Abstract
Background
Beta-trace protein (BTP), measured in serum or plasma, has potential as a novel biomarker for kidney function. Little is known about the genes influencing BTP levels.
Methods
We conducted a genome-wide association study of log-transformed plasma BTP levels in 6720 European Americans (EAs) and replicated the significant associations in 1734 African Americans (AAs) from the Atherosclerosis Risk in Communities (ARIC) study.
Results
We identified a genome-wide significant locus in EA upstream of Prostaglandin D2 synthase (PTGDS), the gene encoding BTP. Each copy of the A allele at rs57024841 was associated with 5% higher BTP levels (P = 1.2 × 10−23). The association at PTGDS was confirmed in AAs (6% higher BTP for each A allele at rs57024841, P = 1.9 × 10−7). The index single nucleotide polymorphisms (SNPs) in EAs and AAs explained ∼1.1% of the log(BTP) variance within each population and explained over 30% of the difference in log(BTP) levels between EAs and AAs. The index SNPs at the PTGDS locus in the two populations were not associated with the estimated glomerular filtration rate (eGFR) or the urine albumin creatinine ratio (P > 0.05). We further tested for the associations of BTP with 16 known loci of the eGFR in EA, and BTP was associated with 3 of 16 tested.
Conclusions
The identification of a novel BTP-specific (non-renal related) locus and the confirmation of several genetic loci of the eGFR with BTP extend our understanding of the metabolism of BTP and inform its use as a kidney filtration biomarker.
Keywords: chronic kidney disease, genome-wide association study, glomerular filtration rate, GWAS, kidney function biomarker
INTRODUCTION
The glomerular filtration rate (GFR) is the most widely accepted index of kidney function. The direct measure of the GFR using an exogenous molecule is an invasive procedure and often impractical in both clinical and research settings [1]. Therefore, it is often estimated using equations based on endogenous biomarker levels, such as serum creatinine (SCr). Discovery and characterization of novel filtration markers, such as beta-trace protein (BTP) and cystatin C, are important for improving the precision and reducing bias in GFR estimation due to non-GFR factors influencing SCr [2]. However, novel filtration markers will have their own non-GFR factors. Advances in human genetics are enabling an unbiased characterization of common genetic variation influencing any novel filtration biomarker.
BTP, measured in serum or plasma, has been found to be more sensitive than SCr in detecting a modest decline in the GFR [3–6]. BTP is a low-molecular weight protein (23–29 kDa) that is freely filtered by the kidney. It has been shown to be associated with risk of kidney failure and mortality in participants with chronic kidney disease (CKD) in the Modification of Diet in Renal Disease and African American Study of Kidney Disease and Hypertension studies [7] and with risk of end-stage renal disease (ESRD) in African Americans (AAs) having hypertensive CKD after controlling for measured GFR [8]. Recently, the Atherosclerosis Risk in Communities (ARIC) population-based cohort also found BTP to have stronger associations with ESRD, cardiovascular disease and mortality than SCr estimates of the GFR [9]. Little is known about with non-kidney-related factors influencing BTP levels. The primary goal of this study was to identify genetic loci of BTP. As a biomarker of kidney function, BTP may be associated with kidney function loci or loci specific to the regulation of BTP. Our secondary goal was to examine the association between BTP and known eGFR loci. Investigation into the genetic associations of BTP can enhance our understanding of the biological mechanisms underlying BTP metabolism and inform its potential use as a kidney function biomarker. We conducted a genome-wide association study (GWAS) of plasma BTP in a large population-based cohort, tested replication of the genome-wide significant results, determined whether this locus was associated with kidney function and finally tested for the associations of BTP with 16 previously identified GFR loci among individuals of European ancestry [10].
METHODS
Study population and phenotype and covariate measures
The ARIC study is a prospective observational cohort study of middle-age adults (baseline age between 45 and 64) in four US communities. Details of the study design were reported previously [11]. Briefly, four visits, each ∼3 years apart, were conducted between 1987 and 1998. The baseline sample included 15 792 participants, ∼12 000 European Americans (EAs) and 4000 AAs.
All biomarkers (BTP, serum and urinary creatinine and urinary albumin) for this study were measured from samples collected at visit 4. BTP was measured from plasma samples using nephelometric technology run on the Dade Behring Nephelometer II (BNII) system (sample reliability coefficient: 0.96 in 381 replicates after the removal of 7 outliers with BTP levels >3 SD). A total of 10 557 observations were available (8269 for EA; 2288 for AAs). Combined with the availability of genotype data and the removal of 20 individuals with the self-reported dialysis status or BTP levels >8 SD, the final sample sizes were 6720 in EA and 1734 in AAs (details in Supplementary Methods section).
SCr was measured using the modified kinetic Jaffe method. eGFRscr was calculated using the CKD-EPI equation [12] after standardizing SCr levels by calibrating to the age- and sex-specific means in the Third National Health and Nutrition Examination Survey (NHANES III) and adding 0.5 [13]. In the calculation of the urine albumin creatinine ratio (UACR), the detectable limit of albumin was determined to be 2 mg/L and that of creatinine was 1 mg/dL. Individuals with non-detectable albumin and creatinine levels were given a value of half of the detectable limit.
Diabetes was defined as fasting glucose ≥126 mg/dL, non-fasting glucose ≥200 mg/dL, self-reported physician diagnosis of diabetes mellitus or the use of oral hypoglycemic medication or insulin. Hypertension was defined as systolic blood pressure ≥140 mmHg, diastolic blood pressure ≥90 mmHg or the use hypertension treatment medication.
Statistical methods
A natural-log transformation was applied to BTP levels because its distribution was right skewed. In the genome-wide and the candidate region association analyses, a linear regression model was used to test for the associations between each SNP and log(BTP) levels controlling for age, gender, center and principal components associated (P < 0.05) with log(BTP). Each SNP assumed an additive genetic effect. To determine the existence of multiple independent signals in each genome-wide significant locus, we conducted linear regression for each SNP adding the index SNP at the locus as an additional covariate. The statistic significant threshold for independent signals was set at 0.05 divided by the number of genotyped SNPs that were relatively independent (variance inflation factor, VIF, <2) within the locus. The VIF was calculated using PLINK v1.07 [14] with a window size of 50 SNPs and defined as 1/(1-R2), where R2 is the multiple correlation coefficient for a SNP being regressed against all other SNPs in a window. The replication P-value threshold in AAs was set at 0.001 (=0.05/45) based on the number of genotyped SNPs with VIF <2 in the imputed region (139.4–140.4 mb on chromosome 9 in build 37 position). Details of genotyping and imputation and principal component generation are reported in the Supplementary Methods section. The genome-wide and candidate region association analyses in EAs and AAs were performed using ProbABEL [15] and PLINK v1.07 [14], respectively. Other analyses were conducted using SAS 9.2 or R.
We tested for the differences in the study participant characteristics between EAs and AAs. (t-tests for age and eGFRscr, Wilcoxon test for BTP levels and Chi-square tests for dichotomous variables). To assess whether the GWAS signals upstream of PTGDS could explain the difference in BTP levels between EAs and AAs, we combined the data of the two cohorts and regressed self-reported race against log(BTP) in three regression models. Model 1 included age, gender and center as covariates. Model 2 added hypertension, diabetes, eGFRscr and common-log-transformed UACR as covariates. Model 3 added one of the index SNPs, rs7019538, which was selected because its association with BTP was similar to the other two index SNPs among EA based on P-value and was the strongest among AAs.
The regression analyses of the BTP index SNPs at the PTGDS locus against eGFRscr and UACR and the regression analyses of 16 eGFR index SNPs against scaled BTP and eGFRscr in EA were conducted controlling for age, gender, center and significant principal component(s). In the regression analyses of the eGFR index SNPs against BTP, we took the reciprocal of BTP and scaled it to have the same mean as eGFRscr, so that the effect size of the SNPs against BTP could be compared with that against eGFRscr. We refer to this as scaled BTP in this manuscript.
RESULTS
Table 1 presents the basic characteristics of our study populations. Compared with EAs, AAs had lower BTP levels, higher estimated GFR based on SCr and more individuals with albuminuria, hypertension or diabetes.
Table 1.
Study participant characteristics
| EAs | AAs | P-valuea | |
|---|---|---|---|
| Overall, n | 6720 | 1734 | |
| Age, mean (SD) | 63.1 (5.6) | 61.9 (5.7) | 2.6E-16 |
| Women, % (n) | 54.8 (3686) | 64.3 (1115) | 1.4E-12 |
| Hypertension, % (n) | 42.2 (2818) | 67.6 (1166) | 3.5E-79 |
| Diabetes, % (n) | 13.7 (916) | 27.5 (469) | 5.3E-43 |
| eGFRscr <60 mL/min/1.73 m2, % (n) | 6.6 (437) | 7.15 (124) | 4.4E-01 |
| eGFRscr, mean (SD) | 82.8 (13.8) | 89.3 (19.2) | 2.9E-39 |
| BTP (mg/L), median (IQR) | 0.69 (0.60, 0.78) | 0.60 (0.51, 0.69) | 1.6E-110 |
| Albuminuriab, % (n) | 6.6 (440) | 15.0 (258) | 1.9E-29 |
| CKDc | 11.8 (773) | 19.1 (328) | 3.4E-15 |
eGFRscr, glomerular filtration rate estimated based on the serum creatinine level using the Chronic Kidney Disease Epidemiology Collaboration equation; CKD, chronic kidney disease; IQR, inter-quartile range.
aThe Chi-square test for categorical variables; t-test for age and eGFRscr; the Wilcoxon test for BTP.
bAlbuminuria defined as urinary albumin creatinine ratio >30.
cCKD defined as eGFRscr <60 mL/min/1.73 m2 or the presence of albuminuria.
The GWAS of BTP levels in EAs was initially conducted using imputed dosage based on a HapMap phase 2 reference panel, and it identified 11 SNPs, representing a single locus, on chromosome 9 that achieved genome-wide significance (P < 5 × 10−8, Supplementary Tables S1 and S2). This genome-wide significant locus was upstream of PTGDS, the gene encoding BTP (rs7040970, beta = 0.04, % variance explained = 0.7%, P = 1.5 × 10−17; Table 2). Figure 1 shows the regional association plot of the PTGDS locus. Supplementary Figure S1 presents a plot of the –log10(P-values) by genomic position, and Supplementary Figure S2 presents the quantile–quantile plot of the GWAS results in the EA samples with a genomic control factor of 1.02. Two other loci showed suggestive significance (P-value < 5 × 10−6). One was near MSTN on chromosome 2 (rs16832277, beta = 0.02, P = 1 × 10−6, MAF = 0.28, variance explained = 0.3%). The other was at GCKR on chromosome 4 (rs1260326, beta = 0.018, P = 2 × 10−6, MAF = 0.41, variance explained = 0.3%), which was a reported eGFR locus [10].
Table 2.
Association of log(BTP) with index SNPs at the PTGDS locus in European and African Americans in ARIC
| SNP | Position (b37) | Coded/non-coded allele | Coded allele frequency | Beta | SE | P-value |
|---|---|---|---|---|---|---|
| EA HapMap imputation | ||||||
| rs7040970 | 139 859 013 | C/T | 0.48 | 0.04 | 0.005 | 1.5E-17 |
| EA 1000 Genomes imputation | ||||||
| rs57024841 | 139 862 633 | A/G | 0.58 | 0.05 | 0.005 | 1.2E-23 |
| AA 1000 Genomes imputation | ||||||
| rs57024841 | 139 862 633 | A/G | 0.34 | 0.06 | 0.01 | 1.9E-07 |
| rs7019538 | 139 861 470 | C/T | 0.21 | 0.06 | 0.01 | 6.8E-08 |
EA, European American; AA, African American; SE, standard error.
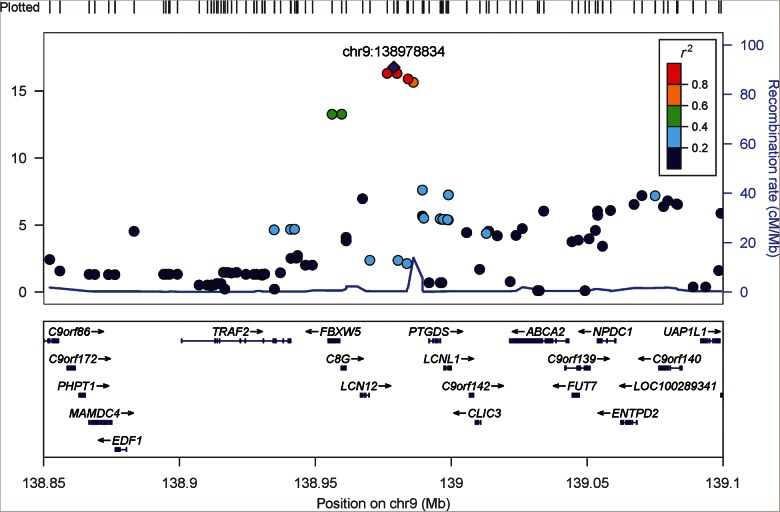
FIGURE 1:
The regional association plot of BTP genome-wide significant locus upstream of PTGDS using genotype dosage imputed from HapMap Phase II reference panel. Build 37 genomic positions are shown on the x-axis. The purple diamond marks the index SNP, rs7040970.
We decided to follow-up on the novel genome-wide significant locus at PTGDS. To finemap the region near PTGDS, we subsequently imputed SNPs across this locus (139.4–140.4 mb in build 37 position) in both EAs and AAs based on 1000 Genomes reference panels [August 2010 release for EA and the Phase I (interim) for AAs]. This 139.4–140.4 mb region was selected because the closest recombination peak was ∼400 kb upstream of the GWAS index SNP rs7040970 (Supplementary Figure S3) and cis-regulatory elements are unlikely to exist far from the protein-coding gene [16]. Using the 1000 Genomes imputed dosage in EAs, the top SNP was rs57024841 (beta = 0.05, P = 1.2 × 10−23; Table 2, Supplementary Table S3). In the regression analysis controlling for this index SNP (rs57024841), we did not detect other significant signals (all SNPs had P > 2.9 × 10−3 > 0.05/28, the number of independent genotyped SNPs in this locus in EAs). The two index SNPs from HapMap phase 2 and 1000 Genomes imputed data (rs7040970 and rs57024841, respectively) were in linkage disequilibrium (LD) (r2 of 0.76 in the 1000 Genomes Interim EUR sample and 0.96 in the ARIC EA samples (Supplementary Table S4).
The replication analysis in AAs using 1000 Genomes imputed dosage at the PTGDS locus detected 64 significant SNPs with P-value below the pre-specified threshold of 0.001. The top SNP was rs7019538 (beta = 0.06, P = 6.8 × 10−8; Table 2 and Supplementary Table S5). After controlling for this top SNP, rs7019538, no additional signals were detected among AAs (P > 1.5 × 10−3). The index SNP identified in EA (rs57024841) was also significant in AAs (beta = 0.06, P = 1.9 × 10−7; Supplementary Table S1).
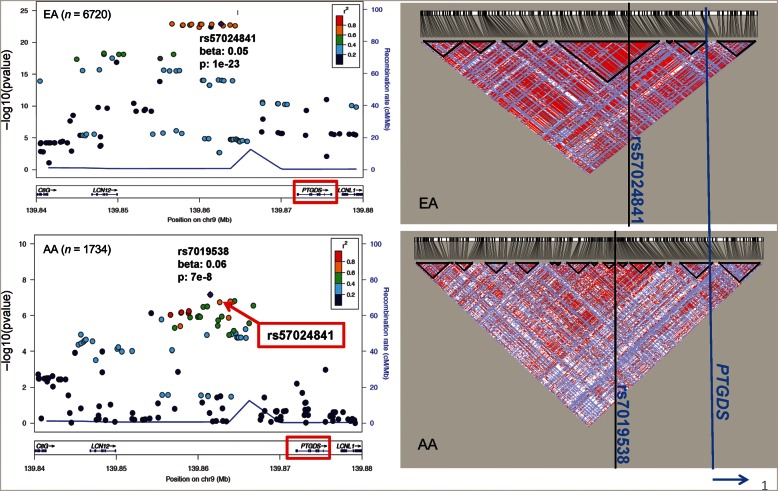
Each of the index SNPs (rs57024841 in EA and rs7019538 in AAs) explained ∼1.1% of the log(BTP) variance in their respective populations (Table 2). The two SNPs were ∼1 kb apart and in high LD within both populations (D' of 1 in 1000 Genomes EUR and AFR populations, Supplementary Table S4). Figure 2 presents the regional association plots in EAs and AAs with their respective LD plots showing more diffused but largely similar LD pattern in AAs compared with the EA samples. As shown in Supplementary Table S1, the two index SNPs had comparable beta estimates within each population (0.05 for rs57024841 and 0.04 for rs7019538 in EA and 0.06 for both SNPs in AAs), indicating that they likely represent the same signal(s). Across the two populations, the coding allele of two index SNPs had lower allele frequencies in AAs than in EAs (0.34 in AAs versus 0.58 in EAs for rs57024841, 0.21 in AAs versus 0.51 in EAs for rs7019538).
FIGURE 2:
Genetic association and LD patterns at the PTGDS locus using imputed dosage from the 1000 Genomes reference panels in EAs and AAs. The Haploview plots on the right show the D' measures between SNPs in the same region as the regional association plots on the left. The positions of the index SNPs and the PTGDS gene are marked with vertical lines in the Haploview plots.
To determine whether the lower allele frequencies of the coding allele of the index SNPs in AAs could partly account for the lower BTP levels in AAs observed between the ARIC EA and AA participants, we performed a combined analysis of the association between self-reported race and log(BTP) levels. In a model controlled for age, sex and center, self-report ‘black’ race was associated with lower BTP levels (beta = −0.10, equivalent to 10% lower in BTP levels, Table 3). In a model controlled for rs57024841 in addition to age, sex, center, diabetes, hypertension, eGFRscr and UACR, the effect estimate of self-reported ‘black’ race was reduced by 38% (Model 3, beta = −0.035) compared with the model without the SNP (Model 2, beta: −0.056, Table 3). Adjusted for these variables, log BTP was not associated with diabetes and hypertension (P > 0.05) and moderately associated with UACR (beta = 0.06 per 10-fold higher UACR) and, as expected, strongly related to eGFRscr (beta = −0.008/1 mL/min/1.73 m2, equivalent to 38% lower BTP levels for 60 mL higher eGFR).
Table 3.
Association of log(BTP) with self-reported race and rs57024841 in combined race data set
| Parameter | Beta | SE | P-value |
|---|---|---|---|
| Model 1: controlled for age, sex, center | |||
| Race (black) | −0.100 | 0.007 | <0.0001 |
| Model 2: controlled for age, sex, center, hypertension, diabetes, eGFRscr and UACR | |||
| Race (black) | −0.056 | 0.0061 | <0.0001 |
| Diabetes | −0.009 | 0.0060 | 0.1234 |
| Hypertension | 0.003 | 0.0045 | 0.5212 |
| eGFRscr | −0.008 | 0.0001 | <0.0001 |
| Log10 (UACR, mg/g) | 0.060 | 0.0036 | <0.0001 |
| Model 3: controlled for age, sex, center, hypertension, diabetes, eGFRscr, UACR and rs7019538 | |||
| Race (black) | −0.035 | 0.0063 | <0.0001 |
| rs57024841 | 0.042 | 0.0034 | <0.0001 |
| Diabetes | −0.010 | 0.0059 | 0.0893 |
| Hypertension | 0.003 | 0.0045 | 0.5707 |
| eGFRscr (mL/min/1.73 m2) | −0.008 | 0.0001 | <0.0001 |
| Log10(UACR, mg/g) | 0.060 | 0.0036 | <0.0001 |
eGFRscr, glomerular filtration rate estimated based on the serum creatinine level using the Chronic Kidney Disease Epidemiology Collaboration equation; UACR, urinary albumin-creatinine ratio.
To determine the potential function of the significant SNPs identified from our GWAS, we interrogated gene expression databases for expression SNP (eSNP) associations with PTGDS gene expression. [17, 18] Two eSNPs downstream of PTGDS were reported to be associated with PTGDS expression [19], but did not explain our GWAS signals upstream of PTGDS (Supplementary Table S6). Since transcription factor binding may affect gene expression and thus protein levels [20], we also queried the data from the ENCODE project [21] for evidence of transcription factor activities at or near the PTDGS locus and found some evidence of transcription factor binding (Supplementary Figure S4).
To assess whether the PTGDS locus may be associated with kidney function, we tested for the association of the BTP index SNPs at the PTGDS locus against eGFRscr and log-transformed UACR in our EA and AA cohorts and found no evidence of associations (P-value >0.05, Supplementary Table S7). Finally, to assess the associations between known eGFR loci [10] and BTP levels, we tested for the association of scaled BTP (for variable definition, see the section ‘Methods’) with the index SNPs at 16 known eGFR loci and compared their association with scaled BTP with their association with eGFRscr. These 16 loci were identified with a total sample size of ∼90 000 [10] from cohorts in the CKDGen consortium. In our study of 6720 individuals, scaled BTP was associated with three loci (GCKR, NAT8 and UMOD) at an alpha level of 0.003 (=0.05/16). The eGFR index SNP at the GCKR locus, rs1260326, was the same SNP that attained suggestive significance in the EA GWAS of log(BTP). Even though only three index SNPs reached statistical significance, 13 of the 16 loci had beta estimates in the same direction as the beta estimates originally reported [10] (binomial test P-value = 0.01), an event that is unlikely due to chance. Based on the same P-value threshold of 0.003, eGFRscr was associated with seven loci with all beta estimates in the same direction as those originally reported [10] (Supplementary Table S8).
DISCUSSION
We identified a genome-wide significant locus of BTP upstream of PTGDS, the gene encoding BTP. The A allele of rs57024841 had a frequency of 58% in EAs compared with 34% in AAs but was associated with a similarly higher level of BTP in the two populations (5 and 6%, respectively). As a result, the PTGDS locus explained over a third of the difference in log(BTP) levels between EAs and AAs in the ARIC study. It accounted for 1.1% of the log(BTP) variance in both EAs and AAs in our sample, and was not associated with either eGFRscr or UACR. However, 13 of the 16 known eGFR loci showed directionally consistent associations with BTP levels, and three loci were statistically significant after a Bonferroni correction. In addition, BTP levels were highly associated with eGFRscr. After adjustment for eGFRscr, BTP was moderately associated with self-reported race and UACR and not associated with hypertension and diabetes.
BTP belongs to the lipocalin protein family and is also known as lipocalin-type prostaglandin (PG) D synthase (L-PGDS). It catalyzes the isomerization of PGH2 as an enzyme [22] and was found to have an independent role in inhibiting astrocyte proliferation [23] and protection against oxidative stress in neural cells [24] in functional experiments. It was first isolated in the cerebrospinal fluid [25] and subsequently shown to be expressed in a variety of tissues, including cells in the glomeruli and the Loop of Henle [26]. After BTP is filtered by the glomeruli, it may be partially reabsorbed and degraded by tubular cells [26] and then excreted in the urine [27–29]. Besides kidney function, little is known about the biological mechanism affecting its level in the blood. White et al., using successive 5′ deletions, identified a core promoter within the 5′ flanking sequence of PTDGS and a thyroid hormone response element (TRE) from 2576 to 2562 bp upstream of PTDGS. There were no SNPs within the 14 bp of the TRE in our imputed data. As reported in the ‘Results’ section, our query of a gene expression database did not find SNP associations with PTGDS expression levels that could explain the GWAS signals. The upstream location of the GWAS signals together with evidence of transcription factor binding in this region from the ENCODE project is consistent with the hypothesis of the existence of distal regulatory elements [30–32]. Further studies are needed to elucidate the biological mechanism underlying the SNP associations at the PTGDS locus with BTP levels.
Although BTP has been studied as a serum or plasma biomarker for kidney function in patients with kidney disease [3, 5, 6, 33–36] and in a community-based cohort [9], relatively little is known about the non-kidney factors influencing BTP levels. In our study of population-based European and African American cohorts, we found the median BTP levels to be ∼0.09 mg/L lower in AA than in their EA counterparts. Moreover, we have identified a genetic locus that is associated with 5–6% differences in BTP levels and accounts for about a third of the racial difference. GFR estimating equations usually require a race coefficient to adjust for race differences in biomarker levels unrelated to kidney function. Self-reported race can be imprecise. If genetic information becomes a routine part of the medical chart, our discovery can be used to account for the largest genetic non-kidney-related individual difference in BTP levels. However, it is also useful to note that this genetic influence is only of moderate size (5 in EAs and 6% in AAs), and our unbiased genome-wide scan suggests that common variants of a larger effect on BTP are unlikely to exist. We also showed that after adjustment for eGFRscr, plasma BTP levels were moderately higher at higher UACR levels.
A limitation of this study is that further functional studies and replication are warranted to fully characterize the causal variant(s) in the novel locus we identified. In contrast, a strength of the present study is the robust, consistent association in both EAs and AAs in a population-based cohort. The GWAS panel spanned the whole genome (∼2.5 million imputed SNPs) and the study sample was large with data collected by uniform methods. BTP was measured with high reliability (reliability coefficient of 0.96).
In conclusion, we discovered a variant upstream of the BTP encoding gene, PTGDS, with a frequency of 58% in EAs, associated with 5% higher levels of BTP and not associated with other measures of kidney function. The frequency of this variant was only 34% in AAs, coding for 6% higher BTP, as a result explaining one-third of the racial difference in log(BTP) levels despite explaining only ∼1.1% of the variance in log(BTP) levels in each ethnic group. Known genetic loci influencing creatinine-based measures of kidney function showed direction consistent associations with BTP levels supporting the role of BTP as a novel filtration marker whose metabolism is known to be different from that of creatinine.
SUPPLEMENTARY DATA
Supplementary data are available online at http://ndt.oxfordjournals.org.
FUNDING
This study was supported by the National Institute of Diabetes and Digestive and Kidney Diseases R01 DK076770–01 Siemens Healthcare Diagnostics provided the reagents and loan of a BNII instrument to conduct the BTP assays. The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201100005C, HHSN268201100006C, HHSN268201100007C, HHSN268201100008C, HHSN268201100009C, HHSN268201100010C, HHSN268201100011C and HHSN268201100012C), R01HL087641, R01HL59367 and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the staff and participants of the ARIC study for their important contributions.
REFERENCES
- 1.K/DOQI. K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis. 2002;39(2 Suppl. 1):S1–266. [PubMed] [Google Scholar]
- 2.Stevens LA, Padala S, Levey AS. Advances in glomerular filtration rate-estimating equations. Curr Opin Nephrol Hypertens. 2010;19:298–307. doi: 10.1097/MNH.0b013e32833893e2. doi:10.1097/MNH.0b013e32833893e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Priem F, Althaus H, Birnbaum M. Beta-trace protein in serum: a new marker of glomerular filtration rate in the creatinine-blind range. Clin Chem. 1999;45:567–568. [PubMed] [Google Scholar]
- 4.Woitas RP, Stoffel-Wagner B, Poege U. Low-molecular weight proteins as markers for glomerular filtration rate. Clin Chem. 2001;47:2179–2180. [PubMed] [Google Scholar]
- 5.Kobata M, Shimizu A, Rinno H. Beta-trace protein, a new marker of GFR, may predict the early prognostic stages of patients with type 2 diabetic nephropathy. J Clin Lab Anal. 2004;18:237–239. doi: 10.1002/jcla.20029. doi:10.1002/jcla.20029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poge U, Gerhardt TM, Stoffel-Wagner B. Beta-trace protein is an alternative marker for glomerular filtration rate in renal transplantation patients. Clin Chem. 2005;51:1531–1533. doi: 10.1373/clinchem.2005.048959. doi:10.1373/clinchem.2005.048959. [DOI] [PubMed] [Google Scholar]
- 7.Tangri N, Inker LA, Tighiouart H. Filtration markers may have prognostic value independent of glomerular filtration rate. J Am Soc Nephrol. 2011;23:351–359. doi: 10.1681/ASN.2011070663. doi:10.1681/ASN.2011070663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhavsar NA, Appel LJ, Kusek JW. Comparison of measured GFR, serum creatinine, cystatin C, and beta-trace protein to predict ESRD in African Americans with hypertensive CKD. Am J Kidney Dis. 2011;58:886–893. doi: 10.1053/j.ajkd.2011.07.018. doi:10.1053/j.ajkd.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Astor BC, Shafi T, Hoogeveen RC. Novel markers of kidney function as predictors of ESRD, cardiovascular disease, and mortality in the general population. Am J Kidney Dis. 2012;59:653–662. doi: 10.1053/j.ajkd.2011.11.042. doi:10.1053/j.ajkd.2011.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kottgen A, Pattaro C, Boger CA. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42:376–384. doi: 10.1038/ng.568. doi:10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.ARIC. The atherosclerosis risk in communities (ARIC) study: design and objectives. The ARIC investigators. Am J Epidemiol. 1989;129:687–702. [PubMed] [Google Scholar]
- 12.Levey AS, Stevens LA, Schmid CH. A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604–612. doi: 10.7326/0003-4819-150-9-200905050-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Selvin E, Manzi J, Stevens LA. Calibration of serum creatinine in the National Health and Nutrition Examination Surveys (NHANES) 1988–1994, 1999–2004. Am J Kidney Dis. 2007;50:918–926. doi: 10.1053/j.ajkd.2007.08.020. doi:10.1053/j.ajkd.2007.08.020. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinf. 2010;11:134. doi: 10.1186/1471-2105-11-134. doi:10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gaffney DJ, Veyrieras JB, Degner JF. Dissecting the regulatory architecture of gene expression QTLs. Genome Biol. 2012;13:R7. doi: 10.1186/gb-2012-13-1-r7. doi:10.1186/gb-2012-13-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. GTEx (Genotype-Tissue Expression) eQTL Database.
- 18. eQTL resources @ the pritchard lab. http://eqtl.uchicago.edu/Home.html. (15 August 2011, date last accessed)
- 19.Zeller T, Wild P, Szymczak S. Genetics and beyond—the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. doi:10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Consortium EP, Birney E, Stamatoyannopoulos JA. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. doi:10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raney BJ, Cline MS, Rosenbloom KR. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. doi:10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Urade Y, Hayaishi O. Biochemical, structural, genetic, physiological, and pathophysiological features of lipocalin-type prostaglandin D synthase. Biochim Biophys Acta. 2000;1482:259–271. doi: 10.1016/s0167-4838(00)00161-8. doi:10.1016/S0167-4838(00)00161-8. [DOI] [PubMed] [Google Scholar]
- 23.Xin X, Huber A, Meyer P. L-PGDS (betatrace protein) inhibits astrocyte proliferation and mitochondrial ATP production in vitro. J Mol Neurosci. 2009;39:366–371. doi: 10.1007/s12031-009-9214-7. doi:10.1007/s12031-009-9214-7. [DOI] [PubMed] [Google Scholar]
- 24.Fukuhara A, Yamada M, Fujimori K. Lipocalin-type prostaglandin D synthase protects against oxidative stress-induced neuronal cell death. Biochem J. 2012;443:75–84. doi: 10.1042/BJ20111889. doi:10.1042/BJ20111889. [DOI] [PubMed] [Google Scholar]
- 25.Saso L, Leone MG, Sorrentino C. Quantification of prostaglandin D synthetase in cerebrospinal fluid: a potential marker for brain tumor. Biochem Mol Biol Int. 1998;46:643–656. doi: 10.1080/15216549800204172. [DOI] [PubMed] [Google Scholar]
- 26.Nagata N, Fujimori K, Okazaki I. De novo synthesis, uptake and proteolytic processing of lipocalin-type prostaglandin D synthase, beta-trace, in the kidneys. FEBS J. 2009;276:7146–7158. doi: 10.1111/j.1742-4658.2009.07426.x. doi:10.1111/j.1742-4658.2009.07426.x. [DOI] [PubMed] [Google Scholar]
- 27.Vynckier LL, Flore KM, Delanghe SE. Urinary beta-trace protein as a new renal tubular marker. Clin Chem. 2009;55:1241–1243. doi: 10.1373/clinchem.2008.119727. doi:10.1373/clinchem.2008.119727. [DOI] [PubMed] [Google Scholar]
- 28.Donadio C. Serum and urinary markers of early impairment of GFR in chronic kidney disease patients: diagnostic accuracy of urinary beta-trace protein. Am J Physiol Renal Physiol. 2010;299:F1407–F1423. doi: 10.1152/ajprenal.00507.2009. doi:10.1152/ajprenal.00507.2009. [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann A, Nimtz M, Conradt HS. Molecular characterization of beta-trace protein in human serum and urine: a potential diagnostic marker for renal diseases. Glycobiology. 1997;7:499–506. doi: 10.1093/glycob/7.4.499. doi:10.1093/glycob/7.4.499. [DOI] [PubMed] [Google Scholar]
- 30.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev Biol. 2010;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. doi:10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. doi:10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maston GA, Evans SK, Green MR. Transcriptional regulatory elements in the human genome. Annu Rev Genomics Hum Genet. 2006;7:29–59. doi: 10.1146/annurev.genom.7.080505.115623. doi:10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 33.Filler G, Priem F, Lepage N. Beta-trace protein, cystatin C, beta(2)-microglobulin, and creatinine compared for detecting impaired glomerular filtration rates in children. Clin Chem. 2002;48:729–736. [PubMed] [Google Scholar]
- 34.Solichova P, Novackova L, Ochmanova R. Assessment of serum beta-trace protein (BTP) measurement in the prediction of glomerular filtration rate. Comparison with serum cystatin C. Biomed Pap Med Fac Univ Palacky Olomouc Czech Repub. 2006;150:83–84. doi: 10.5507/bp.2006.009. doi:10.5507/bp.2006.009. [DOI] [PubMed] [Google Scholar]
- 35.White CA, Akbari A, Doucette S. Estimating GFR using serum beta trace protein: accuracy and validation in kidney transplant and pediatric populations. Kidney Int. 2009;76:784–791. doi: 10.1038/ki.2009.262. doi:10.1038/ki.2009.262. [DOI] [PubMed] [Google Scholar]
- 36.Spanaus KS, Kollerits B, Ritz E. Serum creatinine, cystatin C, and beta-trace protein in diagnostic staging and predicting progression of primary nondiabetic chronic kidney disease. Clin Chem. 2010;56:740–749. doi: 10.1373/clinchem.2009.138826. doi:10.1373/clinchem.2009.138826. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.