Abstract
Background & Aims
Metabolic stress during liver injury enhances autophagy and provokes stellate cell activation, with secretion of scar matrix. Conditions that augment protein synthesis increase demands on the endoplasmic reticulum (ER) folding capacity and trigger the unfolded protein response (UPR) to cope with resulting ER stress. Generation of reactive oxygen species (ROS) is a common feature of hepatic fibrogenesis, and crosstalk between oxidative stress and ER stress has been proposed. The aim of our study was to determine the impact of oxidant and ER stress on stellate cell activation.
Methods
Oxidant stress was induced in hepatic stellate cells using H2O2 in culture or by ethanol feeding in vivo, and the UPR response was analyzed. Because the branch of the UPR mainly affected was IREα, we blocked this pathway in stellate cells and analyzed the fibrogenic response, together with autophagy and downstream MAPK signaling. The Nrf2 antioxidant response was also evaluated in stellate cells under oxidant stress conditions.
Results
H2O2 treatment in culture or ethanol feeding in vivo increased the UPR response based on splicing of XBP1 mRNA, which triggered autophagy. The Nrf2-mediated antioxidant response, as measured by qRT-PCR of its target genes was also induced under ER stress conditions. Conversely, blockade of the IRE1 pathway in stellate cells significantly decreased both their activation and autophagic activity in a p38 MAPK dependent manner, leading to a reduced fibrogenic response.
Conclusions
These data implicate mechanisms underlying protein folding quality control in regulating the fibrogenic response in hepatic stellate cells.
Keywords: liver fibrosis, hepatic stellate cells, autophagy, ER stress, Nrf2
Introduction
Acquisition of a fibrogenic phenotype by resident hepatic stellate cells is a critical event in the liver’s response to injury. This cellular ‘activation’ results in the deposition of collagen and other components of the extracellular matrix that, if injury persists, leads to the progressive replacement of the liver parenchyma by scar and eventual cirrhosis [1, 2]. We have recently demonstrated that up-regulation of autophagy drives the fibrogenic response in stellate cells [3]. However, upstream events that instigate autophagy in stellate cells remain unknown.
Chronic alcohol intake provokes stellate cell activation and initiation of the wound-healing response, and can lead to alcoholic fibrosis. In fact, up to 44% of all deaths from liver disease may be attributable to alcohol [4]. Alcohol metabolism generates ROS, and this oxidant stress is a well characterized inducer of stellate cell activation and collagen secretion [5]. In addition, oxidant stress can result in activation of the Nrf2 transcription factor, a major regulator of the cellular antioxidant response [6]. Upon activation, Nrf2 is liberated from its cytoplasmic inhibitor Keap1, becomes stabilized and accumulates in the nucleus where it binds to the antioxidant response elements (ARE) in the promoter region of several antioxidant genes, including the genes encoding sulfiredoxin-1 (Srxn1) glutathione S-transferase mu subunit (Gstm), NADPH: quinone oxidoreductase 1 (Nqo1), glutamate-cysteine ligase catalytic (Gclc) and modulatory (Gclm) subunits and several peroxiredoxins [7, 8].
As stellate cells are activated and generate more extracellular matrix constituents, this increased protein synthesis imposes a higher demand on the ER folding capacity that may disturb ER homeostasis. In most tissues oxidant stress can alter ER homeostasis and provoke UPR activation [9, 10]. Three transmembrane proteins, normally kept inactive bound to the ER chaperone BIP, can sense perturbation of ER homeostasis: inositol requiring enzyme 1 (IRE1 ), activating transcription factor 6 (ATF-6), and RNA-activated protein kinase (PKR)-like ER kinase (PERK)[11]. IRE1 is the most conserved branch among the three. Its dissociation from BIP allows its trans-phosphorylation, activates its RNase activity, and results in the modification of X-box transcription factor (Xbp1) into its transcriptionally active splice variant (sXbp1)[12]. sXBP1 can translocate to the nucleus and activate specific target genes. The role of the IRE1 -XBP1 pathway in stellate cell activation has not been explored. Because both autophagy and the UPR are induced upon perturbation of cellular homeostasis, we have explored the possibility that they contribute to the fibrogenic response in stellate cells.
Our study sought to determine whether oxidant and ER stress activate autophagy in hepatic stellate cells, thereby stimulating cellular activation and collagen deposition.
Material and methods
In vitro experiments
Mouse hepatic stellate cells were isolated from C57/BL6 wild type mice by enzymatic digestion and Percoll density gradient centrifugation with modifications [13]. The mouse immortalized stellate cell line JS1, previously generated in our laboratory [14], was cultured with Dulbecco’s modified Eagle medium Nutrient Mixture F12 (DMEM-12) containing 10% fetal bovine serum. Immortalized rat hepatic stellate cells from rats fed during 8 months with either control or ethanol Lieber-DeCarli diets were isolated as previously described[15, 16]. Some cells were treated with 10 mM chloroquine (CQ) (Sigma, St. Louis, MO), 20 μM H2O2 (Sigma, St. Louis, MO), 5 μg/ml tunicamycin (Sigma, St. Louis, MO), the p38 MAPK inhibitor SB203580 (Cell Signaling, Boston, MA) or incubated with 10 ng/mL PDGF-BB (BD Biosciences San Jose, CA). An expression plasmid encoding a dominant-negative Nrf2 mutant was used in some studies [17]. Addgene plasmid 20745 (Addgene, Cambridge, MA, USA) coding for DN IRE1 (K599A, a dominant negative kinase mutant), generated by Dr. Fumihiko Urano was also used. Cells were transfected using TransITρ-LT1 Transfection Reagent (Mirus Bio LLC, Madison, WI, USA).
Quantitative Real Time Reverse Transcriptase Polymerase Chain Reaction (q-rt PCR)
Total RNA was extracted using Qiagen mini-columns (Qiagen, Germantown, MD) with an on-column DNase treatment. One microgram of RNA was reverse transcribed using RT complete double pre-primed kit (Clontech, Mountain View, CA). FastStartSYBR Green Master Mix (Roche, Indianapolis, IN) was used for PCR reaction. Samples were analyzed in triplicate in Microsoft Excel and normalized to -actin expression. For spliced Xbp1 detection by standard PCR, the following program was used: (1) 94 C for 4 min, (2) 35 cycles of 94 C for 45s, 63 C for 30s, and 72 C for 30s, (3) 72 C for 10 min. PCR products were separated by agarose gel electrophoresis to resolve the 473 bp (unspliced) and 428 bp (spliced) amplicons.
Immunoblot
Cell lysates were subjected to immunoblot analysis. Membranes were incubated with the following primary antibodies: rabbit anti-LC3 (Sigma, St. Louis, MO), rabbit anti-GAPDH (Sigma, St. Louis, MO), rabbit anti-type I collagen (Rockland Inc., Gilbertsville, PA), rabbit anti-SMA (Billerica, MA), rabbit anti--PDGFR (Santa Cruz, CA.), rabbit anti-MMP2 (Abcam, Cambridge, MA), mouse anti-tubulin (Sigma, St. Louis, MO), rabbit anti-P62 (Enzo, New York, NY), rabbit anti-ATF6 (Santa Cruz, CA.), rabbit anti-ATF4/CREB-2 (Santa Cruz, CA.), mouse anti-P38 (Cell Signaling, Boston, MA), mouse anti-phospho-P38 (Cell Signaling, Boston, MA), rabbit anti-phospho-JNK (Cell Signaling, Boston, MA), rabbit anti-phospho-ERK (Cell Signaling, Boston, MA), rabbit anti-phospho-AKT (Cell Signaling, Boston, MA), rabbit anti-ERK (Cell Signaling, Boston, MA), and rabbit anti-PDI (Cell Signaling, Boston, MA). GCLC and GCLM antibodies were donated by Dr. Terrence Kavanaugh (University of Washington, WA). The reactions were detected with HRP-conjugated secondary antibodies. Blots were developed using ECL detection system (Amersham Pharmacia Biotech, Buckinghamshire, UK) and a Laser4000 (Fujitsu).
GST Activity
GST activity was determined according to the method of Habig et al. [18], with modifications. The reaction was carried out in 0.1 M potassium phosphate, pH 6.5, 10 mM sodium phosphate, pH 7.4, 20 mM GSH, and 20 mM 1-chloro-2, 4-dinitrobenzene dissolved in 96% ethanol in the presence of 5 μL cell lysate (approximately 20 ng protein). The change in absorbance was monitored at 340 nm and 25°C over a 6-minute period. Results are expressed as units of specific activity defined as the amount of the enzyme that produces 1 μmol of conjugated product per minute per milligram of protein.
Statistical Analysis
Results are expressed as the mean and standard error of the mean (SEM). P values (Student two tailed, unpaired t test) of at least three independent determinations were calculated with Microsoft Excel software. Data were considered to be statistically significant at P <0.05.
Results
ROS generation provokes ER stress in hepatic stellate cells
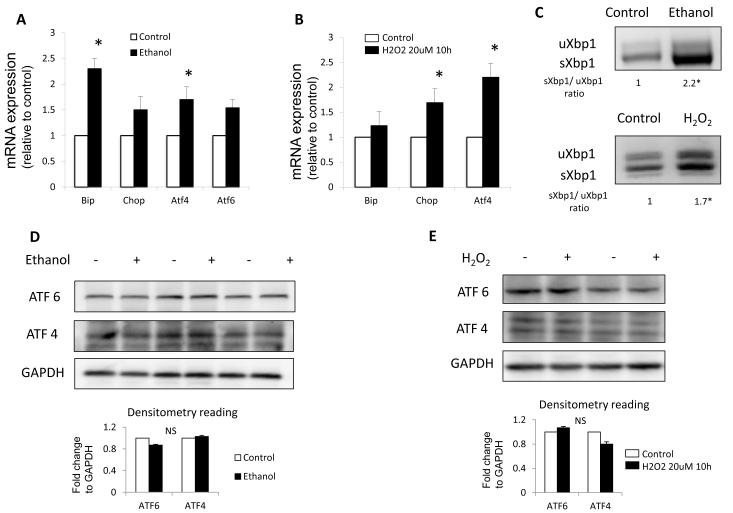
The ER stress response was characterized in stellate cells isolated from rats fed with either control or ethanol-containing (Lieber-DeCarli) diet for eight months. Expression of Bip, Chop, Atf6 and Atf4 mRNAs was increased in stellate cells from ethanol-treated rats (Fig. 1A). Long-term ethanol feeding, however did not change protein levels of either ATF6 or ATF4 as determined by Western blot (Fig. 1D). Stellate cells from ethanol-fed rats had markedly increased splicing of Xbp1 mRNA (Fig. 1C), similar to a previous study of alcohol induced pancreatic damage [10].
Fig. 1. Oxidant stress induces ER stress.
(A, B) quantitative real time RT-PCR analysis of ER stress related genes Bip, Chop, Atf4 and Atf6, normalized to the housekeeping gene -actin in (A) rat stellate cell isolated from ethanol fed rats and (B) JS1 cells treated with H2O2. (C) Standard RT-PCR for Xbp1 showing an increase in the spliced isoform (sXbp1). (D, E) Western blot analysis for ATF6 and ATF4 with GAPDH shown as a loading control. Messenger RNA normalized to the housekeeping gene - actin is expressed as fold change relative to control (*P<0.05; error bars indicate SEM). Protein ratios (normalized to GAPDH) were used to quantify fold change relative to control and are shown in a plot graph below each blot. Data represent the mean value of 3 experiments (*P < .05, NS=Not significant).
To further verify that ROS induce the UPR in stellate cells, we also induced oxidant stress by exposing either JS1 (an immortalized murine hepatic stellate cell line [14]) or primary murine stellate cells to H2O2, a potent pro-oxidant species implicated in fibrogenic stimulation. H2O2 treatment led to an increase in Atf4, Chop, Bip (Fig. 1B) and spliced Xbp1 mRNA levels (Fig. 1C) whereas ATF4 and ATF6 protein expression remained unchanged (Fig. 1E).
Secreted proteins require proper folding to exit the ER, and Protein Disulfide Isomerase (PDI) is one of the essential ER oxidoreductases that catalyzes this reaction in an oxidative state-dependent manner [19]. To examine whether oxidant stress induced PDI expression, we measured PDI levels by Western blot. Indeed, stellate cells from ethanol-fed rats displayed increased PDI levels in response to the oxidizing environment (Fig. S1).
To investigate a potential link between ER stress and fibrogenesis, we treated mouse hepatic stellate cells (JS1) with tunicamycin (TNC), a classical ER stress inducer. Incubation of these cells with TNC (5 g/ml for 6 hours) induced a strong increase in mRNA expression of Col 1 1, Col 1 2, -pdgfr and -sma (Fig. S2A). Next, we extended these findings to a mouse in vivo model with short-term liver injury due to 3 doses of either CCl4 (0.5 L/g body weight) or thioacetamide (TAA) (100 mg/g body weight) every other day. Freshly isolated stellate cells from these injured livers displayed an accumulation of ATF4, ATF6 (Fig. S2B) and spliced Xbp1 mRNA levels, indicative of ER stress signaling, compared to cells from vehicle-treated mice (Fig. S2C).
ER stress triggers autophagy and stellate cell activation
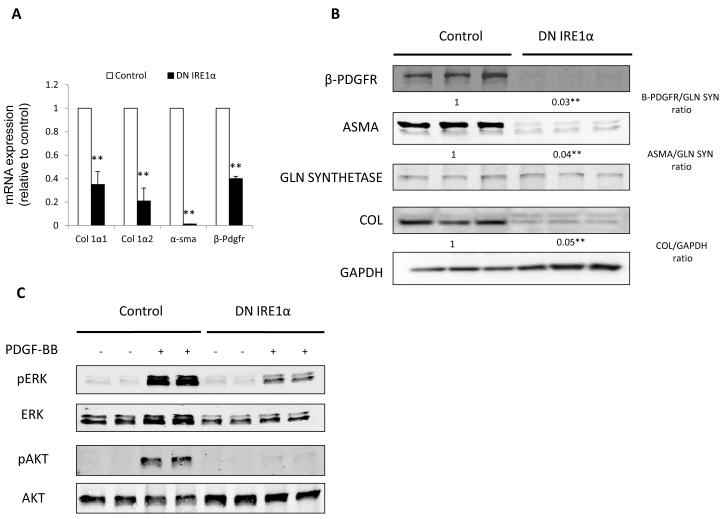
Because our findings primarily implicated IRE1 induction among the UPR branches, we next confirmed its importance in activated stellate cells. To do so, either a plasmid encoding a dominant-negative (DN) mutant of IRE1 (‘DN IRE1 cells’) or an empty vector control (‘Control cells’) was used for generation of stably transfected JS1 cells, as previously reported [20]. Expression of the DN IRE1 led to a significant decrease in the expression of Col 1 1, Col 1 2, -pdgfr and -sma mRNAs (Fig. 2A) and of their corresponding proteins (Fig. 2B), compared to control stellate cells. DN IRE1 cells also showed a decresed response to PDGF-BB-induced activation of the pro-mitogenic and pro-survival Erk and Akt pathways (Fig. 2C) and had impaired migration (data not shown).
Fig. 2. Blocking the IRE1 pathway leads to decreased stellate cell activation and autophagy.
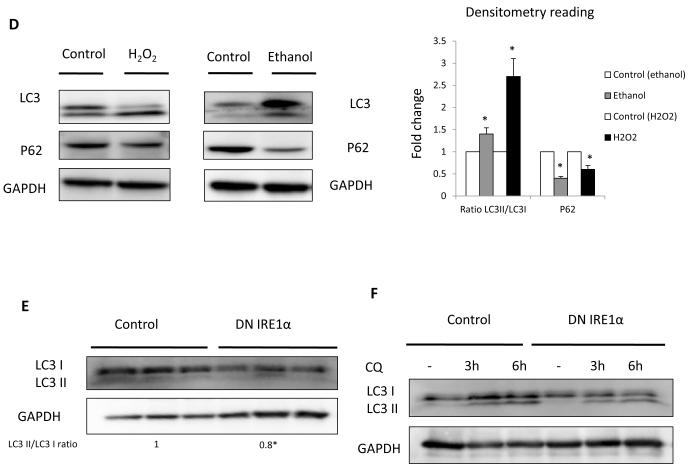
(A) Fibrogenic genes Col 1 1, Col 1 2, -sma and -pdgfr messenger RNA (q-rt RT-PCR) and (B) their protein expression in JS1 cells expressing DNIRE1 or control. (C) Immunoblots showing decreased levels of total and phosphorylated ERK and AKT after stimulation with PDGF-BB in JS1 cells expressing DN IRE1 or control. (D) Immunoblots of stellate cells isolated from ethanol fed rats and JS1 cells treated with H2O2, showing an increase in LC3II conversion (LC3II/LC3I ratio) and decrease of P62. (E) Immunoblots of JS1 cells expressing DNIRE1 or control showing a decrease in LC3 at baseline conditions and (F) after treatment with the lysosomal inhibitor CQ. Messenger RNA normalized to the housekeeping gene -actin is expressed as fold change relative to control (**P<0.001: error bars indicate SEM); the same blot was used as panel B and therefore has the same loading controls shown. Data represent the mean value of at least 3 experiments. Protein ratios (normalized to GAPDH or Gln Synthetase) were used to quantify fold change relative to control and are shown below each blot or as a plot graph (Fig 2D). Data represent the mean value of at least 3 experiments (*P<0.05, **P < .001)
We have recently shown that autophagy is required for stellate cell activation [3], however, it remained critical to uncover upstream events that provoke autophagy and the subsequent fibrogenic response. Intracellular ROS accumulation has been associated with increased autophagy within human hepatocytes, but it is unclear whether a similar link occurs in fibrogenic cells [21]. In addition to UPR activation, stellate cells isolated from rats fed ethanol, and JS1 cells treated with H2O2 demonstrated accumulation of LC3II, the conjugated form of LC3, and decreased levels of the selective autophagy adaptor molecule P62/SQSTM1 as determined by immunoblot, indicating an upregulation of autophagy (Fig. 2D). Blocking the IRE1 pathway was associated with decreased autophagy levels, however, expressed as the accumulation of the conjugated form of LC3 (Fig. 2E). We also measured autophagy flux using the lysosomal inhibitor CQ that leads to accumulation of LC3II by preventing degradation of the cargo by the lysosomal enzimes. Treatment with CQ caused in control cells an increase of LC3II over time as new vacuoles are being formed but cannot be degraded whereas levels remain unchanged in DNIRE1 cells indicating and impairment in autophagy (Fig. 2F). Together, these data support the idea that induction of the UPR response triggers autophagy, which then leads to stellate cell activation
IRE1 regulates autophagy in a p38MAPK dependent manner
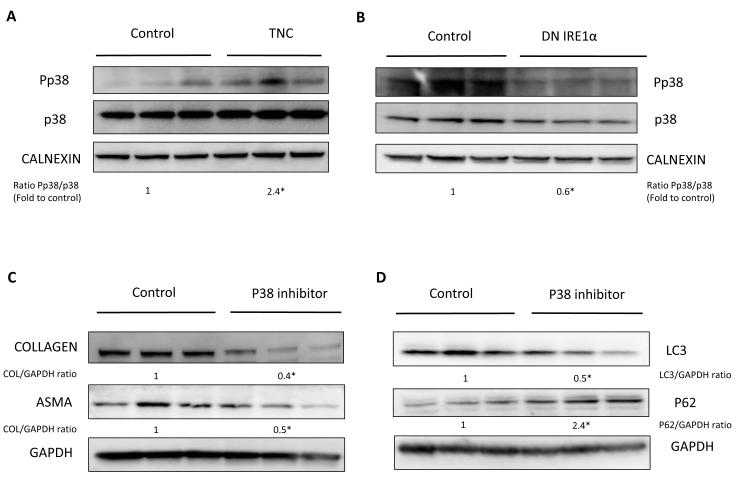
It remained important to identify the mechanism through which IRE1 signaling modulates autophagy. The Mitogen-Activated Protein Kinase (MAPK) pathway has been previously implicated in the regulation of autophagy in different cells, including hepatoma cell lines [22-25]. Therefore, we evaluated whether induction of ER stress was associated with altered activity of the MAPK pathway. Treatment with TNC led to significantly increased levels of phospho-p38 (Fig. 3A), which was abrogated when IRE1 was blocked (Fig. 3B). This effect of TNC was restricted to p38 MAPK, because phospho-JNK levels were not altered after either TNC or DNIRE1 over-expression, and phospho-ERK levels changed only slightly (Fig. S3). To address the prospect that ER stress may provoke fibrogenesis through activation of the p38 MAP kinase pathway, we incubated JS1 cells with the p38 inhibitor, SB203580. Blocking p38 with this compound mimicked the effect of blocking the IRE1 pathway in diminishing fibrogenic activity (Fig. 3C) and decreasing autophagy levels (Fig. 3D).
Fig. 3. IRE1 regulates the p38/MAPK pathway.
(A, B) Western blots for p38 and phospho-p38 from JS1 cells treated with TNC (A) and baseline expression in DN IRE1 cells (B). (C, D) Western blots for COL 1, ASMA, LC3 and P62 from JS1 cells treated with the P38 inhibitor SB 203580. Three different experiments with either calnexin or GAPDH as a loading control are shown. Data represent the mean value of at least 3 experiments. Protein ratios (normalized to calnexin or GAPDH) were used to quantify fold change relative to control and are shown below each blot. Data represent the mean value of at least 3 experiments (*P < 0.05).”
Oxidant and ER stress induce the antioxidant response via Nrf2
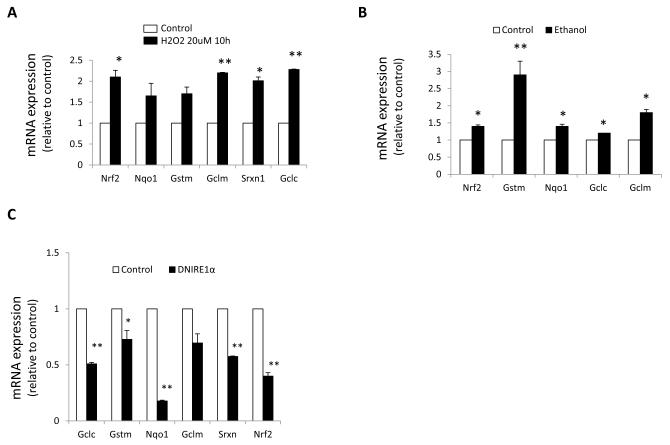
Nrf2 is a key mediator in the defense against oxidative stress in hepatocytes [26], and Xbp1 mRNA is induced in hepatocytes in an Nrf2-dependent fashion during oxidative stress conditions [27]. Moreover tunicamycin has also been shown to induce Nrf2 in pancreatic β cells, suggesting that Nrf2 may be involved in the ER stress response[28]Yet, the specific role of Nrf2 in stellate cells remains unknown. We therefore investigated the contribution of Nrf2 to the defense against oxidative and ER stress in stellate cells isolated from either ethanol-fed rats or a control diet, and in cells exposed to H2O2 (20 M) for 10 hours. Oxidant stress induced the expression of Nrf2 and its target genes, which encode the antioxidant enzymes Gst, Nqo1, Srxn1, Gclc and Gclm (Fig. 4A-B). TNC treatment was also able to induce expression of the Nrf2 targets (data not shown) whereas DNIRE1 cells exhibited a marked reduction in ARE-mediated gene expression and GST activity (Fig. 4C), implicating control of the antioxidant response through the IRE1 pathway.
Fig. 4. Oxidant stress induces the Nrf2-dependent antioxidant response.
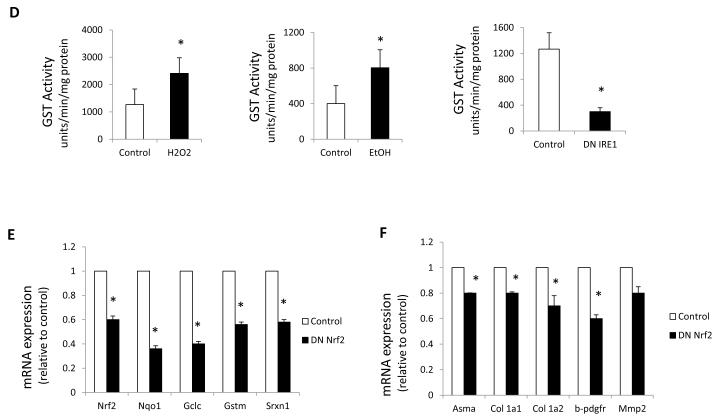
(A-C), Nqo1, Gstm, Gclc, Gclm, Srx and Nrf2 messenger RNA expression assessed by q-rt RT-PCR on (A) JS1 cells treated with H2O2 (B) stellate cells isolated from ethanol fed rats and (C) JS1 cells expressing DN IRE1 .( D) GST activity. (E) Nrf2, Nqo1, Gclc, Gstm Srx messenger RNA levels assessed by q-rt RT-PCR in stellate cells expressing DN NRF2. (F) Col 1 1, Col 1 2, -sma, -pdgfr and Mmp2 messenger RNA levels assessed by q-rt RT-PCR in primary stellate cells expressing DN NRF2. Messenger RNA normalized to the housekeeping gene -actin is expressed as fold change relative to control (*P<0.05, **P<0.001: error bars indicate SEM). Data represent the mean value of at least 3 experiments.
The GST enzyme superfamily is among the most important detoxifying enzymes regulated by Nrf2. These enzymes catalyze the conjugation of reduced glutathione, which neutralizes their electrophilic sites and renders the products more water-soluble. To verify the down-regulation of GST detected by q-rt PCR, we measured total GST activity [18]. Indeed, chronic ethanol feeding or H2O2 treatment increased GST activity in cultured stellate cells, whereas it was decreased in DNIRE1 dells (Fig. 4D).
Glutathione plays a key role in maintaining the intracellular redox balance under oxidative stress conditions. The rate-limiting step of the de novo synthesis of glutathione is catalyzed by the enzyme glutamate-cysteine ligase, which is composed of a catalytic subunit (GCLC) and modifier subunit (GCLM)[29]. To exclude the possibility that the differences found between stellate cells from ethanol-fed rats, or JS1 cells treated with H2O2 were due to glutathione depletion, we evaluated GCLC and GCLM by Western Blot. However, no differences were found, indicating that glutathione depletion was not provoking the changes in Nrf2 expression (Fig. S4).
It remained vital to establish the role of the antioxidant response in stellate cell activation. The stellate cell’s response to injury entails autophagy up-regulation that allows proliferation and acquisition of the fibrogenic phenotype in order to encapsulate the damaged parenchyma. However, perpetuation of this initially protective response becomes detrimental. In fact, selective autophagy inhibition in stellate cells has been suggested as an antifibrotic strategy [3]. ROS upregulate autophagy in an attempt to cope with the injury, but also induce the antioxidant response controlled by Nrf2, however the contribution of each mechanism to the fibrogenic response in stellate cells remained to be elucidated. Moreover, induction of the antioxidant response in stellate cells reportedly promotes myofibroblastic transdifferentiation via the autophagy regulator PI3K [30], suggesting a crosstalk between the two pathways in stellate cells.
To address the contribution of the Nrf2-dependent antioxidant response, we inhibited Nrf2 activity in stellate cells via expression of a dominant-negative Nrf2 mutant (DN Nrf2)[17] (Fig. 4E). Nrf2 down-regulation decreased stellate cells’ fibrogenic potential (Fig. 4F and Fig. S5) without affecting cell survival.
This data support the idea that oxidant injuries induce the Nrf2-dependent antioxidant response in parallel with the induction of the fibrogenic response.
Discussion
Stellate cell activation is a central mechanism underlying liver fibrogenesis and therefore an attractive target for the development of new antifibrotic drugs; however a better understanding of the pathways implicated in the regulation of activation is needed. In agreement with studies in other tissues [31, 32] our findings establish ER stress as a novel intracellular pathway implicated in the fibrogenic response in liver.
Once activated, stellate cells produce and secrete excessive amounts of collagen and extracellular matrix that challenge the cell’s ER folding capacity. Stellate cell activation entails ER stress and UPR activation, in particular IRE1 activation and Xbp1s expression. Therefore, we hypothesize that the IRE1 /Xbp1 pathway may be essential to maintain stellate cell activation. Indeed, blocking the IRE1 pathway in stellate cells decreases their fibrogenic potential.
Our recent work established the role of autophagy as an important driver of stellate cell activation and fibrogenesis [3], however specific triggers of autophagy in this process remained unknown. Here we show that, as reported previously in other organs [9, 10, 31, 32], induction of the UPR up-regulates autophagy. In liver injury, this leads to stellate cell activation and the fibrogenic response. We have also dissected the molecular mechanism of this response under ER stress conditions. Induction of the mitogen-activated protein kinase (MAPK) family is activated during ER stress conditions [33-36], and in particular p38 MAPK plays a role in hepatic [37, 38] and pancreatic [39] stellate cell activation. Our results demonstrate that p38MAPK is activated along with the IRE1 branch of the UPR and modulates fibrogenic gene expression through autophagy. Reduction in phospho-p38 in DNIRE1 cells or after treatment with the p38 inhibitor, SB 203580 not only decreases autophagy but also attenuates fibrogenesis, revealing its contribution to the liver’s fibrotic response. Similar results, supporting the induction of autophagy through p38MAPK under ER stress conditions, have been reported in human fibroblasts from patients with Pompe disease [33].
As oxidant stress frequently accompanies liver fibrogenesis and is a well-described determinant of stellate cell activation and collagen deposition [19, 40], we hypothesized that it may also trigger the UPR. As reported previously in other organs [9, 10], oxidant stress induced by ethanol feeding or H2O2 treatment incites ER stress and induces UPR activation. In stellate cells, this response involves the IRE1 /Xbp1 branch and increases autophagy levels.
In hepatocytes, oxidative stress induces an antioxidant response through the activation of the transcription factor Nrf2 [26]. Nrf2 binds to the ARE in the promoters of genes encoding ROS-detoxifying enzymes and other antioxidant proteins, including Gst, Nqo1, Srxn1, Gclc and Gclm, and Nrf2 itself. In hepatocytes, Nrf2 induction plays a protective role after different types of liver injury [6, 41-43], enhancing hepatocyte survival and regeneration. However, its role in stellate cells has not been adequately examined, and no prior studies have assessed the impact of manipulating Nrf2 activity in this cell type. Moreover, no antioxidant drug has been approved yet for the treatment of hepatic fibrosis. We have discovered that oxidant stress (H2O2 treatment and ethanol feeding) as well as ER stress through the IRE1 pathway induces the expression of Nrf2-dependent detoxifying enzymes and antioxidant proteins in hepatic stellate cells. Previous studies show that compared to quiescent HSC, activated HSC have an increased activation of the ARE signaling pathway[44], which may offer resistance to hepatic stellate cells against oxidative damage during hepatic injury. However this resistance could contribute to their increased proliferation and production of extracellular matrix through stimulation of autophagy. In another study of stellate cells[30] tert-butylhydroquinone (tBHQ), a known inducer of the ARE genes stimulated both an antioxidant response via Nrf2 and myofibroblast transdifferentiation with increased fibrogenic gene expression. Interestingly this effect was abrogated when tBHQ was administered together with the autophagy inhibitor LY294002 [30]. In our current study, blocking the IRE1 pathway strongly reduces fibrogenesis and down-regulates the Nrf2 mediated response. Additionally DNNrf2 cells displayed a decrease mRNA expression of the ARE genes. In aggregate, these data suggest that the activation of the ARE response in stellate cells may confer an increased fibrogenic response through its primary stimulation of autophagy, independent of its impact on oxidant stress.
Altogether, our data support the idea that oxidant stress perturbs ER homeostasis and activates the UPR in stellate cells, contributing to cellular activation. Activation of the IRE1 -Xbp1 pathway triggers both autophagy and the Nrf2-dependent antioxidant response in a p38-dependent manner in an attempt to cope with the injury however its long-term activation may precipitate the fibrogenic response and promote liver fibrosis via autophagy regulation. This evidence provides a rationale to explore the specific blockade of IRE1 in stellate cells as antifibrotic strategy.
Supplementary Material
Acknowledgments
Funding Supported by US National Institutes of Heath grants DK56621, AA020709 and P20AA017067 and 1K05AA018408. Moira Hilscher was supported by a Howard Hughes Medical Institute Medical Student Research Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Friedman SL. Mechanisms of hepatic fibrogenesis. Gastroenterology. 2008;134:1655–1669. doi: 10.1053/j.gastro.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hernandez-Gea V, Ghiassi-Nejad Z, Rozenfeld R, Gordon R, Fiel MI, Yue Z, et al. Autophagy releases lipid that promotes fibrogenesis by activated hepatic stellate cells in mice and in human tissues. Gastroenterology. 2012;142:938–946. doi: 10.1053/j.gastro.2011.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Yoon YH, Yi HY. In: Surveillance report #75: Liver cirrhosis mortality in the United States, 1970-2003. NIAAA, editor. Bethesda, MD: 2003. 2003. [Google Scholar]
- [5].Purohit V, Brenner DA. Mechanisms of alcohol-induced hepatic fibrosis: a summary of the Ron Thurman Symposium. Hepatology. 2006;43:872–878. doi: 10.1002/hep.21107. [DOI] [PubMed] [Google Scholar]
- [6].Gong P, Cederbaum AI. Nrf2 is increased by CYP2E1 in rodent liver and HepG2 cells and protects against oxidative stress caused by CYP2E1. Hepatology. 2006;43:144–153. doi: 10.1002/hep.21004. [DOI] [PubMed] [Google Scholar]
- [7].Keum YS, Han YH, Liew C, Kim JH, Xu C, Yuan X, et al. Induction of heme oxygenase-1 (HO-1) and NAD[P]H: quinone oxidoreductase 1 (NQO1) by a phenolic antioxidant, butylated hydroxyanisole (BHA) and its metabolite, tert-butylhydroquinone (tBHQ) in primary-cultured human and rat hepatocytes. Pharm Res. 2006;23:2586–2594. doi: 10.1007/s11095-006-9094-2. [DOI] [PubMed] [Google Scholar]
- [8].Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. J Biol Chem. 2009;284:13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Chen G, Ma C, Bower KA, Shi X, Ke Z, Luo J. Ethanol promotes endoplasmic reticulum stress-induced neuronal death: involvement of oxidative stress. J Neurosci Res. 2008;86:937–946. doi: 10.1002/jnr.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lugea A, Tischler D, Nguyen J, Gong J, Gukovsky I, French SW, et al. Adaptive unfolded protein response attenuates alcohol-induced pancreatic damage. Gastroenterology. 2011;140:987–997. doi: 10.1053/j.gastro.2010.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Schroder M, Kaufman RJ. The mammalian unfolded protein response. Annu Rev Biochem. 2005;74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- [12].Tirasophon W, Welihinda AA, Kaufman RJ. A stress response pathway from the endoplasmic reticulum to the nucleus requires a novel bifunctional protein kinase/endoribonuclease (Ire1p) in mammalian cells. Genes Dev. 1998;12:1812–1824. doi: 10.1101/gad.12.12.1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Blomhoff R, Berg T. Isolation and cultivation of rat liver stellate cells. Methods Enzymol. 1990;190:58–71. doi: 10.1016/0076-6879(90)90009-p. [DOI] [PubMed] [Google Scholar]
- [14].Guo J, Loke J, Zheng F, Hong F, Yea S, Fukata M, et al. Functional linkage of cirrhosis-predictive single nucleotide polymorphisms of Toll-like receptor 4 to hepatic stellate cell responses. Hepatology. 2009;49:960–968. doi: 10.1002/hep.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Urtasun R, Cubero FJ, Nieto N. Oxidative Stress Modulates KLF6(Full) and Its Splice Variants. Alcohol Clin Exp Res. 2012 doi: 10.1111/j.1530-0277.2012.01798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lieber CS, DeCarli LM. Liquid diet technique of ethanol administration: 1989 update. Alcohol Alcohol. 1989;24:197–211. [PubMed] [Google Scholar]
- [17].auf dem Keller U, Huber M, Beyer TA, Kumin A, Siemes C, Braun S, et al. Nrf transcription factors in keratinocytes are essential for skin tumor prevention but not for wound healing. Mol Cell Biol. 2006;26:3773–3784. doi: 10.1128/MCB.26.10.3773-3784.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Habig WH, Pabst MJ, Jakoby WB. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J Biol Chem. 1974;249:7130–7139. [PubMed] [Google Scholar]
- [19].Shimizu Y, Hendershot LM. Oxidative folding: cellular strategies for dealing with the resultant equimolar production of reactive oxygen species. Antioxid Redox Signal. 2009;11:2317–2331. doi: 10.1089/ars.2009.2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lim MP, Devi LA, Rozenfeld R. Cannabidiol causes activated hepatic stellate cell death through a mechanism of endoplasmic reticulum stress-induced apoptosis. Cell Death Dis. 2011;2:e170. doi: 10.1038/cddis.2011.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Bhogal RH, Weston CJ, Curbishley SM, Adams DH, Afford SC. Autophagy: A cyto-protective mechanism which prevents primary human hepatocyte apoptosis during oxidative stress. Autophagy. 2012;8 doi: 10.4161/auto.19012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Ogata M, Hino S, Saito A, Morikawa K, Kondo S, Kanemoto S, et al. Autophagy is activated for cell survival after endoplasmic reticulum stress. Mol Cell Biol. 2006;26:9220–9231. doi: 10.1128/MCB.01453-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Wang J, Whiteman MW, Lian H, Wang G, Singh A, Huang D, et al. A non-canonical MEK/ERK signaling pathway regulates autophagy via regulating Beclin 1. J Biol Chem. 2009;284:21412–21424. doi: 10.1074/jbc.M109.026013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Xu P, Das M, Reilly J, Davis RJ. JNK regulates FoxO-dependent autophagy in neurons. Genes Dev. 2011;25:310–322. doi: 10.1101/gad.1984311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chen KL, Chang WS, Cheung CH, Lin CC, Huang CC, Yang YN, et al. Targeting cathepsin S induces tumor cell autophagy via the EGFR-ERK signaling pathway. Cancer Lett. 2012;317:89–98. doi: 10.1016/j.canlet.2011.11.015. [DOI] [PubMed] [Google Scholar]
- [26].Xu W, Hellerbrand C, Kohler UA, Bugnon P, Kan YW, Werner S, et al. The Nrf2 transcription factor protects from toxin-induced liver injury and fibrosis. Lab Invest. 2008;88:1068–1078. doi: 10.1038/labinvest.2008.75. [DOI] [PubMed] [Google Scholar]
- [27].Kwak MK, Wakabayashi N, Itoh K, Motohashi H, Yamamoto M, Kensler TW. Modulation of gene expression by cancer chemopreventive dithiolethiones through the Keap1-Nrf2 pathway. Identification of novel gene clusters for cell survival. J Biol Chem. 2003;278:8135–8145. doi: 10.1074/jbc.M211898200. [DOI] [PubMed] [Google Scholar]
- [28].Lee S, Hur EG, Ryoo IG, Jung KA, Kwak J, Kwak MK. Involvement of the Nrf2-proteasome pathway in the endoplasmic reticulum stress response in pancreatic beta-cells. Toxicol Appl Pharmacol. 2012;264:431–438. doi: 10.1016/j.taap.2012.08.021. [DOI] [PubMed] [Google Scholar]
- [29].Wu G, Fang YZ, Yang S, Lupton JR, Turner ND. Glutathione metabolism and its implications for health. J Nutr. 2004;134:489–492. doi: 10.1093/jn/134.3.489. [DOI] [PubMed] [Google Scholar]
- [30].Reichard JF, Petersen DR. Involvement of phosphatidylinositol 3-kinase and extracellular-regulated kinase in hepatic stellate cell antioxidant response and myofibroblastic transdifferentiation. Arch Biochem Biophys. 2006;446:111–118. doi: 10.1016/j.abb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- [31].Lawson WE, Cheng DS, Degryse AL, Tanjore H, Polosukhin VV, Xu XC, et al. Endoplasmic reticulum stress enhances fibrotic remodeling in the lungs. Proc Acad Sci U S A. 2011;108:10562–10567. doi: 10.1073/pnas.1107559108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Baek HA, Kim do S, Park HS, Jang KY, Kang MJ, Lee DG, et al. Involvement of endoplasmic reticulum stress in myofibroblastic differentiation of lung fibroblasts. Am J Respir Cell Mol Biol. 2012;46:731–739. doi: 10.1165/rcmb.2011-0121OC. [DOI] [PubMed] [Google Scholar]
- [33].Shimada Y, Kobayashi H, Kawagoe S, Aoki K, Kaneshiro E, Shimizu H, et al. Endoplasmic reticulum stress induces autophagy through activation of p38 MAPK in fibroblasts from Pompe disease patients carrying c.546G>T mutation. Mol Genet Metab. 2011;104:566–573. doi: 10.1016/j.ymgme.2011.09.005. [DOI] [PubMed] [Google Scholar]
- [34].Kim DS, Kim JH, Lee GH, Kim HT, Lim JM, Chae SW, et al. p38 Mitogenactivated protein kinase is involved in endoplasmic reticulum stress-induced cell death and autophagy in human gingival fibroblasts. Biol Pharm Bull. 2010;33:545–549. doi: 10.1248/bpb.33.545. [DOI] [PubMed] [Google Scholar]
- [35].Kawakami T, Inagi R, Takano H, Sato S, Ingelfinger JR, Fujita T, et al. Endoplasmic reticulum stress induces autophagy in renal proximal tubular cells. Nephrol Dial Transplant. 2009;24:2665–2672. doi: 10.1093/ndt/gfp215. [DOI] [PubMed] [Google Scholar]
- [36].Hamamura K, Goldring MB, Yokota H. Involvement of p38 MAPK in regulation of MMP13 mRNA in chondrocytes in response to surviving stress to endoplasmic reticulum. Arch Oral Biol. 2009;54:279–286. doi: 10.1016/j.archoralbio.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Reeves HL, Dack CL, Peak M, Burt AD, Day CP. Stress-activated protein kinases in the activation of rat hepatic stellate cells in culture. J Hepatol. 2000;32:465–472. doi: 10.1016/s0168-8278(00)80398-0. [DOI] [PubMed] [Google Scholar]
- [38].Furukawa F, Matsuzaki K, Mori S, Tahashi Y, Yoshida K, Sugano Y, et al. p38 MAPK mediates fibrogenic signal through Smad3 phosphorylation in rat myofibroblasts. Hepatology. 2003;38:879–889. doi: 10.1053/jhep.2003.50384. [DOI] [PubMed] [Google Scholar]
- [39].Masamune A, Satoh M, Kikuta K, Sakai Y, Satoh A, Shimosegawa T. Inhibition of p38 mitogen-activated protein kinase blocks activation of rat pancreatic stellate cells. J Pharmacol Exp Ther. 2003;304:8–14. doi: 10.1124/jpet.102.040287. [DOI] [PubMed] [Google Scholar]
- [40].Svegliati-Baroni G, Ridolfi F, Di Sario A, Saccomanno S, Bendia E, Benedetti A, et al. Intracellular signaling pathways involved in acetaldehyde-induced collagen and fibronectin gene expression in human hepatic stellate cells. Hepatology. 2001;33:1130–1140. doi: 10.1053/jhep.2001.23788. [DOI] [PubMed] [Google Scholar]
- [41].Chan K, Han XD, Kan YW. An important function of Nrf2 in combating oxidative stress: detoxification of acetaminophen. Proc Natl Acad Sci U S A. 2001;98:4611–4616. doi: 10.1073/pnas.081082098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tan KP, Yang M, Ito S. Activation of nuclear factor (erythroid-2 like) factor 2 by toxic bile acids provokes adaptive defense responses to enhance cell survival at the emergence of oxidative stress. Mol Pharmacol. 2007;72:1380–1390. doi: 10.1124/mol.107.039370. [DOI] [PubMed] [Google Scholar]
- [43].Beyer TA, Xu W, Teupser D, auf dem Keller U, Bugnon P, Hildt E, et al. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 2008;27:212–223. doi: 10.1038/sj.emboj.7601950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Vasiliou V, Qamar L, Pappa A, Sophos NA, Petersen DR. Involvement of the electrophile responsive element and p53 in the activation of hepatic stellate cells as a response to electrophile menadione. Arch Biochem Biophys. 2003;413:164–171. doi: 10.1016/s0003-9861(03)00095-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.