Abstract
Genetic diversity created by transposable elements is an important source of functional variation upon which selection acts during evolution1–6. Transposable elements are associated with adaptation to temperate climates in Drosophila7, a SINE element is associated with the domestication of small dog breeds from the gray wolf8 and there is evidence that transposable elements were targets of selection during human evolution9. Although the list of examples of transposable elements associated with host gene function continues to grow, proof that transposable elements are causative and not just correlated with functional variation is limited. Here we show that a transposable element (Hopscotch) inserted in a regulatory region of the maize domestication gene, teosinte branched1 (tb1), acts as an enhancer of gene expression and partially explains the increased apical dominance in maize compared to its progenitor, teosinte. Molecular dating indicates that the Hopscotch insertion predates maize domestication by at least 10,000 years, indicating that selection acted on standing variation rather than new mutation.
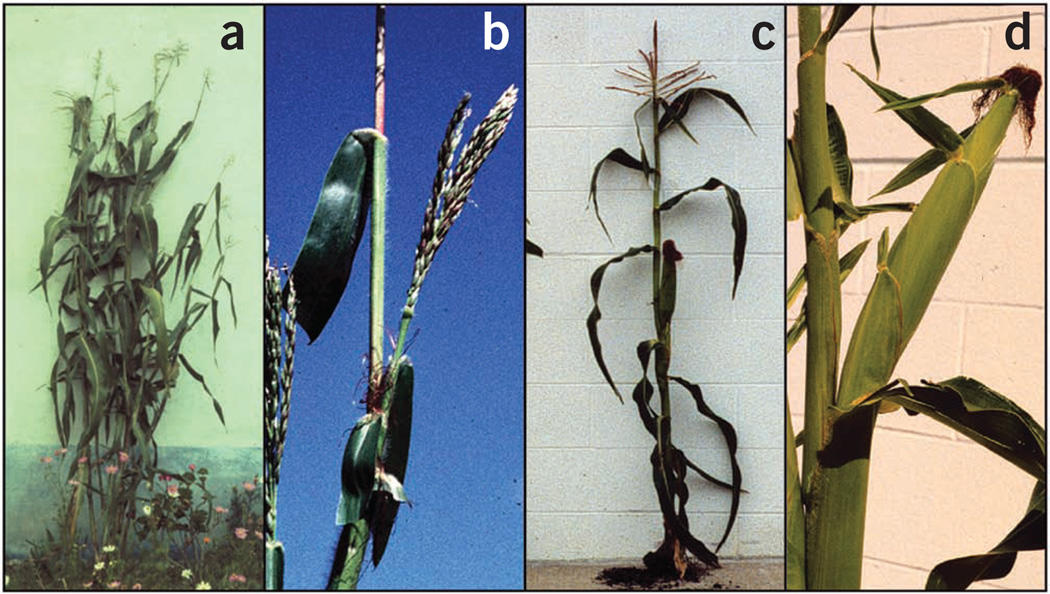
During domestication, maize underwent a dramatic transformation in both plant and inflorescence architecture as compared to its wild progenitor, teosinte10. Like many wild grasses, teosinte has a highly branched architecture (Fig. 1). The main stalk of a teosinte plant has multiple long branches, each tipped by a tassel and bearing many small ears of grain at its nodes. By comparison, the stalk of a modern maize plant has only one or two short branches, each of these tipped by a large, grain-bearing ear. The difference in size of the teosinte and maize ears is substantial. The small ears of teosinte have only 10 or 12 kernels, whereas a single ear of maize can have 300 or more. Overall, maize shows much greater apical dominance, with the development of the branches repressed relative to the development of the main stalk.
Figure 1.
Teosinte and maize plants. (a) Highly branched teosinte plant. (b) Teosinte lateral branch with terminal tassel. (c) Unbranched maize plant. (d) Maize ear shoot (that is, lateral branch).
The teosinte branched1 (tb1) gene corresponds to a quantitative trait locus (QTL)11 that was a major contributor to the increase in apical dominance during maize domestication. tb1 encodes a member of the TCP family of transcriptional regulators12. The TBl protein acts as a repressor of organ growth and thereby contributes to apical dominance by repressing branch outgrowth. Prior research has shown that the maize allele of tb1 is expressed more highly than the teosinte allele, thereby conditioning greater repression of branching13. The regulatory element or ‘control region’ modulating this difference in expression is located between 58.7 kb and 69.5 kb upstream of the tb1 ORF14. Although the region containing the causative factor distinguishing maize and teosinte was narrowed to this ~11-kb interval, the nature of this factor, whether simple or multipartite, and the identity of the exact causative polymorphism(s) have not been elucidated.
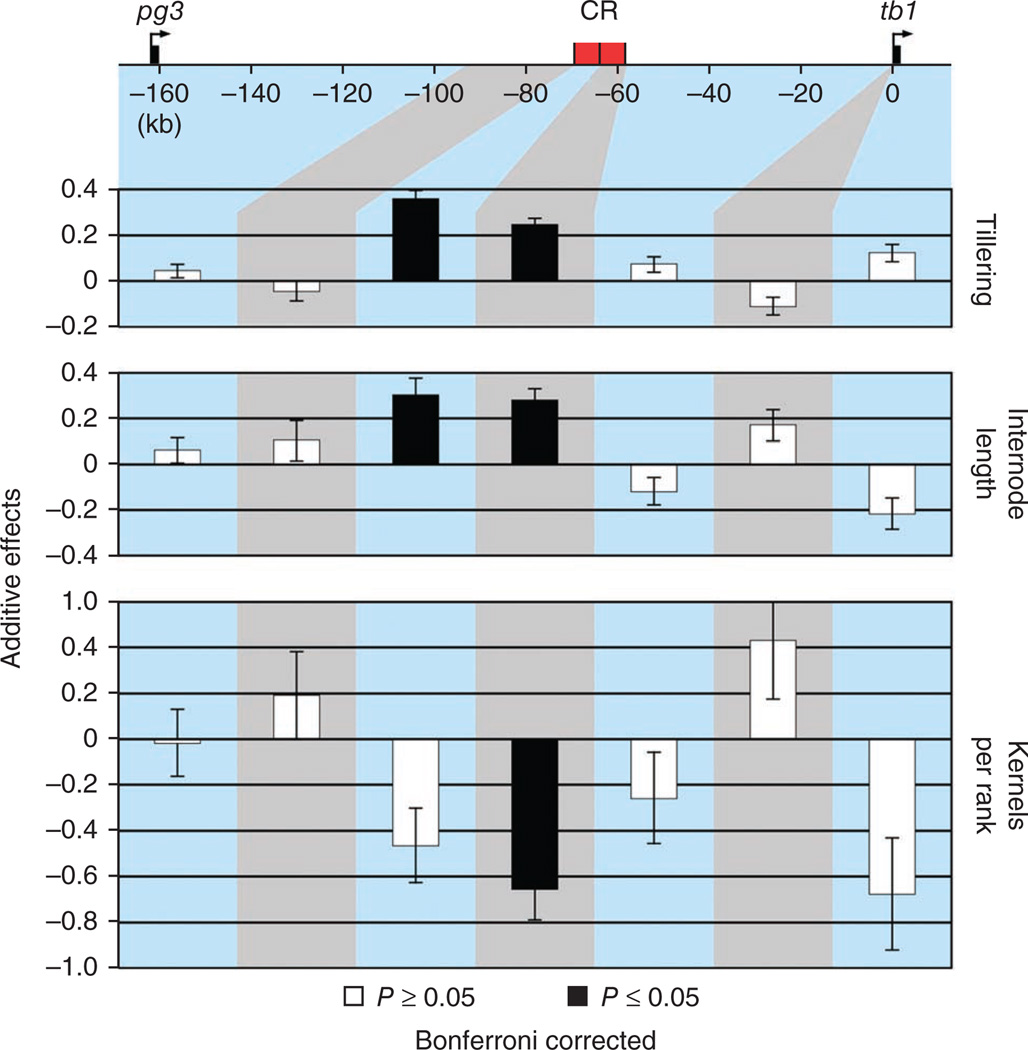
We used genetic fine-mapping to locate the factors influencing phenotype in the control region. We isolated 18 maize-teosinte recombinant chromosomes, each containing a unique teosinte portion of the tb1 genomic region, and we made these 18 recombinant chromosomes isogenic in a common maize inbred background (Supplementary Fig. 1 and Supplementary Tables 1 and 2a). This collection of recombinant chromosomes enabled us to divide the tb1 genomic region into seven intervals based on recombination breakpoints (Supplementary Table 3). The isogenic lines for these recombinant chromosomes were evaluated over four growing seasons, and the phenotypes of more than 5,500 plants were recorded. The resulting data were analyzed using a mixed linear statistical model, enabling us to test each interval for an effect on phenotype. This analysis confirmed that the control region previously described by Clark and colleagues14 is responsible for differences in both plant and ear architecture between maize and teosinte (Fig. 2). Moreover, our data show that the control region is complex, having two independent components affecting phenotype. These two components, which we call the proximal and distal components, are separated by recombination breakpoints located ~63.9 kb upstream of the tb1 ORF. The independent phenotypic effects of the proximal and distal components are readily seen in lines that segregate for only one or the other of these components (Supplementary Fig. 2).
Figure 2.
The phenotypic additive effects for seven intervals across the tb1 genomic region. The horizontal axis represents the tb1 genomic region to scale. Base-pair positions are relative to AGPv2 position 265,745,977 of the maize reference genome sequence. The tb1 ORF and the nearest upstream predicted gene (pg3) are shown. The previously defined control region (CR)14 is shown in red and is divided into its proximal and distal components. Vertical columns represent the additive effects shown with standard error bars for each of the three traits in each of the seven intervals that were tested for an effect on phenotype. Black columns are statistically significant (P (Bonferroni) < 0.05); white bars are not statistically significant (P (Bonferroni) > 0.05).
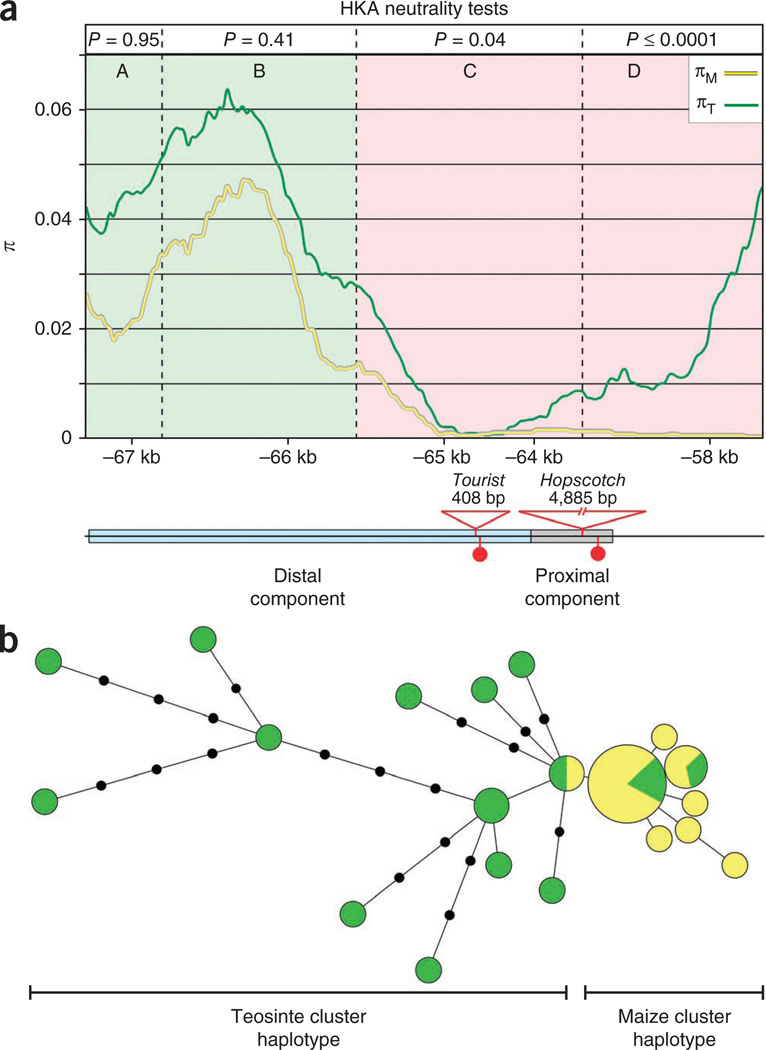
Previous analyses indicated that the tb1 genomic region shows evidence of a selective sweep during domestication that extends from the ORF to −58.6 kb but ends before −93.4 kb15. To better define the extent of the sweep, we performed population-genetic analyses for the region between −57.4 and −67.6 kb using a diverse set of maize and teosinte lines. Nucleotide diversity (π) at −58 kb is high in teosinte but low in maize (Fig. 3a). Between −58 and −65 kb, nucleotide diversity is low in both maize and teosinte but lower in maize. The low diversity for both maize and teosinte in this region suggests that the region is evolving under functional constraint. Beyond 65 kb upstream of the ORF, diversity rises in both maize and teosinte. The rise in nucleotide diversity in maize beyond −65 kb suggests that the selective sweep ends near this point.
Figure 3.
Sequence diversity in maize and teosinte across the control region. (a) Nucleotide diversity across the tb1 upstream control region. Base-pair positions are relative to AGPv2 position 265,745,977 of the maize reference genome sequence. P values correspond to HKA neutrality tests for regions A–D, as defined by the dotted lines. Green shading signifies evidence of neutrality, and pink shading signifies regions of non-neutral evolution. Nucleotide diversity (π) for maize (yellow line) and teosinte (green line) were calculated using a 500-bp sliding window with a 25-bp step. The distal and proximal components of the control region with four fixed sequence differences between the most common maize haplotype and teosinte haplotype are shown below. (b) A minimum spanning tree for the control region with 16 diverse maize and 17 diverse teosinte sequences. Size of the circles for each haplotype group (yellow, maize; green, teosinte) is proportional to the number of individuals within that haplotype.
We applied the HKA test16 to address whether individual segments of the control region show evidence of past selection (Supplementary Table 4). Our results confirm previous findings17 that the region from −65.6 to −67.6 kb (segments A and B in Fig. 3a) does not depart significantly from neutral expectations, but the neutral model can be rejected for the region from −58.8 to −57.4 kb (segment D). We also tested, for the first time, an additional segment (segment C, from −65.6 to −63.7 kb) in the middle of the control region, which our data show departs significantly the neutral model. Prior results15 demonstrated that the sweep extends from −58 kb to the tb1 ORF; thus, overall, the sweep includes approximately 65.6 kb from the control region to the ORF.
Phenotypic fine-mapping with recombinant chromosomes indicated that the factors controlling phenotype lie between 58.7 and 69.5 kb upstream of the ORF. Population genetic analysis indicates that the selective sweep extends only to −65.6 kb. Together, these two sources of information suggest that the causative polymorphism(s) lies between −58.7 and −65.6 kb of the ORF. We looked in greater detail at sequence diversity for maize and teosinte in the ~7-kb segment that these two methods define. A minimum spanning tree for a sample of 16 diverse maize and 17 diverse teosintes in this region revealed two distinct clusters of haplotypes, one composed mostly of maize sequences and the other composed mostly of teosinte sequences (Fig. 3b). We designated these clusters as the maize cluster haplotype (MCH) and the teosinte cluster haplotype (TCH), respectively. There are four fixed differences between the sequences in the maize and teosinte clusters (Fig. 3a). Two of these fixed differences are single-nucleotide polymorphisms (SNPs), and two are large insertions in the maize cluster haplotype relative to the teosinte cluster haplotype. BLAST searches of the insertion sequences revealed that one is a Hopscotch retrotransposon and the other is a Tourist miniature inverted-repeat transposable element (MITE). Of the four fixed differences, Hopscotch and one SNP are in the proximal component, whereas Tourist and the other SNP are in the distal component, as delineated by phenotypic fine-mapping.
To estimate the frequency of the two haplotype groups in maize and teosinte, we assayed 139 additional diverse maize chromosomes and 148 additional diverse teosinte chromosomes (Supplementary Table 5). For this purpose, we used the Hopscotch and Tourist insertions as markers for the haplotype groups (Supplementary Table 2b). The MCH is present in >95% of the maize chromosomes assayed but in <5% of teosinte chromosomes. The fact that the MCH is not fixed in maize suggests either that the initial selective sweep was not complete or that post-domestication gene flow from teosinte to maize has reintroduced the TCH into the maize gene pool. Correspondingly, the presence of the MCH in teosinte may represent either a haplotype variant that existed in teosinte before domestication or post-domestication gene flow from maize into teosinte, which is known to occur18.
Inspection of the sequence alignment of the Hopscotch-Tourist region suggests that the two insertions differ in relative age. The Tourist insertion has accumulated greater nucleotide diversity (π = 0.0054) since insertion, including a pair of sites that fail the four-gamete test, which is indicative of recombination among Tourist sequences. Nucleotide diversity in the Hopscotch insertion is much lower (π = 0.0016) and shows no evidence of past recombination. These observations point to the Hopscotch insertion being more recent than the Tourist. Our sequences do show evidence of recombination between Hopscotch and a SNP in the flanking sequence between the two insertions, likely explaining how the Hopscotch insertion has come to be associated with multiple alleles of the Tourist element.
These nucleotide diversity data allow us to ask whether the Hopscotch insertion arose before or during domestication. Following Thomson et al.19 and Hudson20, we estimate a most recent common ancestor for the Hopscotch alleles at ~28,000 years before present (BP), with a 95% lower bound of ~15,000 BP. A more conservative approach, which counts only singletons and assumes a star phylogeny, yields a slightly lower estimate of ~23,000 BP, with a 95% lower bound of ~13,000 BP. Both estimates conservatively use a relatively high mutation rate21, strongly suggesting that the Hopscotch insertion (and thus, the older Tourist as well) existed as standing genetic variation in the teosinte ancestor of maize. Thus, we conclude that the Hopscotch insertion likely predated domestication by more than 10,000 years and the Tourist insertion by an even greater amount of time.
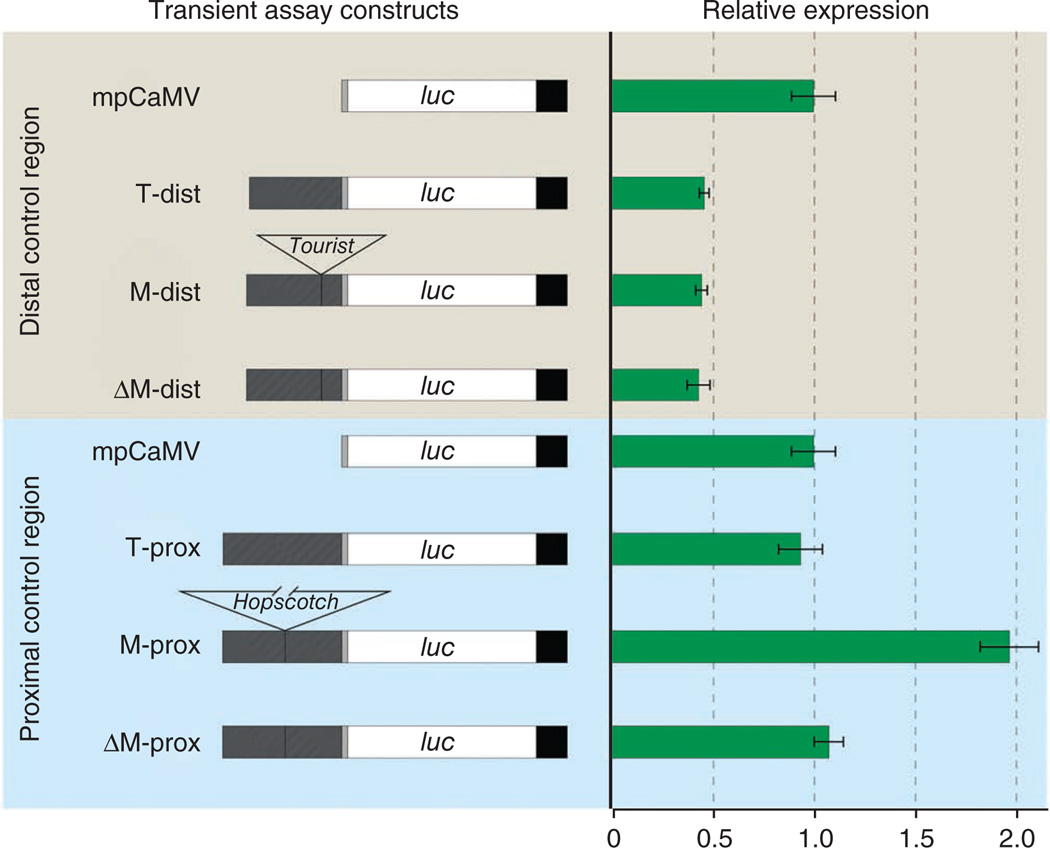
We identified four fixed differences in the portion of the proximal and distal components of the control region that show evidence of selection. We used transient assays in maize leaf protoplasts to test all four differences for effects on gene expression. Maize and teosinte chromosomal segments for the portions of the proximal and distal components with these four differences were cloned into reporter constructs upstream of the minimal promoter of the cauliflower mosaic virus (mpCaMV), the firefly luciferase ORF and the nopaline synthase (NOS) terminator (Fig. 4). Each construct was assayed for luminescence after transformation by electroporation into maize protoplast. The constructs for the distal component contrast the effects of the Tourist insertion plus the single fixed nucleotide substitution that distinguish maize and teosinte. Both the maize and teosinte constructs for the distal component repressed luciferase expression relative to the minimal promoter alone. The maize construct with Tourist excised gave luciferase expression equivalent to the native maize and teosinte constructs and less expression than the minimal promoter alone. These results indicate that this segment is functionally important, acting as a repressor of luciferase expression and, by inference, of tb1 expression in vivo. However, we did not observe any difference between the maize and teosinte constructs as anticipated. One possible cause for the lack of differences in expression between the maize and teosinte constructs might be that additional proteins required to cause these differences are not present in maize leaf protoplast. Another possibility is that the factor affecting phenotype in the distal component lies in the unselected region between −64.8 and −69.5 kb, which is not included in the construct. Nevertheless, the results do indicate that the distal component has a functional element that acts as a repressor. The functional importance of this segment is supported by its low level of nucleotide diversity (Fig. 3a), suggesting a history of purifying selection.
Figure 4.
Constructs and corresponding normalized luciferase expression levels. Transient assays were performed in maize leaf protoplast. Each construct is drawn to scale. The construct backbone consists of the minimal promoter from the cauliflower mosaic virus (mpCaMV, gray box), luciferase ORF (luc, white box) and the nopaline synthase terminator (black box). Portions of the proximal and distal components of the control region (hatched boxes) from maize and teosinte were cloned into restriction sites upstream of the minimal promoter. “Δ” denotes the excision of either the Tourist or Hopscotch element from the maize construct. Horizontal green bars show the normalized mean with s.e.m. for each construct.
The constructs for the proximal component of the control region contrast the effects of the Hopscotch insertion plus a single fixed nucleotide substitution that distinguish maize and teosinte. The construct with the maize sequence including Hopscotch increased expression of the luciferase reporter twofold relative to the teosinte construct for the proximal control region and the minimal promoter alone (Fig. 4). Luciferase expression was returned to the level of the teosinte construct and the minimal promoter construct by deleting the Hopscotch element from the full maize construct. These results indicate that the Hopscotch element enhances luciferase expression and, by inference, tb1 expression in vivo. They also indicate that Hopscotch rather than the fixed SNP difference between maize and teosinte is the causative polymorphism. The observed enhancement of gene expression by the Hopscotch element is consistent with the known higher level of tb1 expression in maize as compared to teosinte.
Our observation of a transposable element providing an enhancer element in tb1 is similar to observations made with globin genes in primates, in which an EVR-9 element was shown to function as a long-distance enhancer of gene expression22. Similarly, in Drosophila, the long terminal repeat of an Accord element acts like an enhancer of Cyp6g1, which metabolizes the pesticide DDT, thereby conferring pesticide resistance23, 24. Over 25 years ago, Barbara McClintock proposed that transposable elements represent a key source of variation for evolution25. Remarkably, a transposable element insertion appears to represent the causal variant for one of the key steps in the domestication of maize, the organism in which McClintock discovered transposable elements.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/naturegenetics/.
ONLINE METHODS
Phenotypic and genotypic data collection
Nine introgression lines generated by Clark and colleagues14 were used for fine-mapping the control region (Supplementary Fig. 1, Supplementary Table 1). Nine additional introgression lines were recovered using the same strategy used by Clark and colleagues14. This entailed backcrossing homozygous introgression lines to W22 and screening individual F2 progeny for crossovers in the introgressed chromosomal segment. Genotyping was accomplished using a set of eight PCR-based indel markers (GS1–GS8, Supplementary Table 2a) tagged with a 5′ HEX or FAM label and then assayed using an ABI 3700 fragment analyzer.
Plants were grown at the University of Wisconsin West Madison Agricultural Research Station (Madison, Wisconsin, USA) during the summers of 2006–2009. F2 seed derived from the cross of each of the homozygous introgression lines to W22 were planted in a completely randomized design using grids with 0.9-m spacing between plants in both dimensions. This spacing minimized the degree to which plants shaded their neighbors. The following three traits were phenotyped: cupules per rank (CUPR, number of cupules in a single rank from base to the tip of the ear), lateral branch length (LBLH, length in centimeters of uppermost lateral branch) and tillering (TILL, the ratio of the sum of tiller heights to plant height). All plants were genotyped individually using a combination of the eight PCR-based markers described above.
Phenotypic data analysis
The tb1 genomic region was divided into 16 intervals based on the recombination breakpoints of the 18 introgression lines. To examine the near-collinearity of these intervals with one another, the CORR procedure of the SAS statistics package (SAS Inc.) was used to calculate the correlation coefficient between intervals. If the correlation between two intervals was high, then the model did not adequately fit the phenotypic effects of the two correlated intervals. Thus, not all intervals can be simultaneously tested in a single model. Only if two intervals showed a correlation coefficient of <0.8 were they included as separate factors in the model. Our final analysis included seven intervals (Supplementary Table 3), which represented the entire tb1 genomic region (Fig. 2). The number of plants included for each trait model were as follows: 5,491 (TILL), 4,591 (LBIL) and 3,499 (CUPR). The MIXED procedure of SAS was used to test each interval for an effect on phenotype. Intervals (1–7) were considered fixed effects, whereas year (2006–2009), the introgression line by ear interaction term and the introgression line by interval interaction terms were treated as random effects. The linear model used was
Yhijkl = µ + ah + bi + cj*dk + cj*ah + ehijkl
where Yhijkl is the trait value for the lth plant with hth intervals from the kth ear of the jth introgression line in the ith year, µ is the overall mean of the experiment, ah is the interval effect, bi is the year effect, cj*dk is the introgression line by ear interaction, cj*ah is the introgression line by interval interaction and ehijkl is the sampling error. The random effects of this full model were subjected to the likelihood ratio test for significance for each trait. Effects that were not significant were dropped from the model on a trait-by-trait basis. All genotype and phenotype data are available at the Panzea website (see URLs).
Nucleotide diversity
A sample of 16 maize landraces made haploid for DNA extraction26 and 17 inbred teosinte lines were used to assay nucleotide diversity in the control region (Supplementary Table 5). Sequencing of PCR fragments for the 33 individuals was performed using standard PCR conditions and Applied Biosystems BigDye kit at the University of Wisconsin Biotechnology Center using Sanger sequencing methods. Initial alignment of nucleotide sequences was performed using ClustalW27 and then finished by hand. Nucleotide diversity (π) was calculated using a 500-bp sliding window with a 25-bp step with a correction for small sample size. Nucleotide sites in the alignment were only used for calculating π if at least ten individuals had ungapped and unambiguous calls from the maize and from the teosinte groups.
Tests for neutrality
The HKA tests16 for neutrality were performed using DnaSP28. Zea diploperennis was used as an outgroup, and its sequence was aligned with that of the 33 individuals used in the nucleotide diversity survey. A set of six previously described neutral loci29 were used as control genes (Supplementary Table 4). For each HKA test, an overall χ2 value was calculated by taking the sum of the individual χ2 values calculated for the six individual neutral loci. These overall χ2 values were then used to obtain overall P values, as shown in Figure 3a.
Minimum spanning tree
The minimum spanning tree was constructed using the same 33 individuals as used in the nucleotide diversity survey (Supplementary Table 5). The alignment of the sequences was trimmed of gaps and missing data and then imported into Arlequin version 3.5 (ref. 30), which was used to define the haplotypes and calculate the minimum spanning tree among haplotypes. Arlequin’s distance matrix output was used in Hapstar31 to draw the minimum spanning tree.
Insertion frequencies
The frequency of Tourist and Hopscotch insertions was calculated using a diverse set of 139 maize chromosomes and 148 teosinte chromosomes (Supplementary Table 5). The frequency of each insertion was assayed using a three-primer PCR reaction (Supplementary Table 2b), which allowed both homozygous and heterozygous individuals to be scored on a 2% agarose gel using standard PCR conditions.
Insertion dating
Initial alignment of nucleotide sequences of the Tourist and Hopscotch elements was performed using ClustalW27 and then finished by hand. Diversity analyses were performed using the “compute” program of the analysis package of libsequence32. Of the 16 maize alleles sequenced, 15 have the Hopscotch insertion. These 15 alleles had 2 insertion or deletions and 16 segregating sites, of which 13 were singleton mutations and 3 were found in 2 sequences. Following equation (3) of Thomson et al.19 and using the mutation rate estimate of Clark et al.21, this gives an age of ~28,000 years BP. If we assume a star phylogeny, we can estimate the time since insertion of Hopscotch as T = S(15 µl)−1, where T is time in generations, S is the number of segregating sites, L is the length of the sequence in base pairs and µ is the per generation mutation rate per base pair. Given 1,524 bp of sequence, 16 segregating sites, a generation time of 1 year and a mutation rate of 3 × 10−8 (ref. 21), this gives an estimate of approximately 23,300 years. Although these assumptions (star phylogeny, mutation rate and ignoring doubletons) are clearly unrealistic, changing any of them leads to an increase in the estimated time of insertion. To estimate confidence intervals around these estimates, we used equation (5) of ref. 20.
Protoplast transient assays
Two reporter constructs were developed for the transient assays. A reporter construct containing the cauliflower mosaic virus (CaMV) 35S minimal promoter33 driving expression of the firefly luciferase gene was used to test control region segments. The second reporter containing the rice actin1 promoter driving expression of the Renilla luciferase gene was used as an internal transformation control. Transient expression assays using maize mesophyll protoplasts were performed following a detailed protocol from the Sheen laboratory (see URLs), with transformation conditions modified as follows. Briefly, 2–4 × 105 freshly isolated protoplasts in 400 µl electroporation buffer were mixed with 50 µl of the plasmids. The protoplast-plasmid mixes were transferred into 0.5-ml cuvettes and electroporated with the Gene Pulser II Electroporation System (Bio-Rad) set at 250 V. Each sample received three pulses of 1.5 ms each with a 20-s pause between the pulses. After electroporation, protoplasts were incubated for 18 h at 25 °C and then harvested. The harvested protoplasts were lysed with CCLR (Cell Culture Lysis Reagent, Promega) and assayed using the Dual-Luciferase Reporter Assay System (Promega) following manufacturer’s instructions. Four to six biological replicates, each with two technical replicates, were assayed per construct.
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Doebley laboratory for technical assistance, especially H. Wang. We also thank A.J. Eckert, K. Thornton, G. Coop and R.A. Cartwright for helpful discussion and J. Holland for statistical advice. This work is supported by US Department of Agriculture Hatch grant MSN101593 and US National Science Foundation grant DBI0820619.
Footnotes
URL. Panzea, http://www.panzea.org/; Sheen laboratory protocols (including transient expression assay using maize mesophyll protoplasts), http://genetics.mgh.harvard.edu/sheenweb/protocols_reg.html.
Accession numbers. Sequence data has been deposited in GenBank with the accession numbers JN205126–JN205158.
Note: Supplementary information is available on the Nature Genetics website.
AUTHOR CONTRIBUTIONS
A.S. and J.D. designed the experiments and wrote the paper. A.S., J.R.-I. and Q.Z. performed population genetic analyses. Genetic mapping, transient assays, sequencing and informatics were done by A.S.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Naito K, et al. Unexpected consequences of a sudden and massive transposon amplification on rice gene expression. Nature. 2009;461:1130–1134. doi: 10.1038/nature08479. [DOI] [PubMed] [Google Scholar]
- 2.Xiao H, Jiang N, Schaffner E, Stockinger EJ, van der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 3.White SE, Habera LF, Wessler SR. Retrotransposons in the flanking regions of normal plant genes: A role for copia-like elements in the evolution of gene structure and expression. Proc. Natl. Acad. Sci. USA. 1994;91:11792–11796. doi: 10.1073/pnas.91.25.11792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bejerano G, et al. A distal enhancer and an ultraconserved exon are derived from a novel retrotransposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 5.Mackay TFC, Lyman RF, Jackson MS. Effects of P element insertions on quantitative traits in Drosophila melanogaster. Genetics. 1992;130:315–332. doi: 10.1093/genetics/130.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Torkamanzehi A, Moran C, Nicholas FW. P element transposition contributes substantial new variation for a quantitative trait in Drosophila melanogaster. Genetics. 1992;131:73–78. doi: 10.1093/genetics/131.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.González J, Karasov TL, Messer PW, Petrov DA. Genome-wide patterns of adaptation to temperate environments associated with transposable elements in Drosophila. PLoS Genet. 2010;6:e1000905. doi: 10.1371/journal.pgen.1000905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray MM, Sutter NB, Ostrander EA, Wayne RK. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:16. doi: 10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britten RJ. Transposable element insertions have strongly affected human evolution. Proc. Natl. Acad. Sci. USA. 2010;107:19945–19948. doi: 10.1073/pnas.1014330107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doebley J. The genetics of maize evolution. Annu. Rev. Genet. 2004;38:37–59. doi: 10.1146/annurev.genet.38.072902.092425. [DOI] [PubMed] [Google Scholar]
- 11.Doebley J, Stec A, Gustus C. Teosinte branched1 and the origin of maize: Evidence for epistasis and the evolution of dominance. Genetics. 1995;141:333–346. doi: 10.1093/genetics/141.1.333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cubas P, Lauter N, Doebley J, Coen E. The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 1999;18:215–222. doi: 10.1046/j.1365-313x.1999.00444.x. [DOI] [PubMed] [Google Scholar]
- 13.Doebley J, Stec A, Hubbard L. The evolution of apical dominance in maize. Nature. 1997;386:485–488. doi: 10.1038/386485a0. [DOI] [PubMed] [Google Scholar]
- 14.Clark RM, Nussbaum Wagler T, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 15.Clark RM, Linton E, Messing J, Doebley JF. Pattern of diversity in the genomic region near the maize domestication gene tb1. Proc. Natl. Acad. Sci. USA. 2004;101:700–707. doi: 10.1073/pnas.2237049100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–159. doi: 10.1093/genetics/116.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Q. Molecular Population Genetics of Maize Regulatory Genes During Maize Evolution. PhD Thesis. University of Wisconsin–Madison; 2006. [Google Scholar]
- 18.Fukunaga K, et al. Genetic diversity and population structure of teosinte. Genetics. 2005;169:2241–2254. doi: 10.1534/genetics.104.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomson R, et al. Recent common ancestry of human Y chromosomes: Evidence from DNA sequence data. Proc. Natl. Acad. Sci. USA. 2000;97:7360–7365. doi: 10.1073/pnas.97.13.7360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hudson RR. The variance of coalescent time estimates from DNA sequences. J. Mol. Evol. 2007;64:702–705. doi: 10.1007/s00239-006-0261-1. [DOI] [PubMed] [Google Scholar]
- 21.Clark RM, Tavare S, Doebley J. Estimating a nucleotide substitution rate for maize from polymorphism at a major domestication locus. Mol. Biol. Evol. 2005;22:2304–2312. doi: 10.1093/molbev/msi228. [DOI] [PubMed] [Google Scholar]
- 22.Pi W, et al. Long-range function of an intergenic retrotransposon. Proc. Natl. Acad. Sci. USA. 2010;107:12992–12997. doi: 10.1073/pnas.1004139107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung H, et al. Cis-regulatory elements in the Accord retrotransposon result in tissue-specific expression of the Drosophila melanogaster insecticide resistance gene Cyp6g1. Genetics. 2007;175:1071–1077. doi: 10.1534/genetics.106.066597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schmidt JM, et al. Copy number variation and transposable elements feature in recent, ongoing adaptation at the Cyp6g1 locus. PLoS Genet. 2010;6:e1000998. doi: 10.1371/journal.pgen.1000998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- 26.Tenaillon MI, et al. Patterns of DNA sequence polymorphism along chromsome 1 of maize (Zea mays ssp. mays L) Proc. Natl. Acad. Sci. USA. 2001;98:9161–9166. doi: 10.1073/pnas.151244298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R, Dna SP. DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19:2496–2497. doi: 10.1093/bioinformatics/btg359. [DOI] [PubMed] [Google Scholar]
- 29.Zhao Q, Weber AL, McMullen MD, Guill K, Doebley J. MADS-box genes of maize: frequent targets of selection during domestication. Genet. Res. (Camb.) 2011;93:65–75. doi: 10.1017/S0016672310000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- 31.Teacher AGF, Griffiths DJ. HapStar: automated haplotype network layout and visualization. Mol. Ecol. Resour. 2011;11:151–153. doi: 10.1111/j.1755-0998.2010.02890.x. [DOI] [PubMed] [Google Scholar]
- 32.Thornton K. Libsequence: a C. class library for evolutionary genetic analysis. Bioinformatics. 2003;19:2325–2327. doi: 10.1093/bioinformatics/btg316. [DOI] [PubMed] [Google Scholar]
- 33.Benfey PN, Chua N. The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science. 1990;250:959–966. doi: 10.1126/science.250.4983.959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.