Abstract
The nuclear envelope (NE) is a specific extension of the endoplasmic reticulum (ER) that wraps around the nucleus and enables the spatial separation of gene transcription and protein translation, one of the signature features of eukaryotes. Rather than being completely closed, the double lipid bilayer of the NE is perforated at sites where the inner and outer nuclear membranes fuse, resulting in circular openings lined with sharply bent membranes. These openings are filled with nuclear pore complexes (NPCs), enormous protein assemblies that facilitate nuclear transport. The scaffold components of the NPC surprisingly share interesting similarities with elements of coat protein complexes, which have general implications for function and evolution of these membrane-coating complexes. Here I discuss, from a structural perspective, what these findings might teach us.
Recent studies have begun to dissect the molecular architecture of nuclear pore complexes. Interestingly, their scaffold components share similarities with elements of coat protein complexes (e.g., COPII).
As discussed in other articles, the endoplasmic reticulum (ER) is a structurally and functionally diverse organelle. The nuclear envelope (NE) is perhaps the most extreme substructure of the ER. It consists of two flat nuclear membrane sheets that are kept apart in a well-defined distance of ∼5 nm and that encapsulate the genetic material of the eukaryotic cell (Stewart et al. 2007; Mekhail and Moazed 2010). The area between the outer nuclear membrane (ONM) and the inner nuclear membrane (INM) is called the perinuclear space (PNS), and it is continuous with the remainder of the ER lumen. The INM and ONM are fused at approximately seven sites per square micrometer (Dultz and Ellenberg 2010), resulting in circular openings with a diameter of ∼100 nm. These openings are the main portals for molecular exchange between the nucleus and the cytoplasm. Traffic is regulated by nuclear pore complexes (NPCs) that fill the portals (Brohawn et al. 2009; Hetzer and Wente 2009; Wente and Rout 2010). In many ways, NPCs are unique structures:
Unlike any other membrane transporter, they have an enormous cargo spectrum ranging from small metabolites and ions to nucleic acids and proteins up to the megadalton sizes of ribosomal subunits and some viral capsids.
Transport is uniquely regulated via a gradient of the small G-protein Ran and its two nucleotide-bound states (Görlich and Kutay 1999).
An aqueous gel made of fiber-like polypeptides that fills the central transport gate is thought to provide a sieve to limit uncontrolled exchange of large molecules (Hülsmann et al. 2012).
The enormous size and complexity of the NPC have long prevented a detailed analysis. Only in recent years have studies begun to dissect the molecular architecture of the NPC, and we are only now beginning to understand the myriad functions of this assembly. Despite the uniqueness of the NPC, its scaffold parts share intriguing structural similarities with vesicle-coating proteins. Based on evolutionary considerations and structure prediction, such similarity was first postulated in 2004 (Devos et al. 2004; Mans et al. 2004). X-ray crystallographic analysis of vesicle coat proteins and the NPC scaffolding units has firmly established specific relationships and has also revealed where there are differences (Brohawn et al. 2009).
THE OVERALL STRUCTURE OF THE NUCLEAR PORE COMPLEX
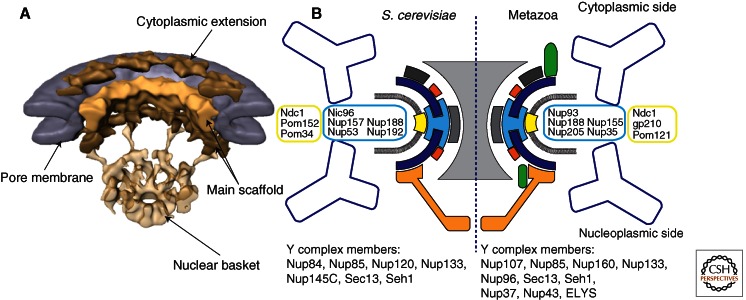
The NPC is a massive protein assembly, composed of about 30 different proteins, called nucleoporins, or nups (Alber et al. 2007; Brohawn et al. 2009; Grossman et al. 2012). In electron micrographs, NPCs appear as hollow cylinders with distinct cytoplasmic and nucleoplasmic faces, and have an overall eightfold rotational symmetry (Goldberg and Allen 1996). The best-resolved NPC images are obtained by cryo-electron tomography (cryo-ET), reaching a resolution of up to 6 nm (Fig. 1) (Beck et al. 2004, 2007; Maimon et al. 2012). In cryo-ET, various building blocks of the NPC can be distinguished, yet individual proteins are not resolved. On the cytoplasmic side of the pore, fiber-like extensions emanate from the central NPC mass. On the nucleoplasmic side, eight filaments protrude and cojoin in a narrow ring, a component named a “fishtrap” or “basket.” The central mass is organized in three electron-dense stacked rings, interspersed with less-dense zones. These stacked rings comprise the basic scaffold nucleoporins. The functionally most significant phenylalanine-glycine (FG)–rich fibrous extensions, which line the central transport gate, make up 20% of the NPC mass and act as the size-exclusion barrier. The FG fibers are invisible in the cryo-ET reconstructions, presumably because of their intrinsic disorder. Overall, NPCs are ∼100 nm in diameter, and in cross section show an hourglass shape. The narrowest constriction, ∼50 nm in the center, roughly defines the upper transport limit of the NPC (Pante and Kann 2002). It is thought that about 500 individual nucleoporin molecules make up one NPC, which amounts to a size of ∼40–60 MDa (Alber et al. 2007). Although the set of about 30 nucleoporins is quite similar between highly divergent eukaryotes (Rout et al. 2000; Cronshaw et al. 2002; Degrasse et al. 2009; Neumann et al. 2010), the height of the NPC from these different species is not very well conserved (Elad et al. 2009). This difference could be the result of a few nucleoporins that are species specific and thus influence the assembly differently. Or alternatively, it may reflect a difference in the copy number of scaffold nups.
Figure 1.
The nuclear pore complex. (A) Cryo-electron tomographic reconstruction of the NPC from Dictyostelium discoideum at ∼6-nm resolution. Cutaway view across the nuclear envelope (NE). The NPC appears organized into three stacked rings that cover the circular openings in the NE. NPCs show eightfold rotational ring symmetry. On the nucleoplasmic side, eight filaments protrude and cojoin in a ring. This resolution reveals the overall dimensions of the NPC and its most prominent features (labeled). (B) Nucleoporins are organized into subcomplexes that are well conserved across divergent eukaryotes. The main NPC scaffold is likely made up of two heteromeric subassemblies, the Y-complex (dark blue) and the Nic96 or Nup93 complex (light blue). The components of these subassemblies have architectural domains, mainly composed of α-helical stack domains and β-propellers. These scaffold complexes are connected to the pore membrane via membrane proteins (yellow). The other subassemblies of the NPC decorate the main scaffold and perform the many functions of the NPC.
NPCs are dynamic assemblies. Although some nucleoporins are stably attached, at least one-third of them dissociate and associate rather rapidly (Rabut et al. 2004). The scaffold structure of the NPC is most likely very stable, because there is virtually no turnover of its components (D’Angelo et al. 2009; Savas et al. 2012). The scaffold may resemble a coat made of approximately 15 nucleoporins that covers the pore membrane and that forms attachment sites for the more mobile, peripheral nucleoporins.
THE SCAFFOLD COMPONENTS OF THE NPC
The principal scaffold of the NPC can be divided into several subcomplexes (Fig. 1B). First, the NPC is anchored to the sharply curved pore membrane via a set of membrane proteins—Ndc1, Pom152, and Pom34—in yeast. (The nomenclature for nucleoporins is confusing because many homologous proteins have different names in yeast versus metazoans; therefore, for simplicity, I exclusively use the yeast nomenclature unless explicitly stated otherwise.) These proteins interact with soluble nucleoporins that line the pore membrane. Two of the soluble scaffold protein subcomplexes likely make up the central, symmetrical core of the NPC. These two subcomplexes are the heteromeric Y-complex and the heteromeric Nic96 complex.
The Y-complex, so named because of its Y-shaped appearance in electron micrographs (Lutzmann et al. 2002; Kampmann and Blobel 2009), is composed of a universally conserved core set of seven proteins: Nup133, Nup84, Nup145C, Nup85, Nup120, Seh1, and Sec13. In some species, up to three additional proteins (Nup37, Nup43, and ELY5/ELYS) extend the Y-complex. Crystal structures of several 40- to 135-kDa fragments of the 575-kDa conserved Y-complex core are available and account for a combined 90% of the structure (Berke et al. 2004; Brohawn et al. 2008; Brohawn and Schwartz 2009; Leksa et al. 2009; Nagy et al. 2009; Whittle and Schwartz 2009). All proteins fold into elongated, stacked α-helical domains or β-propeller domains, or binary combinations thereof. Both fold types are often used in modular protein assemblies; thus, it is fitting that they appear abundantly in the NPC.
The Nic96 complex is composed of at least five nucleoporins: Nic96, Nup188, Nup192, Nup53/59, and Nup157/170 (Nehrbass et al. 1996; Zabel et al. 1996; Franz et al. 2005; Hawryluk-Gara et al. 2008; Makio et al. 2009; Onischenko et al. 2009; Theerthagiri et al. 2010; Amlacher et al. 2011). In Saccharomyces cerevisiae, Nup53 and Nup157 have two mostly redundant orthologs as a result of whole-genome duplication. Similar to the Y-complex, structural studies have revealed that, except for Nup53 (Handa et al. 2006), all Nic96 complex components are also composed of β-propeller or α-helical stack domains (Jeudy and Schwartz 2007; Whittle and Schwartz 2009; Flemming et al. 2011). In contrast to the Y-complex, however, only isolated domains and no assembled complexes have been determined for the Nic96 substructure. Therefore, the three-dimensional (3D) arrangement of the Nic96 complex has not been determined.
Even though one can grossly classify the scaffold nucleoporins into just two folding types, analyzing the individual structures with attention to more detail reveals more significant functional features. Compared with other β-propeller proteins, Sec13 and Seh1 are unusual. Instead of being closed and seven-bladed as most β-propellers are (Chaudhuri et al. 2008; Stirnimann et al. 2010), they are open and six-bladed. The Sec13 and Seh1 interaction partners, Nup145C and Nup85, respectively, insert a seventh blade into the propeller to close it in trans, generating a substantial interface area that helps to stabilize the interaction (Brohawn et al. 2008; Brohawn and Schwartz 2009). In COPII, Sec13 interacts very similarly with Sec31 and Sec16, respectively (Fath et al. 2007; Whittle and Schwartz 2010). So far, this particular interaction type has only been detected in Sec13 and Seh1.
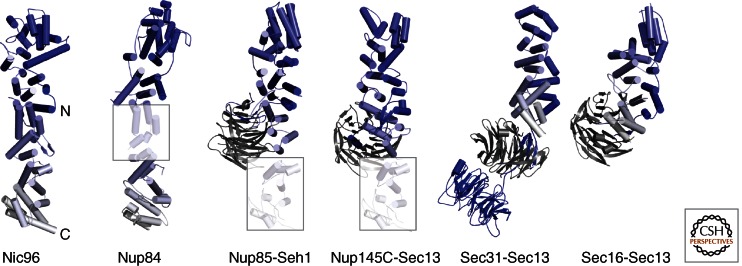
The α-helical stack domains can be classified into several classes based on how well they superpose onto one another. The first such class is the group of nucleoporins that share the ancestral coatomer element ACE1 (Brohawn et al. 2008). ACE1 is a 65-kDa tripartite module of 28 core α-helices. Although helical stacks are typically unidirectional, ACE1 has a characteristic fold-back topology (Fig. 2). The first five α-helices zigzag up on one side of the module, within helices α5–α11 ACE1 folds back, and α-helices α13–α28 zigzag down on the opposite side of α1–α5. The fold-back architecture might equip ACE1 proteins with higher rigidity than the pairwise helical stacking found in many canonical helical repeat proteins.
Figure 2.
The ancestral coatomer element ACE1. Four nucleoporins (Nic96, Nup84, Nup85, Nup145C) share a specific α-helical stack domain with Sec31 and Sec16 of the COPII vesicle system. The six proteins are aligned such that ACE1, shown in a blue-white color gradient from the amino to the carboxyl terminus, is oriented similarly. ACE1 is characterized by a fold-back architecture, which is highly unusual for α-helical stack domains. The 65-kDa ACE1 core has 28 α-helices. In Nup84, Nup85, and Nup145C, parts of the ACE1 domain are homology-modeled (shaded boxes) based on the Nic96 structure. Nup85, Nup145C, Sec31, and Sec16 bind the six-bladed, open β-propeller proteins Seh1 and Sec13 (gray), respectively, by inserting a seventh blade. The orientation relative to the ACE1 domain can vary. Sec31 contains an additional β-propeller at its amino terminus. ACE1 has not yet been found outside of the NPC and COPII, indicating a distinct evolutionary relationship.
Nup133 and Nup157 define a second class of closely related nups (Whittle and Schwartz 2009). They both combine a canonical amino-terminal, closed seven-bladed β-propeller with an extended 60- to 70-kDa α-helical stack domain. Both domains are merely tethered and can likely adopt multiple orientations with respect to each other. Rather than forming a continuous stack of paired helices, the helical stack is organized into several short, consecutive modules (Fig. 3).
Figure 3.
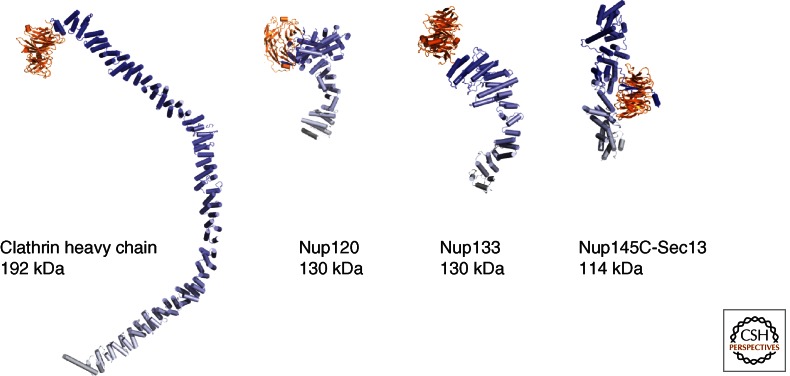
Various combinations of β-propellers with α-helical stack domains. Sequence comparison and structure prediction methods have revealed that many scaffolding proteins in the NPC, in vesicle coats and elsewhere, are built up from β-propeller domains combined, in cis or in trans, with α-helical stacks. Four examples of architectural proteins are given and drawn to scale. Although β-propellers (orange) do not structurally diverge drastically and can generally be superimposed well, α-helical stack domains (in blue–white color gradient) can adopt many shapes.
Nup120 is a unique architectural nucleoporin. It starts with an amino-terminal β-propeller that is interrupted by an α-helical insert between blades 6 and 7 (Leksa et al. 2009). Unlike the Nup133/Nup157 class, the β-propeller is integrated with an irregular α-helical segment to form a compact 80-kDa domain. So far, no other protein outside the NPC has been found to have a similar architecture. Only the carboxy-terminal 40–60 kDa of Nup120 folds into a regular HEAT-repeat structure (Fig. 3) (Bilokapic and Schwartz 2012).
The largest architectural Nups are Nup188 and Nup192. Although crystal structures are not available yet, electron microscopic studies combined with structure prediction suggest that these Nups might form extended, spiraling HEAT-repeat stacks, similar to the nuclear transport receptors (Amlacher et al. 2011; Flemming et al. 2011). Thus, Nup188 and Nup192 are likely to form yet another subclass of α-helical, stack domains.
COMMONALITIES BETWEEN THE NPC SCAFFOLD AND VESICLE COAT PROTEIN COMPLEXES
Although obtaining crystal structures of NPC subcomplexes is necessary for understanding the complete NPC scaffold structure, they alone are insufficient for building an accurate 3D model. Currently two main problems exist: (1) the interactions between subcomplexes are weak and experimentally poorly defined (Alber et al. 2007; Fernandez-Martinez et al. 2012); and (2) fitting subcomplex crystal structures into the cryo-ET structures is not yet possible because of their insufficient resolution (Beck et al. 2007; Maimon et al. 2012). The cryo-ET field is likely to improve resolution such that docking of large subcomplexes should become possible. In addition, single-particle cryo-EM has not been used for NPC components yet, and it should also help bridge the resolution gap between crystallographic and cryo-ET studies.
A major breakthrough was the discovery that the NPC shares surprising similarities with vesicle coat complexes. In the early 2000s, various groups speculated that such a connection might exist, mainly spurred by two observations. First, it was established that the COPII coat component Sec13 (Pryer et al. 1993) is also a bona fide nucleoporin (Siniossoglou et al. 1996). Second, advanced protein structure prediction methods became available and were able to detect phylogenetic relationships between proteins with very low sequence similarity. On that basis, a common origin of various coat complexes was postulated (Devos et al. 2004, 2006; Mans et al. 2004). However, specific relationships between individual proteins were not detected until 2008, when the crystal structure of the Seh1–Nup85 complex was published (Brohawn et al. 2008). Now, it became apparent that four nucleoporins—Nic96, Nup84, Nup85, and Nup145C—all share their 65-kDa ACE1 domain, introduced above, with the COPII component Sec31 (Fig. 2). This is remarkable, given that the sequence identity between these proteins can be as low as 8%–10%. In addition to the shared ACE1 element, the interaction of Nup85 with the Sec13-homolog Seh1, an insertion of a seventh blade into the open, six-bladed β-propeller, is very similar in Nup145C–Sec13 (Brohawn and Schwartz 2009; Nagy et al. 2009) and also in Sec31–Sec13 (Fath et al. 2007). This relationship indicated that these elements of COPII and the NPC likely share a common ancestor. But how does that help to decipher the structure of the NPC? Clues regarding how the NPC might assemble from its subcomplexes come from more advanced knowledge regarding the structure of the COPII coat.
THE CAGE STRUCTURES OF VESICLE COATS
The assembly of COPII and clathrin cages is quite well understood because these systems have been studied more extensively than the NPC and both are much simpler in construction, because they are built from fewer components. Most importantly, both cages can be reassembled in vitro from purified components, such that cryo-EM studies in combination with X-ray crystallography revealed pseudoatomic resolution for both systems (Fig. 4) (Fotin et al. 2004; Stagg et al. 2006; Fath et al. 2007). The COPII system is described in detail in the article by Lord et al. (2013). Here I focus on structural aspects and the comparison with the NPC.
Figure 4.
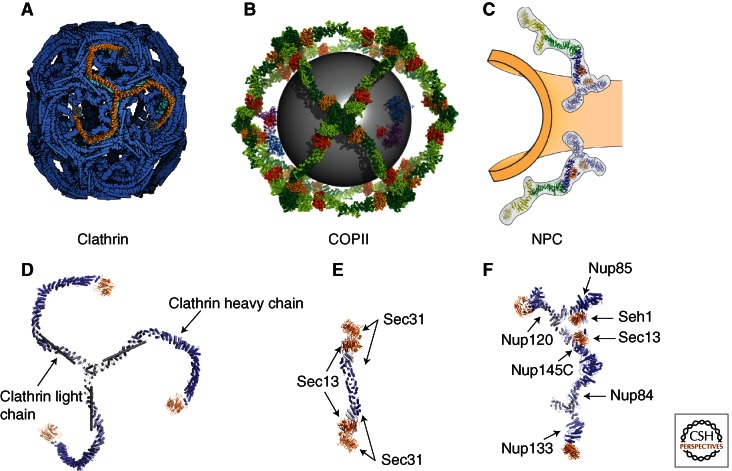
Lattice arrangements of clathrin and COPII coats in comparison with the NPC. (A,B) The simplest cages that can be formed in vitro for clathrin (A) and COPII vesicles (B). In both cases, the cages are built from distinct repeating units. In clathrin, three heavy (orange, blue–white) and three light chains (gray) form a triskelion. In COPII, the heterotetrameric Sec31–13 assembly forms an edge element. The repeating units can be assembled to form larger vesicles as well. (C) The NPC scaffold has not yet been reconstituted in vitro; therefore, the arrangement of its repeating units is still speculative. (D) In clathrin, three heavy (orange, blue–white) and three light chains (gray) form a triskelion. (E) In COPII, the heterotetrameric Sec31–13 assembly forms an edge element. The repeating units can be assembled to form larger vesicles as well. Owing to the close relationship with COPII elements, it is most likely that two Y-complexes (F), the main scaffolding unit, arrange symmetrically along the positive curvature of the pore membrane (Brohawn and Schwartz 2009). Contacts between two Y-complexes might be direct or indirect. Other models have also been suggested (for a discussion, see Brohawn et al. 2009).
In the COPII cage, the outer coat is entirely built from heterotetrameric Sec13–Sec31 units (Stagg et al. 2006; Fath et al. 2007). These form elongated edge elements, capped on both ends with two β-propeller units (Fig. 4). The outer Sec31 β-propeller engages with three neighboring Sec31 β-propellers to form a vertex element. Several cages have been observed, which differ in the number of edge elements per cage (Stagg et al. 2008). To accommodate different numbers of edge elements per coat, the angle between neighboring edge elements changes to adjust for different radii. Some known cargo, like chylomicrons and procollagen fibers, would still not fit into the COPII coats that have been assembled in vitro and need even larger assemblies. Recently, it was shown that monoubiquitinylation of Sec31 might regulate coats and would enable the formation of giant-sized COPII cages (Jin et al. 2012). It should be noted that another architectural factor, Sec16, is also found at ER exit sites (ERESs) (Connerly et al. 2005; Hughes et al. 2009). Sec16 can interact with all COPII components; for example, contact with Sec13 is mediated by an ACE1 domain just as is seen in the interaction between Sec13 and Sec31 (Whittle and Schwartz 2010). Additionally, as seen with Sec31, two Sec16 molecules form homodimers. Why then is Sec16 not found in the assembled COPII coat? In contrast to Sec31, Sec16 does not contain its own β-propeller domain; thus, it presumably cannot form vertices. Although the molecular details of Sec16’s role in COPII are still unclear, it seems most likely that it is a general assembly factor of COPII formation (Zanetti et al. 2012).
The clathrin coat is also constructed from separate outer and inner coats (Fotin et al. 2004; Edeling et al. 2006). The outer coat is built from two proteins: ∼190-kDa clathrin heavy chain (CHC) and ∼25-kDa clathrin light chain (CLC). CHC contains an amino-terminal β-propeller followed by an ∼40-nm-long α-helical zigzag. Three CHCs come together to form a triskelion, the principal building block of the clathrin cage. Even though superficially the combination of architectural elements is similar between CHC and Sec31, the arrangement into a cage is entirely different. Although vertices in COPII are constructed from neighboring β-propellers (Stagg et al. 2006, 2008; Fath et al. 2007), in clathrin, the β-propellers point inward. Instead, clathrin vertices are entirely α-helical. The edges are constructed from the α-helical zigzags of three neighboring CHCs that wrap around each other (Fotin et al. 2004). The 25-kDa CHC is a long helical rod that decorates the outside of the cage edges. The inner coat of clathrin contains heterotetrameric adaptor (AP) complexes (Collins et al. 2002; Heldwein et al. 2004; Owen et al. 2004). In function, these complexes mimic the COPII inner coat, that is, they also connect the membrane with the outer coat, and they are also involved in cargo selection. However, judged by structure and sequence, there is no apparent phylogenetic relationship between the inner coats of COPII and clathrin. As in COPII, however, a small G-protein, Arf-1, helps to recruit the AP complex to the membrane.
COPI, the third well-studied vesicle coat complex, directs Golgi-to-ER retrotransport (Nickel et al. 2002). COPI is entirely built of multiple copies of a heteroheptameric βγδζ-αβ′ε unit (Sahlmüller et al. 2011). The βγδζ heterotetrameric unit has structural and sequence similarities to the AP complex, suggesting that it represents an inner COPI coat. In fact, using structural data from the AP complex, the COPI coat can be modeled with confidence (Yu et al. 2012). A portion of the heterotrimeric αβ′ε unit has now been crystallized (Hsia and Hoelz 2010; Lee and Goldberg 2010). Its makeup of β-propellers and α-helical stacks suggests that it forms an outer coat. Although β-propellers are generally superimposable, α-helical stacks come in many forms, as detailed earlier for the architectural nucleoporins. The α-helical segment in the COPI outer coat bear little resemblance with the α-helical stacks found in COPII and clathrin, which precludes definitive statements regarding their arrangement into a lattice-like cage. So far, COPI vesicles have not been analyzed using single-particle cryo-EM, because of the lack of pure, recombinant COPI coatomer complex. There are speculations as to how similar the COPI outer coat is compared with either COPII or clathrin (Lee and Goldberg 2010; Faini et al. 2012), but they need to be substantiated by further experimental data.
IMPLICATIONS OF VESICLE COAT ASSEMBLIES FOR THE NPC SCAFFOLD
In all vesicle coats, one can distinguish between a membrane-proximal or inner coat, and a membrane-distal or outer coat. To what extent do these principles apply to the NPC scaffold?
As mentioned above, Sec16, Sec31, and four nucleoporins have the ACE1 domain in common, which is not found in either clathrin or COPI. Two of these nucleoporins, Nup85 and Nup145C, also bind open, six-bladed β-propellers just as is true for Sec16 and Sec31. In addition, in the Y-complex, the two neighboring ACE1 proteins Nup84 and Nup145C bind to each other in a way that resembles the ACE1 homotypic Sec16 and Sec31 in the COPII coat (Brohawn and Schwartz 2009). The main difference is that the homotypic Sec16 and Sec31 interface in COPII includes a domain swap, whereas the heterotypic Nup84–Nup145C interaction does not. However, it is not yet clear whether those domain swaps are crystallographic artifacts or whether they are functionally relevant (Whittle and Schwartz 2010). Regardless, it is now evident that COPII and the NPC not only have similar building blocks, but several of these are also connected in related fashion. Therefore, the relationship between the NPC and COPII is closer than its relationship with any other vesicle coat.
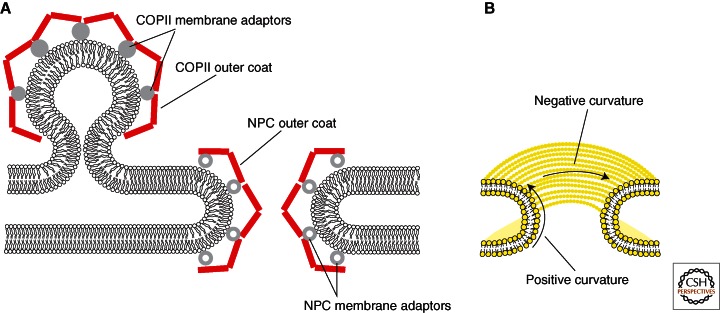
Of course, one has to ask how similar the NPC and vesicle coats can be, given their vastly different tasks and the different membrane geometries they need to stabilize (Fig. 5).
Figure 5.
General considerations regarding the geometry of vesicle coats and the NPC. (A) COPII vesicle budding and NPC formation require stabilization of highly curved ER membranes. Thus, it is conceptually understandable why both systems are related. In NPC and COPII-related outer coat components (red) form a lattice-like arrangement that is connected to the membrane via adaptor proteins, the inner coat (gray spheres). Owing to significantly different task profiles of the two systems, the inner coats appear to be entirely unrelated. (B) Unlike spherical or ellipsoidal vesicle coats, the NPC coat needs to stabilize a membrane that has both positive and negative curvature. Although stabilization of positive curvature is likely similar to the COPII system, the stabilization of negative curvature is not yet understood.
COPI, COPII, and clathrin coats all have in common that they cover spherical or ellipsoidal membrane vesicles, which only show positively curved membranes. In addition, vesicles need to assemble and disassemble rapidly and in a controlled manner. Small GTP-binding proteins, Sar1 in COPII and Arf1 in COPI and clathrin, respectively, regulate vesicle coat assembly in at least a conceptually similar way—membrane bending followed by recruitment of coat components. The NPC coat is different in geometry because the pore membrane has both positive curvature in the direction perpendicular to the INM and ONM sheets, and negative curvature in the horizontal plane of the pore (Antonin and Mattaj 2005). Furthermore, vesicle coats are seamless, whereas the NPC coat needs to terminate at the pore membrane periphery. This is precisely why the lattice model of the NPC, based on the evolutionary relationship with COPII, postulates that two Y-complexes interact with one another such that their respective COPII-related edge elements are aligned along the positive curvature of the pore membrane to form a bracket around the membrane (Brohawn and Schwartz 2009; Brohawn et al. 2009). Nup133, which does not contain an ACE1 domain, is at the nucleoplasmic and cytoplasmic periphery of the bracket, preventing further lattice propagation in those directions.
The propagation of the coats is likely self-guided and similar in all cases. All coats have stable repeating units, separated into inner and outer coat components. Whereas the inner coat components are not reported to interact with their inner coat neighbors, the outer coats all form lattice-like cages. The affinity between outer coat subassemblies is low and is typically not detected in protein–protein interaction studies. Nonetheless, because of the avidity of repeating the same weak interaction many times, stable cages do form. The molecular details of the lattice assembly can be very different, as shown by the contrast between COPII and clathrin assemblies. It seems that β-propellers and α-helical stacks are useful structural elements in all coats, but the way in which they assemble is not uniform. β-propellers can interact with partners in various ways, presumably because so many backbone hydrogen acceptors and donors are surface exposed by design of the domain. Each propeller blade exposes an unsatisfied β-strand at its edge, possibly allowing various, nonsequence-specific interactions. The α-helical stacks and their interactions are also difficult to classify and show an immense spectrum of different assemblies. In the COPII cage, for instance, Sec31 interacts with a neighboring Sec31 ACE1 domain via a small interface, whereas the triskelion legs of clathrin are intertwined over an extended distance (Fig. 4).
The equivalent to an inner coat for the NPC is not yet evident. Sequence comparison between nucleoporins and inner vesicle coat proteins has not revealed any relationship. Other than tethering the outer coat to the membrane, the inner coat of the NPC does not have the same functions as the inner coat of vesicles. First, there is no cargo the NPC inner coat needs to interact with. Second, in contrast to vesicles, NPCs do not assemble and disassemble rapidly. NPCs typically assemble only once during the cell cycle (Antonin et al. 2008; Kutay and Hetzer 2008). In fact, architectural nucleoporins are among the most stable proteins in the cell (D’Angelo et al. 2009; Savas et al. 2012). On the other hand, inner coat proteins of the NPC have potential roles that are irrelevant to vesicle coats. For instance, it is still unclear how INM proteins exactly pass the NPC en route to their destination, after membrane insertion in the ER via the Sec61 channel. If these INM proteins, in fact, stay membrane inserted during NPC passage, it is most likely that they pass through peripheral pore channels between outer coat and pore membrane (Zuleger et al. 2012). Cryo-ET studies of the NPC clearly indicate the existence of peripheral channels ∼8 nm wide (Beck et al. 2007; Maimon et al. 2012). Inner coat proteins could influence the size of these channels.
There are multiple candidate proteins for the inner coat of the NPC. The three transmembrane Nups—Ndc1, Pom152, and Pom34—are by definition involved in anchoring the NPC to the pore membrane. Nup53 might also be an inner coat component, because it interacts with the membrane directly via a well-conserved, carboxy-terminal amphipathic helix (Marelli et al. 2001). Nup53 also binds to the Ndc1–Pom34–Pom152 network (Onischenko et al. 2009). Finally, Nup157, Nup188, and Nup192 are all inner coat candidates as well, because they are at least reported to be close to the membrane or they directly affect transport of INM proteins (Franz et al. 2005; Hawryluk-Gara et al. 2008; Theerthagiri et al. 2010).
CONCLUDING REMARKS
Even though it is now established that vesicle coats and the NPC coat share common features, much remains to be studied. A lattice-like arrangement of repeating building blocks with related architecture is seen in all coats as well as in the NPC, but the molecular details of the two best understood assemblies, the COPII and the clathrin coats, are quite different. For COPI and the NPC, the assembly models are still largely speculative and lack atomic detail. Thus, the exact evolutionary relationship between the known coats, as well as additional systems, like the intraflagellar transport machinery (Jékely and Arendt 2006), remains unclear and probably holds further surprises.
The size variation that is seen in vesicle coats might be regulated differently among the COPI, COPII, and clathrin systems. For the NPC, it is not yet clear whether all pores in a given cell are identical or whether there are different assemblies. In addition, the large size variation between NPCs from different organisms might be the result of assembly from different copy numbers of architectural nups, different sets of nucleoporins, or a combination of both.
Lastly, the sharing of the Sec13 component between COPII and NPC is obviously a strong argument for suggesting common ancestry. However, it could also point to possible communication between these systems. As we become aware of more and more proteins with multiple functions, it might well be that the current view of strictly separate functions for different vesicle-coating systems and the NPC is oversimplified. It has been shown, for example, that COPI components interact with nups during nuclear envelope breakdown (Liu et al. 2003). Thus, the study of vesicle coats and the NPC remain very active fields with still many fundamental biological questions remaining to be answered.
ACKNOWLEDGMENTS
I thank Greg Kabachinski for critically reading the manuscript. This work is supported by National Institutes of Health grant GM077537.
Footnotes
Editors: Susan Ferro-Novick, Tom A. Rapoport, and Randy Schekman
Additional Perspectives on The Endoplasmic Reticulum available at www.cshperspectives.org
REFERENCES
*Reference is also in this collection.
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. 2007. The molecular architecture of the nuclear pore complex. Nature 450: 695–701 [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E 2011. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell 146: 277–289 [DOI] [PubMed] [Google Scholar]
- Antonin W, Mattaj IW 2005. Nuclear pore complexes: Round the bend? Nat Cell Biol 7: 10–12 [DOI] [PubMed] [Google Scholar]
- Antonin W, Ellenberg J, Dultz E 2008. Nuclear pore complex assembly through the cell cycle: Regulation and membrane organization. FEBS Lett 582: 2004–2016 [DOI] [PubMed] [Google Scholar]
- Beck M, Förster F, Ecke M, Plitzko JM, Melchior F, Gerisch G, Baumeister W, Medalia O 2004. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 306: 1387–1390 [DOI] [PubMed] [Google Scholar]
- Beck M, Lucić V, Förster F, Baumeister W, Medalia O 2007. Snapshots of nuclear pore complexes in action captured by cryo-electron tomography. Nature 449: 611–615 [DOI] [PubMed] [Google Scholar]
- Berke IC, Boehmer T, Blobel G, Schwartz TU 2004. Structural and functional analysis of Nup133 domains reveals modular building blocks of the nuclear pore complex. J Cell Biol 167: 591–597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S, Schwartz TU 2012. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci 109: 15241–15246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Schwartz TU 2009. Molecular architecture of the Nup84–Nup145C–Sec13 edge element in the nuclear pore complex lattice. Nat Struct Mol Biol 16: 1173–1177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Leksa NC, Spear ED, Rajashankar KR, Schwartz TU 2008. Structural evidence for common ancestry of the nuclear pore complex and vesicle coats. Science 322: 1369–1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brohawn SG, Partridge JR, Whittle JRR, Schwartz TU 2009. The nuclear pore complex has entered the atomic age. Structure 17: 1156–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhuri I, Söding J, Lupas AN 2008. Evolution of the β-propeller fold. Proteins 71: 795–803 [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ 2002. Molecular architecture and functional model of the endocytic AP2 complex. Cell 109: 523–535 [DOI] [PubMed] [Google Scholar]
- Connerly PL, Esaki M, Montegna EA, Strongin DE, Levi S, Soderholm J, Glick BS 2005. Sec16 is a determinant of transitional ER organization. Curr Biol 15: 1439–1447 [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ 2002. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Raices M, Panowski SH, Hetzer MW 2009. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell 136: 284–295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT 2009. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics 8: 2119–2130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Alber F, Williams R, Chait BT, Sali A, Rout MP 2004. Components of coated vesicles and nuclear pore complexes share a common molecular architecture. PLoS Biol 2: e380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D, Dokudovskaya S, Williams R, Alber F, Eswar N, Chait BT, Rout MP, Sali A 2006. Simple fold composition and modular architecture of the nuclear pore complex. Proc Natl Acad Sci 103: 2172–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dultz E, Ellenberg J 2010. Live imaging of single nuclear pores reveals unique assembly kinetics and mechanism in interphase. J Cell Biol 191: 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edeling MA, Smith C, Owen D 2006. Life of a clathrin coat: Insights from clathrin and AP structures. Nat Rev Mol Cell Biol 7: 32–44 [DOI] [PubMed] [Google Scholar]
- Elad N, Maimon T, Frenkiel-Krispin D, Lim RYH, Medalia O 2009. Structural analysis of the nuclear pore complex by integrated approaches. Curr Opin Struct Biol 19: 226–232 [DOI] [PubMed] [Google Scholar]
- Faini M, Prinz S, Beck R, Schorb M, Riches JD, Bacia K, Brügger B, Wieland FT, Briggs JAG 2012. The structures of COPI-coated vesicles reveal alternate coatomer conformations and interactions. Science 336: 1451–1454 [DOI] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, Goldberg J 2007. Structure and organization of coat proteins in the COPII cage. Cell 129: 1325–1336 [DOI] [PubMed] [Google Scholar]
- Fernandez-Martinez J, Phillips J, Sekedat MD, Diaz-Avalos R, Velazquez-Muriel J, Franke JD, Williams R, Stokes DL, Chait BT, Sali A, et al. 2012. Structure–function mapping of a heptameric module in the nuclear pore complex. J Cell Biol 196: 419–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemming D, Devos DP, Schwarz J, Amlacher S, Lutzmann M, Hurt E 2011. Analysis of the yeast nucleoporin Nup188 reveals a conserved S-like structure with similarity to karyopherins. J Struct Biol 177: 99–105 [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T 2004. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature 432: 573–579 [DOI] [PubMed] [Google Scholar]
- Franz C, Askjaer P, Antonin W, Iglesias CL, Haselmann U, Schelder M, de Marco A, Wilm M, Antony C, Mattaj IW 2005. Nup155 regulates nuclear envelope and nuclear pore complex formation in nematodes and vertebrates. EMBO J 24: 3519–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg MW, Allen TD 1996. The nuclear pore complex and lamina: Three-dimensional structures and interactions determined by field emission in-lens scanning electron microscopy. J Mol Biol 257: 848–865 [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U 1999. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol 15: 607–660 [DOI] [PubMed] [Google Scholar]
- Grossman E, Medalia O, Zwerger M 2012. Functional architecture of the nuclear pore complex. Annu Rev Biophys 41: 557–584 [DOI] [PubMed] [Google Scholar]
- Handa N, Kukimotoniino M, Akasaka R, Kishishita S, Murayama K, Terada T, Inoue M, Kigawa T, Kose S, Imamoto N 2006. The crystal structure of mouse Nup35 reveals atypical RNP motifs and novel homodimerization of the RRM domain. J Mol Biol 363: 114–124 [DOI] [PubMed] [Google Scholar]
- Hawryluk-Gara LA, Platani M, Santarella R, Wozniak RW, Mattaj IW 2008. Nup53 is required for nuclear envelope and nuclear pore complex assembly. Mol Biol Cell 19: 1753–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC 2004. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci 101: 14108–14113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer MW, Wente SR 2009. Border control at the nucleus: Biogenesis and organization of the nuclear membrane and pore complexes. Dev Cell 17: 606–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsia K-C, Hoelz A 2010. Crystal structure of α-COP in complex with ε-COP provides insight into the architecture of the COPI vesicular coat. Proc Natl Acad Sci 107: 11271–11276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes H, Budnik A, Schmidt K, Palmer KJ, Mantell J, Noakes C, Johnson A, Carter DA, Verkade P, Watson P, et al. 2009. Organisation of human ER-exit sites: Requirements for the localisation of Sec16 to transitional ER. J Cell Sci 122: 2924–2934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülsmann BB, Labokha AA, Görlich D 2012. The permeability of reconstituted nuclear pores provides direct evidence for the selective phase model. Cell 150: 738–751 [DOI] [PubMed] [Google Scholar]
- Jékely G, Arendt D 2006. Evolution of intraflagellar transport from coated vesicles and autogenous origin of the eukaryotic cilium. Bioessays 28: 191–198 [DOI] [PubMed] [Google Scholar]
- Jeudy S, Schwartz TU 2007. Crystal structure of nucleoporin Nic96 reveals a novel, intricate helical domain architecture. J Biol Chem 282: 34904–34912 [DOI] [PubMed] [Google Scholar]
- Jin L, Pahuja KB, Wickliffe KE, Gorur A, Baumgärtel C, Schekman R, Rape M 2012. Ubiquitin-dependent regulation of COPII coat size and function. Nature 482: 495–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampmann M, Blobel G 2009. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol 16: 782–788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutay U, Hetzer MW 2008. Reorganization of the nuclear envelope during open mitosis. Curr Opin Cell Biol 20: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Goldberg J 2010. Structure of coatomer cage proteins and the relationship among COPI, COPII, and clathrin vesicle coats. Cell 142: 123–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leksa NC, Brohawn SG, Schwartz TU 2009. The structure of the scaffold nucleoporin Nup120 reveals a new and unexpected domain architecture. Structure 17: 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Prunuske AJ, Fager AM, Ullman KS 2003. The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell 5: 487–498 [DOI] [PubMed] [Google Scholar]
- *.Lord C, Ferro-Novick S, Miller EA 2013. The highly conserved COPII coat complex sorts cargo from the endoplasmic reticulum and targets it to the Golgi. Cold Spring Harb Perspect Biol 10.1101/cshperspect.a013367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E 2002. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J 21: 387–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maimon T, Elad N, Dahan I, Medalia O 2012. The human nuclear pore complex as revealed by cryo-electron tomography. Structure 20: 998–1006 [DOI] [PubMed] [Google Scholar]
- Makio T, Stanton LH, Lin C-C, Goldfarb DS, Weis K, Wozniak RW 2009. The nucleoporins Nup170p and Nup157p are essential for nuclear pore complex assembly. J Cell Biol 185: 459–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mans BJ, Anantharaman V, Aravind L, Koonin EV 2004. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle 3: 1612–1637 [DOI] [PubMed] [Google Scholar]
- Marelli M, Lusk CP, Chan H, Aitchison JD, Wozniak RW 2001. A link between the synthesis of nucleoporins and the biogenesis of the nuclear envelope. J Cell Biol 153: 709–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mekhail K, Moazed D 2010. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol 11: 317–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy V, Hsia K-C, Debler EW, Kampmann M, Davenport AM, Blobel G, Hoelz A 2009. Structure of a trimeric nucleoporin complex reveals alternate oligomerization states. Proc Natl Acad Sci 106: 17693–17698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nehrbass U, Rout MP, Maguire S, Blobel G, Wozniak RW 1996. The yeast nucleoporin Nup188p interacts genetically and physically with the core structures of the nuclear pore complex. J Cell Biol 133: 1153–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann N, Lundin D, Poole AM 2010. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS ONE 5: e13241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickel W, Brügger B, Wieland FT 2002. Vesicular transport: The core machinery of COPI recruitment and budding. J Cell Sci 115: 3235–3240 [DOI] [PubMed] [Google Scholar]
- Onischenko E, Stanton LH, Madrid AS, Kieselbach T, Weis K 2009. Role of the Ndc1 interaction network in yeast nuclear pore complex assembly and maintenance. J Cell Biol 185: 475–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen DJ, Collins BM, Evans PR 2004. Adaptors for clathrin coats: Structure and function. Annu Rev Cell Dev Biol 20: 153–191 [DOI] [PubMed] [Google Scholar]
- Pante N, Kann M 2002. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell 13: 425–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pryer NK, Salama NR, Schekman R, Kaiser CA 1993. Cytosolic Sec13p complex is required for vesicle formation from the endoplasmic reticulum in vitro. J Cell Biol 120: 865–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabut G, Doye V, Ellenberg J 2004. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol 6: 1114–1121 [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT 2000. The yeast nuclear pore complex: Composition, architecture, and transport mechanism. J Cell Biol 148: 635–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlmüller MC, Strating JRPM, Beck R, Eckert P, Popoff V, Haag M, Hellwig A, Berger I, Brügger B, Wieland FT 2011. Recombinant heptameric coatomer complexes: Novel tools to study isoform-specific functions. Traffic 12: 682–692 [DOI] [PubMed] [Google Scholar]
- Savas JN, Toyama BH, Xu T, Yates JR, Hetzer MW 2012. Extremely long-lived nuclear pore proteins in the rat brain. Science 335: 942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC 1996. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell 84: 265–275 [DOI] [PubMed] [Google Scholar]
- Stagg SM, Gürkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE 2006. Structure of the Sec13/31 COPII coat cage. Nature 439: 234–238 [DOI] [PubMed] [Google Scholar]
- Stagg SM, LaPointe P, Razvi A, Gürkan C, Potter CS, Carragher B, Balch WE 2008. Structural basis for cargo regulation of COPII coat assembly. Cell 134: 474–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart CL, Roux KJ, Burke B 2007. Blurring the boundary: The nuclear envelope extends its reach. Science 318: 1408–1412 [DOI] [PubMed] [Google Scholar]
- Stirnimann CU, Petsalaki E, Russell RB, Müller CW 2010. WD40 proteins propel cellular networks. Trends Biochem Sci 35: 565–574 [DOI] [PubMed] [Google Scholar]
- Theerthagiri G, Eisenhardt N, Schwarz H, Antonin W 2010. The nucleoporin Nup188 controls passage of membrane proteins across the nuclear pore complex. J Cell Biol 189: 1129–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Rout MP 2010. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol 2: a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Schwartz TU 2009. Architectural nucleoporins Nup157/170 and Nup133 are structurally related and descend from a second ancestral element. J Biol Chem 284: 28442–28452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JRR, Schwartz TU 2010. Structure of the Sec13–Sec16 edge element, a template for assembly of the COPII vesicle coat. J Cell Biol 190: 347–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Breitman M, Goldberg J 2012. A structure-based mechanism for Arf1-dependent recruitment of coatomer to membranes. Cell 148: 530–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel U, Doye V, Tekotte H, Wepf R, Grandi P, Hurt EC 1996. Nic96p is required for nuclear pore formation and functionally interacts with a novel nucleoporin, Nup188p. J Cell Biol 133: 1141–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti G, Pahuja KB, Studer S, Shim S, Schekman R 2012. COPII and the regulation of protein sorting in mammals. Nat Cell Biol 14: 20–28 [DOI] [PubMed] [Google Scholar]
- Zuleger N, Kerr ARW, Schirmer EC 2012. Many mechanisms, one entrance: Membrane protein translocation into the nucleus. Cell Mol Life Sci 69: 2205–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]