Abstract
In a search for previously unknown genes that are required for lymphocyte development in zebrafish, a retroviral sequence was identified in a subtracted thymus cDNA library and in genomic DNA libraries. The provirus is 11.2 kb and contains intact open reading frames for the gag, pol, and env genes, as well as nearly identical flanking long terminal repeat sequences. As determined by in situ hybridization, the thymus appears to be a major tissue for retroviral expression in both larval and adult fish. Several viral transcripts were found by Northern blotting in the adult thymus. The provirus was found at the same genomic locus in sperm from four fish, suggesting that it is an endogenous retrovirus. Phylogenetic analysis indicates that it is closest to, yet distinct from, the cluster of murine leukemia virus-related retroviruses, suggesting that this virus represents a new group of retroviruses.
Endogenous retroviruses, which have been identified in almost all vertebrate genomes (14), are inherited as cellular genes and passed on to succeeding generations. Although most endogenous retroviruses are defective due to extensive mutations and deletions in their genomes, some are apparently intact (1, 21, 49). Endogenous retroviruses may be transcriptionally active or silent, depending on the differentiation stage of the cell type in which they reside (25, 29, 38, 47), the presence of inducing agents (6, 12, 35, 60), and the genomic locus of viral integration (12, 46). Infectious viral particles can be produced from intact proviruses (4) or generated through genetic recombination involving exogenous and transcriptionally active, either intact or defective, endogenous retroviruses (25, 45).
Genomic structures of endogenous retroviruses have been characterized in chickens, mice, pigs, and humans (20, 21, 33, 41). In fish, fragments derived from endogenous retroviral elements have been reported (14), but they exhibit extensive mutations and deletions and are unlikely to generate functional proteins. To date, the genomes of only a few intact piscine retroviruses—snakehead fish retrovirus (13), walleye dermal sarcoma virus (WDSV) (18) and walleye epidermal hyperplasia virus (WEHV) (28), all of which are exogenous infectious agents—have been cloned. To our knowledge, no full-length fish endogenous retrovirus has yet been identified.
The zebrafish, Danio rerio, has gained recognition in recent years as an excellent system for developmental studies in vertebrates. In a search for previously unknown zebrafish genes that are selectively expressed in the thymus, we have identified an apparently intact endogenous retrovirus, and we report here its full-length genomic structure and sequence. We refer to this full-length endogenous retrovirus as ZFERV and to other related endogenous retroviruses, identified by Southern hybridization with env or long terminal repeat (LTR) probes, as ZFERV-related proviruses. Evidence for expression of ZFERV-related viral transcripts in the thymus, as well as for the presence of ZFERV-related proviruses in the zebrafish genome, is also presented. By phylogenetic analysis, ZFERV is closest to, yet distinct from, murine leukemia virus (MLV)-related retroviruses, suggesting that it may represent a new group of retroviruses.
MATERIALS AND METHODS
Animals.
Zebrafish (D. rerio) of the Tübingen stock were obtained from C. Nüsslein-Volhard (Tübingen, Germany) and maintained at 28°C. Tissues, RNA, and DNA were derived from these fish. The λ phage cDNA library (provided by V. S. Hohman) was derived from outbred zebrafish. Genome Jump DNA libraries (10) and sperm samples derived from AB_Tübingen fish were provided by N. Hopkins, Massachusetts Institute of Technology (MIT), Cambridge. Families of these fish are partially inbred but are not as uniform genetically as strains of inbred mice. The use of animals was in compliance with the guidelines set by the MIT Committee on Animal Care.
Preparation of thymus RNA.
Thymi from adult zebrafish (age 5 to 6 months) were pooled and minced in 15 ml of phosphate-buffered saline (PBS), followed by centrifugation (250 × g, 10 min). The cell pellet was resuspended in 1 ml of trypsin-EDTA (Sigma, St. Louis, Mo.) for 3 min, followed by the addition of 10 ml of PBS. The cell suspension was layered onto 2 ml of Histopaque-1077 (Sigma) in a 17-by-120-mm conical tube and then centrifuged at 150 × g for 10 min; the top cell layer was collected and washed once with PBS. Larval fish (2 or 7 days postfertilization [dpf]) were collected and washed once with PBS. RNA was isolated from thymus cells and from whole larvae with TRIzol reagent (Life Technologies, Grand Island, N.Y.) according to the manufacturer's instructions.
Subtractive hybridization.
RNA (250 ng) from thymus cells from two adult fish and from 250 larval fish at 2 dpf was converted to cDNA and amplified by PCR with the SMART PCR cDNA synthesis kit (Clontech Laboratories, Palo Alto, Calif.). The amplified thymus cDNA was subtracted with that from the larvae according to the manufacturer's instructions for the PCR-Select cDNA subtraction kit (Clontech). The subtracted cDNA was inserted into the TA cloning vector pCRII-TOPO (InVitrogen, Carlsbad, Calif.), and the ligated plasmid was transformed into Escherichia coli strain TOP10F′ (InVitrogen). The insert in each cloned plasmid in 384 independent bacterial colonies was PCR amplified with the nested PCR primers 1 and 2R (Clontech) and resolved in a 2% agarose gel, followed by Southern blotting analysis. Hybridization with 32P-labeled cDNA probes prepared from adult thymus, but not with those prepared from 2-dpf larval fish, identified candidate clones. cRNA probes prepared from these clones were used for in situ hybridization (see below) on 7-day-old fish to identify clones corresponding to transcripts selectively expressed in the thymus at this stage; these clones were sequenced (Tufts Core Facility, Tufts University Medical Center, Boston, Mass.).
Screening of λ phage cDNA library.
To obtain longer cDNA corresponding to the subtracted cDNA sequences, a λ phage library constructed from zebrafish thymus cDNA was screened. Plaques were transferred to nitrocellulose membranes and hybridized to random-primed 32P-labeled DNA probes prepared from the subtracted cDNA clones. Positive plaques were isolated and transduced into E. coli BM25.8 (Clontech) at 31°C for 16 h to convert λ phages into plasmids. Plasmid DNA from selected bacterial colonies was purified, and the sizes of cDNA inserts were estimated by restriction enzyme digestion, followed by agarose gel electrophoresis and ethidium bromide staining. The plasmids containing the longest inserts were sequenced.
Cloning of env.
Thymus RNA (1 μg) was resuspended in 10 μl of H2O containing 0.5 μg oligo(dT)12-18 (Life Technologies), followed by incubation at 65°C for 5 min, followed in turn by the addition of 10 μl of reverse transcription (RT) premix (2 mM deoxynucleoside triphosphate [dNTP], 20 mM dithiothreitol, 2× first-strand buffer, and 200 U of SuperScript II RNase H− reverse transcriptase [Life Technologies]) and incubation at 42°C for 1 h. The RT reaction product was then diluted 10-fold with 1× first-strand buffer. The diluted RT reaction product (1 μl) was added to 19 μl of PCR premix (0.5 μM each for top- and bottom-strand primers, 0.4 mM dNTP, 1× PCR buffer, 1 U of AmpliTag DNA polymerase [Roche Molecular Biochemicals, Mannheim, Germany]), and the sample was incubated at 95°C for 3 min and then at 95°C for 20 s, 50°C for 30 s, and 72°C for 2.5 min for 35 cycles. The primer pairs were 5′-GAAGCATCTAGGCCTGCAGA-3′ (top strand) and 5′-CAGGTGTTAAACCACATCCTGTAC-3′ (bottom strand). The PCR product was diluted 50-fold, and the diluted product (1 μl) was used as a template for the second PCR amplification. The reaction conditions were the same as described above except for a different primer pair: 5′-ATTACTCGAGGGCCACATTCAGGTAATTCTCCTA-3′ (top strand) and 5′-ATTATCTAGATCTCATAAGAGATCACACCATATC-3′ (bottom strand). These primer sequences, located either upstream or downstream of the env coding region, were selected based on the insert sequences of the λ phage plasmids. The PCR product was cloned into the pCRII-TOPO vector, and the insert was sequenced.
Cloning and sequencing of ZFERV.
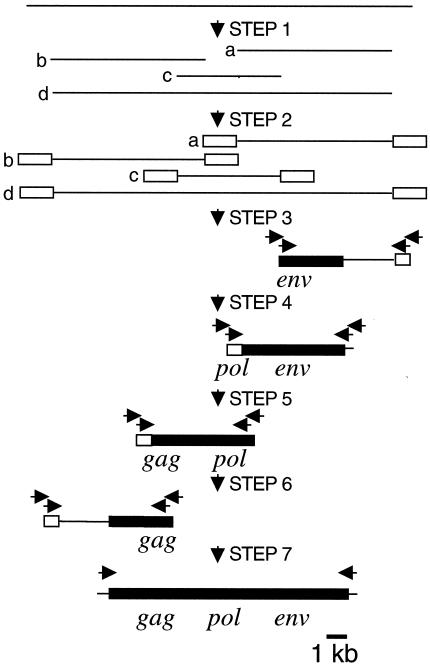
Genome Jump DNA libraries had been generated by digesting genomic DNA with a restriction enzyme, followed by ligation of DNA fragments to an adaptor containing two universal primer sequences for nested PCR (Fig. 1, steps 1 and 2). Nine different restriction enzymes were used to generate a set of nine different libraries.
FIG. 1.
Cloning strategy for ZFERV. The zebrafish Genome Jump DNA libraries had been generated in the Hopkins laboratory by digesting fish genomic DNA with restriction enzymes (step 1), followed by ligating a universal DNA adapter (open boxes) to the ends of DNA fragments (step 2). Each of nine libraries was derived by digestion with a single enzyme. Nested PCR was performed with universal adapter primers and ZFERV-specific primers (indicated as arrows on top of the ZFERV fragments) in conjunction with each of the nine genome jump DNA libraries (steps 3 to 6). The largest PCR fragments (indicated as lines and boxes) among all amplified products generated from the different libraries were sequenced, and ZFERV-specific primers were designed accordingly for PCR amplification in the next step. Finally, various primers containing the derived 5′ and 3′ cellular junction sequences were used in conjunction with the set of libraries to amplify the entire ZFERV segment by PCR (step 7). Filled boxes represent ZFERV sequences and lines are cellular sequences.
To amplify DNA fragments containing genomic sequences downstream to the env gene, top-strand primers corresponding to sequences in the env gene and bottom-strand primers corresponding to the adapter sequence were used (Fig. 1, step 3). The amplified fragments were sequenced, and two different 3′ cellular junction sequences were identified. Next, another set of bottom-strand primers corresponding to one of the 3′ cellular junction sequences was used with the same Genome Jump DNA libraries to amplify DNA fragments containing additional sequence upstream to the 3′ cellular junction (Fig. 1, step 4). Among all DNA fragments amplified from the libraries, the longest one (5.5 kb) was isolated, and the sequence near its 5′ end was obtained.
To obtain genomic sequences upstream to the 5.5-kb genomic DNA fragment, the same strategy was used except that a new set of bottom-strand primers was applied according to the 5′-end sequence of the previously derived largest fragment. Such genomic DNA amplification was performed twice more. Three different 5′ cellular junctions were found (Fig. 1, steps 5 and 6).
To generate the entire ZFERV fragment by PCR, six combinations of primers containing the derived 5′ and 3′ cellular junction sequences were used together with each Genome Jump DNA library (Fig. 1, step 7). For PCR amplification, a 50-μl PCR mixture (1 μg of Genome Jump DNA library, 400 nM each for top- and bottom-strand primers, 400 μM each for dNTP, 1× Expand HF buffer with 1.5 mM MgCl2, 3.5 U of Expand Long Template System enzyme mix [Roche]) was incubated at 95°C for 3 min for 1 cycle, 95°C for 20 s and 68°C for 10 min for 10 cycles, 95°C for 20 s and 68°C for 12 min for 20 cycles, and 68°C for 10 min for 1 cycle. Only one pair of primers (top-strand primer [5′-TCTAAAGGAAAATGAACTTAACAGTTGCGAGTGA-3′] and bottom-strand primer [5′-TATTCGCAATACTCTGTTCAGTTTACTGTACTTTGCTA-3′]), together with EagI-digested Genome Jump DNA library, generated a PCR product (11,249 bp). The PCR product was cloned into the TA cloning vector pCRII-TOPO, and the insert was sequenced from both ends (Biopolymers Laboratories, MIT).
Phylogenetic analysis.
Amino acid sequence from the fifth residue N terminal to the highly conserved Gln residue through the signature motif YXDD of the reverse transcriptase (61) of ZFERV was aligned with sequences corresponding to the same region of representative members of recognized retroviruses. An unrooted phylogenetic tree was constructed with the PHYLIP program (http://evolution.genetics.washington.edu/phylip/phylipweb.html) [7]). The names and GenBank accession numbers of the viruses are listed below.
Preparation of genomic DNA.
To prepare DNA from whole fish, each fish was frozen in liquid N2, ground into powder, and incubated in digestion buffer (100 mM NaCl, 10 mM Tris-HCl [pH 8], 25 mM EDTA, 0.5% sodium dodecyl sulfate, 0.1 mg of proteinase K [Roche]/ml) at 50°C for 16 h. Samples were extracted twice with phenol-chloroform-isoamyl alcohol (25:24:1), followed by ethanol precipitation. DNA was suspended in 100 μl of H2O. To prepare DNA from sperm, semen (5 to 10 μl) from male fish was mixed with 100 μl of digestion buffer and incubated at 50°C for 16 h. The samples were extracted and precipitated as for whole fish. Purified sperm DNA was suspended in 10 μl of H2O.
Detection of ZFERV-related proviruses in genomic DNA. (i) Whole fish.
DNA (10 μg) was digested with EcoRV or SpeI and resolved in 1% agarose gel, followed by Southern blotting with env and LTR probes.
(ii) Sperm.
DNA was amplified by PCR, followed by detection of viral fragments in the PCR products by Southern blotting. For PCR amplification, a 50-μl PCR mixture (1 μl of sperm DNA, 400 nM each for top- and bottom-strand primers, 500 μM each for dNTP, 1× PCR buffer with 2.25 mM MgCl2 and detergents, and 3.5 U of Expand Long Template System enzyme mix) was incubated at 95°C for 3 min for 1 cycle; 94°C for 20 s, 55°C for 30 s, and 72°C for 1 min for 35 cycles; and 72°C for 5 min for 1 cycle. The top-strand primer sequence (5′-TTGCTGCAGCCGAAGGGGATGACGTGAT-3′; nucleotides [nt] 10470 to 10497 of ZFERV) is located within the env gene, and the bottom-strand primer sequence (5′-TATTCGCAATACTCTGTTCAGTTTACTGTACTTTGCTA-3′) is located 73 bp downstream of the 3′ cellular junction of ZFERV. Thus, generation of an 853-bp PCR product containing the putative ZFERV 3′ LTR sequence is expected if the provirus is located in the ZFERV locus. To confirm that the PCR product contains the LTR sequence, 1 μl of the PCR products was resolved in 1% agarose gel, followed by Southern blotting with the LTR probe.
The DNA templates for generating the random-primed 32P-labeled probes were as follows: for the env probe, a 1,929-bp insert consisting of the env coding sequence (see Results) cloned into the pCRII-TOPO vector; and for the LTR probe, a 425-bp fragment extending from the HindIII site of the U3 region to the 3′ end of the R region of ZFERV LTR sequence.
In situ hybridization.
Larval fish (3 to 5 dpf) were fixed with 4% paraformaldehyde in PBS for 2 h at room temperature, followed by gradual dehydration in methanol and rehydration in PBS. Adult fish (3 months old) were fixed with 4% paraformaldehyde in PBS at 4°C for 16 h, followed by gradual dehydration in ethanol and xylene, embedded in melted paraffin and sectioned (2 or 20 μm thick). Sections were dewaxed with xylene, followed by gradual rehydration in H2O. Both larval fish and paraffin sections were treated with 0.3% Triton X-100 for 15 min and with proteinase K (15 μg/ml) for 10 min, followed by incubation in acetylation solution (0.1 M triethanolamine [pH 8], 0.25% [vol/vol] acetic anhydride; Sigma) twice for 5 min each.
For hybridization, samples were incubated with prehybridization buffer (4× SSC [600 mM NaCl, 60 mM sodium citrate; pH 7], 50% [vol/vol] formamide) at 37°C for 10 min, followed by incubation at 42°C for 16 h in hybridization buffer (50% [vol/vol] formamide, 4× SSC, 1× Denhardt solution [200 mg/liter each for Ficoll 400, polyvinylpyrrolidone, and bovine serum albumin], 100 μg of heparin/ml, 0.1% Tween 20, 1 mg of yeast Torula RNA [Sigma]/ml) containing fluorescein- or digoxigenin [DIG]-labeled RNA probe (0.2 μg/ml). After two washes with 0.2× SSC at 60°C, samples were incubated with blocking solution (0.1 M maleic acid [pH 7.6], 150 mM NaCl, 2% blocking reagent [Roche]) for 1 h. Alkaline phosphatase-conjugated anti-DIG or alkaline phosphatase-conjugated anti-fluorescein antibody (Roche) was added to the blocking solution in a ratio of 1:1,000, and incubation continued for two more hours. Samples were washed twice with maleic acid buffer (0.1 M maleic acid [pH 7.6], 150 mM NaCl).
For colorimetric detection, samples containing anti-DIG antibodies were washed once with alkaline phosphatase buffer (0.1 M Tris-HCl [pH 9.5], 0.1 M NaCl, 50 mM MgCl2), followed by staining in buffer containing 1 mM levamisole, 0.9 mg of nitroblue tetrazolium salt and 0.35 mg of BCIP (5-bromo-4-chloro-3-indolylphosphate; Roche)/ml. For fluorescence detection, samples containing anti-fluorescein antibodies were washed once with 0.1 M Tris-HCl (pH 8.2), followed by incubation in Fast Red staining solution (Roche) according to the manufacturer's instructions.
The 1,929-bp fragment containing the env coding sequence in the pCRII-TOPO vector was used as a template for synthesizing sense and antisense env fluorescein- and DIG-labeled cRNA probes. pZr1 (57) containing a 638-bp fragment of rag1 (11) was used to synthesize antisense rag1 DIG-labeled cRNA probe. The cRNA probes were prepared with an RNA labeling kit (Roche) according to the manufacturer's instructions. The sense and antisense env probes (50 ng) were incubated in alkaline hydrolysis buffer (40 mM NaHCO3, 60 mM Na2CO3) at 60°C for 14 min, followed by ethanol precipitation, to generate shorter fragments prior to addition of the hybridization buffer.
Northern blotting.
Thymus RNA (5 μg) from 20 adult fish (5 months old) was electrophoresed in 1% agarose-formaldehyde gel, followed by Northern blotting with the env or LTR probe.
RT-PCR.
RNA (2 μg) from whole larvae (2 and 7 dpf) was incubated in 25 μl of DNase solution (1× RQ1 RNase-free DNase reaction buffer, 2 U of RQ1 RNase-free DNase, 20 U of RNase inhibitor; Promega, Madison, Wis.) at 37°C for 30 min, followed by phenol-chloroform extraction and ethanol precipitation. The RNA pellet was resuspended in 10 μl of H2O containing 0.5 μg oligo(dT)12-18, followed by incubation at 65°C for 5 min, followed by the addition of 10 μl of RT premix (2 mM dNTP, 20 mM DTT, 2× first-strand buffer, 200 U of SuperScript II RNase H− reverse transcriptase) and incubation at 42°C for 1 h. The reaction products were diluted threefold and sixfold with 1× first-strand buffer and were used as the template in PCR to assess unsaturated PCR amplification of cDNA.
For PCR amplification of cDNA, 1 μl of the diluted RT reaction products was added to 19 μl of PCR premix (500 nM each for top- and bottom-strand primers, 400 μM dNTP, 1× PCR buffer, 1 U of AmpliTag DNA polymerase), followed by incubation at 95°C for 3 min and at 95°C for 20 s, 55°C for 30 s, and 72°C for 30 s for 25 cycles. When EF-1α-specific primrs were used, the PCR condition was the same as described above except that 45°C was used for primer annealing and 20 cycles of amplification were applied. The following primer pairs (top strand and bottom strand) corresponding to ZFERV LTR env and gag sequences (GenBank accession number AF503912) were used for PCR amplification: LTR, 5′-GCTGCAGCCGAAGGGGATGACGT-3′ and 5′-CAGGTGTTAAACCACATCCCTGTAC-3′ (positions 10472 to 10494 and positions 10860 to 10836); env, 5′-CATCACTCTAGGGGTAGATGTAGA-3′ and 5′-AATCATGTAATGGAGCGGGTTCAG-3′ (positions 9088 to 9111 and positions 9398 to 9375); and gag, 5′-GTACCTGTGAGGACAGAGACAAGA-3′ and 5′-GTACCCATCTTTTAGTTCTGTCTGACA-3′ (positions 2671 to 2694 and positions 2814 to 2788). The primer pair sequences (top strand and bottom strand) for amplifying EF-1α cDNA were 5′-CTGGTGACAACGTTGGCTTC-3′ and 5′-TGGAACGGTGTGATTGAGGG-3′ (positions 971 to 990 and positions 1473 to 1454 [GenBank accession no. L23807]). A total of 5 μl of PCR products was resolved in 2% agarose gel, followed by ethidium bromide staining.
Nucleotide sequence accession numbers.
The nucleotide sequences of the env gene and of ZFERV have been submitted to GenBank under accession numbers AY075045 and AF503912, respectively. Other viral sequences used for the phylogenetic analysis are from avian leukosis virus (GenBank accession number NC001408), baboon endogenous retrovirus (BAA89659), equine foamy virus (NP054716), feline endogenous retrovirus (FERV; P31792), feline foamy virus (NP056914), feline leukemia virus (NP047255), gibbon ape leukemia virus (AAC80264), human immunodeficiency virus type 1 (NC001802), human T-cell leukemia virus type 1 (NC001436), human T-cell leukemia virus type 2 (NC001488), human foamy virus (NP044280), MLV (P03355), mouse mammary tumor virus (NC001503), porcine endogenous retrovirus (CAC82505), simian foamy virus (NP056803), simian T-cell leukemia virus 2 (NP056907), WDSV (NP045937), WEHV type 1 (WEHV1; AAD30048), and WEHV2 (AAC59311). Retrovirus sequences from birds, reptiles, and amphibians have been described (14).
RESULTS
Identification of retrovirus sequences in a subtracted thymus cDNA library. In a search for genes selectively expressed in the zebrafish thymus, we generated a thymus cDNA library by subtracting cDNA from adult fish thymus with that from 2-day-old larval fish, which have not yet developed T lymphocytes (59). Of 384 clones in the subtracted cDNA library, 43 yielded strong signals after hybridization with cDNA probes prepared from the thymus but not with probes from 2-day-old larval fish. To evaluate expression in the thymus, we performed whole-mount in situ hybridization on 7-day-old fish with cRNA probes prepared from these 43 clones. Probes from 21 clones stained only the thymus, whereas probes from the other 22 clones either stained the entire body or did not stain any tissue (data not shown). These results suggest that the 21 cDNA clones correspond to relatively abundant transcripts expressed in the thymus (at age 7 dpf).
The 21 positive clones were sequenced; eight, including three redundant ones, were nearly identical in sequence to portions of several zebrafish unannotated expressed sequence tags (ESTs; Washington University Zebrafish EST Project). These EST sequences did not contain open reading frames (ORFs). The remaining sequenced clones did not match any EST sequences. All 21 clones were subsequently shown to correspond to different segments of the same or related retroviral sequences (see below).
To identify coding sequences that might be adjacent to these EST sequences, we screened a λ phage library that had been constructed from zebrafish thymus cDNA. Each of the nonredundant cDNA clones that stained only the thymus was used independently as a probe. Two positive plaques containing long cDNA inserts (1.8 and 2.3 kb, respectively) were identified and sequenced. About 600 bp at the 3′ end of the 1.8-kb cDNA fragment and at the 5′ end of the 2.3-kb cDNA fragment were identical in sequence (later identified as a part of the env coding sequence; see below). When combined, the superimposed 3.5-kb sequence showed a 113-bp direct repeat sequence at each end, a structural feature similar to the R sequence of retroviral genomes. Further, the internal sequence contained an ORF encoding a 642-amino-acid residue protein that included a transmembrane domain, as found in retroviral envelope proteins of many vertebrate species.
Since the 3.5-kb sequence was assembled from two independent cDNA fragments, we sought to determine whether transcripts containing an intact env coding sequence are expressed in the thymus. By RT-PCR with thymus RNA, we amplified the entire env fragment (1,929 bp). The fragment was sequenced and found to be identical to the coding sequence of the 3.5-kb superimposed fragment (this env sequence has been deposited in GenBank under the accession number AY075045). Moreover, the entire env fragment was also amplified from zebrafish genomic DNA (data not shown). It seemed possible, therefore, that the env gene is part of an endogenous retrovirus in the zebrafish genome, which we provisionally designated ZFERV (for zebrafish endogenous retrovirus).
Cloning and structure of ZFERV.
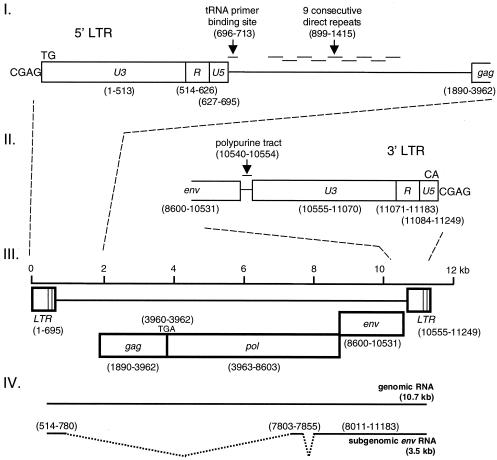
If intact ZFERV is present in the zebrafish genome, it should be possible to clone the whole viral entity from genomic DNA. To this end, we chose the zebrafish Genome Jump DNA libraries (10) over classic λ phage genomic DNA libraries as the source for cloning, mainly because the former is PCR based so that the genome-walking process is directional and more efficient. The strategy to clone ZFERV from zebrafish genomic DNA was to search for junction sequences flanking the LTRs of ZFERV in the zebrafish genome. Primers corresponding to these junction sequences were then applied to PCR amplify the entire ZFERV fragment from zebrafish genomic DNA (Fig. 1). By this approach, a long contiguous retroviral DNA, which consists of 11,249 bp, was cloned and sequenced. No fragments other than ZFERV were amplified by this approach (data not shown). The sequence contains ORFs for the gag, pol, and env genes and flanking LTR sequences (Fig. 2III).
FIG. 2.
Structure of ZFERV genome. The viral genome consists of 11,249 bp and contains ORFs for gag, pol, and env genes and two flanking LTRs (open boxes; panel III). The gag and pol genes are contiguous and are in the same reading frame, whereas env is in a different reading frame. The Gag-Pol polyprotein (2,237 amino acid residues) could be translated from the hypothetical viral genomic RNA (10.7 kb; panel IV) by suppressing the termination codon (TGA) at the junction of the gag and pol genes. The Env protein (643 amino acid residues) could be generated from the spliced subgenomic env RNA (3.5 kb; panel IV). Three sequence blocks (U3, R, and U5) of both LTR segments, as well as features of adjacent sequences, are shown in panels I (for the 5′ LTR) and II (for the 3′ LTR). The numbers shown in parentheses refer to ZFERV DNA (GenBank accession number AF503912).
LTR.
The 5′- and 3′-flanking LTR sequences (nt 1 to 695 and nt 10555 to 11249, respectively) consist of 695 bp each and are 99% identical (Fig. 2I and II). As in other retroviruses, the LTR segments contain three consecutive sequence blocks, from 5′ to 3′, the unique 3′ (U3) sequence, the repeat (R) sequence and the unique 5′ (U5) sequence. Because the middle block, the R sequence (113 bp), is identical to the flanking repeated sequences of the superimposed 3.5-kb cDNA mentioned above, the upstream U3 and the downstream U5 sequences in the ZFERV LTR segment were recognized by comparing the sequences of ZFERV and the 3.5-kb cDNA.
As an integrated provirus, ZFERV also exhibits recognizable features in the U3 and U5 sequences as a result of integration. The 5′ U3 sequence (513 bp) is 3 bp shorter than the 3′ U3 sequence, presumably because of an AAT trinucleotide deletion in the 5′ LTR upon viral integration. Similarly, the 3′ U5 sequence (66 bp) is 3 bp shorter than the 5′ U5 sequence (due to an ATT trinucleotide deletion in the 3′ LTR). At the 5′ and 3′ ends of the LTR segments, the consensus dinucleotide TG/CA inverted repeats are juxtaposed with a duplicated 4-bp cellular sequence (CGAG). Taken together, these apparently symmetric structures suggest that ZFERV follows common rules for retroviruses upon integration (16).
ORFs.
Like all retroviruses, ZFERV contains gag, pol, and env genes (Fig. 2III). ZFERV appears to have the same gene organization as MLV-related retroviruses (62), in which the gag and pol genes are in the same reading frame, whereas the env gene is in another (Fig. 2III). Other ORFs are quite short (≤216 bp), suggesting that they are unlikely to encode functional proteins. The ZFERV gag-pol ORF (nt 1890 to 8603) would encode a 2,237-amino-acid residue Gag-Pol polyprotein, when the internal stop codon (nt 3960 to 3962) between the gag and pol genes is translationally suppressed, as in the case for MLV (62). The env ORF (nt 8600 to 10531) encodes a 643-amino-acid residue Env protein. Interestingly, the ZFERV env ORF has an extra codon relative to the env cDNA fragment amplified by RT-PCR. Since the Genome Jump DNA libraries (10) were prepared from AB × Tübingen fish, whereas the thymus RNA was prepared from Tübingen fish, it is possible that a length polymorphism in env may exist among different fish.
Many protein domains that are conserved among different retroviral genera are also encoded in ZFERV, including the protease, reverse transcriptase, RNase H, and integrase domains of the polymerase (Pol) polyprotein and the transmembrane domain of Env protein. However, the variable Gag polyprotein and the surface domain in Env protein are not similar to these in any known retrovirus, suggesting that ZFERV belongs to a distinct retroviral group.
Other features.
Like other retroviruses (9), the polypurine tract (nt 10540 to 10554) and the tRNA primer-binding site (nt 696 to 713), which serve as primer sites in the process of converting viral RNA to double-stranded DNA, were also found in ZFERV (Fig. 2I and II). The sequence between the 5′ LTR and the gag gene contains nine consecutive direct repeats (nt 899 to 1415), each approximately 60 nt in length (Fig. 2I). As a result, a long 5′-untranslated region (1,376 nt) for viral genomic RNA is expected.
Comparison of the sequence of genomic DNA with that of the superimposed 3.5-kb cDNA encoding the env gene revealed two pairs of RNA splice donor and acceptor sites in the genome (Fig. 2IV). The first exon extends from the beginning (nt 514) of the 5′ R sequence to nt 780; the first intron extends from nt 781 to 7802. The second exon extends from nt 7803 to 7855; the second intron extends from nt 7856 to 8010. The third exon extends from nt 8011 to the end (nt 11183) of the 3′ R sequence. Consequently, the subgenomic RNA lacks most of the sequences between the 5′ LTR and gag, the entire gag, and the majority of the pol gene (Fig. 2IV). The size of this subgenomic RNA is expected to be 3,493 bp plus the size of the poly(A) tail. It is not known whether other species of subgenomic RNA are also generated.
Phylogenetic analysis of ZFERV.
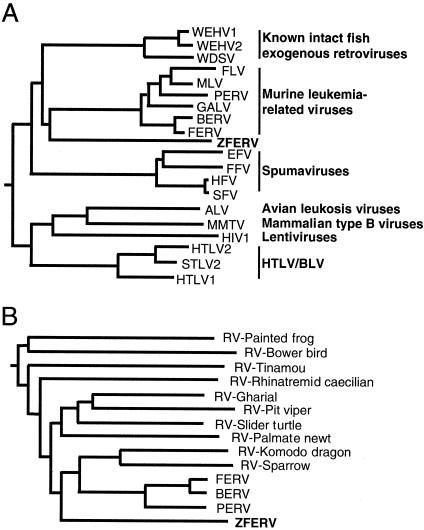
To determine the evolutionary relationships between ZFERV and other retroviruses, we performed a phylogenetic analysis based on an alignment of RT sequences, the most conserved sequence among all retroid elements, including endogenous retroviruses (37, 61). Sequences of the same RT region from representative members of recognized retroviral genera and from three previously identified exogenous walleye fish retroviruses (18, 28) were used in the alignment.
As shown in Fig. 3A, the analysis places ZFERV closest to the cluster of MLV-related viruses (43 to 44% identity) and to the group of exogenous walleye fish retroviruses (42 to 44% identity) and more distant to other retroviral genera (28 to 35% identity). In contrast, the percent identity between any two viruses within the group of MLV-related viruses is at least 82%. Similarly, the percent identity between WDSV and WEHV1 is 82%. Therefore, although ZFERV shares a common ancestor with MLV-related retroviruses and walleye fish retroviruses, it appears that it belongs to a distinct group.
FIG. 3.
Phylogenetic relationship between ZFERV and other retroviruses. (A) The ZFERV RT conserved region, consisting of 161 amino acid residues, was aligned with sequences from the same region of representative members of recognized retroviral genera. The phylogenetic tree was constructed with the PHYLIP program (7). (B) The same RT regions of endogenous retroviral elements from birds (bower bird and sparrow), reptiles (pit viper, Komodo dragon, gharial, and slider turtle), amphibians (palmate newt and rhinatremid caecilian) (14), and mammals (FERV, baboon endogenous retrovirus [BERV], and porcine endogenous retrovirus [PERV]) were also compared. The GenBank accession numbers of these viruses are given in Materials and Methods. Abbreviations: ALV, avian leukosis virus; EFV, equine foamy virus; FFV, feline foamy virus; FLV, feline leukemia virus; GALV, gibbon ape leukemia virus; HIV1, human immunodeficiency virus type 1; HTLV1, human T-cell leukemia virus type 1; HTLV2, human T-cell leukemia virus type 2; HFV, human foamy virus; MMTV, mouse mammary tumor virus; SFV, simian foamy virus; STLV2, simian T-cell leukemia virus 2; RV, retrovirus.
We also compared ZFERV with other endogenous retroviral elements from birds, reptiles, and amphibians (14). As shown in Fig. 3B, ZFERV is not closely related (<40% identity) to any of these. This result further supports the notion that ZFERV belongs to a distinct group of retroviruses.
ZFERV is an endogenous retrovirus in zebrafish.
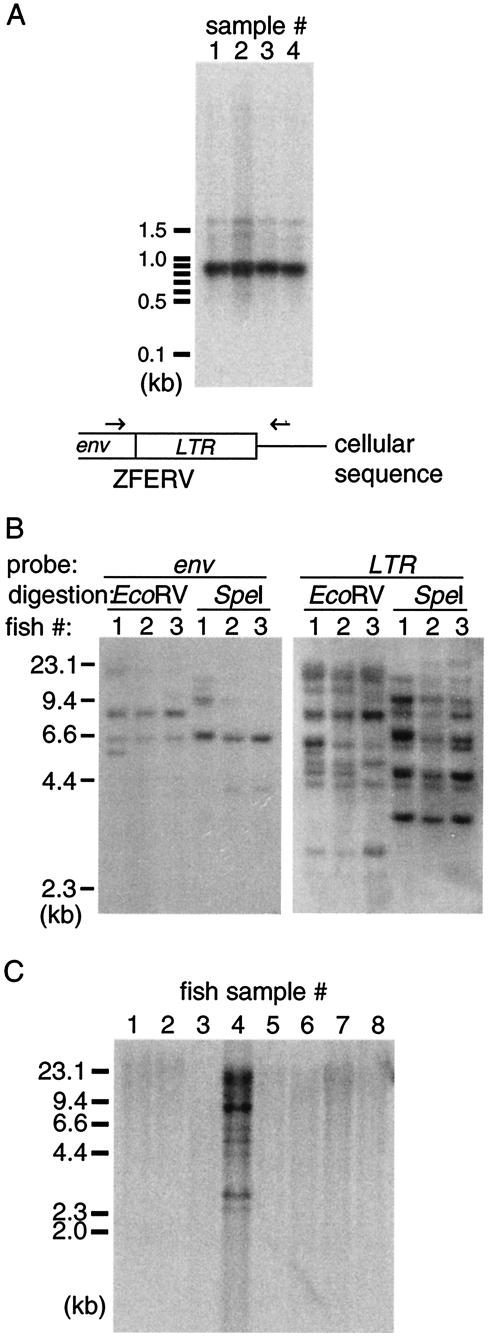
An endogenous retrovirus remains in the same genetic locus in every cell and is transmitted through germ cells to the next generation. To establish that ZFERV is indeed an endogenous virus, we performed Southern blotting to detect ZFERV in genomic DNA from sperm and from whole fish. Several individual fish samples were examined for the localization of ZFERV in the genome.
To detect ZFERV in sperm DNA, we amplified a fragment containing the putative ZFERV 3′ LTR segment from four sperm DNA samples by PCR; one primer was located in the env gene, and the other was located in the 3′ cellular sequence (determined previously in the process of cloning ZFERV). The presence of the LTR sequence in the amplified fragment was verified by Southern blotting. As shown in Fig. 4A, a predominant band, located between 800 and 900 bp (the expected size is 853 bp), was detected in every sperm DNA sample, indicating that genomic DNA from zebrafish germ cells does contain ZFERV sequences. In addition, since one of the primer sequences is located in the downstream cellular sequence, this result suggests that ZFERV, like cellular genes, resides in the same genetic locus in different fish.
FIG.4.
ZFERV is an endogenous retrovirus. (A) Sperm DNA from each of four zebrafish was used as a template in PCR to detect the provirus in the putative ZFERV locus. The top-strand primer sequence, located within the ZFERV env gene, and the bottom-strand primer sequence, located in the cellular sequence downstream of ZFERV, were used in PCR (arrows in the diagram). The products were resolved in a 1% agarose gel, followed by Southern blotting with the LTR probe. (B) Genomic DNA (10 μg) from three individual zebrafish (3 months old) was digested with either EcoRV or SpeI, resolved by 1% agarose gel electrophoresis, and subjected to Southern blotting with probes for the env gene (left panel) or for LTR segments (right panel). (C) Genomic DNA (10 μg) from different fish species was digested with EcoRV, resolved by 1% agarose gel electrophoresis, and subjected to Southern blotting with the LTR probe. Lane 1, sturgeon (Acipenser baerc); lane 2, bowfin (Amia calva); lane 3, Atlantic trout (Salmo salar); lane 4, zebrafish (Danio rerio); lane 5, Danio albolineatus; lane 6, Danio kerri; lane 7, Danio nigrofasciatus; lane 8, Danio shanensis.
In another experiment, genomic DNA from three fish was digested with restriction enzymes, followed by Southern blotting with the env and LTR probes. EcoRV and SpeI restriction enzymes were chosen so that the entire env gene is contained within a single DNA fragment, since neither restriction site is present in the env gene. As shown in Fig. 4B, two to four bands were detected with the env probe, suggesting that no more than four copies of ZFERV exist in the zebrafish genome. Consistent with the results obtained with sperm DNA (Fig. 4A), the darkest bands hybridizing to the env probe (∼8 kb in EcoRV- and ∼6.5 kb in SpeI-digested samples) are the same size in DNA from each fish, suggesting that these ZFERV-related proviruses are located in the same genetic locus. The other env-positive bands with lower intensities may result from copies with truncated env genes. However, it is also possible that EcoRV and SpeI sites are present in the env gene of these copies due to restriction fragment length polymorphism. Eight to ten bands were detected with the LTR probe in each digested genomic DNA, suggesting that some provirus entities are probably defective or recombinant since they contain the same LTR segments but lack the env gene (e.g., “solo LTR” [51]).
Most of the LTR-positive bands are consistent in size among different fish, but some bands were detected in the genome of only one fish. These results suggest that most ZFERV-related proviruses, intact or defective, reside in the same respective genomic loci in different fish but that some fish either contain different copy numbers of ZFERV or exhibit polymorphism in viral and/or cellular sequences.
ZFERV may be limited to zebrafish (Danio rerio).
The genomes of several other fish species were also examined for the presence of ZFERV. As shown in Fig. 4C, none of these other fish, including several closely related Danio species, was found to contain ZFERV (lanes 1 to 3 and lanes 5 to 8), suggesting that ZFERV may be limited to the zebrafish.
Thymic expression of ZFERV.
To determine in which tissues ZFERV-related transcripts are expressed, larval fish and sections from adult fish were subjected to in situ hybridization with probes specific for the ZFERV env gene. In 5-day-old fish, the thymus appeared to be the only tissue with detectable staining, suggesting that the thymus is a major tissue for viral RNA expression at this developmental stage (Fig. 5A). In sections of adult fish, the thymus was also the most strongly stained tissue compared to others on the same sections (Fig. 5B to D).
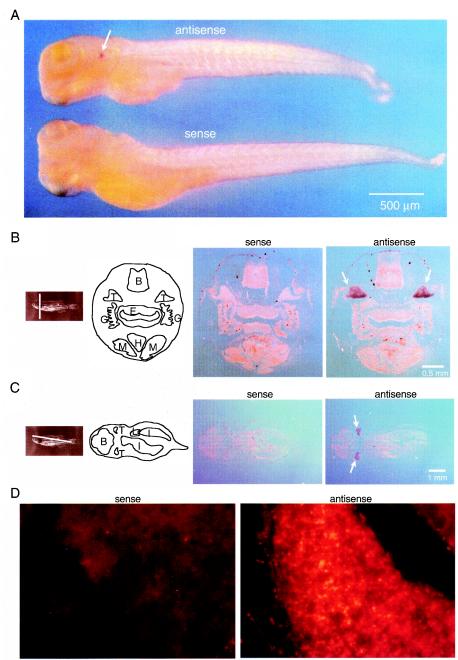
FIG.5.
Expression of ZFERV-related RNA in thymus. (A) Larval fish at 5 dpf were fixed and subjected to whole mount in situ hybridization with a DIG-labeled sense or antisense probe for the env gene, followed by colorimetric detection. (B and C) Contiguous sections (20 μm thick) of 3-month-old fish (right two panels) were subjected to in situ hybridization with probes as described in panel A. The section planes and main organs are indicated. Abbreviations: B, brain; E, esophagus; G, gill; H, heart; I, intestine; L, liver; M, muscle; T, thymus. The bilateral thymi hybridizing to the antisense probe are highlighted with arrows. (D) Sections (2 μm thick) of the thymus from 3-month-old fish were hybridized to fluorescein-labeled sense (left panel) or antisense (right panel) env probe, followed by fluorescence detection with the alkaline phosphatase-Fast Red system.
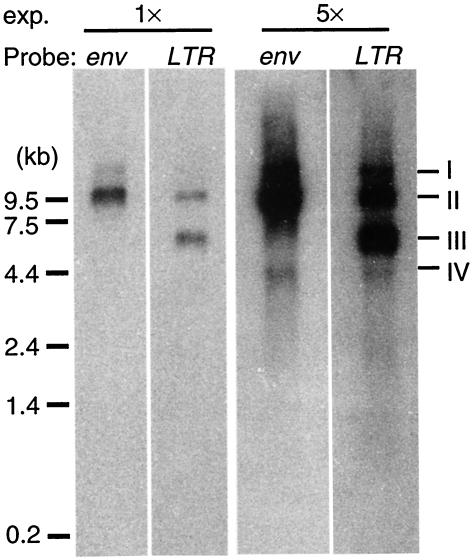
To determine the sizes of different ZFERV-related transcripts, RNA isolated from adult thymus was subjected to Northern blotting with probes for the ZFERV env gene and for the LTR segments. As shown in Fig. 6, an RNA species migrating at ca. 9.5 kb was detected by both probes (band II), presumably corresponding to the full-length genomic RNA (10.7 kb, Fig. 2A). Longer exposure of the same blot showed a weaker band at about 4 kb (band IV, right panel), presumably corresponding to the env subgenomic RNA (3.5 kb, Fig. 2A). In addition, an RNA species at about 6 kb (band III) hybridized strongly to the LTR probe. We do not know whether this is an early termination of the full-length genomic RNA or whether it represents a transcript from other proviruses with LTR sequences similar to that of ZFERV in the fish genome. A more slowly migrating band, estimated to be about 15 kb (band I), was also detected by both probes, possibly corresponding to a readthrough transcript that does not terminate at the regular polyadenylation site of the viral genomic RNA. Although there is uncertainty regarding the identity of bands I and III, the sizes of bands II and IV are consistent with those of the putative genomic and subgenomic RNA predicted from the genome structure of ZFERV and from the sequenced cDNA clones (Fig. 2A).
FIG. 6.
Northern blotting showing various ZFERV-related transcripts. Total RNA (5 μg) from thymocytes (pooled from 20 adult fish) was resolved by 1% agarose gel electrophoresis, followed by Northern hybridization with probes for env gene or LTR segment. Short and fivefold-longer exposures (1× and 5×) are indicated, the longer exposure revealing the 3.5-kb subgenomic RNA. The major bands are indicated on the right (bands I to IV).
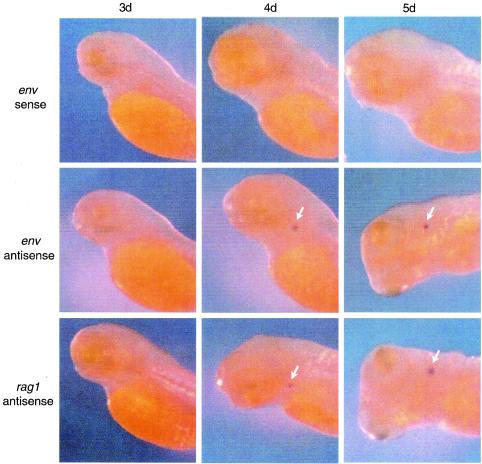
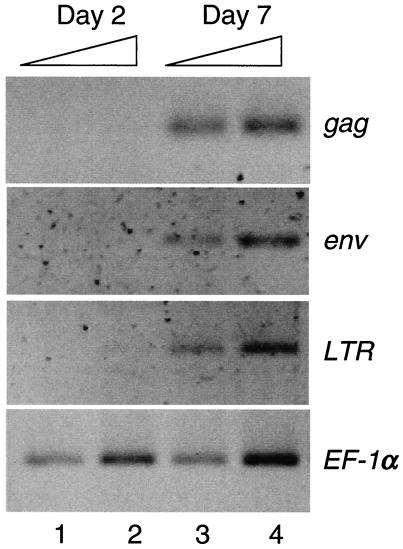
Since the thymus appears to be the major tissue in larval fish for viral RNA expression, we wanted to determine when ZFERV-related transcripts are detected during thymus development. As a hallmark of early thymus development, rag1 gene expression, which is first detected in the thymus at about 92 hpf (59), was used for comparison. As shown in Fig. 7, ZFERV-related RNA expression in the thymus was first detected with the antisense env probe by day 4, at the same time as expression of rag1. Higher expression levels were observed for both rag1 and the env at day 5, but there was no staining with either probe at day 3. No staining was observed at any stage with the sense env probe. These results indicate that expression of ZFERV-related transcripts begins at 4 dpf. Consistently, results from RT-PCR analysis also showed that 7-day-old larval fish contain ZFERV-related transcripts, whereas no viral transcripts were detected in 2-day-old fish (Fig. 8).
FIG. 7.
Temporal expression of ZFERV-related proviruses and rag1. Larval fish at stages of 3, 4, and 5 dpf were fixed and subject to whole-mount in situ hybridization with DIG-labeled probes for env or rag1 gene, followed by colorimetric detection. The thymus is highlighted with an arrow.
FIG. 8.
Detection of ZFERV-related transcripts in larval fish by RT-PCR. Total RNA from 2-day-old (lanes 1 and 2) and 7-day-old (lanes 3 and 4) larval fish were isolated and amplified by RT-PCR with primers specific for each indicated region of the virus. Onefold (lanes 1 and 3) and twofold (lanes 2 and 4) amounts of RT reaction mixtures were used as a template in PCRs. PCR products of the housekeeping gene EF-1α served as an internal control for equal RNA loading from both 2- and 7-day-old fish.
DISCUSSION
Properties of ZFERV.
We have shown that an intact endogenous retrovirus, ZFERV, is present in the zebrafish genome. Several other teleost species were also examined for the presence of ZFERV in their genomes (Fig. 4C). These included more distantly related species such as Atlantic trout, sturgeon, and bowfin and close relatives of zebrafish such as other Danio species. ZFERV was not found in any of these fish, suggesting that it may be limited to zebrafish. It appears that ZFERV is widely distributed among zebrafish, because the zebrafish EST and genomic databases (Washington University Zebrafish EST Project and Sanger Center Zebrafish Genome Sequencing Project, respectively) contain many ZFERV-related sequences from several lines: AB, SJD, and Tübingen.
We amplified the entire ZFERV fragment by PCR with primers corresponding to the 5′ and the 3′ cellular-viral junction sequences. When the products were analyzed by gel electrophoresis, no smaller fragment was seen, suggesting that no solo LTR has the same flanking sequences. An identical 5′ junction sequence was found in the zebrafish genome sequence database (trace file number zfishC-a1368f09.p1c [Sanger Center Zebrafish Genome Sequencing Project]). Further, the same 3′ cellular-viral junction sequence was detected in each of four individual sperm DNA samples (Fig. 4A). The darkest bands hybridizing to the env probe by Southern blotting (Fig. 4B) appear to be the only ones that are the same size in DNA from each fish and probably correspond to ZFERV. Other bands with lower densities may be derived from truncated ZFERV-related copies. Taken together, these data strongly suggest that at least one copy of ZFERV is present in every zebrafish at the same genomic locus. The additional, nonconserved bands may be related to restriction fragment length polymorphism in these fish. It is also possible that exogenous infection with ZFERV or retrotransposition contributes to the variability noted.
When compared to proviral MLV (8.9 kb; GenBank accession number AF033811), ZFERV has a relatively large genome (11.2 kb), mainly due to the following features. (i) Long U3 segments in the LTRs. Since the 5′ U3 segment serves as viral promoter and contains many potential transcription factor binding sites (see below), a longer sequence suggests a more complex gene regulation for ZFERV. (ii) Nine consecutive direct repeats between the 5′ LTR and gag. The retrovirus packaging signal, ψ element, usually resides between the 5′ LTR and gag (3). We speculate that these repeats may form stem-loop structures, which serve as a signal for encapsidation. (iii) An extended pol gene that could encode an additional protein domain at its 3′ end. The sequence near the C terminus of the presumable Gag-Pol polyprotein is homologous to the phosphoesterase domain found in macrohistone 2 and in Appr-1′-p processing enzyme (36, 42).
Another intriguing feature of ZFERV is the double splicing used to generate the subgenomic env RNA (Fig. 2). These splicing events join three exons, the last containing the env ORF. The fragments derived from pol do not contain translation initiation codons and therefore could not contribute to the Env protein sequence.
Phylogenetic analysis places ZFERV closest to the MLV-related retroviruses and to the exogenous piscine retroviruses (Fig. 3A). When genomic structures are compared, however, ZFERV is more similar to MLV since both genomes consist of the same number of ORFs, whereas exogenous piscine retroviruses contain several extra ORFs (18, 28). Herniou et al. (14) have characterized many endogenous retroviral elements from a wide spectrum of vertebrates, including mammals, reptiles, amphibians and fish. However, none of these retroviral sequences is >40% identical to ZFERV in the conserved RT region (Fig. 3B). Therefore, ZFERV appears to be sufficiently distinct from all known vertebrate endogenous retroviruses to represent a new group of retroviruses.
Expression of ZFERV.
In the initial screening of the subtracted thymus cDNA library, we chose clones with the strongest hybridization signals for further analysis. Of these, 21 showed selective expression in the thymus. Unexpectedly, all 21 were related to ZFERV. Evidently, ZFERV transcripts are particularly abundant among transcripts found in the adult zebrafish thymus but not in the 2-day-old larval fish.
In addition to the transcripts presumably corresponding to the full-length genomic and subgenomic env RNA, two additional ZFERV-related RNA species were identified in the thymus (Fig. 6). It is possible that these transcripts are generated from ZFERV by aberrant RNA termination or from other defective ZFERV-related proviruses in the fish genome. Another possibility is that they are generated from recombinant retroviruses derived from ZFERV, a scenario similar to that for mink cell focus-forming (MCF) MLV in AKR mice (23, 25). The latter possibility is supported by our finding that a cDNA clone from the subtracted thymus cDNA library contains a partial ZFERV gag segment combined with a non-ZFERV DNA segment that encodes a protein homologous to many retroviral protease proteins (data not shown). This recombinant sequence matches one of the sequences in the zebrafish EST database (accession number AW232029; Washington University Zebrafish EST project). Therefore, additional retroviral transcripts, which are related to ZFERV but not exactly the same as the ZFERV RNA, are also expressed in zebrafish. To understand the complexity of viral expression, a series of probes for different ZFERV segments and other non-ZFERV viral segments may be required to detect these transcripts and reconstruct their detailed structures.
The thymus was found to be the major tissue for expression of ZFERV in larval fish, and a high level of viral RNA expression persists in the thymus of adult fish. A previous study had shown that the adult thymus is filled with lymphocytes, which appear as groups of packed small cells interspersed between nonlymphoid cells (59). Consistent with this observation, staining with the env probe revealed that the thymus was packed with small cells having the appearance of small lymphocytes (Fig. 5D). A search of the zebrafish EST database for ZFERV-related sequences revealed a large number of EST sequences identical to parts of ZFERV. Unexpectedly, the mRNA sources of these EST clones were derived from a variety of tissue types, including the brain, kidney, olfactory rosettes, fin regenerates, retina, and heart. Since these tissues are from anatomically discrete organs and many of them are distant from the thymus, it seems unlikely that they are contaminated with thymic tissue.
There appear to be several possibilities to account for the presence of ZFERV transcripts in tissues other than the thymus. Viral transcripts may indeed be expressed, but at a low level, in these tissues. A number of potential transcription factor binding sites can be identified in the viral LTR, suggesting that ZFERV is expressed in a wide range of cell types. These sites could bind lymphoid-specific (Ikaros and E47 [8, 44]), myeloid-specific (AML and MZF [39, 48]), and hematopoietic-specific (GATA [22]) factors, as well as more general factors (STAT, NF-κB, C/EBP, AP-1/ATF, and Oct-1 [19, 30, 50, 53, 54]). Different cell types may contain limited amounts and numbers of these factors generating a low level of viral transcripts. These transcription factors may be expressed simultaneously in the thymus, synergistically activating the viral promoter to generate a relatively high level of ZFERV transcripts.
Another possibility is that mobile white blood cells from the thymus, such as T lymphocytes, may migrate to and reside in other tissues. In zebrafish, the intermediate cell mass (ICM), the first hematopoietic tissue in the embryo, is observed at approximately 24 hpf (2). The thymic primordium appears at 60 hpf, and it is colonized by immature lymphoblasts by 65 hpf (58). By 92 hpf, cells of the T lineage, presumably pre-T cells, expressing the recombination activating genes rag1 and rag2, are detected in the thymus (59). The same temporal expression patterns of rag1 and ZFERV-related proviruses in larval fish thymus suggest that activation of ZFERV may be subject to the developmental program of lymphopoiesis. Although circulation and homing of mature T cells may contribute to the ZFERV-related transcripts in various tissue types, ZFERV is probably not activated in cells of the erythroid lineage. The evidence for this is that ZFERV-related transcripts were not detected by in situ hybridization or by RT-PCR in larval fish before 4 dpf, when circulating erythrocytes are already prevalent (5, 58). Additional evidence is that radioactive cDNA probes prepared from mRNA of adult red blood cells did not hybridize with the subtracted thymus cDNA clones containing ZFERV fragments in Southern blotting analyses (data not shown). The definitive cell types that express substantial amount of viral transcripts remain to be determined.
Finally, it is possible that viral particles, if produced, may circulate and spread among different tissues. Endogenous retroviruses capable of generating exogenous viruses in mammals have been reported (31, 32, 34). In mice harboring AKR-type MLV, there is a good correlation between intact AKR viral genomes in cellular DNA and the capacity of the cells to release infectious AKR-type MLV (34). Unlike ancient remnant endogenous retroviruses that contain numerous sequence deletions and frameshifts in their genomes (33, 56), the ZFERV genomic structure is essentially intact and contains three long ORFs for gag, pol, and env genes; abundant viral transcripts are expressed and processed correctly. Therefore, ZFERV may have the potential for generating viral progeny. In addition, the presence of a number of ZFERV-related copies in the fish genome could also be the source of viral transcripts and provide a reservoir for generating recombinant retroviruses. Such a case has been well studied in AKR mice harboring various intact and defective endogenous retroviruses. These studies indicate that expression of these endogenous retroviruses may eventually lead to the generation of leukemogenic recombinant MCF MLV (15, 17, 23-25). Since cells expressing various retroviral transcripts concurrently are more likely to generate recombinant viruses (25), the thymus in which multiple viral transcripts are coexpressed (Fig. 6), may be the potential source for such viral particles. It is also possible that a tissue other than the thymus is the primary site for ZFERV expression and that the resulting viral particles subsequently infect a permissive site, perhaps the thymus, for replication.
Endogenous retroviruses exist in almost all vertebrate genomes (14). It has been estimated that ca. 10% of the mouse and human genomes, respectively, consist of endogenous retroviral DNA (27, 52, 55). In humans, endogenous retroviruses have been thought to perform a variety of physiological roles, from being essential to placental development (and therefore critical to human survival) to tumorigenesis and induction of autoimmune diseases (26, 29, 38, 40, 43). The potential risks involving activation of endogenous retroviruses in xenotransplantation and in gene therapies are still being evaluated (33). The interaction between the host immune system and endogenous retroviruses is complex and largely unknown in most experimental systems. Given the powerful genetic and molecular tools, together with the biological features of this model system, the zebrafish may provide an alternative approach to studying these questions.
Acknowledgments
We thank S.-Y. Lin, R. Nissen, and N. Hopkins for providing the zebrafish Genome Jump DNA libraries; V. S. Hohman for the λ phage zebrafish thymus cDNA library; G. W. Warr for fish genomic DNA samples; and D. M. Parichy for Danio fish samples. We also thank S. P. Bradley, J. Chen, N. Danilova, H. N. Eisen, C. Esau, Q. Ge, F. B. Gertler, J. J. Loureiro, S. A. Menzies, F. Sacher, and L. Van Parijs for advice and N. Hopkins, L. S. Lerman, S. Schlesinger, M. Schlesinger, and A. Yesilaltay for helpful comments on the manuscript.
This study was supported in part by grant 2RO1 AI08054 from the National Institutes of Health.
REFERENCES
- 1.Akiyoshi, D. E., M. Denaro, H. Zhu, J. L. Greenstein, P. Banerjee, and J. A. Fishman. 1998. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J. Virol. 72:4503-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Al-Adhame, M. A., and Y. W. Kunz. 1977. Ontogenesis of haematopoietic sites in Brachydanio rerio (Hamilton-Buchanan) (Teleostei). Dev. Growth Differ. 19:171-179. [DOI] [PubMed] [Google Scholar]
- 3.Berkowitz, R., J. Fisher, and S. P. Goff. 1996. RNA packaging. Curr. Top. Microbiol. Immunol. 214:177-218. [DOI] [PubMed] [Google Scholar]
- 4.Bonham, L., G. Wolgamot, and A. D. Miller. 1997. Molecular cloning of Mus dunni endogenous virus: an unusual retrovirus in a new murine viral interference group with a wide host range. J. Virol. 71:4663-4670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Detrich, W. H., M. W. Kieran, F. Y. Chan, L. M. Barone, K. Yee, J. A. Runstadler, S. Pratt, D. Ransom, and L. I. Zon. 1995. Intraembryonic hematopoietic cell migration during vertebrate development. Proc. Natl. Acad. Sci. USA 92:10713-10717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Emanoil-Ravier, R., G. Mercier, M. Canivet, M. Garcette, J. Lasneret, F. Peronnet, M. Best-Belpomme, and J. Peries. 1988. Dexamethasone stimulates expression of transposable type A intracisternal retroviruslike genes in mouse (Mus musculus) cells. J. Virol. 62:3867-3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1989. PHYLIP-phylogeny inference package. Cladistics 5:164-166. [Google Scholar]
- 8.Georgopoulos, K., M. Bigby, J.-H. Wang, A. Molnar, P. Wu, S. Winandy, and A. Sharpe. 1994. The Ikaros gene is required for the development of all lymphoid lineages. Cell 79:143-156. [DOI] [PubMed] [Google Scholar]
- 9.Gilboa, E., S. W. Mitra, S. Goff, and D. Baltimore. 1979. A detailed model of reverse transcription and tests of crucial aspects. Cell 18:93-100. [DOI] [PubMed] [Google Scholar]
- 10.Golling, G., A. Amsterdam, Z. Sun, M. Antonelli, E. Maldonado, W. Chen, S. Burgess, M. Haldi, K. Artzt, S. Farrington, S.-Y. Lin, R. M. Nissen, and N. Hopkins. 2002. Insertional mutagenesis in zebrafish rapidly identifies genes essential for early vertebrate development. Nat. Genet. 31:135-140. [DOI] [PubMed] [Google Scholar]
- 11.Greenhalgh, P., and L. A. Steiner. 1995. Recombination activating gene 1 (Rag-1) in zebrafish and shark. Immunogenetics 41:54-55. [DOI] [PubMed] [Google Scholar]
- 12.Groudine, M., R. Eisenman, and H. Weintraub. 1981. Chromatin structure of endogenous retroviral genes and activation by an inhibitor of DNA methylation. Nature 292:311-317. [DOI] [PubMed] [Google Scholar]
- 13.Hart, D., G. N. Frerichs, A. Rambaut, and D. E. Onions. 1996. Complete nucleotide sequence and transcriptional analysis of the snakehead fish retrovirus. J. Virol. 70:3606-3616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herniou, E., J. Martin, K. Miller, J. Cook, M. Wilkinson, and M. Tristem. 1998. Retroviral diversity and distribution in vertebrates. J. Virol. 72:5955-5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herr, W. 1984. Nucleotide sequence of AKV murine leukemia virus. J. Virol. 49:471-478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hindmarsh, P., and J. Leis. 1999. Retroviral DNA integration. Microbiol. Mol. Biol. Rev. 63:836-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoggan, M. D., C. E. Buckler, J. F. Sears, W. P. Rowe, and M. A. Martin. 1983. Organization and stability of endogenous xenotropic murine leukemia virus proviral DNA in mouse genomes. J. Virol. 45:473-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holzschu, D. L., D. Martineau, S. K. Fodor, V. M. Vogt, P. R. Bowser, and J. W. Casey. 1995. Nucleotide sequence and protein analysis of a complex piscine retrovirus, walleye dermal sarcoma virus. J. Virol. 69:5320-5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Horwitz, B. H., M. L. Scott, S. R. Cherry, R. T. Bronson, and D. Baltimore. 1997. Failure of lymphopoiesis after adoptive transfer of NF-κB-deficient fetal liver cells. Immunity 6:765-772. [DOI] [PubMed] [Google Scholar]
- 20.Humphries, E. H., M. L. Danhof, and I. Hlozanek. 1984. Characterization of endogenous viral loci in five lines of white leghorn chickens. Virology 135:125-138. [DOI] [PubMed] [Google Scholar]
- 21.Jenkins, N. A., N. G. Copeland, B. A. Taylor, and B. K. Lee. 1982. Organization, distribution and stability of endogenous ecotropic murine leukemia virus DNA in chromosomes of Mus musculus. J. Virol. 43:26-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joulin, V., D. Bouries, J. F. Eleouet, M. C. Labastie, S. Chretien, M. G. Mattei, and P. H. Romeo. 1991. A T-cell specific TCRδ DNA binding protein is a member of the human GATA family. EMBO J. 10:1809-1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khan, A. F., F. Laigret, and C. P. Rodi. 1987. Expression of mink cell focus-forming murine leukemia virus-related transcripts in AKR mice. J. Virol. 61:876-882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan, A. S. 1984. Nucleotide sequence analysis establishes the role of endogenous murine leukemia virus DNA segments in formation of recombinant mink cell focus-forming murine leukemia viruses. J. Virol. 50:864-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laigret, F., R. Repaske, K. Boulukos, A. B. Rabson, and A. S. Khan. 1988. Potential progenitor sequences of mink cell focus-forming (MCF) murine leukemia viruses: ecotropic, xenotropic, and MCF-related viral RNAs are detected concurrently in thymus tissues of AKR mice. J. Virol. 62:376-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lan, M. S., A. Mason, R. Coutant, Q. Y. Chen, A. Vargas, J. Rao, R. Gomez, S. Chalew, R. Garry, and N. K. Maclaren. 1998. HERV-K10s and immune-mediated (type I) diabetes. Cell 95:14-16. [DOI] [PubMed] [Google Scholar]
- 27.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 28.LaPierre, L. A., D. L. Holzschu, G. A. Wooster, P. R. Bowser, and J. W. Casey. 1998. Two closely related but distinct retroviruses are associated with walleye discrete epidermal hyperplasia. J. Virol. 72:3484-3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsson, E., and G. Andersson. 1998. Beneficial role of human endogenous retroviruses: facts and hypotheses. Scand. J. Immunol. 48:329-338. [DOI] [PubMed] [Google Scholar]
- 30.Lekstrom-Himes, J., and K. G. Xanthopoulos. 1998. Biological role of the CCAAT/enhancer-binding protein family of transcription factors. J. Biol. Chem. 273:28545-28548. [DOI] [PubMed] [Google Scholar]
- 31.Levy, J. A. 1973. Xenotropic viruses: murine leukemia virusses associated with NIH Swiss, NZB, and other mouse strains. Science 182:1151-1153. [DOI] [PubMed] [Google Scholar]
- 32.Lower, R., K. Boller, B. Hasenmaier, C. Korbmacher, N. Muller-Lantzsch, J. Lower, and R. Kurth. 1993. Identification of human endogenous retroviruses with complex mRNA expression and particle formation. Proc. Natl. Acad. Sci. USA 90:4480-4484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lower, R., J. Lower, and R. Kurth. 1996. The viruses in all of us: characteristics and biological significance of human endogenous retroviruses. Proc. Natl. Acad. Sci. USA 93:5177-5184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowy, D. R., S. K. Chattopadhyay, N. M. Teich, W. P. Rowe, and A. S. Levine. 1974. AKR murine leukemia virus genome: frequency of sequences in DNA of high-, low-, and non-virus yielding mouse strains. Proc. Natl. Acad. Sci. USA 71:3555-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lowy, D. R., W. P. Rowe, N. Teich, and J. W. Hartley. 1971. Murine leukemia virus: high-frequency activation in vitro by 5-iododeoxyuridine. Science 174:155-156. [DOI] [PubMed] [Google Scholar]
- 36.Martzen, M. R., S. M. McCraith, S. L. Spinelli, F. M. Torres, S. Fields, E. J. Grayhack, and E. M. Phizicky. 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286:1153-1155. [DOI] [PubMed] [Google Scholar]
- 37.McClure, M. A., M. S. Johnson, D. F. Feng, and R. F. Doolittle. 1988. Sequence comparisons of retroviral proteins: relative rates of change and general phylogeny. Proc. Natl. Acad. Sci. USA 85:2469-2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mi, S., X. Lee, X. Li, G. M. Veldman, H. Finnerty, L. Racie, E. LaVallie, X. Y. Tang, P. Edouard, S. Howes, J. C. Keith, Jr., and J. M. McCoy. 2000. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 403:785-789. [DOI] [PubMed] [Google Scholar]
- 39.Morris, J. F., R. Hromas, and F. J. Rauscher III. 1994. Characterization of the DNA-binding properties of the myeloid zinc finger protein MZF1: two independent DNA-binding domains recognize two DNA consensus sequences with a common G-rich core. Mol. Cell. Biol. 14:1786-1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murphy, V. J., L. C. Harrison, W. A. Rudert, P. Luppi, M. Trucco, A. Fierabracci, P. A. Biro, and G. F. Bottazzo. 1998. Retroviral superantigens and type I diabetes mellitus. Cell 95:9-11. [DOI] [PubMed] [Google Scholar]
- 41.Patience, C., W. M. Switzer, Y. Takeuchi, D. J. Griffiths, M. E. Goward, W. Heneine, J. P. Stoye, and R. A. Weiss. 2001. Multiple groups of novel retroviral genomes in pigs and related species. J. Virol. 75:2771-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pehron, J. R., and R. N. Fuji. 1998. Evolutionary conservation of histone macroH2A subtypes and domains. Nucleic Acids Res. 26:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Perron, H., J. A. Garson, F. Bedin, F. Beseme, G. Paranhos-Baccala, F. Komurian-Pradel, F. Mallet, P. W. Tuke, C. Voisset, J. L. Blond, B. Lalande, J. M. Seigneurin, B. Mandrand, et al. 1997. Molecular identification of a novel retrovirus repeatedly isolated from patients with multiple sclerosis. Proc. Natl. Acad. Sci. USA 94:7583-7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schlissel, M., A. Voronova, and D. Baltimore. 1991. Helix-loop-helix transcription factor E47 activates germ-line immunoglobulin heavy-chain gene transcription and rearrangement in a pre-T-cell line. Genes Dev. 5:1367-1376. [DOI] [PubMed] [Google Scholar]
- 45.Stoye, J. P., and J. M. Coffin. 1987. The four classes of endogenous murine leukemia virus: structural relationships and potential for recombination. J. Virol. 61:2659-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stoye, J. P., and C. Moroni. 1983. Endogenous retrovirus expression in stimulated murine lymphocytes. Identification of a new locus controlling mitogen induction of a defective virus. J. Exp. Med. 157:1660-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takayama, Y., M. A. O'Mara, K. Spilsbury, R. Thwaite, P. B. Rowe, and G. Symonds. 1991. Stage-specific expression of intracisternal A-particle sequences in murine myelomonocytic leukemia cell lines and normal myelomonocytic differentiation. J. Virol. 65:2149-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka, T., K. Tanaka, S. Ogawa, M. Kurokawa, K. Mitani, J. Nishida, Y. Shibata, Y. Yazaki, and H. Hirai. 1995. An acute myleoid leukemia gene, AML1, regulates hemopoietic myeloid cell differentiation and transcriptional activation antagonistically by two alternative spliced forms. EMBO J. 14:341-350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tonjes, R. R., F. Czauderna, and R. Kurth. 1999. Genome-wide screening, cloning, chromosomal assignment, and expression of full-length human endogenous retrovirus type K. J. Virol. 73:9187-9195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ullmann, K. S., W. M. Flanagan, C. A. Edwards, and G. R. Crabtree. 1991. Activation of early gene expression in T lymphocytes by Oct-1 and an inducible protein, OAP40. Science 254:558-562. [DOI] [PubMed] [Google Scholar]
- 51.Varmus, H. E., N. Quintrell, and S. Ortiz. 1981. Retroviruses as mutagens: insertion and excision of a nontransforming provirus alter expression of a resident transforming provirus. Cell 25:23-36. [DOI] [PubMed] [Google Scholar]
- 52.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 53.Wang, Z. Q., C. Ovitt, A. E. Grigoriadis, U. Mohle-Steinlein, U. Ruther, and E. F. Wagner. 1992. Bone and haematopoietic defects in mice lacking c-fos. Nature 360:741-745. [DOI] [PubMed] [Google Scholar]
- 54.Ward, A. C., I. Touw, and A. Yoshimura. 2000. The Jak-Stat pathway in normal and perturbed hematopoiesis. Blood 95:19-29. [PubMed] [Google Scholar]
- 55.Waterston, R. H., K. Lindblad-Toh, E. Birney, J. Rogers, J. F. Abril, P. Agarwal, R. Agarwala, R. Ainscough, M. Alexandersson, P. An, S. E. Antonarakis, J. Attwood, R. Baertsch, J. Bailey, K. Barlow, S. Beck, E. Berry, B. Birren, T. Bloom, P. Bork, M. Botcherby, N. Bray, M. R. Brent, D. G. Brown, S. D. Brown, C. Bult, J. Burton, J. Butler, R. D. Campbell, P. Carninci, S. Cawley, F. Chiaromonte, A. T. Chinwalla, D. M. Church, M. Clamp, C. Clee, F. S. Collins, L. L. Cook, R. R. Copley, A. Coulson, O. Couronne, J. Cuff, V. Curwen, T. Cutts, M. Daly, R. David, J. Davies, K. D. Delehaunty, J. Deri, E. T. Dermitzakis, C. Dewey, N. J. Dickens, M. Diekhans, S. Dodge, I. Dubchak, D. M. Dunn, S. R. Eddy, L. Elnitski, R. D. Emes, P. Eswara, E. Eyras, A. Felsenfeld, G. A. Fewell, P. Flicek, K. Foley, W. N. Frankel, L. A. Fulton, R. S. Fulton, T. S. Furey, D. Gage, R. A. Gibbs, G. Glusman, S. Gnerre, N. Goldman, L. Goodstadt, D. Grafham, T. A. Graves, E. D. Green, S. Gregory, R. Guigo, M. Guyer, R. C. Hardison, D. Haussler, Y. Hayashizaki, L. W. Hillier, A. Hinrichs, W. Hlavina, T. Holzer, F. Hsu, A. Hua, T. Hubbard, A. Hunt, I. Jackson, D. B. Jaffe, L. S. Johnson, M. Jones, T. A. Jones, A. Joy, M. Kamal, E. K. Karlsson, et al. 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420:520-562. [DOI] [PubMed] [Google Scholar]
- 56.Weinberg, R. A. 1980. Origins and roles of endogenous retroviruses. Cell 22:643-644. [DOI] [PubMed] [Google Scholar]
- 57.Willett, C. E., J. J. Cherry, and L. A. Steiner. 1997. Characterization and expression of the recombination activating genes (rag1 and rag2) of zebrafish. Immunogenetics 45:394-404. [DOI] [PubMed] [Google Scholar]
- 58.Willett, C. E., A. Cortes, A. Zuasti, and A. G. Zapata. 1999. Early hematopoiesis and developing lymphoid organs in the zebrafish. Dev. Dyn. 214:323-336. [DOI] [PubMed] [Google Scholar]
- 59.Willett, C. E., A. G. Zapata, N. Hopkins, and L. A. Steiner. 1997. Expression of zebrafish rag genes during early development identifies the thymus. Dev. Biol. 182:331-341. [DOI] [PubMed] [Google Scholar]
- 60.Wolgamot, G., and A. D. Miller. 1999. Replication of Mus dunni endogenous retrovirus depends on promoter activation followed by enhancer multimerization. J. Virol. 73:9803-9809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiong, Y., and T. H. Eickbush. 1990. Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9:3353-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yoshinaka, Y., I. Katoh, T. D. Copeland, and S. J. Oroszlan. 1985. Murine leukemia virus protease is encoded by the gag-pol gene and is synthesized through suppression of an amber termination codon. Proc. Natl. Acad. Sci. USA 82:1618-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]