Abstract
Cancer-causing mutations disrupt coordinated, precise programs of gene expression that govern cell growth and differentiation. Microarray-based gene-expression profiling (GEP) is a powerful tool to globally analyze these changes to study cancer biology and clinical behavior. Despite overwhelming genomic chaos in multiple myeloma (MM), expression patterns within tumor samples are remarkably stable and reproducible. Unique expression patterns associated with recurrent chromosomal translocations and ploidy changes defined molecular classes with differing clinical features and outcomes. Combined molecular techniques also dissected two distinct, reproducible forms of hyperdiploid disease and have molecularly defined MM with high risk for poor clinical outcome. GEP is now used to risk-stratify patients with newly diagnosed MM. Groups with high-risk features are evident in all GEP-defined MM classes, and GEP studies of serial samples showed that risk increases over time, with relapsed disease showing dramatic GEP shifts toward a signature of poor outcomes. This suggests a common mechanism of disease evolution and potentially reflects preferential expansion of therapy-resistant cells. Correlating GEP-defined disease class and risk with outcomes of therapeutic regimens reveals class– specific benefits for individual agents, as well as mechanistic insights into drug sensitivity and resistance. Here, we review modern genomics contributions to understanding MM pathogenesis, prognosis, and therapy.
Keywords: gene-expression profiling, array comparative hybridization, multiple myeloma, classification, prognosis
Multiple myeloma: the disease
Multiple myeloma (MM) is a plasma-cell dyscrasia that homes to and expands in the bone marrow, in which it causes a constellation of disease manifestations that includes osteolytic lesions because of osteoblast inactivation and osteoclast activation, anemia, and immunosuppression because of loss of normal hematopoietic stem cell function and end-organ damage because of excessive monoclonal immunoglobulin secretion;1 increased bone-marrow angiogenesis is also frequently observed.2 MM presents with a common histological diagnosis, but it displays enormous genomic complexity as well as marked variations in clinical characteristics and patient survival. To advance treatments, clinical outcome data must be interpreted within the framework of genetic entities, which has proved useful in leukemia and lymphoma,3–11 and over the past 10 years has contributed to advances in treatment and survival of patients with MM.
For many years, investigations into the molecular lesions driving initiation and progression of MM languished, in part because of the enormously complex karyotypes typically seen in this malignancy. In fact, MM has cytogenetic features more similar to tumors of epithelial origin than to hematological malignancies. Whereas most leukemias and lymphomas present with single chromosomal translocations, karyotypes of myeloma cells from newly diagnosed disease have an average of seven different structural and/or numeric chromosomal abnormalities. This genomic chaos, along with the rarity of the disease, made it difficult to perform the comprehensive correlative studies necessary to identify and better understand the abnormalities involved in initiation and/or progression of the disease and to distinguish nonspecific bystander effects of chromosome instability. Indeed, the first link between a recurrent chromosome abnormality and prognosis was observed only 10 years ago, when deletions of chromosome 13 were associated with aggressive clinical course.12
The advent of new technologies, such as interphase fluorescence in situ hybridization (FISH), spectral karyotyping, comparative genomic hybridization, single nucleotide polymorphism genotyping, and gene-expression profiling (GEP), has provided the necessary tools to study MM in unprecedented detail. Combining these approaches with maturing technologies, such as high-throughput proteomics, microRNA profiling, and whole-genome sequencing, broadens the spectrum of molecular variables that can be tested, but also poses immense bioinformatics challenges to integrate the massive complexity of these high-dimensional datasets to improve management of MM. This review focuses on the use of GEP of primary disease to classify the disease, define risk, and elucidate underlying mechanisms that are beginning to change clinical decision making and inform drug design.
Studying the complexities of the transcriptome
It is likely that each of the six hallmarks of cancer, outlined in the Hanahan–Weinberg model,13 ultimately causes or is related to reproducible changes in the expression of subsets of genes within clonal tumor cells and that these patterns are unique and specific to each malignancy. This hypothesis was difficult to test, however, until the completion of the human-genome project14, 15 and the development of high-throughput tools capable of analyzing the activities of all genes simultaneously.16 It is now believed that the human genome consists of approximately 25000 mRNA-encoding genes, and this complexity is increased by post-transcriptional modifications, such as alternative splicing.
In the mid-1990s, Brown and coworkers developed a system that used DNA microarrays to monitor the expression levels of thousands of genes in parallel,16–18 which paved the way for tools that revolutionized molecular biology. The system worked similar to reverse northern blots: cloned DNA fragments immobilized on a solid matrix were used simultaneously to probe mRNA pools from a control source and from the tumor or other tissue of interest, each labeled with a different fluorescent dye (e.g. Cy5 and Cy3). Building on this concept, more advanced high-density oligonucleotide microarrays capable of unprecedented levels of sensitivity and throughput was developed using photolithography and solid-phase chemistry. Now in the industry standard, these whole-genome high-density oligonucleotide microarrays contain hundreds of thousands of oligonucleotide probes, packed at extremely high densities.19 The probes are designed to maximize sensitivity, specificity, and reproducibility, which allows consistent discrimination between specific and background signals and between closely related target sequences.20 Using microarrays for GEP generates large amounts of complex data, demanding equally complex analyses. Indeed, GEP analysis has evolved into a field of its own and in many ways represents a central node in translational research; a comprehensive review of the principles and tools used to analyze microarray data was recently published.21 Here, we focus on the specific use of microarray profiling in MM, a research that has exploded over the past 10 years.
Microarray technology was first used to study cancer in 1996,22 and De Vos et al.23 were the first to use GEP to study MM in 2001. In these early experiments, human myeloma cell lines and plasma-cell leukemia samples were analyzed on small-scale, filter-based cDNA arrays to identify genes involved in intercellular signaling. In spite of its small scale, this study revealed that key signaling molecules within the Wnt pathway were altered in MM. Subsequently, Stewart et al. used a combination of high-throughput DNA sequencing and microarrays on cells pooled from several cases of plasma-cell leukemia to establish a comprehensive list of genes expressed in MM.24
Microarray profiling of MM
On account of the heterogeneous nature of MM growth within the bone marrow, with variable percentages of tumor in a given site as low as 5%, molecular profiling of unfractionated bone-marrow aspirates complicates interpretation of results. To overcome this limitation, researchers have used various means of cell enrichment of plasma cells from bone-marrow aspirates. Plasma cells typically make up <1% of the cells in healthy human bone marrow, so isolation of sufficient numbers of plasma cells from healthy human marrow made large-scale GEP experiments an impractical endeavor for most laboratories. To isolate sufficient numbers of cells for GEP, two different but complementary specialized methodologies were developed. Zhan et al.25 used automated immunomagnetic bead sorting of plasma cells from large-volume bone-marrow aspirates using a monoclonal antibody, BB4, raised against syndecan-1/CD138; this technique routinely has isolated highly homogeneous populations of healthy plasma cells from both bone marrow and tonsil.26 To create a source of polyclonal plasma cells from healthy donors, Tarte et al.27 developed a method for in vitro differentiation of peripheral blood B cells. Global GEP of polyclonal plasma cells and healthy bone-marrow plasma cells derived from immunomagnetic sorting has revealed strong similarities, but also distinct and reproducible differences between the two populations and myeloma cells,27, 28 suggesting that polyclonal plasma cells may not fully recapitulate the molecular biology of a bone-marrow plasma cell.
Early studies made several contributions to understanding the molecular basis of MM by comparing gene-expression profiles of CD138-enriched plasma cells from the bone marrow of healthy donors and patients with monoclonal gammopathy of unknown significance (MGUS), newly diagnosed MM, and end-stage MM.25 These studies uncovered potential clues to the molecular pathogenesis of MM—disease-specific changes in gene expression. Myeloma plasma cells can be clearly distinguished from those of healthy donors based on expression of approximately 120 of 6800 genes analyzed. Unsupervised clustering of these early global gene-expression data showed that MM could be divided into four distinct molecular subgroups, MM1–MM4, with MM1 being more similar to MGUS and MM4 being related to myeloma cell lines. The MM4 group also had a higher incidence of cytogenetic abnormalities (CAs) and high serum levels of β-2-microglobulin, clinical features historically linked to poor prognosis. Consistent with these data, genes distinguishing MM4 from the other groups were related to cell proliferation. More advanced microarray technologies and larger sample sizes have now further divided MM into seven disease classes (discussed below).
These results provided the first evidence that MM is likely numerous molecular entities that presumably use different molecular mechanisms to get to a tumor with a common histology, which has enormous clinical implications. First, the high resolution of molecular classifications allows retrospective evaluation of class-specific efficacy of current therapeutic regimens, which is exceedingly important when designing clinical trials. For example, a new drug might not show a significant effect on a given end point when considering MM as a whole, but the results might be dramatically different if the end point is examined in the context of a particular molecular classification of MM, which might include only 5% of the overall population. Second, identifying the genes whose expression is driving these classes can inform the use of existing agents that might not have been considered and can direct development of new class-specific drugs.
To provide insights into the molecular characterization of plasma-cell dyscrasias and to investigate the contributions of specific genetic lesions to the biological and clinical heterogeneity of MM, Mattioli et al. compared the GEP of plasma cells isolated from 7 cases of MGUS, 39 of MM, and 6 of plasma-cell leukemia. MM was heterogeneous at the transcriptional level, whereas MGUS was distinguished from plasma-cell leukemias and the majority of MM cases by differential expression of genes involved in DNA metabolism and proliferation. The clustering of MM cases was mainly driven by the presence of one of five recurrent translocations involving the immunoglobulin heavy-chain (IGH) locus.29 For example, overexpression of CCND2 and genes involved in cell-adhesion pathways was observed in cases with t(14;16) and t(14;20), whereas upregulated genes showed apoptosis-related functions in cases with t(4;14). The peculiar finding in cases with t(11;14) was downregulation of the α-subunit of the interleukin-6 receptor (IL6R). Finally, cancer-testis antigens were specifically expressed in a subgroup of patients characterized by aggressive clinical evolution of MM.30
To further decipher the differences between malignant and normal plasma cells, Jourdan et al.31 recently focused on 58 genes linked with extrinsic and intrinsic apoptotic pathways, caspases, and inhibitor of apoptosis proteins and found B-cell differentiation was associated with change in the expression of pro-apoptotic and anti-apoptotic genes with TRAIL being upregulated, whereas FAS, APAF1, and BNIP3 were down-regulated in MM cells compared with normal bone-marrow plasma cells.
GEP reveals interactions between MM cells and bone-marrow microenvironment
The bone-marrow microenvironment (BMME) has a critical function in MM cell proliferation, apoptosis, migration, and drug resistance.32 MM cells in turn disrupt normal bone-marrow homeostasis leading to bone destruction and impaired hemato-poiesis. Clarifying these interactions between the BMME and MM cells is a prerequisite necessary to completely understand MM initiation and progression. GEP has been used to reveal the molecular basis of these interactions.
Corre et al.33 studied bone-marrow mesenchymal stem cells, the only long-lived cells of the BMME, by GEP in patients with MM, patients with MGUS, and healthy subjects. Among the 145 distinct genes differentially expressed in MM and normal bone-marrow mesenchymal stem cells, 46% were classified in the ‘tumor microenvironment’ category according to gene ontology annotations. Genes overexpressed in MM bone-marrow mesenchymal stem cells included known myeloma growth factors such as IL-6, amphiregulin, and IL-1β, angiogenic factors, and proteins involved in bone disease such as DKK1. Pérez-Andrés et al.34 showed different expression profiles of molecules involved in the interaction with the immunological BMME across MGUS, plasma-cell leukemia, and MM.
MM cells have an affinity for bone, in which they cause osteolytic lesions.35 Comparing the GEP of MM cells from the MM patients with and without focal bone lesions, Tian et al.36 identified DKK1, a Wnt signaling antagonist, as a key gene that was secreted by MM cells and regulates homeostatsis of bone lysis and formation.37
Growing evidence indicates a function for increased angiogenesis in MM progression, and markers of angiogenesis have prognostic potential.2,38,39 Vacca et al.40 used GEP to show an overall induction of VEGF, FGF-2, HGF-SF, IGF-1, and IGF-binding protein 3 genes in the endothelial cells derived from MM and MGUS patients compared with human umbilical vein endothelial cell. Hedvat et al.41 compared GEP of MM cells, plasma-cell leukaemia plasma cells, and extramedullary plasmacytoma cells (EPC) and identified several angiogenesis-related genes upregulated in extramedullary plasmacytoma cells, which could confer malignant plasma cells with the ability to grow outside the normal bone-marrow environment. Comparing GEP of genetically identical twin samples, Munshi et al.42 observed increased levels of expression of angiogenesis-related interleukin-8 (5-fold) and angiopoietin-1 (5.8-fold) transcripts in MM cells versus healthy twin PCs. Hose et al.43 assessed the expression of 402 angiogenesis-associated genes by GEP in 466 samples, including CD138-purified myeloma cells. They found that although MM cells did not show a significantly higher median number of expressed pro- or anti-angiogenic genes, 97% of MM cell samples aberrantly express at least one of the angiogenic factors: HGF, IL-15, ANG, APRIL, CTGF, or TGFA.
Heparanase (HPSE) degrades Syndecan-1 and, therefore, enhances tumor cell metastasis,44 and may have an important function in regulating the growth and progression of MM.45 Using GEP, Mahtouk et al.46 found that HPSE was expressed mainly in the bone-marrow environment from polymorphonuclear cells, T cells, monocytes, and osteoclasts. Bret et al.47 investigated 100 genes involved in synthesis of heparan sulfate (HS) and chondroitin sulfate (CS) chains, which covalently bind to syndecan-1 and are the bioactive components of syndecan-1 in MM cells. Among 100 genes involved in synthesis of HS and CS, 16 were significantly different between normal and malignant plasma cells. Nine of these genes, EXT2, CHSY3, CSGALNACT1, HS3ST2, HS2ST1, CHST11, CSGALNACT2, HPSE and SULF2, encode proteins involved in glycosaminogly-can-chain synthesis or modifications. GEP data also revealed that syndecan-1 (SDC1) is essential for MM cell growth activity of EGF-family ligands in MM.48
BAFF and APRIL promote survival and growth of MM cells.49,50 BMME was the main source of BAFF and APRIL in MM patients.51 TACI is the receptor of BAFF/APRIL and its expression level varies in MM patients and is a good indicator of a BAFF-binding receptor. Using GEP, Moreaux et al.51 found that MM cells with high TACI displayed a mature plasma-cell gene signature, indicating dependence on the bone-marrow environment. In contrast, MM cells with low TACI exhibited a gene signature of plasmablasts, suggesting an attenuated dependence on the bone-marrow environment.
To investigate the molecular consequences of myeloma cell interactions with osteoclasts, Ge et al.52 exploited an ex vivo culture system and GEP to identify genes whose expression was consistently altered in osteoclasts by coculture with myeloma cells. They identified fibroblast activation protein (FAP), a serin protease, as one of 28 genes significantly overexpressed in cocultured osteoclasts and suggested FAP as a critical microenvironmental factor and a potential therapeutic target for MM.
GEP reveals universal event in MM is cyclin D dysregulation
Genomic profiling in a large cohort of primary disease revealed that dysregulated expression of cyclin D might be a universal event in myelomagenesis. Relative to plasma cells from the bone marrow of healthy donors, myeloma plasma cells exhibit increased and/or dysregulated expression of either CCND1, CCND2, or CCND3.53 IGH-mediated translocations can directly activate CCND1 (11q13)54 or CCND3 (6p21);55 MAF (16q23)- or MAFB (20q11)-activating translocations lead to their transactivation of adhesion molecules and CCND2, which is elevated in t(4;14)-positive tumors.56 Biallelic dysregulation of CCND1 occurs in nearly 40% of tumors, most of which are hyperdiploid.53 Elevated levels of CCND2 and the absence of IGH translocation spikes characterize a novel form of MM discovered through GEP of primary disease (termed ‘low bone,’ discussed below);57 interestingly, elevated expression of CCND2 is not an adverse prognostic factor in this setting.58
Validated molecular classification of MM
Using a supervised classification approach that uses prior knowledge of the disease, Bergsagel and Kuehl29 developed a classification schema based on GEP spikes of the five recurrent translocations, specific trisomies, and expression of cyclin D genes. Reducing the complexity of the microarray from over 50 000 probes to <30 genes, eight translocation/cyclin D (TC) groups were identified. These were termed the 11q13/TC1, 6p21/TC2, 4p16/TC3, maf/TC4, D1/TC5, D1+D2/TC6, D2/TC7, and none/TC8 classes.53 The authors proposed that these genetic entities are defined by early, perhaps initiating, oncogenic events. The classes exhibited significant, uniform differences in global gene-expression profiles and clinical features, such as prevalence of bone disease, frequency distribution at relapse, and progression to extramedullary tumor growth.53
Agnelli et al.59 used this GEP class-prediction model on purified plasma cells from 50 MM cases. The TC1, TC2, TC4, and TC5 groups were characterized by 112 probe sets, but TC3 samples showed heterogeneous phenotypes and no gene biomarkers. The TC2 group, with extra copies of the CCND1 locus and no IGH translocations or 13q deletion, was characterized by overexpression of genes involved in regulation of protein translation. The failure to validate all TC classes is likely related to the small sample size. Another possibility is that the TC classification is not robust because several of the classes are difficult to classify using this approach; new, different methods of classification are required when dealing with large datasets.
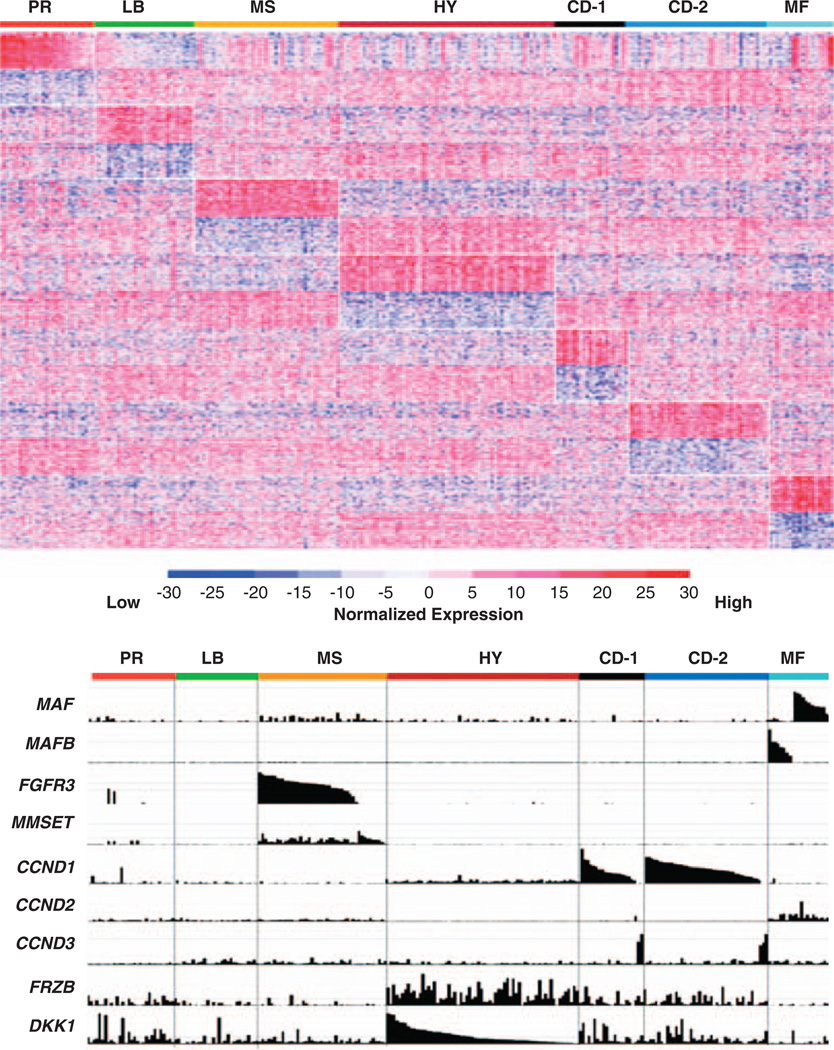
Complementing the supervised approach, unsupervised hierarchical clustering allows samples to self-organize based on underlying correlations in gene-expression patterns (Figure 1). Using a training set of 351 MM cases and a test set of nearly 200 newly diagnosed MM cases, Zhan et al.57 divided MM into seven different reproducible classes (Table 1). These molecular classes, largely consistent with the TC classification, are strongly influenced by distinct gene-expression profiles associated with known genetic lesions, including hyperdiploidy, translocations, cell proliferation, and tumor cell interactions with the BMME.57
Figure 1.
Classes are characterized by unique GEP patterns. (upper panel) A supervised clustergram of the expression of 700 genes (50 SAM-defined overexpressed and 50 underexpressed genes from each of the 7 classes) across 256 newly diagnosed cases. Genes are indicated along the vertical axis and samples on the horizontal axis. The normalized expression value for each gene is indicated by a color, with red representing high expression and blue representing low expression. (lower panel) The Affymetrix gene-expression signal (expression level: vertical axis) for the mRNA of MAF MAFB, FGFR3, MMSET, CCND1, CCND2, CCND3, FRZB, and DKK1, within classes presented in the upper panel, are indicated. The normalized expression level for each gene across the samples is given by the height of each bar. Note that spiked expression of CCND1, MAF and MAFB, and FGFR3 and MMSET is strongly correlated with specific subgroup designations. Also note that cases retaining the MMSET spike, but lacking FGFR3 spikes maintain similar cluster designation, and MAF and MAFB spikes cluster in the same subgroups. Several MMSET and CCND1 spikes cases are evident in the PR class. CCND3 expression is mutually exclusive of CCND1 expression. Although overexpressed in the HY subgroup, FRZB and DKK1 are significantly underexpressed in LB and MF. Figures reproduced with permission from Blood.
Table 1.
Characteristics of validated molecular classes as defined by unsupervised hierarchical clustering
| Molecular subtype |
%of newly diagnosed patientsa |
Genetic characteristics | Characteristic genes elevated in class |
Risk | Features |
|---|---|---|---|---|---|
| MS(MMSET) | 17 | t(4;14) |
FGFR3, MMSET CCND2, IL6R |
High | Overexpression MMSET and FGFR3’ FGFR3 not evident in ∼30%; bone disease is rare |
| MF(MAF/MAFB) | 6 | t(14;16) or t(14;20) |
MAF or MAFB CCND2, IL6R |
High/Moderate | Elevated expression of CCND2; bone disease rare; low DKK1; High NF-kB signature; low TNF-a induced gene TNFAIP8. |
| CD-1 (CCND1 or CCND3) |
6 | t(11;14) or t(6;14) |
CCND1 or CCND3 |
Low | Few cases express CCND2 in absence of CCND1 or CCND3; can have high DKK1 |
| CD-2 (CCND1 or CCND3) with CD20 expression |
12 | t(11;14) or t(6;14) |
CCND1 or CCND3, CD20, VPREB3 |
Low | Few cases express CCND2 in absence of CCND1 or CCND3 |
| HY (HYperdiploid) |
31 | Typical trisomies +3, +5, +7, +9, +11, +15, +19; |
GNG11, DKK1 FRZB |
Moderate | Low-ectopic expression of CCND1; del13 and gain of 1q are rare; high expression of interferon-induced genes |
| LB (Low Bone disease) |
12 | Typical HY trisomies; exception is frequent del13, gain of 1q, rare gain of 11 |
CCND2, CST6 ARHE, IL6R |
Low | Expression of CCND2; low level DKK1 FRZB,CCR2 HIF1A, SMAD1; low expression of interferon-induced genes |
| PR (PRoliferation) | 10 | Made up of all subgroups |
CCNB1, CCNB2, PCNA, MKI67 TOP2A, TYMS |
High | Overexpression of 1q genes; evolves from other groups |
Approximately 13% of newly diagnosed cases were not classified. Unclassified samples typically derived from bone-marrow aspirates containing low-bone marrow plasmacytosis. This so-called MY subgroup exhibits superior survival relative to other groups. CD138-purified cells from these cases have GEP of signatures consistent with contamination of preparations with the myeloid, T-cell, B-cell, and plasma-cell lineages. Group consists of all molecular classes described above as spikes and class-specific GEP features are evident in most. Bold and underlined values indicates the source of abbreviations.
Four (MF, MS, CD-1, and CD-2) of the seven subclasses are characterized by hyperelevated expression that results from recurrent chromosomal translocations present in approximately 40% of MM, which occur as a result of errors in switch recombination and/or somatic hypermutation.60 These translocations cause normally silent genes to become juxtaposed with powerful immunoglobulin enhancer elements, resulting in expression ‘spikes’ readily detectable in microarray studies. Hyperdiploid (HY) is characterized by low-level ectopic expression of CCND1 and overexpression of genes mapping to the odd-numbered chromosomes that typically exhibit trisomy in MM. The LB (low-bone disease) class, characterized by a low incidence of magnetic resonance imaging (MRI)-defined bone lesions, expresses high levels of CCND2 and a unique constellation of genes, including endothelin-1/EDN1. Unique, reproducible gene-expression programs can define at least six molecular entities; the seventh class of MM, PR (proliferation), is not related to a primary genetic lesion, but to high expression levels of proliferation-associated genes. This class likely consists of the other classes, but underlying features are masked by expression of proliferation genes. The 700 most differentially expressed genes across seven molecular classes can be found in Supplementary Tables 2 and 3 in Ref.57. In the following section, we highlight subsets of these 700 genes thought to be significant in class-specific disease pathogenesis.
MS class
The t(4;14)(p16;q32) translocation characterizes the MMSET (MS) class of MM, which is a high-risk entity that predicts poor prognosis.61 The t(4;14)(p16;q32) reciprocal translocation results in hyperactivation of both FGFR3 and MMSET/WHSC1 genes.62 All t(4;14)-positive disease expresses elevated levels of MMSET, but in about 30% of these cases, expression of FGFR3 is lost.61,63 As loss of FGFR3 expression is the only obvious GEP difference between these two types of t(4;14)-positive MM, it seems that MMSET has a central function in driving downstream transcriptional events in the MS class. Furthermore, 25% of MM cases in other classes also exhibit upregulation of MMSET, supporting its importance in MM pathogenesis.64 In a comparison of cases with and without t(4;14), GEP studies identified 127 genes as differentially expressed,64 including MMSET and CCND2. Notable genes overexpressed in the MS class, relative to other classes, encode N-cadherin/CDH2, cadherin-family member desmoglein2/DSG2, Wnt receptors FZ2 and FZD8, and B-cell oncogene PBX1. Underexpressed genes with potential relevance encode adhesion molecules ICAM4, cadherin 7/CDH7, and transcription factor PAX5.
Functional studies suggest that MMSET has a function in cell proliferation by decreasing cell viability and cell-cycle progression, possibly acting through desmoglein 2 (DSG2), a cell-surface cadherin molecule involved in cell adhesion and perhaps drug resistance. Thus, desmoglein 2 may be a therapeutic target for MS class MM.65
MF class
Accounting for approximately 6% of cases, the MAF/MAFB (MF) class of MM is characterized by the t(14;16)(q32;q23) and t(14;20)(q32;q11) translocations, which result in activation of c-MAF and MAFB proto-oncogenes, respectively. Cases lacking characteristic c-MAF or MAFB spikes can be classified as MF, suggesting that other genes of the MAF family may be activated in these cases or that low ectopic expression of either gene is sufficient to drive this classification. Although translocations involving c-MAF are seen in <5% of MM cases, c-MAF expression is elevated in myeloma cell lines lacking the translocations and in up to 50% of primary samples.56 These data strongly suggest that c-MAF expression may be activated by other mechanisms and attest to the importance of this family of transcription factors in MM pathogenesis. The NF-kB gene-expression signature in the MF class is significantly higher than in the other classes, with the exception of the LB class (see below).66 It is perhaps noteworthy that TNF-induced TNFAIP8 is the gene most significantly underexpressed in the MF class, relative to the other disease classes. Clinically, the MF class has relatively low incidence of bone lesions and, consistent with this, has low expression of DKK1, a Wnt antagonist produced by myeloma cells and associated with bone disease.36
Although mutually exclusive, MAF- and MAFB-activating translocations induce a common gene-expression signature, suggesting that ectopic expression of the MAF family of transcription factors results in dysregulation of common downstream targets. In GEP studies aimed at identifying MAFB targets in MM, 284 transcripts were modulated—14 were common to c-MAF and some had functional relationships with MAFB.67 Additional genes uniquely overexpressed in the MF class that represent known and putative targets of these transcription factors include NUAK1/ARK5,68 NTRK2, ARID5A, SMARCA1, TLR4, SPP1, and G6MB6.
CX3CR1 and ITGB7 are overexpressed in the MF class,69 consistent with the report that CCND2, CX3CR1, and ITGB7 are targets of the c-MAF transcription factor.56 c-MAF–driven expression of ITGB7 enhanced adhesion of myeloma cells to bone-marrow stroma and increased production of VEGF in these cells, suggesting that MAF can drive myeloma cell adhesion to the extracellular matrix and cellular stroma, resulting in drug resistance and angiogenesis. As expression of ITGB7 is also elevated in other MM classes, it could be a critical-adhesion molecule in MM that can be activated through multiple mechanisms. Expression of CCND2 is elevated in other disease classes, but is highest in the MF class. Hurt and coworkers showed that c-MAF transactivated the CCND2 promoter and enhanced MM proliferation, whereas dominant inhibition of c-MAF blocked tumor formation in immunodeficient mice, which highlights the potential significance of this transcription factor in myelomagenesis and the recognized association between MM and activation of one the three CCND genes. The MAF/CCND2 pathway might also interact with therapeutic strategies as it was regulated by CS1,70 a cell-surface glycoprotein to which a therapeutic monoclonal antibody reacts.71
CD-1 and CD-2 classes
The t(11;14)(q13;q32) and t(6;14)(p21;q32) translocations, characteristics of the CD-1 and CD-2 classes, directly activate expression of CCND1 and CCND3, respectively. Tumors with CCND1 and CCND3 spikes have gene-expression profiles that cluster membership, suggesting that activation of the two cyclin D orthologs results in dysregulation of common downstream transcriptional programs.
Nevertheless, CCND1 and CCND3 spikes are associated with two distinct, nonoverlapping gene-expression signatures that were used to distinguish the CD-1 and CD-2 classes. CD-2 is characterized by elevated expression of CD20/MS4A1, VPREB3, and PAX5—genes expressed in B cells, but normally extinguished in terminally differentiated plasma cells. Of note, CD20 mRNA and protein levels are correlated,57 but cells expressing elevated PAX5 mRNA do not express the protein.72 Unlike CD-2, CD-1 lacks expression of CD59 (potent inhibitor of complement membrane-attack complex), notch-like protein NOTCH2NL, and notch target gene HES1. CD-1 is characterized by overexpression of KLHL4 (a transcription factor), INHBE, FYN proto-oncogene, CEBPB/NF-IL6, and EVER1 and EVER2, two cytoplasmic proteins that colocalize with calnexin, an integral membrane protein of the endoplasmic reticulum (ER).57
HY class
Hyperdiploid MM is characterized by trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19, and 21. The trisomies are one of two central genetic pathways in development of MM, and this type of disease was earlier shown to have a distinct gene-expression signature.29 The HY signature is present in nearly 50% of cases and is associated with hyperdiploid karyotypes in >90% of these; however, the signature is also observed in cases that are not identified as hyperdiploid by flow-cytometry analyses. Such cases may arise through a similar initiating genetic mechanism (trisomies of odd chromosomes) with clonal evolution that results in DNA loss from other chromosomes, resulting in a DNA complement that is essentially diploid. Indeed, recent array-based comparative genomic hybridization (aCGH) data revealed that all molecular classes have at least a partial complement of the common trisomies (our unpublished data).
Genes uniquely overexpressed in the HY class encode guanine nucleotide-binding protein, gamma 11/GNG11, Trail/ TNFSF10, Wnt signaling antagonists FRZB/SFRP3 and DKK1, and MIP1-α chemokine receptor/CCR5. Overexpression of several interferon-induced genes, including OAS2, IFI27, and IFI35, is also characteristic of this class. Genes significantly underexpressed in the HY class, relative to the other classes, included CD52 and genes mapping to chromosome 1q—TAGLN2, CKS1B, and OPN3—whose overexpression has been linked to poor survival.73 Consistent with these results, integration of aCGH and GEP studies has shown that HY disease rarely exhibits gain of 1q (manuscript submitted). These data are also consistent with aCGH studies that defined disease clusters based on copy-number variation.74 These studies revealed that the classical hyperdiploid trisomies defined a specific subset of MM that lacks gains of 1q and deletion of chromosome 13.
LB class
A novel class of MM, LB is characterized by low incidence of MRI-defined focal bone lesions and lacks evidence of translocation spikes or HY gene-expression features. Consistent with the absence of MRI-defined focal lesions in the LB class, recent studies integrating positron emission tomography imaging with GEP also revealed that the LB class is uniquely inversely correlated with F18-fluorodeoxyglucose positron emission tomography-defined focal lesion number and intensity.75
LB disease is distinguished by overexpression of endothelin 1/ EDN1, a soluble factor that is secreted by prostate cancers and that causes osteoblastic metastases of prostate cancer. Interestingly, purified EDN1 induces osteoblast differentiation through suppression of Wnt/β-catenin signaling antagonist DKK1,76, 77 and the LB class is characterized by significantly lower expression of DKK1, suggesting that EDN1 may downregulate DKK1 in myeloma cells. LB is also associated with high-expression levels of IL6R; the MS and MF classes that also have low DKK1 and lower incidence of MRI-defined focal bone lesions, relative to HY, CD-1, and CD-2 classes, share this feature. In contrast to LB, the HY class rarely expresses IL6R and expresses high levels of DKK1 and EDN1 decoy receptor EDNRB. These data suggest a potential connection between EDN1 signaling and DKK1 production and bone disease in MM. It is also noteworthy that relative to the other classes, the LB class expresses higher levels of the apoptosis inducing gene BIK and the notch target gene and transcriptional repressor HES5 and lower levels of the chemokine receptor CCR2, hypoxia-induced Factor 1-a (HIF1A), and SMAD1, a transcription factor that mediates BMP signaling.
PR class
The PR class is characterized by overexpression of numerous genes related to cell-cycle progression and cell proliferation, including CCNB2, CCNB1, MCM2, CDCA2, BUB1, CDC2, and TYMS, and also cancer-testis antigen genes, including MAGEA6, MAGEA3, GAGE1, and GAGE4. Plasma cells from all MM classes have a higher gene-expression–defined proliferation index than plasma cells from healthy donors, but that of the PR class is similar to myeloma cell lines and is significantly higher than non-PR classes. Metaphase CAs, a surrogate for cell proliferation, are present in an extraordinarily high percentage of cases in the PR class. The PR class is associated with poorer survival than other classes, and both hyperdiploid and nonhyperdiploid cases are equally common, with and without concomitant translocation spikes, suggesting that hyperdiploid versus nonhyperdiploid status is likely not sufficient as a sole parameter for risk assessment. Indeed, those cases with both PR signature and hyperdiploidy are at higher risk than those with the HY signature. Several factors—overexpression of proliferation-associated genes in all non-PR classes, the presence of expression spikes in the PR class, and a shift to the PR class on disease progression—suggest that the PR class is driven by a transformation event secondary to underlying primary genetic lesions.
Classification outliers
In the study by Zhan et al., about 25% of newly diagnosed cases could not be classified because gene-expression signatures of myeloid/lymphoid-lineage cells and/or polyclonal plasma cells predominated and, similar to the PR signature, prevented unsupervised classification. The presence of translocation spikes in these cases supports the idea that this is not a unique class of MM. This contamination signature seems to hold important clinical implications because patients with this signature have lower levels of bone-marrow plasmacytosis, lower incidence of CAs, low β-2-microglobulin and creatinine, and better event-free survival (EFS) and overall survival (OS) than those without the signature.57
Using GEP to dissect specific genetic features of MM
GEP and hyperdiploid disease
GEP has been used to further dissect hyperdiploid and nonhyperdiploid disease. A combination of FISH and GEP showed that differential expression of genes involved in protein biosynthesis, transcriptional machinery, and oxidative phosphorylation distinguished the two types of disease.78 Of 204 genes upregulated in hyperdiploid disease, the majority mapped to the hyperdiploid chromosomes, and 29% of genes upregulated in nonhyperdiploid disease mapped to chromosome 16q;78 these findings were validated in independent datasets.78 Consistent with earlier studies,74 hyperdiploid MM was further divided into two distinct molecular and transcriptional entities, one characterized by trisomy 11 and another lacking this feature, but harboring chromosome 1q gains and chromosome 13 deletes.78
Chng et al.79 used GEP to show that hyperdiploid MM is primarily defined by a protein biosynthesis signature driven by a gene-dosage mechanism and to identify four independently validated patient clusters within hyperdiploid MM. One prominent cluster was characterized by cancer-testis antigen, proliferation-associated genes, and higher median plasma-cell labeling index; these patients experienced much shorter survival times than those in the other three clusters. Genes involved in tumor necrosis factor α/TNF-α and NF-κB signaling and anti-apoptosis characterized another cluster, and these patients had better responses to bortezomib than those in other clusters. This hyperdiploid disease cluster is probably the same disease entity as the hyperdiploid disease characterized by gain of 1q, lack of trisomy 11, and deletion of chromosome 13, as well as the LB class of MM. Studies integrating aCGH and GEP revealed that the LB class is primarily composed of this novel type of hyperdiploid disease,80 and the LB class is significantly associated with increased NF-kB activation.66
GEP and chromosome 13 deletion
A cohort of MM cases followed for 9 years was used to evaluate the prognostic implications of all individual CAs. Among all CAs and standard prognostic factors examined before therapy, only nonhyperdiploid and deletion of chromosome 13 (del13), alone or in combination, were associated with shortest EFS and OS.81 A combination of metaphase cytogenetics (to identify CAs), GEP, and interphase FISH (to identify del13) on 146 patient samples showed that overexpression of cell-cycle genes distinguished disease with CA from that without CA; this was especially evident in cases lacking FISH-defined del13.81 Interphase FISH evidence of del13 was significantly associated with reduced expression of a subset of genes mapping to chromosome 13, including RB1. The authors proposed that haploinsufficiency of genes mapping to chromosome 13, as well as significant upregulation of IGF-1R (insulin-like growth factor receptor), may have an amplifying effect on expression of cell-cycle genes, providing a molecular explanation for the dire outcome of patients with del13 compared with those with CA, but lacking del13.
Historically, del13 has been associated with an unfavorable prognosis, but increasing data indicate that its prognostic relevance must be related to the presence of other molecular features. Studies using FISH to detect del13, combined with GEP, on highly purified plasma cells from 80 patients newly diagnosed with MM identified 67 differentially expressed genes, all of which were downregulated in disease with del13. Of these, 44 mapped to chromosome 13, 7 to chromosome 11, and 3 to chromosome 19. del13-positive and -negative cases were differentiated by modulations in global gene expression; in particular, FISH-defined del13 was associated with upregulation of genes mapping to 1q21–1q42 and downregulation of genes mapping to 19p and most of chromosome 11.82
GEP and gains of chromosome 1q
Found in up to 45% of patients, abnormalities of chromosome 1 are among the most frequent chromosomal alterations in MM;83 the short arm is most often associated with deletions and the long arm with amplifications.74 Gains/amplification of 1q21 increases the risk of MM progression, and incidence of the amplification is higher in relapsed than in newly diagnosed MM.83,84 GEP studies comparing MM with and without 1q gains85 identified 61 genes that distinguished the two groups. In cases with 1q gains, 41 of the 43 upregulated genes mapped to 1q12–q44, whereas most of the 18 downregulated genes were localized to chromosomes 13q (7/18) and 11 (6/18). These data suggest that cases with 1q gains typically also harbor del13 and lack trisomy of chromosome 11, features consistent with the LB class. Integrating GEP and DNA copy-number variation data, several independent studies revealed that numerous 1q genes are copy-number sensitive in MM.74,86
Gains of 1q are associated with upregulation of genes involved in intracellular protein transport; prominent in the list were COPA and ARF1, which have functions in vesicle-mediated transport from the ER to the Golgi, and RABIF and RAB3GAP2, which are related to the Rab GTPases that regulate membrane-vesicle transport. These findings may also partially account for increased expression of genes-encoding proteins involved in energy-production pathways. Genes downregulated in cases with 1q gains include three genes involved in protein translation (RPLP2, RPL21, and FAU), which is potentially significant because recent studies suggest that survival of B-cell malignancies (including MM) may be highly dependent on ER– Golgi protein transport, thus targeting this process may be a novel therapeutic strategy.87
Cases with 1q gains also showed significantly modulated expression of genes involved in ER stress-induced responses, including upregulation of CLN3 (chaperone gene), UBAP2L and UBE2Q1 (ubiquitin cycle), PSMD4 (proteasome degradation), and CASP4 (initiates apoptosis in response to ER stress).85 As ER stress-induced apoptosis can have an important function in malignant cells’ sensitivity to certain drugs, including bortezomib, these studies suggest that a better understanding of ER stress-induced responses may contribute to important new treatment strategies.85
GEP and deletion of 17p13/TP53
A high-risk feature in MM is deletion of 17p13 presumably leads to loss of heterozygosity of TP53,88 a tumor suppressor gene that transcriptionally regulates cell-cycle progression and apoptosis to modulate cellular responses to DNA damage. Xiong et al.89 found that low expression of TP53, seen in approximately 10% of newly diagnosed patients, is highly correlated with FISH-defined TP53 deletion and inferior clinical outcome and is an independent risk factor. Only a few of the 122 known p53 target genes were highly correlated with TP53 expression in primary myeloma cells. GEP after ectopic expression of TP53 in four TP53-null-cell lines identified 85 significantly differentially expressed genes—50 upregulated and 35 downregulated. Using these 85 putative target genes, unsupervised hierarchical clustering of myeloma-cell samples from 351 newly diagnosed and 90 relapsed patients revealed two major subgroups that strongly correlated with TP53 expression and survival. These data suggest that loss of TP53 expression in MM confers high risk and probably results in deregulation of a novel set of p53 target genes specific to MM and perhaps unique to different cell lineages.89
Integrating GEP and high-resolution DNA copy-number analyses
Cigudosa et al.,90 Gutierrez et al.,91 and Avet-Loiseau et al.92 first applied traditional comparative genomic hybridization approaches93 to expand our knowledge about chromosome instability and copy-number changes in MM. Recently, developed aCGH, similar to GEP, allows simultaneous, high-resolution investigation of copy-number alterations across the entire genome.94–96 GEP and high-resolution analysis of recurrent copy-number alterations defined 87 discrete minimal common regions within recurrent, highly focal copy-number alterations;74 unsupervised classification using nonnegative matrix factorization uncovered four subtypes of disease and two subtypes of hyperdiploid disease. One hyperdiploid subtype had classic features—trisomies of chromosomes 3, 5, 7, 9, 11, 15, 19, and 21—whereas the other, associated with inferior survival,74 lacked trisomies of chromosomes 7 and 11 and also harbored deletion of chromosome 13 and gains of 1q. Another subtype was characterized by amplification of 1q21 and deletion of 1p, suggesting a relationship between this group and GEP-defined high risk. Indeed, a recent analysis of 92 additional cases revealed that copy-number alterations in chromosome 1q and 1p are highly correlated with gene-expression changes that are strongly correlated with risk of death from disease progression, gene-expression–based proliferation index, and 70/17-gene model of high risk (discussed below, GEP and risk stratification).80
High-density single nucleotide polymorphism microarrays of genomic DNA in the KMS-26 myeloma cell line showed that an amplicon within an unstable chromosomal region (17p11.2-p12) was characterized by a large number of low-copy repeats that mediate deletion and duplication in several genomic disorders and amplifications in solid tumors. Combining this data with GEP and FISH mapping narrowed the region of interest to the TNFRSF13B/TACI gene, an important receptor in B-cell development.97
In a FISH study of over 800MM cases, deletion of 16q was identified in 19.5% of cases, was associated with poor outcome, was an independent prognostic marker, and conferred additional adverse survival in cases with t(4;14) and/or del(17p).98 GEP and single nucleotide polymorphism-mapping arrays revealed loss of heterozygosity at 16q12, mapping near CYLD, and at 16q23, near WWOX. Cases with low expression of CYLD, a negative regulator of the NF-kB pathway, defined a ‘low-CYLD signature.’ Cases with 16q loss of heterozygosity or t(14;16) had significantly reduced expression of WWOX, a tumor suppressor gene involved in apoptosis that lies at the t(14;16) break point.98
GEP and risk stratification
Although most cases of MM initially respond to treatment, a subset exhibits resistance to therapy from the outset, and most will develop resistance over time. Therefore, long-term survival in patients with MM can vary considerably, and it is difficult to predict outcome based on current laboratory tests. High-risk MM is routinely defined by laboratory parameters alone or in combinations as in the Durie–Salmon staging system99 and International Staging System (ISS).100 A staging system based on cell morphology, the Bartl grade, has also been developed,101 and the presence of abnormal metaphase or interphase genetics,102 high plasma-cell labeling index,103 and a recently defined flow-cytometry–based test on minimal residual disease are also used.104 Importantly, the molecular mechanisms by which cells develop resistance are not yet known, but understanding the mechanisms underlying the disease escape from initial drug responsiveness will contribute to more robust prognostic strategies.
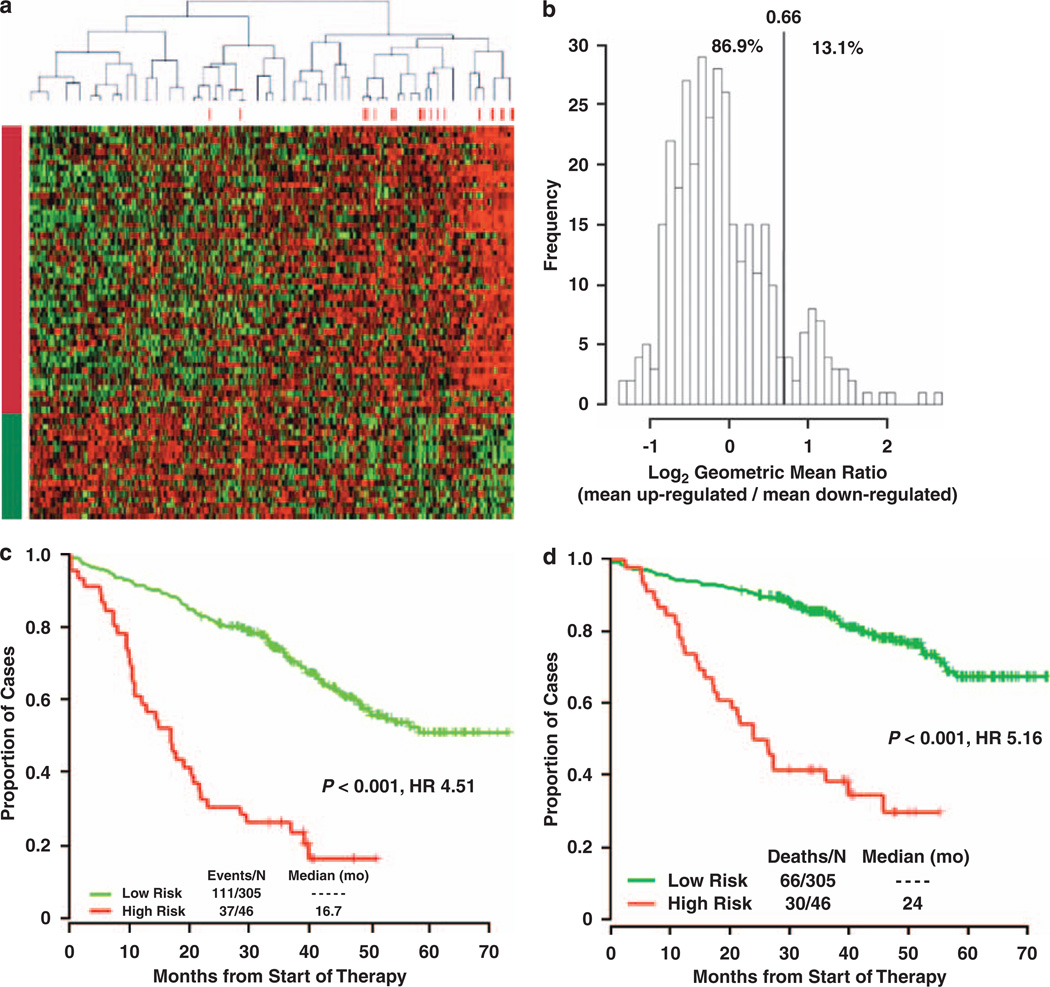
To determine whether GEP might provide a better measure of risk stratification, microarray data was correlated with outcome in two independent cohorts, permitting identification and validation of a high-risk gene-expression signature present in approximately 15% of newly diagnosed disease73 (Figure 2). The high-risk signature is evident in a subset of all molecular classes and negatively influences regardless of class—for example low-risk MS disease fares much better than high-risk MS disease. The ‘70/17-gene model’ of high risk is based on expression patterns of 70 genes, reducible to 17 genes—predominately increased expression of genes from the q arm and reduced expression of genes from the p arm of chromosome 173—which was confirmed by whole-genome microarrays and high-resolution comparative genomic hybridization (our unpublished data).
Figure 2.
A GEP-based 70-gene score can define high-risk myeloma. (a) Heat map of the 70 genes illustrate remarkably similar expression patterns in CD138+ selected tumor cells among 351 newly diagnosed patients. Red bars above the patient columns denote patients with disease-related deaths at the time of analysis. The 51 genes in rows designated by the red bar on the left (top rows; upregulated) identified patients in the upper quartile of expression at high risk for early disease-related death. The 19 gene rows designated by the green bar (downregulated), identified patients in the lower quartile of expression at high risk of early disease-related death. (b) Frequencies of the risk score defined as the log2 geometric mean ratio of the 51 quartile 4 genes and 19 quartile 1 genes. This self-normalizing expression ratio has a marked bimodal distribution, consistent with the upper/lower quartile log-rank differential expression analysis, which was designed to detect genes that define a single high-risk group (13.1%) with an extreme expression distribution. Interpreted as an up/downregulation ratio on the log2-scale, higher values are associated with poor outcome. The vertical line shows the high-risk versus low-risk cutoff for the log2-scale ratio determined by K-means clustering: the percentage of samples below and above the cutoff is also shown. Kaplan–Meier estimates of EFS (c) and OS (d) in low-risk myeloma (green) and high-risk myeloma (red) showed inferior 5-year actuarial probabilities of EFS (18% versus 60%, P<0.001; HR = 4.51) and OS (28% versus 78%, P <0.001; HR=5.16) in the 13.1% patients with a high-risk signature. Reproduced with Permission from Blood.
When subjected to multivariate analysis including the ISS and a gene-expression–based proliferation index, the 70/17-gene model remained a significant predictor of outcome. Improving on risk stratification provided by the ISS, Mulligan et al.105 used U133A microarray data to develop response and survival classifiers for relapsed disease treated with single-agent bortezomib or high-dose dexamethasone that were significantly associated with outcome; a modified version of the 70/17-gene model also predicted poor outcome in relapsed disease.106 U133A data from newly diagnosed disease validated the 70/17-gene model, but also showed that the t(4;14) translocation remained a significant variable for poor outcome.107 In addition to its ability to predict the outcome of newly diagnosed MM patients, a recent study showed that the 70-gene model was also an independent and the most significant prognostic factor in an analysis of post-relapse survival in relapsing MM.108
Decaux et al.109 recently used a custom cDNA microarray to define a 15-gene model of high risk related to cell proliferation, with a hyperdiploid signature being related to a better survival. Multivariate analysis comparing the 70/17-gene model with the 15-gene model revealed that the 70/17-gene model was significant in all datasets tested, but the 15-gene model was significant in bortezomib trials only. These data, together with unpublished studies, suggest that the 70/17-gene model captures more outcome variability than models or indexes of cell proliferation, considerably improving standard measures; however, it must be noted that the R2 is only ∼30%.
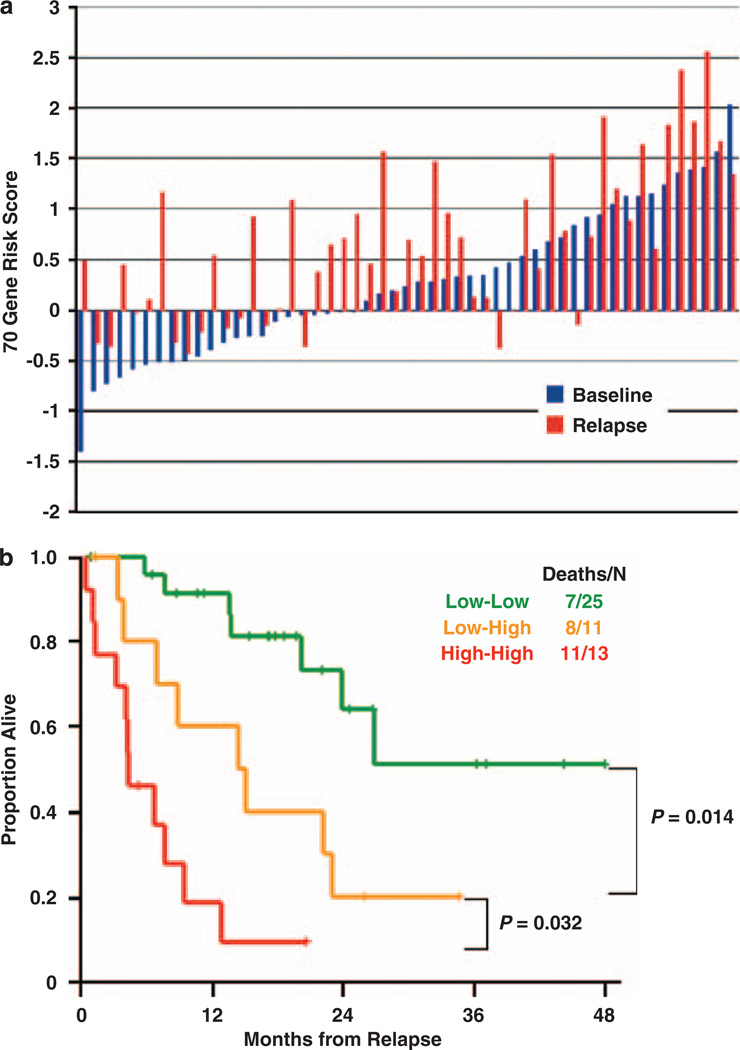
Conversion of GEP low to high risk at relapse
GEP on 71 paired diagnostic and relapse samples indicated increased 70/17-gene model scores in 80% of cases, and this conversion from low to high risk severely impacted post-relapse survival in 14 of 24 cases (58%) (Figure 3). This quantifiable increase in the high-risk score over time, combined with the report that percentages of cells with gains of chromosome 1q increased over time,73 suggests expansion of a dominant clone with survival and/or proliferation advantages. The almost universal increase in this risk score during disease evolution suggests that evaluating minimal residual disease may benefit from monitoring this traceable molecular signature, in combination with flow-cytometry–based surrogate. An urgent task is to determine whether a specific baseline GEP signature can prospectively identify which low-risk cases will convert to high risk at relapse.
Figure 3.
70-gene risk score can increase in relapsed relative to newly diagnosed disease and an increase predicts poor post-relapse survival. (a) The 70-gene risk score in paired diagnostic (blue) and relapse (red) samples of 51 patients. The gene expression risk score is indicated to the left. Sample pairs are order from left to right based on lowest baseline score. (b) Kaplan–Meier plot of post-relapse survival of the three groups defined by low risk both at diagnosis and relapse (low-low), low risk at diagnosis and high risk at relapse (low-high), and high risk at both time points (high-high). Reproduced with Permission from Blood.
GEP and the centrosome index
The mechanisms underlying aneuploidy in MM are unclear, but centrosome amplification has been implicated as the cause of chromosomal instability in a variety of tumors and may be involved in MM. Immunofluorescence staining detected centrosome amplification in 67% of monoclonal gammopathies.110 A GEP-based centrosome index (CI) was created from gene expression of centrosome proteins, which correlated well with immunofluorescence-detected centrosome amplification. High CI (>4) was associated with poor prognostic genetic features and was an independent prognostic factor in a small cohort of heterogeneously treated cases of MM.110 Prognostic significance of the CI was subsequently validated in two large cohorts of patients entered into clinical trials, showing that a high CI is a powerful independent prognostic factor in both newly diagnosed and relapsed patients, whether treated by intensive therapy or novel agents.111 Human myeloma cell lines with higher CIs are more responsive to treatment with a novel aurora kinase inhibitor, suggesting the potential for aurora kinases as novel therapeutic targets for patients with poor prognoses.111
GEP and CD200
Elevated expression of CD200 is an additional prognostic marker that emerged from GEP studies as a high-risk feature in MM.112 CD200 is a membrane glycoprotein that imparts an immunoregulatory signal through CD200R, leading to suppression of T-cell–mediated immune responses.113 CD200 expression was predictive for EFS independent of ISS stage or β-2-microglobin serum levels,112 but it has not yet been validated in independent datasets or evaluated in the context of OS and all molecular subtypes and models. As failure of immune surveillance may account for MM progression, CD200 modulation might be an important adjunct to cellular immunotherapy. As a cell-surface protein, CD200 is a potential target for monoclonal antibody therapy, but such strategies will have to consider off-target effects and the critical function of this molecule in immune regulation.
Using GEP to understand signaling in disease and its response to therapy
GEP and cancer-testis antigen expression in MM
Cancer-testis antigens are expressed in testis and malignant tumors, but rarely in nongametogenic tissues, which makes them attractive targets for cancer vaccination approaches. GEP studies determined that patients newly diagnosed with MM expressed variable numbers of cancer-testis genes (98% expressed at least one, 86% at least two, and 70% at least three) and that expression of six or more cancer-testis genes was associated with shorter EFS.114 Ten cancer-testis genes are desirable for circumventing tumor escape mechanisms, and GEP could be useful in identifying which antigens should be used to vaccinate a given patient.114 Additional independent studies confirmed the association of cancer-testis antigen expression and poor outcome in patients with MM.115,116 Global GEP studies showed that cancer-testis antigen NY-ESO-1 could be an ideal tumor target antigen for immunotherapy of patients with MM. NY-ESO-1 expression was higher in tumor cells from patients with CAs than in those with no CAs,117 and NY-ESO-1 expression was significantly higher in cases of relapsing MM, especially in patients with CA.
GEP and IGF signaling
IGF-1R have been implicated in cancer pathophysiology, and IGF-1R is universally expressed in various cells of hematologic malignancies (i.e. MM, lymphoma, leukemia) and solid tumors. Specific in vitro inhibition of IGF-1R with neutralizing antibody, antagonistic peptide, or selective kinase inhibitor (NVP-ADW742) has activity against diverse tumor-cell types (particularly MM), even those resistant to conventional therapies. Global transcriptional profiles also delineated pleiotropic antiproliferative/pro-apoptotic molecular sequelae— specifically, modulated intracellular concentrations of key components of these pathways, including Akt, Raf, and IKK. Therapies with NVP-ADW742 or GSK1904529A, alone or in combination with cytotoxic chemotherapy, had significant antitumor activity in an orthotopic xenograft MM model, providing in vivo proof of principle for therapeutic use of selective IGF-1R inhibitors.118,119 Sprynski et al.120 showed that an IGF-1 autocrine loop promoted survival in CD45-negative MM cell lines, whereas CD45-positive cells required addition of either IL-6 or IGF-1. GEP analysis in primary disease revealed that elevated expression of IGF-1R and IL6R conferred an adverse prognosis. High expression of both IGF-1R and IL6R is seen in the MS class, but elevated IGF-1R expression is also seen in non-MS classes and is associated with a poor prognosis. Combining IGF-1–targeted therapy with anti-IL-6 therapy could be promising in the subset of patients with myeloma cells that express IGF-1R.
Pharmacogenomics of short-term in vivo exposure can reveal drug efficacies and mechanisms of action
Mechanisms of cancer cell resistance to chemotherapy are poorly understood, and efficacy measures typically rely on clinical outcome data; however, GEP studies potentially can delineate mechanisms and identify novel strategies for avoiding or overcoming drug resistance, which is a major hurdle for MM therapy and cure. Marton et al.121 and Gray et al.122 were the first to use microarrays to discover targets and effects of therapeutic agents in yeast, and Cheok et al.123 first revealed gene-expression patterns in drug responses of human cancer.
GEP has been used to study drug responses of MM cells in vitro. It was first used to identify genes regulated by two FDA-approved drugs, dexamethasone and bortezomib, and genes that may confer drug resistance in MM cells.124–126 Numerous other studies have since been reported. The Dana–Farber group used GEP to identify the transcriptional signatures of a bunch of distinct drugs in MM celles, such as 2-methoxyestradiol (2ME2), an estrogen derivative,127 two histone deacetylase inhibitors, SAHA and VPA,128, 129 IGF-1R) inhibitors,118 a IL-6 receptor inhibitor, Sant7,130 an inhibitor of Wnt signaling pathway, PKF115–584,131 ribonucleotide reductase inhibitor, Didox,132 and Atiprimod, a oral agent with anti-inflammatory properties.133 Using GEP, the transcriptional signatures of cellular responses to 5-aza-2′-deoxycytidine and trichostatin,134 a HSP90 inhibitor, 17-AAG,135 and a marine-derived compound, Zalypsis,136 have recently been described.
Recently, GEP has been successfully used to study drug response of MM patients in vivo. By comparing GEP of myeloma cells before and 48 h after single-agent therapy with dexamethasone, thalidomide, or lenalidomide, Burington et al. found that genes differentially expressed after therapy were prognostic for EFS and OS and were enriched for genes involved in oxidative stress reactions and actin cytoskeleton rearrangements.137 Remarkably, gene expression altered by thalidomide in newly diagnosed disease and associated with subsequent survival was also altered by lenalidomide, a thalidomide analogue, and the changes were associated with EFS in a salvage trial of patients with relapsed disease.137 This finding strongly suggests that these genes are powerful biomarkers, and the similar acute gene-expression responses to two related chemotherapeutic agents may provide important insights into the drugs’ potential mechanism(s) of action. These results also highlight the similar acute molecular responses to chemotherapies in both primary and refractory diseases.
GEP studies after therapy with proteasome inhibitor bortezomib in 142 newly diagnosed symptomatic MM cases identified 113 genes with significantly altered expression—predominately downregulated proteasome genes—seen in tumor cells from 76% of patients.138 The post-bortezomib gene-expression signature was associated with a 3-year survival estimate of >80%, which dramatically contrasts a median survival of <24 months in those with activated proteasome genes.138 Multivariate analysis showed that the post-bortezomib score was an independent predictor of outcome that alone accounted for >50% of outcome variability.138 These data implied that the activation status of proteasome genes in tumor cells after short-term proteasome inhibition is associated with significant outcome differences in patients with MM receiving polychemotherapy that includes bortezomib.138
Conclusions
Using high-throughput genomic analyses and data-mining techniques, a complete landscape of MM molecular pathogenesis is emerging, and powerful validated prognostic models have been developed. Unsupervised clustering of GEP data of large patient cohorts also have revealed that MM heterogeneity can be accurately cataloged and disease classes defined. Importantly, the improved survival observed in specific classes through the use of new treatments, such as thalidomide and bortezomib, buttress the concept of personalized treatment approaches.
Large-scale gene-expression data and large cohorts of uniformly treated patients with long follow-up times have provided more precise and independent prognostic models for stratifying patients with MM. Investigating GEP changes between baseline and relapse has shed light on the mechanisms underlying MM progression and the nearly universal development of multidrug-resistant MM. Pharmacogenomics studies comparing gene-expression profiles at diagnosis and after short-term single-agent therapy have identified genes associated with drug responses, contributing to mechanistic understanding. This is critical for improving existing therapies with personalized treatments and for informing research and discovery of new therapeutics.
It is well known that MM growth and survival are highly dependent on interactions with the BMME, and GEP has uncovered many molecular details of these interactions, which might prove to be the Achilles heels of MM. A prominent example was the use of GEP and MRI imaging of bone to learn that myeloma cells aberrantly synthesize DKK1, a potent inhibitor of Wnt/β-catenin signaling, which is required for osteoblast differentiation and function. These data provided a potential underlying mechanism for the unique MM osteolytic bone disease characterized by complete loss of osteoblast function. An inhibitor of DKK1 is in early Phase I/II clinical trials, attesting to the translational potential of GEP studies. GEP of whole-bone biopsies that contain tumor cells and all the accessory cells are also beginning to reveal details of MM pathogenesis and potential therapeutic targets.
After 10 years of applying GEP to thousands of patient samples in numerous institutions, GEP is emerging from the research laboratory as a clinical tool with the potential for transforming routine management of MM. Although the majority of MM patients can anticipate long-term disease control through a variety of treatment approaches, patients with molecularly defined high-risk disease do not benefit from current approaches. To address this, clinical trials designed to reduce toxicities in low-risk disease and to test new treatment strategies in high-risk disease are underway. When routinely available, molecular-based classification and risk stratification will meet their potential to shift strategies for MM treatment and cure.
We have made every effort to reference as much of the literature on the molecular characterization of myeloma as possible. However, because of size limitations and the rapidly growing body of work, it is likely that we have failed to include some of the works of our coworkers. We apologize for this oversight.
Acknowledgements
This work was supported by the National Cancer Institute (grants CA55819-09 and CA97513-01), the Lebow Fund to Cure Myeloma, and the Nancy and Stephen Grand Fund. The manuscript was edited by the Office of Grants and Scientific Publications, University of Arkansas for Medical Sciences.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- 1.Barlogie B, Shaughnessy J, Epstein J, Sanderson R, Anaissie E, Walker R, et al. Plasma cell myeloma. In: Lichtman MA, Beutler E, Kaushansky K, Kipps TJ, Seligsohn U, Prchal J, editors. Williams Hematology. 7 edn. New York: McGraw-Hill Professional; 2005. pp. 1501–1533. [Google Scholar]
- 2.Ribatti D, Nico B, Vacca A. Importance of the bone marrow microenvironment in inducing the angiogenic response in multiple myeloma. Oncogene. 2006;25:4257–4266. doi: 10.1038/sj.onc.1209456. [DOI] [PubMed] [Google Scholar]
- 3.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 4.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, et al. Diffuse large B-cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 5.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–197. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 6.Ross ME, Zhou X, Song G, Shurtleff SA, Girtman K, Williams WK, et al. Classification of pediatric acute lymphoblastic leukemia by gene expression profiling. Blood. 2003;102:2951–2959. doi: 10.1182/blood-2003-01-0338. [DOI] [PubMed] [Google Scholar]
- 7.Bullinger L, Dohner K, Bair E, Frohling S, Schlenk RF, Tibshirani R, et al. Use of gene-expression profiling to identify prognostic subclasses in adult acute myeloid leukemia. N Engl J Med. 2004;350:1605–1616. doi: 10.1056/NEJMoa031046. [DOI] [PubMed] [Google Scholar]
- 8.Valk PJ, Verhaak RG, Beijen MA, Erpelinck CA, Barjesteh van Waalwijk van Doorn-Khosrovani S, Boer JM, et al. Prognostically useful gene-expression profiles in acute myeloid leukemia. N Engl J Med. 2004;350:1617–1628. doi: 10.1056/NEJMoa040465. [DOI] [PubMed] [Google Scholar]
- 9.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, et al. Prediction of survival in diffuse large-B-cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 10.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N Engl J Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 11.Dave SS, Fu K, Wright GW, Lam LT, Kluin P, Boerma EJ, et al. Molecular diagnosis of Burkitt′s lymphoma. N Engl J Med. 2006;354:2431–2442. doi: 10.1056/NEJMoa055759. [DOI] [PubMed] [Google Scholar]
- 12.Tricot G, Barlogie B, Jagannath S, Bracy D, Mattox S, Vesole DH, et al. Poor prognosis in multiple myeloma is associated only with partial or complete deletions of chromosome 13 or abnormalities involving 11q and not with other karyotype abnormalities. Blood. 1995;86:4250–4256. [PubMed] [Google Scholar]
- 13.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 14.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. Science. Vol. 291. New York, NY: 2001. The sequence of the human genome; pp. 1304–1351. [DOI] [PubMed] [Google Scholar]
- 15.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 16.Schena M, Shalon D, Davis RW, Brown PO. Science. Vol. 270. New York, NY: 1995. Quantitative monitoring of gene expression patterns with a complementary DNA microarray; pp. 467–470. [DOI] [PubMed] [Google Scholar]
- 17.Shalon D, Smith SJ, Brown PO. A DNA microarray system for analyzing complex DNA samples using two-color fluorescent probe hybridization. Genome Res. 1996;6:639–645. doi: 10.1101/gr.6.7.639. [DOI] [PubMed] [Google Scholar]
- 18.Schena M, Shalon D, Heller R, Chai A, Brown PO, Davis RW. Parallel human genome analysis: microarray-based expression monitoring of 1000 genes. Proc Natl Acad Sci USA. 1996;93:10614–10619. doi: 10.1073/pnas.93.20.10614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fodor SP, Read JL, Pirrung MC, Stryer L, Lu AT, Solas D. Science. Vol. 251. New York, NY: 1991. Light-directed, spatially addressable parallel chemical synthesis; pp. 767–773. [DOI] [PubMed] [Google Scholar]
- 20.Lipshutz RJ, Fodor SP, Gingeras TR, Lockhart DJ. High density synthetic oligonucleotide arrays. Nat Genet. 1999;21(1 Suppl):20–24. doi: 10.1038/4447. [DOI] [PubMed] [Google Scholar]
- 21.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med. 2006;354:2463–2472. doi: 10.1056/NEJMra042342. [DOI] [PubMed] [Google Scholar]
- 22.DeRisi J, Penland L, Brown PO, Bittner ML, Meltzer PS, Ray M, et al. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 23.De Vos J, Couderc G, Tarte K, Jourdan M, Requirand G, Delteil MC, et al. Identifying intercellular signaling genes expressed in malignant plasma cells by using complementary DNA arrays. Blood. 2001;98:771–780. doi: 10.1182/blood.v98.3.771. [DOI] [PubMed] [Google Scholar]
- 24.Claudio JO, Masih-Khan E, Tang H, Goncalves J, Voralia M, Li ZH, et al. A molecular compendium of genes expressed in multiple myeloma. Blood. 2002;100:2175–2186. doi: 10.1182/blood-2002-01-0008. [DOI] [PubMed] [Google Scholar]
- 25.Zhan F, Hardin J, Kordsmeier B, Bumm K, Zheng M, Tian E, et al. Global gene expression profiling of multiple myeloma, monoclonal gammopathy of undetermined significance, and normal bone marrow plasma cells. Blood. 2002;99:1745–1757. doi: 10.1182/blood.v99.5.1745. [DOI] [PubMed] [Google Scholar]
- 26.Zhan F, Tian E, Bumm K, Smith R, Barlogie B, Shaughnessy J., Jr Gene expression profiling of human plasma cell differentiation and classification of multiple myeloma based on similarities to distinct stages of late-stage B-cell development. Blood. 2003;101:1128–1140. doi: 10.1182/blood-2002-06-1737. [DOI] [PubMed] [Google Scholar]
- 27.Tarte K, De Vos J, Thykjaer T, Zhan F, Fiol G, Costes V, et al. Generation of polyclonal plasmablasts from peripheral blood B cells: a normal counterpart of malignant plasmablasts. Blood. 2002;100:1113–1122. [PubMed] [Google Scholar]
- 28.Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102:592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 29.Bergsagel PL, Kuehl WM. Molecular pathogenesis and a consequent classification of multiple myeloma. J Clin Oncol. 2005;23:6333–6338. doi: 10.1200/JCO.2005.05.021. [DOI] [PubMed] [Google Scholar]
- 30.Mattioli M, Agnelli L, Fabris S, Baldini L, Morabito F, Bicciato S, et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene. 2005;24:2461–2473. doi: 10.1038/sj.onc.1208447. [DOI] [PubMed] [Google Scholar]
- 31.Jourdan M, Reme T, Goldschmidt H, Fiol G, Pantesco V, De Vos J, et al. Gene expression of anti- and pro-apoptotic proteins in malignant and normal plasma cells. Br J Haematol. 2009;145:45–58. doi: 10.1111/j.1365-2141.2008.07562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dalton WS. The tumor microenvironment: focus on myeloma. Cancer Treat Rev. 2003;29(Suppl 1):11–19. doi: 10.1016/s0305-7372(03)00077-x. [DOI] [PubMed] [Google Scholar]
- 33.Corre J, Mahtouk K, Attal M, Gadelorge M, Huynh A, Fleury-Cappellesso S, et al. Bone marrow mesenchymal stem cells are abnormal in multiple myeloma. Leukemia. 2007;21:1079–1088. doi: 10.1038/sj.leu.2404621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perez-Andres M, Almeida J, Martin-Ayuso M, Moro MJ, Martin-Nunez G, Galende J, et al. Clonal plasma cells from monoclonal gammopathy of undetermined significance, multiple myeloma and plasma cell leukemia show different expression profiles of molecules involved in the interaction with the immunological bone marrow microenvironment. Leukemia. 2005;19:449–455. doi: 10.1038/sj.leu.2403647. [DOI] [PubMed] [Google Scholar]
- 35.Mundy GR. Metastasis to bone: causes, consequences and therapeutic opportunities. Nat Rev. 2002;2:584–593. doi: 10.1038/nrc867. [DOI] [PubMed] [Google Scholar]
- 36.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 37.Pinzone JJ, Hall BM, Thudi NK, Vonau M, Qiang YW, Rosol TJ, et al. The role of Dickkopf-1 in bone development, homeostasis, and disease. Blood. 2009;113:517–525. doi: 10.1182/blood-2008-03-145169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jakob C, Sterz J, Zavrski I, Heider U, Kleeberg L, Fleissner C, et al. Angiogenesis in multiple myeloma. Eur J Cancer. 2006;42:1581–1590. doi: 10.1016/j.ejca.2006.02.017. [DOI] [PubMed] [Google Scholar]
- 39.Kumar S, Gertz MA, Dispenzieri A, Lacy MQ, Wellik LA, Fonseca R, et al. Prognostic value of bone marrow angiogenesis in patients with multiple myeloma undergoing high-dose therapy. Bone Marrow Transplant. 2004;34:235–239. doi: 10.1038/sj.bmt.1704555. [DOI] [PubMed] [Google Scholar]
- 40.Vacca A, Scavelli C, Montefusco V, Di Pietro G, Neri A, Mattioli M, et al. Thalidomide downregulates angiogenic genes in bone marrow endothelial cells of patients with active multiple myeloma. J Clin Oncol. 2005;23:5334–5346. doi: 10.1200/JCO.2005.03.723. [DOI] [PubMed] [Google Scholar]
- 41.Hedvat CV, Comenzo RL, Teruya-Feldstein J, Olshen AB, Ely SA, Osman, et al. Insights into extramedullary tumour cell growth revealed by expression profiling of human plasmacytomas and multiple myeloma. Br J Haematol. 2003;122:728–744. doi: 10.1046/j.1365-2141.2003.04481.x. [DOI] [PubMed] [Google Scholar]
- 42.Munshi NC, Hideshima T, Carrasco D, Shammas M, Auclair D, Davies F, et al. Identification of genes modulated in multiple myeloma using genetically identical twin samples. Blood. 2004;103:1799–1806. doi: 10.1182/blood-2003-02-0402. [DOI] [PubMed] [Google Scholar]
- 43.Hose D, Moreaux J, Meissner T, Seckinger A, Goldschmidt H, Benner A, et al. Induction of angiogenesis by normal and malignant plasma cells. Blood. 2009;114:128–143. doi: 10.1182/blood-2008-10-184226. [DOI] [PubMed] [Google Scholar]
- 44.Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, et al. Heparanase degrades syndecan-1 and perlecan heparan sulfate: functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 45.Kelly T, Miao HQ, Yang Y, Navarro E, Kussie P, Huang Y, et al. High heparanase activity in multiple myeloma is associated with elevated microvessel density. Cancer Res. 2003;63:8749–8756. [PubMed] [Google Scholar]
- 46.Mahtouk K, Hose D, Raynaud P, Hundemer M, Jourdan M, Jourdan E, et al. Heparanase influences expression and shedding of syndecan-1, and its expression by the bone marrow environment is a bad prognostic factor in multiple myeloma. Blood. 2007;109:4914–4923. doi: 10.1182/blood-2006-08-043232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bret C, Hose D, Reme T, Sprynski AC, Mahtouk K, Schved JF, et al. Expression of genes encoding for proteins involved in heparan sulphate and chondroitin sulphate chain synthesis and modification in normal and malignant plasma cells. Br J Haematol. 2009;145:350–368. doi: 10.1111/j.1365-2141.2009.07633.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mahtouk K, Cremer FW, Reme T, Jourdan M, Baudard M, Moreaux J, et al. Heparan sulphate proteoglycans are essential for the myeloma cell growth activity of EGF-family ligands in multiple myeloma. Oncogene. 2006;25:7180–7191. doi: 10.1038/sj.onc.1209699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moreaux J, Legouffe E, Jourdan E, Quittet P, Reme T, Lugagne C, et al. BAFF and APRIL protect myeloma cells from apoptosis induced by interleukin 6 deprivation and dexamethasone. Blood. 2004;103:3148–3157. doi: 10.1182/blood-2003-06-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Novak AJ, Darce JR, Arendt BK, Harder B, Henderson K, Kindsvogel W, et al. Expression of BCMA, TACI, and BAFF-R in multiple myeloma: a mechanism for growth and survival. Blood. 2004;103:689–694. doi: 10.1182/blood-2003-06-2043. [DOI] [PubMed] [Google Scholar]
- 51.Moreaux J, Cremer FW, Reme T, Raab M, Mahtouk K, Kaukel P, et al. The level of TACI gene expression in myeloma cells is associated with a signature of microenvironment dependence versus a plasmablastic signature. Blood. 2005;106:1021–1030. doi: 10.1182/blood-2004-11-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ge Y, Zhan F, Barlogie B, Epstein J, Shaughnessy J, Jr, Yaccoby S. Fibroblast activation protein (FAP) is upregulated in myelomatous bone and supports myeloma cell survival. Br J Haematol. 2006;133:83–92. doi: 10.1111/j.1365-2141.2006.05976.x. [DOI] [PubMed] [Google Scholar]
- 53.Bergsagel PL, Kuehl WM, Zhan F, Sawyer J, Barlogie B, Shaughnessy J., Jr Cyclin D dysregulation: an early and unifying pathogenic event in multiple myeloma. Blood. 2005;106:296–303. doi: 10.1182/blood-2005-01-0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chesi M, Bergsagel PL, Brents LA, Smith CM, Gerhard DS, Kuehl WM. Dysregulation of cyclin D1 by translocation into an IgH gamma switch region in two multiple myeloma cell lines. Blood. 1996;88:674–681. [PubMed] [Google Scholar]
- 55.Shaughnessy J, Jr, Gabrea A, Qi Y, Brents L, Zhan F, Tian E, et al. Cyclin D3 at 6p21 is dysregulated by recurrent chromosomal translocations to immunoglobulin loci in multiple myeloma. Blood. 2001;98:217–223. doi: 10.1182/blood.v98.1.217. [DOI] [PubMed] [Google Scholar]
- 56.Hurt EM, Wiestner A, Rosenwald A, Shaffer AL, Campo E, Grogan T, et al. Overexpression of c-maf is a frequent oncogenic event in multiple myeloma that promotes proliferation and pathological interactions with bone marrow stroma. Cancer Cell. 2004;5:191–199. doi: 10.1016/s1535-6108(04)00019-4. [DOI] [PubMed] [Google Scholar]
- 57.Zhan F, Huang Y, Colla S, Stewart JP, Hanamura I, Gupta S, et al. The molecular classification of multiple myeloma. Blood. 2006;108:2020–2028. doi: 10.1182/blood-2005-11-013458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanamura I, Huang Y, Zhan F, Barlogie B, Shaughnessy J. Prognostic value of cyclin D2 mRNA expression in newly diagnosed multiple myeloma treated with high-dose chemotherapy and tandem autologous stem cell transplantations. Leukemia. 2006;20:1288–1290. doi: 10.1038/sj.leu.2404253. [DOI] [PubMed] [Google Scholar]
- 59.Agnelli L, Bicciato S, Mattioli M, Fabris S, Intini D, Verdelli D, et al. Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23:7296–7306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- 60.Kuehl WM, Bergsagel PL. Multiple myeloma: evolving genetic events and host interactions. Nat Rev. 2002;2:175–187. doi: 10.1038/nrc746. [DOI] [PubMed] [Google Scholar]
- 61.Keats JJ, Reiman T, Maxwell CA, Taylor BJ, Larratt LM, Mant MJ, et al. In multiple myeloma, t(4;14)(p16;q32) is an adverse prognostic factor irrespective of FGFR3 expression. Blood. 2003;101:1520–1529. doi: 10.1182/blood-2002-06-1675. [DOI] [PubMed] [Google Scholar]
- 62.Chesi M, Nardini E, Lim RS, Smith KD, Kuehl WM, Bergsagel PL. The t(4;14) translocation in myeloma dysregulates both FGFR3 and a novel gene, MMSET, resulting in IgH/MMSET hybrid transcripts. Blood. 1998;92:3025–3034. [PubMed] [Google Scholar]
- 63.Santra M, Zhan F, Tian E, Barlogie B, Shaughnessy J., Jr A subset of multiple myeloma harboring the t(4;14)(p16;q32) translocation lacks FGFR3 expression but maintains an IGH/MMSET fusion transcript. Blood. 2003;101:2374–2376. doi: 10.1182/blood-2002-09-2801. [DOI] [PubMed] [Google Scholar]
- 64.Dring AM, Davies FE, Fenton JA, Roddam PL, Scott K, Gonzalez D, et al. A global expression-based analysis of the consequences of the t(4;14) translocation in myeloma. Clin Cancer Res. 2004;10:5692–5701. doi: 10.1158/1078-0432.CCR-04-0467. [DOI] [PubMed] [Google Scholar]
- 65.Brito JL, Walker B, Jenner M, Dickens NJ, Brown NJ, Ross FM, et al. MMSET deregulation affects cell cycle progression and adhesion regulons in t(4;14) myeloma plasma cells. Haematologica. 2009;94:78–86. doi: 10.3324/haematol.13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, et al. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell. 2007;12:115–130. doi: 10.1016/j.ccr.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.van Stralen E, van de Wetering M, Agnelli L, Neri A, Clevers HC, Bast BJ. Identification of primary MAFB target genes in multiple myeloma. Exp Hematol. 2009;37:78–86. doi: 10.1016/j.exphem.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 68.Suzuki A, Iida S, Kato-Uranishi M, Tajima E, Zhan F, Hanamura I, et al. ARK5 is transcriptionally regulated by the large-MAF family and mediates IGF-1-induced cell invasion in multiple myeloma: ARK5 as a new molecular determinant of malignant multiple myeloma. Oncogene. 2005;24:6936–6944. doi: 10.1038/sj.onc.1208844. [DOI] [PubMed] [Google Scholar]
- 69.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tai YT, Soydan E, Song W, Fulciniti M, Kim K, Hong F, et al. CS1 promotes multiple myeloma cell adhesion, clonogenic growth, and tumorigenicity via c-maf-mediated interactions with bone marrow stromal cells. Blood. 2009;113:4309–4318. doi: 10.1182/blood-2008-10-183772. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 71.Tai YT, Dillon M, Song W, Leiba M, Li XF, Burger P, et al. Anti-CS1 humanized monoclonal antibody HuLuc63 inhibits myeloma cell adhesion and induces antibody-dependent cellular cytotoxicity in the bone marrow milieu. Blood. 2008;112:1329–1337. doi: 10.1182/blood-2007-08-107292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lin P, Mahdavy M, Zhan F, Zhang HZ, Katz RL, Shaughnessy JD. Expression of PAX5 in CD20-positive multiple myeloma assessed by immunohistochemistry and oligonucleotide microarray. Mod Pathol. 2004;17:1217–1222. doi: 10.1038/modpathol.3800169. [DOI] [PubMed] [Google Scholar]
- 73.Shaughnessy JD, Jr, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- 74.Carrasco DR, Tonon G, Huang Y, Zhang Y, Sinha R, Feng B, et al. High-resolution genomic profiles define distinct clinico-pathogenetic subgroups of multiple myeloma patients. Cancer Cell. 2006;9:313–325. doi: 10.1016/j.ccr.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 75.Bartel TB, Haessler J, Brown TL, Shaughnessy JD, Jr, van Rhee F, Anaissie E, et al. F18-fluorodeoxyglucose positron emission tomography in the context of other imaging techniques and prognostic factors in multiple myeloma. Blood. 2009 doi: 10.1182/blood-2009-03-213280. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yin JJ, Mohammad KS, Kakonen SM, Harris S, Wu-Wong JR, Wessale JL, et al. A causal role for endothelin-1 in the pathogenesis of osteoblastic bone metastases. Proc Natl Acad Sci USA. 2003;100:10954–10959. doi: 10.1073/pnas.1830978100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Clines GA, Mohammad KS, Bao Y, Stephens OW, Suva LJ, Shaughnessy JD., Jr . Mol Endocrinol. Vol. 21. Baltimore, Md: 2007. Dickkopf homolog 1 mediates endothelin-1-stimulated new bone formation; pp. 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Agnelli L, Fabris S, Bicciato S, Basso D, Baldini L, Morabito F, et al. Upregulation of translational machinery and distinct genetic subgroups characterise hyperdiploidy in multiple myeloma. Br J Haematol. 2007;136:565–573. doi: 10.1111/j.1365-2141.2006.06467.x. [DOI] [PubMed] [Google Scholar]
- 79.Chng WJ, Kumar S, Vanwier S, Ahmann G, Price-Troska T, Henderson K, et al. Molecular dissection of hyperdiploid multiple myeloma by gene expression profiling. Cancer Res. 2007;67:2982–2989. doi: 10.1158/0008-5472.CAN-06-4046. [DOI] [PubMed] [Google Scholar]
- 80.Zhou Y, Barlogie B, Herman D, Stephens O, Tian E, Williams D, et al. Integration of DNA copy number and gene expression alteration reveal novel insights into the molecular pathogenesis and prognosis of multiple myeloma. Blood (ASH Annual Meeting Abstracts) 2008;12:250. [Google Scholar]
- 81.Shaughnessy J, Jacobson J, Sawyer J, McCoy J, Fassas A, Zhan F, et al. Continuous absence of metaphase-defined cytogenetic abnormalities, especially of chromosome 13 and hypodiploidy, ensures long-term survival in multiple myeloma treated with total therapy I: interpretation in the context of global gene expression. Blood. 2003;101:3849–3856. doi: 10.1182/blood-2002-09-2873. [DOI] [PubMed] [Google Scholar]
- 82.Agnelli L, Bicciato S, Fabris S, Baldini L, Morabito F, Intini D, et al. Integrative genomic analysis reveals distinct transcriptional and genetic features associated with chromosome 13 deletion in multiple myeloma. Haematologica. 2007;92:56–65. doi: 10.3324/haematol.10414. [DOI] [PubMed] [Google Scholar]
- 83.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–1732. doi: 10.1182/blood-2006-03-009910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rosinol L, Carrio A, Blade J, Queralt R, Aymerich M, Cibeira MT, et al. Comparative genomic hybridisation identifies two variants of smoldering multiple myeloma. Br J Haematol. 2005;130:729–732. doi: 10.1111/j.1365-2141.2005.05673.x. [DOI] [PubMed] [Google Scholar]
- 85.Fabris S, Ronchetti D, Agnelli L, Baldini L, Morabito F, Bicciato S, et al. Transcriptional features of multiple myeloma patients with chromosome 1q gain. Leukemia. 2007;21:1113–1116. doi: 10.1038/sj.leu.2404616. [DOI] [PubMed] [Google Scholar]
- 86.Agnelli L, Mosca L, Fabris S, Lionetti M, Andronache A, Kwee I, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer. 2009;48:603–614. doi: 10.1002/gcc.20668. [DOI] [PubMed] [Google Scholar]
- 87.Carew JS, Nawrocki ST, Krupnik YV, Dunner K, Jr, McConkey DJ, Keating MJ, et al. Targeting endoplasmic reticulum protein transport: a novel strategy to kill malignant B cells and overcome fludarabine resistance in CLL. Blood. 2006;107:222–231. doi: 10.1182/blood-2005-05-1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chng WJ, Price-Troska T, Gonzalez-Paz N, Van Wier S, Jacobus S, Blood E, et al. Clinical significance of TP53 mutation in myeloma. Leukemia. 2007;21:582–584. doi: 10.1038/sj.leu.2404524. [DOI] [PubMed] [Google Scholar]
- 89.Xiong W, Wu X, Starnes S, Johnson SK, Haessler J, Wang S, et al. An analysis of the clinical and biological significance of TP53 loss and the identification of potential novel transcriptional targets of TP53 in multiple myeloma. Blood. 2008;112:4235–4246. doi: 10.1182/blood-2007-10-119123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cigudosa JC, Rao PH, Calasanz MJ, Odero MD, Michaeli J, Jhanwar SC, et al. Characterization of nonrandom chromosomal gains and losses in multiple myeloma by comparative genomic hybridization. Blood. 1998;91:3007–3010. [PubMed] [Google Scholar]
- 91.Gutierrez NC, Garcia JL, Hernandez JM, Lumbreras E, Castella-nos M, Rasillo A, et al. Prognostic and biologic significance of chromosomal imbalances assessed by comparative genomic hybridization in multiple myeloma. Blood. 2004;104:2661–2666. doi: 10.1182/blood-2004-04-1319. [DOI] [PubMed] [Google Scholar]
- 92.Avet-Loiseau H, Andree-Ashley LE, Moore D, II, Mellerin MP, Feusner J, Bataille R, et al. Molecular cytogenetic abnormalities in multiple myeloma and plasma cell leukemia measured using comparative genomic hybridization. Genes Chromosomes Cancer. 1997;19:124–133. doi: 10.1002/(sici)1098-2264(199706)19:2<124::aid-gcc8>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 93.Houldsworth J, Chaganti RS. Comparative genomic hybridization: an overview. Am J Pathol. 1994;145:1253–1260. [PMC free article] [PubMed] [Google Scholar]
- 94.Barrett MT, Scheffer A, Ben-Dor A, Sampas N, Lipson D, Kincaid R, et al. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. Proc Natl Acad Sci USA. 2004;101:17765–17770. doi: 10.1073/pnas.0407979101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pollack JR, Perou CM, Alizadeh AA, Eisen MB, Pergamenschikov A, Williams CF, et al. Genome-wide analysis of DNA copy-number changes using cDNA microarrays. Nat Genet. 1999;23:41–46. doi: 10.1038/12640. [DOI] [PubMed] [Google Scholar]
- 96.Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, et al. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. [DOI] [PubMed] [Google Scholar]
- 97.Fabris S, Todoerti K, Mosca L, Agnelli L, Intini D, Lionetti M, et al. Molecular and transcriptional characterization of the novel 17p11.2-p12 amplicon in multiple myeloma. Genes Chromosomes Cancer. 2007;46:1109–1118. doi: 10.1002/gcc.20494. [DOI] [PubMed] [Google Scholar]
- 98.Jenner MW, Leone PE, Walker BA, Ross FM, Johnson DC, Gonzalez D, et al. Gene mapping and expression analysis of 16q loss of heterozygosity identifies WWOX and CYLD as being important in determining clinical outcome in multiple myeloma. Blood. 2007;110:3291–3300. doi: 10.1182/blood-2007-02-075069. [DOI] [PubMed] [Google Scholar]
- 99.Salmon SE, Durie BG. Clinical staging and new therapeutic approaches in multiple myeloma. Recent Results Cancer Res. 1978;65:12–20. doi: 10.1007/978-3-642-81249-1_3. [DOI] [PubMed] [Google Scholar]
- 100.Greipp PR, San Miguel J, Durie BG, Crowley JJ, Barlogie B, Blade J, et al. International staging system for multiple myeloma. J Clin Oncol. 2005;23:3412–3420. doi: 10.1200/JCO.2005.04.242. [DOI] [PubMed] [Google Scholar]
- 101.Bartl R. Histologic classification and staging of multiple myeloma. Hematol Oncol. 1988;6:107–113. doi: 10.1002/hon.2900060209. [DOI] [PubMed] [Google Scholar]
- 102.Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- 103.Greipp PR, Kumar S. Plasma cell labeling index. Methods Mol Med. 2005;113:25–35. doi: 10.1385/1-59259-916-8:25. [DOI] [PubMed] [Google Scholar]
- 104.Paiva B, Vidriales MB, Cervero J, Mateo G, Perez JJ, Montalban MA, et al. Multiparameter flow cytometric remission is the most relevant prognostic factor for multiple myeloma patients who undergo autologous stem cell transplantation. Blood. 2008;112:4017–4023. doi: 10.1182/blood-2008-05-159624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mulligan G, Mitsiades C, Bryant B, Zhan F, Chng WJ, Roels S, et al. Gene expression profiling and correlation with outcome in clinical trials of the proteasome inhibitor bortezomib. Blood. 2007;109:3177–3188. doi: 10.1182/blood-2006-09-044974. [DOI] [PubMed] [Google Scholar]
- 106.Zhan F, Barlogie B, Mulligan G, Shaughnessy JD, Jr, Bryant B. High-risk myeloma: a gene expression based risk-stratification model for newly diagnosed multiple myeloma treated with high-dose therapy is predictive of outcome in relapsed disease treated with single-agent bortezomib or high-dose dexamethasone. Blood. 2008;111:968–969. doi: 10.1182/blood-2007-10-119321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Chng WJ, Kuehl WM, Bergsagel PL, Fonseca R. Translocation t(4;14) retains prognostic significance even in the setting of high-risk molecular signature. Leukemia. 2008;22:459–461. doi: 10.1038/sj.leu.2404934. [DOI] [PubMed] [Google Scholar]
- 108.Nair B, Shaughnessy JD, Jr, Zhou Y, Astrid-Cartron M, Qu P, van Rhee F, et al. Gene expression profiling of plasma cells at myeloma relapse from total therapy 2 predicts subsequent survival. Blood. 2009;113:6572–6575. doi: 10.1182/blood-2009-02-207803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Decaux O, Lode L, Magrangeas F, Charbonnel C, Gouraud W, Jezequel P, et al. Prediction of survival in multiple myeloma based on gene expression profiles reveals cell cycle and chromosomal instability signatures in high-risk patients and hyperdiploid signatures in low-risk patients: a study of the intergroupe francophone du myelome. J Clin Oncol. 2008;26:4798–4805. doi: 10.1200/JCO.2007.13.8545. [DOI] [PubMed] [Google Scholar]
- 110.Chng WJ, Ahmann GJ, Henderson K, Santana-Davila R, Greipp PR, Gertz MA, et al. Clinical implication of centrosome amplification in plasma cell neoplasm. Blood. 2006;107:3669–3675. doi: 10.1182/blood-2005-09-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chng WJ, Braggio E, Mulligan G, Bryant B, Remstein E, Valdez R, et al. The centrosome index is a powerful prognostic marker in myeloma and identifies a cohort of patients that might benefit from aurora kinase inhibition. Blood. 2008;111:1603–1609. doi: 10.1182/blood-2007-06-097774. [DOI] [PubMed] [Google Scholar]
- 112.Moreaux J, Hose D, Reme T, Jourdan E, Hundemer M, Legouffe E, et al. CD200 is a new prognostic factor in multiple myeloma. Blood. 2006;108:4194–4197. doi: 10.1182/blood-2006-06-029355. [DOI] [PubMed] [Google Scholar]
- 113.Gorczynski RM, Lee L, Boudakov I. Augmented Induction of CD4+CD25+ Treg using monoclonal antibodies to CD200R. Transplantation. 2005;79:1180–1183. [PubMed] [Google Scholar]
- 114.Condomines M, Hose D, Raynaud P, Hundemer M, De Vos J, Baudard M, et al. Cancer/testis genes in multiple myeloma: expression patterns and prognosis value determined by microarray analysis. J Immunol. 2007;178:3307–3315. doi: 10.4049/jimmunol.178.5.3307. [DOI] [PubMed] [Google Scholar]
- 115.Andrade VC, Vettore AL, Regis Silva MR, Felix RS, Almeida MS, de Carvalho F, et al. Frequency and prognostic relevance of cancer testis antigen 45 expression in multiple myeloma. Exp Hematol. 2009;37:446–449. doi: 10.1016/j.exphem.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 116.Atanackovic D, Luetkens T, Hildebrandt Y, Arfsten J, Bartels K, Horn C, et al. Longitudinal analysis and prognostic effect of cancer-testis antigen expression in multiple myeloma. Clin Cancer Res. 2009;15:1343–1352. doi: 10.1158/1078-0432.CCR-08-0989. [DOI] [PubMed] [Google Scholar]
- 117.van Rhee F, Szmania SM, Zhan F, Gupta SK, Pomtree M, Lin P, et al. NY-ESO-1 is highly expressed in poor-prognosis multiple myeloma and induces spontaneous humoral and cellular immune responses. Blood. 2005;105:3939–3944. doi: 10.1182/blood-2004-09-3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell. 2004;5:221–230. doi: 10.1016/s1535-6108(04)00050-9. [DOI] [PubMed] [Google Scholar]
- 119.Sabbatini P, Rowand JL, Groy A, Korenchuk S, Liu Q, Atkins C, et al. Antitumor activity of GSK1904529A, a small-molecule inhibitor of the insulin-like growth factor-I receptor tyrosine kinase. Clin Cancer Res. 2009;15:3058–3067. doi: 10.1158/1078-0432.CCR-08-2530. [DOI] [PubMed] [Google Scholar]
- 120.Sprynski AC, Hose D, Caillot L, Reme T, Shaughnessy J, Barlogie B, et al. The role of IGF-1 as a major growth factor for myeloma cell lines and the prognostic relevance of the expression of its receptor. Blood. 2009;113:4614–4626. doi: 10.1182/blood-2008-07-170464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Marton MJ, DeRisi JL, Bennett HA, Iyer VR, Meyer MR, Roberts CJ, et al. Drug target validation and identification of secondary drug target effects using DNA microarrays. Nat Med. 1998;4:1293–1301. doi: 10.1038/3282. [DOI] [PubMed] [Google Scholar]
- 122.Gray NS, Wodicka L, Thunnissen AM, Norman TC, Kwon S, Espinoza FH, et al. Science. Vol. 281. New York, NY: 1998. Exploiting chemical libraries, structure, and genomics in the search for kinase inhibitors; pp. 533–538. [DOI] [PubMed] [Google Scholar]
- 123.Cheok MH, Yang W, Pui CH, Downing JR, Cheng C, Naeve CW, et al. Treatment-specific changes in gene expression discriminate in vivo drug response in human leukemia cells. Nat Genet. 2003;34:85–90. doi: 10.1038/ng1151. [DOI] [PubMed] [Google Scholar]
- 124.Chauhan D, Auclair D, Robinson EK, Hideshima T, Li G, Podar K, et al. Identification of genes regulated by dexamethasone in multiple myeloma cells using oligonucleotide arrays. Oncogene. 2002;21:1346–1358. doi: 10.1038/sj.onc.1205205. [DOI] [PubMed] [Google Scholar]
- 125.Mitsiades N, Mitsiades CS, Poulaki V, Chauhan D, Fanourakis G, Gu X, et al. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc Natl Acad Sci USA. 2002;99:14374–14379. doi: 10.1073/pnas.202445099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Mitsiades N, Mitsiades CS, Richardson PG, Poulaki V, Tai YT, Chauhan D, et al. The proteasome inhibitor PS-341 potentiates sensitivity of multiple myeloma cells to conventional chemotherapeutic agents: therapeutic applications. Blood. 2003;101:2377–2380. doi: 10.1182/blood-2002-06-1768. [DOI] [PubMed] [Google Scholar]
- 127.Chauhan D, Li G, Auclair D, Hideshima T, Richardson P, Podar K, et al. Identification of genes regulated by 2-methoxyestradiol (2ME2) in multiple myeloma cells using oligonucleotide arrays. Blood. 2003;101:3606–3614. doi: 10.1182/blood-2002-10-3146. [DOI] [PubMed] [Google Scholar]
- 128.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101:540–545. doi: 10.1073/pnas.2536759100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Neri P, Tagliaferri P, Di Martino MT, Calimeri T, Amodio N, Bulotta A, et al. In vivo anti-myeloma activity and modulation of gene expression profile induced by valproic acid, a histone deacetylase inhibitor. Br J Haematol. 2008;143:520–531. doi: 10.1111/j.1365-2141.2008.07387.x. [DOI] [PubMed] [Google Scholar]
- 130.Tassone P, Neri P, Burger R, Savino R, Shammas M, Catley L, et al. Combination therapy with interleukin-6 receptor superantagonist Sant7 and dexamethasone induces antitumor effects in a novel SCID-hu in vivo model of human multiple myeloma. Clin Cancer Res. 2005;11:4251–4258. doi: 10.1158/1078-0432.CCR-04-2611. [DOI] [PubMed] [Google Scholar]
- 131.Sukhdeo K, Mani M, Zhang Y, Dutta J, Yasui H, Rooney MD, et al. Targeting the beta-catenin/TCF transcriptional complex in the treatment of multiple myeloma. Proc Natl Acad Sci USA. 2007;104:7516–7521. doi: 10.1073/pnas.0610299104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Raje N, Kumar S, Hideshima T, Ishitsuka K, Yasui H, Chhetri S, et al. Didox, a ribonucleotide reductase inhibitor, induces apoptosis and inhibits DNA repair in multiple myeloma cells. Br J Haematol. 2006;135:52–61. doi: 10.1111/j.1365-2141.2006.06261.x. [DOI] [PubMed] [Google Scholar]
- 133.Neri P, Tassone P, Shammas M, Yasui H, Schipani E, Batchu RB, et al. Biological pathways and in vivo antitumor activity induced by Atiprimod in myeloma. Leukemia. 2007;21:2519–2526. doi: 10.1038/sj.leu.2404912. [DOI] [PubMed] [Google Scholar]
- 134.Heller G, Schmidt WM, Ziegler B, Holzer S, Mullauer L, Bilban M, et al. Genome-wide transcriptional response to 5-aza-2′-deoxycytidine and trichostatin a in multiple myeloma cells. Cancer Res. 2008;68:44–54. doi: 10.1158/0008-5472.CAN-07-2531. [DOI] [PubMed] [Google Scholar]
- 135.Duus J, Bahar HI, Venkataraman G, Ozpuyan F, Izban KF, Al-Masri H, et al. Analysis of expression of heat shock protein-90 (HSP90) and the effects of HSP90 inhibitor (17-AAG) in multiple myeloma. Leuk Lymphoma. 2006;47:1369–1378. doi: 10.1080/10428190500472123. [DOI] [PubMed] [Google Scholar]
- 136.Ocio EM, Maiso P, Chen X, Garayoa M, Alvarez-Fernandez S, San-Segundo L, et al. Zalypsis: a novel marine-derived compound with potent antimyeloma activity that reveals high sensitivity of malignant plasma cells to DNA double-strand breaks. Blood. 2009;113:3781–3791. doi: 10.1182/blood-2008-09-177774. [DOI] [PubMed] [Google Scholar]
- 137.Burington B, Barlogie B, Zhan F, Crowley J, Shaughnessy JD., Jr Tumor cell gene expression changes following short-term in vivo exposure to single agent chemotherapeutics are related to survival in multiple myeloma. Clin Cancer Res. 2008;14:4821–4829. doi: 10.1158/1078-0432.CCR-07-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shaughnessy JD, Jr, Qu P, Edmondson P, Herman D, Zhou Y, Tian E, et al. Changes in the expression of proteasome genes in tumore cells following short-term proteasome inhibitor therapy predicts survival in multiple myeloma treated with bortezomib-containing multi-agent chemotherapy. Blood (ASH Annual Meeting Abstracts) 2008;12:733. [Google Scholar]