Abstract
We describe a novel knowledge-based protein-ligand scoring function that employs a new definition for the reference state, allowing us to relate a statistical potential to a Lennard-Jones (LJ) potential. In this way, the LJ potential parameters were generated from protein-ligand complex structural data contained in the PDB. Forty-nine types of atomic pairwise interactions were derived using this method, which we call the knowledge-based and empirical combined scoring algorithm (KECSA). Two validation benchmarks were introduced to test the performance of KECSA. The first validation benchmark included two test sets that address the training-set and enthalpy/entropy of KECSA The second validation benchmark suite included two large-scale and five small-scale test sets to compare the reproducibility of KECSA with respect to two empirical score functions previously developed in our laboratory (LISA and LISA+), as well as to other well-known scoring methods. Validation results illustrate that KECSA shows improved performance in all test sets when compared with other scoring methods especially in its ability to minimize the RMSE. LISA and LISA+ displayed similar performance using the correlation coefficient and Kendall τ as the metric of quality for some of the small test sets. Further pathways for improvement are discussed which would KECSA more sensitive to subtle changes in ligand structure.
Keywords: Knowledge-based scoring function, Empirical scoring function, Potential of mean force
Introduction
Knowledge-based protein-ligand scoring functions1–18, building on the idea of potential of mean force (PMF),19 are derived from structural information regarding protein-ligand complexes. Their pairwise interaction parameters are directly converted from the frequency of occurrence of given atom pairs contained in a large database of complexes. The concept of the potential of mean force can be illustrated by a simple fluid system of N particles whose positions are r1… rN.
The average potential ω(n)(r1… rN) is expressed as:
| (1) |
where g(n) is called a correlation function. β=1/kBT and kB is the Boltzmann constant and T is the system temperature. Hence the mean potential of the system with N particles is strictly the potential that gives the average force over all the configurations of the n+1…N particles acting on a particle at any fixed configuration keeping the 1…n particles fixed. The mean potential can be described as follows:
| (2) |
where U is the total potential energy of the system. Described by Sippl and others,1–5 the average potential is expressed as Equation 3 for the special case of a system with an observed particle number of n=2, as is the case herein (pairwise atoms from the protein and ligand).
| (3) |
where g(2)(r) is the pair distribution function, ρij(r) is the number density for the atom pairs of types i and j observed in the known protein structures and ρ*ij(r) is the number density of the corresponding pair in a reference state. In order to obtain the pure interaction potential between atoms, a reference state is required to remove the contribution of the ideal-gas state potential. So, in the reference state, the system of particles is like an ideal-gas state defined by fundamental statistical mechanics, in which particles would be evenly distributed in the binding site. Equation 3 can also be expressed as:
| (4) |
where nij(r) and n*ij(r) are numbers of atom pairs of type i and j, respectively, at distance r for the observed structures and the reference state.
In potential of mean force methods, the number of the corresponding pairs in the reference state cannot be exactly obtained for protein-ligand systems due to the effects of connectivity, excluded volume, composition, etc.6 Therefore, the pairwise interaction potential cannot be accurately calculated. Nonetheless, this idea of PMF scoring has advantages over empirical scoring, because it directly relates pairwise interaction to structural data instead of fitting to known binding affinity data. Additionally, the PMF is more efficient than force field scoring due to the avoidance of higher expense computations. Our intent is to introduce a new concept of the reference state, in order to relate the statistical potential to atomic pairwise interaction potential. Hence the atomic pairwise interaction model can be parameterized exclusively from structural data instead of binding data or quantum calculations.
Methods and Results
Construction of traditional statistical potentials starts by collecting structural information from large numbers of protein-ligand complexes, in order to simulate a "mean force" state in which the protein-ligand atomic pairwise radial distribution arises from all possible interactions in the binding site. Various reference states have been designed to remove the non-interacting energy from the "mean force" state in order to correlate the pairwise radial distribution to the interaction potential between selected atoms of a specific atom type i, j with all other atoms in the protein-ligand binding site.
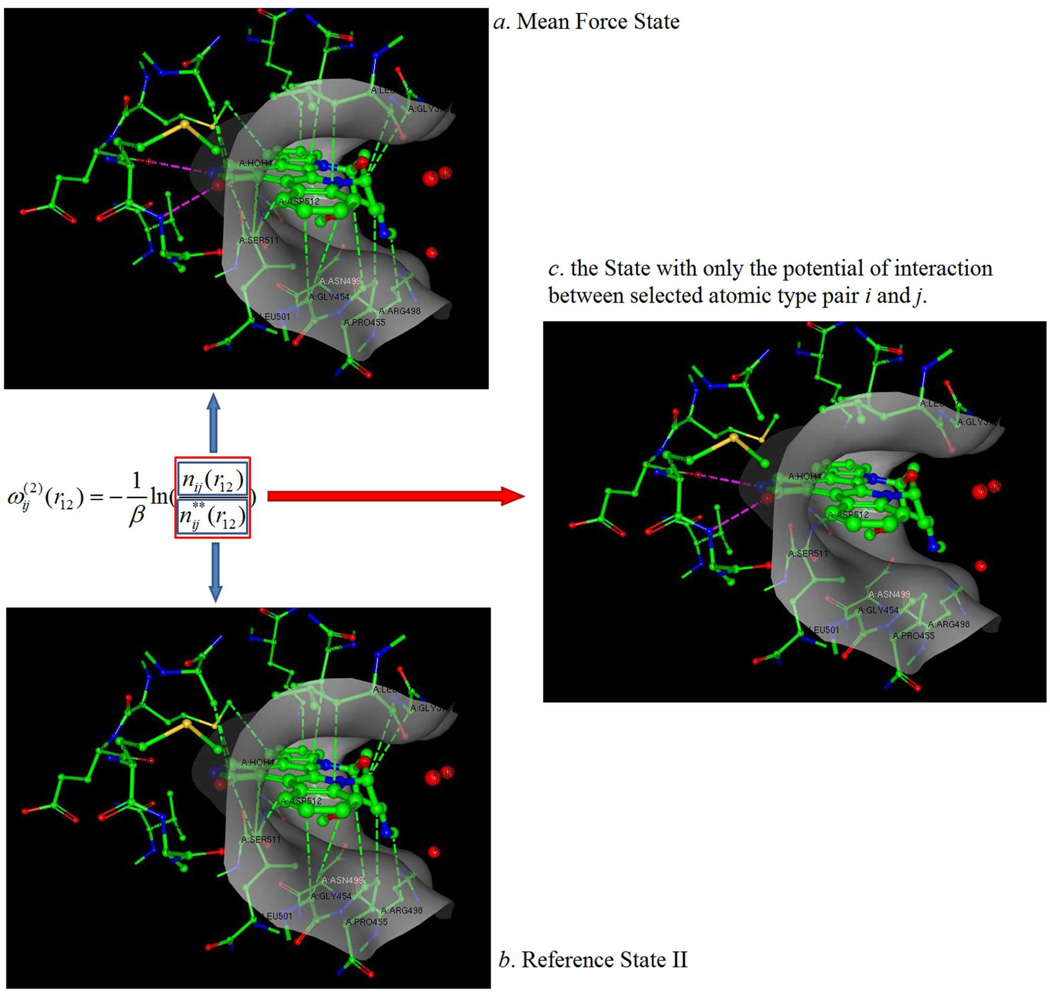
Our goal is to equate the statistical potential to the Lennard-Jones potential for each pairwise interaction. However, the LJ potential reflects pairwise interactions between two types of atoms, while a statistical potential is an average potential contributed by all atoms within the binding region. In this case, when trying to equate the statistical potential to a pairwise interaction potential, we need to remove all interactions except the pairwise interaction between atoms of type i and j in the binding region by defining a new reference state (denominator) in the PMF model. Unlike the traditional reference state, in which the selected atom pairs i and j are at an infinite separation where the interaction energy is zero (as in the ideal gas state), within this new reference state (which we will call reference state II), a system of particles is under an average force contributed by all atoms in the binding region excluding the interaction force between the selected atom pairs i and j. In other words, the only difference between the mean force state and the reference state II is that the latter state does not contain the pairwise interaction potential between the selected atoms of type i and j. Figure 1 provides a graphic illustration of the KECSA statistical potential model.
Figure 1.
A protein-ligand structural illustration (using PDBID 1xbc) of how the KECSA statistical potential is modeled. The protein binding site is shown as a grey surface with the ligand located within the binding site surrounded by protein residues which it makes contacts with. The pink dashed lines indicate interactions between certain atom pair types i and j, (i.e. carbonyl oxygen with amine nitrogens in this example) which are defined as "selected interactions" in this manuscript. Green dashed lines indicate all other non-covalent interactions between the protein and ligand atoms in the binding pocket, defined as "background interactions". (a) In the mean force state, the system is filled with all types of interactions. (b) The reference state II contains all the background interactions. (c) Removing all the background interactions from total interactions results in a state with only the selected interactions for each i and j combination.
When equated to the LJ potential, the statistical potential can be expressed as:
| (5) |
where σ is the distance at which the inter-particle potential is zero and ε is the well depth. The exponents for the repulsive term and attractive term are α and β, respectively. We derive the exponents instead of assigning “typical” exponent values (i.e., 12–6), because (1) the repulsive and attractive forces change with different types of pairwise interactions and (2) Eij(r) in Equation 5 includes both van der Waals and electrostatic interactions, which means the LJ-potential formula on the right hand side of Equation 5 accounts for two components:
| (6) |
The reason we use the LJ formula on the left hand side of Equation 6 instead of partitioning them into van der Waals and electrostatic potentials is that the LJ potential reaches 0 at σ and R, while reaching its minimum value when r is . Based on these properties, equations can be derived in order to determine the unknown parameters.
In Equation 5, and nij(r) are the number of protein-ligand atomic pairwise interactions in the bin (r, r+Δr), with the volume 4πraΔr in reference state II and in the training set that mimics the mean force state, respectively. Δr is defined as 0.005Å. We introduce a to-be-determined parameter a for the shell volume because of the inaccessible volume present in protein-ligand systems, and because of the deviation of nij(r) in the training set from the "perfect" pairwise number. Hence, the expectation is the parameter a will adopt values other than 2.
The central issue in the KECSA model construction is to build up the radial distribution function of the selected atom pairs in reference state II. A way in which to do this is to measure the similarity of the reference state II with two known states: the mean force state, and ideal gas state. Then build the radial distribution function with information collected from these two states. In reference state II, the radial distribution of a certain atom pair i and j is associated with a certain "background interaction" which is related to the total number of selected atom pairs Nij. Because the "background interaction" potential contains all atom pairwise interactions in the binding site excluding the "selected interactions" between atom pairs i and j, the difference in energy between the mean force state and reference state II for each atom pair type depends on the total number of the selected atom pairs Nij and the total number of atom pairwise interactions N found in the binding site. The "background interaction" energy approaches the "mean force" state energy as becomes smaller, while the energy difference increases when becomes larger. Hence we decide to make the "background interaction" potential, as well as the radial distribution function, a function of . The modeling of the atom pairwise number distribution function in reference state II starts from the two extreme situations for : (1) When Nij approaches zero (Nij →0), the background energy ≈ the "mean force" state energy, resulting in . (2) When Nij approaches to N (Nij →N), the background energy ≈ the ideal-gas energy, resulting in resembling an ideal gas state radial distribution. The radial distribution function for an ideal gas state is defined as implying that the number of the selected atom pairs i and j is evenly distributed in the binding site which has an average volume V. The average V of protein-ligand binding site is given as , with the same to-be-determined parameter a as introduced above. Hence, the two extreme situations for reference state II can be defined as:
| (7) |
| (8) |
At certain distance r, is a function of with a range from to nij(r). In addition, with Nij tending towards 0 or N, the reference state II would be more similar to the mean force state or the ideal gas state, respectively. Hence, is defined as a weighted combination of both the ideal gas state and the mean force state radial distribution functions. Due to the fact that the integral of from 0 to R (cutoff distance where the atomic interaction is regarded as zero) is Nij, a linear combination (Equation 9) of the weighted radial distribution functions for both the ideal gas and mean force state meets all the necessary conditions:
| (9) |
In this way the new reference state is designed as state intermediate between the ideal gas and the mean force state. At a certain distance between the atom pairs of type i and j, the total energy of reference state II , or the "background interaction" energy is:
| (10) |
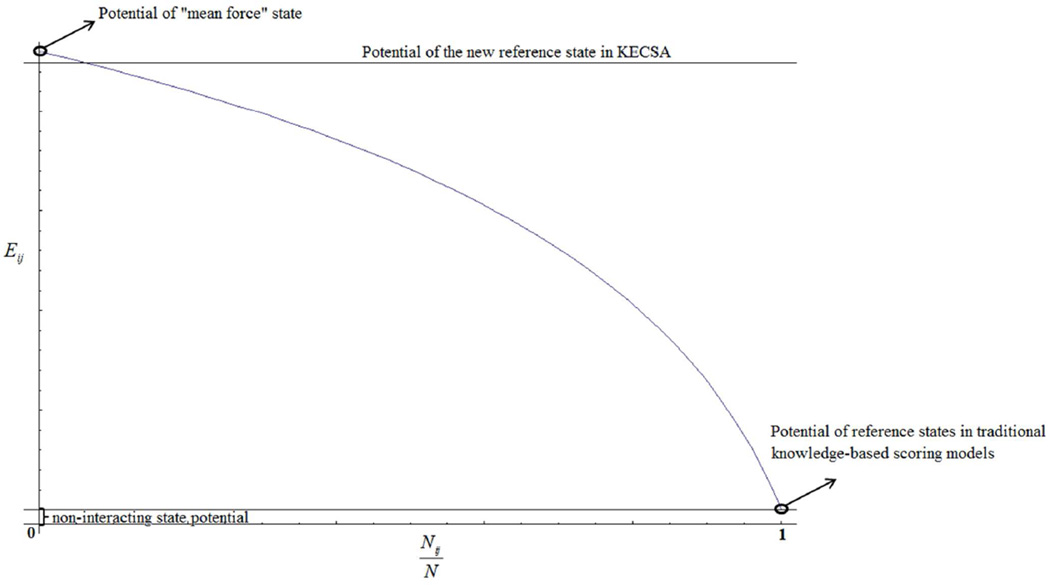
A plot illustrating the relationship between the number fraction and the new reference state potential is shown in Figure 2, to better illustrate the differences in energy between the different states.
Figure 2.
Ratio of the observed atom pairs to the total interacting atom pairs vs. the new reference state II potential.
Combining Equations 5 and 9 we obtain:
| (11) |
Using and using the fact that the Lennard-Jones potential is zero at rij = σ and at rij=R, we arrive at:
| (12) |
| (13) |
In addition, the LJ potential reaches its minimum value when r is . So the first derivative (D) of the statistical energy term with respect to r is zero at .
| (14) |
To simplify the resultant expressions the factor is given as η.
| (15) |
Simplifying Equations 12, 13 and 15 yields:
| (16) |
| (17) |
and
| (18) |
Although we don't know the values of α and β yet, we do know that the value of η is unique for each combination of α and β. Supplementary Table 1 lists all η values for each integer combination of α and β from 2-1 to 15-14. Different η values will be chosen for every pairwise interaction, to satisfy the well depth distance at ησ (e.g., σ is 21/6σ or rij* (well-depth at the minimum) for the 12-6 potential).
In order to find the a, σ and η values within Equations 16–18, we still need to determine the value of R, the cutoff distance. We introduce a nonlinear programming method to find a reasonable R for each pairwise interaction type instead of assigning a fixed R value. Ideally, R should be as large as possible since the LJ potential approaches 0 when the distance approaches infinity. Meanwhile, for any r between σ and R, the potential value is below 0. Here we use the following inequality constraint in our nonlinear programming approach:
| (19) |
which can be simplified as:
| (20) |
With the goal of maximizing the value of R, coupled with the three constraint equations (Equations 16–18) and an inequality constraint (Equation 20), a, σ and η can be determined. The values of η obtained in this way can then be compared with the η values in supplementary Table 1, in order to determine the closest α and β pair. Inserting these values into Equation 11 we can calculate all of the corresponding ε values.
One important issue in the parameterization of KECSA is that the LJ potential parameters for each type of pairwise interaction should be independent of the other types of interactions, instead of being dependent. Derivation of a, σ, α, β and R comes from Equations 16–18 and 20, none of which contains the total interaction number N. This indicates that for each type of pairwise interaction, the average volume , the distances at which the LJ potential reaches zero and has a minimum , the relative strength of the repulsive and attractive forces in the LJ potential (α and β), and the long-range cutoff distance (R) are independently derived in KECSA. The only issue lies in the derivation of the ε values from Equation 11, where the probability of occurrence is included in the calculation. In order to avoid relative energies generated for each interaction type based on their probability of occurrence in protein-ligand binding sites, we used a normalized for each interaction type. Thereby the number fractions for all interaction types are identical.
In the present work, all pairwise interactions among 18 atom types (listed in Table 1) were examined resulting in 49 significant interaction types being identified. The remaining interaction types were abandoned or merged into similar interaction types due of the paucity of data to fit to or because they are randomly distributed across the observed distance range. The chosen interaction types included 38 van der Waals and 11 hydrogen bonding interaction types. In this case, all interaction types share the same probability of occurrence in protein-ligand binding sites. Equation 11 can be rewritten as follows in order to generate the ε values. All derived parameters are listed in Table 2.
| (21) |
Table 1.
List of selected atom types.
| Atom Type | Description |
|---|---|
| C3 | sp3 hybridized carbon |
| C2 | sp2 hybridized carbon |
| Car | aromatic carbon |
| C1 | sp hybridized carbon |
| N4 | positively charged nitrogen |
| Nam | amide nitrogen |
| N3 | sp3 hybridized nitrogen |
| Nar | nitrogen aromatic |
| N2 | sp2 hybridized nitrogen |
| Npl3 | trigonal planar nitrogen |
| O3 | sp3 hybridized oxygen |
| O2 | sp2 hybridized oxygen |
| S | sulfur |
| P | phosphorus |
| F | fluorine |
| Cl | chlorine |
| Br | bromine |
| I | iodine |
Table 2.
Parameters for all 49 pairwise potentials.
| interaction type |
C2C2 | C2Car | C2N2 | C2N3 | C2N4 | C2Nam | C2Nar | C2Npl3 | C2O2 | C2O3 |
|---|---|---|---|---|---|---|---|---|---|---|
| σ | 4.145 | 3.630 | 3.450 | 3.285 | 3.215 | 3.505 | 3.575 | 3.505 | 3.370 | 3.135 |
| a | 3.375 | 2.224 | 3.085 | 2.810 | 3.089 | 4.296 | 2.662 | 2.273 | 3.298 | 2.992 |
| R | 5.900 | 6.535 | 4.755 | 4.220 | 4.235 | 4.265 | 5.390 | 6.485 | 4.430 | 5.345 |
| ε | 0.091 | 0.041 | 0.388 | 0.035 | 0.133 | 1.003 | 0.296 | 0.769 | 1.735 | 0.071 |
| LJ model | 12-5 | 11-1 | 10-9 | 12-8 | 14-12 | 15-6 | 12-11 | 15-14 | 12-11 | 13-4 |
| interaction type |
C2S | C3C2 | C3C3 | C3Car | C3N2 | C3N3 | C3N4 | C3Nam | C3Nar | C3Npl3 |
| σ | 4.350 | 3.940 | 4.290 | 3.850 | 3.580 | 3.650 | 4.570 | 4.470 | 3.455 | 3.815 |
| a | 2.505 | 3.049 | 2.759 | 2.237 | 2.404 | 1.759 | 2.988 | 3.581 | 2.990 | 2.347 |
| R | 6.425 | 6.210 | 6.840 | 6.775 | 6.130 | 6.945 | 6.850 | 6.165 | 5.435 | 6.160 |
| ε | 0.387 | 0.085 | 0.364 | 0.454 | 0.053 | 0.123 | 0.022 | 0.071 | 0.067 | 0.129 |
| LJ model | 12-11 | 14-3 | 5-4 | 5-3 | 15-9 | 13-12 | 12-7 | 12-7 | 12-9 | 4-3 |
| interaction type |
C3O2 | C3O3 | C3S | CarCar | CarN2 | CarN3 | CarN4 | CarNam | CarNar | CarNpl3 |
| σ | 3.200 | 3.325 | 3.940 | 3.700 | 3.600 | 3.700 | 4.360 | 3.720 | 3.565 | 3.665 |
| a | 2.742 | 3.164 | 1.965 | 1.898 | 2.079 | 2.032 | 1.089 | 3.655 | 1.389 | 1.736 |
| R | 4.515 | 5.650 | 6.630 | 6.855 | 6.440 | 6.845 | 6.980 | 6.030 | 6.865 | 6.675 |
| ε | 0.343 | 0.038 | 0.016 | 0.249 | 0.013 | 0.005 | 0.056 | 0.279 | 0.206 | 0.016 |
| LJ model | 9-6 | 13-7 | 14-1 | 4-3 | 11-1 | 8-1 | 9-5 | 14-13 | 15-14 | 15-6 |
| interaction type |
CarO2 | CarO3 | CarS | N2O2HB | N2O3HB | N3O2HB | N3O3HB | NamO2HB | NamO3HB | Npl3O2HB |
| σ | 3.430 | 3.690 | 3.920 | 2.640 | 2.670 | 2.550 | 2.605 | 2.610 | 2.625 | 2.585 |
| a | 2.840 | 2.204 | 1.627 | 2.056 | 2.365 | 0.989 | 1.788 | 2.057 | 3.475 | 1.377 |
| R | 6.600 | 6.505 | 6.975 | 6.420 | 6.465 | 6.745 | 4.585 | 4.765 | 4.160 | 4.995 |
| ε | 0.120 | 0.030 | 0.050 | 0.062 | 0.036 | 0.196 | 0.217 | 1.700 | 0.172 | 0.219 |
| LJ model | 12-10 | 6-2 | 9-5 | 15-8 | 15-5 | 14-10 | 13-9 | 12-10 | 11-8 | 13-8 |
| interaction type |
Npl3O3HB | O2N2 | O2Nam | O2Nar | O2O2 | O3N2HB | O3O2HB | O3O2 | O3O3HB | |
| σ | 2.635 | 2.570 | 4.125 | 3.380 | 3.065 | 2.510 | 2.445 | 3.365 | 2.080 | |
| a | 1.899 | 2.397 | 2.784 | 2.292 | 2.767 | 1.345 | 1.998 | 3.250 | 2.408 | |
| R | 6.755 | 6.845 | 6.065 | 6.070 | 6.055 | 4.395 | 6.065 | 6.480 | 6.990 | |
| ε | 0.272 | 0.010 | 0.008 | 0.073 | 0.034 | 0.116 | 2.002 | 0.024 | 0.038 | |
| LJ model | 15-12 | 7-1 | 13-3 | 11-7 | 4-1 | 15-8 | 14-13 | 3-2 | 11-3 | |
With all of the enthalpy terms determined in the analytical manner described above, the entropy terms are then decided upon in an empirical manner. Structural information such as the number of rotatable bonds, number of double and aromatic bonds, molecular mass, counts of carbon/oxygen/nitrogen atoms, buried surface area, etc. were collected for all ligands contained in the training set. The selection of entropy terms is based on their contribution to our linear regression model, whose 95% confidence interval should not include 0. Finally, 9 entropy terms are selected: number of rotatable bonds in the ligand, the molecular mass of the ligand, number of aromatic bonds in the ligand, number of oxygen atoms in the ligand, number of nitrogen atoms in the ligand, the nonpolar buried surface area, total buried surface area, the ratio of the nonpolar buried surface and total ligand surface area and, finally, the ratio of the total buried surface area and the total ligand surface area. The PDBbind v2010 data set21,22 including 5054 protein-ligand complexes is chosen as the training set for the parameterization of the enthalpy terms. We chose 1982 protein-ligand complexes found in the PDBbind v2011 refined data set as the training set for the selection and parameterization of the entropy terms.
Model Validation and Discussion
I) Validation to Detect Over-Fitting and Ligand-Size Dependence
Several validation benchmarks were introduced to test the performance of KECSA. The first benchmark included two test sets in order to examine the dependence of KECSA on the training set. Because the entropy term was obtained by fitting to experimental binding free energies care was taken to ensure that the resultant model was not over-fit. First, a leave-one-out cross validation was used against the training set, which includes 1982 protein-ligand complexes used in the KECSA entropy term parameterization. Comparison of the Pearson correlation coefficient r, RMSE (root-mean-square error) and Kendall τ between the training set and the leave-one-out cross validation are shown in Table 4. The three statistical measures all showed small differences between the training set and the leave-one-out prediction, indicating that the KECSA entropy model was properly built.
Table 4.
Leave-one-out cross validation of KECSA.
| Pearson's r | RMSE(kcal/mol) | Kendall τ | |
|---|---|---|---|
| Training | 0.601 | 2.20 | 0.442 |
| Leave-One-Out | |||
| Calculation | 0.594 | 2.22 | 0.437 |
Second, we introduced a test set including 1934 protein-ligand complexes chosen from the PDBbind v2011 dataset, with no overlap with the training set used above. From the total of 6051 protein-ligand complexes with binding affinity data in the PDBbind v2011 dataset, complexes forthis test set were selected following four criteria: (1) Crystal structures of all selected complexes had X-ray resolutions ≤ 2.5Å. (2) Only complexes with pKi or pKd values distributed between 2 to 8 were selected, mimicking what might be found in a virtual screening database or a pharmaceutically relevant ligand database. (3) Only complexes with molecular weights (MWs) distributed from 80 to 800 were selected, to avoid ligand size-dependent prediction results. (4) Complexes used in the KECSA entropy term training set were excluded.
The second test set was used to verify the robustness of KECSA against and external dataset as well as investigating the contributions of the enthalpy and entropy terms in KECSA’s binding affinity prediction. In addition, because the entropy term in KECSA was modeled as a linear combination of several ligand properties including ligand size/mass information this test demonstrated that KECSA was not ligand size-dependent. We split the KECSA scoring function and used both LJ potential and entropy terms for binding affinity prediction and then compared with the full KECSA scoring function. We also calculated the correlation coefficient between the experimental pKi or pKd with ligand MW. The predictions and ranking results are listed in Table 5.
Table 5.
Statistical results for KECSA, KECSA LJ, KECSA entropy and Ligand MW correlated with experimental binding affinity.
| Pearson's r | Kendall τ | |
|---|---|---|
| KECSA Scoring | ||
| Function | 0.590 | 0.404 |
| LJ Potentials in KECSA | 0.509 | 0.352 |
| Entropy in KECSA | 0.521 | 0.349 |
| Ligand Molecular Weight | 0.381 | 0.272 |
KECSA produces a Pearson's r of 0.590 and a Kendall τ of 0.404. Comparison to the training set result (a Pearson's r of 0.610 and a Kendall τ of 0.442) indicates that KECSA gives robust predictions against an unknown binding affinity dataset. When just using the ligand MW for binding affinity prediction, the Pearson's r drops by 0.128 and the Kendall τ drops by 0.08 when compared to the LJ potential only prediction, while Pearson's r drops by 0.140 and the Kendall τ drops by 0.08 when compared to the entropy term prediction. The drop in prediction is even greater when compared to the full KECSA score function. This result suggests that the KECSA prediction is minimally affected by MW considerations. The LJ potential and entropy only predictions are quite similar, but lower than the full KECSA prediction (see Table 5). None of the two independent parts, enthalpy or entropy, shows a significant performance over the other, suggesting that both enthalpy and entropy play important roles in the KECSA scoring function. RMSE is not listed for comparison because the LJ potential is generated from a statistical potential while entropy is derived by fitting to pKd or pKi values, which results in different scales making comparison difficult.
II) Validation Benchmark for Comparison with LISA, LISA+ and Other Scoring Methods
KECSA is the second-generation scoring function we have developed after LISA23 and LISA+.24 The latter two were developed for the fast calculation of protein-ligand binding affinity (pKd and pKi), and were successful in the SAMPL3 challenge (first rank for all scoring methods), which was a blind test for docking and scoring methods.24 Comparisons between KECSA and other scoring methods including LISA and LISA+ are necessary for further understanding its performance. The second validation benchmark consists of two large-scale test sets and four smaller test sets for comparison of KECSA with LISA/LISA+, as well as one small scale test set (Wang’s test set25) with 100 diverse protein-ligand complexes for comparison of KECSA and several other well-known scoring methods.
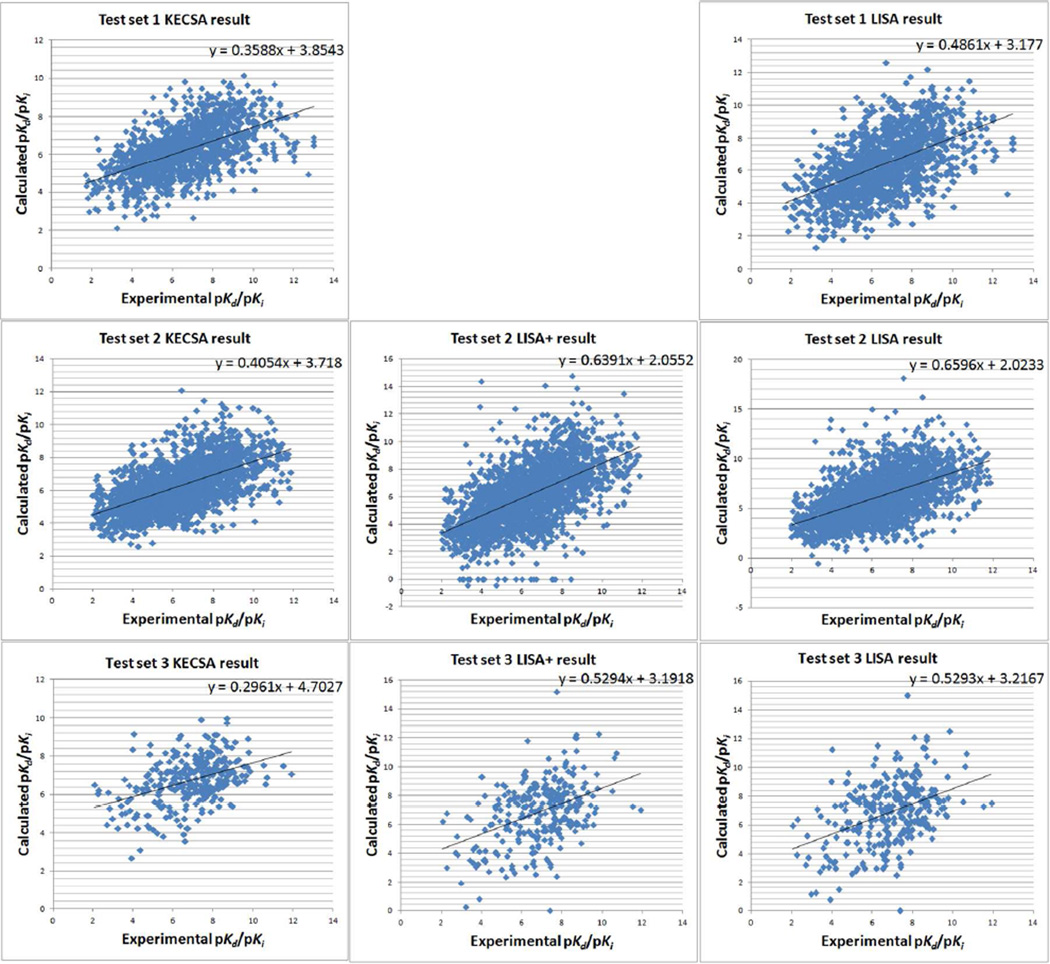
First, two large scale test sets both with more than 1000 complexes were introduced to KECSA, LISA and LISA+. The first test set contained 1399 complexes from the PDBbind v2010 database, which was previously used for LISA validation. KECSA reproduces a Pearson correlation coefficient r of 0.553, an RMSE of 2.46kcal/mol and a Kendall τ of 0.401, while LISA reproduces a Pearson correlation coefficient r of 0.534, an RMSE of 2.65kcal/mol and a Kendall τ of 0.378. LISA+ was trained based on this data set, so it was excluded from this validation benchmark. A larger test set was applied for all three scoring functions, including 2456 protein-ligand complexes from the PDBbind v2011 refined data set, of out which 290 complexes had Zn-ligand binding. For those 2166 non-metal containing complexes, KECSA gets an r of 0.589, an RMSE of 2.31kcal/mol and a Kendall τ of 0.429, while LISA gets an r of 0.542, an RMSE of 3.06kcal/mol and a Kendall τ of 0.397, LISA+ yields an r of 0.572, an RMSE of 2.81kcal/mol and a Kendall τ of 0.419. For those complexes with Zn-ligand binding, KECSA has an r of 0.415, an RMSE of 2.33kcal/mol and a Kendall τ of 0.267, LISA has an r of 0.409, an RMSE of 3.08kcal/mol and a Kendall τ of 0.252, and LISA+ has an r of 0.420, an RMSE of 3.00kcal/mol and a Kendall τ of 0.257. Calculation and statistical results of KECSA, LISA+ and LISA for all large-scale validation studies are shown in Table 6 and Figure 3.
Table 6.
Statistical results of KECSA, LISA+ and LISA from large-scale validation studies.
| KECSA | LISA+ | LISA | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Test set 1a |
Test set 2b |
Test set 3c |
Test set 1 |
Test set 2 |
Test set 3 |
Test set 1 |
Test set 2 |
Test set 3 |
|
| Correlation | |||||||||
| Coefficient | 0.553 | 0.589 | 0.415 | - | 0.572 | 0.420 | 0.534 | 0.542 | 0.409 |
| RMSE | |||||||||
| (kcal/mol) | 2.46 | 2.31 | 2.33 | - | 2.81 | 3.00 | 2.65 | 3.06 | 3.08 |
| Kendall τ | 0.401 | 0.429 | 0.267 | - | 0.419 | 0.257 | 0.378 | 0.397 | 0.252 |
Test set 1 contains 1399 complexes from PDBbind v2010 database.
Test set 2 contains 2166 non-metal complexes from PDBbind v2011 refined data set.
Test set 3 contains 290 metalloprotein-ligand complexes from PDBbind v2011 refined data set.
Figure 3.
Plot of KECSA, LISA+ and LISA calculated pKd or pKi vs. Experimental pKd or pKi in obtained in validation studies.
In the large-scale test, KECSA yields a better prediction than our two first-generation scoring functions. It produces better predicted results based on RMSE and more reliable binding affinity ranking based on Kendall τ compared with the other two scoring methods. LISA+ can compete with KECSA based on correlation coefficient, and even achieves better r in the subset of complexes with Zn-ligand binding. We believe that the improvement of LISA+ compared with LISA is because the complexes in the training set are categorized based on ligand properties (mass and hydrophobicity) and different models are trained for each category. This proves that a multi-model scheme can improve the predictive ability of empirical scoring functions.
Our validation studies indicate an improvement from LISA to KECSA. Introducing PMF theory for non-bonding interaction modeling shows its advantages over simply fitting to binding affinity data. However, metal-ligand binding prediction remains a challenge for classical-mechanics based or statistical scoring methods. KECSA improves the binding affinity predicting ability mostly in RMSE for this subgroup of complexes. However, although KECSA shows improvement in both correlation coefficient r and RMSE, predictions in the low and high binding regions are poor. Seen from the linear regression functions in Figure 3 the slopes of LISA and LISA+ generated data vs. experimental data are 0.66 and 0.64, while that for KECSA is 0.41; the intercepts for LISA and LISA+ are 2.02 and 2.06, while that for KECSA result is 3.72. The reason is that 3314 complexes, which comprised 65.3% of the whole training set, are in the mid-binding region (pKd or pKi between 4 and 8). Hence the scoring function tends to overestimate binding affinity of the low-binding region while underestimating that of the high-binding region if there is no significant decrease or increase in contact number for the protein-ligand complexes from these two regions. We used a scaling procedure to help improve the prediction of binding affinity in the high and low binding regions. The KECSA generated results were fit to a linear model to reproduce the PDBbind v2011 refined data set pKd values. So we get:
| (22) |
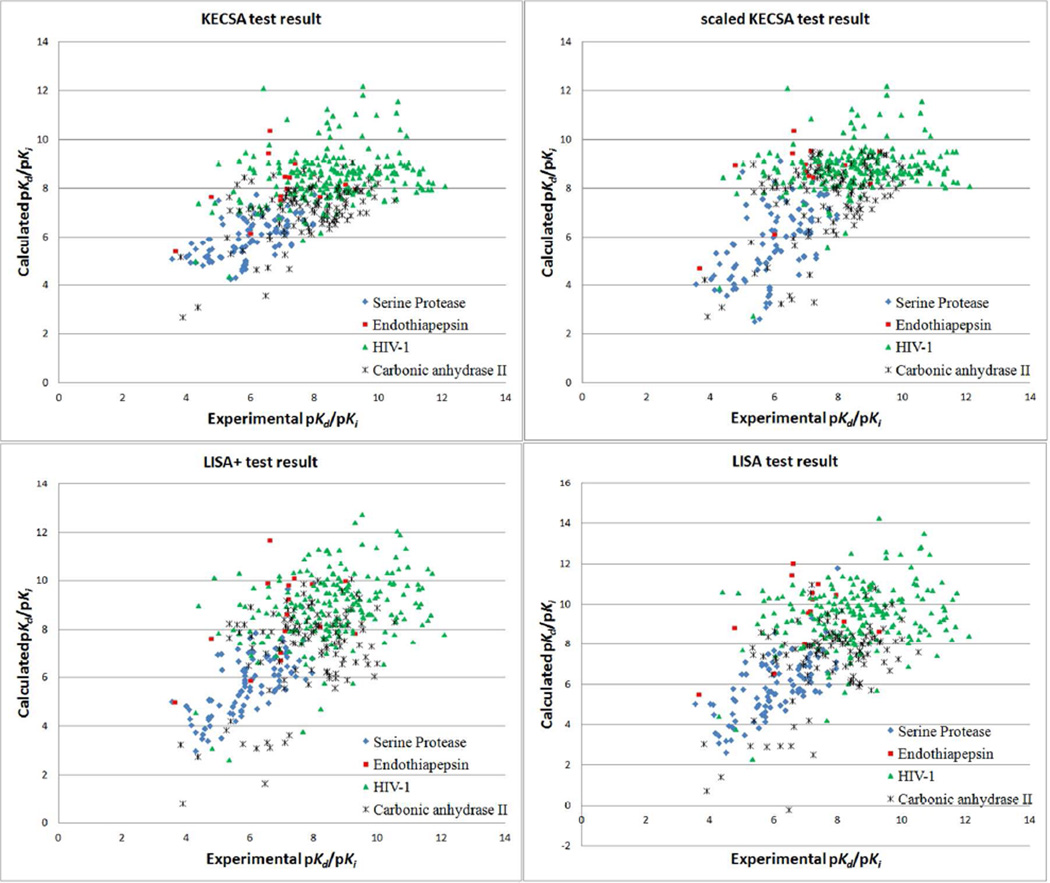
Next, four test sets containing 427 protein-ligand complexes from four protein families was examined. The list of complexes, their families and binding constants are given in Supplementary Table 2. Statistical and calculation results are shown in Table 7 and Figure 4. KECSA improves the RMSE for all test sets, indicating that KECSA makes a more robust prediction with respect to experimental binding affinity data than do LISA and LISA+. However, LISA and LISA+ both give a better correlation coefficient and Kendall τ for the serine protease, endothiapepsin and HIV-1 protease test sets, showing that they perform better in binding affinity ranking in these small test sets. Results for each test set were carefully examined. The serine protease test set contains many complexes with low-mass ligands, while most ligands are relatively larger in the endothiapepsin test set. Statistical results of the three scoring methods' predictions are similar in these two test sets: KECSA generates a better RMSE, while LISA and LISA+ yielded both a better correlation coefficient and Kendall τ. In the serine protease test set, 11 out of 96 protein-ligand complexes had small ligands with molecular mass lower than 200 Daltons. KECSA prediction overestimates most of their pKd or pKi values (Supplementary Table 2), while LISA and LISA+ to some degree underestimates these values. Hence, KECSA decreases the binding affinity differences between these low-binding complexes and other high-binding complexes and LISA/LISA+ increases these differences. The implication is that LISA and especially LISA+ differentiate the binding affinity values of the complexes better in this test set and give higher r and τ values, but they have worse RMSEs. In the endothiapepsin test set, on the other hand, all complexes have ligands with molecular mass higher than 500 Daltons. LISA and LISA+ overestimated binding affinities with larger errors compared with KECSA, while distinguishing the complexes better, generating a better linear correlation towards the experimental data. These test results suggest that LISA and LISA+ are more sensitive to ligand mass changes, while KECSA makes more precise prediction with smaller error, but has difficulty in ranking complexes with similar pKds or pKis. The HIV-1 protease test set is a significant challenge for all three scoring methods. Most of the complexes have high-mass ligands and high binding affinity. Because the ligands in this test set have similar structures binding affinity predictions from all three scoring methods are not able to rank these complexes. LISA+ does better in correlation coefficient and ranking than others, which indicates that training scoring functions for different complex categories based on ligand or binding pocket properties may help scoring methods improve their ability to identify subtle changes in ligand structure. The carbonic anhydrase II test set includes 100 out of 110 complexes with Zn chelation, and contains more polar and charged interactions. KECSA demonstrates better performance in the correlation coefficient, RMSE and Kendall τ, pointing to its advantage over LISA and LISA+ on reproducing binding affinity data of complexes with more hydrophilic interactions and metal chelation. For the reproduction of the binding affinity for the four combined test sets all three scoring methods give a similar correlation coefficient r, while LISA+ has a small advantage. KECSA does better in both RMSE and Kendall τ. For all four test sets, scaling of the KECSA score does help to improve the slope of calculated data vs. experimental data, but does not improve the statistical tests. Overall, KECSA give better RMSE values and better binding affinity ranking of the complexes belonging to different protein families.
Table 7.
Statistical results of KECSA, scaled KECSA, LISA+ and LISA from four small test sets.
| Serine protease |
Endothiapepsin | KECSA HIV-1 protease |
Carbonic anhydrase II |
All | Serine protease |
Endothiapepsin | Scaled KECSA HIV-1 protease |
Carbonic anhydrase II |
All | |
|---|---|---|---|---|---|---|---|---|---|---|
| Correlation | ||||||||||
| Coefficient | 0.568 | 0.441 | 0.244 | 0.495 | 0.591 | 0.568 | 0.547 | 0.245 | 0.487 | 0.607 |
| RMSE | ||||||||||
| (kcal/mol) | 0.894 | 1.700 | 1.678 | 1.405 | 1.466 | 1.299 | 1.987 | 1.677 | 1.478 | 1.562 |
| Kendall τ | 0.423 | 0.202 | 0.139 | 0.246 | 0.420 | 0.421 | 0.067 | 0.123 | 0.228 | 0.390 |
| Serine protease |
Endothiapepsin | LISA+ HIV-1 protease |
Carbonic anhydrase II |
All | Serine protease |
Endothiapepsin | LISA HIV-1 protease |
Carbonic anhydrase II |
All | |
| Correlation | ||||||||||
| Coefficient | 0.692 | 0.484 | 0.334 | 0.427 | 0.603 | 0.645 | 0.496 | 0.256 | 0.492 | 0.586 |
| RMSE | 0.970 | 2.171 | 1.781 | 1.814 | 1.661 | 1.131 | 2.880 | 2.089 | 1.821 | 1.884 |
| Kendall τ | 0.508 | 0.353 | 0.195 | 0.147 | 0.407 | 0.479 | 0.269 | 0.118 | 0.228 | 0.392 |
Figure 4.
Plot of KECSA, scaled KECSA, LISA+ and LISA calculated pKd or pKi vs. Experimental pKd or pKi in four small test sets.
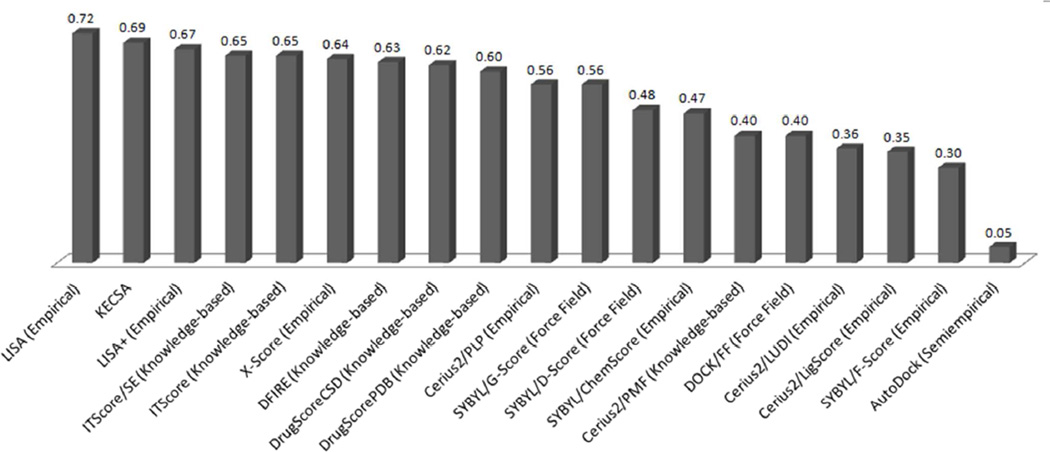
For the last test set, we introduced Wang’s test set25 with 100 diverse protein-ligand complexes. The purpose was to compare binding affinity prediction ability of KECSA not only with LISA and LISA+, but also with other well-known scoring functions. We obtain Pearson's r = 0.69, RMSE = 2.25 kcal/mol using KECSA, compared with Pearson's r = 0.72, RMSE = 2.32 kcal/mol using LISA, Pearson's r = 0.67, RMSE = 2.80 kcal/mol using LISA+. This result coincides with the conclusion gained from the second validation benchmark, that among these three scoring functions, KECSA prediction has the smallest RMSE. Pearson correlation coefficients of KECSA together with other score functions are presented in Figure 5, showing its performance on this test set.
Figure 5.
With the test set built by Wang,25 binding affinity comparison was done for KECSA, LISA, LISA+ and several other well-known scoring functions, ITScore/SE,18 ITScore,17 X-Score,26 DFIRE,27 DrugScoreCSD,12 DrugScorePDB,11 Cerius2/PLP,28,29 SYBYL/G-Score,30 SYBYL/D-Score,31 SYBYL/ChemScore,32 Cerius2/PMF,8 DOCK/FF,31 Cerius2/LUDI,33,34 Cerius2/LigScore,35 SYBYL/F-Score,36 AutoDock.37
Conclusion and Outlook
Based on atom pairwise interactions, interaction enthalpy terms in KECSA were parameterized by combining PMF theory with the Lennard-Jones potential, without fitting to any binding affinity data. This procedure parameterizes the LJ potential with neither QM calculations nor binding affinity data, hence lowering the computational expense while improving the prediction accuracy relative to empirical scoring functions. Generally, KECSA improves the binding affinity RMSE, when compared to LISA and LISA+, especially for complexes dominated by polar and charged interactions. With respect to ranking predictions, KECSA better distinguishes complexes in the large-scale test sets. KECSA yields the lowest RMSE values illustrated by its superior performance in all test sets for this measure of quality. It is less responsive, however, to minor structural changes of the ligand or binding pocket, reducing its ability to rank complexes from the same or similar protein families. In the KECSA model, the solvent accessible surface area is introduced to describe the desolvation effect and entropy terms were empirically modeled. Since we have formulated and parameterized the enthalpy component of non-covalent interactions with this alternative method, an interesting possibility is to use similar procedures to build a force field model solely based on experimental structural data. In this way, the desolvation and entropy terms can also be included instead of using empirical models. We believe that more accurate and effective scoring methods can be developed using this concept.
Our group has constructed an in-house docking program where KECSA (as well as LISA and LISA+) is employed as the scoring module in this program. KECSA's ability to score docking poses and distinguish native poses from decoys will be further evaluated and refined in future work using this docking module.
Supplementary Material
Table 3.
Entropy parameters and their 95% confidence intervals
| parameter | 95% confidence | interval | |
|---|---|---|---|
| enthalpy | 0.0928 | 0.0650 | 0.1206 |
| number of rotatable bonds | 0.0900 | 0.0601 | 0.1200 |
| molecular mass | −0.0170 | −0.0191 | −0.0149 |
| N_number | 0.2455 | 0.1838 | 0.3072 |
| O_number | 0.3131 | 0.2528 | 0.3733 |
| Number of aromatic bonds | 0.0359 | 0.0130 | 0.0588 |
| nonpolar buried surface area | 0.0152 | 0.0047 | 0.0257 |
| total buried surface area | −0.0089 | −0.0167 | −0.0012 |
| nonpolar buried surface | |||
| area/total surface area | −4.1454 | −6.9496 | −1.3412 |
| total buried surface area/total | |||
| surface area | −6.2438 | −8.1509 | −4.3368 |
Acknowledgments
We would like to thank the NIH (GM044974 and GM066859) for supporting the present research. Dr. Michael Weaver is also acknowledged for numerous helpful discussions.
Footnotes
Supporting Information
Supporting Information is available including the Lennard-Jones potential models parameter η values and Predicted pKd or pKi vs. experimental pKd or pKi for our test sets. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Sippl MJ. Calculation of conformational ensembles from potentials of mean force. J. Mol. Biol. 1990;213:859–883. doi: 10.1016/s0022-2836(05)80269-4. [DOI] [PubMed] [Google Scholar]
- 2.Miyazawa S, Jernigan RL. Estimation of effective interresidue contact energies from protein crystal structures: quasi-chemical approximation. Macromolecules. 1985;18:534–552. [Google Scholar]
- 3.Hendlich M, Lackner P, Weitckus S, Floeckner H, Froschauer R, Gottsbacher K, Casari G, Sippl MJ. Identification of native protein folds amongst a large number of incorrect models. The calculation of low energy conformations from potentials of mean force. J. Mol. Biol. 1990;216:167–180. doi: 10.1016/S0022-2836(05)80068-3. [DOI] [PubMed] [Google Scholar]
- 4.Jones DT, Taylor WR, Thornton JM. A new approach to protein fold recognition. Nature. 1992;358:86–89. doi: 10.1038/358086a0. [DOI] [PubMed] [Google Scholar]
- 5.Thomas PD, Dill KA. An iterative method for extracting energy-like quantities from protein structures. Proc. Natl. Acad. Sci. USA. 1996;93:11628–11633. doi: 10.1073/pnas.93.21.11628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thomas PD, Dill KA. Statistical potentials extracted from protein structures: How accurate are they? J. Mol. Biol. 1996;257:457–469. doi: 10.1006/jmbi.1996.0175. [DOI] [PubMed] [Google Scholar]
- 7.Lu H, Skolnick J. A Distance-Dependent Atomic Knowledge-Based Potential for Improved Protein Structure Selection. Proteins: Struct., Funct., Genet. 2001;44:223–232. doi: 10.1002/prot.1087. [DOI] [PubMed] [Google Scholar]
- 8.Muegge I, Martin YC. A General and Fast Scoring Function for Protein-Ligand Interactions: A Simplified Potential Approach. J. Med. Chem. 1999;42:791–804. doi: 10.1021/jm980536j. [DOI] [PubMed] [Google Scholar]
- 9.Muegge I. A knowledge-based scoring function for protein-ligand interactions: Probing the reference state. Perspect. Drug Discovery Des. 2000;20:99–14. [Google Scholar]
- 10.Muegge I. Effect of ligand volume correction on PMF scoring. J. Comput. Chem. 2001;22:418–425. [Google Scholar]
- 11.Gohlke H, Hendlich M, Klebe G. Knowledge-based scoring function to predict protein-ligand interactions. J. Mol. Biol. 2000;295:337–356. doi: 10.1006/jmbi.1999.3371. [DOI] [PubMed] [Google Scholar]
- 12.Velec HFG, Gohlke H, Klebe G. DrugScore(CSD)-knowledge-based scoring function derived from small molecule crystal data with superior recognition rate of near-native ligand poses and better affinity prediction. J. Med. Chem. 2005;48(20):6296–6303. doi: 10.1021/jm050436v. [DOI] [PubMed] [Google Scholar]
- 13.DeWitte RS, Shakhnovich EI. SMoG: de Novo design method based on simple, fast, and accutate free energy estimate. 1. Methodology and supporting evidence. J. Am. Chem. Soc. 1996;118:11733–11744. [Google Scholar]
- 14.Ishchenko AV, Shakhnovich EI. Small molecule growth 2001 (SMoG2001): An improved knowledge-based scoring function for protein-ligand interactions. J. Med. Chem. 2002;45:2770–2780. doi: 10.1021/jm0105833. [DOI] [PubMed] [Google Scholar]
- 15.Mitchell JBO, Laskowski RA, Alex A, Thornton JM. BLEEP–potential of mean force describing protein–ligand interactions: I. Generating potential. J. Comput. Chem. 1999;20:1165–1176. [Google Scholar]
- 16.Mitchell JBO, Laskowski RA, Alex A, Forster MJ, Thornton JM. BLEEP - Potential of mean force describing protein-ligand interactions: II. Calculation of binding energies and comparison with experimental data. J. Comput. Chem. 1999;20(11):1177–1185. [Google Scholar]
- 17.Huang S-Y, Zou X. An iterative knowledge-based scoring function to predict protein-ligand interactions: II. Validation of the scoring function. J. Comput. Chem. 2006;27:1876–1882. doi: 10.1002/jcc.20505. [DOI] [PubMed] [Google Scholar]
- 18.Huang S-Y, Zou X. Inclusion of Solvation and Entropy in the Knowledge-Based Scoring Function for Protein-Ligand Interactions. J. Chem. Inf. Model. 2010;50:262–273. doi: 10.1021/ci9002987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirkwood JG. Statistical Mechanics of fluid Mixtures. J. Chem. Phys. 1935;3:300–313. [Google Scholar]
- 20.Fan H, Schneidman-Duhovny D, Irwin JJ, Dong G, Shoichet BK, Sali A. Statistical potential for modeling and ranking of protein-ligand interactions. J. Chem. Inf. Model. 2011;51(12):3078–3092. doi: 10.1021/ci200377u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang R, Fang X, Lu Y, Wang S. The PDBbind Database: Collection of Binding Affinities for Protein-Ligand Complexes with KnownThree-Dimensional Structures. J. Med. Chem. 2004;47:2977–2980. doi: 10.1021/jm030580l. [DOI] [PubMed] [Google Scholar]
- 22.Wang R, Fang X, Lu Y, Yang C-Y, Wang S. The PDBbind Database: Methodologies and Updates. J. Med. Chem. 2005;48:4111–4119. doi: 10.1021/jm048957q. [DOI] [PubMed] [Google Scholar]
- 23.Zheng Z, Merz KM. Ligand Identification Scoring Algorithm (LISA) J. Chem. Inf. Model. 2011;51:1296–1306. doi: 10.1021/ci2000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson ML, Faver JC, Ucisik MN, Dashti DS, Zheng Z, Merz KM. Prediction of trypsin/molecular fragment binding affinities by free energy decomposition and empirical scores. J Comput Aided Mol Des. 2012;26(5):647–659. doi: 10.1007/s10822-012-9567-9. [DOI] [PubMed] [Google Scholar]
- 25.Wang R, Lu Y, Wang S. Comparative Evaluation of 11 Scoring Functions for Molecular Docking. J. Med. Chem. 2003;46:2287–2303. doi: 10.1021/jm0203783. [DOI] [PubMed] [Google Scholar]
- 26.Wang R, Lai L, Wang S. Further development and validation of empirical scoring functions for structure-based binding affinity prediction. J. Comput.-Aided Mol. Des. 2002;16:11–16. doi: 10.1023/a:1016357811882. [DOI] [PubMed] [Google Scholar]
- 27.Zhang C, Liu S, Zhu Q, Zhou Y. A Knowledge-Based Energy Function for Protein-Ligand, Protein-Protein, and Protein-DNA Complexes. J. Med. Chem. 2005;48:2325–2335. doi: 10.1021/jm049314d. [DOI] [PubMed] [Google Scholar]
- 28.Gehlhaar DK, Verkhivker GM, Rejto PA, Sherman CJ, Fogel DB, Freer ST. Molecular recognition of the inhibitor AG-1343 by HIV-1 Protease: Conformationally flexible docking by evolutionary programming. Chem. Biol. 1995;2:317–324. doi: 10.1016/1074-5521(95)90050-0. [DOI] [PubMed] [Google Scholar]
- 29.Gehlhaar DK, Bouzida D, Rejto PA. In: Rational Drug Design: Novel Methodology and Practical Applications. Parrill L, Reddy MR, editors. Vol.719. Washington, DC: American Chemical Society; 1999. pp. 292–211. [Google Scholar]
- 30.Jones G, Willett P, Glen RC, Leach AR, Talor R. Development and validation of a genetic algorithm for flexible docking. J. Mol. Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- 31.Meng EC, Shoichet BK, Kuntz ID. Automated docking with grid-based energy approach to macromolecule-ligand interactions. J. Comput. Chem. 1992;13:505–524. [Google Scholar]
- 32.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. Empirical scoring functions: I. The development of a fast empirical scoring function to estimate the binding affinity of ligands in receptor complexes. J. Comput.-Aided Mol. Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 33.Böhm HJ. The development of a simple empirical scoring function to estimate the binding constant for a protein-ligand complex of known three-dimensional structure. J. Comput.-Aided Mol. Des. 1994;8:243–256. doi: 10.1007/BF00126743. [DOI] [PubMed] [Google Scholar]
- 34.Böhm HJ. Prediction of binding constants of ptotein ligands: A fast method for the polarization of hits obtained from de novo design or 3D database search programs. J. Comput.-Aided Mol. Des. 1998;12:309–323. doi: 10.1023/a:1007999920146. [DOI] [PubMed] [Google Scholar]
- 35.CERIUS2 LigandFit User Manual. San Diego, CA: Accelrys Inc; 2000. pp. 3–48. [Google Scholar]
- 36.Rarey M, Kramer B, Lengauer T, Klebe G. A fast flexible docking method using an incremental construction algorithm. J. Mol. Biol. 1996;261:470–489. doi: 10.1006/jmbi.1996.0477. [DOI] [PubMed] [Google Scholar]
- 37.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J. Comput. Chem. 1998;19:1639–1662. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.