Abstract
Background
The Fra-1/AP-1 transcription factor regulates the expression of genes controlling various processes including migration, invasion, and survival as well as extracellular remodeling. We recently demonstrated that loss of Fra-1 leads to exacerbated bleomycin-induced pulmonary fibrosis, accompanied by enhanced expression of various inflammatory and fibrotic genes. To better understand the molecular mechanisms by which Fra-1 confers protection during bleomycin-induced lung injury, genome-wide mRNA expression profiling was performed.
Results
We found that Fra-1 regulates gene expression programs that include: 1) several cytokines and chemokines involved in inflammation, 2) several genes involved in the extracellular remodeling and cell adhesion, and 3) several genes involved in programmed cell death.
Conclusion
Loss of Fra-1 leads to the enhanced expression of genes regulating inflammation and immune responses and decreased the expression of genes involved in apoptosis, suggesting that this transcription factor distinctly modulates early pro-fibrotic cellular responses.
Background
Pulmonary fibrosis is a chronic, progressive, and usually untreatable group of chronic disorders and appears to be regulated by complex cellular processes [1]. In animal models, a single intratracheal administration of bleomycin induces an inflammatory response that is characterized by leukocyte infiltration, apoptosis, fibroblast proliferation, matrix metalloproteinase (MMP)/tissue inhibitor of metalloproteinase (TIMP) imbalance, and an increase in interstitial collagen content [2,3] that can culminate in the development of pulmonary lesions similar to those observed in human interstitial pulmonary fibrosis (IPF) [4]. However, the exact mechanisms underlying pulmonary fibrosis remain unclear.
AP-1 is a dimeric transcription factor, mainly comprised of the Jun (c-Jun, Jun-B, Jun-D), Fos (c-Fos, Fos-B, Fra-1, Fra-2), and ATF (ATF1-4) families of b-ZIP transcription factors. AP-1 binds to the TPA response element (TRE, also known as the AP-1 site) and regulates target gene expression in response to various pro-oxidants and toxicants. These gene products mediate (mitigate or promote) oxidative stress and inflammatory responses, as well as cell growth and tumorigenesis [5]. The promoters of many inflammatory response genes, especially those encoding cytokines and chemokines, have functional AP-1 binding sites [6]. Fra-1 regulates gene expression involved in various processes such as cell growth and cell death and regulates the expression of genes controlling tissue/cell remodeling, such as MMP-1, MMP-2, and MMP-9, mainly at the transcriptional level [7-11]. We have recently shown that Fra-1-deficient (Fra-1∆/∆) mice are more susceptible than wild-type (Fra-1+/+) mice to bleomycin-induced fibrosis [12], suggesting that this transcription factor is involved in the regulation of complex genetic networks to maintain cellular homeostasis during bleomycin-induced lung inflammation, injury, and repair processes. Based on these results, we hypothesized that accelerated inflammation and fibrosis observed in Fra-1∆/∆ mice are caused by enhanced inflammatory and fibrotic gene expression. To test this hypothesis and to better understand the mechanisms by which the Fra-1 transcription factor confers pulmonary protection, we have performed microarray analysis to examine the changes in gene expression in the lungs of Fra-1∆/∆ mice after treatment with bleomycin. In the present study, we have evaluated changes in early inflammatory and pro-fibrotic gene expression after 5 days of bleomycin treatment. Our mRNA expression profiling demonstrated increased expression of genes involved in inflammation and immune responses and decreased levels of apoptotic genes in Fra-1∆/∆ mice, suggesting that the Fra-1 transcription factor dampens the development of late fibrotic injury by modulating the early pro-fibrotic responses.
Results and discussion
Genes that encode cytokines, chemokines, and their receptors
The set of genes that was differentially expressed between PBS-treated Fra-1+/+ and Fra-1∆/∆ mice was studied in order to identify those genes for which a genotypic difference in expression exists. We found that the loss of Fra-1 led to an up-regulation of chemokine (C-X-C motif) ligand 13 (Cxcl13, 1.7-fold) and interleukin 1 alpha (Il1a, 1.7-fold) expression. Similarly, we found a down-regulation of chemokine (C-C motif) ligand 5 (Ccl5, -1.6-fold) and chemokine (C-X-C motif) ligand 9 (Cxcl9, -2.7-fold) in Fra-1∆/∆ mice when compared to Fra-1+/+ mice (Table 1). Next, we compared the differentially up-regulated genes between bleomycin-treated Fra-1∆/∆ and Fra-1+/+ mice (Table 2). The genes that showed a -fold change of ≥1.7 were selected for analysis. Interestingly, our data suggested that the lack of Fra-1 leads to up-regulation of cytokines and chemokines in response to bleomycin, including interleukin-1 alpha (Il1a, 1.87-fold), interleukin-2 receptor alpha chain (Il2ra, 2.15-fold), interleukin 2 receptor, beta chain (Il2rb, 1.69-fold), interleukin 6 (Il6, 2.23-fold), chemokine (C-C motif) ligand 8 (Ccl8, 2.27-fold), and C-X-C motif chemokine 10 (Cxcl10, 2.54-fold), whereas Fra-1+/+ mice showed an up-regulation of interleukin 1 receptor, type II (Il1r2, 2.27-fold). These results suggest that Fra-1 signaling controls the expression of some of the genes that are involved in fibrosis. For instance, expression of interleukin 6, a cytokine that promotes greater inflammation and fibrosis [13], was significantly higher in bleomycin-treated Fra-1∆/∆ mice (2.23-fold) than in Fra-1+/+ mice (Table 2). Also, we noted that Fra-1+/+ mice showed an increased expression of Il1r2 (Il1r2, 2.27-fold) in response to bleomycin as compared to Fra-1∆/∆ mice. Interleukin 1 (Il1), a principal pro-inflammatory cytokine that includes two ligands (Il1α and Il1β) and two cell surface receptors namely Il1r1 and Il1r2. Several reports suggest that binding of Il1 to Il1r1 ultimately leads to the activation of several genes, including those encoding cyclooxygenase, nitricoxide synthase, cell adhesion molecules and cytokines and chemokines [14,15]. More importantly, in mouse models, exogenous administration of recombinant Il1β induced high degree of bleomycin-induced fibrosis, and specific blockade of Il1r1 markedly reduced bleomycin-induced inflammation [16,17]. Due to the lack of a cytoplasmic-signaling domain for Il1r2, this receptor mainly acts as a decoy receptor to prevent Il1-mediated biological responses [18]. Many anti-inflammatory mediators enhance the expression and release of Il1r2 to induce anti-Il1 pathway [19-21]. The increased Il1r2 expression in Fra-1+/+ but not in Fra-1∆/∆ mice suggests that Fra-1 controls bleomycin-induced inflammation by augmenting the expression of anti-inflammatory genes. In the bleomycin-induced fibrosis model, extensive neovascularization has often been observed to follow the airways and sites of injury [22]. Presence/absence of ERL motif in CXC chemokines dictates their angiogenic property [23]. The amount of Cxcl10 chemokine in the lungs has been directly correlated with the degree of fibrosis [24]. Administration of Cxcl10 to bleomycin-treated mice attenuates pulmonary fibrosis in part due to reduced angiogenesis [25]. However, we found that bleomycin-induced Fra-1∆/∆ mice showed a 2.54-fold increase in Cxcl10 when compared to Fra-1+/+ mice (Table 2).
Table 1.
Differentially expressed genes in lung tissues of Fra-1∆/∆and Fra-1+/+ mice
| Affymetrix ID | Symbol | Gene title | KOC vs. WTC (fold change) |
|---|---|---|---|
| Cytokine and chemokines |
|
|
|
| 10487588 |
Il1a |
Interleukin 1 alpha |
1.7 |
| 10523359 |
Cxcl13 |
Chemokine (C-X-C motif) ligand 13 |
1.7 |
| 10389207 |
Ccl5 |
Chemokine (C-C motif) ligand 5 |
−1.6 |
| 10531407 |
Cxcl9 |
Chemokine (C-X-C motif) ligand 9 |
−2.7 |
| Inflammation |
|
|
|
| 10545569 |
Reg3g |
Regenerating islet-derived 3 gamma |
7 |
| 10402390 |
Serpina1b |
Serine preptidase inhibitor, clade A, member 1B |
4.61 |
| 10535559 |
Baiap2l1 |
BAI1-associated protein 2-like 1 |
3.9 |
| 10349648 |
Ctse |
Cathepsin E |
3.61 |
| 10402409 |
Serpina1e |
Serine peptidase inhibitor, clade A, member 1E |
2.47 |
| 10563597 |
Saa3 |
Serum amyloid A 3 |
1.82 |
| 10362138 |
Vnn1 |
Vanin 1 |
1.72 |
| 10398117 |
Bdkrb2 |
Bradykinin receptor B2 |
1.69 |
| 10481627 |
Lcn2 |
Lipocalin 2 |
1.4 |
| 10372652 |
Lyz1 |
Lysozyme 1 |
1.31 |
| 10444824 |
H2-Q6 |
Histocompatibility 2, Q region locus 6 |
−1.66 |
| 10497356 |
Sirpb1a |
Signal-regulatory protein beta 1A |
−1.72 |
| 10574149 |
Nlrc5 |
NLR family, CARD domain containing 5 |
−1.78 |
| 10398069 |
Serpina3m |
Serine peptidase inhibitor, clade A, member 3 M |
−2.32 |
| Cell adhesion molecule |
|
|
|
| 10395553 |
Nrcam |
Neuron-glia-CAM-related cell adhesion molecule |
1.95 |
| 10523717 |
Spp1 |
Secreted phosphoprotein 1 |
−1.81 |
| Transcription |
|
|
|
| 10521537 |
Cytl1 |
Cytokine-like 1 |
2.02 |
| 10598409 |
Tcfe3 |
Transcription factor E3 |
1.78 |
| 10583312 |
Taf1d |
TATA box binding protein (Tbp)-associated factor, RNA polymerase I, D |
−1.63 |
| 10399725 | Sox11 | SRY-box containing gene 11 | −1.96 |
Table 2.
Differentially expressed genes induced by bleomycin in lung tissues of Fra-1+/+ and Fra-1∆/∆mice
| Affymetrix ID | Symbol | Gene title | KOT vs. WTT (fold change) |
|---|---|---|---|
| Cytokine, chemokines, and their receptors |
|
|
|
| 10531415 |
Cxcl10 |
C-X-C motif chemokine 10 |
2.54 |
| 10379535 |
Ccl8 |
Chemokine (C-C motif) ligand 8 |
2.27 |
| 10520452 |
Il6 |
Interleukin 6 |
2.23 |
| 10469278 |
Il2ra |
Interleukin-2 receptor alpha chain |
2.15 |
| 10487588 |
Il1a |
Interleukin-1 alpha |
1.87 |
| 10430344 |
Il2rb |
Interleukin 2 receptor, beta chain |
1.69 |
| 10523128 |
Ppbp |
Chemokine (C-X-C motif) ligand 7 |
–2.17 |
| 10345752 |
Il1r2 |
Interleukin 1 receptor, type II |
−2.27 |
| Inflammation |
|
|
|
| 10531126 |
Igj |
Immunoglobulin joining chain |
17.25 |
| 10496555 |
Gbp1 |
Guanylate binding protein 1 |
7.88 |
| 10403069 |
Igh-6 |
Immunoglobulin heavy constant mu |
7.43 |
| 10545569 |
Reg3g |
Regenerating islet-derived 3 gamma |
4.61 |
| 10502801 |
H28 |
Histocompatibility 28 |
3.3 |
| 10364542 |
CFD |
Complement factor D |
3.09 |
| 10563602 |
Saa4 |
Serum amyloid A 4 |
2.81 |
| 10576757 |
Fcer2a |
Fc receptor, IgE, low affinity II, alpha polypeptide |
2.41 |
| 10466172 |
Ms4a1 |
Membrane-spanning 4-domains, subfamily A, member 1 |
2.39 |
| 10562169 |
Hamp |
Hepcidin antimicrobial peptide |
2.38 |
| 10551025 |
Cd79a |
CD79A antigen |
2.11 |
| 10531987 |
Gbp4 |
Guanylate binding protein 4 |
2.11 |
| 10437224 |
Mx2 |
Myxovirus (influenza virus) resistance 2 |
2.1 |
| 10441233 |
Mx1 |
Interferon-induced GTP-binding protein Mx1 |
1.96 |
| 10500677 |
CD2 |
CD2 antigen |
1.85 |
| 10450675 |
H2-T24 |
Histocompatibility 2, T region locus 24 |
1.81 |
| 10451287 |
Isg15 |
ISG15 ubiquitin-like modifier |
1.81 |
| 10531994 |
Mpa2l |
Guanylate binding protein 6 |
1.79 |
| 10398121 |
Bdkrb1 |
Bradykinin receptor, beta 1 |
1.76 |
| 10399710 |
Rsad2 |
Radical S-adenosyl methionine domain containing 2 |
1.75 |
| 10435982 |
Btla |
B and T lymphocyte associated |
1.74 |
| 10468898 |
Lax1 |
Lymphocyte transmembrane adaptor 1 |
1.71 |
| 10547894 |
CD4 |
CD4 antigen |
1.7 |
| 10444821 |
H2-Q8 |
Histocompatibility 2, Q region locus 8 |
1.69 |
| 10444236 |
H2-DMb2 |
Histocompatibility 2, class II, locus Mb2 |
1.67 |
| 10372652 |
Lyz1 |
Lysozyme 1 |
1.66 |
| 10574098 |
Nlrc5 |
NOD-like receptor C5 |
1.66 |
| 10601385 |
Tlr13 |
Toll-like receptor 13 |
−1.66 |
| 10560242 |
C5ar1 |
Complement component 5a receptor 1 |
−1.72 |
| 10541614 |
Clec4d |
C-type lectin domain family 4, member d |
−1.78 |
| 10347335 |
Slc11a1 |
Solute carrier family 11, member 1 |
−1.85 |
| 10416837 |
Irg1 |
Immunoresponsive gene 1 |
−2.08 |
| 10493831 |
S100a8 |
S100 calcium binding protein A8 |
−2.63 |
| 10349648 |
Ctse |
Cathepsin E |
−2.77 |
| Cell adhesion molecule |
|
|
|
| 10433172 |
Glycam1 |
Glycosylation dependent cell adhesion molecule 1 |
1.98 |
| 10500677 |
CD2 |
CD2 antigen |
1.85 |
| 10450675 |
H2-T24 |
Histocompatibility 2, T region locus 24 |
1.81 |
| 10547894 |
CD4 |
CD4 antigen |
1.7 |
| 10444821 |
H2-Q8 |
Histocompatibility 2, Q region locus 8 |
1.69 |
| 10444236 |
H2-DMb2 |
Histocompatibility 2, class II, locus Mb2 |
1.67 |
| 10562720 |
Siglece |
Sialic acid binding Ig-like lectin E |
−1.78 |
| 10557862 |
Itgam |
Integrin alpha M |
−2.04 |
| Transcription |
|
|
|
| 10404389 |
Irf4 |
Interferon regulatory factor 4 (TF) |
2.06 |
| 10585276 |
Pou2af1 |
POU class 2 associating factor 1 |
2.09 |
| 10562812 |
Spib |
Spi-B transcription factor (Spi-1/PU.1 related) (TF) |
1.83 |
| 10360406 |
Ifi205 |
interferon activated gene 205 |
1.65 |
| 10390691 |
Nr1d1 |
Nuclear receptor subfamily 1, group D, member 1 (TF) |
1.7 |
| 10460585 | Fosl1 | Fos-like antigen 1 | −1.85 |
We then analyzed genes that are uniquely expressed in either Fra-1+/+ (see Additional file 1: Table S1) or Fra-1∆/∆ mice (see Additional file 2: Table S2) after bleomycin treatment. The Fra-1∆/∆ mice showed an up-regulation of chemokine (C-C motif) ligand 19 (Ccl19, 2.07-fold), macrophage-derived chemokine (Ccl22, 2.42-fold), chemokine (C motif) ligand 1 (Xcl1, 2.15-fold), chemokine (C-X-C motif) ligand 11 (Cxcl11, 2.53-fold), chemokine (C-C motif) receptor 4 (Ccr4, 1.75-fold), chemokine (C-C motif) receptor 8 (Ccr8, 2.13-fold) and chemokine (C-C motif) receptor 9 (Ccr9, 2.63-fold) (see Additional file 2: Table S2), whereas Fra-1+/+ mice showed an up-regulation of chemokine (C-X-C motif) receptor 2 (Cxcr2, 2.83-fold), interleukin 1 family, member 9 (Il1f9, 2.43-fold), chemokine (C-X-C motif) ligand 2 (Cxcl2, 3.78-fold), colony stimulating factor 2 receptor, alpha, low-affinity (Csf2ra, 2.02-fold) and interleukin 1 receptor-like 2 (Il1rl2, 1.85-fold) (see Additional file 1: Table S1). Of particular interest is the Ccl22 chemokine, which has been recognized as a Th2 chemokine, and its involvement in the pathophysiology of pulmonary fibrosis has been documented. Belperio et al. have demonstrated that Ccl22 and its receptor, Ccr4, are overexpressed in a mouse model of belomycin-induced fibrosis [26]. Ccl22 and Ccr4 levels are also increased in patients with IPF, and their expression has been detected on epithelium and macrophages, respectively [27]. Neutralization of Ccl22 and Ccr4 has been shown to lead to a significant reduction in lung inflammation during bleomycin-induced fibrosis [26,28]. Interestingly, our results here showed that the expression of Ccl22 and Ccr4 was significantly increased (2.42- and 1.75-fold, respectively) in bleomycin-treated Fra-1∆/∆ mice when compared to Fra-1+/+ mice.
Next, we analyzed genes that are uniquely down-regulated in bleomycin-treated Fra-1∆/∆ mice (see Additional file 2: Table S2), which included chemokine (C-C motif) receptor-like 2 (Ccrl2, -1.75-fold), bone morphogenetic protein 2 (Bmp2, -1.78-fold), and bone morphogenetic protein 3 (Bmp3, -1.78-fold). Similarly, Fra-1+/+ also showed uniquely down-regulated genes, including chemokine (C-X-C motif) receptor 5 (Cxcr5, -2.38-fold), chemokine (C-C motif) ligand 21A (Ccl21a, -1.72-fold), chemokine (C-C motif) ligand 27a (Ccl27a, -1.75-fold) and ciliary neurotrophic factor receptor (Cntfr, -1.75-fold) (see Additional file 1: Table S1). Taken together, the results of the present study have revealed that genetic disruption of Fra-1 differentially regulates a number of cytokines and chemokines in response to bleomycin, indicating a potential role for Fra-1 in cytokine and chemokine signaling during bleomycin-induced acute lung injury.
Genes encoding proteins that are involved in the inflammatory response
In Fra-1∆/∆ mice treated with vehicle, we found up-regulation of some genes involved in the inflammatory response (Table 1) when compared to their Fra-1+/+ counterparts. These genes included regenerating islet-derived 3 gamma (Reg3g, 7.00-fold), cathepsin E (ctse, 3.61-fold), serine peptidase inhibitor, clade A, member 1B (Serpina1b, 4.61-fold) and serum amyloid A3 (saa3, 1.82-fold). On the other hand, the lack of Fra-1 led to the down-regulation of the expression of a few genes, including serine peptidase inhibitor, clade A, member 3 M (Serpina3m, -2.32-fold) and NLR family, CARD domain containing 5 (Nlrc5, -1.78-fold). Matrix metalloproteases (MMPs) play key roles in tissue repair and remodelling; but recent studies indicate a prominent role for the lysosomal proteinases, such as cathepsins, in the extracellular remodelling [29,30]. BAL fluid and extracellular space contain secreted cathepsins and their activities are controlled by endogenous inhibitors of cathepsins [31,32]. Likewise, serine peptidases are implicated in various biological processes such as wound healing and they also contribute to the development of pulmonary fibrosis and acute lung injury [33,34]. Endogenous serine peptidase inhibitors regulate the activities of serine peptidases. Imbalance in the activities of proteinases/peptidases and endogenous proteinases/peptidases inhibitors may contribute to deregulated protein degradation and resulting in the initiation of lung injury [35]. Serum amyloid A is an acute phase protein, induced by several inflammatory mediators and its serum level is elevated in various conditions like COPD [36], bronchial carcinoma [37], and cardiovascular disease [38]. Thus, it is likely that Fra-1 distinctly regulates proteinases/peptidases and their inhibitor’s expression to maintain lung homeostasis.
When we compared the gene expression patterns of Fra-1∆/∆ mice and their Fra-1+/+ littermates following bleomycin treatment, it was evident that there were many more up-regulated genes than down-regulated genes involved in inflammation in the lungs of Fra-1∆/∆ mice when compared to their Fra-1+/+counterparts (Table 2). We identified differential expression of genes that belong to the immunoglobulin family, specifically immunoglobulin joining chain (Igj, 17.25-fold) and immunoglobulin heavy constant mu (Igh-6, 7.43-fold) in Fra-1∆/∆ mice treated with bleomycin (Table 2). It has previously been shown that immunoglobulin concentrations are increased in immune disorders, such as rheumatoid arthritis [39], inflammatory bowel disease [40], and some respiratory disorders including asthma [41], cystic fibrosis [42], and idiopathic pulmonary fibrosis [43]. The up-regulation of immunoglobulin genes was also accompanied by an up-regulation of several genes involved in antigen presentation and antigen binding. This was true for some of the major histocompatibility genes, including histocompatibility 28 (H28, 3.3-fold), histocompatibility 2, class II, locus Mb2 (H2-DMb2, 1.67-fold), histocompatibility 2, T region locus 24 (H2-T24, 1.81-fold), and histocompatibility 2, Q region locus 8 (H2-Q8, 1.69-fold) (Table 2). Complement components such as complement factor D (CFD, 3.09-fold), and antigens such as CD79A antigen (Cd79a, 2.11-fold), CD2 antigen (CD2, 1.85-fold), and CD4 antigen (CD4, 1.70-fold) were differentially up-regulated in Fra-1∆/∆ mice (Table 2). The expression of other inflammatory genes, including regenerating islet-derived 3 gamma (Reg3g, 4.61-fold) and serum amyloid A 4 (Saa4, 2.81-fold), was also differentially up-regulated in Fra-1∆/∆ mice treated with bleomycin (Table 2). The Reg3g and Saa4 genes have now been consistently associated with pulmonary fibrosis and chronic inflammation [36,44]. On the other hand, we also noticed down-regulation of some genes in Fra-1∆/∆ mice, such as solute carrier family 11, member 1 (Slc11a1, -1.85-fold), S100 calcium binding protein A8 (S100a8, -2.63-fold), and cathepsin E (Ctse, -2.77-fold) (Table 2), when compared to Fra-1+/+ mice. Again, we analyzed genes that are uniquely expressed in either Fra-1∆/∆ (see Additional file 2: Table S2) or Fra-1+/+ (see Additional file 1: Table S1) mice in response to bleomycin. The results showed that major histocompatibility molecules such as histocompatibility 2, Q region locus 6 (H2-Q6, 2.35-fold), MHC class I like protein GS10 (H2-gs10, 2.12-fold), and complement components such as complement component 4B (C4b, 1.78-fold) were uniquely up-regulated in Fra-1∆/∆ mice treated with bleomycin (see Additional file 2: Table S2). In contrast, we found down-regulation of immunoglobulin heavy constant mu (Igh-6, -8.33-fold), histocompatibility 2, O region beta locus (H2-Ob, -3.12-fold), Cd226 antigen (Cd226, -1.75-fold), and complement receptor 2 (Cr2, -2.22-fold) in Fra-1+/+ mice after bleomycin treatment. Our previous study demonstrated that Fra-1∆/∆ mice showed increased levels of inflammation after bleomycin treatment [12]. Thus, our present data suggest that deregulation of the expression of immune response genes in Fra-1∆/∆ mice is the likely cause of the increased lung inflammation in Fra-1∆/∆ mice.
Genes that encode extracellular matrix and cell adhesion molecules
The unique gene expression pattern in Fra-1+/+ mice (see Additional file 1: Table S1) treated with bleomycin suggested an increase in the expression of genes that encode extracellular matrix, such as collagen, type IV, alpha 1 (Col4a1, 1.96-fold), collagen, type IV, alpha 2 (Col4a2, 1.76-fold), collagen, type VI, alpha 1 (Col6a1, 1.70-fold), collagen, type VI, alpha 2 (Col6a2, 1.85-fold), collagen, type VI, alpha 3 (Col6a3, 1.98-fold), collagen, type XV, alpha 1 (Col15a1, 1.79-fold), a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 1 (Adamts1, 1.94-fold), and a disintegrin-like and metallopeptidase (reprolysin type) with thrombospondin type 1 motif, 2 (Adamts2, 2.07-fold). In contrast, Fra-1∆/∆ mice showed increased expression levels of laminin, alpha 1 (Lama1, 1.75-fold).
Among the various collagens, types I and III collagens are the most widely distributed in both airways and parenchymal structures [45]. To maintain normal structural properties of lung, the controlled distribution of these proteins is vital and their inappropriate accumulation in fibrotic lungs has been reported [46,47]. In contrast, the physiologic functions and abnormal deposition patterns of other collagens in the lung fibrosis are poorly understood. Our recent study showed an increased expression of the TGF-β1 and type-1-collagen genes in response to bleomycin at the end of 14 and 31 days of bleomycin treatment and demonstrated the presence of increased fibrosis in Fra-1∆/∆ mice [12]. However, we did not observe any differences in the expression levels of type I collagen or type III collagen, nor did we observe altered TGF-β1 gene expression in either genotype at the end of 5 days bleomycin treatment. It has been reported that excessive synthesis and deposition of ECM proteins is a general tissue response to an unresolved chronic inflammation [48]. Hence, we speculate that the persistence of increased inflammation in Fra-1∆/∆ mice is driven by the loss of Fra-1, while higher levels of fibrotic gene expression in the fibrotic stage (14 days) may contribute to the excessive deposition of ECM and disease severity seen in Fra-1∆/∆ mice. This point needs to be addressed in order to better understand the mechanisms underlying the increased fibrosis in Fra-1∆/∆ mice.
We found that a few genes known to be involved in general cell adhesion are affected by bleomycin treatment in Fra-1∆/∆ mice (Table 2). Glycosylation-dependent cell adhesion molecule 1 (Glycam1, 1.98-fold), cd2 antigen (Cd2, 1.85-fold), sialic acid binding Ig-like lectin E (Siglece, -1.78-fold), and integrin alpha M (Itgam, -2.04-fold) were differentially expressed in Fra-1∆/∆ mice treated with bleomycin when compared to Fra-1+/+ mice. In general, the expression of intercellular adhesion molecules is increased by inflammatory signals, which facilitates lymphocytes for higher adhesion and permeation into inflamed tissues. It has been shown that the glycam1 molecule is strongly expressed in inflammatory processes in order to modulate leukocyte trafficking [49]. Sialic acids present on the surface of all mammalian cells and play important roles in physiological and pathological processes, and their expression has been reported to decrease during immune cell activation [50]. Consistent with these findings, we also noted an up-regulation in the expression of glycam1 and a loss of siglece in Fra-1∆/∆ mice when compared to Fra-1+/+mice.
We then analyzed the genes that are uniquely expressed in Fra-1∆/∆ (see Additional file 2: Table S2) or Fra-1+/+ mice (see Additional file 1: Table S1). Fra-1∆/∆ mice showed unique expression of genes that included major histocompatibility molecules such as histocompatibility 2, Q region locus 6 (H2-Q6, 2.35-fold), histocompatibility 2, T region locus 22 (H2-T22, 1.81-fold), histocompatibility 2, K1 region (H2-K1, 1.7-fold), laminin alpha 1 (Lama 1, 1.75-fold), and heparanase (Hpse, 1.98-fold). Fra-1+/+ mice showed unique expression of mesothelin (Msln, 2.16-fold), neuron-glia-CAM-related cell adhesion molecule (Nrcam, 2.1-fold), and a disintegrin and metallopeptidase domain 12 (Adam 12, 2.1-fold). Heparanase is an endoglucuronidase that cleaves heparan sulfate (HS) chains, resulting in HS fragments of 10 to 20 sugar units [51]. Overexpression of heparanase has been reported in numerous tumors, where it regulates angiogenesis and metastasis [52]. Furthermore, the ADAM gene family is associated with proteolytic, cell-cell, and cell-matrix interaction-promoting activities. Several investigations have shown a functional role for ADAMS in collagen deposition in cells and in lungs in which the ADAM gene was knocked down, thus revealing the functional dysregulation of this gene family in the lung fibrosis [53-55]. Thus, an alteration in the expression of cell adhesion molecules may represent another potential mechanism by which more fibrosis can occur in Fra-1∆/∆ mice.
Fra-1-regulated genes involved in programmed cell death
The process of programmed cell death is known to play a major role in maintaining many biological processes, and inappropriate apoptosis can lead to disease conditions, either because cells experience an inappropriately prolonged survival or they die prematurely [56,57]. Studies using genetic models have demonstrated both cooperative and antagonistic roles of AP-1 family of proteins in modulating cell death in response to a variety of pro-apoptotic stimuli. For example, c-Jun-/- mouse embryonic fibroblasts (MEFs) and liver cells show increased levels of oxidative stress and apoptosis [58]. Likewise, c-Fos also participates in both pro- and anti-apoptotic activities. For example, c-Fos-/- MEFs undergo apoptosis when cultured in vitro and also display an increased susceptibility to UV-induced cell death [59]. Overexpression of Fra-1 also inhibits proliferation, induces apoptosis, and reduces the tumorigenicity of c6 glioma cells [60]. Consistent with a role for Fra-1 in apoptosis, we recently found that mouse embryonic fibroblasts lacking Fra-1 show an increased resistance to oxidant-induced cell death [61]. Fra-1 appears to uniquely up-regulate some genes modulating apoptosis in Fra-1+/+ mice (see Additional file 1: Table S1), including paternally expressed 3 (Peg3, 2.55-fold), the tumor necrosis factor receptor superfamily, member 10b (Tnfrsf10b, 2.03-fold), AXL receptor tyrosine kinase (Axl, 1.73-fold), Eph receptor A2 (Epha2, 1.7-fold), zinc finger matrin type 3 (Zmat 3, 1.76-fold), solute carrier family 40 (iron-regulated transporter, member 1) (Slc40a1, 1.70-fold), and EGL nine homolog 3 (Egln3, 1.81-fold). Similarly, Fra-1∆/∆ mice showed (see Additional file 2: Table S2) up-regulation of glutamate-cysteine ligase, catalytic subunit (GCLC, 1.70-fold) and down-regulation of lectin, galactose binding, soluble 12 (Lgals 12, -1.78-fold), Eph receptor A7 (Epha 7, -1.72-fold), and arachidonate 12-lipoxygenase (Alox12, -1.69-fold). It has been reported that kaempferol exerts an anti-oxidative and anti-apoptotic effects in HEI-OC1 cells treated with cisplatin through enhancing GCLC expression [62]. Consistent with this observation, we found that GCLC was induced in Fra-1∆/∆ mice. Our gene expression results from Fra-1∆/∆ mice are therefore in good agreement with the observation that inappropriate apoptosis can lead to exaggerated lung fibrosis.
Validation of microarray data
Amongst several genes that were significantly affected by bleomycin, we randomly selected 17 genes according to the microarray results to confirm their differential expression by qRT-PCR. We confirmed that bleomycin treatment significantly induced the expression of Il1a, Irf4, Reg3g, and Ccr4 and reduced the expression of S100a8 in Fra-1∆/∆ mice when compared to similarly treated Fra-1+/+ mice (Table 3). These results confirmed the expression patterns of the microarrays. Next, we analyzed the genes that were uniquely expressed in both genotypes. The results revealed that Fbln2 expression was significantly higher in Fra-1+/+ mice treated with bleomycin than in the vehicle-treated control group (Table 4); however, there was no difference observed in Fra-1∆/∆ mice. Similarly, Fra-1∆/∆ mice treated with bleomycin also showed significantly increased expression of Hpse, Gclc, Runx3, Xcl1, and Cxcl11 when compared to the vehicle-treated group, but no differences were observed with their wild-type counterparts. These results further confirmed the expression patterns of the microarrays. According to the qRT-PCR results, there was a tendency for increased expression of Ccr8 and Ccl22 in the Fra-1∆/∆ mice treated with bleomycin as compared to the vehicle-treated mice, but the differences were not significant. Similarly, wild-type mice treated with bleomycin showed greater expression of Col4a1 and Tnfrsf10b than did the vehicle controls. While these qRT-PCR results agree in general direction with the trends measured by microarray, the results did not reach statistical significance. Comparison of microarray and qRT-PCR results revealed discordance in 2 of the 17 genes selected. In the case of Marco, microarray analysis revealed no change in Marco expression in Fra-1+/+ mice but a significant increase in bleomycin-treated (3.08 ± 0.11) Fra-1+/+ mice when compared to vehicle-treated mice, as assayed by qRT-PCR. In the second case, the microarray results showed no change in the expression of Snai2 in Fra-1+/+ mice, but the real-time RT-PCR results indicated a significant decrease (0.37± 0.15) in expression in these mice. The discordance in these two genes may be explained by the lower sensitivity of the microarrays.
Table 3.
Validation of differentially expressed genes induced by bleomycin in lung tissues from both Fra-1+/+ and Fra-1∆/∆ mice
|
Affymetrix ID |
Symbol |
Gene title |
Micro array |
qRT-PCR |
|---|---|---|---|---|
| |
|
|
KOT vs. WTT |
KOT vs. WTT |
| Fold change | Fold change | |||
| 10487588 |
Il1a |
Interleukin 1 alpha |
1.87 |
3.40 ± 1.68* |
| 10404389 |
Irf4 |
Interferon regulatory factor 4 (TF) |
2.06 |
3.29 ± 1.08* |
| 10597420 |
Ccr4 |
C-C chemokine receptor type 4 |
1.78 |
1.68 ± 0.18* |
| 10545569 |
Reg3g |
Regenerating islet-derived 3 gamma |
4.61 |
6.89 ± 0.86* |
| 10493831 | S100a8 | S100 calcium binding protein A8 | −2.63 | -3.57±1.70* |
Data are presented as mean ± SD (n=3-4). *P<0.05, WTT vs KOT after bleomycin treatment.
Table 4.
Validation of uniquely expressed genes induced by bleomycin in lung tissues from both Fra-1+/+ and Fra-1∆/∆ mice
|
Affymetrix ID |
Symbol |
Gene title |
Micro array |
qRT-PCR |
||
|---|---|---|---|---|---|---|
| |
|
|
WTT vs. WTC |
KOT vs. KOC |
WTT vs. WTC |
KOT vs. KOC |
| Fold change | Fold change | Fold change | Fold change | |||
| 10540085 |
Fbln2 |
Fibulin 2 |
2.28 |
ND |
2.35 ± 0.27* |
ND |
| 10531737 |
Hpse |
Heparanase |
ND |
1.98 |
ND |
1.57 ± 0.10* |
| 10587266 |
Gclc |
Glutamate-cysteine ligase, catalytic subunit |
ND |
1.70 |
ND |
1.58 ± 0.27* |
| 10509030 |
Runx3 |
Run related transcription factor 3 |
ND |
2.02 |
ND |
1.30 ± 0.14* |
| 10359697 |
Xcl1 |
Chemokine (C motif) ligand 1 |
ND |
2.15 |
ND |
2.57 ± 0.48* |
| 10531420 |
Cxcl11 |
Chemokine (C-X-C motif) ligand 11 |
ND |
2.53 |
ND |
4.72 ± 2.33* |
| 10590242 |
Ccr8 |
Chemokine (C-C motif) receptor 8 |
ND |
2.13 |
ND |
1.58 ± 0.00NS |
| 10574213 |
Ccl22 |
Macrophage-derived chemokine |
ND |
2.42 |
ND |
1.73 ± 0.54NS |
| 10576973 |
Col4a1 |
Collagen, type IV, alpha 1 |
1.96 |
ND |
1.83 ± 1.08NS |
ND |
| 10416230 |
Tnfrsf10b |
Tumor necrosis factor receptor superfamily, member 10b |
2.03 |
ND |
2.30 ± 1.09NS |
ND |
| 10357261 |
Marco |
Macrophage receptor with collagenous structure |
ND |
2.03 |
3.08 ± 0.11*† |
2.23 ± 0.21* |
| 10433776 | Snai2 | Snail homolog 2 (Drosophila) | ND | -1.92 | -2.23*† | -3.50 ± 1.95* |
Data are presented as mean ± SD (n=3-4). *P<0.05, PBS vs bleomycin of same genotype. NS means non significant. † Result did not agree with microarray result.
Analysis of selected microarray genes at different time points after bleomycin treatment
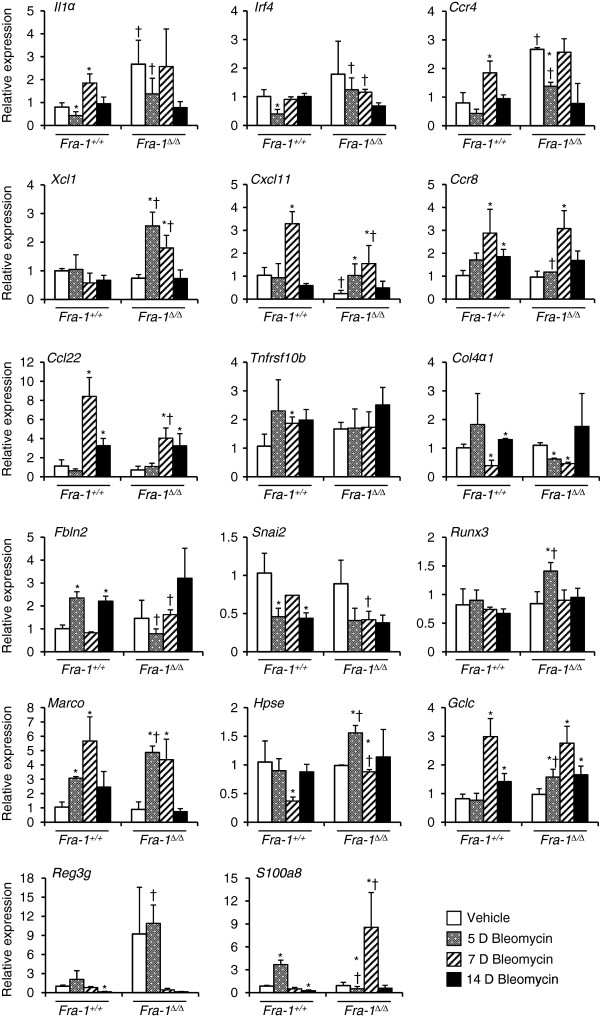
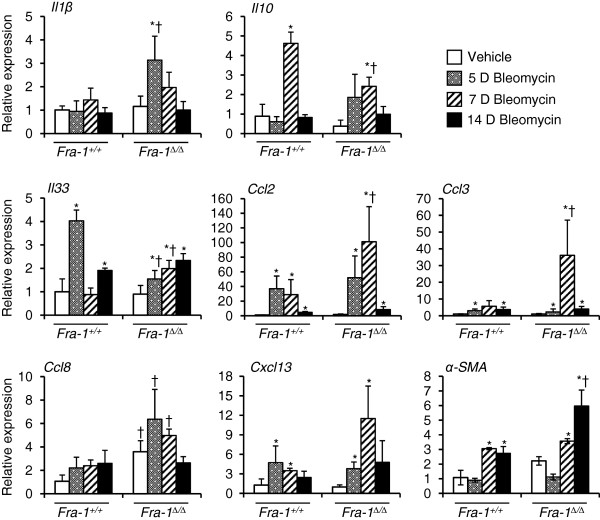
To identify the time course of gene induction by bleomycin, we analyzed samples at different time points for selected genes. All the genes that were used for microarray validation were also used for our analysis of temporal patterns of gene expression (Figure 1). The results revealed that most of the genes continued to show no significant differences between Fra-1+/+ and Fra-1∆/∆ at 7 and 14 days after bleomycin treatment. However, some of the genes were predominantly up-regulated (Xcl1 and S100a8) or down-regulated (Cxcl11 and Ccl22) in Fra-1∆/∆ mice only at 7 days following bleomycin treatment, when compared to the vehicle-treated control and Fra-1+/+ mice. We were further interested in the delayed response of some of the genes involved in lung fibrosis that did not exhibit significant differences in microarray results at 5 days (Figure 2). The results revealed a significant difference in the induction of C-C chemokines such as Ccl2, Ccl3, and Ccl8 between Fra-1+/+ and Fra-1∆/∆ mice: Fra-1∆/∆ mice showed significantly elevated expression at 7 days following bleomycin treatment when compared to wild-type mice. Interestingly, excessive production of Ccl2, Ccl3, and Ccl8 has been shown to aid the development of pulmonary fibrosis [63,64]. In addition to the up-regulation of Ccl2, Ccl3, and Ccl8, we observed a marked down-regulation of Il10 and Il33 in bleomycin-treated Fra-1∆/∆ mice. Previous studies have reported that in vivo IL-10 gene delivery before [65] and after bleomycin [66] administration suppresses the development of pulmonary fibrosis. Finally, fibroblast transdifferentiation has been shown to contribute to the pathology of pulmonary fibrosis. Therefore, we analyzed the expression of α-SMA, a marker for myofibroblasts. The results revealed that Fra-1∆/∆ mice treated with bleomycin had a significantly higher expression of α-SMA than did vehicle-treated control or Fra-1+/+ mice at 14 days. This result further supports the enhanced susceptibility of Fra-1-null mice to bleomycin-induced lung fibrosis.
Figure 1.
Validation of the differences in mRNA expression between Fra-1+/+and Fra-1Δ/Δ mice for selected microarray genes at different time points after bleomycin treatment. Lung mRNA abundance was determined by quantitative real-time RT-PCR. The graphs represent the fold change over vehicle treated Fra-1+/+ controls after normalization with the expression of GAPDH. Results are mean ± SD for 3-4 mice in each group. *p<0.05, PBS vs bleomycin; †p<0.05, Fra-1Δ/Δ vs Fra-1+/+ mice.
Figure 2.
Analysis of some gene expression between Fra-1+/+and Fra-1Δ/Δ mice at different time points after bleomycin treatment. Lung mRNA abundance was determined by quantitative real-time RT-PCR. The graphs represent the fold change over vehicle treated Fra-1+/+ controls after normalization with the expression of GAPDH. Results are mean ± SD for 3-4 mice in each group. *p<0.05, PBS vs bleomycin; †p<0.05, Fra-1Δ/Δ vs Fra-1+/+ mice.
Conclusion
The factors that contribute to the pathogenesis of pulmonary fibrosis include persistent inflammation, generation of pro-inflammatory, pro-fibrotic and angiogenic mediators, alveolar epithelial cell injury, fibroblast differentiation, and poor apoptotic activity of the myofibroblasts. These deregulated cellular processes eventually lead to excessive deposition of extracellular collagen and pathological fibrosis [48,67,68]. The present mRNA expression profiling analysis has revealed an important role for Fra-1 in regulating components of complex regulatory networks controlling the lung injury and fibrosis. We found that Fra-1∆/∆ mice displayed some of the factors that contribute to pulmonary fibrosis, such as increased expression of pro-inflammatory genes and decreased expression of genes involving in apoptotic process during bleomycin treatment. Thus, we propose that strategies enhancing Fra-1 functions may represent a promising approach to mitigate pulmonary fibrosis.
Methods
Mice
Conventional deletion of Fra-1 is embryonic lethal due to extra-embryonic tissue defects [69]. The mice bearing Fra-1 “floxed” allele [69] (hereafter referred as Fra-1FF mice) were obtained from Erwin F. Wagner (Spanish National Cancer Research Centre, Madrid, Spain). These mice are maintained on a mixed (C57BL6/129) background. Meox2 (Sox2)-Cre transgenic mice (C57BL6/129), in which Cre expression specifically restricted in embryo but not in extra-embryonic tissues, were obtained from the Jackson Labs. Meox2 Cre mice were crossed to Fra-1F/F mice [70], in order to obtain Fra-1F/F-Meox2-Cre mice as described earlier [12]. Fra-1F/F mice with and without Cre are hereafter referred to as Fra-1∆/∆ and Fra-1+/+ genotypes, respectively.
Bleomycin treatment
Bleomycin (0.075U) (APP Pharmaceuticals, LLC, Schaumburg, IL, USA) diluted in 30 μL of PBS was intratracheally administered to mice (n=3) (10-14 weeks old) as described previously [71]. Mice treated with PBS (n=3) served as controls. All experiments were conducted under a protocol approved by the institutional animal care use committee of the University of Illinois at Chicago. At the end of 5 days treatment, the left lungs were frozen immediately in RNAlater (Ambion) for subsequent microarray and qRT-PCR analysis.
RNA isolation and labeling
Total RNA was isolated from Fra-1+/+ and Fra-1∆/∆ mice administered with PBS and bleomycin using Qiagen RNeasy micro kit (Cat no. 74004). RNA concentration and purity was determined before gene expression profiling using the Affymetrix MoGene 1.0ST v1 Array (gene array) (Affymetrix, Inc., Santa Clara, California). The microarray labeling, hybridization and processing was performed at the University of Illinois Research Resource Center according to the manufacturer’s protocol. Microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus database (Accession number: GSE43695).
Microarray data analysis
The raw probe signal intensities were quantile normalized over all samples, summarized with the robust multi-array average (RMA) algorithm [72] and log2 transformed with a median polish, using the Affymetrix Power Tools. We considered a transcript cluster (gene-level) to be reliably expressed in a sample if the Affymetrix implemented DABG (detection above ground) p-value was less than 0.05. We used local-pooled-error (LPE) estimates and robust statistical tests [73] for evaluating significance of each gene’s differential expression in a comparison (e.g., wild-type vs. wild-type with bleomycin treatment). The LPE estimation was shown to be powerful and effective in case of a small number of replicate arrays [73]. False discovery rate (FDR) was controlled at 1% using the LPE library for the R Statistical Package [74].
Pathway analysis
We searched for any enriched pathways and biological processes among the differential genes in each comparison relative to the genes covered on the gene expression profiling platform using the NIH/DAVID (The Database for Annotation, Visualization and Integrated Discovery) [75]. Particularly, the following databases were interrogated: KEGG (Kyoto Encyclopedia of Genes and Genomes) [76] and GO (Gene Ontology) [77]. A minimum of 5 genes and the Benjamini corrected p-value less than 0.01 were used to call significantly enriched pathways or biological processes. There are distinct temporal phases during bleomycin-induced lung injury and fibrosis. To dissect the differential gene expression during bleomycin-induced initial lung injury, we have analyzed the gene expression profiles in Fra-1+/+ and Fra-1∆/∆ mice given PBS or bleomycin. We then compared the gene expression profiles in various categories: (1) differentially expressed genes in the lung tissue of Fra-1∆/∆ mice vs. Fra-1+/+ mice (Table 1), (2) differentially expressed genes induced by bleomycin in the lung tissue of Fra-1+/+ vs. Fra-1∆/∆ (Table 2), (3) unique gene expression induced by bleomycin in the lung tissue of Fra-1+/+ mice (see Additional file 1: Table S1), and (4) unique gene expression induced by bleomycin in the lung tissue of Fra-1∆/∆ mice (see Additional file 2: Table S2). The resulting gene lists were divided into several categories based on functional analysis in order to dissect Fra-1-dependent and -independent transcriptional programs.
Validation of microarray analysis
Total RNA (1 μg) was reverse transcribed using qScript cDNA super mix (Quanta Biosciences, Inc. Cat no. 95048-100). qRT-PCR was performed using fluorogenic SYBR Green and detection system (Applied Biosystems). PCR was performed using primers listed in (Additional file 3: Table S3). For Gclc, Marco and α-SMA, TaqMan gene expression assays were purchased from Applied Biosystems (Foster City, CA). The cycle threshold (CT) values for each gene were normalized to that of GAPDH, and the relative value for PBS treated Fra-1+/+ was set as one arbitrary unit (AU). Values are shown as mean ± SD, with n=3-4 for each experimental condition. Student’s T test was used and p ≤ 0.05 was considered significant.
Competing interests
The author’s declare that they have no competing interests.
Authors’ contributions
SR and SPR were involved in the conception, delineation of hypotheses, and design of the study, as well as the analysis and interpretation of data. SR performed the experiments. NMR and WZ participated in bioinformatics analysis. SR and SPR wrote the manuscript. All the authors read and approved the final manuscript.
Supplementary Material
Several unique genes expression induced by bleomycin in lung tissues of Fra-1+/+ mice.
Several unique genes expression induced by bleomycin in lung tissues of Fra-1∆/∆ mice.
Sequences of sybr green mouse primers used for qRT-PCR.
Contributor Information
Subbiah Rajasekaran, Email: rsubbiah@uic.edu.
Narsa M Reddy, Email: nmreddy@uic.edu.
Wei Zhang, Email: weizhan1@uic.edu.
Sekhar P Reddy, Email: sreddy03@uic.edu.
Acknowledgments
We acknowledge the Core Genomics Facility at UIC for performing microarray analysis on service charge basis. We thank Dr. Erwin F. Wagner (Spanish National Cancer Research Centre, Madrid, Spain) for providing the mice bearing a “Floxed” Fra-1 allele. This study was supported by grants from the NIH (RO1 ES11863, RO1 HL66109, and R21 ES18998) and Flight Attendents Medical Research Institute (FAMRI) to SPR. We thank Debbie McClellan for her help in editing the manuscript.
References
- Wynn TA. Integrating mechanisms of pulmonary fibrosis. J Exp Med. 2011;208(7):1339–1350. doi: 10.1084/jem.20110551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aso Y, Yoneda K, Kikkawa Y. Morphologic and biochemical study of pulmonary changes induced by bleomycin in mice. Lab Invest. 1976;35(6):558–568. [PubMed] [Google Scholar]
- Schrier DJ, Kunkel RG, Phan SH. The role of strain variation in murine bleomycin-induced pulmonary fibrosis. Am Rev Respir Dis. 1983;127(1):63–66. doi: 10.1164/arrd.1983.127.1.63. [DOI] [PubMed] [Google Scholar]
- Crystal RG, Fulmer JD, Roberts WC, Moss ML, Line BR, Reynolds HY. Idiopathic pulmonary fibrosis. Clinical, histologic, radiographic, physiologic, scintigraphic, cytologic, and biochemical aspects. Ann Intern Med. 1976;85(6):769–788. doi: 10.7326/0003-4819-85-6-769. [DOI] [PubMed] [Google Scholar]
- Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer. 2003;3(11):859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- Rahman I, MacNee W. Role of transcription factors in inflammatory lung diseases. Thorax. 1998;53(7):601–612. doi: 10.1136/thx.53.7.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adiseshaiah P, Vaz M, Machireddy N, Kalvakolanu DV, Reddy SP. A Fra-1-dependent, matrix metalloproteinase driven EGFR activation promotes human lung epithelial cell motility and invasion. J Cell Physiol. 2008;216(2):405–412. doi: 10.1002/jcp.21410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincenti MP. The matrix metalloproteinase (MMP) and tissue inhibitor of metalloproteinase (TIMP) genes. Transcriptional and posttranscriptional regulation, signal transduction and cell-type-specific expression. Methods Mol Biol. 2001;151:121–148. doi: 10.1385/1-59259-046-2:121. [DOI] [PubMed] [Google Scholar]
- Westermarck J, Kahari VM. Regulation of matrix metalloproteinase expression in tumor invasion. FASEB J. 1999;13(8):781–792. [PubMed] [Google Scholar]
- Tower GB, Coon CI, Belguise K, Chalbos D, Brinckerhoff CE. Fra-1 targets the AP-1 site/2G single nucleotide polymorphism (ETS site) in the MMP-1 promoter. Eur J Biochem. 2003;270(20):4216–4225. doi: 10.1046/j.1432-1033.2003.03821.x. [DOI] [PubMed] [Google Scholar]
- Ho HH, Antoniv TT, Ji JD, Ivashkiv LB. Lipopolysaccharide-induced expression of matrix metalloproteinases in human monocytes is suppressed by IFN-gamma via superinduction of ATF-3 and suppression of AP-1. J Immunol. 2008;181(7):5089–5097. doi: 10.4049/jimmunol.181.7.5089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajasekaran S, Vaz M, Reddy SP. Fra-1/AP-1 transcription factor negatively regulates pulmonary fibrosis In vivo. PLoS One. 2012;7(7):e41611. doi: 10.1371/journal.pone.0041611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito F, Tasaka S, Inoue K, Miyamoto K, Nakano Y, Ogawa Y, Yamada W, Shiraishi Y, Hasegawa N, Fujishima S. et al. Role of interleukin-6 in bleomycin-induced lung inflammatory changes in mice. Am J Respir Cell Mol Biol. 2008;38(5):566–571. doi: 10.1165/rcmb.2007-0299OC. [DOI] [PubMed] [Google Scholar]
- Murphy JE, Robert C, Kupper TS. Interleukin-1 and cutaneous inflammation: a crucial link between innate and acquired immunity. J Invest Dermatol. 2000;114(3):602–608. doi: 10.1046/j.1523-1747.2000.00917.x. [DOI] [PubMed] [Google Scholar]
- Ghosh S, May MJ, Kopp EB. NF-kappa B and Rel proteins: evolutionarily conserved mediators of immune responses. Annu Rev Immunol. 1998;16:225–260. doi: 10.1146/annurev.immunol.16.1.225. [DOI] [PubMed] [Google Scholar]
- Wilson MS, Madala SK, Ramalingam TR, Gochuico BR, Rosas IO, Cheever AW, Wynn TA. Bleomycin and IL-1beta-mediated pulmonary fibrosis is IL-17A dependent. J Exp Med. 2010;207(3):535–552. doi: 10.1084/jem.20092121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasse P, Mary C, Guenon I, Noulin N, Charron S, Schnyder-Candrian S, Schnyder B, Akira S, Quesniaux VF, Lagente V. et al. IL-1R1/MyD88 signaling and the inflammasome are essential in pulmonary inflammation and fibrosis in mice. J Clin Invest. 2007;117(12):3786–3799. doi: 10.1172/JCI32285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahan CJ, Slack JL, Mosley B, Cosman D, Lupton SD, Brunton LL, Grubin CE, Wignall JM, Jenkins NA, Brannan CI. et al. A novel IL-1 receptor, cloned from B cells by mammalian expression, is expressed in many cell types. EMBO J. 1991;10(10):2821–2832. doi: 10.1002/j.1460-2075.1991.tb07831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F, Re F, Muzio M, Bertini R, Polentarutti N, Sironi M, Giri JG, Dower SK, Sims JE, Mantovani A. Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science. 1993;261(5120):472–475. doi: 10.1126/science.8332913. [DOI] [PubMed] [Google Scholar]
- Re F, Muzio M, De Rossi M, Polentarutti N, Giri JG, Mantovani A, Colotta F. The type II “receptor” as a decoy target for interleukin 1 in polymorphonuclear leukocytes: characterization of induction by dexamethasone and ligand binding properties of the released decoy receptor. J Exp Med. 1994;179(2):739–743. doi: 10.1084/jem.179.2.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colotta F, Re F, Muzio M, Polentarutti N, Minty A, Caput D, Ferrara P, Mantovani A. Interleukin-13 induces expression and release of interleukin-1 decoy receptor in human polymorphonuclear cells. J Biol Chem. 1994;269(17):12403–12406. [PubMed] [Google Scholar]
- Keane MP, Belperio JA, Moore TA, Moore BB, Arenberg DA, Smith RE, Burdick MD, Kunkel SL, Strieter RM. Neutralization of the CXC chemokine, macrophage inflammatory protein-2, attenuates bleomycin-induced pulmonary fibrosis. J Immunol. 1999;162(9):5511–5518. [PubMed] [Google Scholar]
- Strieter RM, Polverini PJ, Kunkel SL, Arenberg DA, Burdick MD, Kasper J, Dzuiba J, Van Damme J, Walz A, Marriott D. et al. The functional role of the ELR motif in CXC chemokine-mediated angiogenesis. J Biol Chem. 1995;270(45):27348–27357. doi: 10.1074/jbc.270.45.27348. [DOI] [PubMed] [Google Scholar]
- Tager AM, Kradin RL, LaCamera P, Bercury SD, Campanella GS, Leary CP, Polosukhin V, Zhao LH, Sakamoto H, Blackwell TS. et al. Inhibition of pulmonary fibrosis by the chemokine IP-10/CXCL10. Am J Respir Cell Mol Biol. 2004;31(4):395–404. doi: 10.1165/rcmb.2004-0175OC. [DOI] [PubMed] [Google Scholar]
- Keane MP, Belperio JA, Arenberg DA, Burdick MD, Xu ZJ, Xue YY, Strieter RM. IFN-gamma-inducible protein-10 attenuates bleomycin-induced pulmonary fibrosis via inhibition of angiogenesis. J Immunol. 1999;163(10):5686–5692. [PubMed] [Google Scholar]
- Belperio JA, Dy M, Murray L, Burdick MD, Xue YY, Strieter RM, Keane MP. The role of the Th2 CC chemokine ligand CCL17 in pulmonary fibrosis. J Immunol. 2004;173(7):4692–4698. doi: 10.4049/jimmunol.173.7.4692. [DOI] [PubMed] [Google Scholar]
- Yogo Y, Fujishima S, Inoue T, Saito F, Shiomi T, Yamaguchi K, Ishizaka A. Macrophage derived chemokine (CCL22), thymus and activation-regulated chemokine (CCL17), and CCR4 in idiopathic pulmonary fibrosis. Respir Res. 2009;10:80. doi: 10.1186/1465-9921-10-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo G, O’Connor EC, Kunkel SL, Hogaboam CM. A novel mechanism for CCR4 in the regulation of macrophage activation in bleomycin-induced pulmonary fibrosis. Am J Pathol. 2008;172(5):1209–1221. doi: 10.2353/ajpath.2008.070832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts V, van der Zee E, Creemers L, Beertsen W. Phagocytosis and intracellular digestion of collagen, its role in turnover and remodelling. Histochem J. 1996;28(4):229–245. doi: 10.1007/BF02409011. [DOI] [PubMed] [Google Scholar]
- Bienkowski RS, Gotkin MG. Control of collagen deposition in mammalian lung. Proc Soc Exp Biol Med. 1995;209(2):118–140. doi: 10.3181/00379727-209-43886a. [DOI] [PubMed] [Google Scholar]
- Wolters PJ, Chapman HA. Importance of lysosomal cysteine proteases in lung disease. Respir Res. 2000;1(3):170–177. doi: 10.1186/rr29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeyabu K, Betsuyaku T, Nishimura M, Yoshioka A, Tanino M, Miyamoto K, Kawakami Y. Cysteine proteinases and cystatin C in bronchoalveolar lavage fluid from subjects with subclinical emphysema. Eur Respir J. 1998;12(5):1033–1039. doi: 10.1183/09031936.98.12051033. [DOI] [PubMed] [Google Scholar]
- Hernandez-Rodriguez NA, Cambrey AD, Harrison NK, Chambers RC, Gray AJ, Southcott AM, duBois RM, Black CM, Scully MF, McAnulty RJ. et al. Role of thrombin in pulmonary fibrosis. Lancet. 1995;346(8982):1071–1073. doi: 10.1016/S0140-6736(95)91744-6. [DOI] [PubMed] [Google Scholar]
- Hoffmann H, Siebeck M, Spannagl M, Weis M, Geiger R, Jochum M, Fritz H. Effect of recombinant hirudin, a specific inhibitor of thrombin, on endotoxin-induced intravascular coagulation and acute lung injury in pigs. Am Rev Respir Dis. 1990;142(4):782–788. doi: 10.1164/ajrccm/142.4.782. [DOI] [PubMed] [Google Scholar]
- Koslowski R, Knoch KP, Wenzel KW. Proteinases and proteinase inhibitors during the development of pulmonary fibrosis in rat. Clin Chim Acta. 1998;271(1):45–56. doi: 10.1016/S0009-8981(97)00228-3. [DOI] [PubMed] [Google Scholar]
- Bozinovski S, Hutchinson A, Thompson M, Macgregor L, Black J, Giannakis E, Karlsson AS, Silvestrini R, Smallwood D, Vlahos R. et al. Serum amyloid a is a biomarker of acute exacerbations of chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2008;177(3):269–278. doi: 10.1164/rccm.200705-678OC. [DOI] [PubMed] [Google Scholar]
- Nel AE, Strachan AF, Welke HE, de Beer FC. Acute phase response in bronchiectasis and bronchus carcinoma. Respiration. 1984;45(4):406–410. doi: 10.1159/000194647. [DOI] [PubMed] [Google Scholar]
- Sin DD, Man SF. Why are patients with chronic obstructive pulmonary disease at increased risk of cardiovascular diseases? The potential role of systemic inflammation in chronic obstructive pulmonary disease. Circulation. 2003;107(11):1514–1519. doi: 10.1161/01.CIR.0000056767.69054.B3. [DOI] [PubMed] [Google Scholar]
- Kormelink TG, Tekstra J, Thurlings RM, Boumans MH, Vos K, Tak PP, Bijlsma JW, Lafeber FP, Redegeld FA, van Roon JA. Decrease in immunoglobulin free light chains in patients with rheumatoid arthritis upon rituximab (anti-CD20) treatment correlates with decrease in disease activity. Ann Rheum Dis. 2010;69(12):2137–2144. doi: 10.1136/ard.2009.126441. [DOI] [PubMed] [Google Scholar]
- Rijnierse A, Redegeld FA, Blokhuis BR, Van der Heijden MW, Te Velde AA, Pronk I, Hommes DW, Nijkamp FP, Koster AS, Kraneveld AD. Ig-free light chains play a crucial role in murine mast cell-dependent colitis and are associated with human inflammatory bowel diseases. J Immunol. 2010;185(1):653–659. doi: 10.4049/jimmunol.0901129. [DOI] [PubMed] [Google Scholar]
- Kraneveld AD, Kool M, van Houwelingen AH, Roholl P, Solomon A, Postma DS, Nijkamp FP, Redegeld FA. Elicitation of allergic asthma by immunoglobulin free light chains. Proc Natl Acad Sci USA. 2005;102(5):1578–1583. doi: 10.1073/pnas.0406808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornick DB, Fick RB Jr. The immunoglobulin G subclass composition of immune complexes in cystic fibrosis. Implications for the pathogenesis of the Pseudomonas lung lesion. J Clin Invest. 1990;86(4):1285–1292. doi: 10.1172/JCI114836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot Kormelink T, Pardo A, Knipping K, Buendia-Roldan I, Garcia-de-Alba C, Blokhuis BR, Selman M, Redegeld FA. Immunoglobulin free light chains are increased in hypersensitivity pneumonitis and idiopathic pulmonary fibrosis. PLoS One. 2011;6(9):e25392. doi: 10.1371/journal.pone.0025392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckgraefe BK, Crimmins DL, Landt V, Houchen C, Anant S, Porche-Sorbet R, Ladenson JH. Expression of the regenerating gene family in inflammatory bowel disease mucosa: Reg Ialpha upregulation, processing, and antiapoptotic activity. J Investig Med. 2002;50(6):421–434. doi: 10.2310/6650.2002.32518. [DOI] [PubMed] [Google Scholar]
- Shahzeidi S, Mulier B, de Crombrugghe B, Jeffery PK, McAnulty RJ, Laurent GJ. Enhanced type III collagen gene expression during bleomycin induced lung fibrosis. Thorax. 1993;48(6):622–628. doi: 10.1136/thx.48.6.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghu G, Striker LJ, Hudson LD, Striker GE. Extracellular matrix in normal and fibrotic human lungs. Am Rev Respir Dis. 1985;131(2):281–289. doi: 10.1164/arrd.1985.131.2.281. [DOI] [PubMed] [Google Scholar]
- Zhang K, Gharaee-Kermani M, McGarry B, Phan SH. In situ hybridization analysis of rat lung alpha 1(I) and alpha 2(I) collagen gene expression in pulmonary fibrosis induced by endotracheal bleomycin injection. Lab Invest. 1994;70(2):192–202. [PubMed] [Google Scholar]
- Wynn TA. Common and unique mechanisms regulate fibrosis in various fibroproliferative diseases. J Clin Invest. 2007;117(3):524–529. doi: 10.1172/JCI31487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leone M, Garcin F, Chaabane W, Boutiere-Albanese B, Albanese J, Dignat-Georges F, Martin C. Activation of adhesion molecules in patients with septic shock. Ann Fr Anesth Reanim. 2003;22(8):721–729. doi: 10.1016/S0750-7658(03)00327-7. [DOI] [PubMed] [Google Scholar]
- Chang YC, Uchiyama S, Varki A, Nizet V. Leukocyte inflammatory responses provoked by pneumococcal sialidase. MBio. 2012;3(1):e00220–11. doi: 10.1128/mBio.00220-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y, Elkin M, Aingorn H, Atzmon R, Ishai-Michaeli R, Bitan M, Pappo O, Peretz T, Michal I. et al. Mammalian heparanase: gene cloning, expression and function in tumor progression and metastasis. Nat Med. 1999;5(7):793–802. doi: 10.1038/10518. [DOI] [PubMed] [Google Scholar]
- Vlodavsky I, Friedmann Y. Molecular properties and involvement of heparanase in cancer metastasis and angiogenesis. J Clin Invest. 2001;108(3):341–347. doi: 10.1172/JCI13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita J, Takeuchi T, Dobashi N, Ohtsuki Y, Tokuda M, Takahara J. Detection of anti-ADAM 10 antibody in serum of a patient with pulmonary fibrosis associated with dermatomyositis. Ann Rheum Dis. 1999;58(12):770–772. doi: 10.1136/ard.58.12.770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongepier H, Boezen HM, Dijkstra A, Howard TD, Vonk JM, Koppelman GH, Zheng SL, Meyers DA, Bleecker ER, Postma DS. Polymorphisms of the ADAM33 gene are associated with accelerated lung function decline in asthma. Clin Exp Allergy. 2004;34(5):757–760. doi: 10.1111/j.1365-2222.2004.1938.x. [DOI] [PubMed] [Google Scholar]
- Keating DT, Sadlier DM, Patricelli A, Smith SM, Walls D, Egan JJ, Doran PP. Microarray identifies ADAM family members as key responders to TGF-beta1 in alveolar epithelial cells. Respir Res. 2006;7:114. doi: 10.1186/1465-9921-7-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matute-Bello G, Martin TR. Science review: apoptosis in acute lung injury. Crit Care. 2003;7(5):355–358. doi: 10.1186/cc1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwano K, Hagimoto N, Nakanishi Y. The role of apoptosis in pulmonary fibrosis. Histol Histopathol. 2004;19(3):867–881. doi: 10.14670/HH-19.867. [DOI] [PubMed] [Google Scholar]
- Wisdom R, Johnson RS, Moore C. c-Jun regulates cell cycle progression and apoptosis by distinct mechanisms. EMBO J. 1999;18(1):188–197. doi: 10.1093/emboj/18.1.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, Baumann B, Cotten M, Angel P, Wagner EF. Fos is an essential component of the mammalian UV response. EMBO J. 1995;14(21):5338–5349. doi: 10.1002/j.1460-2075.1995.tb00218.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirsat NV, Shaikh SA. Overexpression of the immediate early gene fra-1 inhibits proliferation, induces apoptosis, and reduces tumourigenicity of c6 glioma cells. Exp Cell Res. 2003;291(1):91–100. doi: 10.1016/S0014-4827(03)00346-X. [DOI] [PubMed] [Google Scholar]
- Vaz M, Machireddy N, Irving A, Potteti HR, Chevalier K, Kalvakolanu D, Reddy SP. Oxidant-induced cell death and Nrf2-dependent antioxidative response are controlled by Fra-1/AP-1. Mol Cell Biol. 2012;32(9):1694–1709. doi: 10.1128/MCB.06390-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SS, Choi BM, Chen XY, Zhu RZ, Kim Y, So H, Park R, Sung M, Kim BR. Kaempferol suppresses cisplatin-induced apoptosis via inductions of heme oxygenase-1 and glutamate-cysteine ligase catalytic subunit in HEI-OC1 cell. Pharm Res. 2010;27(2):235–245. doi: 10.1007/s11095-009-0003-3. [DOI] [PubMed] [Google Scholar]
- Takada Y, Gresh L, Bozec A, Ikeda E, Kamiya K, Watanabe M, Kobayashi K, Asano K, Toyama Y, Wagner EF. et al. Interstitial lung disease induced by gefitinib and toll-like receptor ligands is mediated by Fra-1. Oncogene. 2011;30(36):3821–3832. doi: 10.1038/onc.2011.101. [DOI] [PubMed] [Google Scholar]
- Ishida Y, Kimura A, Kondo T, Hayashi T, Ueno M, Takakura N, Matsushima K, Mukaida N. Essential roles of the CC chemokine ligand 3-CC chemokine receptor 5 axis in bleomycin-induced pulmonary fibrosis through regulation of macrophage and fibrocyte infiltration. Am J Pathol. 2007;170(3):843–854. doi: 10.2353/ajpath.2007.051213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai T, Abe K, Matsuoka H, Yoshida M, Mori M, Goya S, Kida H, Nishino K, Osaki T, Tachibana I. et al. Introduction of the interleukin-10 gene into mice inhibited bleomycin-induced lung injury in vivo. Am J Physiol Lung Cell Mol Physiol. 2000;278(5):L914–922. doi: 10.1152/ajplung.2000.278.5.L914. [DOI] [PubMed] [Google Scholar]
- Nakagome K, Dohi M, Okunishi K, Tanaka R, Miyazaki J, Yamamoto K. In vivo IL-10 gene delivery attenuates bleomycin induced pulmonary fibrosis by inhibiting the production and activation of TGF-beta in the lung. Thorax. 2006;61(10):886–894. doi: 10.1136/thx.2005.056317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn TA. Fibrotic disease and the T(H)1/T(H)2 paradigm. Nat Rev Immunol. 2004;4(8):583–594. doi: 10.1038/nri1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasek JJ, Gabbiani G, Hinz B, Chaponnier C, Brown RA. Myofibroblasts and mechano-regulation of connective tissue remodelling. Nat Rev Mol Cell Biol. 2002;3(5):349–363. doi: 10.1038/nrm809. [DOI] [PubMed] [Google Scholar]
- Eferl R, Hoebertz A, Schilling AF, Rath M, Karreth F, Kenner L, Amling M, Wagner EF. The Fos-related antigen Fra-1 is an activator of bone matrix formation. EMBO J. 2004;23(14):2789–2799. doi: 10.1038/sj.emboj.7600282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Soriano P. Epiblast-restricted Cre expression in MORE mice: a tool to distinguish embryonic vs. extra-embryonic gene function. Genesis. 2000;26(2):113–115. doi: 10.1002/(SICI)1526-968X(200002)26:2<113::AID-GENE3>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Eitzman DT, McCoy RD, Zheng X, Fay WP, Shen T, Ginsburg D, Simon RH. Bleomycin-induced pulmonary fibrosis in transgenic mice that either lack or overexpress the murine plasminogen activator inhibitor-1 gene. J Clin Invest. 1996;97(1):232–237. doi: 10.1172/JCI118396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Jain N, Thatte J, Braciale T, Ley K, O’Connell M, Lee JK. Local-pooled-error test for identifying differentially expressed genes with a small number of replicated microarrays. Bioinformatics. 2003;19(15):1945–1951. doi: 10.1093/bioinformatics/btg264. [DOI] [PubMed] [Google Scholar]
- Team RDC, editor. A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2005. [Google Scholar]
- da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Kawashima S, Okuno Y, Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32(Database issue):D277–280. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Several unique genes expression induced by bleomycin in lung tissues of Fra-1+/+ mice.
Several unique genes expression induced by bleomycin in lung tissues of Fra-1∆/∆ mice.
Sequences of sybr green mouse primers used for qRT-PCR.