Abstract
Photoconvertible fluorescent proteins, such as Kaede, can be switched irreversibly from their native color to a new one. This property can be exploited to visualize de novo mRNA translation, because newly synthesized proteins can be distinguished from preexisting ones by their color. In this protocol, Kaede cDNA linked to the 3′ untranslated region (UTR) of β-actin is delivered into cells fated to become the retina by injection into Xenopus blastomeres. Brief exposure (6–10 s) to UV light (350–410 nm) of Kaede-positive retinal axons/growth cones efficiently converts Kaede from its native green fluorescence to red. The reappearance of the green signal reports the synthesis of new Kaede protein. This approach can be used to investigate the spatiotemporal control of translation of specific mRNAs in response to external stimuli and to test the efficiency of full-length versus mutant UTRs. The 3-d protocol can be adapted for broad use with other photoactivatable fluorescent proteins.
INTRODUCTION
Protein synthesis can be triggered in spatially restricted cytoplasmic compartments in neuronal processes (dendrites and axons) by external stimuli1,2. This provides an adaptable mechanism for regulating local cellular responses and requires mRNA localization. Direct visualization of protein synthesis over a short timescale (min) can yield valuable information about local protein dynamics in response to external triggers. Fluorescent reporters are essential tools in live cell imaging to study protein dynamics such as trafficking and turnover. For the study of regulated protein synthesis, the regulatory elements of the mRNAs of interest are added to the coding sequence of the fluorescent reporter to recapitulate the endogenous control of protein translation. Under regulated translational control, the changes in the fluorescence intensity of the reporter protein over time can be monitored and used to determine the changes in protein level. Most of the mRNA regulatory elements are located within the UTRs 5′ and 3′ to the coding sequence. Whereas 5′UTRs contain different regulatory elements such as the m7GpppG cap, secondary stem-loop structures, upstream open reading frames, upstream AUG codons and internal ribosome entry sites to initiate and inhibit translation, 3′UTRs have diverse regulatory elements including a poly(A)-tail, cytoplasmic polyadenylation elements and the hexanucleotide signal to control translation3. Previous studies have used the 3′UTRs of mRNAs such as the Ca2+/calmodulin-dependent kinase II-α subunit, tau, EphA2, RhoA and β-actin to control the expression of fluorescence reporters to test local protein synthesis in both dendritic and axonal neurons4–8. In β-actin mRNA, for example, the 3′UTR is particularly important because it functions not only in translation regulation, but also in mRNA localization. The localization of β-actin mRNA has been shown to require a cis-acting element located in the 3′UTR called the zipcode9,10. This sequence can target the linked mRNAs to specific cytoplasmic compartments in axons and fibroblasts8,10,11. Moreover, evidence shows that the zipcode in the 3′UTR of β-actin mRNA is important for the spatiotemporal control of β-actin translation12.
Classically, simple fluorescent reporters such as green fluorescent protein (GFP) have been used to study protein synthesis in neurons by monitoring the changes in the fluorescence intensity4,5,13. This approach can be enhanced by the photobleaching technique to optically isolate the soma from the neurite as well as fluorescence recovery after photobleaching (FRAP) for easier detection of changes in the fluorescence signal relative to the basal level after photobleaching (owing to the higher relative change compared with a lower starting level)4. With the development of properties of fluorescent reporters such as photoactivation, photoconversion and time-dependent spectral changes14–16, new methods to study protein synthesis in neurons have emerged over the past 5 years. For example, Flanagan and colleagues6 used a Fluorescent Timer protein that changes color with time to demonstrate local protein synthesis in axonal growth cones in vivo. More recently, our group and Jan and colleagues7,17 have used a photoconvertible fluorescent protein, called Kaede, to reveal directly local synthesis of new protein driven by the 3′UTRs of β-actin in retinal growth cones and the Kv1.1 channel in hippocampal dendrites. These new methods offer several advantages over FRAP. First, fluorescent proteins that switch their spectral emission allow the possibility of distinguishing between new and preexisting proteins18 and are, therefore, useful for visualizing the synthesis of new proteins. The ability to monitor the preexisting protein simultaneously provides extra information, such as degradation and diffusion of the protein, which cannot be obtained by using FRAP. This information is potentially useful for correcting the artifacts caused by degradation and diffusion, as well for providing internal controls for the translation of the new protein. Second, the time required for the irradiation of a specific wavelength of light to induce photoconversion is in the scale of seconds, and the wavelength used for photoconversion may be different from the observation wavelength. This is in contrast with photobleaching in FRAP, where the observation wavelength is used, and usually complete photobleaching requires a longer time to achieve, which can lead to phototoxic effects due to the energy release after the chromophore excitation. When fluorophores are excited, they produce reactive oxygen species that react with proteins, nucleic acids and fluorophores leading to photobleaching19–21 and, at the same time, cell cycle arrest or cell death22.
Kaede, meaning ‘maple’ in Japanese, is a photoconvertible fluorescent protein cloned from the stony coral Trachyphyllia geoffroyi15. Upon UV or violet light illumination (350–410 nm), Kaede protein changes from its native green form (Kaede-green) to a photoconverted red form (Kaede-red), like maple leaves in autumn. As this change occurs through cleavage of the peptide backbone, the photoconversion is stable15,23. His62 in the Kaede protein is essential for this photoinduced cleavage. Upon UV irradiation, the cleavage occurs at the Nα–Cα bond of the His62 residue. The subsequent loss of a proton from His62-Cβ forms a double bond between His62-Cα and Cβ, leading to the extension of the π-conjugation and, thus, a new red-emitting chromophore23. Under normal aerobic conditions, photoconverted Kaede has been shown to be irreversible after dark exposure or strong illumination at 570 nm (ref. 15). Moreover, studies using fluorescence correlation spectroscopy demonstrate that the photoconversion reaction, indeed, leads to the formation of a new chromophore24. One of the advantages of using Kaede is the large separation between the peak wavelengths required for efficient photoconversion and subsequent observation. Specifically, the excitation wavelengths (470 nm, with emission filters 510WB40 and 575ALP for green and red fluorescence, respectively) used for observation do not induce further photoconversion15. The stable photoconversion permits detection of newly synthesized protein by visualizing the return of Kaede-green, while retaining the ability to track the dynamics of the preexisting Kaede-red protein. Therefore, the use of Kaede permits quantitative measurements to be made of both preexisting and new protein by analyzing the Kaede-red and Kaede-green signals after photoconversion.
Kaede has been expressed successfully in mammalian, avian, amphibian and fish cells, demonstrating the efficient maturation of the chromophore at a wide range of temperatures7,17,18,25,26. Although the maturation time of Kaede has not been accurately measured, the most rapid maturation of a fluorescent protein reported to date, Venus, has a time constant of about 2 min at 37 °C (refs. 27,28). Therefore, there is likely a time lag between completion of translation and the detection of fluorescence expression resulting in the speed of de novo synthesis being underestimated. It should be noted that Kaede is an obligate tetramer and has a tendency to form aggregates when fused to other proteins15, rendering it less suitable for fusion protein studies. Therefore, instead of constructing a Kaede fusion protein, the reporter alone should be built under the control of the 3′UTR of interest. To extend the application of photoconvertible proteins to fusion protein studies, monomeric variants have been developed. EosFP from stony coral Lobophyllia hemprichii has been mutated to generate a monomeric version called mEosFP that forms a chromophore at temperatures below 30 °C (ref. 29). For the application of fusion proteins in bacterial and mammalian cells, dendGFP (cloned from octocoral Dendronephthya sp.) has been mutated to produce a monomeric variant, Dendra, that efficiently matures at 37 °C (ref. 30).
Different transfection techniques have been used to express fluorescent reporter constructs in neuronal cells, including microinjection, viral infection, lipofection and electroporation4,6,8,13,17,31. In Xenopus laevis, microinjection can be used to introduce DNA, mRNA and oligonucleotides, and the procedure can be performed at the single-cell stage or later, which gives the advantage of being able to transfect a large amount of cells32. Unlike other transfection techniques that require uptake of injected material through the membrane, which results in a mosaic expression pattern in fewer cells, microinjection introduces injected material directly into the cells, especially in early-stage embryos. The large size of the fertilized egg (1.4 mm diameter) and subsequent blastomeres allows the direct injection of a large volume (up to 20 nl) of DNA/mRNA, resulting in long-lasting and high levels of expression in a large number of progeny cells. Microinjection targeted to cells fated to give rise to the eyes results in high levels of transgene expression in retinal neurons, including retinal ganglion cells (RGC). Using this method, transgene-expressing eyes can be collected for in vitro culture to study local protein synthesis in retinal ganglion axons. Injected mRNAs are stable up to the neurula stage of development, and injected DNA can persist for up to 8 months33,34. As eye primodia develop only after stage 24, to study local protein synthesis in retinal axons beyond this stage, cDNA constructs were injected. The earliest stage to perform other transfection techniques for eye expression is stage 15, which does not provide sufficient time to highly express the injected DNA for screening at stage 24 to prepare primary neuronal cultures35–37. Although electroporation of RNA at stage 21 can also be used to express Kaede in the eyes, microinjection has the advantage of transfecting a larger number of cells. Therefore, microinjection is the method of choice for these experiments.
In a previous study, we used a cDNA construct of Kaede linked to the β-actin 3′UTR to observe the synthesis of new protein driven by the β-actin 3′UTR in Xenopus retinal growth cones7. Using this method, the translational regulatory functions in the UTRs of specific mRNAs can be tested. The subcloning strategy can be found in the supplementary methods section of the original paper (http://www.nature.com/neuro/journal/v9/n10/extref/nn1775-S8.pdf). In this protocol, detailed procedures of how to obtain transgene-expressing primary neurons by microinjection and analyze them with live imaging are described (Fig. 1). These procedures are divided into four parts: (i) microinjection, (ii) tissue culture, (iii) photoconversion and live imaging and (iv) data analysis. With suitable adaptations, this protocol can be applied to other studies such as cell tracking and protein trafficking in neurons. For example, tracking of a photoconverted red target (e.g., growth cone) in a green background can be performed using time-lapse imaging after photoconversion of a particular cell expressing Kaede. These procedures can be adapted to other fluorescent proteins such as Dendra.
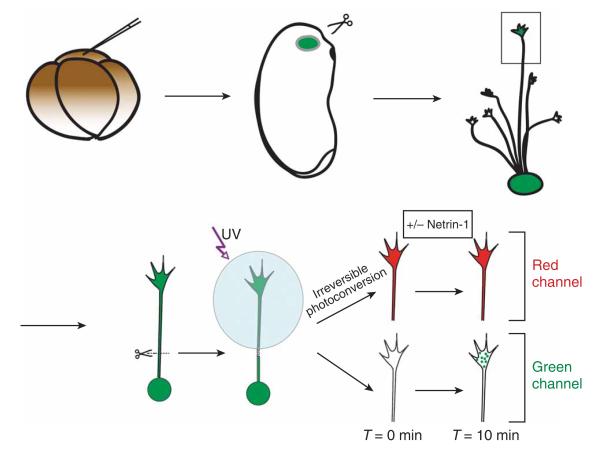
Figure 1.
A schematic diagram showing the experimental design of the protocol. A cDNA construct of Kaede is injected into the blastomeres fated to become the eyes at the four-cell stage. Embryos showing expression of Kaede-green in the eyes are selected and dissected for overnight in vitro tissue culture. Retinal growth cones expressing Kaede-green are selected and severed from the soma. Kaede-green protein is photoconverted to Kaede-red by brief UV illumination. After bath application of netrin-1 or control vehicle, the recovery of the newly synthesized Kaede-green protein is monitored over time by time-lapse imaging. (Adapted from a figure in ref. 7.)
MATERIALS
REAGENTS
Xenopus embryos (see ref. 38 for details)
 : One should observe government regulations concerning the use of animals in scientific procedures.
: One should observe government regulations concerning the use of animals in scientific procedures.Experimental constructs for injections, for example, Kaede-β-actin-3′UTR plasmid (made according to ref. 7)
Control constructs for injections, for example, to perform control experiments for the translation regulation of the 3′UTR, inject a construct with a mutated 3′UTR
Chloroform (Sigma, C2432)
Sylgard (Dow Corning, 240-1673921)
10× modified Barth’s Solution (MBS; see REAGENT SETUP)
Cysteine (Fluka, 30090)
Ficoll (Sigma, F4375)
Antibiotic–antimycotic (100×), liquid (10,000 U ml−1 penicillin, 10,000 μg ml−1 streptomycin and 25 μg ml−1 amphotericin B as fungizone, PSF) (GIBCO, 15240-062).
L15 medium (GIBCO, 11415)
3-Aminobenzoic acid ethylester methanesulfonate salt, MS222 (Sigma, A5040).
Mineral oil (Sigma, M8410)
Ethanol (Riedel-deHaën, 32221).
Poly-l-lysine (Sigma, P1274-25MG)
Laminin-1 (Sigma, L2020-1MG).
Cycloheximide (Sigma, C7698)
Albumin from bovine serum, BSA (Sigma, A2153)
Recombinant netrin-1 (made according to ref. 39)
EQUIPMENT
60-mm Petri dish (Greiner Bio-One, 628160)
Nitex mesh 1.5 mm × 1.5 mm (Sefar, 03-310/45).
Needle puller (World Precision Instruments, PUL-1)
Borosilicate glass capillary tubing, 1.0 mm outer diameter ×0.5 mm inner diameter, 100 mm length (Frederick Haer & Co., no. 27-30-1)
Forceps, no. 3 and no. 5 (Fine Science Tools)
150-mm Petri dish (Falcon, 351058)
Blu-tac
35-mm Petri dish (Falcon, 353001)
0.22-μm vaccum-driven filtration system (Millipore, SCGPU02RE)
Stereomicroscope, 0.8–5× Stemi SV6)
Eyepiece with graticule (Zeiss, W-Pl 10X/23, 455044)
Microloader, 20 μl (Eppendorf, 5424956.003)
Micropipette holder (World Precision Instruments)
Manual micromanipulator (Fine Science Tools)
Microinjector (Harvard Apparatus, PLI-100)
Disposable Pasteur pipette, 3ml (ELKay, 127-P503-000)
Hood (EdgeGard, Laminar flow 3252)
Glass-bottomed dish (MatTek, P50G-1.5-14-F)
Fluorescence dissecting microscope (Leica, MZ FLIII; ebq 100 mercury lamp)
0.15-mm insect pin (Fine Science Tools)
Pin holder (Fine Science Tools, 26018-17)
27G × 3/4 inch needle (TERUMO, NN2719R)
Incubator (Cole Parmer, ECHO Therm)
Inverted microscope (Nikon, Eclipse TE2000-U)
Fluorescence filters for UV, blue and green excitations (Nikon, UV-2E/C; B-2E/C; G-2A)
Fluorescence lamp house (Nikon, LH-M100CB-1)
Super high-pressure mercury lamp power supply (Nikon, C-SHG1)
100× objective, Plan Apochromat, numerical aperture (NA) = 1.40, phase contrast, dark contrast middle type (DM), ∞/0.17, working distance = 0.13 mm, oil immersion (Nikon)
20× objective, Plan Fluor, NA = 0.50, phase contrast, lower contrast type (DLL), ∞/0.17, working distance = 2.1 mm (Nikon)
IEEE 1394 digital charged-couple device camera (Hamamatsu, C4742-80-12AG)
Shutter system (Orbit, CS100K IM)
OpenLab software (Improvision)
ImageJ software (National Institutes of Health)
Excel software (Microsoft)
REAGENT SETUP
10× MBS
88 mM NaCl, 1 mM KCl, 2.4 mM NaHCO3, 10 mM HEPES, 820 μM MgSO4, 330 μM Ca(NO3)2, 410 μM CaCl2, pH 7.5.
0.1× MBS for microinjection
Freshly prepare 1 liter of the solution by diluting 10 ml of 10× MBS to 1 liter of water. The pH value of the concentrated buffer lowers after dilution. Add 1 M NaOH drop by drop, stirring at the same time, to adjust the pH to 7.5.
0.1× MBS for tissue culture
Prepare 250 ml of the solution with 1% (vol/vol) PSF. Adjust the pH to 7.5 by adding 1 M NaOH and filter the solution with a 0.22-μm filtration system.
2% (wt/vol) l-cysteine
Freshly prepare 50 ml of the solution on the day of injection by adding 1 g of l-cysteine to 1× MBS. Adjust the solution to pH 8 by adding 5 M NaOH.  Cysteine solution has to be freshly prepared, and the correct pH level is critical to effective dejellying.
Cysteine solution has to be freshly prepared, and the correct pH level is critical to effective dejellying.
4% (wt/vol) Ficoll
Prepare 250 ml of the solution by mixing 10 g of Ficoll into 0.1× MBS with 1% (vol/vol) PSF. Adjust the pH to 7.5 by adding 1 M NaOH. This solution is stable in alkaline and neutral solutions and takes at least 30 min to dissolve completely with stirring. Prepare it up to 2 weeks in advance and store it at 4 °C.
Culture medium
Under sterile conditions, prepare 250 ml of 60% (vol/vol) L15 with 1% (vol/vol) PSF. Adjust the pH to 7.6–7.8 and filter the solution with a 0.22-μm filtration system. Store at 4 °C.  The pH of the solution decreases with prolonged storage (1 week). The altered pH adversely affects the outgrowth of the culture. Aliquot the culture medium for storage at 4 °C for a maximum of 2 weeks.
The pH of the solution decreases with prolonged storage (1 week). The altered pH adversely affects the outgrowth of the culture. Aliquot the culture medium for storage at 4 °C for a maximum of 2 weeks.
0.04% (wt/vol) MS222
Under sterile conditions, prepare 50 ml of the solution in 1× MBS. Adjust pH to 7.5 with 1 M NaOH. Filter the solution with a 0.22-μm filtration system.  The solution is unstable. Freshly prepare it each time and keep it protected from light at 4 °C.
The solution is unstable. Freshly prepare it each time and keep it protected from light at 4 °C.
EQUIPMENT SETUP
Inverted fluorescence microscope
Set up an epifluorescence microscope. Equip a brightfield inverted microscope with phase contrast objectives and condenser annuli. Use a mercury arc lamp as illumination sources and install filter sets for UV, blue and green excitations.
The specifications of the filter blocks used are as detailed in the following table. Alternative imaging setups suitable for DAPI, GFP and rhodamine can also be used. For details of the excitation and emission spectra of Kaede, please see ref. 15.
| Excitation filter wavelengths (nm) |
Dichromatic mirror cut-on wavelength (nm) |
Barrier filter wavelengths (nm) |
|
|---|---|---|---|
| UV | 360/40 (340–380) | 400 (longpass) | 460/50 (435–485) |
| Blue | 480/30 (465–495) | 505 (longpass) | 535/40 (515–555) |
| Green | 535/50 (510–560) | 565 (longpass) | 590 (longpass) |
Connect the microscope to a cooled CCD digital camera with high resolution (1.37 million pixels) and high quantum efficiency (18,000 electrons) and low noise (6 electrons) for low light level imaging. Use imaging software (e.g., OpenLab) for image acquisition. An optional shutter system can be connected for easy control of the illumination. See EQUIPMENT for the actual equipment used.
Alternatively, set up an inverted confocal microscope equipped with standard diode (or UV), Argon and Helium–Neon lasers for excitation at wavelengths of 405 nm (or 364 nm), 488 nm and 543 nm, respectively.
Gridded dish
This is used to hold and align embryos in place for injection. Cut a circle of 56 mm diameter from the nitex mesh and put it into a 60-mm Petri dish. Secure the nitex mesh by putting four drops of chloroform in the corners to melt the plastic together. Allow it to dry before use. The gridded dishes can be cleaned with water and reused.  Chloroform is toxic. Handle with care in fume hood.
Chloroform is toxic. Handle with care in fume hood.  The grid has to be securely attached to the Petri dish to hold embryos in place to avoid unwanted movement of embryos during injection. The bottom of the Petri dishes has to be flat.
The grid has to be securely attached to the Petri dish to hold embryos in place to avoid unwanted movement of embryos during injection. The bottom of the Petri dishes has to be flat.
Micropipette
Use the micropipette puller to modify the shape of the glass capillaries. The pipette puller breaks a capillary into halves with a long thin tip (Fig. 2, panels a,b). The optimal outer diameter of the tip is 18–20 μm. To obtain this, measure the diameter of the tip under the microscope. Hold the blunt end of the micropipette with one hand. Break off about 300 μm from the tip with a pair of no. 5 forceps at a tilted angle, ideally, to obtain a tapered end for easy penetration into the cells. Store the micropipettes carefully in a container so as not to damage the tips (e.g., 150-mm Petri dish with blu-tac at the bottom to hold the middle of each micropipette individually and securely).
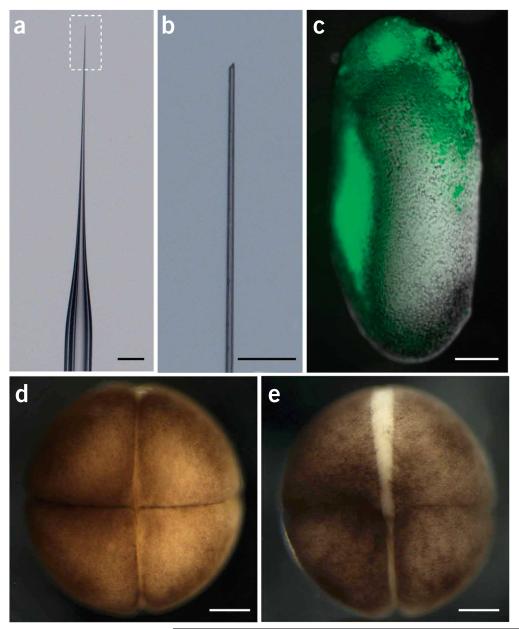
Figure 2.
Images of micropipettes and Xenopus embryos. (a) Modification of the shape of the micropipette. (b) Higher magnification of the boxed area in a showing the tapered shape of the tip. (c) A stage-24 embryo expressing Kaede-green in the eye. (d) A good-quality embryo has tight cell junctions. (e) A poor-quality embryo has white gaps between cells. Scale bars, 1 mm in a and 200 μm in b–e.
Sylgard dish
Mix Sylgard according to the manufacturer’s manual and half-fill a 35-mm Petri dish with the mixture. Leave the mixture to set at 60 °C overnight.
PROCEDURE
Microinjection
1| After fertilization, monitor the development of the embryos closely to catch the right time for dejellying. The embryos can be dejellied (see Step 6) at the four-cell stage, just less than 2 h after fertilization. The protocol for dejellying and microinjection described here are similar to methods described in refs. 32,38.
 All procedures are performed at room temperature (20–21 °C), unless specified.
All procedures are performed at room temperature (20–21 °C), unless specified.
2| While monitoring the embryos periodically, calibrate the volume of injection as follows. Aim to inject 5 nl per cell at the four- or eight-cell stage, but reduce the volume for injection at later stages. Inject a maximum of 500 pg of Kaede DNA or 1 ng of RNA construct and adjust the concentration accordingly.
 The larger the amount of construct injected, the more toxic it is to the embryos. Therefore, it is important to keep quantities of injected material to a minimum.
The larger the amount of construct injected, the more toxic it is to the embryos. Therefore, it is important to keep quantities of injected material to a minimum.
 Injection of DNA results in a more mosaic expression. The constructs are transcribed only after the mid-blastula transition and last for a longer time. It is suitable for translation studies beyond neurula stage (see INTRODUCTION). Injection of RNA results in earlier onset and more homogeneous expression of the transgene. Although the protein expression from RNA injection persists beyond stage 40, injected RNAs are usually degraded before the neurula stage. Therefore, to study translation, RNA injection is only suitable for studies with embryos before neurula stage.
Injection of DNA results in a more mosaic expression. The constructs are transcribed only after the mid-blastula transition and last for a longer time. It is suitable for translation studies beyond neurula stage (see INTRODUCTION). Injection of RNA results in earlier onset and more homogeneous expression of the transgene. Although the protein expression from RNA injection persists beyond stage 40, injected RNAs are usually degraded before the neurula stage. Therefore, to study translation, RNA injection is only suitable for studies with embryos before neurula stage.
3| Fill the micropipette with 3–5 μl injection solution. To back-fill the micropipette, use a microloader; this option is faster, but may introduce bubbles in the micropipette more easily. Alternatively, fill the micropipette by suction using the microinjector. This option prevents air bubble formation, but it takes a long time (about 20 min) to fill the required volume.
4| Insert the micropipette into the micropipette holder attached to the micromanipulator. Position the micromanipulator to one side of the microscope. Lower the filled micropipette into a Petri dish filled with mineral oil until the tip is just immersed. Apply pressure with the microinjector. A small spherical drop will form at the pipette tip in the oil.
5| Calibrate the volume of the drop to 5 nl per pulse by adjusting the pressure and duration of injection, which vary considerably depending on the micropipette. The diameter of a 5-nl sphere is 214 μm. Alternatively, inject a drop formed by 2 pulses into the oil and calibrate the sphere to 270 μm diameter. A guideline to the PLI-100 Microinjector settings is that an injection pressure of 15–60 PSI requires injection times of 10–70 ms. After calibration, remove the micropipette from the mineral oil. Put aside the dish of mineral oil.
6| For injections at stages between the 4-cell stage and the 32-cell stage, dejelly embryos when the second division is finished. At the four-cell stage (2 h postfertilization), remove the buffer in the Petri dish holding the embryos and replace it with 5 ml of 2% (wt/vol) cysteine (pH 7.8–8.0). Make sure that the solution covers the embryos completely. The embryos can also be dejellied at the two-cell stage (1.5 h postfertilization). However, early dejellying is likely to increase the death rate.
 There is about 15 min before the cells divide at the four-cell stage. Perform Steps 6–11 quickly to obtain the embryos at the desired stage.
There is about 15 min before the cells divide at the four-cell stage. Perform Steps 6–11 quickly to obtain the embryos at the desired stage.
 Do not add cysteine when cells are dividing. This will damage the embryos.
Do not add cysteine when cells are dividing. This will damage the embryos.
7| Leave the cysteine for 3–5 min and swirl the dish slightly until the embryos become less sticky. Cysteine is a reducing agent that breaks disulfide bonds of mucopolysaccharides, resulting in fragmentation of the jelly coat.
 Do not leave the embryos in cysteine for any longer than required. Cysteine damages the embryos after the jelly coat is removed. It is advisable to monitor the embryos during the process. Proceed to the next step immediately when the fragmented jelly coats can be seen in the solution.
Do not leave the embryos in cysteine for any longer than required. Cysteine damages the embryos after the jelly coat is removed. It is advisable to monitor the embryos during the process. Proceed to the next step immediately when the fragmented jelly coats can be seen in the solution.
8| Once the embryos swirl around freely, pour them into a 250-ml beaker with about 100 ml of 0.1× MBS. Decant the solution quickly and rinse the embryos five more times with 100 ml of 0.1× MBS to remove trace amounts of cysteine. Pour the cleaned embryos into a clean Petri dish.
9| Pour just enough 4% (wt/vol) Ficoll into the gridded dishes to cover the bottom. Tap the dish on the bench several times to get rid of the bubbles formed in the grid. Alternatively, scrape the grid with a transfer pipette to remove bubbles. Fill the dish halfway with 4% (wt/vol) Ficoll. Injections are performed in 4% (wt/vol) Ficoll to prevent leakage from the injection site.
10| Select 50 healthy, dejellied embryos and transfer them into the gridded dish using a 3-ml transfer pipette. Transfer some healthy embryos to a separate dish; these serve as uninjected controls. Uninjected controls provide information of wild-type viability after dejellying.
 Select only good-quality embryos to increase the survival rate. Good-quality embryos have deep junctions between cells. A white gap between cells is an indication of embryos of inferior quality, which are more likely to die, although sometimes these embryos get themselves repaired after a while (Fig. 2d and e).
Select only good-quality embryos to increase the survival rate. Good-quality embryos have deep junctions between cells. A white gap between cells is an indication of embryos of inferior quality, which are more likely to die, although sometimes these embryos get themselves repaired after a while (Fig. 2d and e).
11| Align and orient the embryos gently with a pair of coarse forceps under the microscope so that the animal side is up and the dorsal side is to the right (i.e., two smaller and lighter cells on the right). The animal dorsal cells will give rise to the eyes40. The alignment of the embryos is easiest at the four-cell stage. It is also possible to do it at the eight-cell stage.
At stages beyond this point, it is more difficult to distinguish the appropriate orientation.
 After aligning the embryos, avoid moving the dish. If it has to be moved, do it slowly and gently so that the embryos stay in their aligned position.
After aligning the embryos, avoid moving the dish. If it has to be moved, do it slowly and gently so that the embryos stay in their aligned position.
12| Inject 5 nl (1 pulse) of the solution into the cytoplasm of one or both animal dorsal cells at the four-cell stage (maximum 10 nl). Push the micropipette through the cell membrane and withdraw it slightly until the cell surface is flat and only the tip is inside the cell. Make a pulse to inject the solution, and remove the micropipette quickly. This withdrawal motion before injection loosens the micropipette to allow fast removal of the micropipette from the cell to prevent backflow. Continue to the next embryo by moving the dish.
 Check the flow of the micropipette between each (row of) injection because the tissue may block the micropipette. To do this, make a pulse in the 4% (wt/vol) Ficoll; the injection solution can be seen leaving the micropipette due to the different densities of the solutions. If the injected material contains a colored dye, a colored liquid sphere can be visualized within the mineral oil during the calibration, and the colored liquid can be seen in the 4% (wt/vol) Ficoll when checking the flow of the micropipette. However, due to the pigmentation of the embryos, the dye is barely visible when injected into the blastomeres.
Check the flow of the micropipette between each (row of) injection because the tissue may block the micropipette. To do this, make a pulse in the 4% (wt/vol) Ficoll; the injection solution can be seen leaving the micropipette due to the different densities of the solutions. If the injected material contains a colored dye, a colored liquid sphere can be visualized within the mineral oil during the calibration, and the colored liquid can be seen in the 4% (wt/vol) Ficoll when checking the flow of the micropipette. However, due to the pigmentation of the embryos, the dye is barely visible when injected into the blastomeres.
 Inject appropriate control constructs to separate batches of embryos to perform control experiments at this stage.
Inject appropriate control constructs to separate batches of embryos to perform control experiments at this stage.
13| Keep the embryos in 4% (wt/vol) Ficoll for about 1 h after injection until the puncture wound is healed. The higher density of the solution collapses the vitelline space and increases the pressure on the embryo to prevent leakage of cytoplasm. Do not raise the embryos in 4% (wt/vol) Ficoll because this often causes developmental defects.
14| Rinse the injected embryos three times with 0.1× MBS. Gene expression from injected DNA constructs is usually detected within 6 h, whereas gene expression from injected RNA constructs is generally observed sooner.
 Protect the embryos from natural light as it can convert the color of Kaede15.
Protect the embryos from natural light as it can convert the color of Kaede15.
15| Raise the embryos in 0.1× MBS at 18–21 °C overnight until they reach stage 24 to perform tissue culture. Meanwhile, proceed with dish coating (see below).
Tissue culture
16| Disinfect the tissue culture hood by UV illumination for 30 min and clean the worktop and all the apparatus with 70% (vol/vol) ethanol. Flame all the metal tools including forceps and dissection pins.
 Perform Steps 17–25 (except 19) under sterile conditions.
Perform Steps 17–25 (except 19) under sterile conditions.
17| Coat ten glass-bottomed dishes with 10 μg ml−1 poly-l-lysine overnight at room temperature.
18| Remove poly-l-lysine and allow the dish to dry. Coat the dish with 10 μg ml−1 laminin-1 in L15 for 1–3 h.
19| In the meantime, check the fluorescence signal of the Kaede protein in the embryos under a fluorescence dissecting microscope with blue excitation light and select embryos that have strong expression in the eyes (Fig. 2c).
 Do not expose the embryos to UV light to avoid converting the Kaede-green protein to Kaede-red.
Do not expose the embryos to UV light to avoid converting the Kaede-green protein to Kaede-red.
20| Remove laminin-1 from the glass-bottomed dish and rinse the dish with culture medium once. Add 400 μl of culture medium into the glass bottom of the dish. The solution forms a convex meniscus.
21| Remove the vitelline membrane of the selected embryos with forceps. Rinse the embryos three times in 0.1× MBS containing 1% (vol/vol) PSF and then anesthetize them with 0.04% (wt/vol) MS222. Secure the embryos with dissection pins in a Sylgard dish containing 1:1 culture medium and MS222.
22| Remove the skin covering the eyes and dissect the eye primordia of the embryos using 0.15-mm dissecting pins. Keep the eyes intact for the culture to avoid non-RGC sending processes outside the eyes on the glass-bottomed dish. If explants have to be used, cut the eyes into 3–4 pieces with a 27G needle or dissection pins.
23| Fill a P200 pipette with 100 μl of culture medium in the pipette tip. Expel a small amount of the solution from the tip and then collect the dissected eyes in the half-filled pipette tip.
 Drawing the eyes into an empty pipette tip will cause the eyes to stick to the wall of the pipette tip.
Drawing the eyes into an empty pipette tip will cause the eyes to stick to the wall of the pipette tip.
24| Wash the eyes three times in culture medium and place 3–5 eyes or 8–10 explants with the RGC side down on each glass-bottomed culture dish with culture medium. Space out the tissues in the middle of the culture dish to allow sufficient space for the axons to grow without touching each other.
 For experiments where guidance cues will be added, only one growth cone can be used in each dish. It is therefore sensible to put just as many eyes in each culture dish that will result in one Kaede-expressing growth cone and prepare more dishes.
For experiments where guidance cues will be added, only one growth cone can be used in each dish. It is therefore sensible to put just as many eyes in each culture dish that will result in one Kaede-expressing growth cone and prepare more dishes.
25| Leave the culture dishes in the hood for at least 30 min to allow the eye tissues to attach to the glass before moving them. Culture the eye tissues for 14 h in a 20 °C incubator. See also ref. 38 for information about tissue culture.
Imaging
26| Transfer the glass-bottomed dish to the inverted microscope for live imaging. To avoid forming a convex meniscus that interferes with the optics due to refraction, remove excess culture medium until the surface is flat. The minimal volume required to keep the culture completely under the medium is 200 μl and it is optimal to reduce the amount of reagent needed for stimulation.
27| Using phase optics, visualize the growth cones at ×100 magnification and locate those that are healthy. A healthy growth cone should have several motile filopodia and have a core width of about 5 μm.
28| Using a blue excitation filter set, illuminate the growth cones and select one that expresses Kaede protein. Use neutral density filters to attenuate the intensity of light to minimize phototoxicity and bleaching of the fluorescence signal. Reduce the size of the aperture so that only the center of the field of view is illuminated. This prevents bleaching of the surrounding fluorescence signal. A shutter system is an optional recommendation for convenient control of illumination.
29| Under the 20× objective, remove the cell body of the growth cone using an insect pin held by a pin holder. This can be done either by hand or by mounting the pin holder to the micromanipulator. When cutting the neurite by hand, rest both hands on the stage of the microscope. It also helps to use one hand to hold the pin and the other to stabilize it. If the micromanipulator is used, a glass micropipette can also be used instead of an insect pin. Scrape/touch the bottom of the dish to cut the neurite by moving the pin back and forth. The cut should be made near the cell body end to obtain a long neurite, because growth cones with long neurites survive better than the ones with short neurites. A 20× (NA = 0.50) objective provides a good field of view with sufficient magnification and resolution to observe the morphology for cutting.
 It is very tricky to make a clean cut because the neurites are elastic and they stick to the dissection pins.
It is very tricky to make a clean cut because the neurites are elastic and they stick to the dissection pins.
As a result, the neurites are often dragged and pulling other tissue in the culture dish. Dip the pin into 0.1% (wt/vol) BSA in medium before cutting to minimize the chances of sticking. Despite this precaution, cutting the neurite is still very difficult and, unfortunately, it is successful less than half of the time.
30| After axotomy, assess the health of the severed neurite, preferably at a high magnification (×40 or above).
A healthy neurite is straight and continuous with small branches. An unhealthy neurite is ‘beaded’ with multiple swellings along its length.
31| Under the 100× objective capture a phase image and fluorescent images of the severed growth cone with blue and green excitations. These are the images before photoconversion. The following image acquisition settings are guidelines only; the end user should determine the parameters depending on the expression of the construct and the actual equipment used.
To start with, take 12-bit images using exposure time 300 ms, neutral density ND4, gain 255, offset 0 and binning 0, and adjust accordingly. The dynamic range of the CCD camera used is 2,250:1. In certain models of the inverted microscope, the built-in ×1.5 intermediate magnification dial can be used to increase the magnification. Therefore, a 60× objective can be used to replace the 100× objective. If the microscope is equipped with a photomask reticle, use it to aid respositioning the growth cone to the original position after photoconversion.
32| Switch to the UV filter set. Use neutral density filters ND 12 to attenuate the intensity of the UV excitation light to 3% transmission to protect the growth cone from phototoxicity. While observing the growth cone, expose it to the UV excitation light for 6–10 s and photoconvert Kaede-green to Kaede-red18. Turn on the bright-field light source at the same time if necessary. Move the stage to expose the adjacent segment and repeat the illumination until the severed end of the neurite is reached.
 The required exposure time and UV excitation intensity vary depending on the magnification and the NA of the objective lens used. For each new set of experiments, determine the suitable intensity of the UV excitation by testing the photoconversion with the lowest UV excitation and increasing it until the photoconversion is achieved within 10 s with no sign of phototoxicity of the growth cone. Growth cones damaged by phototoxicity can be distinguished by the cessation of motility, the withdrawal of filopodia and the beaded appearance of neurites due to the rapid formation of large swellings. To minimize phototoxicity of the growth cone and the neurite, always minimize the amount of UV excitation light used.
The required exposure time and UV excitation intensity vary depending on the magnification and the NA of the objective lens used. For each new set of experiments, determine the suitable intensity of the UV excitation by testing the photoconversion with the lowest UV excitation and increasing it until the photoconversion is achieved within 10 s with no sign of phototoxicity of the growth cone. Growth cones damaged by phototoxicity can be distinguished by the cessation of motility, the withdrawal of filopodia and the beaded appearance of neurites due to the rapid formation of large swellings. To minimize phototoxicity of the growth cone and the neurite, always minimize the amount of UV excitation light used.
33| Switch to the blue excitation filter to check if the green Kaede protein signal has already been converted. If a trace amount of green is still visible, repeat Step 32 once. To check the efficiency of the photoconversion, define a region of interest (ROI) within the growth cone and measure the fluorescence intensity of Kaede-green in the selected region before proceeding to the next step. This value should be similar to the surrounding background area (±10%).
 Repeated UV irradiation to the growth cone increases the chance of impairing its health.
Repeated UV irradiation to the growth cone increases the chance of impairing its health.
34| Return the growth cone to the same position as in Step 31. Capture the first frame of the phase image and fluorescence images with blue and green excitations (time point, −1 min). Add netrin-1 into the bath carefully after 1 min, and capture the phase and fluorescence images again. These images are designated as time point 0 min. The fluorescence intensity level of the Kaede-green signal is very low after photoconversion and is barely detectable by eye. A highly sensitive cooled-CCD camera is required to detect low levels of light for imaging.
 Do not touch the culture dish or the microscope stage when adding reagents. Be careful to keep the growth cone in the same position when imaging for easy comparison from this point until the end of the experiment.
Do not touch the culture dish or the microscope stage when adding reagents. Be careful to keep the growth cone in the same position when imaging for easy comparison from this point until the end of the experiment.
35| Capture images every 2.5 or 5 min for at least 30 min. The severed growth cone can survive for up to 2.5 h.  Save digital images for later analysis.
Save digital images for later analysis.
36| Repeat Steps 26–35 for each experimental group. For experimental groups where translation is inhibited, remember to add protein synthesis inhibitor, for example, cycloheximide (25 μM), into the medium bath just before photoconversion (before Step 32) to test netrin-1-induced translation responses. Guidance cues may stimulate post-translational processing of premature Kaede such as torsional adjustments, dehydration and oxidation for fluorophore formation, resulting in the reappearance of Kaede-green without synthesizing new protein. Therefore, protein synthesis inhibitor is added before photoconversion to minimize the possibility of including signals from conformation maturation of preexisting protein. These results are used to provide evidence that the recovery of Kaede-green signal is dependent on local translation.
37| Repeat Steps 26–35 for appropriate controls. For example, in a separate set of cultures, add control vehicle to perform a control experiment for the netrin-1-induced translation driven by the 3′UTR. To perform control experiments for the translation regulation of the 3′UTR, use growth cones from embryos injected with a construct of mutated 3′UTR.
Analysis of the fluorescence intensity of images
38| Perform quantitative immunofluorescence measurement using imaging software, such as ImageJ (free download from http://rsb.info.nih.gov/ij/download.html) or OpenLab. Define the ROI on the phase image by tracing the outline of the growth cone manually using ‘Freehand Tool’. Although there are some common functions in different software programs that outline objects by detecting the threshold of the intensity on the image, most of them are not able to detect the outline of the growth cone, possibly due to the complexity of the morphology. Measure the mean intensity (expressed as pixels per unit area) of the corresponding fluorescence image with the same outline. As the growth cone moves and changes shape over time, modify the ROI on each phase image at different time points for measurement of the corresponding fluorescence image. Measure the background intensity level in the surrounding area of the growth cone of each fluorescence image for background correction.
39| Plot data as percentage change of fluorescence intensity (F1–F0)/F0, where F0 is the fluorescence intensity at 0 min and F1 is that of the subsequent time points.
Day 1
Steps 1–5: 15 min
Steps 6–8: 10 min
Steps 9–11: 5 min
Step 12, variable: injecting 50 embryos takes 10–15 min.
Step 13: 0.5–2 h
Step 14: 5 min
Step 15: 1 d
Step 16: 45 min
Step 17: 5 min
Day 2
Step 18: 1–3 h
Step 19: 15–30 min
Steps 20–24: 1–2 h
Step 25: 14–20 h
Day 3
Steps 26–28: 15 min
Steps 29–30: 10 min
Steps 31–37: 45 min to 3 h
Steps 38–39, variable: tracing of each frame takes 1–2 min
Troubleshooting advice can be found in Table 1.
TABLE 1.
Troubleshooting table.
| Step | Problem | Possible reason | Solution |
|---|---|---|---|
| 5 | The drop formed for calibration is too small |
Incorrect magnification is used for calibration |
Make sure that the correct magnification is used |
| The opening of the micropipette is too small |
Break the tip of the micropipette a little further |
||
| A string of bubbles come out from the micropipette and do not form one large drop |
The opening of the micropipette is too small |
Break the tip of the micropipette a little further |
|
| 8 | Jelly coat is not removed after incubating with cysteine solution |
The pH of the cysteine solution is incorrect |
Adjust the pH of the cysteine to 8 |
| Embryos die after dejellying | Embryos are not completely divided during dejellying |
Turn the embryos over to the vegetal view to ensure that the cell division is completed |
|
| Embryo quality is not good enough | Use embryos laid by another frog | ||
| 11 and 12 | The micropipette sticks to the embryos. Or, embryos stick to each other |
Jelly coat is not completely removed | Leave cysteine slightly longer until embryos swirl freely before washing it off |
| 34 | Growth cones collapse during time-lapse imaging |
The amount of excitation light is too strong |
Attenuate the amount of excitation light with neutral density filters |
ANTICIPATED RESULTS
The survival rate of the embryos after dejellying depends on the embryo quality and varies from 0% to over 90%. The mechanical damage induced by microinjection can be assessed by comparing injected embryos (with water or control constructs) to dejellied uninjected embryos. Microinjection usually increases the abnormalities or the death rate of the dejellied embryos by about 10%. After injection, the expression of the Kaede construct linked to the β-actin 3′UTR can be detected by stage 10. Kaede construct is expected to be detected in over 80% of surviving embryos. At stage 24, the expression of the construct can be detected in the eyes (Fig. 2c). Tissue cultures from these eyes give rise to growth cones that express the construct. Usually only less than 5% of these growth cones express detectable Kaede. The use of a camera with high sensitivity allows time-lapse imaging of the fluorescent signals of the growth cones. The Kaede-green signal is intense and the Kaede-red is undetectable/low (200–400 pixels) before UV-induced photoconversion (Fig. 3a–f). After UV illumination, the two signals are reversed, that is, the Kaede-green is undetectable/low (200–400 pixels) and the Kaede-red is intense. The level of autofluorescence in the nontransfected neurons is similar to the signal detected in the green channel after photoconversion and in the red channel before photoconversion—around 200–400 pixels, which is ±10% of the surrounding background. In the case that Kaede is driven by the 3′UTR of β-actin, the Kaede-green signal increases with time after stimulation with netrin-1 (within minutes), demonstrating local protein synthesis in severed growth cones7 (Fig. 3g–l). In the control conditions, a decrease in the fluorescence intensity is detected over time, suggesting photobleaching and/or protein degradation during the assay. Therefore, the measured value of fluorescence intensity is lower than the actual increase that results from local protein synthesis in the axonal growth cone. Consistent with this, there is a small decrease (less than 15%) in the red signal over time, which is accounted for by photobleaching and/or protein degradation. To distinguish the difference between photobleaching and degradation, one can do the experiment in the presence of protein degradation inhibitors. In experiments where intact neurons are used and only the growth cones are photoconverted, recovery of the Kaede-green signal is observed within 1 min, implying protein diffusion. Diffusion of proteins from the soma results in overestimation of the amount of newly synthesized protein. Therefore, it is essential to sever the axons from the cell bodies (Step 29). On the other hand, diffusion of proteins away from the growth cone may lead to underestimation of the amount of protein synthesized. Plotting the change of the fluorescence intensity of Kaede-green at each time point relative to that of time 0 min shows the rate of protein synthesis as seen by the recovery of the Kaede-green protein in isolated growth cones (Fig. 3m).
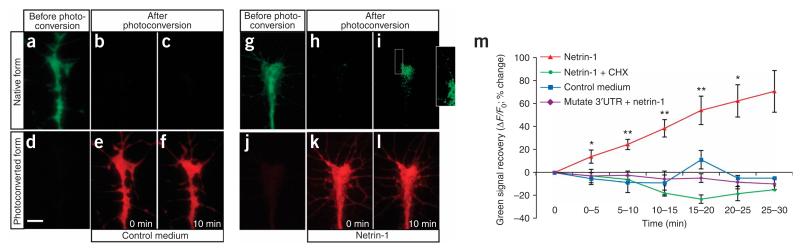
Figure 3.
Typical results of netrin-1-induced synthesis of Kaede driven by β-actin 3′UTR. (a–l) Time-lapse images showing growth cones expressing Kaede driven by β-actin 3′UTR. (a,d) Before photoconversion, the native Kaede-green signal is intense and the Kaede-red signal is low. (b,e) After photoconversion, the Kaede-green signal is low and Kaede-red is intense. (c,f) Bath application of control medium does not change either Kaede-green or Kaede-red. (i,l) Bath application of netrin-1 induces Kaede-green recovery in 10 min, but does not change Kaede-red. (i, boxed area and inset) Recovery is also observed in some filopodia. The exposure gain for the inset picture is increased for visualization of filopodia. (m) Graph showing that the recovery of Kaede-green signal is stimulated by netrin-1, but not control medium. The increase is not observed in growth cones expressing a mutated construct and is blocked by CHX. Data are presented as percentage change of the fluorescence intensity (F) over time. CHX, cycloheximide. Mutated 3′UTR, Kaede construct linked to a mutated β-actin 3′UTR. *P<0.05, **P<0.01, Mann–Whitney test. Error bars, s.e.m. Scale bar, 5 μm. (g–m are adapted from a figure in ref. 7).
ACKNOWLEDGMENTS
We thank M. Spira for suggesting the use of Kaede. We also thank M. Agathocleous, J. Falk, L. Leung, A. Lin and F. van Horck for critical reading of the manuscript. This work was supported by a Croucher Scholarship (K.-M.L.) and a Wellcome Trust Programme Grant (C.E.H.).
References
- 1.Piper M, Holt C. RNA translation in axons. Annu. Rev. Cell. Dev. Biol. 2004;20:505–523. doi: 10.1146/annurev.cellbio.20.010403.111746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steward O, Schuman EM. Compartmentalized synthesis and degradation of proteins in neurons. Neuron. 2003;40:347–359. doi: 10.1016/s0896-6273(03)00635-4. [DOI] [PubMed] [Google Scholar]
- 3.Wilkie GS, Dickson KS, Gray NK. Regulation of mRNA translation by 5′- and 3′-UTR-binding factors. Trends Biochem. Sci. 2003;28:182–188. doi: 10.1016/S0968-0004(03)00051-3. [DOI] [PubMed] [Google Scholar]
- 4.Aakalu G, Smith WB, Nguyen N, Jiang C, Schuman EM. Dynamic visualization of local protein synthesis in hippocampal neurons. Neuron. 2001;30:489–502. doi: 10.1016/s0896-6273(01)00295-1. [DOI] [PubMed] [Google Scholar]
- 5.Aronov S, Aranda G, Behar L, Ginzburg I. Visualization of translated tau protein in the axons of neuronal P19 cells and characterization of tau RNP granules. J. Cell Sci. 2002;115:3817–3827. doi: 10.1242/jcs.00058. [DOI] [PubMed] [Google Scholar]
- 6.Brittis PA, Lu Q, Flanagan JG. Axonal protein synthesis provides a mechanism for localized regulation at an intermediate target. Cell. 2002;110:223–235. doi: 10.1016/s0092-8674(02)00813-9. [DOI] [PubMed] [Google Scholar]
- 7.Leung KM, et al. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu KY, et al. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kislauskis EH, Li Z, Singer RH, Taneja KL. Isoform-specific 3′-untranslated sequences sort alpha-cardiac and beta-cytoplasmic actin messenger RNAs to different cytoplasmic compartments. J. Cell Biol. 1993;123:165–172. doi: 10.1083/jcb.123.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kislauskis EH, Zhu X, Singer RH. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J. Cell Biol. 1994;127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bassell GJ, et al. Sorting of beta-actin mRNA and protein to neurites and growth cones in culture. J. Neurosci. 1998;18:251–265. doi: 10.1523/JNEUROSCI.18-01-00251.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huttelmaier S, et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 13.Job C, Eberwine J. Identification of sites for exponential translation in living dendrites. Proc. Natl. Acad. Sci. USA. 2001;98:13037–13042. doi: 10.1073/pnas.231485698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Terskikh A, et al. ‘Fluorescent timer’: protein that changes color with time. Science. 2000;290:1585–1588. doi: 10.1126/science.290.5496.1585. [DOI] [PubMed] [Google Scholar]
- 15.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc. Natl. Acad. Sci. USA. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lukyanov KA, Chudakov DM, Lukyanov S, Verkhusha VV. Innovation: photoactivatable fluorescent proteins. Nat. Rev. Mol. Cell Biol. 2005;6:885–891. doi: 10.1038/nrm1741. [DOI] [PubMed] [Google Scholar]
- 17.Raab-Graham KF, Haddick PC, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- 18.Hatta K, Tsujii H, Omura T. Cell tracking using a photoconvertible fluorescent protein. Nat. Protoc. 2006;1:960–967. doi: 10.1038/nprot.2006.96. [DOI] [PubMed] [Google Scholar]
- 19.Bernas T, Zarebski M, Dobrucki JW, Cook PR. Minimizing photobleaching during confocal microscopy of fluorescent probes bound to chromatin: role of anoxia and photon flux. J. Microsc. 2004;215:281–296. doi: 10.1111/j.0022-2720.2004.01377.x. [DOI] [PubMed] [Google Scholar]
- 20.Song L, Varma CA, Verhoeven JW, Tanke HJ. Influence of the triplet excited state on the photobleaching kinetics of fluorescein in microscopy. Biophys. J. 1996;70:2959–2968. doi: 10.1016/S0006-3495(96)79866-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song L, van Gijlswijk RP, Young IT, Tanke HJ. Influence of fluorochrome labeling density on the photobleaching kinetics of fluorescein in microscopy. Cytometry. 1997;27:213–223. [PubMed] [Google Scholar]
- 22.Dixit R, Cyr R. Cell damage and reactive oxygen species production induced by fluorescence microscopy: effect on mitosis and guidelines for non-invasive fluorescence microscopy. Plant J. 2003;36:280–290. doi: 10.1046/j.1365-313x.2003.01868.x. [DOI] [PubMed] [Google Scholar]
- 23.Mizuno H, et al. Photo-induced peptide cleavage in the green-to-red conversion of a fluorescent protein. Mol. Cell. 2003;12:1051–1058. doi: 10.1016/s1097-2765(03)00393-9. [DOI] [PubMed] [Google Scholar]
- 24.Dittrich PS, Schafer SP, Schwille P. Characterization of the photoconversion on reaction of the fluorescent protein Kaede on the single-molecule level. Biophys. J. 2005;89:3446–3455. doi: 10.1529/biophysj.105.061713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sato T, Takahoko M, Okamoto H. HuC:Kaede, a useful tool to label neural morphologies in networks in vivo. Genesis. 2006;44:136–142. doi: 10.1002/gene.20196. [DOI] [PubMed] [Google Scholar]
- 26.Stark DA, Kulesa PM. An in vivo comparison of photoactivatable fluorescent proteins in an avian embryo model. Dev. Dyn. 2007;236:1583–1594. doi: 10.1002/dvdy.21174. [DOI] [PubMed] [Google Scholar]
- 27.Nagai T, et al. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 28.Wachter RM. Chromogenic cross-link formation in green fluorescent protein. Acc. Chem. Res. 2007;40:120–127. doi: 10.1021/ar040086r. [DOI] [PubMed] [Google Scholar]
- 29.Nienhaus GU, et al. Photoconvertible fluorescent protein EosFP: biophysical properties and cell biology applications. Photochem Photobiol. 2006;82:351–358. doi: 10.1562/2005-05-19-RA-533. [DOI] [PubMed] [Google Scholar]
- 30.Gurskaya NG, et al. Engineering of a monomeric green-to-red photoactivatable fluorescent protein induced by blue light. Nat. Biotechnol. 2006;24:461–465. doi: 10.1038/nbt1191. [DOI] [PubMed] [Google Scholar]
- 31.Sahly I, Erez H, Khoutorsky A, Shapira E, Spira ME. Effective expression of the green fluorescent fusion proteins in cultured Aplysia neurons. J. Neurosci. Methods. 2003;126:111–117. doi: 10.1016/s0165-0270(03)00072-4. [DOI] [PubMed] [Google Scholar]
- 32.Vize PD, Melton DA, Hemmati-Brivanlou A, Harland RM. Assays for gene function in developing Xenopus embryos. Methods Cell Biol. 1991;36:367–387. doi: 10.1016/s0091-679x(08)60288-5. [DOI] [PubMed] [Google Scholar]
- 33.Etkin LD, Pearman B. Distribution, expression and germ line transmission of exogenous DNA sequences following microinjection into Xenopus laevis eggs. Development. 1987;99:15–23. doi: 10.1242/dev.99.1.15. [DOI] [PubMed] [Google Scholar]
- 34.Harland R, Misher L. Stability of RNA in developing Xenopus embryos and identification of a destabilizing sequence in TFIIIA messenger RNA. Development. 1988;102:837–852. doi: 10.1242/dev.102.4.837. [DOI] [PubMed] [Google Scholar]
- 35.Holt CE, Garlick N, Cornel E. Lipofection of cDNAs in the embryonic vertebrate central nervous system. Neuron. 1990;4:203. doi: 10.1016/0896-6273(90)90095-w. [DOI] [PubMed] [Google Scholar]
- 36.Ohnuma S, Mann F, Boy S, Perron M, Harris WA. Lipofection strategy for the study of Xenopus retinal development. Methods. 2002;28:411–419. doi: 10.1016/s1046-2023(02)00260-8. [DOI] [PubMed] [Google Scholar]
- 37.Nieuwkoop PD, Faber J. The Normal Table of Xenopus laevis (Daudin) Garland Publishing Inc.; New York: 1994. [Google Scholar]
- 38.Sive HL, Grainger RM, Harland R. Early Development of Xenopus Laevis: a Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor; New York: 1998. [Google Scholar]
- 39.Shirasaki R, Mirzayan C, Tessier-Lavigne M, Murakami F. Guidance of circumferentially growing axons by netrin-dependent and -independent floor plate chemotropism in the vertebrate brain. Neuron. 1996;17:1079–1088. doi: 10.1016/s0896-6273(00)80241-x. [DOI] [PubMed] [Google Scholar]
- 40.Huang S, Moody SA. The retinal fate of Xenopus cleavage stage progenitors is dependent upon blastomere position and competence: studies of normal and regulated clones. J. Neurosci. 1993;13:3193–3210. doi: 10.1523/JNEUROSCI.13-08-03193.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]