Abstract
Background
The oncogenic microRNAs (miRNAs) miR-21 and miR-31 negatively regulate tumor-suppressor genes. Their potential as serum biomarkers has not been determined in human colorectal cancer (CRC).
Methods
To determine whether miR-21 and miR-31 are secretory miRNAs, we screened expression in medium from 2 CRC cell lines, which was followed by serum analysis from 12 CRC patients and 12 control subjects. We validated expression of candidate miRNAs in serum samples from an independent cohort of 186 CRC patients, 60 postoperative patients, 43 advanced adenoma patients, and 53 control subjects. We analyzed miR-21 expression in 166 matched primary CRC tissues to determine whether serum miRNAs reflect expression in CRC. Patient survival analyses were performed by Kaplan–Meier analyses and Cox regression models. All statistical tests were two-sided.
Results
Although miR-21 was secreted from CRC cell lines and upregulated in serum of CRC patients, no statistically significant differences were observed in serum miR-31 expression between CRC patients and control subjects. In the validation cohort, miR-21 levels were statistically significantly elevated in preoperative serum from patients with adenomas (P < .001) and CRCs (P < .001). Importantly, miR-21 expression dropped in postoperative serum from patients who underwent curative surgery (P < .001). Serum miR-21 levels robustly distinguished adenoma (area under the curve [AUC] = 0.813; 95% confidence interval [CI] = 0.691 to 0.910) and CRC (AUC = 0.919; 95% CI = 0.867 to 0.958) patients from control subjects. High miR-21 expression in serum and tissue was statistically significantly associated with tumor size, distant metastasis, and poor survival. Moreover, serum miR-21 was an independent prognostic marker for CRC (hazard ratio = 4.12; 95% CI = 1.10 to 15.4; P = .03).
Conclusions
Serum miR-21 is a promising biomarker for the early detection and prognosis of CRC.
Colorectal cancer (CRC) is a leading cause of cancer-related death worldwide. In the United States, CRC is the third most common cancer, with more than 143000 new cases and more than 52000 deaths each year (1). Several CRC screening tests, including fecal occult-blood testing and colonoscopy, have been available for years (2) and have aided in reducing the mortality associated with this disease (3–5). However, compliance with these screening tests has been far from adequate. Patients with metastatic disease frequently receive expensive cytotoxic chemotherapeutic regimens coupled with targeted monoclonal antibodies but with relatively modest benefits (6). Without a priori knowledge of which patients will experience tumor recurrence, there is inevitable overtreatment with agents associated with toxic side effects (7). These limitations underscore the need for novel biomarkers, particularly noninvasive biomarkers in serum or plasma, for diagnosis, prognosis, and prediction of response to chemotherapy.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that play a central role in the regulation of mRNA expression (8). The discovery that miRNA expression is frequently dysregulated in a cancer-specific manner provides an opportunity to develop these RNAs as biomarkers for cancer detection (9). Although most previous studies on miRNA expression have been performed on tissue specimens, some studies have shown diagnostic and prognostic potential for circulating miRNAs (10–14) because tumor-derived miRNAs can be present in blood and appear to be stably protected from endogenous ribonuclease activity in the circulation (15). Nonetheless, it is unclear whether expression profiles of circulating miRNAs reflect miRNA profiles of tumor tissues and to the best of our knowledge, no systematic investigation of the relationship between miRNA profiles in body fluids vs matched primary CRCs has thus far been undertaken. This is critical because increased expression of circulating miRNAs could be indicative of miRNAs secreted from a tumor, raising the overall diagnostic specificity of the biomarker.
MiR-21 is an oncogenic miRNA that modulates the expression of multiple cancer-related target genes such as PTEN, TPM1, and PDCD and has been shown to be overexpressed in various human tumors (16–18). In addition, miR-21 expression is upregulated in CRC tissues, is elevated during tumor progression, and is also associated with poor survival and response to chemotherapy (19–22). The clinical significance of circulating miR-21 levels in CRC remains unclear at this time. Although an earlier study was unable to use plasma miR-21 as a biomarker because of low levels of detection using a direct amplification method (10), a more recent study demonstrated statistically significantly elevated plasma miR-21 expression in CRC patients using TaqMan-based approaches (23). On the other hand, miR-31 is another miRNA frequently overexpressed in CRC tissues and has been shown to be associated with tumor prognosis (19,24). Additionally, both miR-21 and miR-31 are frequently upregulated, even in premalignant lesions such as colonic adenomas, which are the target lesions of CRC screening (25–27). In light of these observations, we hypothesized that these two miRNAs might be good candidates for exploration as circulating biomarkers for the early detection and prognosis of CRC, assuming that the expression pattern for these miRNAs in serum mirrors that in the neoplastic tissues.
We have systematically investigated the expression of miR-21 and miR-31 in a two-phase study. In the first phase, we determined whether cultured CRC cells secrete these miRNAs into the culture medium, establishing their secretory potential. We then performed quantitative analyses of these miRNAs in a subset of serum samples from CRC patients and healthy control subjects to determine the feasibility of their detection in the circulation. In the second phase, using a large validation cohort comprised of matched serum and tissue samples from patients with colorectal neoplasia and serum from healthy control subjects, we evaluated the clinical significance of these miRNAs as potential biomarkers for diagnosis and prognosis of CRC patients.
Methods
Study Design
This study included analysis of 568 serum and tissue specimens that were obtained from healthy volunteers and consecutively enrolled patients with colorectal adenomas and cancers at the Mie University Medical Hospital, Japan, between January 1, 2005 and December 31, 2010. This study was designed as an initial screening phase and a subsequent validation phase. In the screening phase, oncogenic miR-21 and miR-31 were selected (16–18,24,28), and their expression was measured using TaqMan-based quantitative reverse transcription polymerase chain reaction (qRT-PCR) using cell culture medium and matched serum and tissue samples. To determine the secretory potential of these miRNAs, two CRC cell lines, HCT116 and SW620, were cultured and a fraction of the culture medium was collected at 0, 24, and 48 hours after the initial seeding of cells in 10-cm dishes. In addition, a small set of preoperative serum samples were collected from 12 CRC patients and from 12 sex- and age-matched healthy subjects as control subjects. To further assess the specificity of miRNA expression in serum, CRC and adjacent normal tissues were analyzed from eight of the 12 CRC patients, from whom both matched normal and neoplastic tissues were available.
In the validation phase, changes in miRNA expression patterns in serum and tissues from CRC patients were validated in a large, independent cohort of patients where preoperative sera (n = 186) and matched surgical tissues (n = 166) were collected from a pool of 200 CRC patients. Additionally, postoperative sera (day 7 postoperation) were collected from an independent set of 60 patients from whom matching preoperative sera were available to determine whether miRNA expression was altered subsequent to tumor resection. To better appreciate the diagnostic utility of these miRNAs, sera from 43 patients with advanced adenomas and 53 healthy control subjects were also collected. The serum specimens from healthy subjects were sex- and age-matched and each volunteer had a negative colonoscopic examination and no prior diagnosis of any other malignancy. The Tumor Node Metastasis (TNM) staging system from the American Joint Committee on Cancer was used for classification of pathological tumor staging of CRC (29). All CRC patients who underwent surgery were followed up for tumor recurrence at regular intervals for up to 5 years. During each annual hospital visit, all patients underwent a chest x-ray, colonoscopy, and abdominal computerized tomography. Survival time was calculated from the date of diagnosis to the date of death or last date of follow-up. Patients treated with radiotherapy or chemotherapy before surgery were excluded from the study. Patients with stage III/IV disease received 5-fluorouracil–based chemotherapy, whereas no adjuvant therapy was given to stage I/ II CRC patients.
Ethics Statement
Both serum- and tissue-based specimen collection and studies were approved by the institutional review boards of Mie University Hospital, Japan, and Baylor University Medical Center, Dallas, Texas. All participants provided written consent and indicated willingness to donate their blood and tissue samples for research.
RNA isolation and Quantitative Reverse-Transcription Polymerase Chain Reaction
MiRNA extraction from serum and culture media samples was performed with miRNeasy RNA isolation Kits (Qiagen, Valencia, CA), whereas miRNA extraction from Formalin-fixed and paraffin-embedded (FFPE) samples was performed using RecoverAll Total Nucleic Acid Isolation Kits (Ambion Inc, Austin, TX). TaqMan miRNA qRT-PCR (Applied Biosystems, Foster City, CA) were used to detect and quantify miRNA expression using the 2–ΔCt method (for details, refer to Supplementary Methods, available online).
Statistical Analyses
Results were expressed as mean ± standard deviation. Mann–Whitney U and Kruskal–Wallis analyses of variance were used to evaluate statistical differences in serum or tissue miRNA expression between unpaired groups and multiple comparison groups, respectively. The Wilcoxon test was used to compare miR-21 expression in paired serum samples obtained before surgical tumor resection and 7 days after surgical tumor resection. The Spearman correlation test was used to examine correlation between miRNA expression in serum and matched CRC tissues. Receiver operating characteristic (ROC) analysis was performed to determine the diagnostic performance of miR-21 expression levels in distinguishing patients with colorectal adenomas or cancers from the healthy control subjects. Sensitivity against 100% minus specificity was plotted at each cutoff threshold, and the area under the curve (AUC) values that reflect the probability of correctly identifying adenoma or CRC patients from control subjects were computed. The optimal cutoff thresholds for diagnosis were obtained by Youden index (30). In brief, the optimal cutoff threshold values were determined at the point on the ROC curve at which Youden’s index (sensitivity + [100% − specificity]) was maximal. By using these optimal cutoff values, sensitivity, specificity, and positive and negative predictive values were calculated.
To validate the accuracy estimates of ROC curves and optimal cutoff threshold values and to adjust for optimism bias in discriminating CRC or adenoma patients from control subjects, the bootstrap bias correction and accelerated (BCa) bootstrap methods were performed (31). In general, there are no standard recommended methods for adjusting bias. However, we selected the BCa bootstrap method because it adjusts for both bias and skewness in the bootstrap distribution of data. For this analysis, we randomly included data from the original serum samples, followed by sensitivity and specificity determination for various cutoff thresholds. This process was repeated 1000 times, and resultant mean values (95% confidence interval [CI]) for sensitivity and specificity were computed. In addition, ROC convex hull (ROCCH) curves were generated using approaches that allowed the hull segment to be viewed as being generated by a tie between the plots of sensitivity and 100% minus specificity for various cutoffs from the original data and adjusted data by BCa bootstrap methods. Thereafter, the AUCs of ROCCH curves were calculated by Trapezoidal rule. Finally, a two-sided z test was used to compare the AUCs of two ROC curves (32). A multivariable logistic regression model was used to calculate odds ratios (ORs) for age- and sex-adjusted cases associated with CRC or adenoma according to serum miRNA levels.
We estimated that 154 patients were needed to achieve 80% power to substantiate more than 20% differences in prognostic outcome, a number that was much smaller than our cohort of 200 CRC patients. Survival analyses were performed using the Kaplan–Meier method, and the differences in survival were examined using log-rank tests. ROC curves were established to discriminate the patients with or without death, and the Youden index (30) was used to determine the optimal cutoff threshold of serum or tissue miRNAs levels to predict the overall survival. Cox’s proportional hazard regression analyses were used to estimate hazard ratios (HRs) of death according to serum and tumor miRNA levels, unadjusted and adjusted for potential confounding factors for death, including age, sex, pathological differentiation, T stage, N stage, M stage, and serum carcinoembryonic antigen (CEA) levels. Assumptions of proportionality were confirmed for the Cox proportional hazards analyses by generating Kaplan−Meier survival curves (eg, high- vs low-expression groups) and by ensuring that the two curves did not intersect each other. All P values are two-sided; P less than or equal to .05 was considered statistically significant. All statistical analyses were carried out using Medcalc version 12.3.0 (Broekstraat 52, 9030; Mariakerke, Belgium).
Results
MiR-21 Expression in CRC Cell Culture Medium
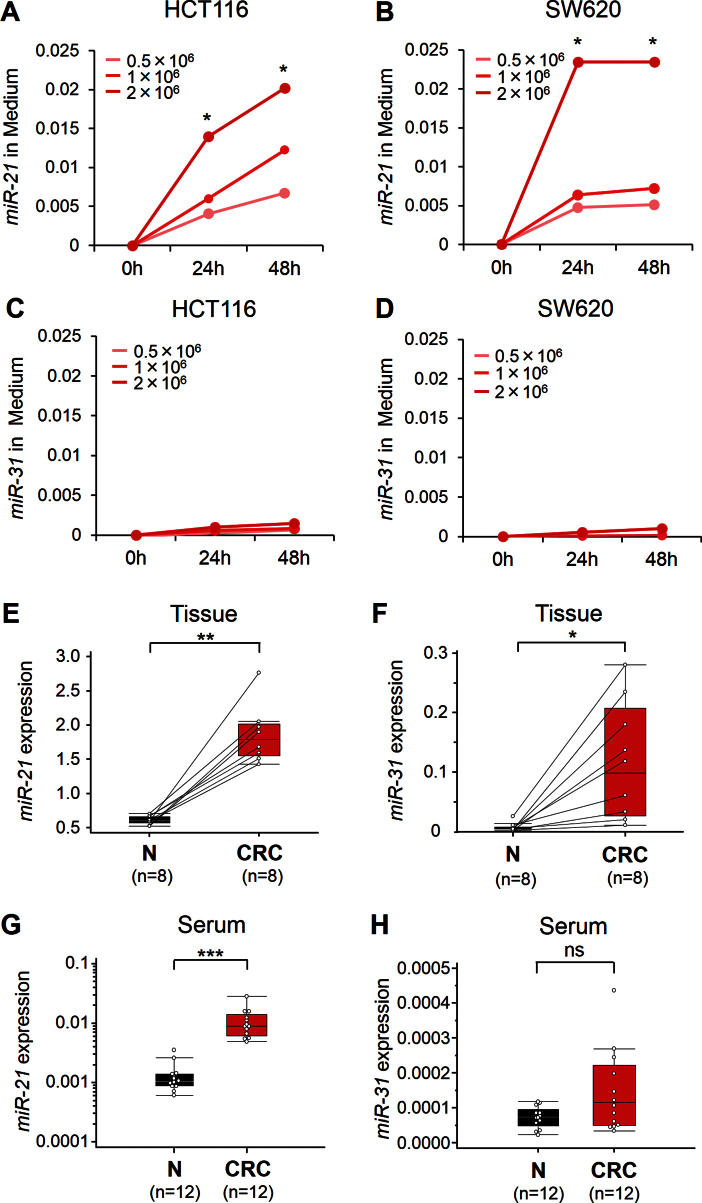
In this initial experiment, we determined whether miR-21 and miR-31 act as secretory miRNAs and are excreted into the culture media by HCT116 and SW620 CRC cell lines. We observed that miR-21 expression in the culture media from both cell lines increased with time (24 and 48 hours; P < .05) and with increasing numbers of tumor cells (P < .05) (Figure 1A, HCT116; Figure 1B, SW620). However, miR-31 expression levels did not reveal statistically significant changes in either cell line (Figure 1C, HCT116; Figure 1D, SW620), suggesting that miR-21 but not miR-31 is a secretory miRNA.
Figure 1.
Expression of miR-21 and miR-31 in culture media of colorectal cancer (CRC) cell lines (HCT116 and SW620). MiR-21 levels in media of both HCT116 A) and SW620 B) increased with increased cell counts (0.5–2×106 cells/well) and longer incubation intervals, whereas miR-31 levels did not change in either cell line C, D). The y-axis represents relative expression of miR-21 and miR-31 normalized to cel-miR-39. Initial screening for miR-21 and miR-31 expression in the screening phase, using a small subset of tissue and serum specimens from CRC patients, was done. Box plots are shown for miR-21 expression E) and miR-31 expression F) levels in primary tumor tissues (CRC) and adjacent normal mucosa (N) from eight CRC patients. Box plots are shown for serum levels of miR-21 G) and miR-31 H) in mucosa from normal control subjects (N; n = 12) and CRC patients (n = 12). Boxes represent interquartile range, and the horizontal line across each box indicates median value. The y-axis represents relative expression of miR-21 and miR-31, and data were normalized to cel-miR-39 and miR-16 expression in sera and tissue, respectively. Statistical analysis was performed using two-sided Wilcoxon and Mann–Whitney U tests. *P < .05; **P < .01; ***P < .001; ns, not statistically significant.
Tissue and Serum miR-21 Expression During Screening Phase
In the screening phase of the study, miR-21 and miR-31 expression were determined in a small set of eight CRCs and the adjacent normal mucosa. Both miR-21 and miR-31 levels were statistically significantly elevated in CRC tissues compared with adjacent normal control tissues (miR-21: P < .01; miR-31: P < .05) (Figures 1E and 1F). We next examined the feasibility of detecting the expression of circulating miR-21 and miR-31 in 24 serum samples from CRC patients (n = 12) and healthy control subjects (n = 12). We noted that miR-21 levels were statistically significantly elevated in the sera of CRC patients (P < .001) (Figure 1G), whereas no statistically significant differences were noted in serum miR-31 expression between CRC patients and control subjects (P > .05) (Figure 1H). Based on our result that only miR-21 acts a secretory miRNA, we focused the rest of our study on miR-21 for further assessment of its efficacy as a diagnostic and prognostic biomarker in patients with colorectal neoplasia.
Tissue and Serum miR-21 Expression During the Validation Phase
Patient Characteristics.
The clinicopathological and other patient characteristics are summarized in Supplementary Table 1 (available online). There were no statistically significant differences observed in age between healthy control subjects (mean = 64 years; standard deviation [SD] = 12.9 years) and patients with adenomas (mean = 66 years; SD = 9.8 years) or CRCs (mean = 67 years; SD = 7.5 years; P > .05). Likewise, there were no sex differences between different groups, with 27 men and 26 women in the control group, 30 men and 13 women in the adenoma group, and 106 men and 80 women in the CRC group (P > .05). The median follow-up time period for CRC patients was 44 months (range = 2–84 months).
Serum miR-21 Expression in Patients With Colorectal Adenomas and Cancers.
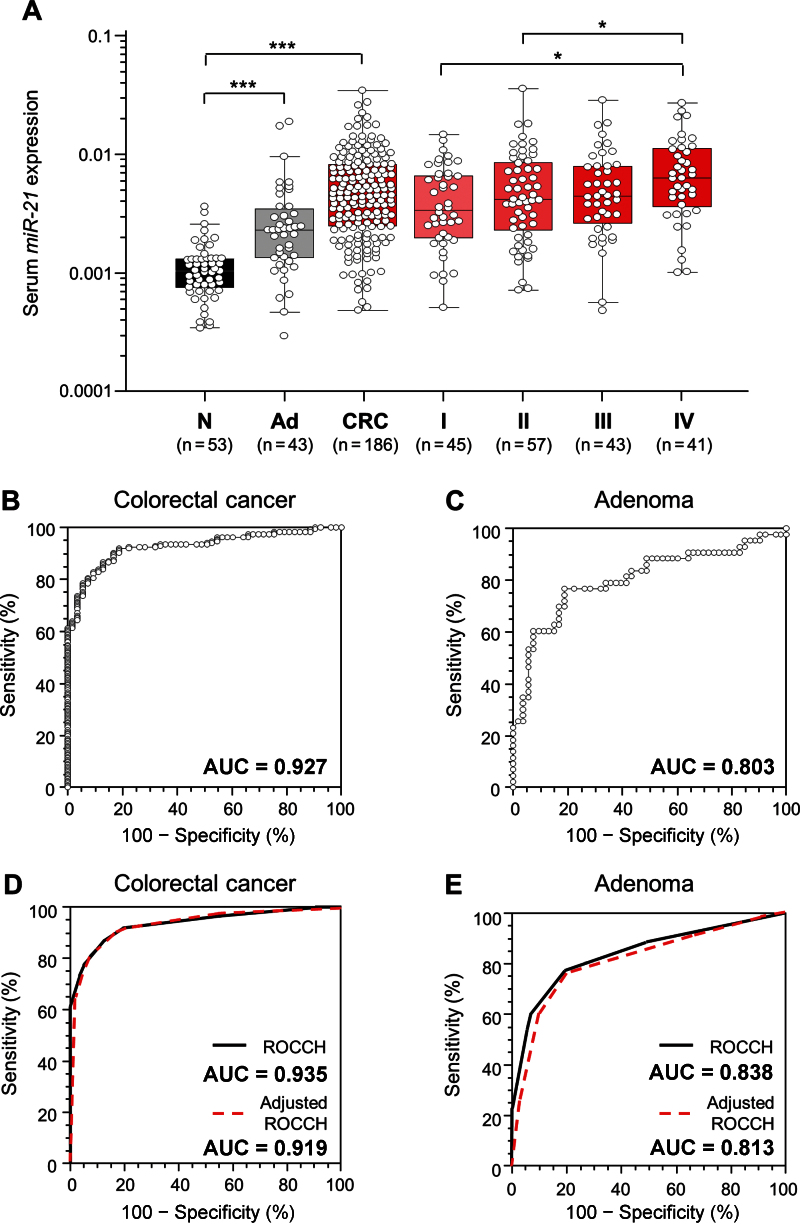
To evaluate the diagnostic potential of miR-21, a total of 282 serum samples, including those from patients with CRC (n = 186), patients with adenomatous polyps (n = 43), and normal control subjects (n = 53) were examined. In comparison with healthy control subjects, the expression levels of serum miR-21 demonstrated a stepwise increase in patients with adenomatous polyps (P < .001) (Figure 2A) and CRC (P < .001) (Figure 2A). Furthermore, when all CRC patients were segregated based upon TNM stage, the gradual increase in serum miR-21 levels was clearly discernible, with statistically significantly higher expression levels in stage IV patients compared with stage I or II patients (both P < .05) (Figure 2A).
Figure 2.
MiR-21 expression levels in serum samples (n = 282). A) Box plots represent serum miR-21 levels in healthy control subjects (N; n = 53) and patients with adenomatous polyps (Ad; n = 43) and different Tumor Node Metastasis (TNM) stages (I, II, III, and IV) of colorectal cancer (CRC) (n = 186). The y-axis (log10 scale) represents relative expression of miR-21 normalized to cel-miR-39. Boxes represent the interquartile range, and the horizontal line across each box indicates median values. Statistically significant differences were determined using the Mann–Whitney U test and Kruskal–Wallis tests. *P < .05; ***P < .001. Receiver operating characteristics (ROC) curve analysis using serum miR-21 for distinguishing patients with colorectal neoplasms from normal control subjects was performed. B) Serum miR-21 yielded an area under the curve (AUC) value of 0.927 (95% confidence interval [CI] = 0.89 to 0.96), with 82.8% sensitivity and 90.6% specificity in distinguishing CRC from normal control subjects. C) Serum miR-21 yielded AUC values of 0.803 (95% CI = 0.71 to 0.88) with 76.8% sensitivity and 81.1% specificity in discriminating adenomas from normal control subjects. ROC convex hull (ROCCH) curve analysis using raw data and bootstrap bias correction and accelerated (BCa) bootstrap bias-corrected data for distinguishing patients with colorectal neoplasms from normal control subjects was performed. D) AUC values derived from ROCCH curves of original and BCa bootstrap-corrected samples were 0.935 (95% CI = 0.812 to 0.982) and 0.919 (95% CI = 0.867 to 0.958), respectively, in distinguishing CRC patients from normal control subjects. E) AUC values derived from ROCCH curves of original and BCa bootstrap-corrected samples were 0.838 (95% CI = 0.619 to 0.964) and 0.813 (95% CI = 0.691 to 0.910), respectively, in distinguishing adenoma patients from normal control subjects.
Next, we generated ROC curves to assess the potential usefulness of serum miR-21 as a noninvasive biomarker for the early diagnosis of colorectal neoplasia. Our ROC analyses revealed that serum miR-21 levels were robust in discriminating patients with CRC from control subjects, with an AUC value of 0.927 (95% CI = 0.886 to 0.956) (Figure 2B). Using a cutoff value of 0.0019, the sensitivity, specificity, and positive and negative predictive values were 82.8%, 90.6%, 96.3%, and 60.8%, respectively, to identify a patient with CRC (Supplementary Table 2, available online). Even more important from a screening perspective, serum miR-21 levels could reliably differentiate patients with advanced adenomatous polyps from healthy control subjects, as evidenced by an AUC value of 0.803 (95% CI = 0.669 to 0.869) (Figure 2C). With a cutoff value of 0.0013, the sensitivity, specificity, and positive and negative predictive values were 76.8 % and 81.1%, 76.7%, and 81.1%, respectively (Supplementary Table 2, available online).
To validate the accuracy estimates of ROC curves and optimal cutoff values for discriminating patients with colorectal adenoma or cancer from healthy control subjects, we performed an internal validation by BCa bootstrap methods. The results obtained with the original and the BCa bootstrap samples were in good agreement (Supplementary Tables 2 and 3, available online). With serum miR-21 at 0.0013 (95% CI = 0.0009 to 0.00134), the sensitivity and specificity were 91.9% and 81.1%, respectively, to identify a patient with CRC, and the sensitivity and specificity were 81.13% and 76.74%, respectively, for a patient with colorectal adenoma using a cutoff value of 0.0013 (95% CI = 0.0010 to 0.00134) (Supplementary Table 3, available online).
ROCCH curves with BCa bootstrap bias correction data in both CRC patients vs control subjects and adenoma patients vs control subjects were very similar to those without bias correction (Figure 2, D and E). In addition, AUC values obtained from ROCCH analysis from the original and bootstrap bias-corrected samples for identifying a patient with CRC were non-statistically significant (original AUC = 0.935, 95% CI = 0.812 to 0.982; BCa corrected AUC = 0.919, 95% CI = 0.867 to 0.958; P = .80) (Supplementary Table 4, available online). In a similar manner, no statistically significant differences were observed in ROCCH-derived AUC values for discriminating between two samples with colorectal adenomas (original AUC = 0.838, 95% CI = 0.619 to 0.964; BCa bootstrap AUC = 0.813, 95% CI = 0.691 to 0.910; P = .84) (Supplementary Table 4, available online).
In addition, multivariable logistic regression analyses revealed that serum miR-21 could be used as a potential diagnostic biomarker for the identification of patients with CRC or adenomas after adjustment for patients’ age and sex (P < .001 for both) (Table 1). The odds ratio for patients with serum miR-21 was 43.3 (95% CI = 17.53 to 107.13), and the odds ratio for patients with adenomas was 6.62 (95% CI = 2.63 to 16.88) (Table 1).
Table 1.
Multivariable logistic analyses for serum miR-21 levels and various diagnostic factors in patients with colorectal cancer (CRC) and adenomas*
| Variables | OR (95% CI) | P |
|---|---|---|
| CRC patients vs control subjects | ||
| Age, >67 y vs ≤67 y† | 1.69 (0.73 to 3.92) | .22 |
| Sex, female vs male | 1.81 (0.78 to 4.22) | .17 |
| miR-21 in serum, >0.0019 vs ≤0.0019‡ | 43.3 (17.53 to 107.13) | <.001 |
| Adenoma patients vs control subjects | ||
| Age, >67 y vs ≤67 y† | 0.54 (0.21 to 1.39) | .20 |
| Sex, female vs male | 1.32 (0.53 to 3.34) | .55 |
| miR-21 in serum, >0.0013 vs ≤0.0013‡ | 6.62 (2.63 to 16.88) | <.001 |
* CI = confidence interval; OR = odds ratio.
† Median age was 67 years.
‡ The cutoff values of serum miR-21 in CRC patients vs control subjects and adenoma patients vs control subjects were derived by receive operating characteristic curves with Youden’s index.
Correlation Between Serum and Tissue miR-21 Expression in CRC Patients.
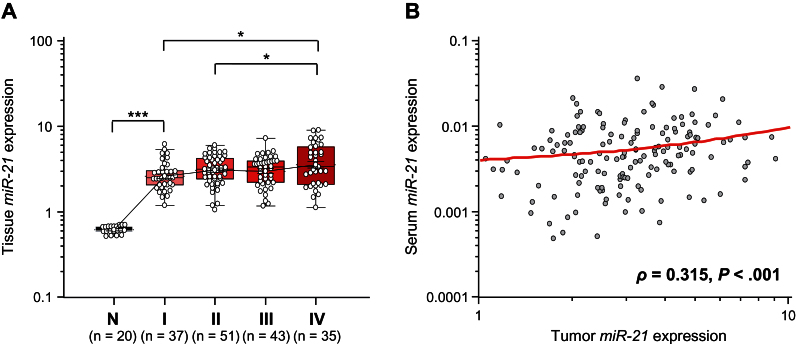
Next, we asked whether miR-21 expression in matching tumor tissues and serum samples correlated with any of the other clinicopathological data. Table 2 illustrates that miR-21 levels in both tumor tissues and matched serum were statistically significantly higher in CRC patients with larger tumor size (P = .01 and P = .004, respectively) and those with distant metastases (P = .02 and P = .01, respectively). Tissue levels of miR-21 expression associated with CRC clinical stage (stage I vs IV: P < .05; stage II vs IV: P < .05) (Figure 3A). However, miR-21 levels in both tumor tissues and matched serum samples did not correlate with specific tumor location within the colorectum.
Table 2.
Clinical significance of miR-21 expression in serum and tissue specimens from colorectacl cancer patients*
| Variables | Serum miR-21 expression, mean ± SD (n = 186) | P | Tissue miR-21 expression, mean ± SD (n = 166) | P |
|---|---|---|---|---|
| Age, y† | ||||
| ≤67 | 0.0062±0.0047 (n = 88) | .22 | 3.16±1.46 (n = 80) | .07 |
| >67 | 0.0059±0.0052 (n = 98) | 3.64±1.73 (n = 86) | ||
| Sex | ||||
| Male | 0.0059±0.0053 (n = 106) | .30 | 3.23±1.46 (n = 100) | .36 |
| Female | 0.0062±0.0044 (n = 80) | 3.52±1.72 (n = 66) | ||
| Tumor location | ||||
| Right side | 0.0073±0.0065 (n = 59) | .16 | 3.38±1.45 (n = 56) | .70 |
| Left side | 0.0058±0.0048 (n = 127) | 3.33±1.57 (n = 110) | ||
| Tumor type | ||||
| Colon cancer | 0.0063±0.0055 (n = 110) | .89 | 3.50±1.51 (n = 101) | .07 |
| Rectal cancer | 0.0062±0.0054 (n = 76) | 3.11±1.52 (n = 65) | ||
| Tumor size, cm‡ | ||||
| ≤5 | 0.0048±0.0035 (n = 91) | .004 | 3.13±1.47 (n = 78) | .01 |
| >5 | 0.0070±0.0056 (n = 95) | 3.67±1.71 (n = 88) | ||
| Serosal invasion | ||||
| Negative | 0.0051±0.0041 (n = 58) | .08 | 2.82±1.37 (n = 47) | .004 |
| Positive | 0.0064±0.0052 (n = 128) | 3.64±1.66 (n = 119) | ||
| Lymphatic invasion | ||||
| Negative | 0.0058±0.0041 (n = 46) | .83 | 3.04±1.52 (n = 39) | .02 |
| Positive | 0.0061±0.0051 (n = 140) | 3.53±1.64 (n = 127) | ||
| Venous invasion | ||||
| Negative | 0.0058±0.0045 (n = 108) | .53 | 3.36±1.53 (n = 91) | .60 |
| Positive | 0.0064±0.0054 (n = 78) | 3.47±1.73 (n = 75) | ||
| Lymph node metastasis | ||||
| Negative | 0.0055±0.0042 (n = 106) | .12 | 3.24±1.48 (n = 92) | .11 |
| Positive | 0.0068±0.0057 (n = 80) | 3.62±1.76 (n = 74) | ||
| Distant metastasis | ||||
| Negative | 0.0055±0.0045 (n = 145) | .01 | 3.21±1.37 (n = 131) | .02 |
| Positive | 0.0078±0.0060 (n = 41) | 4.18±2.19 (n = 35) | ||
* SD = standard deviation.
† The median age was 67 years.
‡ The median tumor size was 5 cm.
Figure 3.
Validation of miR-21 expression in matched tissue samples (n = 186). A) Box plots illustrating tissue miR-21 levels in different Tumor Node Metastasis (TNM) stages (I,II,III, and IV) of colorectal cancers (CRCs) (n = 166) and adjacent normal mucosa (N; n = 20). The y-axis (log10 scale) represents relative expression of miR-21 normalized to miR-16 in tissue samples. Boxes represent the interquartile range, and the horizontal line across each box indicates the median value. Statistically significant differences were determined using the Mann–Whitney U test and Kruskal–Wallis tests. *P < .05: ***P < .001. B) Scatter plots showing the correlation between relative expression of miR-21 levels in serum (y-axis: log10 scale) and matched tumor tissues (x-axis: log10 scale) obtained from 154 CRC patients. A positive correlation was found by Spearman correlation (ρ = 0.315; 95% CI = 0.17 to 0.45; P < .001).
To further enhance the specificity of our assay and validate that circulating miR-21 expression accurately reflects concentrations found in CRC tissues, we determined the relationship between miR-21 levels in primary CRC tissues and matched serum from individual CRC patients. Interestingly, we observed a statistically significantly positive correlation between miR-21 expression in primary CRC lesions and matched serum samples from these patients (ρ = 0.315; 95% CI = 0.17 to 0.45; P < .001; Spearman’s correlation analysis) (Figure 3B).
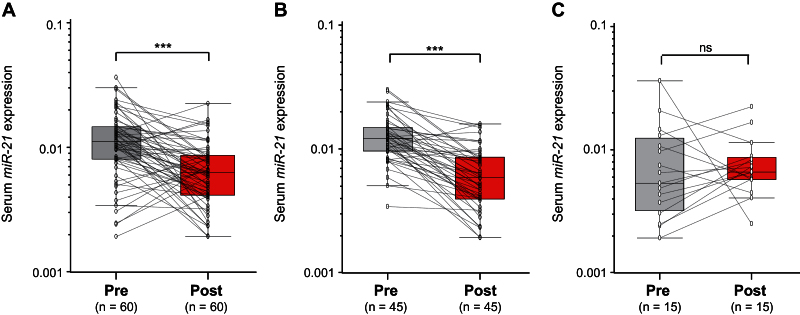
Thereafter, we analyzed paired pre- and postoperative serum samples in a subset of 60 CRC patients who underwent surgical resection of their tumors. In the 60 CRC patients, 45 underwent potentially curative resection, whereas 15 had multiple hepatic metastases and underwent primary resection to prevent bleeding and bowel obstruction (noncurative resection). It was interesting to note that serum levels of miR-21 statistically significantly plummeted after surgery in the same subset of patients (P < .001) (Figure 4A). Furthermore, when data were analyzed based on potentially curative vs noncurative surgeries, postoperative reductions in serum miR-21 levels occurred exclusively among patients with potentially curative surgeries (P < .001) (Figure 4B). Contrariwise, no statistically significant differences were observed in miR-21 levels before or after surgery in patients with noncurative resections (P = .72) (Figure 4C). Collectively, these data underscore the importance of serum miR-21 expression as a highly specific biomarker for the diagnosis of colorectal neoplasia.
Figure 4.
Alterations in serum miR-21 expression levels in patients with colorectal cancer (CRC) before surgery (Pre) and 7 days after postsurgical removal of primary tumors (Post). A) Comparison of serum miR-21 levels from all CRC patients (n = 60). B) Comparison of serum miR-21 levels in 45 CRC patients who underwent potentially curative surgeries. C) Comparison of serum miR-21 levels in 15 CRC patients who underwent noncurative surgeries. The y-axis (log10 scale) represents relative expression of miR-21 normalized to cel-miR-39. Boxes represent the interquartile range, and the horizontal line across each box indicates the median value. Statistically significant differences were determined using the Wilcoxon test. ***P < .001; ns, not statistically significant.
Association of Serum miR-21 Expression With Survival in Patients With CRC.
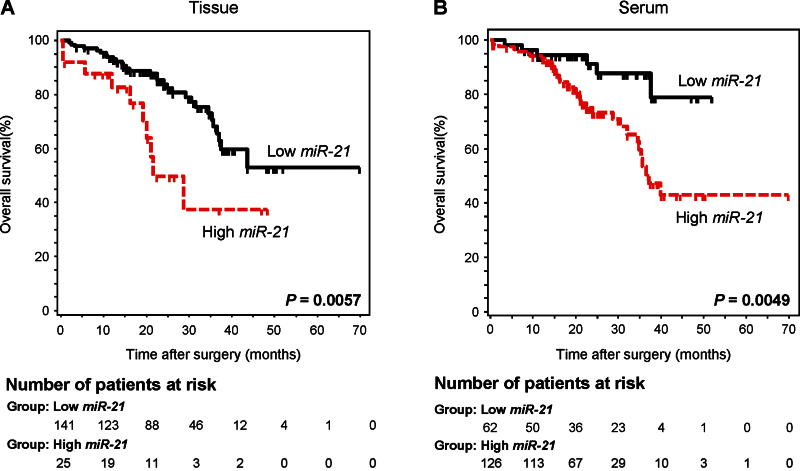
To further evaluate whether serum miR-21 levels can serve as a predictor of patient outcome, we performed Kaplan–Meier survival analysis. As anticipated, patients with higher levels of miR-21 in the tumor tissues had statistically significantly worse overall survival (P = .006; log-rank test) (Figure 5A). Moreover, a similar pattern of increased miR-21 concentrations associated with statistically significantly decreased overall survival was observed when the analysis used serum miR-21 expression levels (P = .005; log-rank test) (Figure 5B).
Figure 5.
Kaplan–Meier survival analysis in colorectal cancer (CRC) patients based upon miR-21 expression in primary tumors and matched serum samples. A) The overall survival rate in CRC patients with high miR-21 expression in tumor tissue (n = 25) was statistically significantly lower than that for those with low miR-21 expression (n = 141) (>3.7 vs ≤3.7; P = .006; log-rank test). B) The overall survival rate in CRC patients with high serum miR-21 expression (n = 126) was statistically significantly lower than that for those with low serum miR-21 expression (n = 62) (>0.003 vs ≤.003; P = .005; log-rank test). Cutoff values for miR-21 expression in serum and primary tumor tissues were determined from the receiver operating characteristic curves by using Youden’s index. The number of patients at risk are shown below each curve at various timepoints.
Furthermore, Cox proportional hazard regression analyses revealed that in the univariate analysis, poor prognosis in CRC patients was associated with high levels of miR-21 in both tumor and serum (P = .01 and P = .003, respectively), high levels of carcinoembryonic antigen (CEA > 5ng/mL; P = .001), high T stage (T3/T4; P = .002), lymph node metastasis (P < .001), poorly differentiated tumors (P = .04), and distant metastasis (P < .001) (Table 3). More important, multivariable analysis demonstrated that high levels of serum miR-21, but not high concentrations of miR-21 in tumor tissues or high CEA levels, served as an independent prognostic marker for indicating overall survival in CRC patients (HR = 4.12; 95% CI = 1.10 to 15.4; P = .03).
Table 3.
Univariate and multivariable analyses of factors predictive of poor overall survival in colorectal cancer patients*
| Variables | Univariate† | Multivariable‡ | ||
|---|---|---|---|---|
| HR (95% CI) | P | HR (95% CI) | P | |
| Age#, >67 y vs ≤67 y | 0.72 (0.43 to 1.37) | .37 | 1.23 (0.57 to 2.67) | .59 |
| Sex, female vs male | 1.02 (0.56 to 1.86) | .92 | 2.07 (0.86 to 4.96) | .10 |
| Pathological T, T3/4 vs T1/2 | 8.97 (2.19 to 36.70) | .002 | 2.38 (0.30 to 29.77) | .31 |
| Pathology, poor diff. vs diff. | 2.26 (1.05 to 4.84) | .04 | 2.39 (0.57 to 9.92) | .23 |
| Lymph node metastasis, yes vs no | 17.1 (6.18 to 47.80) | <.001 | 6.83 (1.69 to 28.36) | .008 |
| Distant metastasis, yes vs no | 35.6 (14.40 to 86.60) | <.001 | 21.7 (5.92 to 79.99) | <.001 |
| CEA§, >5ng/mL vs ≤5ng/mL | 4.84 (2.15 to 10.89) | .001 | 1.03 (0.33 to 3.22) | .94 |
| miR-21 in tissue¶, >3.7 vs ≤3.7 | 2.66 (1.29 to 5.45) | .01 | 0.59 (0.21 to 1.63) | .31 |
| miR-21 in serum¶, >0.0031 vs ≤0.0031 | 3.25 (1.36 to 7.73) | .003 | 4.12 (1.10 to 15.40) | .03 |
* CEA = carcinoembryonic antigen; CI = confidence interval; diff. = differentiation; HR = hazard ratio.
# The median age was 67 years.
§ Cutoff value for CEA is 5ng/mL (as per American Association of Clinical Oncology recommendations).
¶ Cutoff values of miR-21 in tissue and serum are derived from receiver operating characteristic curve with Youden’s index.
† Univariate analysis was performed using clinical data available from 200 patients, serum miR-21 data from 186 patients, and tissue miR-21 data from 166 colorectal cancer patients, respectively..
‡ Multivariable analysis was performed using data from 153 colorectal cancer patients from whom matched matching data were available for all clinico-pathological factors as well as serum and tissue miR-21 expression results.
Discussion
Most approaches aimed at the identification of diagnostic or prognostic miRNAs in the circulation have focused on miRNAs that are frequently overexpressed (12,15,33). However, relatively few miRNAs that are abundantly expressed in cancer cells have been detectable in the circulation (34), and nearly 30% of the released miRNAs do not mirror the cellular expression profiles originating in specific cancer cells (35).These reports underscore the need for more careful evaluation of criteria depending upon whether these miRNAs are retained in the cell or are released into the systemic circulation to provide a robust substrate for a noninvasive diagnostic strategy.
Data from our initial screening phase revealed two important observations. First, miR-21 levels in cell culture medium increased statistically significantly in a time- and cell number–dependent manner, establishing the secretory nature of this miRNA for development as a noninvasive biomarker of the early detection of colorectal neoplasia. Second, a statistically significant correlation was observed in miR-21 expression between matching serum and tissues from a small subset of CRC patients, validating the specificity of miR-21 expression in the circulation. Increased miR-21 expression was thereafter successfully validated in a large, independent set of matching serum and tissue samples. Although our observation of increased miR-21 expression in CRC tissues is consistent with previous reports (19–22), our results are the first to demonstrate that high levels of miR-21 in both primary CRC tissues and matched serum samples are associated with large tumor size, distant metastasis, and advanced TNM stage. Another interesting feature of our study was the existence of a statistically significant correlation between miR-21 expression in primary lesions and those in sera. The fact that the circulating miR-21 in serum of CRC patients is likely produced by the CRCs was further strengthened by our observation of a statistically significant drop in serum miR-21 expression in postoperative serum vis-à-vis preoperative samples after potentially curative surgery in patients with CRC.
This study is the first to demonstrate the potential role of serum miR-21 in the early detection of colorectal neoplasia. This is supported by the markedly high AUC values of 0.919 derived from comparisons between CRC patients and healthy control subjects (sensitivity = 91.9%; specificity = 81.1%). Even more important from a CRC screening perspective, serum miR-21 expression levels demonstrated AUC values of 0.813, associated with 81.1% sensitivity and 76.7% specificity, for discerning patients with adenomatous polyps from healthy control subjects, an observation that is novel and perhaps represents the highest levels of sensitivity and specificity for any serum biomarker aimed at the identification of colorectal adenomas in a noninvasive manner. In addition, the odds ratio for case subjects with high levels of miR-21 expression being associated with CRC was 43.3 (95% CI = 17.53 to 107.13), which is an extraordinary performance for a noninvasive biomarker compared with recently reported data for other circulating miRNAs (OR = 18.7) (11) or a positive first guaiac FOBT (OR = 7.6) (36). Another important feature of our study was that we observed detectable levels of serum miR-21 in premalignant adenomas, even though these lesions are not fully vascularized. Thus, our study highlights that miR-21 expression can be exploited not only as a promising noninvasive biomarker for early detection of CRC but also for the identification of clinically meaningful adenomas—a critical target lesion for any CRC screening strategy.
Another interesting finding of our study is that miR-21 expression in serum also serves as a prognostic biomarker for CRC. Our results are consistent with previous studies that have demonstrated the potential of tissue miR-21 as a prognostic and predictive marker for response to chemotherapy in CRC (20,22,37). However, our findings that high levels of serum (rather than tissue) miR-21 indicate a poor prognosis in patients with CRC is an important step forward in the identification of a noninvasive biomarker for this disease. Furthermore, the multivariable Cox proportional hazards model illustrated that high expression of serum miR-21 was an independent prognostic variable, whereas the prognostic values of miR-21 expression in tumors and CEA levels were statistically significantly compromised by other clinical factors. Therefore, serum levels of miR-21 might not only diagnose neoplasia but also help predict metastases or tumor recurrence with higher accuracy than miR-21 expression in the tumor tissue.
This study clearly highlights that the relative expression of serum miR-21 was statistically significantly different between case patients (for both adenomas and cancers) and control subjects. Unlike tissue or cellular miRNAs, at present there is no consensus housekeeping miRNA for normalizing the expression of circulating miRNAs (38,39); therefore, measurement of relative expression levels of circulating miRNAs has been a common approach in published studies. In spite of this limitation, we circumvented this concern using two approaches. First, we used the same amount of starting serum (200 μL) from each patient for every quantitation. Second, to further ensure the technical aspects of our assay, including variability in serum RNA extraction and polymerase chain reaction amplification efficiencies, we used the recently described approach for normalization of experimental miRNA data using spiked-in synthetic, nonhuman mature miRNA from Caenorhabditis elegans (40). Although our method of quantifying relative expression [2–ΔCt; ΔCt = Ct (miRNA of interest) − Ct (cel-miR-39 [this miRNA used as a normalizer for sample to sample variation]) of serum miRNAs was quite robust, absolute quantitation of serum miR-21 expression levels may further improve the translation of these data into a clinically viable diagnostic test for the early detection of colorectal neoplasia in the immediate future.
Although our current assay may become a promising screening tool for colorectal neoplasia, we acknowledge two potential limitations of using miR-21 as a single biomarker for the early detection of colorectal neoplasia. First, circulating expression of miR-21 has been described in many solid cancers besides CRC, including glioblastoma, pancreatic cancer, and breast cancer, underscoring the need for being vigilant about organ and disease specificity while investigating miR-21 as solitary biomarker for CRC. As a consequence, it might be challenging to differentiate whether circulating miR-21 expression is specifically associated with CRC itself or if this is a common phenomenon that manifests during progression of any cancer as a result of perturbations in the host immune response (41). In this study, we have addressed this issue by demonstrating that a statistically significant correlation existed between tissue and serum levels of miR-21 across all stages of colorectal neoplasia and that miR-21 levels fell after surgical removal of the colorectal tumors in a subset of patients, highlighting the specificity of miR-21 as a specific biomarker for colorectal neoplasia. Second, although it is highly unlikely to have a substantial impact, use of serum miR-21 expression levels as a diagnostic and prognostic biomarker must be validated in diverse ethnic populations because the clinical materials analyzed in this study were solely from patients of Japanese origin.
In conclusion, our results provide compelling evidence for the potential usefulness of serum miR-21 as a noninvasive screening and prognostic tool in patients with colorectal neoplasia, a concept that can be incorporated into routine clinical practice in the not-so-distant future pending validation in large-scale prospective trials.
Funding
This work was supported by grants R01 CA72851 and CA129286 from the National Cancer Institute, National Institutes of Health, and funds from the Baylor Research Institute to CRB and AG. This study was also supported by a pilot project grant from the Charles A. Sammons Cancer Center, Baylor University Medical Center, to AG.
Supplementary Material
The study sponsor had no role in the design of the study; the collection, analysis, and interpretation of the data; the writing of the manuscript; and the decision to submit the manuscript for publication. The author contributions are as follows: study concept and design (YT, AG); provision of samples (TN, KT, YI, MK); acquisition of data (YT, AG); analysis and interpretation of data (YT, MT, KH, AG); statistical analysis (YT, MT, KH, AG); drafting of the manuscript (YT, MT, CRB, AG).
References
- 1. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2012. CA Cancer J Clin. 2012;62(1):10–29 [DOI] [PubMed] [Google Scholar]
- 2. Weizman AV, Nguyen GC. Colon cancer screening in 2010: an up-date. Minerva Gastroenterol Dietol. 2010;56(2):181–188 [PubMed] [Google Scholar]
- 3. Hewitson P, Glasziou P, Watson E, Towler B, Irwig L. Cochrane systematic review of colorectal cancer screening using the fecal occult blood test (hemoccult): an update. Am J Gastroenterol. 2008;103(6):1541–1549 [DOI] [PubMed] [Google Scholar]
- 4. Walsh JM, Terdiman JP. Colorectal cancer screening: scientific review. JAMA. 2003;289(10):1288–1296 [DOI] [PubMed] [Google Scholar]
- 5. Baxter NN, Warren JL, Barrett MJ, Stukel TA, Doria-Rose VP. Association between colonoscopy and colorectal cancer mortality in a US cohort according to site of cancer and colonoscopist specialty. J Clin Oncol. 2012;30(21):2664–2669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Halama N, Herrmann C, Jaeger D, Herrmann T. Treatment with cetuximab, bevacizumab and irinotecan in heavily pretreated patients with metastasized colorectal cancer. Anticancer Res. 2008;28(6B):4111–4115 [PubMed] [Google Scholar]
- 7. Meropol NJ, Schulman KA. Cost of cancer care: issues and implications. J Clin Oncol. 2007;25(2):180–186 [DOI] [PubMed] [Google Scholar]
- 8. Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post- transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9(2):102–114 [DOI] [PubMed] [Google Scholar]
- 9. van Kouwenhove M, Kedde M, Agami R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat Rev Cancer. 2011;11(9):644–656 [DOI] [PubMed] [Google Scholar]
- 10. Pu XX, Huang GL, Guo HQ, et al. Circulating miR-221 directly amplified from plasma is a potential diagnostic and prognostic marker of colorectal cancer and is correlated with p53 expression. J Gastroenterol Hepatol. 2010;25(10):1674–1680 [DOI] [PubMed] [Google Scholar]
- 11. Ng EK, Chong WW, Jin H, et al. Differential expression of microRNAs in plasma of patients with colorectal cancer: a potential marker for colorectal cancer screening. Gut. 2009;58(10):1375–1381 [DOI] [PubMed] [Google Scholar]
- 12. Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–126 [DOI] [PubMed] [Google Scholar]
- 13. Cheng H, Zhang L, Cogdell DE, et al. Circulating plasma MiR-141 is a novel biomarker for metastatic colon cancer and predicts poor prognosis. PLoS One. 2010;6(3):e17745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wang LG, Gu J. Serum microRNA-29a is a promising novel marker for early detection of colorectal liver metastasis. Cancer Epidemiol. 2012;36(1)e61–e67 [DOI] [PubMed] [Google Scholar]
- 15. Mitchell PS, Parkin RK, Kroh EM, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–10518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1). J Biol Chem. 2007;282(19):14328–14336 [DOI] [PubMed] [Google Scholar]
- 18. Asangani IA, Rasheed SA, Nikolova DA, et al. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–2136 [DOI] [PubMed] [Google Scholar]
- 19. Slaby O, Svoboda M, Fabian P, et al. Altered expression of miR-21, miR-31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5–6): 397–402 [DOI] [PubMed] [Google Scholar]
- 20. Shibuya H, Iinuma H, Shimada R, Horiuchi A, Watanabe T. Clinicopathological and prognostic value of microRNA-21 and microRNA-155 in colorectal cancer. Oncology. 2010;79(3–4):313–320 [DOI] [PubMed] [Google Scholar]
- 21. Chang KH, Miller N, Kheirelseid EA, et al. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol. 2011;37(7):597–603 [DOI] [PubMed] [Google Scholar]
- 22. Schetter AJ, Leung SY, Sohn JJ, et al. MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA. 2008;299(4):425–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kanaan Z, Rai SN, Eichenberger MR, et al. Plasma MiR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3): 544–551 [DOI] [PubMed] [Google Scholar]
- 24. Wang CJ, Zhou ZG, Wang L, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26(1):27–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oberg AL, French AJ, Sarver AL, et al. miRNA expression in colon polyps provides evidence for a multihit model of colon cancer. PLoS One. 2011;6(6):e20465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Fassan M, Pizzi M, Giacomelli L, et al. PDCD4 nuclear loss inversely correlates with miR-21 levels in colon carcinogenesis. Virchows Arch. 2011;458(4):413–419 [DOI] [PubMed] [Google Scholar]
- 27. Cekaite L, Rantala JK, Bruun J, et al. MiR-9, -31, and -182 deregulation promote proliferation and tumor cell survival in colon cancer. Neoplasia. 2012;14(9):868–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cottonham CL, Kaneko S, Xu L. miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J Biol Chem. 2008;285(46):35293–35302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Edge SB, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A, eds. AJCC Cancer Staging Manual. 7th ed. New York: Springer; 2010. [Google Scholar]
- 30. Ruopp MD, Perkins NJ, Whitcomb BW, Schisterman EF. Youden index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom J. 2008;50(3):419–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Efron B. Better bootstrap confidence intervals. J Am Stat Associ. 1987;82 (397): 171–185 [Google Scholar]
- 32. Hanley JA, McNeil BJ. A method of comparing the areas under receiver operating characteristic curves derived from the same cases. Radiology. 1983;148(3):839–843 [DOI] [PubMed] [Google Scholar]
- 33. Skog J, Wurdinger T, van Rijn S, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008;10(12):1470–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chen TS, Lai RC, Lee MM, Choo AB, Lee CN, Lim SK. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010;38(1):215–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pigati L, Yaddanapudi SC, Iyengar R, et al. Selective release of microRNA species from normal and malignant mammary epithelial cells. PLoS One. 2010;5(10):e13515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fraser CG, Matthew CM, Mowat NA, Wilson JA, Carey FA, Steele RJ. Immunochemical testing of individuals positive for guaiac faecal occult blood test in a screening programme for colorectal cancer: an observational study. Lancet Oncol. 2006;7(2):127–131 [DOI] [PubMed] [Google Scholar]
- 37. Kulda V, Pesta M, Topolcan O, et al. Relevance of miR-21 and miR-143 expression in tissue samples of colorectal carcinoma and its liver metastases. Cancer Genet Cytogenet. 2010;200(2):154–160 [DOI] [PubMed] [Google Scholar]
- 38. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zen K, Zhang CY. Circulating microRNAs: a novel class of biomarkers to diagnose and monitor human cancers. Med Res Rev. 2012;32(2):326–348 [DOI] [PubMed] [Google Scholar]
- 40. Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods. 2010;50(4):298–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiao J, Luo X, Lin H, et al. MicroRNA miR-133 represses HERG K+ channel expression contributing to QT prolongation in diabetic hearts. J Biol Chem. 2011;286(32):28656 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.