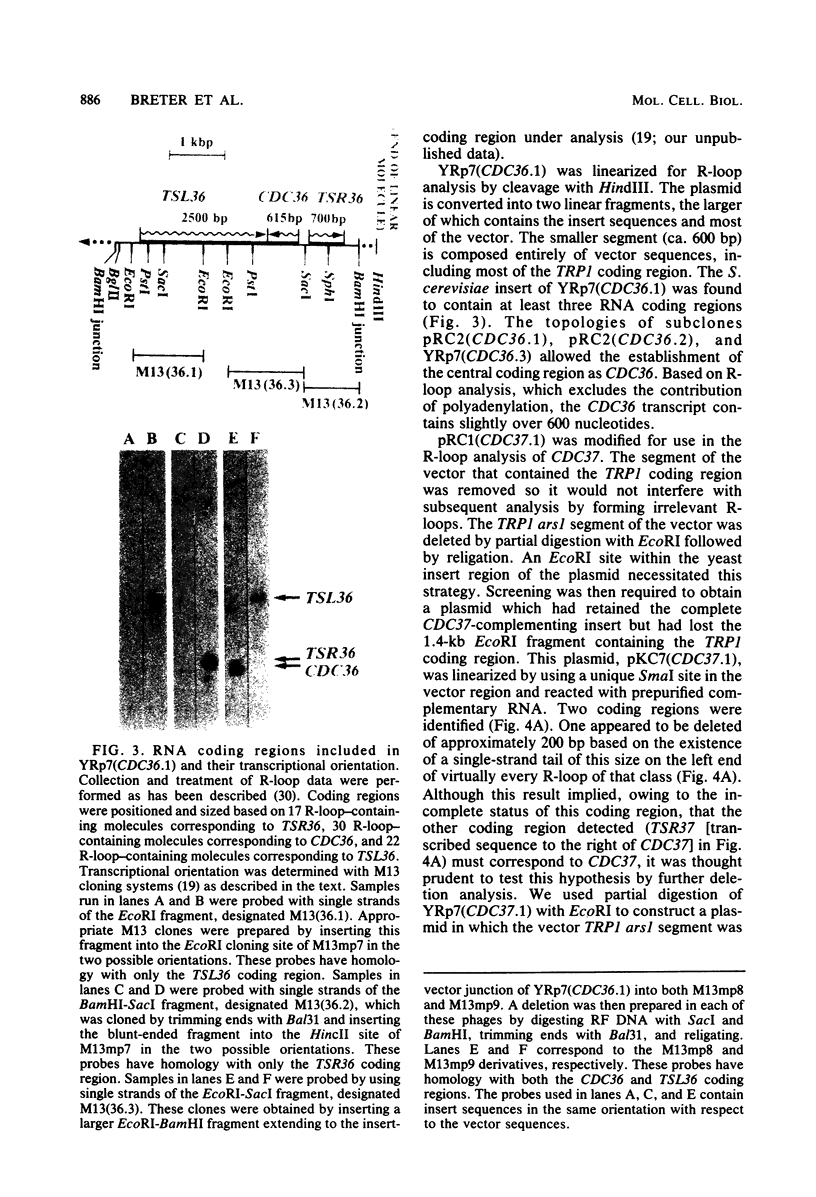
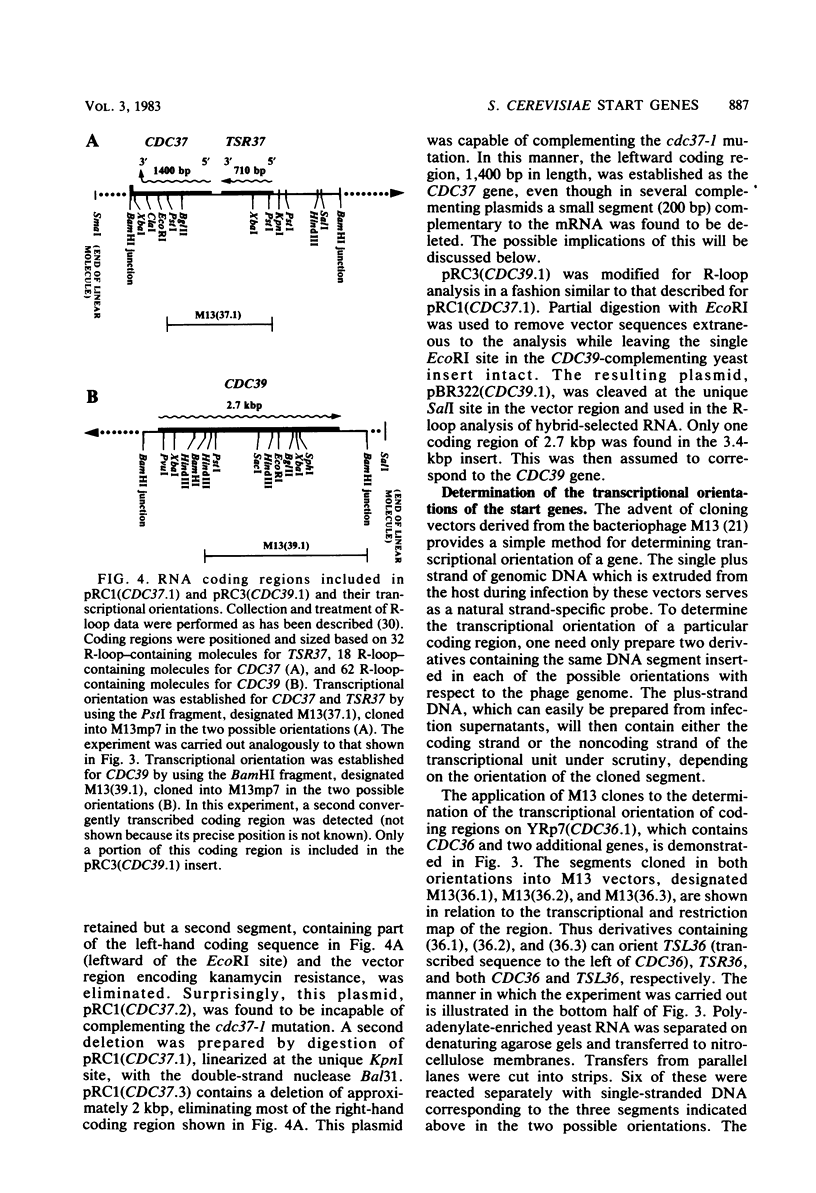
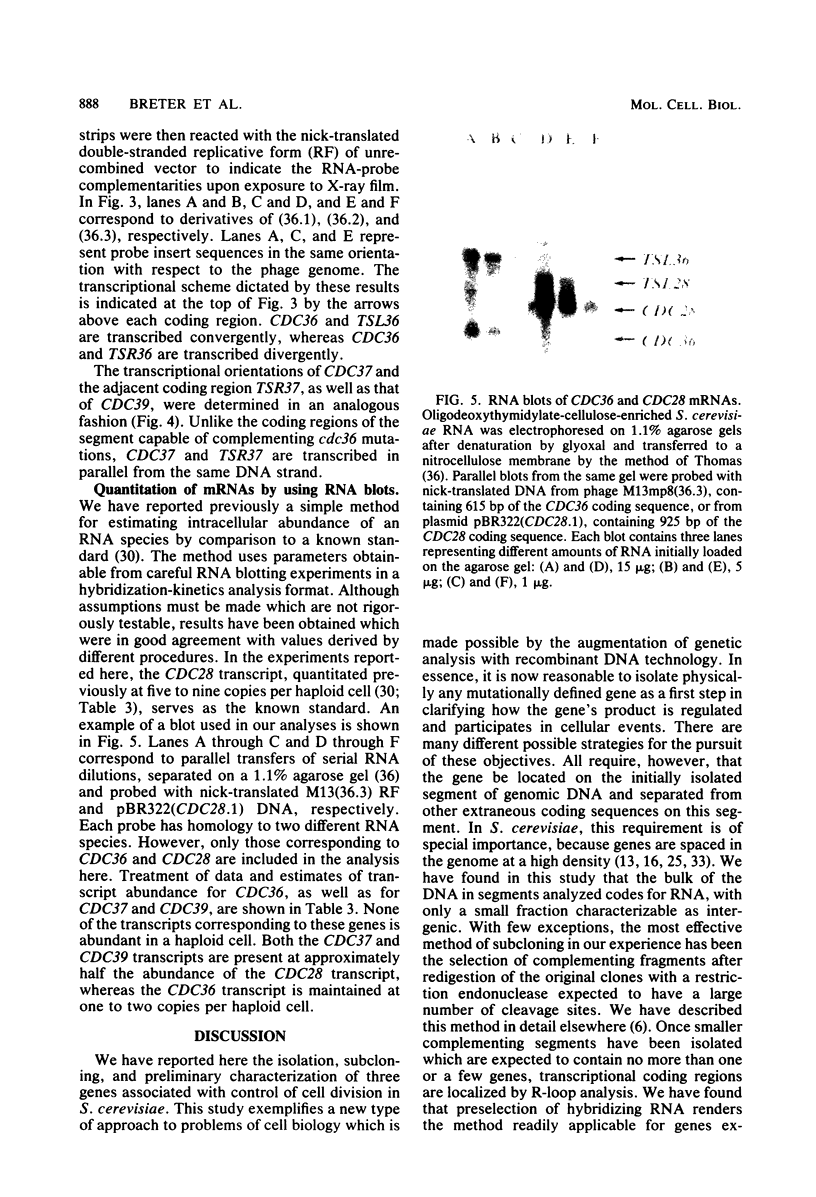
Abstract
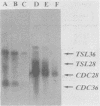
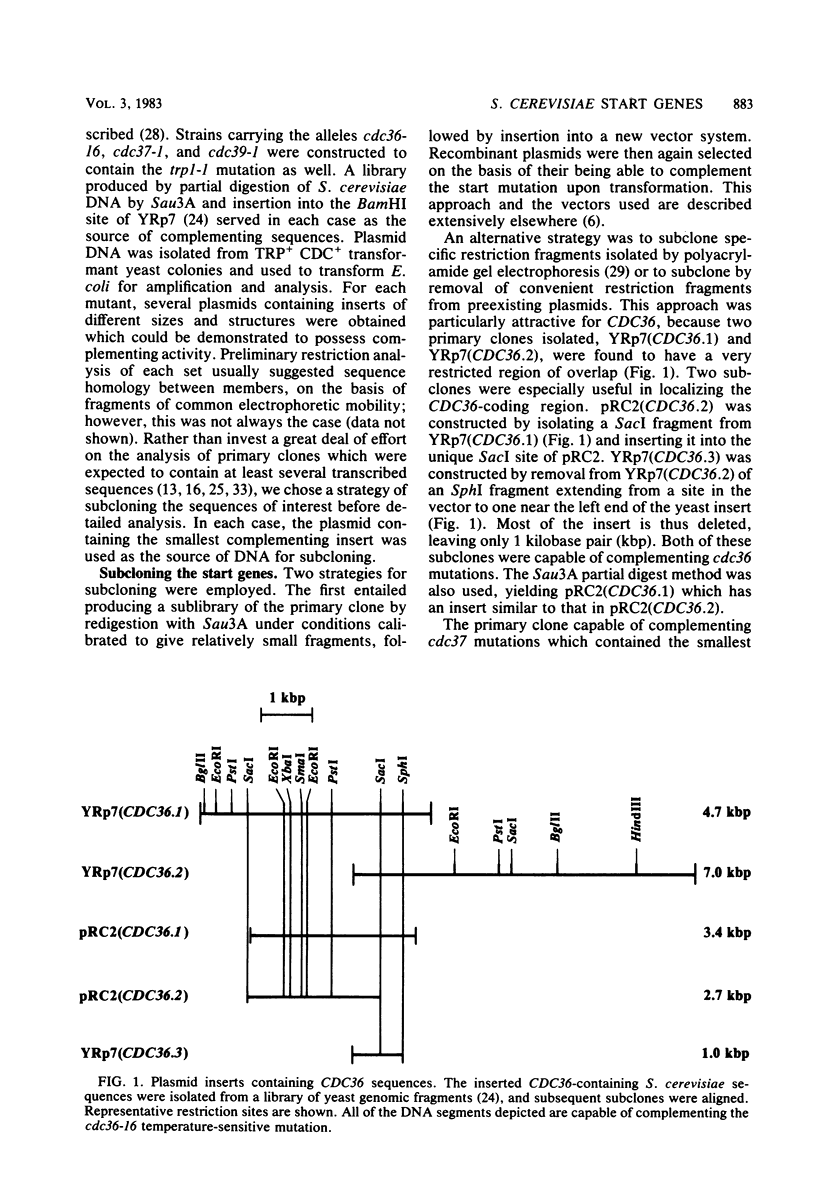
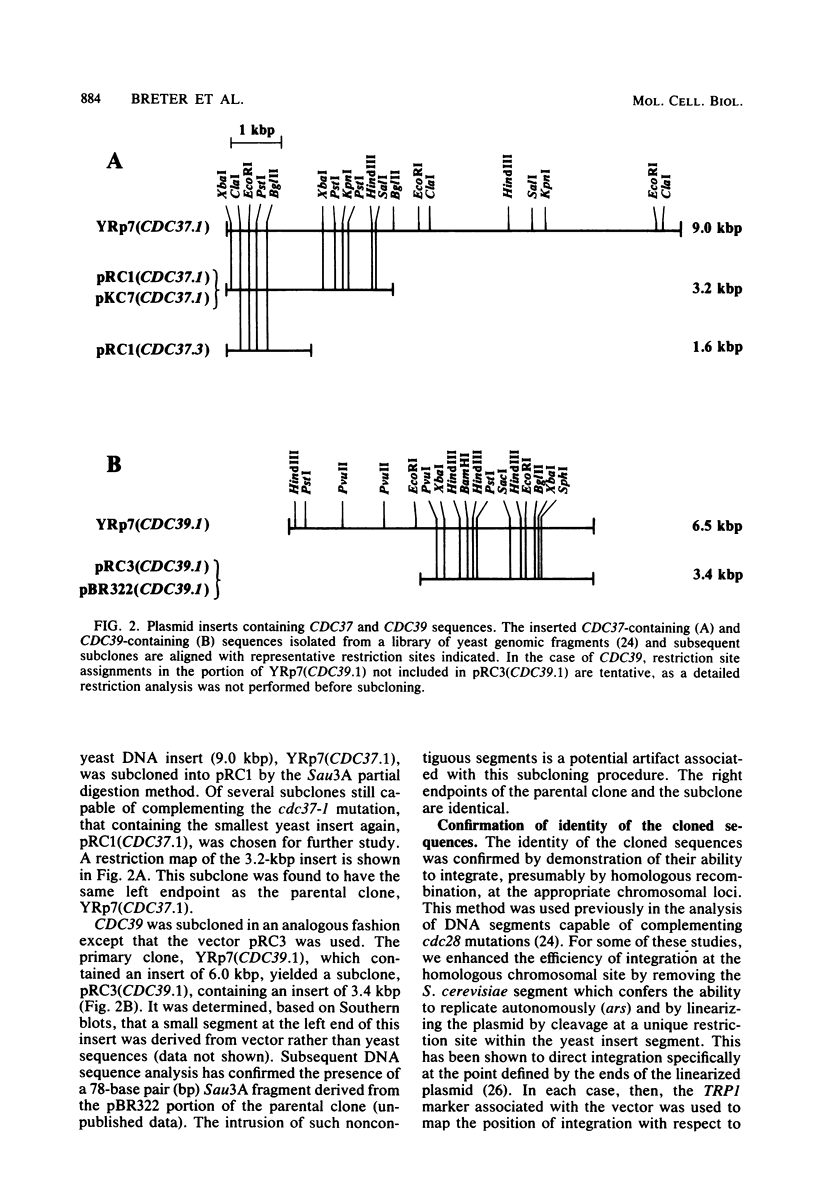
The genes CDC36, CDC37, and CDC39, thought to function in the cell division control process in Saccharomyces cerevisiae, were isolated from a recombinant plasmid library prepared by partial digestion of S. cerevisiae genomic DNA with Sau3A and insertion into the S. cerevisiae-Escherichia coli shuttle vector YRp7. In each case, S. cerevisiae DNA sequences were identified which could complement mutant alleles of the gene in question and which could direct integration of a plasmid at the chromosomal location known to correspond to that gene. Complementing DNA segments were subcloned to remove extraneous coding regions. The coding regions corresponding to CDC36, CDC37, and CDC39 were then identified and localized by R-loop analysis. The estimated sizes of the three coding regions were 615, 1,400, and 2,700 base pairs, respectively. Transcriptional orientation of the coding regions was established by using M13 vectors to prepare strand-specific probes followed by hybridization to blots of electrophoresed S. cerevisiae mRNA. The intracellular steady-state abundance of the mRNA species corresponding to the genes was estimated by comparing hybridization signals on RNA blots to that of a previously determined standard, the cell cycle start gene CDC28. The quantities calculated for the three mRNA species were low, ranging from 1.5 +/- 1 copies per haploid cell for the CDC36 mRNA to 3.1 +/- 1.5 and 4.6 +/- 2 copies per haploid cell for the CDC37 and CDC39 mRNAs, respectively. The CDC28 mRNA had been previously estimated at 7.0 +/- 2 copies per cell.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beggs J. D. Transformation of yeast by a replicating hybrid plasmid. Nature. 1978 Sep 14;275(5676):104–109. doi: 10.1038/275104a0. [DOI] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Bücking-Throm E., Duntze W., Hartwell L. H., Manney T. R. Reversible arrest of haploid yeast cells in the initiation of DNA synthesis by a diffusible sex factor. Exp Cell Res. 1973 Jan;76(1):99–110. doi: 10.1016/0014-4827(73)90424-2. [DOI] [PubMed] [Google Scholar]
- Ferguson J., Groppe J. C., Reed S. I. Construction and characterization of three yeast-Escherichia coli shuttle vectors designed for rapid subcloning of yeast genes on small DNA fragments. Gene. 1981 Dec;16(1-3):191–197. doi: 10.1016/0378-1119(81)90075-5. [DOI] [PubMed] [Google Scholar]
- Goldberg M. L., Lifton R. P., Stark G. R., Williams J. G. Isolation of specific RNA's using DNA covalently linked to diazobenzyloxymethyl cellulose or paper. Methods Enzymol. 1979;68:206–220. doi: 10.1016/0076-6879(79)68016-3. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Culotti J., Pringle J. R., Reid B. J. Genetic control of the cell division cycle in yeast. Science. 1974 Jan 11;183(4120):46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967 May;93(5):1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H. Sequential function of gene products relative to DNA synthesis in the yeast cell cycle. J Mol Biol. 1976 Jul 15;104(4):803–817. doi: 10.1016/0022-2836(76)90183-2. [DOI] [PubMed] [Google Scholar]
- Hartwell L. H., Unger M. W. Unequal division in Saccharomyces cerevisiae and its implications for the control of cell division. J Cell Biol. 1977 Nov;75(2 Pt 1):422–435. doi: 10.1083/jcb.75.2.422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hereford L. M., Hartwell L. H. Sequential gene function in the initiation of Saccharomyces cerevisiae DNA synthesis. J Mol Biol. 1974 Apr 15;84(3):445–461. doi: 10.1016/0022-2836(74)90451-3. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Osley M. A., Ludwig T. R., 2nd, McLaughlin C. S. Cell-cycle regulation of yeast histone mRNA. Cell. 1981 May;24(2):367–375. doi: 10.1016/0092-8674(81)90326-3. [DOI] [PubMed] [Google Scholar]
- Hereford L. M., Rosbash M. Number and distribution of polyadenylated RNA sequences in yeast. Cell. 1977 Mar;10(3):453–462. doi: 10.1016/0092-8674(77)90032-0. [DOI] [PubMed] [Google Scholar]
- Hereford L., Fahrner K., Woolford J., Jr, Rosbash M., Kaback D. B. Isolation of yeast histone genes H2A and H2B. Cell. 1979 Dec;18(4):1261–1271. doi: 10.1016/0092-8674(79)90237-x. [DOI] [PubMed] [Google Scholar]
- Hinnen A., Hicks J. B., Fink G. R. Transformation of yeast. Proc Natl Acad Sci U S A. 1978 Apr;75(4):1929–1933. doi: 10.1073/pnas.75.4.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston G. C., Pringle J. R., Hartwell L. H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp Cell Res. 1977 Mar 1;105(1):79–98. doi: 10.1016/0014-4827(77)90154-9. [DOI] [PubMed] [Google Scholar]
- Kaback D. B., Angerer L. M., Davidson N. Improved methods for the formation and stabilization of R-loops. Nucleic Acids Res. 1979 Jun 11;6(7):2499–2317. doi: 10.1093/nar/6.7.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleid D. G., Yansura D., Small B., Dowbenko D., Moore D. M., Grubman M. J., McKercher P. D., Morgan D. O., Robertson B. H., Bachrach H. L. Cloned viral protein vaccine for foot-and-mouth disease: responses in cattle and swine. Science. 1981 Dec 4;214(4525):1125–1129. doi: 10.1126/science.6272395. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortimer R. K., Schild D. Genetic map of Saccharomyces cerevisiae. Microbiol Rev. 1980 Dec;44(4):519–571. doi: 10.1128/mr.44.4.519-571.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Reed S. I. Isolation of genes by complementation in yeast: molecular cloning of a cell-cycle gene. Proc Natl Acad Sci U S A. 1980 Apr;77(4):2119–2123. doi: 10.1073/pnas.77.4.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. A., Tatchell K., Hall B. D., Astell C., Smith M. A position effect in the control of transcription at yeast mating type loci. Nature. 1981 Jan 22;289(5795):244–250. doi: 10.1038/289244a0. [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I., Ferguson J., Groppe J. C. Preliminary characterization of the transcriptional and translational products of the Saccharomyces cerevisiae cell division cycle gene CDC28. Mol Cell Biol. 1982 Apr;2(4):412–425. doi: 10.1128/mcb.2.4.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed S. I. Preparation of product-specific antisera by gene fusion: antibodies specific for the product of the yeast cell-division-cycle gene CDC28. Gene. 1982 Dec;20(2):255–265. doi: 10.1016/0378-1119(82)90044-0. [DOI] [PubMed] [Google Scholar]
- Reed S. I. The selection of S. cerevisiae mutants defective in the start event of cell division. Genetics. 1980 Jul;95(3):561–577. doi: 10.1093/genetics/95.3.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Shuman H. A., Silhavy T. J., Beckwith J. R. Labeling of proteins with beta-galactosidase by gene fusion. Identification of a cytoplasmic membrane component of the Escherichia coli maltose transport system. J Biol Chem. 1980 Jan 10;255(1):168–174. [PubMed] [Google Scholar]
- St John T. P., Davis R. W. Isolation of galactose-inducible DNA sequences from Saccharomyces cerevisiae by differential plaque filter hybridization. Cell. 1979 Feb;16(2):443–452. doi: 10.1016/0092-8674(79)90020-5. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas M., White R. L., Davis R. W. Hybridization of RNA to double-stranded DNA: formation of R-loops. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2294–2298. doi: 10.1073/pnas.73.7.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]