Abstract
As the limitations of antiretroviral drug therapy, such as toxicity and resistance, become evident, interest in alternative therapeutic approaches for human immunodeficiency virus (HIV) infection is growing. We developed the first gene therapeutic strategy targeting entry of a broad range of HIV type 1 (HIV-1) variants. Infection was inhibited at the level of membrane fusion by retroviral expression of a membrane-anchored peptide derived from the second heptad repeat of the HIV-1 gp41 transmembrane glycoprotein. To achieve maximal expression and antiviral activity, the peptide itself, the scaffold for presentation of the peptide on the cell surface, and the retroviral vector backbone were optimized. This optimized construct effectively inhibited virus replication in cell lines and primary blood lymphocytes. The membrane-anchored C-peptide was also shown to bind to free gp41 N peptides, suggesting that membrane-anchored antiviral C peptides have a mode of action similar to that of free gp41 C peptides. Preclinical toxicity and efficacy studies of this antiviral vector have been completed, and clinical trials are in preparation.
One priority in AIDS research is to develop novel, effective, and less-toxic therapies. Although gene therapy is an interesting option, initial clinical trials have been disappointing (15, 28). One major problem for gene therapy of human immunodeficiency virus (HIV) infection has been that in the patient only a minor fraction of the >1011 target cells for HIV can be protected by an antiviral gene. Overall reduction of viral load and therapeutic success therefore depend critically on in vivo selection of the gene-protected cells. We argue that in this situation genes that block HIV prior to integration of the provirus have an advantage over late inhibitory genes that suppress production of viral RNA and protein (transdominant Rev and Tat, RRE decoys). Although early inhibitory genes are expected to lead to an accumulation of noninfected gene-protected cells, late inhibitors allow the provirus to integrate and are thus expected to mediate selection of cells containing a suppressed HIV provirus. This accumulation of HIV-infected cells counteracts the antiviral activity of the therapeutic gene. Mathematical models predict that late-acting antiviral genes only lead to an overall reduction of viral load if they have a very high inhibitory activity (D. Von Laer, S. Hasselmann, and K. Hasselmann, unpublished data). Surprisingly, very few early inhibitors for gene therapy of HIV-1 infection are available. These either have a low antiviral activity, such as the intracellular single-chain variable fragments against reverse transcriptase or integrase (17) or only act on R5-tropic virus by inhibiting CCR5 expression (7, 24). We have therefore concentrated on the development of an effective and broadly active early inhibitory gene.
The C peptides (C36 = DP-178 = T-20 and C34), which are derived from the C-terminal heptad repeat of the HIV-1 transmembrane glycoprotein gp41, inhibit HIV type 1 (HIV-1) entry at the level of membrane fusion by interacting with the trimeric coiled coil structure formed by the N-terminal heptad repeat of gp41 (3, 26, 27). gp41 mediates fusion of the viral and cellular membranes and binding of C peptides is thought to lock gp41 in a fusion-incompetent state. One C peptide (T-20 [enfurvirtide]) was shown to be highly effective in clinical trials. However, the lack of oral bioavailability, the high production costs, and the rapid emergence of resistant viruses still hinder broad application (14, 25). To overcome these problems, the C36 peptide was engineered for expression on the cell membrane, leading to a high local concentration of peptide at the site of action (12). Surface expression was achieved by fusing an N-terminal signal peptide and a C-terminal scaffold consisting of a hinge and a membrane anchor to the antiviral peptide C36. This membrane-anchored peptide was expressed from a retroviral vector (M87) and had good antiviral activity in cell lines. Further testing in our laboratory, however, failed to show reproducible inhibition in primary lymphocytes (unpublished data). In the present study, we now have developed a retroviral vector expressing a membrane-anchored antiviral peptide that was highly effective also in primary cells and had minimal potential immunogenicity and no detectable toxicity. To our knowledge, this is the first antiviral gene that effectively inhibits entry of a broad range of different HIV-1 isolates in cell lines and primary cells, thus representing an interesting candidate for clinical applications.
MATERIALS AND METHODS
Cell lines, primary lymphocyte culture, and virus isolates.
PM-1 cells were kindly provided by Buchacher and coworkers (19), and Phoenix packaging cells was provided by G. P. Nolan (9). Peripheral blood mononuclear cells (PBMC) were obtained from healthy donors by separation through a Ficoll gradient (PAA, Cölbe, Germany) and stimulated with OKT3 (10 ng/ml) and interleukin-2 (100 U/ml) as described previously (16). All virus isolates were grown on PBMC. Their identity was verified by analysis of coreceptor usage and sequencing of the env. The virus titers were determined by a p24 focus-forming assay on U87 cells as described previously (4).
Generation of retroviral vectors.
The vectors MP71EGFP and M87-Ineo have been described previously (12, 23). The vector M87/C46-Ineo was created by an overlapping PCR: the 5′ part of the transgene was amplified with the primers M87-F1241 (5′CCTACATCGTGACCTGGGAAG-3′) and M87N+1R (5′-ATTGTTAATTTCTCTGTCCCACTCCATCCAAGATCTGGCACCTCCAAGGG -3′). The remaining 3′ section was amplified with the primers M87N+2F (5′-TCTTGGATGGAGTGGGACAGAGAAATTAACAA TTACACTAGCTTAATACACTCC-3′) and M87-R1911 (5′-GTGCGTCTAAGTTACGGGAAG-3′). The PCR products were combined and extended to full-length with the primers SqM87-f2 (5′-TTATCCAGCCCTCACTCCTTC-3′) and M87-R1911. The product was then cloned into the retroviral vector M87-Ineo by using BstXI and SalI.
The synthetic gene encoding for the optimized membrane-anchored peptide was obtained from Geneart (Regensburg, Germany) and amplified by PCR with the primers M171-F632-BstXI (5′-AATTACCACCGCGGTGGGACTCACTATAGGGCGAATTG-3′) and M171-R1116-SalI (5′-AATTAGTCGACGCTGGAGCTCTCCGGATCAG-3′). The product was introduced into the retroviral vector M87-Ineo by using BstXI and SalI, generating the vector M87o-Ineo. The vector was digested with BspEI, blunted, and HindIII digested, and the wPRE cassette was introduced, substituting it for the IRES-neo cassette. One ATG in the 5′ region of the wPRE was removed by site-directed mutagenesis with the QuikChange kit (Stratagene, Amsterdam, The Netherlands) and the primers M176-S-PRE-qcF1910 (5′-CGTCGACCTGCAGGCTAGCAAGCTGGGCTGCAG-3′) and M176-S-PRE-qcR1910 (5′-CTGCAGCCCAGCTTGCTAGCCTGCAGGTCGACG-3′). This ATG starts a 166-amino-acid open reading frame. The DNA fragment with the open reading frame encoding the membrane-anchored peptide and the wPRE was obtained by AgeI/HindIII digestion and then inserted into the optimized retroviral vector MP71 (11) to obtain M87o. The RRE decoy was added by HindIII digestion of M87o and ligated with the hybridized primers RRE-HindIII-F (5′-AGCTTCACTATGGGCGCAGTGTCATTGACGCTGACGGTACAGGCCA-3′) and RRE-HindIII-R (5′-AGCTCTGGCCTGTACCGTCAGCGTCAATGACACTGCGCCCATAGTGA-3′). The IRES-neo cassette was obtained from M87-Ineo with HindIII and cloned into the HindIII site of M87o to generate M87o-Ineo.
Introduction of T-20-resistant mutations in HIV-1HXB2 env.
Mutations within the HIVHXB2 env were introduced by site-directed mutagenesis with the QuikChange mutagenesis kit (Stratagene) and the primers M104-SIM-F (5′-GCCAGACAATTATTGTCTAGTATAATGCAGCAGCAGAACAATTTG-3′) and M104-SIM-R (5′-CAAATTGTTCTGCTGCTGCATTATACTAGACAATAATTGTCTGGC-3′) for HxB2resI-Env and M104-DTV-F (5′-GCCAGACAATTATTGTCTGATACAGTGCAGCAGCAGAACAATTTG-3′) and M104-DTV-R (5′-CAAATTGTTCTGCTGCTGCACTGTATCAGACAATAATTGTCTGGC-3′) for HxB2resII-Env into the plasmid pSG-HxB2 (10).
Generation of cell lines expressing membrane-anchored peptides.
Retroviral vectors were packaged by transfection of Phoenix packaging cells, and supernatants were used for transduction at low multiplicities of infection (MOIs) to omit multiple vector integrations. PBMC were transduced on days 3, 4, and 5 of culture as described elsewhere (16). PM-1 cells were transduced once.
PM-1 cells transduced with vectors containing a neomycin resistance gene (neoR) were selected with G418 for 10 days with 1 mg of G418/ml. PM-1 cells transduced with M87o or M87oRRE (not containing neoR) were stained with the human monoclonal antibody 2F5 directed against a motif in the C peptide of gp41 (kindly provided by H. Katinger) as described previously and sorted on a fluorescence-activated cell sorter (FACScalibur; BD, Heidelberg, Germany) (12). Staining with 2F5 was shown to be specific by competition assays with the free C36 peptide (data not shown).
Inhibition of C36-resistant HIV-1 strains.
After G418 selection, PM-1/MP1neo, PM-1/M87-Ineo, and M87/C46-Ineo were used for single-round infection with a lentiviral vector expressing enhanced green fluorescent protein (EGFP) in the position of nef. This vector was pseudotyped with the HIV Env proteins HxB2, HxB2resI, or HxB2resII and as a control with the vesicular stomatitis virus G protein. The lentiviral vector supernatants were produced by transient transfection of 293T as described previously (10). The titers of the different lentiviral pseudotyped vectors on PM-1 were 1.5 × 103 for the vesicular stomatitis virus G pseudotype, 7.4 × 103 for the HxB2 pseudotype, 3 × 102 for the HxB2mtI and 2 × 102 for the HxB2mtII. The transduction efficacy of the lentiviral vector in the different cell lines was monitored by flow cytometric analysis of EGFP expression and given relative to the efficacy in the control cell line PM-1/MP1neo. The vector supernatants were used at a dilution that gave between 1 and 10% marking in the control cells, since <10% marking a linear relation between vector input and GFP positivity was seen (data not shown). A total of 105 cells per sample were analyzed by fluorescence-activated cell sorting (FACS) to allow accurate analysis of marking even in the inhibited cultures with fewer than 1% GFP-positive cells.
Inhibition of different HIV-1 strains by membrane-anchored peptides.
The different PM-1 cell lines and PBMC cultures were infected with HIV-1 strains at MOIs of 0.01 to 0.0001. HIV p24 antigen was measured in the culture supernatants collected at different time points by enzyme-linked immunosorbent assay (ELISA) (Innogenetics, Heiden, Germany).
Cell-cell fusion assay.
Cell-cell fusion was performed essentially as described previously (20). In brief, ca. 5 × 106 PM-1 target cells were labeled with a 30 μM concentration of the cytoplasmic marker 7-amino-4-chloromethylcoumarin (CMAC). As effector cells, the cell line 293T was cotransfected with the plasmid p202 expressing gp160 of the T-tropic HxB2 strain of HIV-1 and the rev-plasmid pcRev. A total of 5 × 106 cells/ml of the transfected cells were labeled with 1.3 μM calcein AM. Next, 105 effector cells and 105 fluorescence-labeled target cells were mixed in HEPES-buffered Dulbecco modified Eagle medium (pH 7.2) supplemented with 1 mg of bovine serum albumin/ml, followed by incubation for the indicated periods at 37°C in 8-chamber glass slides. Fusion was measured by visual microscope examination and counting of cell contacts that produced cell fusion with mixing of dyes. Per sample, between 100 and 120 cell contacts were counted. The assay was performed in duplicates.
Binding of N peptides.
To investigate the functional expression of C46 on the surface by M87oRRE, the binding of the following two N-peptides was determined by flow cytometry: N36, representing the N-terminal heptad region of gp41, and IZN17, consisting of the 17 aa of the N heptad region and the artificial 24-aa prepeptide IZm fused to the N terminus (5). N-biotinylated N36, IZN17, and the IZm prepeptide were purchased from Jerini, Berlin, Germany. Circular dichroism (CD) experiments revealed a molar helicity of 97% for IZN17 and 68% for N36. A total of 2 × 105 PM-1/M87oRRE cells were incubated with 10 μg of N36 or IZN17 in phosphate-buffered saline for 1 h at 37°C. After a single washing, cells were incubated with phosphate-buffered saline-1.5% formaldehyde for 10 min at 4°C, followed by an additional washing step. Steptavidin-phycoerythrin (Dianova, Hamburg, Germany) was used for detection. The IZm prepeptide was used as a control to exclude that binding of IZN17 to C46 was not exerted by the prepeptide alone. Untransduced PM1 cells served as controls.
RESULTS
The antiviral peptide.
Two antiviral C peptides, C36 and C46, both expressed as membrane-anchored peptides, were compared for antiviral activity. C36 (also known as DP-178 or T-20) is a 36-aa peptide corresponding to aa 638 to 673 of HIVHXB2 gp41. The C46 peptide contains 10 additional amino acids at the N terminus, thus spanning aa 628 to 673 of gp41. In previous reports, peptides containing these additional 10 N-terminal amino acids (but lacking C-terminal amino acids present in C36, such as C34 [T-1249]) were shown to effectively inhibit HIV isolates resistant to C36 (8). The retroviral vector M87-Ineo expresses membrane-anchored C36 and the neomycin resistance gene. In M87/C46-Ineo the C36 peptide in M87-Ineo was replaced by the C46 peptide (Fig. 1A and Table 1). For both peptides, the signal peptide of the low-affinity nerve growth factor receptor (LNGFR) directed translocation into the endoplasmic reticulum, and the murine immunoglobulin G2 (IgG2) hinge fused to the LNGFR membrane-spanning domain served as a scaffold that anchored the peptide to the cell surface (Fig. 1 and Table 1).
FIG. 1.
Basic design of retroviral vectors. (A) Retroviral vectors with anti-HIV activity code for a protein that consists of the following modules: a signal peptide to direct translocation into the endoplasmic reticulum (SP), the antiviral C peptide (C46), a hinge (H), and an MSD. Some vectors coexpress the neomycin resistance gene via a poliovirus internal ribosome entry site as a marker or express an RRE decoy or a wPRE element in the 3′ UTR. (B) Amino acid sequence of the membrane-anchored antiviral peptide encoded by M87o. LTR, long terminal repeat.
TABLE 1.
Design of antiviral vectors derived from M87 and of control vectorsa
| Vector | LTR | Leader | SP | C peptide | H (hinge) | MSD | Marker gene | 3′ UTR |
|---|---|---|---|---|---|---|---|---|
| MP1-neo | MPSV | 1 | None | None | None | None | neoR | |
| MP71-EGFP | MPSV | 1 | None | None | None | None | EGFP | wPRE |
| M87-Ineo | MPSV | 1 | dLNGFR | C36 | Murine IgG2 | LNGFR | PolioIRES + neoR | |
| M87/C46-Ineo | MPSV | 1 | dLNGFR | C46 | Murine IgG2 | LNGFR | PolioIRES + neoR | |
| M87o-Ineo | MPSV | 71 | dLNGFR | C46 | Human IgG2 | CD34 | PolioIRES + neoR | |
| M87o | MPSV | 71 | dLNGFR | C46 | Human IgG2 | CD34 | None | wPRE |
| M87o-RRE | MPSV | 71 | dLNGFR | C46 | Human IgG2 | CD34 | None | RRE, wPRE |
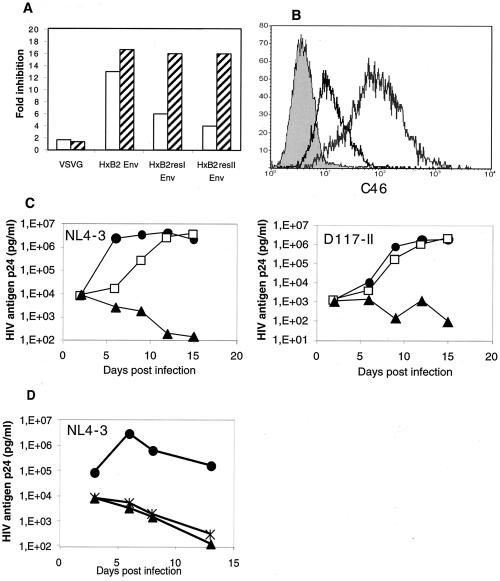
PM-1 cells were transduced with M87-Ineo, M87/C46-Ineo, or MP1neo containing the resistance gene only as a control. Two independent cultures were initiated for each vector. Cells were selected and stained for surface expression of C36 or C46 with the human monoclonal antibody 2F5 detecting an epitope (ELDKWA) present in both peptides (2, 21). The selected bulk cultures were >95% pure, and the level of C36 and C46 peptide expression did not differ significantly. Single-round infections of the G418-resistant PM-1 bulk cultures were then performed with a replication-incompetent lentiviral vector. The lentiviral vector expressing EGFP as a marker gene was packaged with the envelope glycoprotein of HIVHXB2 (10). In addition, two mutations were introduced into the HxB2 env (gp41) sequence that have been shown to confer resistance to the C36 peptide. The GIV motif in the N-terminal heptad repeat was exchanged for SIM and DTV, generating the HxB2resI and HxB2resII mutants, respectively (22). The lentiviral vector pseudotyped with the G protein of the vesicular stomatitis virus served as a negative control. The results are shown in Fig. 2A. For the wild-type HxB2env, entry was inhibited in C36- and C46-expressing cells with approximately equal efficiencies of 13- and 17-fold inhibition, respectively. As expected, inhibition of the C36-resistant HIV-1HXB2 envelopes was less efficient in the C36-expressing cells, with a six- and fourfold inhibition for the HxB2resI and HxB2resII mutants, respectively. In contrast, entry was inhibited with the same efficacy for wild-type and mutant env's in C46-expressing cells. In conclusion, the C46 peptide was found to have full antiviral activity against C36-resistant HIV-1 variants and was therefore selected for further vector development.
FIG. 2.
Optimizing the antiviral vector. (A) PM-1 cells expressing the control vector or antiviral vector M87-Ineo (□) or M87/C46-Ineo (▨) were transduced with a lentiviral vector expressing EGFP pseudotyped with the indicated viral envelope glycoprotein. In HxB2resI and HxB2resII, mutations that confer resistance to C36 (T-20) have been introduced. The inhibitory activity (fold inhibition) is represented by the ratio of the transduction efficiency for PM-1 cells expressing the control vector relative to the cells expressing the antiviral vector. The vector supernatants were used at a dilution that gave between 1 and 10% marking in the control cells, since in this range a linear relation between vector input and marking was observed (data not shown). A total of 105 cells per sample were analyzed by FACS to allow accurate analysis of marking even in the inhibited cultures with fewer than 1% GFP-positive cells. The mean values of two independent selected bulk cultures per vector are given. (B) Wild-type PM-1 (shaded curve) and PM-1 cells expressing M87-Ineo (black line) or M87o-Ineo (gray line) were stained with the human monoclonal antibody 2F5 that detects an epitope in the antiviral peptides and a phycoerythrin-conjugated goat-anti-human serum followed by flow cytometric analysis. (C) G418-selected bulk cultures of PM-1 expressing the control vector MP1neo (•) M87-Ineo (□) or M87o-Ineo (▴) were infected with the T-tropic isolate HIV-1NL4-3 or the dualtropic primary isolate D117-II at an MOI of 0.01 or 0.0001, respectively. HIV-1 p24 antigen production into the supernatant was monitored by ELISA. Experiments were performed in triplicate. (D) M87o and M87oRRE have comparable antiviral activity. PM-1 cells were transduced with M87o, M87oRRE, orMPIN and then sorted for expression of C46 or mock sorted (MP1N) by FACS. Purified bulk cultures were infected with NL4-3 at an MOI of 0.01. HIV-1 production was quantified as described in the text. The assays were performed in triplicates. Mean values are shown. Symbols: •, MP1neo; ✠, M87oRRE; ▴, M87o.
Improvement of the scaffold and expression of the membrane-anchored peptide.
Five modifications were introduced into the basic construct M87-Ineo. (i) To reduce potential immunogenicity, the murine IgG2 hinge was replaced by the human IgG2 hinge (H). (ii) To improve vector safety, the membrane-spanning domain (MSD) of LNGFR was exchanged for the MSD of human tCD34. Recent reports have suggested that LNGFR could have contributed to a leukemia observed after retroviral gene transfer in mice (18). In addition, tCD34 mediates more stable anchoring in the membrane than the LNGFR MSD. The amino acid sequence of the optimized M87o protein containing C46 and the two exchanged modules H and MSD is shown in Fig. 1B. (iii) To increase the expression level, the DNA sequence coding for the M87o protein was adapted for human codon usage and cloned into an optimized retroviral vector backbone (13, 23) (Table 1). Expression and antiviral activity of the optimized vector M87o-Ineo were compared to the basic construct M87-Ineo in G418 selected bulk cultures by flow cytometry. The optimized construct M87o-Ineo showed an ∼10-fold-higher expression than the basic construct M87-Ineo (Fig. 2B).
In addition, inhibition of the replication of the HIV-1 isolates NL4-3 and D117-II was analyzed. Although replication of both isolates was not completely inhibited in PM-1 cultures expressing M87-Ineo, only a low level of virus replication was detected in cells expressing the optimized vector M87o-Ineo (Fig. 2C).
(iv) To further reduce immunogenicity, the neo gene was eliminated from the vector, generating the monocistronic vector M87o. (v) In addition, a 41-bp sequence derived from the HIV-1 RRE element was added in the 3′-untranslated region (UTR) of the vector as a second antiviral principle, generating the vector M87oRRE. Expression of this RRE sequence was shown to inhibit HIV replication since it acts as an RNA decoy (1). Cells were transduced with M87o and M87oRRE. The level of surface expression of C46 was comparable for both vectors. C46-positive cells were sorted and challenged with the HIV strains NL4-3 (X4) and D117-II (X4R5, primary isolate). Nearly complete inhibition of HIV replication was observed for both M87o and M87oRRE-transduced cultures. Under these conditions, no improvement of antiviral potency by addition of the RRE element was observed (Fig. 2D). Nevertheless, the M87oRRE vector was chosen for further preclinical testing, since the short RRE sequence is not expected to have adverse effects and may reduce the emergence of resistant virus.
Antiviral activity of M87oRRE.
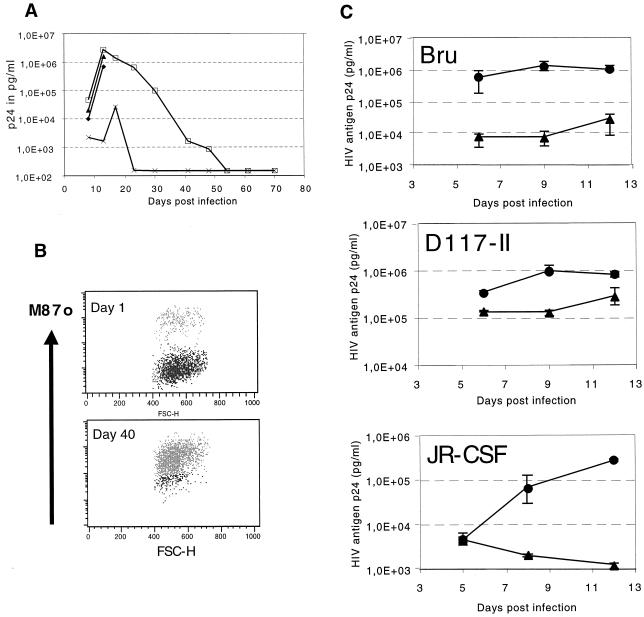
Treatment of HIV-1 infection with antiviral genes generally relies on the selective advantage of the gene-modified cells that are expected to accumulate to therapeutically effective levels over time in the patient. To test whether the M87oRRE vector can confer such a selective advantage, a mixture of vector-positive and -negative cells was challenged with HIV-1NL4-3. PM-1 cells were transduced with M87oRRE and sorted for expression of C46. The sorted bulk cultures were 97% positive. These sorted cells were mixed with wild-type PM-1 to generate a 17% C46-positive culture. The following cells served as positive and negative controls: PM-1 sorted for expression of C46 (>95% positive), wild-type PM-1, and a mixture of wild-type PM-1 with FACS-purified cells transduced with a vector expressing EGFP (20% EGFP positive). The results are shown in Fig. 3A. Wild-type PM-1 and the cells expressing EGFP showed a strong HIV-induced cytopathic effect (CPE), and all cells were dead by day 12 postinfection. In contrast, PM-1 cells sorted for the expression of C46 showed no viral CPE, and the 17% C46-positive culture only showed a transient CPE around day 10 and then grew normally for the rest of the experiment. Maximum virus production was seen in the control cultures on day 12. A short small peak of virus production was observed in the C46-positive culture on day 16, but no virus production was observed thereafter. Interestingly, the 17% C46-positive cells produced an amount of virus approximately equal to that produced by the control cultures on day 12. Thereafter, however, virus production dropped continuously to become undetectable on day 55. Intracellular p24 staining showed no expression of HIV gag (data not shown), and even coculture with wild-type PM-1 cells failed to recover virus from these cultures. Flow cytometric analysis on day 40 showed that C46-positive cells had accumulated from 17 to >95% (Fig. 3B). Uninfected control cells cultured in parallel remained 17% C46 positive. These data clearly show that the M87o-RRE vector can confer an effective selective advantage to gene-modified cells in the presence of HIV replication.
FIG. 3.
Antiviral activity of M87oRRE. (A) Wild-type PM-1 (♦), PM-1 transduced with an EGFP vector and FACS purified for EGFP expression and then mixed 1 to 5 with wild-type PM-1(▴), PM-1 transduced with M87oRRE and FACS purified for C46 expression (×), and FACS-purified M87oRRE mixed with wild-type PM-1 to generate a 17% C46-positive culture (□) were infected with HIV-1NL4-3 at an MOI of 0.001. p24 secretion into the supernatant was monitored by an ELISA. (B) A culture that was initially 17% C46 positive was stained for expression of C46 on days 1 and 40 with the human monoclonal antibody 2F5. Cells on day 40 were 98% positive for C46. (C) PBMC from healthy donors were transduced with M87oRRE(▴) or mock transduced (•), sorted for expression of C46, and infected with the indicated HIV-1 isolates. p24 production was monitored by ELISA.
Inhibition of HIV replication in primary cells.
To test the antiviral activity in primary cells, primary blood lymphocytes from healthy donors were transduced with M87oRRE and M87o on days 3, 4, and 5 of culture; sorted for C46 expression on day 7; and challenged with HIV-1 between days 9 and 11. Sorted cultures were between 76 and 91% positive. As a control, mock-transduced cells were used. Several virus isolates and PBMC from different donors were tested. A representative experiment for the three HIV-isolates Bru (X4), JR-CSF (R5), and D117-II (X4R5) is shown. HIV replication was inhibited by 1 to 2 logs in PBMC expressing M87oRRE. In this experiment, JR-CSF seemed to be more susceptible to M87oRRE. This was not, however, observed for all donors. As in the cell lines, no difference was seen in the inhibitory activity of M87o and M87oRRE in PBMC (data not shown).
Mechanism of action of M87o.
The current model is that the antiviral C peptides derived from the HIV-1 gp41 second heptad repeat interact with the trimeric coiled coil of gp41 formed by the first (N-terminal) gp41 heptad repeat. gp41 is consequently locked into a fusion-incompetent state, and entry of HIV-1 into the target cell is inhibited (6). We tested whether membrane-anchored C46 acts like the free C peptides. Initially, we tested whether membrane-anchored C46 can bind to the coiled-coil structure formed by N-peptides derived from the first heptad repeat of gp41. Two biotinylated N peptides were used: N36 and IZN17. The latter is a peptide containing an N-terminal artificial helix sequence (IZm) fused to a 17-aa sequence derived from the gp41 first heptad repeat (5). IZm was shown to induce a nearly entire helical structure of the N peptide. PM-1 cells expressing M87oRRE and control PM-1 were incubated with the biotinylated peptides and then stained with streptavidin-phycoerythrin. Flow cytometric analysis showed that the N peptides specifically bound to cells expressing C46, whereas a control peptide did not (Fig. 4). This indicates that membrane-anchored C46 can interact with the coiled-coil domain of gp41.
FIG. 4.
Binding of gp41 N peptides to a membrane-anchored C peptide. Wild-type PM-1 cells (shaded curve) and FACS-purified PM-1 cells expressing M87oRRE (solid line) were incubated with the biotinylated gp41 N peptide IZN17 (A) or N36 (B) or the 24-aa control prepeptide IZm (C) and then stained with streptavidin-phycoerythrin. The peptides and staining procedure are described in Materials and Methods.
In addition, the proposed mode of action for C peptides would predict that infection is blocked at the level of membrane fusion. We therefore tested the fusion of wild-type PM-1 cells, PM-1/M87-Ineo, and PM-1/M87o-Ineo cells with 293T cells expressing the HIV-1HXB2 envelope. Fusion was inhibited transiently ∼2-fold in cells expressing the basic construct and more sustained at a level of ∼4-fold in cells expressing the optimized construct M87o-Ineo (Fig. 5). These data suggest that membrane-anchored antiviral C peptides have a mode of action similar to that of the free C peptides.
FIG. 5.
Inhibition of membrane fusion. The fusion kinetics of PM-1 cells expressing the indicated vectors with 293T expressing the HIV-1 envelope glycoprotein were analyzed. The percentage of cell contacts that led to fusion is given for each time point. A total of 100 to 120 cell contacts were counted per time point and sample. The assay was performed in duplicates. Symbols: •, MP1neo; □, M87-Ineo; ▴, M87o-Ineo.
DISCUSSION
In the present study, the retroviral vector M87oRRE that expresses a membrane-anchored fusion inhibitory peptide derived from the C-terminal heptad repeat of gp41 was shown to effectively inhibit HIV-1 entry. To develop this highly active antiviral vector, all modules of the vector M87-Ineo described previously had to be individually optimized (12).
M87oRRE had an >10-fold-higher expression level and antiviral activity than the basic construct M87-Ineo. Furthermore, the optimized vector effectively suppressed virus replication of different isolates in primary cells, which was not reproducibly seen for the basic construct. Additional experiments have shown that the enhanced antiviral potency is mostly due to the higher level of transgene expression by the optimized construct (unpublished data).
In addition, M87oRRE has less potential immunogenicity than the basic construct, since the neoR gene was removed and the murine IgG2 hinge was humanized. However, immunogenicity of the C46 sequence and the fusion domains between the four modules of the membrane-anchored peptide (SP, C46, H, and MSD; Fig. 1) cannot be excluded. It is difficult to predict whether such an immune reaction would impair the therapeutic effect of M87oRRE, and this must be tested in future clinical trials.
As in any antiviral therapy, resistance is a major issue also for M87oRRE. Three steps were taken to minimize the possibility that resistant strains might emerge. An RRE element was added in the 3′ UTR of M87o. The RRE sequence can act as an RRE decoy and thereby inhibit production of HIV-1 genomic RNA. Although the antiviral activity of M87o was not improved by the addition of the RRE element, this element was retained in the vector that underwent further preclinical testing since it is not expected to have toxic side effects and, as a second antiviral principle, could reduce the chance for the development of M87o-resistant virus strains in a clinical setting. To verify this hypothesis, current studies aim at testing the activity of the M87oRRE vector against M87o-resistant virus strains. However, despite repeated attempts, we have not yet been able to generate M87o-resistant virus. In addition, the chance for resistance was reduced by using the elongated C46 peptide that is known to interact with a highly conserved pocket of gp41. Lastly, the high and stable expression of the antiviral peptide reduces the chance for resistance, which is more likely under a suboptimal dose of the antiviral peptide (25).
Several observations clearly indicate that the mode of action of membrane-anchored C46 is the same as for the free gp41 C peptides. C peptides have been shown to interact with gp41 and thereby inhibit membrane fusion during virus entry. Likewise, the membrane-anchored peptide was found to block early steps of virus replication in single-round infection assays (Fig. 2). In addition, membrane fusion mediated by HIV Env was effectively inhibited. For different antiviral constructs, the level of fusion inhibition (Fig. 5) was found to correlate with the level of inhibition of virus replication (Fig. 3). Finally, the membrane-anchored peptide bound to free N peptides that are known to form the trimeric coiled-coil structure of gp41 (Fig. 4).
The vector M87oRRE has several qualities that could be crucial for therapeutic success. The major advantage over previous antiviral genes is that viral infection is blocked at the level of virus entry. Thus, infection of the cell is effectively prevented. Previous genes mostly target viral RNA or protein synthesis and therefore require suppression of the integrated provirus for the total life span of the cell. In addition, the gene-modified cells that carry a suppressed provirus are expected to accumulate in the patient, which would counteract the antiviral effect of the gene. Late inhibitors therefore are expected to require a much stronger and sustained antiviral activity than early inhibitors to be therapeutically effective. In addition, the membrane-anchored peptides are located exactly at their site of action, which is the membrane of the T helper cells, thus minimizing potential adverse effects at distant and therapeutically irrelevant locations. Furthermore, the membrane-anchored peptides are encoded by a relatively small open reading frame, thus allowing for the expression of additional antiviral genes in the same vector. Finally, gene modification with M87oRRE can be easily detected by flow cytometry with the antibody 2F5, so that a marker gene is dispensable.
Extensive preclinical toxicity studies have been completed in mice and rhesus macaques for M87oRRE without any detectable side effects or immunogenicity (unpublished data). This lack of toxicity and the high antiviral efficacy thus justify further development of M87oRRE and in particular the initiation of clinical trials.
Acknowledgments
The p202 expression plasmid for HXB2 env was kindly provided by Feng Yang (Whitehead Institute for Biomedical Research). We thank Sascha Döring and Frank Vorpahl for excellent technical assistance.
This study was supported by Fresenius AG and EU grant QLK2-CT 2000-01040 (HIVComTher).
REFERENCES
- 1.Bauer, G., P. Valdez, K. Kearns, I. Bahner, S. F. Wen, J. A. Zaia, and D. B. Kohn. 1997. Inhibition of human immunodeficiency virus-1 (HIV-1) replication after transduction of granulocyte colony-stimulating factor-mobilized CD34+ cells from HIV-1-infected donors using retroviral vectors containing anti-HIV-1 genes. Blood 89:2259-2267. [PubMed] [Google Scholar]
- 2.Buchacher, A., R. Predl, K. Strutzenberger, W. Steinfellner, A. Trkola, M. Purtscher, G. Gruber, C. Tauer, F. Steindl, and A. Jungbauer. 1994. Generation of human monoclonal antibodies against HIV-1 proteins; electrofusion and Epstein-Barr virus transformation for peripheral blood lymphocyte immortalization. AIDS Res. Hum. Retrovir. 10:359-369. [DOI] [PubMed] [Google Scholar]
- 3.Chan, D. C., D. Fass, J. M. Berger, and P. S. Kim. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263-273. [DOI] [PubMed] [Google Scholar]
- 4.Clapham, P. R., A. McKnight, and R. A. Weiss. 1992. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J. Virol. 66:3531-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert, D. M., and P. S. Kim. 2001. Design of potent inhibitors of HIV-1 entry from the gp41 N-peptide region. Proc. Natl. Acad. Sci. USA 98:11187-11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eckert, D. M., and P. S. Kim. 2001. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70:777-810. [DOI] [PubMed] [Google Scholar]
- 7.Feng, Y., M. Leavitt, R. Tritz, E. Duarte, D. Kang, M. Mamounas, P. Gilles, F. Wong-Staal, S. Kennedy, J. Merson, M. Yu, and J. R. Barber. 2000. Inhibition of CCR5-dependent HIV-1 infection by hairpin ribozyme gene therapy against CC-chemokine receptor 5. Virol. 276:271-278. [DOI] [PubMed] [Google Scholar]
- 8.Greenberg, M. L., D. Davison, L. Jin, S. Mosier, T. Melby, P. Sista, R. Demasi, D. Miralles, N. Cammack, and T. J. Matthews. 2002. In vitro antiviral activity of T-1249. Antiviral Ther. 7:S10. [Google Scholar]
- 9.Grignani, F., T. Kinsella, A. Mencarelli, M. Valtieri, D. Riganelli, L. Lanfrancone, C. Peschle, G. P. Nolan, and P. G. Pelicci. 1998. High-efficiency gene transfer and selection of human hematopoietic progenitor cells with a hybrid EBV/retroviral vector expressing the green fluorescence protein. Cancer Res. 58:14-19. [PubMed] [Google Scholar]
- 10.He, J., Y. Chen, M. Farzan, H. Choe, A. Ohagen, S. Gartner, J. Busciglio, X. Yang, W. Hofmann, W. Newman, C. R. Mackay, J. Sodroski, and D. Gabuzda. 1997. CCR3 and CCR5 are coreceptors for HIV-1 infection of microglia. Nature 385:645-649. [DOI] [PubMed] [Google Scholar]
- 11.Hildinger, M., K. L. Abel, W. Ostertag, and C. Baum. 1999. Design of 5′ untranslated sequences in retroviral vectors developed for medical use. J. Virol. 73:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hildinger, M., M. T. Dittmar, P. Schult-Dietrich, B. Fehse, B. S. Schnierle, S. Thaler, G. Stiegler, R. Welker, and D. von Laer. 2001. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J. Virol. 75:3038-3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildinger, M., H. G. Eckert, A. J. Schilz, J. John, W. Ostertag, and C. Baum. 1998. FMEV vectors: both retroviral long terminal repeat and leader are important for high expression in transduced hematopoietic cells. Gene Ther. 5:1575-1579. [DOI] [PubMed] [Google Scholar]
- 14.Kilby, J. M., S. Hopkins, T. M. Venetta, B. DiMassimo, G. A. Cloud, J. Y. Lee, L. Alldredge, E. Hunter, D. Lambert, D. Bolognesi, T. Matthews, M. R. Johnson, M. A. Nowak, G. M. Shaw, and M. S. Saag. 1998. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4:1302-1307. [DOI] [PubMed] [Google Scholar]
- 15.Kohn, D. B., G. Bauer, C. R. Rice, J. C. Rothschild, D. A. Carbonaro, P. Valdez, Q. Hao, C. Zhou, I. Bahner, K. Kearns, K. Brody, S. Fox, E. Haden, K. Wilson, C. Salata, C. Dolan, C. Wetter, E. Aguilar-Cordova, J. Church, T. C. Lee, B. A. Sullenger, H. F. Gallardo, G. E. Ungers, and E. Gilboa. 1999. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34+ cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood 94:368-371. [PubMed] [Google Scholar]
- 16.Kuhlcke, K., F. A. Ayuk, Z. Li, C. Lindemann, A. Schilz, U. M. Schade, A. A. Fauser, A. R. Zander, H. G. Eckert, and B. Fehse. 2000. Retroviral transduction of T lymphocytes for suicide gene therapy in allogeneic stem cell transplantation. Bone Marrow Transplant. 26:S96-S98. [DOI] [PubMed] [Google Scholar]
- 17.Levy-Mintz, P., L. Duan, H. Zhang, B. Hu, G. Dornadula, M. Zhu, J. Kulkosky, D. Bizub-Bender, A. M. Skalka, and R. J. Pomerantz. 1996. Intracellular expression of single-chain variable fragments to inhibit early stages of the viral life cycle by targeting human immunodeficiency virus type 1 integrase. J. Virol. 70:8821-8832. (Erratum, J. Virol. 72:3505-3506, 1998.) [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 18.Li, Z., J. Dullmann, B. Schiedlmeier, M. Schmidt, C. von Kalle, J. Meyer, M. Forster, C. Stocking, A. Wahlers, O. Frank, W. Ostertag, K. Kuhlcke, H. G. Eckert, B. Fehse, and C. Baum. 2002. Murine leukemia induced by retroviral gene marking. Science 296:497. [DOI] [PubMed] [Google Scholar]
- 19.Lusso, P., F. Cocchi, C. Balotta, P. D. Markham, A. Louie, P. Farci, R. Pal, R. C. Gallo, and M. S. J. Reitz. 1995. Growth of macrophage-tropic and primary human immunodeficiency virus type 1 (HIV-1) isolates in a unique CD4+ T-cell clone (PM-1): failure to downregulate CD4 and to interfere with cell-line-tropic HIV-1. J. Virol. 69:3712-3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Melikyan, G. B., R. M. Markosyan, H. Hemmati, M. K. Delmedico, D. M. Lambert, and F. S. Cohen. 2000. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J. Cell Biol. 151:413-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Purtscher, M., A. Trkola, A. Grassauer, P. M. Schulz, A. Klima, S. Dopper, G. Gruber, A. Buchacher, T. Muster, and H. Katinger. 1996. Restricted antigenic variability of the epitope recognized by the neutralizing gp41 antibody 2F5. AIDS 10:587-593. [DOI] [PubMed] [Google Scholar]
- 22.Rimsky, L. T., D. C. Shugars, and T. J. Matthews. 1998. Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72:986-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schambach, A., H. Wodrich, M. Hildinger, J. Bohne, H. G. Krausslich, and C. Baum. 2000. Context dependence of different modules for posttranscriptional enhancement of gene expression from retroviral vectors. Mol. Ther. 2:435-445. [DOI] [PubMed] [Google Scholar]
- 24.Steinberger, P., J. Andris-Widhopf, B. Buhler, B. E. Torbett, and C. F. Barbas III. 2000. Functional deletion of the CCR5 receptor by intracellular immunization produces cells that are refractory to CCR5-dependent HIV-1 infection and cell fusion. Proc. Natl. Acad. Sci. USA 97:805-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wei, X., J. M. Decker, H. Liu, Z. Zhang, R. B. Arani, J. M. Kilby, M. S. Saag, X. Wu, G. M. Shaw, and J. C. Kappes. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896-1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weissenhorn, W., A. Dessen, S. C. Harrison, J. J. Skehel, and D. C. Wiley. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426-430. [DOI] [PubMed] [Google Scholar]
- 27.Wild, C. T., D. C. Shugars, T. K. Greenwell, C. B. McDanal, and T. J. Matthews. 1994. Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. USA 91:9770-9774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woffendin, C., U. Ranga, Z. Yang, L. Xu, and G. J. Nabel. 1996. Expression of a protective gene-prolongs survival of T cells in human immunodeficiency virus-infected patients. Proc. Natl. Acad. Sci. USA 93:2889-2894. [DOI] [PMC free article] [PubMed] [Google Scholar]