Abstract
A series of inhibitors of acetylcholinesterase (AChE) have been screened by back-scattering interferometry (BSI). Enzyme levels as low as 100 pM (22,000 molecules of AChE) can be detected. This method can be used to screen for mixed AChE inhibitors, agents that have shown high efficacy against Alzheimer's disease, by detecting dual-binding interactions.
Keywords: detection method, interferometry, acetylcholinesterase, high-throughput screening, enzyme assay
In recent years, the emphasis of drug discovery and optimization has shifted from traditional methods to rapid microfluidic screening to minimize cost and maximize output. Back-scattering interferometry (BSI),[1] a technique which quantifies refractive index (RI) changes arising from intermolecular binding interactions, is a novel biosensing platform that may be useful in drug discovery. Acetylcholinesterase (AChE), a widely studied serine hydrolase that plays pivotal roles in Alzheimer’s disease (AD),[2] inflammatory processes,[3] and nerve-agent poisoning,[4] is an interesting system with which to study the use of BSI for the rapid detection of drug candidates. The search for potent AChE inhibitors (AChEI) has been driven largely by the need for an effective AD treatment, and is based on the long-standing cholinergic hypothesis[2] and the more recent amyloid hypothesis.[5] It is well-established that AChE accelerates amyloid-beta (Ab) peptide deposition, a process which may be mediated by an interaction between plaque precursors and the peripheral anionic site (PAS) of the enzyme.[6] Recent drug discovery efforts have focused on the design of AChEIs that are able to interact with the PAS to target both cholinergic and noncholinergic AD pathologies.[7] Seminal work in this field has led to the discovery of dual-binding inhibitors, such as bisgalantamine[8] and pseudo-irreversible carbamate inhibitors.[9] Given the large number of studies on several diverse classes of AChEIs, this system is a widely accepted benchmark for the development and testing of novel screening techniques.
Traditional methods used to detect interactions with AChE focus on the indirect measurement of substrate procedures.[10] To explore the utility of BSI to detect AChEIs, several known and novel inhibitors with diverse potencies and inhibitory mechanisms have been screened. Herein, we show that BSI can: 1) quantify AChE–inhibitor interactions in the absence of substrate (S); 2) detect signal changes at enzyme concentrations lower than previously reported techniques; 3) sense and deconvolute multiple binding events; and 4) discern between different types of inhibitors by comparing BSI binding data with half-maximal inhibitory concentrations (IC50).
The straightforward setup of BSI utilizes a multipass configuration,[1] where the refraction of monochromatic light within a microfluidic channel generates an interference fringe pattern. Because one of the surfaces of the channel is curved, the light deviates from its original path and hence refracts several times within the channel prior to exiting.[1f] The resulting high-contrast fringes are then reflected off a mirror and directed onto a charge-coupled device array camera, which acts as the detector. This unique optical train increases the effective pathlength and allows for the analysis of pM to µM receptor and ligand concentrations for an optical probe volume of 360 pL.[1a,f] The high-sensitivity interference fringe pattern allows the quantification of RI changes based on phase shifts of the back-scattered laser beam. Several studies have shown that these shifts correlate well with ligand–receptor binding interactions and can result from conformational changes, solvation/desolvation, changes in dipole moments, and polarizability.[1] Because RI changes may arise from molecular interactions unrelated to target binding, ligand and receptor blanks must be prepared to ensure that the measured changes reflect an actual binding event. Furthermore, a temperature-controlled stage is employed to avoid minor RI shifts arising from temperature fluctuations. From the BSI-generated data, an equilibrium dissociation constant (KD) can be obtained by fitting the average signal shift generated at each concentration to an exponential rise to max equation.
BSI can be used to measure binding interactions in free solution without the need for surface immobilization, specialized reagents, or fluorescent probes, thus providing a clear advantage over techniques such as isothermal titration calorimetry (ITC),[11] surface plasmon resonance (SPR),[12] and fluorometric assays.[13] Furthermore, cosolvents, such as DMSO[1b] and acetonitrile,[1c] can be used without assay interference.
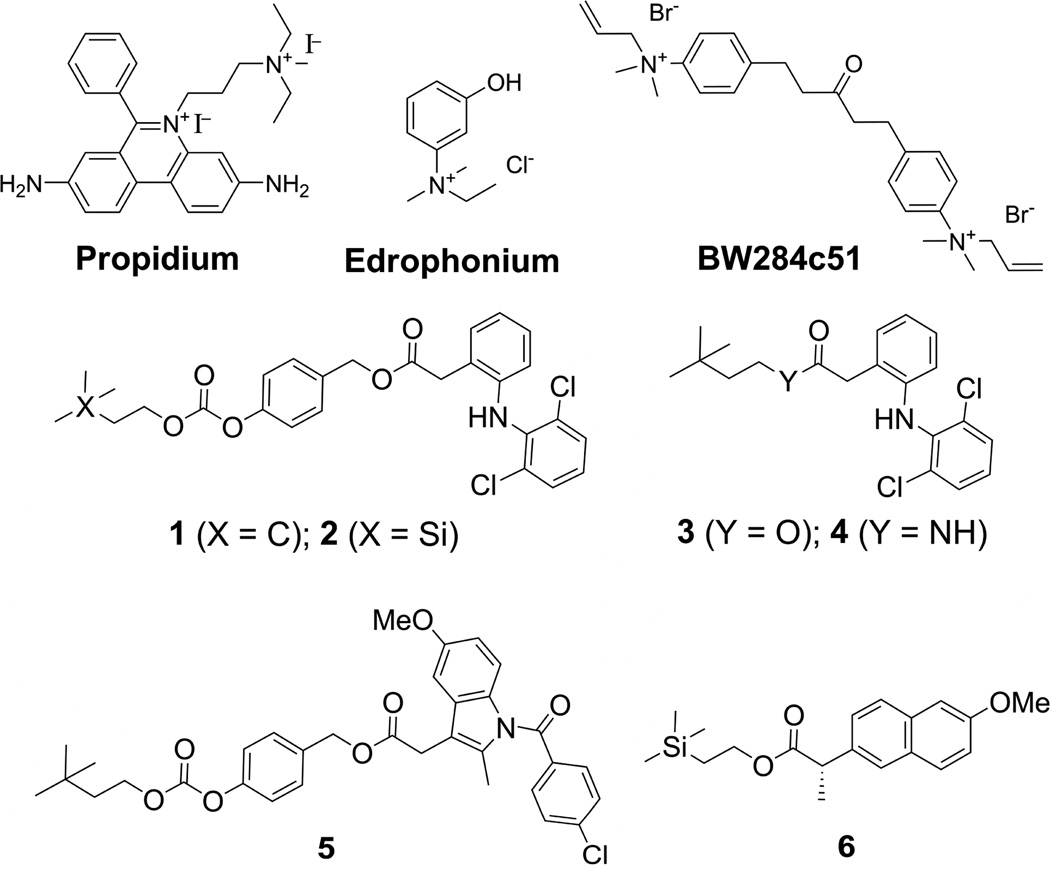
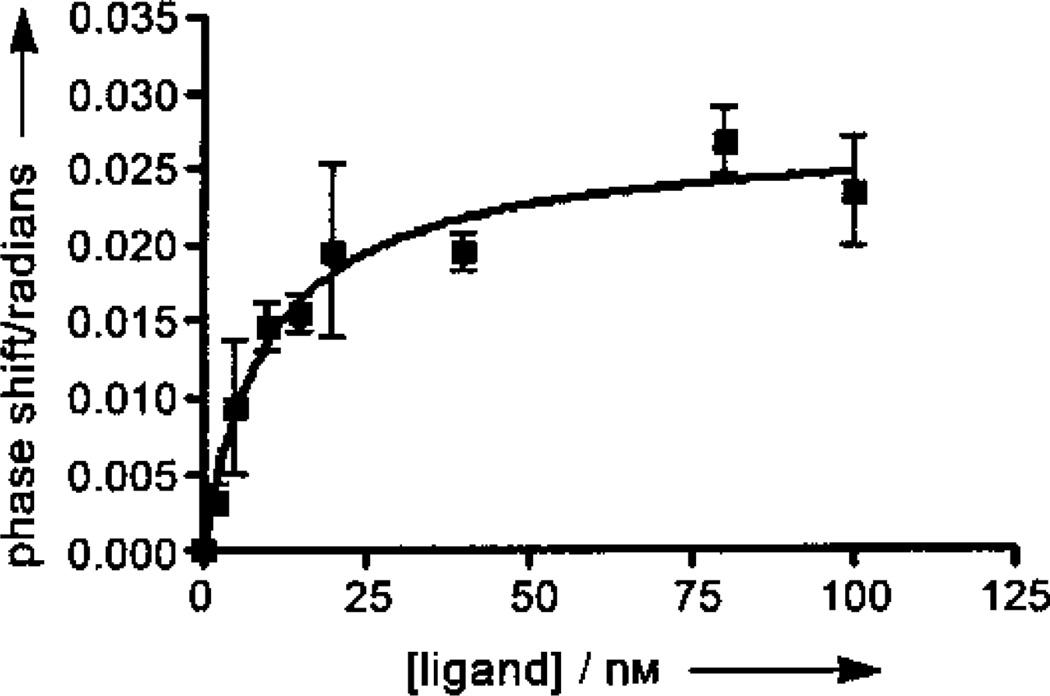
We first wanted to probe the utility of BSI to screen AChEIs using a set of known and novel AChEIs (Scheme 1). Edrophonium, a competitive inhibitor, propidium, a noncompetitive inhibitor, and the mixed inhibitors, 1,5-bis(4-allyldimethylammoniumphenyl) pentan-3-one dibromide (BW284c51) and galantamine, were the standards used to validate BSI as a method for the detection of AChEIs. Physostigmine,[14] a pseudo-irreversible carbamate AChEI used for the treatment of AD, was additionally explored to determine whether BSI could be used to detect a range of inhibitor types. It is worth noting that carbamates are not only effective anticholinesterases,[9] they have also been extensively investigated as inhibitors of other serine hydrolases, such as fatty acid amide hydrolase.[15] A binding curve for the high affinity ligand, BW284c51, is shown in Figure 1. An enzyme concentration of 72 nm with varying ligand concentrations (1–100 nm) resulted in signal shifts of about 0.02 radians and a KD that is consistent with previously reported values. KD values for other known inhibitors also are in the range of literature values (Table 1).
Scheme 1.
Structures of AchEIs screened using BSI.
Figure 1.
BSI binding curve of signal versus concentration of BW284c51 ligand. Signal shifts of the back-scattered laser for equilibrated samples of AChE (72 nM) with ligand (2, 5, 10, 20, 40, 80, and 100 nM) in PBS were measured. Each data point represents the average of at least five trials and the error bars shown indicate the full value of the standard error of the measurement.
Table 1.
Reported Ki and experimental KD values for known AchEIs.
| Ligand | Ki [mM] | BSI-KD [mM] | References |
|---|---|---|---|
| Edrophonium | 1.5–3.8 | 1.27 ± 0.4 | [16, 17] |
| Physostigmine | 0.013[a] –0.11 | 0.020 ± 0.009 | [18] |
| Galantamine | 0.20–0.61 | 1.23 ± 0.5 | [19, 8a] |
| Propidium | 0.63–1.5 | 0.64 ± 0.09 | [16, 20] |
| BW284c51 | 0.0032–0.008 | 0.0084 ± 0.002 | [21, 22] |
Value listed is an IC50 value.
Besides KD (or inhibitor dissociation constants Ki) values, another common measurement of inhibitor potency is an IC50 value. While KD/Ki determinations require complex graphical methods,[23] IC50 values are easily determined and are better suited for the screening of large compound libraries. However, IC50 measurements can vary depending on experimental conditions, such as substrate and enzyme concentrations.[24a] Furthermore, cases exist where IC50 values differ from KD values by up to three orders of magnitude, a trend which usually occurs with complex inhibitory mechanisms.[24]
We next decided to explore the utility of BSI for the investigation and mechanistic characterization of novel noncompetitive AChEIs by comparing IC50 values (determined via Ellman’s method[10a]) to BSI-generated KD values (Table 2). Interestingly, while the IC50 value for inhibitor 3 correlates well with the BSI KD values for ligands 1, 2, and 5 are 5- to 10-fold lower than their corresponding IC50 values. These results were originally perplexing, because noncompetitive inhibitors traditionally exhibit KD values close in magnitude to their IC50 values.
Table 2.
Correlation of BSI KD values with anticholinesterase activities.
| Ligand | Kies/IC50 (µM)[a] | BSI KD (µM) |
|---|---|---|
| 1 | 0.51 ± 0.02 | 0.11 ± 0.02 |
| 2 | 1.36 ± 0.1 | 0.12 ± 0.04 |
| 3 | 2.69 ± 0.1 | 2.18 ± 0.4 |
| 4 | 6.34 ± 0.5 | 0.97 ± 0.57[b] |
| 5 | 2.29 ± 0.9 | 0.21 ± 0.07 |
| 6 | 13.9 ± 0.3 | Dual-binding |
Previously reported IC50 values against AchE from Electrophorus electricus.[25]
KD value is for second binding event for dual-binding ligand.
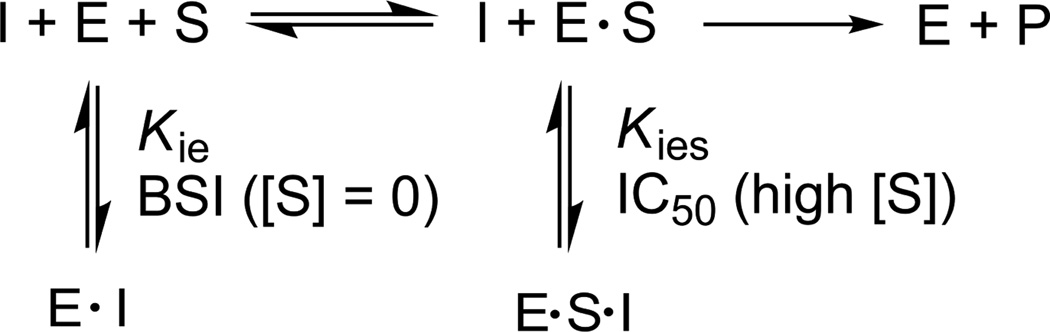
The above KD/IC50 relationship can be rationalized by considering the Cheng–Prusoff equation.[23, 26] For noncompetitive and mixed inhibition, the IC50 is equal to the inhibitor’s affinity for the enzyme–substrate complex (Kies) in the presence of high substrate concentrations (that is, traditional Ellman assay conditions). Conversely, BSI measurements are performed in the absence of substrate and are an indication of a given ligand’s affinity for the enzyme alone (KD or Kie). This relationship is depicted in Scheme 2.
Scheme 2.
Relationship between BSI KD (Kie) and Ellman assay IC50 for noncompetitive and mixed inhibition.[23]
With the Cheng–Prusoff relationship in mind, we revisited our KD/IC50 correlation for ligands 1, 2, 3, and 5. When the IC50 is measured at high [S], the portion of the ligand with affinity for the active site is not able to compete with the substrate. However, under conditions used for BSI, these particular ligands are free to interact with both AChE sites, thus resulting in higher binding affinities (lower KD values). These data, then, suggest that ligands 1, 2, and 5 are able to bind to both the active and peripheral sites in the absence of competing substrate. On the other hand, 3 has affinity for the PAS alone, as the affinity of this ligand for the enzyme is unaffected by substrate. This result is profound because it suggests that by measuring IC50 values at high [S] along with BSI KD values (two easily determined parameters), valuable insight is gained into the relative affinities that a mixed inhibitor has for the active and peripheral sites of AChE. Furthermore, these data indicate that if an IC50 value is equal to the BSI-generated KD, the inhibitor is acting by a true noncompetitive or an uncompetitive mechanism and thus has affinity for the PAS alone. Conversely, if the IC50 is greater than the KD, it implies that the compound interacts with the active site by a competitive or mixed interaction.
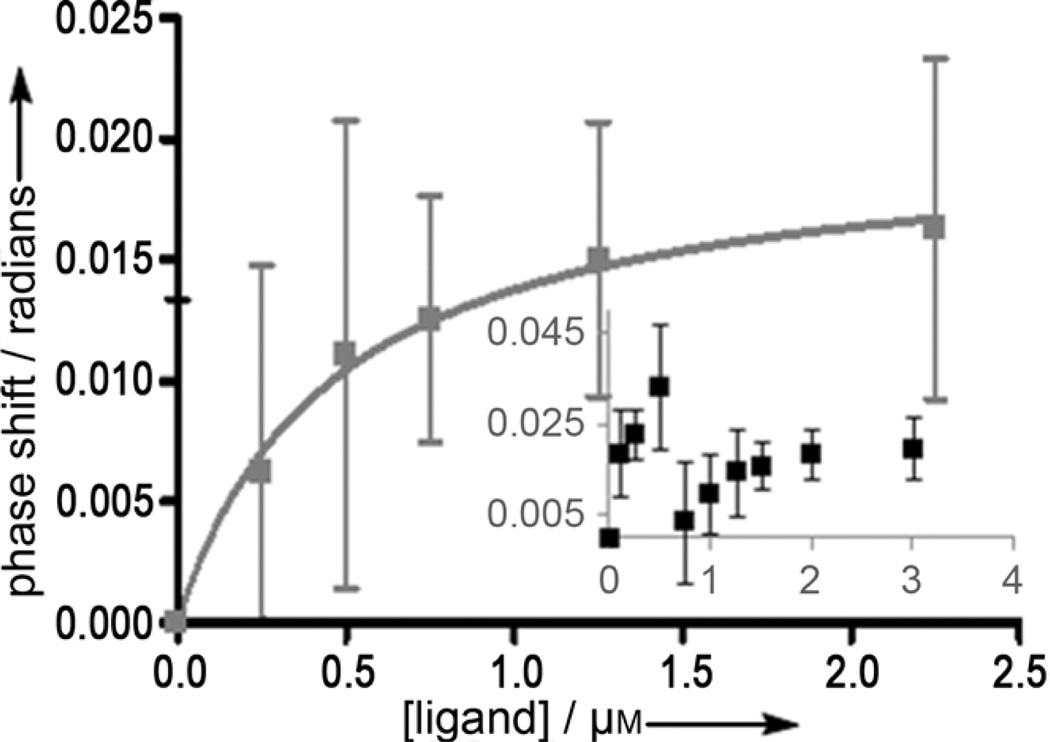
Two separate and distinct saturation curves were observed for inhibitors 4 and 6 (Figure 2 and Supporting Information, Figure S7, respectively). For ligand 4, the dual-binding curve is shown in the graph inset of while the fit of the second binding event is shown in the main graph. While the first KD of 4 could not be accurately determined because of the close proximity to the second curve, the KD of the second curve was calculated as 0.97 ± 0.57 µm. To calculate this dissociation constant, the ligand concentrations were normalized to treat the second curve as an isolated binding event (Figure 2).
Figure 2.
Inset: BSI dual-binding curve of signal versus concentration of 4. Signal shifts of the back-scattered laser for equilibrated samples of AChE (6.9 µM) with ligand (0.1, 0.25, 0.5, 0.75, 1.0, 1.25, 1.5, 2.0, and 3.0 µM) in PBS/methanol (9:1) were measured. The main graph depicts the fit of the second binding curve. To fit this curve, the second curve was normalized and treated as an isolated binding event. In both graphs, each data point is the average of at least five trials, and the error bars shown indicate the full value of the standard error of measurement.
We were initially surprised that the signal shift approached zero prior to the second binding event for both dual-binding ligands. It is important to note, however, that RI shifts may not always occur in the same direction. Rather, the transition to a zero signal shift simply is a measureable change in the optical properties of the system. Once the first site of AChE is saturated and the second site starts to be occupied by ligand, enough water molecules could be displaced to change the RI of the bulk solution (causing the signal to change and the phase shift to approach zero). Putting this result into context, the active-site gorge of AChE contains a considerable amount of easily displaced water molecules, about half of which are in a region neighboring the active site.[27] Perhaps once the active site is occupied, water molecules are displaced to facilitate ligand binding to the PAS. In addition to displaced water molecules, a conformational shift of the enzyme may occur upon saturation of the active site to allow PAS binding.[28] This result suggest that BSI has the potential to screen for mixed AChEIs, compounds which have shown high efficacy against AD.[5] Furthermore, with further optimization of this method, both the potency and the type of inhibition could be determined using BSI and may eliminate the need for enzyme kinetics and fluorescence displacement assays, which are labor-intensive and prone to error.[24]
As indicated by the IC50 and dual-binding data, BSI has the ability to distinguish between true noncompetitive and mixed inhibition. This is important because of the well established role of the PAS in AD. Blocking this site by noncompetitive inhibition has proven very effective at treating both cholinergic and non-cholinergic AD symptoms. Seminal work by Andrisano et al. revealed that the true PAS inhibitor, propidium, attenuates Ab aggregation by 82%, while mixed inhibitors only provide moderate suppression (22–30%). Competitive inhibitors, such as edrophonium, have no effect on AChE-induced Ab aggregation, indicating that affinity for the PAS is required for the observed aggregation suppression.[5] By discerning between true noncompetitive and competitive (or mixed) inhibitory mechanisms, BSI may be useful as part of a rapid screen for therapeutically important PAS inhibitors.
We next determined the detection limit of BSI for AChE sensing. At the lowest limit, concentrations of 100 pm can be detected, which equates to an astonishing 22000 molecules of enzyme with an optical probe volume of 360 pL. This detection level approaches and, in many cases, surpasses the sensitivity of recently reported AChE detection methods, such as chemiluminescent dioxetane probes,[10b] the aggregation-induced emission (AIE) of tetraphenylethylenes (TPE),[10c,d] cyano-substituted poly(p-phenylenevinylene) (PPV) probes[10e] and other fluorescence assays,[10f,g] and electrochemical detection methods involving gold nanoparticles[10h] (Table 3). Unlike many of these techniques, BSI does not require substrate analogues, specialized probes, or laborious procedures.
Table 3.
Comparison of the limit of detection (LOD) of BSI to previously reported AChE detection methods.
| Technique | ATCh (Y/N) |
LOD [mUmL−1] |
LOD [moles] |
Ref. |
|---|---|---|---|---|
| BSI | N | 24 | 3.6 × 10−20 | – |
| Ellman assay | Y | – | 1.85 × 10−18 | [10a,b] |
| Chemiluminescence | Y | 30–40 | 2.5 × 10−19 | [10b] |
| AIE of TPE | Y | 5 | – | [10c] |
| AIE of TPE | N | 500 | – | [10d] |
| Cyano-PPV Fluor. | N | 12,500 | – | [10e] |
| Conjugated polymer Fluor. | N | 50 | – | [10f] |
| Laser-induced Fluor. | Y | 75 | – | [10g] |
| Electrochemical AuNP detection | N | 1000 | – | [10h] |
Such low-level detection of AChE, an enzyme whose price ranges from hundreds to thousands of dollars per mg, is also very cost-effective. The study of AChE mutants will therefore be more accessible with BSI, as the preparation and isolation of large quantities of enzyme are not required. Furthermore, one of the drawbacks with the use of the Ellman assay is nonenzymatic hydrolysis of acetylthiocholine (ATCh), an analogue of acetylcholine (ACh), which can result in misleading cholinesterase activities. This effect has been reported in the investigation of nucleophilic oximes.[29] Spurious signals may presumably be observed with techniques relying on the quantification of ATCh hydrolysis (or a derivative thereof; Table 3). As a substrate is not required to generate a BSI signal, this is a useful method to screen diverse compound libraries without the potential for false signals.
Any intermolecular interactions which generate a RI change of at least 10−8 units can be detected by BSI.[1e] There are, however, some limitations to the current platform. For novel and insufficiently characterized receptors, appropriate control experiments are critical to correct for such events as enzyme aggregation or denaturation. Furthermore, care should be taken during the interpretation of KD values, as they may not correlate directly with physiological action or inhibitory activity. Finally, using BSI data alone, the site of inhibition cannot be determined (unless the ligand is a mixed inhibitor, as shown with AChE ligands 4 and 6).
In summary, BSI provides a highly sensitive method for the detection of anticholinesterases with a range of potencies and inhibition types. Unlike previous methods, BSI does not require the use of substrate (or a derivative thereof) for signal generation, thus avoiding the possibility of false signals. Using the multipass configuration of BSI, a detection limit of 3.6 × 10−5 fmol of AChE was obtained for an optical probe volume of 360 pL, a sensitivity which surpasses that of previously described colorimetric, fluorescent, chemiluminescent, and electrochemical techniques.[10] Two distinct binding interactions of inhibitors 4 and 6 were also detected in one experiment, demonstrating that BSI is well-suited for the rapid screening for dual-binding AChEIs. Finally, by comparing IC50 values generated at high [S] to BSI KD values, therapeutically important PAS inhibitors can be screened. While the focus of the present study is on AChE–small molecule interactions, the approach described herein should be applicable to other enzyme–inhibitor interactions and thus may be useful to screen libraries for ligands of orphan receptors. In a broader sense, BSI has the potential to track events other than target binding, such as metal complexation, liposome formation, and intermolecular interactions of medically and biologically relevant molecules.
Experimental Section
Acetylcholinesterase (Type VI-S from Electrophorus electricus), 1,5- bis(4-allyldimethylammoniumphenyl)pentan-3-one dibromide, propidium diiodide, and phosphate-buffered saline (PBS) packets were purchased from Sigma–Aldrich (Saint Louis, MO). Methanol was obtained from EMD Chemicals (Gibbstown, NJ) and 2(2-ethoxyethoxy) ethanol was purchased from Acros Organics (Geel, Belgium). Edrophonium chloride was purchased from MP Biomedicals (Solon, Ohio). PBS was prepared with MilliQ water (Millipore, Billerica, MA), and both PBS and the cosolvent were filtered using 0.2 µm membrane filters prior to use. AChE was dialyzed against PBS (3 × 300 mL) using regenerated cellulose dialysis tubing with a molecular weight cutoff of 3500 Da (Fisher, Hampton, NH). Inhibitors 1–3, 5, and 6 were synthesized as previously described.[25] For the synthesis of ligand 4, see the Supporting Information.
The previously reported instrumental setup contains a helium–neon (HeNe) laser (λ= 632.8 nm) directed onto a poly(dimethylsiloxane) (PDMS) chip containing a semicircular microfluidic channel (90 µm wide, 40 µm deep, cross-sectional area of 2.9 nm2).[1] The HeNe laser and temperature controller were allowed to equilibrate for at least 1 h before experiments were run. Chip silanization was performed using the procedure described in the Supporting Information. The microfluidic channels were rinsed with MilliQ water and PBS prior to each run. Owing to the high lipophilicty of many of the AChEIs, BSI experiments were run in PBS with either 10% methanol or 5% 2(2-ethoxyethoxy)ethanol as a cosolvent. The AChE concentration was held constant (0.025–9.5 µm) while the ligand concentrations were varied based either on the IC50 value determined by Ellman’s method or based on previously determined literature Ki values. Enzyme and ligand blanks were additionally prepared in order to correct for concentration-dependent RI changes. Significant RI shifts arising from changes in analyte concentration were subtracted out from the overall BSI-generated signal for that particular concentration. AChE:ligand samples were prepared in advance and allowed to equilibrate at 48C for at least 3 h prior to each experiment. The signal was measured for 45 to 60 s at 25 °C. Once the data were collected for each ligand concentration, the resulting binding curve was fit to a one site binding hyperbola function using GraphPad Prism (Version 4).
Supplementary Material
Acknowledgements
We thank Dr. Esther Pesciotta for her initial contributions to this work. We are grateful for support from the National Science Foundation (CHE-0848788) and the National Institutes of Health (U54AR055073).
References
- 1.a) Bornhop DJ, Latham JC, Kussrow A, Markov DA, Jones RD, Sorensen HS. Science. 2007;317:1732–1736. doi: 10.1126/science.1146559. [DOI] [PubMed] [Google Scholar]; b) Morcos EF, Kussrow A, Enders C, Bornhop D. Electrophoresis. 2010;31:3691–3695. doi: 10.1002/elps.201000389. [DOI] [PMC free article] [PubMed] [Google Scholar]; c) Pesciotta EN, Bornhop DJ, Flowers RA., II Org. Lett. 2011;13:2654–2657. doi: 10.1021/ol200757a. [DOI] [PubMed] [Google Scholar]; d Kussrow A, Enders CS, Castro AR, Cox DL, Ballard RC, Bornhop DJ. Analyst. 2010;135:1535–1537. doi: 10.1039/c0an00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]; e Markov D, Begari D, Bornhop DJ. Anal. Chem. 2002;74:5438–5441. doi: 10.1021/ac020403c. [DOI] [PubMed] [Google Scholar]; f) Swinney K, Markov D, Bornhop DJ. Anal. Chem. 2000;72:2690–2695. doi: 10.1021/ac000261r. [DOI] [PubMed] [Google Scholar]
- 2.Bartus RT, Dean RL, III, Beer B, Lippa AS. Science. 1982;217:408–414. doi: 10.1126/science.7046051. [DOI] [PubMed] [Google Scholar]
- 3.Wang H, Yu M, Ochani M, Amella CA, Tanovic M, Susarla S, Li JH, Wang H, Yang H, Ulloa L, Al-Abed Y, Czura CJ, Tracey KJ. Nature. 2003;421:384–388. doi: 10.1038/nature01339. [DOI] [PubMed] [Google Scholar]
- 4.Somani SM, editor. Chemical Warfare Agents. London: Academic Press; 1992. [Google Scholar]
- 5.a) De Ferrari GV, Canales MA, Shin I, Weiner LM, Silman I, Inestrosa NC. Biochemistry. 2001;40:10447–10457. doi: 10.1021/bi0101392. [DOI] [PubMed] [Google Scholar]; b) Bartolini M, Bertucci C, Cavrini V, Andrisano V. Biochem. Pharmacol. 2003;65:407–416. doi: 10.1016/s0006-2952(02)01514-9. [DOI] [PubMed] [Google Scholar]
- 6.Inestrosa NC, Alvarez A, Perez CA, Moreno RD, Vicente M, Linker C, Casanueva OI, Soto C, Garrido J. Neuron. 1996;16:881–891. doi: 10.1016/s0896-6273(00)80108-7. [DOI] [PubMed] [Google Scholar]
- 7.a) del Monte-Millan M, Garcia-Palomero E, Valenzuela R, Usan P, de Austria C, Munoz-Ruiz P, Rubio L, Dorronsoro I, Martinez A, Medina M. J. Mol. Neurosci. 2006;30:85–88. doi: 10.1385/JMN:30:1:85. [DOI] [PubMed] [Google Scholar]; b) Castro A, Martinez A. Curr. Pharm. Des. 2006;12:4377–4387. doi: 10.2174/138161206778792985. [DOI] [PubMed] [Google Scholar]; c) Munoz-Ruiz P, Rubio L, Garcia-Palomero E, Dorronsoro I, del Monte-Millan M, Valenzuela R, Usan P, de Austria C, Bartolini M, Andrisano V, Bidon-Chanal A, Orozco M, Luque FJ, Medina M, Martinez A. J. Med. Chem. 2005;48:7223–7233. doi: 10.1021/jm0503289. [DOI] [PubMed] [Google Scholar]; d) Reyes AE, Chacon MA, Dinamarca MC, Cerpa W, Morgan C, Inestrosa NC. Am. J. Pathol. 2004;164:2163–2174. doi: 10.1016/s0002-9440(10)63774-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Greenblatt HM, Guillou C, Guenard D, Argaman A, Botti S, Badet B, Thal C, Silman I, Sussman JL. J. Am. Chem. Soc. 2004;126:15405–15411. doi: 10.1021/ja0466154. [DOI] [PubMed] [Google Scholar]; b) Mary A, Renko DZ, Guillou C, Thal C. Bioorg. Med. Chem. 1998;6:1835–1850. doi: 10.1016/s0968-0896(98)00133-3. [DOI] [PubMed] [Google Scholar]
- 9.a) Greig NH, Pei XF, Soncrant TT, Ingram DK, Brossi A. Med. Res. Rev. 1995;15:3–31. doi: 10.1002/med.2610150103. [DOI] [PubMed] [Google Scholar]; b) Zhu XD, Cuadra G, Brufani M, Maggi T, Pagella PG, Williams E, Giacobini E. J. Neurosci. Res. 1996;43:120–126. doi: 10.1002/jnr.490430116. [DOI] [PubMed] [Google Scholar]
- 10.a) Ellman GL, Courtney KD, Andres V, Jr, Feather-Stone RM. Biochem. Pharmacol. 1961;7:88–95. doi: 10.1016/0006-2952(61)90145-9. [DOI] [PubMed] [Google Scholar]; b) Sabelle S, Renard PY, Pecorella K, de Suzzoni-Dezard S, Creminon C, Grassi J, Mioskowski C. J. Am. Chem. Soc. 2002;124:4874–4880. doi: 10.1021/ja0171299. [DOI] [PubMed] [Google Scholar]; c) Peng L, Zhang G, Zhang D, Xiang J, Zhao R, Wang Y, Zhu D. Org. Lett. 2009;11:4014–4017. doi: 10.1021/ol9016723. [DOI] [PubMed] [Google Scholar]; d) Wang M, Gu X, Zhang G, Zhang D, Zhu D. Anal. Chem. 2009;81:4444–4449. doi: 10.1021/ac9002722. [DOI] [PubMed] [Google Scholar]; e) Zhang W, Zhu L, Qin J, Yang C. J Phys Chem. B. 2011;115:12059–12064. doi: 10.1021/jp206930v. [DOI] [PubMed] [Google Scholar]; f) Feng F, Tang Y, Wang S, Li Y, Zhu D. Angew. Chem. 2007;119:8028–8032. Angew. Chem. Int. Ed. 2007, 46, 7882-7886; [Google Scholar]; g) Heleg-Shabtai V, Gratziany N, Liron Z. Anal. Chim. Acta. 2006;571:228–234. doi: 10.1016/j.aca.2006.04.087. [DOI] [PubMed] [Google Scholar]; h) Zhao W, Sun SX, Xu JJ, Chen HY, Cao XJ, Guan XH. Anal. Chem. 2008;80:3769–3776. doi: 10.1021/ac702395c. [DOI] [PubMed] [Google Scholar]
- 11.Velazquez Campoy A, Freire E. Biophys. Chem. 2005;115:115–124. doi: 10.1016/j.bpc.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 12.Boozer C, Kim G, Cong S, Guan H, Londergan T. Curr. Opin. Biotechnol. 2006;17:400–405. doi: 10.1016/j.copbio.2006.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Hemmila I. Clin. Chem. 1985;31:359–370. [PubMed] [Google Scholar]
- 14.Triggle DJ, Mitchell JM, Filler R. CNS Drug Rev. 1998;4:87–136. [Google Scholar]
- 15.a) Cravatt BF, Lichtman AH. Curr. Opin. Chem. Biol. 2003;7:469–475. doi: 10.1016/s1367-5931(03)00079-6. [DOI] [PubMed] [Google Scholar]; b) Kathuria S, Gaetani S, Fegley D, Valino F, Duranti A, Tontini A, Mor M, Tarzia G, La Rana G, Calignano A, Giustino A, Tattoli M, Palmery M, Cuomo V, Piomelli D. Nat. Med. 2003;9:76–81. doi: 10.1038/nm803. [DOI] [PubMed] [Google Scholar]
- 16.Shafferman A, Velan B, Ordentlich A, Kronman C, Grosfeld H, Leitner M, Flashner Y, Cohen S, Barak D, Ariel N. EMBO J. 1992;11:3561–3568. doi: 10.1002/j.1460-2075.1992.tb05439.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schalk I, Ehret-Sabatier L, Le Feuvre Y, Bon S, Massoulie J, Goeldner M. Mol. Pharmacol. 1995;48:1063–1067. [PubMed] [Google Scholar]
- 18.a) Bartolini M, Cavrini V, Andrisano V. J. Chromatogr. A. 2007;1144:102–110. doi: 10.1016/j.chroma.2006.11.029. [DOI] [PubMed] [Google Scholar]; b) Jennings NA, Pezzementi L, Lawrence AL, Watts SA. Comp. Biochem. Physiol. Part B. 2008;149:401–409. doi: 10.1016/j.cbpb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Loizzo MR, Tundis R, Menichini F, Menichini F. Curr. Med. Chem. 2008;15:1209–1228. doi: 10.2174/092986708784310422. [DOI] [PubMed] [Google Scholar]
- 20.Camps P, Cusack B, Mallender WD, ElAchab RE, Morral J, Munoz-Torrero D, Rosenberry TL. Mol. Pharmacol. 2000;57:409–417. [PubMed] [Google Scholar]
- 21.Velan B, Kronman C, Ordentlich A, Flashner Y, Leitner M, Cohen S, Shafferman A. Biochem. J. 1993;296:649–656. doi: 10.1042/bj2960649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchot P, Khelif A, Ji YH, Mansuelle P, Bougis PE. J. Biol. Chem. 1993;268:12458–12467. [PubMed] [Google Scholar]
- 23.Burlingham BT, Widlanski TS. J. Chem. Educ. 2003;80:214–218. [Google Scholar]
- 24.a) Cer RZ, Mudunuri U, Stephens R, Lebeda FJ. NucleicAcids Res. 2009;37:W441–W445. doi: 10.1093/nar/gkp253. [DOI] [PMC free article] [PubMed] [Google Scholar]; b) Taylor P, Lappi S. Biochemistry. 1975;14:1989–1997. doi: 10.1021/bi00680a029. [DOI] [PubMed] [Google Scholar]; c) Rosenberry TL, Sonoda LK, Dekat SE, Cusack B, Johnson JL. Biochemistry. 2008;47:13056–13063. doi: 10.1021/bi8015197. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Tseng SJ, Hsu JP. J. Theor. Biol. 1990;145:457–464. doi: 10.1016/s0022-5193(05)80481-3. [DOI] [PubMed] [Google Scholar]; e) Lam P, Jadhav P, Eyermann C, Hodge C, Ru Y, Bacheler L, Meek J, Otto M, Rayner M, Wong Y, et al. Science. 1994;263:380–384. doi: 10.1126/science.8278812. [DOI] [PubMed] [Google Scholar]; f) Prusakiewicz JJ, Duggan KC, Rouzer CA, Marnett LJ. Biochemistry. 2009;48:7353–7355. doi: 10.1021/bi900999z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young S, Fabio K, Guillon C, Mohanta P, Halton TA, Heck DE, Flowers RA, II, Laskin JD, Heindel ND. Bioorg. Med. Chem. Lett. 2010;20:2987–2990. doi: 10.1016/j.bmcl.2010.02.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng Y, Prusoff WH. Biochem. Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 27.Koellner G, Kryger G, Millard CB, Silman I, Sussman JL, Steiner T. J. Mol. Biol. 2000;296:713–735. doi: 10.1006/jmbi.1999.3468. [DOI] [PubMed] [Google Scholar]
- 28.a) Bourne Y, Radic´ Z, Sulzenbacher G, Kim E, Taylor P, Marchot P. J. Biol. Chem. 2006;281:29256–29267. doi: 10.1074/jbc.M603018200. [DOI] [PubMed] [Google Scholar]; b) Bourne Y, Taylor P, Radic´ Z, Marchot P. EMBO J. 2003;22:1–12. doi: 10.1093/emboj/cdg005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.a) Sinko G, Calic M, Bosak A, Kovarik Z. Anal. Biochem. 2007;370:223–227. doi: 10.1016/j.ab.2007.07.023. [DOI] [PubMed] [Google Scholar]; b) Sakurada K, Ikegaya H, Ohta H, Akutsu T, Takatori T. Toxicol. Lett. 2006;166:255–260. doi: 10.1016/j.toxlet.2006.07.339. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.