencing data. Haplotypes with frequencies >2% are listed in Table IV in the online-only Data Supplement. Haplotype designations were based on the amino acid sequence of the encoded allozyme, with *2, *3, *4, *5, *7, *8, and *9 denoting variants that encoded Ser3, Arg91, Leu146, Ile388, Ala499, Ser520, and Asn521 variant amino acid sequences, respectively. The *10 designation was used for a Gly477, Asn521 double variant. Gly477 would otherwise have been designated *6, but we did not find this variant independently, but rather only in combination with Asn521. The WT amino acid sequence was designated *1. Letter designations were then added on the basis of descending haplotype frequencies, beginning with data for AA subjects (Table IV in the online-only Data Supplement).
NPR3 Protein Expression Levels
Expression constructs were created for the NPR3 WT and the 9 variant receptors encoded by the nsSNPs that we observed to determine the possible effect of the nsSNPs on protein levels. These 10 NPR3 expression constructs were transfected into HEK293 cells to ensure that mammalian posttranslation modification and protein degradation systems would be present. Immunoreactive protein levels were then determined by quantitative Western blot analysis (Figure 2A). The levels of NPR3 immunoreactive protein as a percentage of WT are shown graphically in Figure 2B. Four of the variants: Cys3 (38±8% of WT; P=3.4×10–6), Arg146 (20±6% of WT; P=2.17×10–7), Val499 (55±11% of WT; P=0.0001), and Ser477+521Asp (53±14% of WT; P=0.0002) showed at least a 40% decrease in protein levels as compared with WT, and the Ser477 allozyme was decreased to 64±18% of the WT (P=0.003). The allozyme with the least protein was the Arg146 variant which was decreased to 20±6% of WT protein. There was no statistically significant difference in immunoreactive protein compared with WT for the other 4 variants: Pro91 (95±38% of WT; P=0.791), Val388 (109±26% of WT; P=0.447), Arg520 (92±23% of WT; P=0.484), and Asp521 (98±18% of WT; P=0.865).
Figure 2.
Natriuretic peptide receptor-3 (NPR3) functional genomics. A, Western blot analysis showing NPR3 wild-type (WT) protein expression as compared with variant NPR3 protein expression for constructs with nonsynonymous single-nucleotide polymorphisms expressed in HEK293 cells. B, Levels of NPR3 immunoreactive protein as a percentage of WT. Each bar represents the average of 3 independent transfections (mean±SEM). All values are corrected for transfection efficiency. *P<0.05 and **P<0.001 when compared with the WT value.
NPR3 Arg146 Protein Degradation
In an effort to understand mechanisms responsible for the decreased protein expression that we observed for variant NPR3 receptors, the variant with the lowest level of protein expression, Arg146, was selected for further study. The decrease in Arg146 immunoreactive protein could be due to several mechanisms, including mRNA instability, decreased rate of synthesis or increased rate of degradation. Degradation especially of receptor proteins could be mediated by lysosomal proteases. However, the most common mechanism responsible for decreased levels of protein in association with nsSNPs is accelerated degradation involving the ubiquitin-proteasome system and autophagy.24,25 Therefore, we set out to test the hypothesis that accelerated degradation involving the ubiquitin-proteasome system and autophagy might explain, at least in part, the decreased level of Arg146 NPR3.
To determine the possible roles of proteasome- and autophagy-mediated degradation, the effect of the proteasome inhibitor MG132 and autophagy inhibitor 3MA on WT and Arg146 expression was determined. Figure 3 shows graphically that, after treatment with MG132, the average level of change for Arg146 protein (34% increase) was not significantly different from that of the change in WT receptor, indicating that proteasome-mediated degradation did not seem to play a major role in variant receptor degradation. However, after 3MA treatment, the changes in protein levels for WT (304% increase; P=4×10–6) and Arg146 (892% increase; P=9×10–6) were 4- and 10-fold higher, respectively, when compared with untreated WT or untreated Arg146-transfected cells, indicating the importance of autophagy in the degradation of NPR3 (Figure 3). A representative Western blot showing these results after normalizing for the expression of β-galactosidase as a control for transfection efficiency and for actin as an internal control is shown in Figure 3B. Our experiments demonstrated that autophagy-mediated degradation and to a lesser extent proteasomes seem to play an important role in degradation of both the Arg146 variant and the WT NPR3 because both were greatly increased by 3MA, but more so for Arg146 than WT (P=8×10–4), indicating enhanced degradation of the variant receptor. To ensure reproducibility of the observation that autophagy plays an important role, not only in the degradation of the Arg146 variant but also in the other variants that have a significant decrease in protein expression (Cys3, Val499, Ser477+521Asp, and Ser477), we determined the effect of MG132 and 3MA on the degradation of these variant proteins, with similar results showing autophagy and, to a lesser extent proteasome-mediated degradation playing a major role (Figure IV in the online-only Data Supplement). These observations are compatible with the concept that autophagy is the principal process involved in the turnover of plasma membrane–associated receptors.26 However, it is possible that these variant allozymes demonstrate decreased protein expression not because of enhanced degradation but because of the SNPs resulting in alteration of the secondary mRNA structure.27
Figure 3.
Natriuretic peptide receptor-3 (NPR3) degradation by autophagy and proteasome-mediated processes. A, Levels of Arg146 NPR3 receptor immunoreactive protein expressed as a percentage of untreated wild type (WT) or Arg146 expressed in HEK293 cells after exposure to MG132, a proteasome inhibitor, or 3-methyladenosine (3MA), an autophagy inhibitor. Each value is the mean±SEM of 3 independent experiments. B, Representative Western blot analysis of WT and Arg146 NPR3 receptor immunoreactive protein after treatment of HEK293 cells with MG132 or 3MA. NT indicates nontreated cells.
We next used the NPR3 X-ray crystal structures that are currently available to determine whether the Arg146 variant might compromise the structural integrity of the NPR3 protein. The Arg146 variant was selected because we could model its effect on NPR3 protein structure by X-ray crystallography because this was the only variant with reduced protein expression that was included in the crystal structures that are currently available.
Structural Analysis of NPR3 Variants
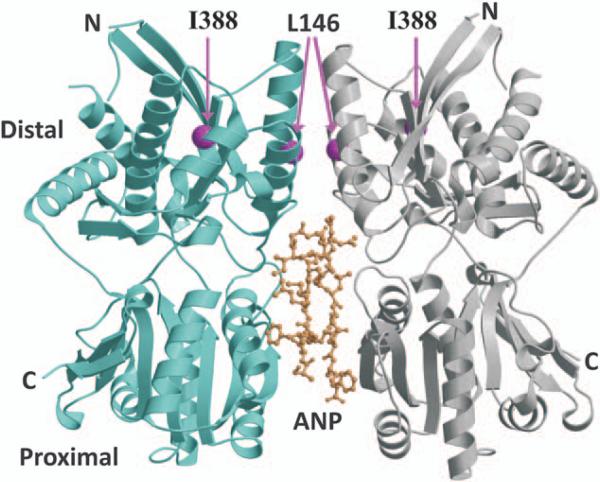
All NPR3 crystal structures currently available include only the ligand-binding ECD.3,11 The ECD has a bilobal protein fold consisting of membrane-distal and membrane-proximal domains (Figure 4). In both free and ligand-bound forms, the ECD is dimeric, with the membrane-distal domains forming the dimer interface. Of the NPR3 variants identified during our resequencing study, Ala(499)Val, Ser(520)Arg, and Asn(521) Asp are located in the transmembrane and cytoplasmic domains, for which there are no experimentally determined molecular structures. Of the remaining variants, Ser(3)Cys, Arg(91)Pro, and Gly(477)Ser alter residues that are missing in the ECD crystal structures. Therefore, structural analysis was only possible for the Leu(146)Arg and Ile(388)Val variants.
Figure 4.
Natriuretic peptide receptor-3 (NPR3) extracellular domain structure and location of 2 variant sites. The crystal structure of atrial natriuretic peptide (ANP)-bound NPR3 (11) includes 2 copies of the extracellular domain (cyan and gray backbone ribbons with labeled N and C termini) and 1 ANP ligand (yellow ball-and-stick structure) bound primarily to the 2 membrane-proximal domains. The Leu146 and Ile388 residues are located in the membrane-distal domain (pink spheres).
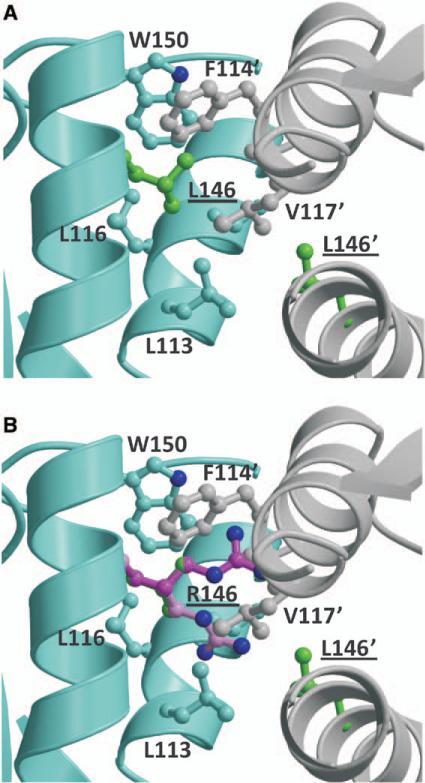
Both Leu146 and Ile388 are located in the membrane-distal domain, which does not change conformation during ligand binding (Figure 4). The Leu146 side chain is buried in a hydrophobic environment and interacts with several hydrophobic residues, both within the same monomer and across the dimer interface (Figure 5A). Computational substitution with the much longer Arg146 side chain introduces several sterically unfavorable short contacts (Figure 5B). All Arg146 conformations result in unfavorable intermolecular contacts that can be relieved only by significant alteration of the dimer interface. In addition, the Arg146 substitution introduces a positive charge into a hydrophobic environment, which would be energetically unfavorable. Therefore, molecular modeling suggests that the Leu(146)Arg substitution is incompatible with the WT NPR3 ECD structure and would either destabilize the protein or generate an altered conformation that, in turn, would likely affect function. Ile388 is located in a small surface helix. The conservative Ile(388)Val substitution results in the loss of a terminal methyl group in the hydrophobic side chain. The resulting small gap could be easily accommodated by small local shifts in the helix and loop and is not predicted to have significant structural consequences. Therefore, the results of structural modeling of these 2 NPR3 variants were compatible with the results of our functional genomic studies, with the Arg146 variant showing large differences from WT in protein expression, whereas the Val388 variant was no different from WT (Figure 2B).
Figure 5.
Structural modeling of the natriuretic peptide receptor-3 (NPR3) Leu146Arg variant. A, The Leu146 side chain (green ball-and-stick) is surrounded by hydrophobic residues from both monomers in the dimeric NPR3 crystal structure (side chains shown in cyan and gray ball-and-stick). B, The modeled Arg146 variant side chain in the cyan monomer is shown in 2 possible conformations (ball-and-stick with dark and light pink carbon atoms, superimposed on green Leu146 of wild-type structure), both of which sterically clash with neighboring hydrophobic residues.
Discussion
This is the first study to systematically resequence NPR3, a gene encoding an important protein in the natriuretic peptide pathway, in 3 different ethnic populations, resulting in the identification of 50 novel genetic variants, of which 7 were novel nsSNPs. In addition, we determined the functional significance of these nonsynonymous SNPs by demonstrating their effect on protein expression, characterized degradation mechanisms for the WT and the genetic variant with the least protein expression, that is, Arg146, and compared those results with structural modeling predictions.
DNA sequence variation in or near the NPR3 gene has been reported to be associated with a series of cardiovascular phenotypes, especially phenotypes related to blood pressure regulation. The rs2270915 (G/A) nonsynonymous SNP located in exon 8 NPR3 gene that was identified during our resequencing study was associated with differences in systolic blood pressure and was replicated in 2 large populations with diabetes mellitus (z score=–3.109; P=0.002).9 Furthermore, subjects homozygous for the WT (AA) exhibited a greater reduction in systolic blood pressure with reduction in dietary salt intake than did G carriers.
Other genetic variants in or near NPR3 have also been identified by GWAS to be significantly associated with hypertension. For example, in 19 414 subjects from the Asian Genetic Epidemiology Network Blood Pressure (AGEN-BP) consortium, a SNP (rs1173766, C/T) on chromosome 5p13.3 in the 3′-FR of NPR3 was identified as genome-wide significant for its association with both systolic (β=0.63; P=1.9×10–8) and diastolic blood pressure (β=0.36; P=1.2×10–7), a finding replicated by de novo genotyping in 10 461 Japanese subjects.7 The rs1173766 SNP (NCBI36 chromosomal location 32840285) is ≈50 000 bp downstream from the 3′-untranslated region of the NPR3 gene. Because we did not resequence this far downstream, we did not identify this SNP in our study. Genetic variation (rs7726475, A/G) in the 5′-FR of the NPR3 gene has also been significantly associated with interindividual variation in both systolic (β=2.00; P=8.85×10–7) and diastolic (β=1.04; P=3.64×10–6) blood pressure in AAs by the Candidate Gene Association Resource (CARe) Consortium and in 5 other studies involving AAs.28 This SNP (NCBI36 chromosomal location 32611671) was located ≈130 000 bp upstream from the 5′-untranslated of NPR3 and was not covered by our resequencing experiment because we resequenced up to ≈2000 bp in the 5′-FR of the gene, and the chromosomal location of the first variant identified in this region was at position 32746115. The International Consortium for Blood Pressure Genome-Wide Association Studies involving 200 000 subjects of European descent also identified a SNP (rs1173771, G/A) in the 3′-FR of NPR3 as significantly associated with both systolic (β=0.50; P=1.8×10–16) and diastolic blood pressure (β=0.26; P=9.1×10–12).21 In a study involving 1164 patients undergoing coronary artery bypass grafting, 4 NPR3 SNPs (rs700923, rs16890196, rs765199, and rs700926) were associated with the development of postoperative left ventricular dysfunction (odds ratio, 3.89–4.28; P=0.007–0.034).29
The association of SNPs in or near NPR3 with blood pressure regulation and cardiovascular disease highlights the importance of resequencing this gene to identify novel and rare variants, especially in the coding region that may have functional implications. GWAS can identify common variants. However, for complex diseases, such as hypertension, the effect size of these associations, although reaching genome-wide significance, is often small.30 Resequencing as performed in this study and increasingly with the application of next generation DNA sequencing could make it possible to identify rare variants with larger effects, as demonstrated in a Framingham study of subjects with rare mutations of genes (SLC12A3, SLC12A1, and KCNJ1) involved with renal sodium chloride reabsorption, which were associated with a 60% reduction in the risk of hypertension at the age of 60 years.31 The functionally significant variants in this study were rare. However, rare variants are being increasingly recognized as potentially important for providing insight into the pathophysiology of disease states.32
NPR3 plays an important role in the clearance of natriuretic peptides that modulate blood pressure by their effect on vascular tone as well as renal salt and water excretion.33 Natriuretic peptides have a prolonged half-life in a NPR3 knockout mouse model, with augmentation of their blood pressure lowering and diuretic effects.34 Hence, genetic variation in NPR3 could result in variation of blood pressure and hypertension risk, especially as a result of nsSNPS, such as the Arg146 variant, which significantly alters protein expression. Genetic variation in other components of the natriuretic peptide system, such as the T2238C variant in NPPA (atrial natriuretic precursor A), can be associated with alteration in the cardiovascular effects of antihypertensive drugs, such as chlorthalidone and amlodipine.35 Genetic variation in the gene encoding membrane metallo-endopeptidase or neutral endopeptidase can also affect the ability of the encoded protein to metabolize natriuretic peptides by altering its enzymatic activity, with potential implications for drugs that are neutral endopeptidase substrates, such as human recombinant brain natriuretic peptide or neutral endopeptidase inhibitors, for which it serves as a target.36 The effect of sequence variation in NPR3 on natriuretic peptide levels and the efficacy of anti-hypertensive therapy are presently unknown. However, before performing such translational studies, the range and extent of NPR3 genetic variation need to be characterized in healthy individuals, a major goal of the present study.
In summary, in the present study, we defined genetic variation in NPR3 in 3 ethnic populations by resequencing NPR3, resulting in the identification of 105 polymorphisms, of which 50 were novel, including 8 nsSNPs, 7 were novel. The possible functional significance of the nsSNPs was tested by determining protein expression of the variant allozymes in a mammalian cell line. Four of the variant receptors showed decreases in protein levels to <60% of WT, with the Arg146 variant displaying the lowest level of protein expression. The decreased protein expression of the Arg146 variant was compatible with the results of molecular modeling performed with the presently available X-ray crystallography structures for NPR3. Structural modeling indicated that substitution of Leu146 with Arg146 could alter protein folding or stability. The results of these resequencing and functional genomic studies could improve our understanding of the clinical implications of these genetic variants on cardiovascular disease risk and the effect of drugs that mediate their effect via the natriuretic peptide system.
Supplementary Material
CLINICAL PERSPECTIVE.
The natriuretic peptides play an important role in the pathophysiology of hypertension and heart failure. Circulating natriuretic peptides are cleared by natriuretic peptide receptor-3 (NPR3), and NPR3 also mediates the antiproliferative effects of natriuretic peptides. Although common genetic variation in NPR3 has been associated with hypertension and obesity in previous studies, comprehensive resequencing of the gene in different ethnic populations has not been performed. We set out to identify and functionally characterize variation in the exons, splice junctions, and flanking regions of NPR3 in 96 European American, 96 African American, and 96 Han Chinese American healthy subjects to provide fundamental information that could be expanded to study genetic variation in disease states and in drug response phenotypes. We identified 105 polymorphisms, 50 of which were novel, including 8 nonsynonymous single-nucleotide polymorphisms, 7 were novel. To determine the functional significance of the nonsynonymous single-nucleotide polymorphisms, HEK293 cells were transfected with expression constructs for wild type and variant allozymes; and recombinant proteins were measured by quantitative Western blot analysis. The Arg146 variant allozyme was the most significant, expression being 20% of wild-type protein, primarily because of autophagy-dependent degradation. NPR3 structural modeling confirmed that the Arg146 variant could result in protein misfolding or instability. The genetic variation characterized by these resequencing and functional genomic studies could play an important role in the clearance of natriuretic peptides and, therefore, improve our understanding of the implications of these genetic variants on disease risk and the effect of drugs that mediate their effect via the natriuretic peptide system.
Acknowledgments
Sources of Funding
This work was supported in part by HL 84904 (Heart Failure Clinical Research Network; Dr Pereira), a Marie Ingalls Cardiovascular Career Development Award (Dr Pereira), CTSA grant No. UL1 TR000135 (Dr Pereira), U19 GM61388 (The Pharmacogenomics Research Network; Dr Weinshilboum), R01 GM28157 (Dr Weinshilboum), R01 CA132780 (Dr Weinshilboum), U01 HG005137 (Dr Weinshilboum), and a PhRMA Foundation “Center of Excellence in Clinical Pharmacology” Award (Dr Weinshilboum).
Footnotes
Disclosures
None.
References
- 1.Boerrigter G, Costello-Boerrigter LC, Burnett JC., Jr Natriuretic peptides in the diagnosis and management of chronic heart failure. Heart Fail Clin. 2009;5:501–514. doi: 10.1016/j.hfc.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rademaker MT, Charles CJ, Kosoglou T, Protter AA, Espiner EA, Nicholls MG, et al. Clearance receptors and endopeptidase: equal role in natriuretic peptide metabolism in heart failure. Am J Physiol. 1997;273(5 pt 2):H2372–H2379. doi: 10.1152/ajpheart.1997.273.5.H2372. [DOI] [PubMed] [Google Scholar]
- 3.He X-l, Chow D-c, Martick MM, Christopher Garcia K. Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone. Science. 2001;293:1657–1662. doi: 10.1126/science.1062246. [DOI] [PubMed] [Google Scholar]
- 4.Potter LR. Natriuretic peptide metabolism, clearance and degradation. FEBS J. 2011;278:1808–1817. doi: 10.1111/j.1742-4658.2011.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Murthy KS, Makhlouf GM. Identification of the G protein-activating domain of the natriuretic peptide clearance receptor (NPR-C). J Biol Chem. 1999;274:17587–17592. doi: 10.1074/jbc.274.25.17587. [DOI] [PubMed] [Google Scholar]
- 6.Estrada K, Krawczak M, Schreiber S, van Duijn K, Stolk L, van Meurs JB, et al. A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum Mol Genet. 2009;18:3516–3524. doi: 10.1093/hmg/ddp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato N, Takeuchi F, Tabara Y, Kelly TN, Go MJ, Sim X, et al. Meta-analysis of genome-wide association studies identifies common variants associated with blood pressure variation in east Asians. Nat Genet. 2011;43:531–538. doi: 10.1038/ng.834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sarzani R, Strazzullo P, Salvi F, Iacone R, Pietrucci F, Siani A, et al. Natriuretic peptide clearance receptor alleles and susceptibility to abdominal adiposity. Obes Res. 2004;12:351–356. doi: 10.1038/oby.2004.44. [DOI] [PubMed] [Google Scholar]
- 9.Saulnier PJ, Roussel R, Halimi JM, Lebrec J, Dardari D, Maimaitiming S, et al. SURDIAGENE. DIAB2NEPHROGENE. DIABHYCAR study groups Impact of natriuretic peptide clearance receptor (NPR3) gene variants on blood pressure in type 2 diabetes. Diabetes Care. 2011;34:1199–1204. doi: 10.2337/dc10-2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The International Consortium for Blood Pressure Genome-Wide Association Studies Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478:103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He XL, Dukkipati A, Garcia KC. Structural determinants of natriuretic peptide receptor specificity and degeneracy. J Mol Biol. 2006;361:698–714. doi: 10.1016/j.jmb.2006.06.060. [DOI] [PubMed] [Google Scholar]
- 12.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(pt 12 Pt 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 13.Kraulis PJ. Molscript: a program to produce both detailed and schematic plots of protein structures. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 14.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Meth Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 15.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartl DL, Clark AG. Principles of Population Genetics. Sinauer Associates, Inc; Sunderland, MA: 1997. pp. 96–106. [Google Scholar]
- 17.Hedrick PW. Genetics of Populations. Jones and Bartlett Publishers; Sundbury, MA: 2000. [Google Scholar]
- 18.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 19.Schaid DJ, Rowland CM, Tines DE, Jacobson RM, Poland GA. Score tests for association between traits and haplotypes when linkage phase is ambiguous. Am J Hum Genet. 2002;70:425–434. doi: 10.1086/338688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou H, Murthy KS. Identification of the G protein-activating sequence of the single-transmembrane natriuretic peptide receptor C (NPR-C). Am J Physiol, Cell Physiol. 2003;284:C1255–C1261. doi: 10.1152/ajpcell.00520.2002. [DOI] [PubMed] [Google Scholar]
- 21.Consortium GP, Durbin RM, Abecasis GR, Altshuler DL, Auton A, Brooks LD, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Genet. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 23.Roden DM, Altman RB, Benowitz NL, Flockhart DA, Giacomini KM, Johnson JA, et al. Pharmacogenetics Research Network Pharmacogenomics: challenges and opportunities. Ann Intern Med. 2006;145:749–757. doi: 10.7326/0003-4819-145-10-200611210-00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang L, Yee VC, Weinshilboum RM. Aggresome formation and pharmacogenetics: sulfotransferase 1A3 as a model system. Biochem Biophys Res Commun. 2004;325:426–433. doi: 10.1016/j.bbrc.2004.10.040. [DOI] [PubMed] [Google Scholar]
- 25.Reinstein E, Ciechanover A. Narrative review: protein degradation and human diseases: the ubiquitin connection. Ann Intern Med. 2006;145:676–684. doi: 10.7326/0003-4819-145-9-200611070-00010. [DOI] [PubMed] [Google Scholar]
- 26.Clague MJ, Urbé S. Ubiquitin: same molecule, different degradation pathways. Cell. 2010;143:682–685. doi: 10.1016/j.cell.2010.11.012. [DOI] [PubMed] [Google Scholar]
- 27.Nackley AG, Shabalina SA, Tchivileva IE, Satterfield K, Korchynskyi O, Makarov SS, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–1933. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 28.Zhu X, Young JH, Fox E, Keating BJ, Franceschini N, Kang S, et al. Combined admixture mapping and association analysis identifies a novel blood pressure genetic locus on 5p13: contributions from the CARe consortium. Hum Mol Genet. 2011;20:2285–2295. doi: 10.1093/hmg/ddr113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox AA, Collard CD, Shernan SK, Seidman CE, Seidman JG, Liu KY, et al. Natriuretic peptide system gene variants are associated with ventricular dysfunction after coronary artery bypass grafting. Anesthesiology. 2009;110:738–747. doi: 10.1097/aln.0b013e31819c7496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coffman TM. Under pressure: the search for the essential mechanisms of hypertension. Nat Med. 2011;17:1402–1409. doi: 10.1038/nm.2541. [DOI] [PubMed] [Google Scholar]
- 31.Ji W, Foo JN, O'Roak BJ, Zhao H, Larson MG, Simon DB, et al. Rare independent mutations in renal salt handling genes contribute to blood pressure variation. Nat Genet. 2008;40:592–599. doi: 10.1038/ng.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rubattu S, Sciarretta S, Morriello A, Calvieri C, Battistoni A, Volpe M. NPR-C: a component of the natriuretic peptide family with implications in human diseases. J Mol Med. 2010;88:889–897. doi: 10.1007/s00109-010-0641-2. [DOI] [PubMed] [Google Scholar]
- 34.Matsukawa N, Grzesik WJ, Takahashi N, Pandey KN, Pang S, Yamauchi M, et al. The natriuretic peptide clearance receptor locally modulates the physiological effects of the natriuretic peptide system. Proc Natl Acad Sci USA. 1999;96:7403–7408. doi: 10.1073/pnas.96.13.7403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch AI, Boerwinkle E, Davis BR, Ford CE, Eckfeldt JH, Leiendecker-Foster C, et al. Pharmacogenetic association of the NPPA T2238C genetic variant with cardiovascular disease outcomes in patients with hypertension. JAMA. 2008;299:296–307. doi: 10.1001/jama.299.3.296. [DOI] [PubMed] [Google Scholar]
- 36.Pereira NL, Aksoy P, Moon I, Peng Y, Redfield MM, Burnett JC, Jr, et al. Natriuretic peptide pharmacogenetics: membrane metal-lo-endopeptidase (MME): common gene sequence variation, functional characterization and degradation. J Mol Cell Cardiol. 2010;49:864–874. doi: 10.1016/j.yjmcc.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.