Abstract
The Hha and YdgT proteins are suggested to modulate the expression of horizontally acquired genes by interacting with H-NS and StpA, which play central roles in the transcriptional silencing of such genes. However, it is also possible that Hha/YdgT repress gene expression independently of H-NS/StpA, as we have not fully understood the molecular mechanism through which Hha/YdgT modulate H-NS/StpA activity. To gain further insight into the basic functions of Hha/YdgT, we analysed the impact of hha/ydgT double inactivation on the transcriptome profile of Escherichia coli K-12, and compared the effects with that of hns/stpA double inactivation. In addition, we examined the effects of hha/ydgT inactivation on the chromosomal binding of H-NS, and conversely the effects of hns/stpA inactivation on the chromosomal binding of Hha. Our results demonstrated that the chromosomal binding of Hha requires H-NS/StpA, and is necessary for the repression of a subset of genes in the H-NS/StpA regulon. Furthermore, the distribution of H-NS binding around Hha/YdgT-dependent and -independent genes suggests that Hha/YdgT proteins modulate formation of the H-NS/StpA-DNA complex.
Keywords: Escherichia coli, Hha, YdgT, H-NS, StpA
1. Introduction
The H-NS protein is a major nucleoid component and is conserved amongst γ-proteobacteria. Recent studies have revealed that H-NS plays a central role in transcriptional silencing of horizontally acquired genes1–5 by binding to the nucleation sites in AT-rich sequences and forming higher-order nucleo-protein complexes.6–9 Escherichia coli and related enteric bacteria possess four H-NS homologues, such as H-NS, StpA, Hha and YdgT,10 and their mutual interactions have been demonstrated.11–13
Both H-NS and StpA comprise two domains, with the N-terminal domain involved in dimerization and oligomerization and the C-terminal domain involved in DNA binding.14 Inactivation of hns was found to result in de-repression of hundreds of genes in E. coli and Salmonella.2,4,14,15 In contrast, stpA inactivation is not associated with any notable phenotype under standard growth conditions in E. coli, probably because its function is compensated by H-NS. Conversely, it has been suggested that StpA partially compensates hns inactivation.16–18 Also, stpA transcription is induced in an hns mutant, and growth impairment of the hns/stpA double mutant is severer than that of the hns single mutant.19,20 Recently, we reported that the StpA-binding regions on the E. coli K-12 chromosome essentially overlap with those of H-NS, and that StpA binding was reduced to one-third in the hns mutant, but H-NS binding was unaffected by stpA inactivation.21 Thus, StpA is considered to be a molecular back-up of H-NS in E. coli.13,19–21
Hha structurally resembles the N-terminal domain of H-NS,10 and its expression is sensitive to osmolality.22 Hha was first identified as a repressor of the haemolysin operon (hlyCABD) in E. coli 5K.23–25 In Salmonella enterica, Hha negatively regulates hilA and rtsA, each encoding activator of virulence genes.26–29 Furthermore, the Ler regulator of the esc operon in E. coli O157:H7 is negatively regulated by Hha.30 These virulence genes are under the control of H-NS, and Hha is generally believed to modulate the DNA-binding and nucleoid-organizing properties of H-NS.10,24,25 Gel retardation analysis has shown that Hha binds to the regulatory regions of the rtsA and the hilA gene.26,29 Till now, functional studies of Hha have been hampered by the presence of its paralog, YdgT.10 Transcription of ydgT is induced by hha inactivation, while its overexpression was shown to repress the hly operon in the hha mutant,11 suggesting that the functions of Hha and YdgT may overlap. Furthermore, simultaneous inactivation of hha and ydgT in S. enterica induced numerous genes located in AT-rich, horizontally acquired DNA sequences, many of which are reported to be targets of H-NS.31,32
Taken together, these data indicate that Hha/YdgT function with H-NS/StpA to regulate the expression of horizontally acquired genes. The lack of clear DNA-binding domains in Hha and YdgT strongly suggests that they interact with H-NS/StpA to confer their regulatory activity. However, it remains possible that Hha/YdgT may repress gene expression independently of H-NS/StpA. In addition, the molecular mechanism through which Hha/YdgT modulate the activity of H-NS/StpA in a subset of genes in the H-NS/StpA regulon remains to be clarified. Hha/YdgT are conserved in non-pathogenic E. coli, and Hha is reportedly involved in regulating htrA expression to promote survival at high temperatures,33 and in regulating biofilm formation in E. coli K-12.34
To gain further insight into the basic function of Hha/YdgT, we compared the transcriptome profiles of hha/ydgT double-inactivation mutants with those of hns/stpA double-inactivation mutants in E. coli K-12. In addition, we examined the effects of hha/ydgT inactivation on the chromosomal binding of H-NS, and the effects of hns/stpA inactivation on the chromosomal binding of Hha as shown below.
2. Materials and methods
2.1. Bacterial strains, plasmids and media
The E. coli K-12 strains (strain W3110 and its derivatives) and plasmids used in this study are listed in Supplementary Table S1. The construction of mutant strains and the media used in this study are detailed in Supplementary Methods.
2.2. Transcriptome analysis
The detailed procedures for RNA purification, probe preparation and data acquisition using the Affymetrix E. coli genome 2.0 array are described in Supplementary Methods. The raw data (CEL format) from transcriptome experiments have been deposited in ArrayExpress under accession number E-MEXP-3811.
2.3. Profiling chromosome binding of H-NS and Hha
H-NS and Hha binding profiles were determined using a slight modification of the previously described ChIP-chip21 and ChAP-chip methods.35,36 The detailed procedures for these analysis were described in Supplementary Methods. The raw data (CEL format) from the ChIP-chip and the ChAP-chip experiments have been deposited in ArrayExpress under accession numbers E-MEXP-3812, E-MEXP-3813, respectively.
3. Results and discussion
3.1. Transcriptome analysis of hha/ydgT and hns/stpA double mutants of E. coli K-12
To avoid compensatory effects of the paralogous proteins, we created hha/ydgT and hns/stpA double mutants of E. coli K-12, and compared their transcriptome profiles with that of wild-type. Also, we used cells cultivated under high osmolality condition (LB medium containing 0.3 M NaCl) in which Hha regulated the expression of proteins in E. coli.37 Escherichia coli cells were grown aerobically and the expression level of each gene was assessed using an Affymetrix E. coli genome 2.0 array. Up- or down-regulation in the mutant was judged by the difference in the expression level of >4- or <0.25-fold (>2- or <−2-fold in log2 scale) compared with that of wild-type, with a false discovery rate (FDR) of <0.1.
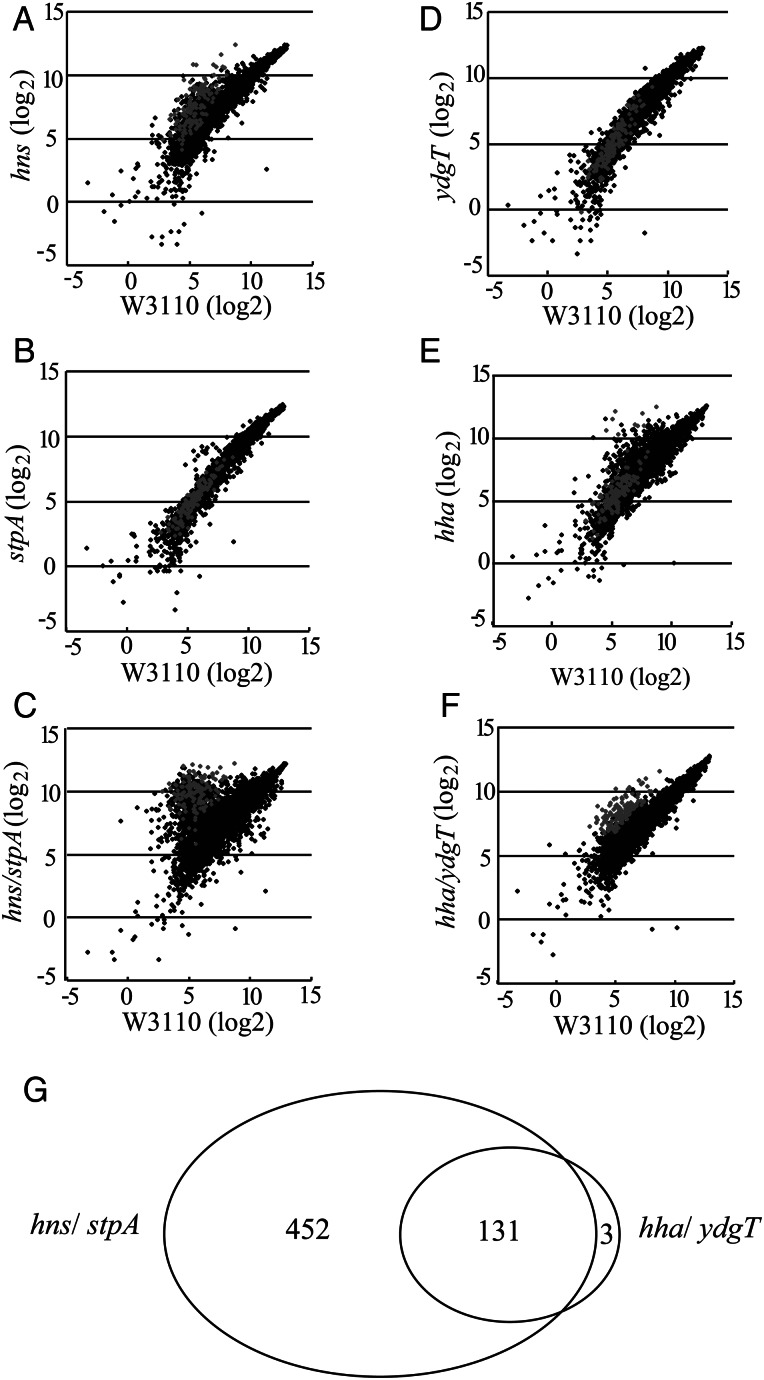
The single inactivation of hns had a significant impact on the transcriptome, resulting in up-regulation of 172 genes and down-regulation of one gene (Fig. 1A and Supplementary Table S2). Up-regulation of gene expression in the hns mutant could reflect indirect effects of down-regulation of the hns homologues. However, we believe that this is unlikely, since hns inactivation up-regulated the expression level of stpA, hha and ydgT by 1.98-, 1.41- and 2.03- (log2) fold, respectively, compared with that in wild-type (Supplementary Table S3). In contrast, the single inactivation of stpA had a far less significant effect (Fig. 1B).
Figure 1.
Transcriptome analysis of mutant cells. (A–F) Log-scale scatter plots (log2) of the transcriptional intensities of each gene in hns (A), stpA (B), hns/stpA (C), ydgT (D), hha (E) and hha/ydgT (F) mutant cells (vertical axis) compared with those in wild-type cells (horizontal axis). The average signal intensities from two independent experiments using each strain are plotted. Genes up-regulated in hha/ydgT mutant cells are shown as grey dots. (G) Venn diagram indicates the number of shared and unique genes up-regulated in hha/ydgT and hns/stpA mutant cells.
The hns/stpA double mutant showed further alterations in the transcriptome profile, with up-regulation of 583 genes, 167 of which were included in the 172 genes up-regulated in the hns single mutant, and down-regulation of 86 genes compared with wild-type (Fig. 1C and Supplementary Table S4). Notably, the up-regulation of hha and ydgT seen in the hns mutant also occurred in the hns/stpA double mutant (Supplementary Table S3), indicating that the up-regulation observed in the hns/stpA mutant was not caused by the down-regulation of hha and/or ydgT. Of the genes up-regulated by hns inactivation, 69% (119 of 172) have been proposed to be horizontally acquired.38,39 Similarly, 62% (363 of 583) of the genes up-regulated in the double mutant were horizontally acquired.
The single inactivation of ydgT had a much less effect, up-regulating only one gene (yhjX) and down-regulating six genes (yfiD, sraA, pdhR, tke1, yifE and yejG; Fig. 1D). In contrast, single hha inactivation up-regulated 113 genes and down-regulated 8 genes (Fig. 1E and Supplementary Table S5). In addition, it moderately up-regulated ydgT [1.48- (log2) fold], but did not cause significant changes in hns and stpA (FDR > 0.1; Supplementary Table S3), indicating that the observed up-regulation was a direct consequence of the hha inactivation. The genes up-regulated in the hha mutant included those related to osmotic (e.g. osmY, kdpA and kdpC) and carbon-starvation (e.g. csiD and rmf) stresses, which is consistent with the previous finding that genes related to these stresses were induced in hha mutant cells.34 The hha/ydgT double inactivation showed transcriptional alterations in a similar number of genes, with 134 genes up-regulated and 5 genes down-regulated (Fig. 1F and Supplementary Table S6). Consistent with the previous observation in S. enterica,32 most of the genes that were up-regulated in hha/ydgT-inactivated cells (108 of 134) are believed to be horizontally acquired (Supplementary Table S6). No statistically significant change was detected in the expression of hns or stpA in the hha/ydgT mutant (Supplementary Table S3).
Only 12 genes were commonly up-regulated in the hha and hha/ydgT mutants (Fig. 1E, grey dots; Supplementary Tables S5 and S6). Furthermore, although most of the genes that were up-regulated in the hha/ydgT mutant were also up-regulated in the hns/stpA mutant (see below), only 47 of the 113 genes that were up-regulated in the hha single mutant were up-regulated in the hns/stpA mutant (Supplementary Tables S4 and S5). These results suggest that the mechanism responsible for the up-regulation of genes in the single hha mutant differs from that in the hha/ydgT double mutant, although we did not further explore the molecular mechanism causing this phenotype in the hha mutant.
In contrast to the altered transcriptome profile in the hha mutant, 131 of the 134 genes that were up-regulated in the hha/ydgT double mutant were also up-regulated in the hns/stpA double mutant (Fig. 1G, Venn diagram; Fig. 1C, grey dots; Supplementary Tables S4 and S6). Of the three genes that were significantly up-regulated only in the hha/ydgT mutant, two were probably also up-regulated in the hns/stpA mutant, though at a low signal intensity (fimZ) or low induction level (yhjA). Thus, only ycgX appears to be regulated by Hha/YdgT and not by H-NS/StpA. Taken together, these results indicate that Hha and YdgT are additionally required for the repression of a subset of genes that are repressed by H-NS and StpA in E. coli K-12.
3.2. H-NS binding profiles are not altered by hha/ydgT inactivation
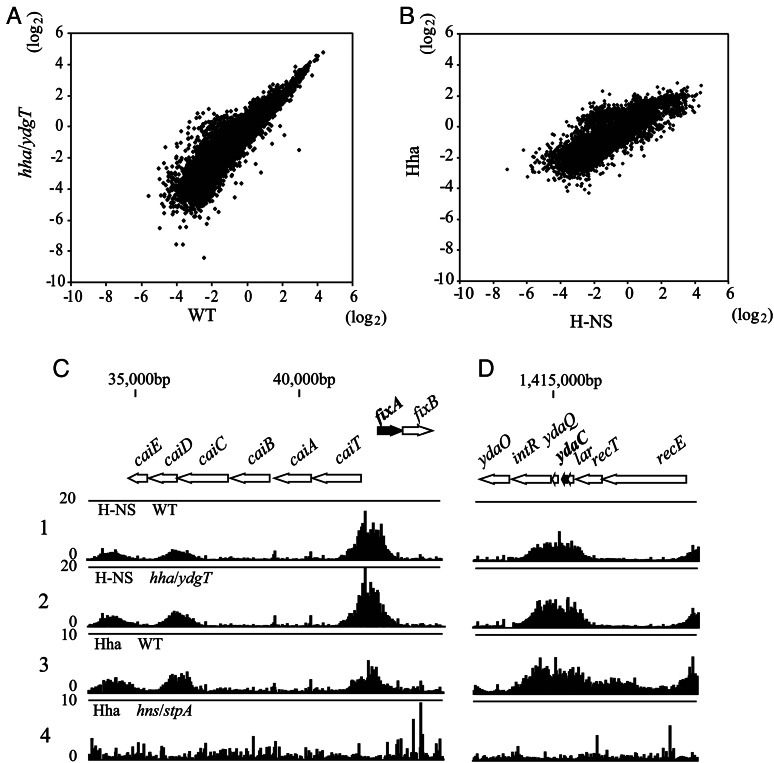
To investigate whether Hha/YdgT modulate the DNA-binding activity of H-NS/StpA, we examined effects of hha/ydgT inactivation on the chromosomal binding of H-NS. To this end, we introduced the hha/ydgT mutation into a strain in which H-NS was tagged with 3× Flag and placed under the control of the native promoter.5,21 Wild-type and hha/ydgT mutant cells expressing H-NS-3× Flag were cultivated in LB (+0.3 M NaCl) medium, formaldehyde treated to crosslink H-NS-Flag with the chromosomal DNA, and then harvested and subjected to immunoprecipitation of H-NS–DNA complexes. The DNA fragments co-purified with H-NS were mapped onto the chromosome by using a custom Affymetrix tiling array. As shown in Figure 2A, scatter plots of the H-NS binding signals in both strains showed a genome-wide positive correlation (correlation coefficient = 0.91). H-NS-binding regions, 417 regions in wild-type and 530 in the hha/ydgT mutant, were reproducibly observed in duplicate ChIP-chip analyses (Supplementary Fig. S1 and Table S7). Moreover, 96% (400 of 417) of the binding regions in wild-type overlapped with those in the hha/ydgT mutant (Supplementary Table S7).
Figure 2.
Impact of hha/ydgT double inactivation on H-NS bindings and the genome-wide correlation of H-NS and Hha bindings. (A and B) Log scatter plots (log2) of the average signal intensities of H-NS binding signals from two independent experiments using wild-type (W3110) and hha/ydgT mutant cells (A), and H-NS binding signals in wild-type (W3110) cells and Hha binding signals in W3110 pQEHha cells (B). (C and D) Typical examples of H-NS-binding profiles in W3110 (lane 1) and W3110 hha::Km ydgT::Cm (lane 2), and Hha-binding profiles in W3110 pQE80Hha (lane 3) and W3110 hns::Km stpA::Cm pQE80Hha (lane 4) are shown with the binding signal of each probe mapped to the corresponding position in the E. coli chromosome. The binding intensity (vertical axis) was determined as the relative ratio of the signal intensity for the hybridization of labelled DNA fragments prepared from the ChIP (ChAP) versus Sup fractions in each experiment. The Hha binding signals were low throughout the genome of hns/stpA mutant cells (lane 4), and the background spike signals were enhanced when we conferred a signal average of 500 during signal intensity normalization prior to calculating the binding intensity. Shown are the H-NS and Hha binding profiles in the vicinity of a gene (fixA; black thick arrow) that was up-regulated only in hns/stpA mutant cells (C), or the H-NS- and Hha-binding profiles in the vicinity to a gene (ydaC) that was up-regulated in both hns/stpA and hha/ydgT mutant cells (D).
Although many of the H-NS-binding regions found in wild-type were segmented in the hha/ydgT mutant and H-NS-binding signals were slightly stronger in the hha/ydgT mutant than in wild-type, 84% (444 of 530) of the regions overlapped with those in wild-type (Supplementary Table S7) and weak binding signals (below the threshold) were observed in wild-type in the remaining regions as well (Supplementary Fig. S1). In addition, there was no significant difference between the effect of Hha/YdgT inactivation on H-NS binding in the vicinity of genes up-regulated only in hns/stpA mutant (Fig. 2C) and of those up-regulated in both hha/ydgT and hns/stpA mutants (Fig. 2D). Thus, we concluded that the hha/ydgT mutation had little effect on the DNA-binding property of H-NS.
3.3. Hha associates with the chromosome in an H-NS/StpA-dependent manner
To examine whether Hha/YdgT interact with target genes independently of H-NS/StpA, we analysed Hha binding on the chromosome in wild-type and the hns/stpA mutant. As the expression level of Hha from the native promoter was expected to be low, to obtain clear chromosomal binding profile by the ChIP-chip analysis,12 cells of wild-type and the hns/stpA mutant expressing 6× histidine-tagged Hha (6 × His-Hha) from a multi-copy plasmid were cultivated in LB (+0.3 M NaCl) medium and subjected to a modified ChIP-chip analysis (ChAP-chip analysis) in which His-tagged Hha–DNA complexes were affinity purified using nickel affinity resin. Western blot analysis of cell extracts with an anti-His antibody indicated that the expression level of 6 × His-Hha in wild-type and hns/stpA mutant cells was similar (data not shown). Therefore, the binding signals of Hha consistently overlapped with those of H-NS in wild-type (Fig. 2C and D lane 3; Supplementary Fig. S1), while the Hha-binding signals seen in wild-type disappeared in the hns/stpA mutant (Fig. 2C and D lane 4; Supplementary Fig. S1). Scatter plots of Hha- and H-NS-binding signals (Fig. 2B) support the genome-wide positive correlation of H-NS- and Hha-binding signals (correlation coefficient = 0.77). Seventy-two per cent (302 of 417) of the H-NS-binding regions were found to overlap with the Hha-binding regions, and conversely 74% (352 of 476) of the Hha-binding regions overlapped with the H-NS-binding regions (Supplementary Table S8). This lesser level of overlap between the Hha- and H-NS-binding regions may be due to the low binding signals against the high background seen in the ChAP-chip analysis of Hha binding, which may reflect its indirect interaction with the chromosomal DNA. It should also be noted that Hha binding exhibited no preference towards the genes up-regulated in hns/stpA and those up-regulated in both hha/ydgT and hns/stpA under the experimental conditions (compare lane 3 in panels Fig. 2C and D). However, it is possible that the reduced amount of Hha/YdgT expressed from the native promoter form a limited amount of H-NS/StpA-Hha/YdgT complexes that recognize the specific regions. Further analysis is needed to clarify the possibility.
Only five regions were found to be possible Hha-binding regions in the hns/stpA mutant (Supplementary Table S9), although the signals observed in the lacI and hha genes may probably be due to contamination from the multi-copy plasmid harbouring lacI and hha. Therefore, H-NS/StpA-independent chromosomal binding of Hha appear to be possible only in the intergenic region of ygjI–ygjH and the coding region of fecA. We concluded that the binding of Hha to the chromosome essentially depends on H-NS/StpA.
3.4. Hha/YdgT support transcriptional repression by H-NS/StpA bound to the coding sequences
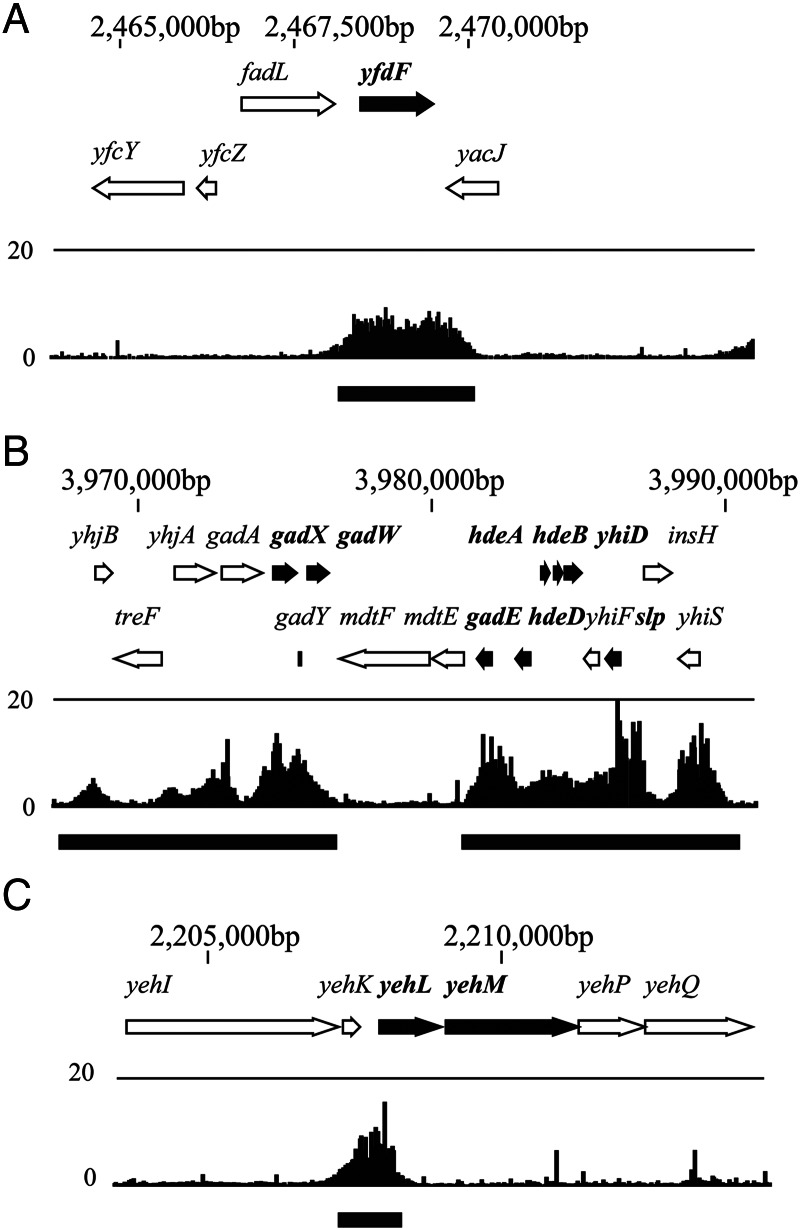
It has been suggested that the binding of H-NS is biased to intergenic regions, where H-NS plays a regulatory role, while its binding to coding regions may be linked to chromosome organization in E. coli.40 Our inspection of H-NS-binding signals in and around the genes that are co-repressed by Hha/YdgT and H-NS/StpA suggest that the coding sequences of Hha/YdgT-dependent genes tend to be bound by H-NS (Fig. 3A). Furthermore, such H-NS binding often extended into multiple coding sequences (Fig. 3B). In contrast, the H-NS binding seemed to be localized in the intergenic regions of the genes exhibiting Hha/YdgT-independent up-regulation (Fig. 3C).
Figure 3.
H-NS binding signals for genes up-regulated in the hns/stpA mutant alone or in both hns/stpA and hha/ydgT mutants. Figures represent the typical H-NS binding signals classified as ‘coding (single)’ [in the vicinity of gene (yfdF; the thick black arrow) that were up-regulated in both hns/stpA and hha/ydgT mutants] (A), ‘coding (multiple)’ [in the vicinity of genes (yhiD, hdeB, hdeA, hdeD, slp, gadE, gadW and gadX) that were up-regulated in both hns/stpA and hha/ydgT mutants] (B), and ‘intergenic’ [in the vicinity of genes (yehL and yehM) that were up-regulated only in hns/stpA mutants] (C). The H-NS-binding regions (concatenated) are shown by black lines at the bottom of the figures.
To perform genome-wide evaluation of these phenomena, we visually inspected the H-NS-binding regions in wild-type into two classes: those localized to intergenic regions (‘intergenic’) and those extending to coding sequences (‘coding’) by concatenating the H-NS-binding regions with the corresponding intergenic regions or the (successive) coding sequences. A half of the concatenated H-NS-binding regions was classified as ‘intergenic’, as their peaks were located at intergenic sequences (Supplementary Fig. S2A and B), while the remaining half was classified as ‘coding’ (106 regions; Table 1), as their peaks were located at coding sequences or their H-NS-binding signals evenly covered coding sequences (Supplementary Fig. S2C and D). About a half of the ‘coding’ extended into multiple coding sequences (Supplementary Fig. S2E and F; Table 1). Ten regions were classified as both ‘coding’ and ‘intergenic’, as they included the intergenic sequences of one gene and the coding sequences of another gene(s) (for example, see Supplementary Fig. S3D).
Table 1.
Number of H-NS-binding regions in ‘intergenic’ and ‘coding’ sequences
| Location of H-NS-binding regions | Number of regions (un)associated with H-NS/StpA-repressed genes |
Total |
||||
|---|---|---|---|---|---|---|
| Unassociated |
Associated |
|||||
| Localized to intergenic regions | 33 | 73 | 106 | |||
| Covering coding sequencesa | 17 | 89 | 106 | |||
| Single | Multiple | Single | Multiple | Single | Multiple | |
| 15 | 2 | 27 | 62 | 42 | 64 | |
| Total | 50 | 162 | 212 | |||
aH-NS-binding regions covering the coding sequence(s) of single and multiple gene(s) are indicated as ‘Single’ and ‘Multiple’, respectively.
We then examined the H-NS-binding regions in and around the genes that were up-regulated in the hns/stpA mutant, and found that those regions were classified as 73 ‘intergenic’ and 89 ‘coding’ (Table 1). Subsequently, we evaluated the relationship between the localization of H-NS-binding signals and the ability of the genes to undergo Hha/YdgT-dependent or -independent repression by H-NS/StpA (Table 2). A majority of the genes up-regulated in both hha/ydgT and hns/stpA mutants was localized in and around H-NS-binding regions classified as ‘coding’ (108 genes; 88%; Supplementary Fig. S3), including those covering multiple coding sequences. Hha/YdgT-independent genes were more enriched (136 genes; 31%) in the genes that were regulated by binding of H-NS/StpA to their intergenic regions than Hha/YdgT-dependent genes (11 genes; 9%), although many Hha/YdgT-independent genes were also found in ‘coding’ H-NS-binding regions (243 genes; 55%). These results suggest that Hha/YdgT are required for efficient transcriptional repression by H-NS/StpA bound to coding sequences.
Table 2.
The Hha/YdgT-dependent and -independent genes up-regulated in the hns/stpA mutant, and their distribution in and around the ‘intergenic’ and ‘coding’ H-NS-binding regions
| Genes up-regulated in hns/stpA mutanta | Genes with intergenic H-NS binding | Genes with coding H-NS binding |
Genes without H-NS-binding regionsb | |
|---|---|---|---|---|
| Hha/YdgT-independent (436 genes) | 136 (31%)c | 243 (55%) | 57 (13%) | |
| Single | Multiple | |||
| 25 (6%) | 218 (50%) | |||
| Hha/YdgT-dependent (122 genes) | 11 (9%)d | 108 (88%) | 3 (2%) | |
| Single | Multiple | |||
| 12 (10%) | 96 (79%) | |||
aTwenty-five genes that were found to be up-regulated in the hns/stpA mutant cells (as detected by transcriptome analysis using the E. coli genome 2.0 array) were not annotated in the genome sequence of E. coli K-12 strain W3110, which was used to design the custom tiling chip for ChIP-chip analysis. We evaluated the correlation of transcriptome alteration and H-NS binding profiles according to the W3110 annotation, and analysed the locations of the 436 genes up-regulated only in the hns/stpA mutant and the 122 genes up-regulated in both the hns/stpA and hha/ydgT mutants. Genes located in or around the H-NS-binding regions were classified as ‘coding’ or ‘intergenic’, respectively.
bSum of the genes that were not located in or around the H-NS-binding regions but showed up-regulation in the hns/stpA mutant alone or in both the hns/stpA and hha/ydgT mutants.
cNumber and percent with respect to the 436 genes up-regulated only in the hns/stpA mutant.
dNumber and percent with respect to the 122 genes up-regulated in both the hns/stpA and hha/ydgT mutants.
3.5. Possible mechanism of transcriptional repression by H-NS and Hha
A number of mechanisms have been proposed to explain the inhibition of transcriptional initiation or elongation by H-NS.10,14,41,42 H-NS binds to two distinct sites and forms a bridge structure to block the binding of RNA polymerase (RNAP) to a promoter43 or trap RNAP in an open complex at a promoter, thereby inhibiting transcriptional elongation.44,45 Others have shown that the binding of H-NS upstream of a regulatory region interferes with either promoter clearance or progression of RNAP,46 and H-NS has been shown to constrain local DNA topology to regulate the promoter activity of supercoiling-sensitive genes.47–49 Mutations impairing the ability of H-NS and its homologue of Pseudomonas aeruginosa termed MvaT to form higher-order oligomers have been shown to reduce their abilities to repress transcription of the E. coli proU and P. aeruginosa cupA fimbria genes.18,50,51 The formation of a DNA–protein filament through cooperative polymerization of MvaT or H-NS along DNA and of protein bridges to constrain DNA loops through the interaction of the DNA–protein filaments were suggested to be important for transcriptional repression.7,52 The direct interaction of Hha and C-terminally truncated H-NS (H-NS64) was previously demonstrated by NMR and fluorescence anisotropy.53,54 In the absence of Hha, H-NS64 formed only dimers even at a high concentration; in the presence of Hha, however, H-NS64 formed high-molecular-mass Hha-(H-NS64) hetero-oligomers.53 We speculate that Hha enhances the higher-order oligomerization of H-NS/StpA and contributes to the formation of DNA–protein filaments and DNA loops, which often include the coding sequences of target genes. The 5.5 protein of phage T7 binds to the oligomerization domain of H-NS to inhibit the oligomerization of H-NS and the repression of more than 200 genes, although the DNA binding of H-NS is not abolished.55 Up-regulation of genes observed in the hha/ydgT mutant might be similar to that caused by the T7 5.5 protein. Even though H-NS binding is not greatly affected in hha/ydgT mutant cells, the oligomerization of H-NS/StpA might be reduced, resulting in the up-regulation of the genes that are normally repressed by oligomerized H-NS/StpA. However, we do not yet fully understand the mechanism underlying the enhancement of H-NS/StpA oligomerization by Hha/YdgT and the repression of transcription by oligomerized H-NS/StpA. Future work is needed to elucidate the co-repression activity of Hha/YdgT and H-NS/StpA.
Supplementary data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by JSPS KAKENHI [grant numbers 23241062 (to N.O.) and 23580114 (to T.O.)].
Supplementary Material
Acknowledgements
We thank the editor and reviewers for valuable comments to revise the manuscript.
Footnotes
Edited by Dr Katsumi Isono
References
- 1.Dillon S.C., Cameron A.D., Hokamp K., Lucchini S., Hinton J.C., Dorman C.J. Genome-wide analysis of the H-NS and Sfh regulatory networks in Salmonella Typhimurium identifies a plasmid-encoded transcription silencing mechanism. Mol. Microbiol. 2010;76:1250–65. doi: 10.1111/j.1365-2958.2010.07173.x. [DOI] [PubMed] [Google Scholar]
- 2.Lucchini S., Rowley G., Goldberg M.D., Hurd D., Harrison M., Hinton J.C. H-NS mediates the silencing of laterally acquired genes in bacteria. PLoS Pathog. 2006;2:e81. doi: 10.1371/journal.ppat.0020081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Navarre W.W., McClelland M., Libby S.J., Fang F.C. Silencing of xenogeneic DNA by H-NS-facilitation of lateral gene transfer in bacteria by a defense system that recognizes foreign DNA. Genes Dev. 2007;21:1456–71. doi: 10.1101/gad.1543107. [DOI] [PubMed] [Google Scholar]
- 4.Navarre W.W., Porwollik S., Wang Y., et al. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science. 2006;313:236–8. doi: 10.1126/science.1128794. [DOI] [PubMed] [Google Scholar]
- 5.Oshima T., Ishikawa S., Kurokawa K., Aiba H.,, Ogasawara N. Escherichia coli histone-like protein H-NS preferentially binds to horizontally acquired DNA in association with RNA polymerase. DNA Res. 2006;13:141–53. doi: 10.1093/dnares/dsl009. [DOI] [PubMed] [Google Scholar]
- 6.Bouffartigues E., Buckle M., Badaut C., Travers A., Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007;14:441–8. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 7.Lang B., Blot N., Bouffartigues E., et al. High-affinity DNA binding sites for H-NS provide a molecular basis for selective silencing within proteobacterial genomes. Nucleic Acids Res. 2007;35:6330–7. doi: 10.1093/nar/gkm712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rimsky S. Structure of the histone-like protein H-NS and its role in regulation and genome superstructure. Curr. Opin. Microbiol. 2004;7:109–14. doi: 10.1016/j.mib.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 9.Rimsky S., Zuber F., Buckle M., Buc H. A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 2001;42:1311–23. doi: 10.1046/j.1365-2958.2001.02706.x. [DOI] [PubMed] [Google Scholar]
- 10.Madrid C., Balsalobre C., Garcia J., Juarez A. The novel Hha/YmoA family of nucleoid-associated proteins: use of structural mimicry to modulate the activity of the H-NS family of proteins. Mol. Microbiol. 2007;63:7–14. doi: 10.1111/j.1365-2958.2006.05497.x. [DOI] [PubMed] [Google Scholar]
- 11.Paytubi S., Madrid C., Forns N., et al. YdgT, the Hha paralogue in Escherichia coli, forms heteromeric complexes with H-NS and StpA. Mol. Microbiol. 2004;54:251–63. doi: 10.1111/j.1365-2958.2004.04268.x. [DOI] [PubMed] [Google Scholar]
- 12.Nieto J.M., Madrid C., Miquelay E., Parra J.L., Rodriguez S.,, Juarez A. Evidence for direct protein-protein interaction between members of the enterobacterial Hha/YmoA and H-NS families of proteins. J. Bacteriol. 2002;184:629–35. doi: 10.1128/JB.184.3.629-635.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johansson J., Uhlin B.E. Differential protease-mediated turnover of H-NS and StpA revealed by a mutation altering protein stability and stationary-phase survival of Escherichia coli. Proc. Natl Acad. Sci. USA. 1999;96:10776–81. doi: 10.1073/pnas.96.19.10776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorman C.J. H-NS: a universal regulator for a dynamic genome. Nat. Rev. 2004;2:391–400. doi: 10.1038/nrmicro883. [DOI] [PubMed] [Google Scholar]
- 15.Hommais F., Krin E., Laurent-Winter C., et al. Large-scale monitoring of pleiotropic regulation of gene expression by the prokaryotic nucleoid-associated protein, H-NS. Mol. Microbiol. 2001;40:20–36. doi: 10.1046/j.1365-2958.2001.02358.x. [DOI] [PubMed] [Google Scholar]
- 16.Deighan P., Free A.,, Dorman C.J. A role for the Escherichia coli H-NS-like protein StpA in OmpF porin expression through modulation of micF RNA stability. Mol. Microbiol. 2000;38:126–39. doi: 10.1046/j.1365-2958.2000.02120.x. [DOI] [PubMed] [Google Scholar]
- 17.Shi X.,, Bennett G.N. Plasmids bearing hfq and the hns-like gene stpA complement hns mutants in modulating arginine decarboxylase gene expression in Escherichia coli. J. Bacteriol. 1994;176:6769–75. doi: 10.1128/jb.176.21.6769-6775.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Williams R.M., Rimsky S.,, Buc H. Probing the structure, function, and interactions of the Escherichia coli H-NS and StpA proteins by using dominant negative derivatives. J. Bacteriol. 1996;178:4335–43. doi: 10.1128/jb.178.15.4335-4343.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sonden B.,, Uhlin B.E. Coordinated and differential expression of histone-like proteins in Escherichia coli: regulation and function of the H-NS analog StpA. EMBO J. 1996;15:4970–80. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang A., Rimsky S., Reaban M.E., Buc H.,, Belfort M. Escherichia coli protein analogs StpA and H-NS: regulatory loops, similar and disparate effects on nucleic acid dynamics. EMBO J. 1996;15:1340–9. [PMC free article] [PubMed] [Google Scholar]
- 21.Uyar E., Kurokawa K., Yoshimura M., Ishikawa S., Ogasawara N., Oshima T. Differential binding profiles of StpA in wild-type and h-ns mutant cells: a comparative analysis of cooperative partners by chromatin immunoprecipitation-microarray analysis. J. Bacteriol. 2009;191:2388–91. doi: 10.1128/JB.01594-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mourino M., Balsalobre C., Madrid C., et al. Osmolarity modulates the expression of the Hha protein from Escherichia coli. FEMS Microbiol. Lett. 1998;160:225–9. doi: 10.1111/j.1574-6968.1998.tb12915.x. [DOI] [PubMed] [Google Scholar]
- 23.Godessart N., Munoa F.J., Regue M.,, Juarez A. Chromosomal mutations that increase the production of a plasmid-encoded haemolysin in Escherichia coli. J. Gen. Microbiol. 1988;134:2779–87. doi: 10.1099/00221287-134-10-2779. [DOI] [PubMed] [Google Scholar]
- 24.Nieto J.M., Madrid C., Prenafeta A., et al. Expression of the hemolysin operon in Escherichia coli is modulated by a nucleoid-protein complex that includes the proteins Hha and H-NS. Mol. Gen. Genet. 2000;263:349–58. doi: 10.1007/s004380051178. [DOI] [PubMed] [Google Scholar]
- 25.Madrid C., Nieto J.M., Paytubi S., Falconi M., Gualerzi C.O.,, Juarez A. Temperature- and H-NS-dependent regulation of a plasmid-encoded virulence operon expressing Escherichia coli hemolysin. J. Bacteriol. 2002;184:5058–66. doi: 10.1128/JB.184.18.5058-5066.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olekhnovich I.N., Kadner R.J. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 2007;189:6882–90. doi: 10.1128/JB.00905-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silphaduang U., Mascarenhas M., Karmali M., Coombes B.K. Repression of intracellular virulence factors in Salmonella by the Hha and YdgT nucleoid-associated proteins. J. Bacteriol. 2007;189:3669–73. doi: 10.1128/JB.00002-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahlen T.F., Wilson R.L., Boddicker J.D., Jones B.D. Hha is a negative modulator of transcription of hilA, the Salmonella enterica serovar Typhimurium invasion gene transcriptional activator. J. Bacteriol. 2001;183:6620–9. doi: 10.1128/JB.183.22.6620-6629.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olekhnovich I.N., Kadner R.J. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enteric. J. Mol. Biol. 2006;357:373–86. doi: 10.1016/j.jmb.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 30.Sharma V.K., Zuerner R.L. Role of hha and ler in transcriptional regulation of the esp operon of enterohemorrhagic Escherichia coli O157:H7. J. Bacteriol. 2004;186:7290–301. doi: 10.1128/JB.186.21.7290-7301.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vivero A., Banos R.C., Mariscotti J.F., et al. Modulation of horizontally acquired genes by the Hha-YdgT proteins in Salmonella enterica serovar Typhimurium. J. Bacteriol. 2008;190:1152–6. doi: 10.1128/JB.01206-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banos R.C., Vivero A., Aznar S., et al. Differential regulation of horizontally acquired and core genome genes by the bacterial modulator H-NS. PLoS Genet. 2009;5:e1000513. doi: 10.1371/journal.pgen.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forns N., Juarez A., Madrid C. Osmoregulation of the HtrA (DegP) protease of Escherichia coli: an Hha-H-NS complex represses HtrA expression at low osmolarity. FEMS Microbiol. Lett. 2005;251:75–80. doi: 10.1016/j.femsle.2005.07.027. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Contreras R., Zhang X.S., Kim Y., Wood T.K. Protein translation and cell death: the role of rare tRNAs in biofilm formation and in activating dormant phage killer genes. PloS One. 2008;3:e2394. doi: 10.1371/journal.pone.0002394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chumsakul O., Takahashi H., Oshima T., et al. Genome-wide binding profiles of the Bacillus subtilis transition state regulator AbrB and its homolog Abh reveals their interactive role in transcriptional regulation. Nucleic Acids Res. 2011;39:414–28. doi: 10.1093/nar/gkq780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa S., Ogura Y., Yoshimura M., et al. Distribution of stable DnaA-binding sites on the Bacillus subtilis genome detected using a modified ChIP-chip method. DNA Res. 2007;14:155–68. doi: 10.1093/dnares/dsm017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Balsalobre C., Johansson J., Uhlin B.E., Juarez A.,, Munoa F.J. Alterations in protein expression caused by the hha mutation in Escherichia coli: influence of growth medium osmolarity. J. Bacteriol. 1999;181:3018–24. doi: 10.1128/jb.181.10.3018-3024.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence J.G.,, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc. Natl Acad. Sci. USA. 1998;95:9413–7. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura Y., Itoh T., Matsuda H., Gojobori T. Biased biological functions of horizontally transferred genes in prokaryotic genomes. Nat. Genet. 2004;36:760–6. doi: 10.1038/ng1381. [DOI] [PubMed] [Google Scholar]
- 40.Grainger D.C., Hurd D., Goldberg M.D., Busby S.J. Association of nucleoid proteins with coding and non-coding segments of the Escherichia coli genome. Nucleic Acids Res. 2006;34:4642–52. doi: 10.1093/nar/gkl542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ali S.S., Xia B., Liu J., Navarre W.W. Silencing of foreign DNA in bacteria. Curr. Opin. Microbiol. 2012;15:175–81. doi: 10.1016/j.mib.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 42.Fang F.C., Rimsky S. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 2008;11:113–20. doi: 10.1016/j.mib.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prosseda G., Falconi M., Giangrossi M., Gualerzi C.O., Micheli G., Colonna B. The virF promoter in Shigella: more than just a curved DNA stretch. Mol. Microbiol. 2004;51:523–37. doi: 10.1046/j.1365-2958.2003.03848.x. [DOI] [PubMed] [Google Scholar]
- 44.Shin M., Song M., Rhee J.H., et al. DNA looping-mediated repression by histone-like protein H-NS: specific requirement of Eσ70 as a cofactor for looping. Genes Dev. 2005;19:2388–98. doi: 10.1101/gad.1316305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dame R.T., Wyman C., Wurm R., Wagner R.,, Goosen N. Structural basis for H-NS-mediated trapping of RNA polymerase in the open initiation complex at the rrnB P1. J. Biol. Chem. 2002;277:2146–50. doi: 10.1074/jbc.C100603200. [DOI] [PubMed] [Google Scholar]
- 46.Schroder O.,, Wagner R. The bacterial DNA-binding protein H-NS represses ribosomal RNA transcription by trapping RNA polymerase in the initiation complex. J. Mol. Biol. 2000;298:737–48. doi: 10.1006/jmbi.2000.3708. [DOI] [PubMed] [Google Scholar]
- 47.Mojica F.J.,, Higgins C.F. In vivo supercoiling of plasmid and chromosomal DNA in an Escherichia coli hns mutant. J. Bacteriol. 1997;179:3528–33. doi: 10.1128/jb.179.11.3528-3533.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Owen-Hughes T.A., Pavitt G.D., Santos D.S., et al. The chromatin-associated protein H-NS interacts with curved DNA to influence DNA topology and gene expression. Cell. 1992;71:255–65. doi: 10.1016/0092-8674(92)90354-f. [DOI] [PubMed] [Google Scholar]
- 49.Blot N., Mavathur R., Geertz M., Travers A.,, Muskhelishvili G. Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 2006;7:710–5. doi: 10.1038/sj.embor.7400729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Castang S.,, Dove S.L. High-order oligomerization is required for the function of the H-NS family member MvaT in Pseudomonas aeruginosa. Mol. Microbiol. 2010;78:916–31. doi: 10.1111/j.1365-2958.2010.07378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Badaut C., Williams R., Arluison V., et al. The degree of oligomerization of the H-NS nucleoid structuring protein is related to specific binding to DNA. J. Biol. Chem. 2002;277:41657–66. doi: 10.1074/jbc.M206037200. [DOI] [PubMed] [Google Scholar]
- 52.Winardhi R.S., Fu W., Castang S., Li Y., Dove S.L.,, Yan J. Higher order oligomerization is required for H-NS family member MvaT to form gene-silencing nucleoprotein filament. Nucleic Acids Res. 2012;40:8942–52. doi: 10.1093/nar/gks669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Garcia J., Cordeiro T.N., Nieto J.M., Pons I., Juarez A.,, Pons M. Interaction between the bacterial nucleoid associated proteins Hha and H-NS involves a conformational change of Hha. Biochem. J. 2005;388:755–62. doi: 10.1042/BJ20050002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garcia J., Madrid C., Juarez A.,, Pons M. New roles for key residues in helices H1 and H2 of the Escherichia coli H-NS N-terminal domain: H-NS dimer stabilization and Hha binding. J. Mol. Biol. 2006;359:679–89. doi: 10.1016/j.jmb.2006.03.059. [DOI] [PubMed] [Google Scholar]
- 55.Ali S.S., Beckett E., Bae S.J., Navarre W.W. The 5.5 protein of phage T7 inhibits H-NS through interactions with the central oligomerization domain. J. Bacteriol. 2011;193:4881–92. doi: 10.1128/JB.05198-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.