Abstract
Homologous recombination is essential for productive DNA replication particularly under stress conditions. We previously demonstrated a stress-induced recruitment of Rad51 to mitochondria and a critical need for its activity in the maintenance of mitochondrial DNA (mtDNA) copy number. Using the human osteosarcoma cell line U20S, we show in the present study that recruitment of Rad51 to mitochondria under stress conditions requires ongoing mtDNA replication.. Additionally, Rad51 levels in mitochondria increase in cells recovering from mtDNA depletion. Our findings highlight an important new role for Rad51 in supporting mtDNA replication, and further promote the idea that recombination is indispensable for sustaining DNA synthesis under conditions of replication stress.
Keywords: Rad51, mtDNA replication, mtDNA copy number, oxidative stress, replication restart
1. Introduction
The recombinase activity of the Rad51 protein forms the catalytic core of the homologous recombination (HR) pathway for the repair of DNA double strand breaks (DSBs). In addition to its traditional role in DSB repair, it is well established that Rad51-mediated HR plays a critical role in facilitating efficient genomic DNA replication in mammalian cells, particularly under DNA damaging stress conditions (Arnaudeau et al., 2001; Groth et al., 2012; Lundin et al., 2002; Lundin et al., 2003; Petermann and Helleday, 2010; Saintigny et al., 2001; Saleh-Gohari et al., 2005). The collapse of a DNA replication fork results in the generation of a one-ended DSB. With the absence of a second end to mediate direct ligation, Rad51-mediated HR is required for repair and restart of replication (Arnaudeau et al., 2001; Roseaulin et al., 2008; Saintigny et al., 2001; Saleh-Gohari et al., 2005). It is now thought that repair of replication-associated DNA damage is the primary role of HR in mammalian cells (Petermann and Helleday, 2010). Indeed, in vertebrate cells, Rad51 is regulated such that its activity is highest during the S and G2 cell cycle phases when the bulk of DNA replication occurs and sister chromatids are present (Hartlerode et al., 2011; Rothkamm et al., 2003).
In addition to nuclear DNA, a 16.5 KB genome is maintained in the mitochondrial matrix. This circular genome is present in thousands of copies per cell with copy number being highly regulated in a tissue specific manner to provide for the energetic needs of particular cell types (Jeng et al., 2008; Mineri et al., 2009; Pohjoismaki et al., 2010). Given its proximity to the site of oxidative phosphorylation, mtDNA is routinely subjected to oxidative stress leading to numerous types of DNA damage. Recent studies have provided an increased appreciation for the ability of mitochondria to repair its DNA using various repair pathways including long and short patch base excision repair as well as mismatch repair (Liu and Demple, 2010). We have recently shown that Rad51 localizes to human mitochondria, and levels increase significantly in response to oxidative stress (Sage et al., 2010). Additionally, we found that Rad51 physically interacts with mtDNA and supports maintenance of mtDNA copy number under stress conditions. Oxidative stress of this type largely generates mtDNA damage in the form of single strand breaks and abasic sites (Shokolenko et al., 2009). These lesions block the progression of DNA polymerases leading to stalled or collapsed replication forks, a classic substrate for Rad51 directed repair and replication restart (Petermann and Helleday, 2010). Indeed, in a recent report Rad51 was implicated in the repair of secondary, replication-associated nuclear DSBs following ionizing radiation (IR) treatment (Groth et al., 2012). In the present study we explore how recruitment of Rad51 to mitochondria may be influenced by ongoing mtDNA replication, and how this may affect mtDNA copy number.
2. Materials and methods
2.1 Cell culture, treatment of cells with oxidative stress, and siRNA transfection
U20S cells (ATCC #HTB-96) were grown at 37 °C in McCoy’s 5A medium (Invitrogen) with 5% CO2, supplemented with 10% fetal bovine serum and antibiotics. Cell number was routinely determined by staining with trypan blue and counting using a haemocytometer. For oxidative stress, glucose oxidase (GO; Sigma G7141) stock solutions were prepared prior to each experiment and added at the indicated concentrations. Cells were washed once with non-supplemented medium that was then replaced with non-supplemented medium containing the indicated concentration of GO. To inhibit DNA replication, aphidicolin (Sigma) was prepared in DMSO and added directly to the culture medium to a final concentration of 3 μM, dideoxycytidine (ddC; Sigma) was prepared in water and added to a final concentration of 20 μM, and ethidium bromide (EtBr; Sigma) was prepared in water and added to a final concentration of 50 ng/mL. Rad51 protein was depleted by treating cells with a pool of siRNAs (10 nM final, Dharmacon L-003530-00) delivered using Dharmafect1. A pool of nonsilencing siRNAs was used as the negative control.
2.2 Bromodeoxyuridine labeling of cells for immunofluorescent detection of nascent DNA
U20S cells were seeded onto glass cover slips prior to the addition of ddC, aphidicolin, or EtBr. Cells were treated for 2 h with inhibitor prior to the addition of 5-bromo-2′-deoxyuridine (BrdU; BD Biosciences) to 10 μg/mL for 2 h. Thirty minutes prior to the end of labeling, cells were treated with 5 nM MitoTracker Deep Red (Invitrogen). Cells were cross-linked by the addition of 4% formaldehyde at room temperature for 10 min. After washing cells twice with PBS, DNA was denatured by incubating cells in 2N HCl for 10 min, followed by pH neutralization by washing twice with 200 mM Tris-HCl pH 8.0. Cells were permeabilized by incubation in ice-cold methanol for 10 min. Cells were incubated for 1 h in blocking buffer [3% FBS and 0.5% Triton X100 in PBS] with rocking. Cells were incubated with a 1:500 dilution of mouse anti-BrdU antibody (Roche) in wash buffer [1% FBS and 0.5% Triton X100 in PBS] for 2 h with rocking, followed by 3, 5 min washes. Cells were then incubated with a 1:500 dilution of Alexa Fluor 488-conjugated goat anti-mouse antibody (Invitrogen) in wash buffer for 1 h with rocking followed by 6 × 5 min washes. Finally cover slips were mounted onto glass slides with VECTASHIELD mounting medium containing DAPI (Vector Labs). Images were captured with an Axioskop fluorescence microscope fitted with the Axiocam MRm camera and accompanying software (Zeiss).
2.3 Subcellular fractionation and immunoblotting
Intact mitochondria were isolated from cells as previously described (Sage et al., 2010). Briefly, following a gentle lysis using mitochondrial isolation buffer [10 mM Tris-HCl, pH 7.8, 0.2 mM EDTA, 250 mM sucrose, 150 mM KCl, 0.15% digitonin (Sigma) and a protease inhibitor cocktail (Roche)], cells were subjected to a series of centrifugation steps to isolate a crude mitochondrial pellet. This pellet was washed 4 times with mitochondrial isolation buffer containing high salt (1 M KCl) prior to solubilization of the mitochondrial membrane to yield the mitochondrial protein fraction. Protein concentrations were determined using the BCA Protein Assay Kit (Pierce), and samples were prepared by boiling in 2x Laemmli sample buffer (Sigma). Samples were run on pre-cast 4–12% Bis-Tris gels (Invitrogen), and immunoblotted as previously described (Sage et al., 2010) using mouse anti-Rad51 (Millipore), mouse anti-ATP synthase (BD Transduction Laboratories), rabbit anti-TFAM (Abcam), mouse anti-GAPDH (Millipore), or rabbit anti-PCNA (Abcam) antibodies. Blots were then incubated with chemiluminescent visualizer (Denville) and developed using an LAS-400 imaging instrument (Fuji).
2.4 Determination of mtDNA copy number
Total cellular DNA was isolated using the QIAamp DNA Mini Kit (Qiagen), and DNA concentrations were determined spectrophotometrically. Equal amounts of total cellular DNA were assayed by quantitative PCR as previously described (Sage et al., 2010), with changes in mtDNA copy number quantified relative to the 18S rRNA gene.
3. Results and discussion
3.1 Inhibition of post-DNA damage mtDNA replication prevents recruitment of Rad51 to mitochondria
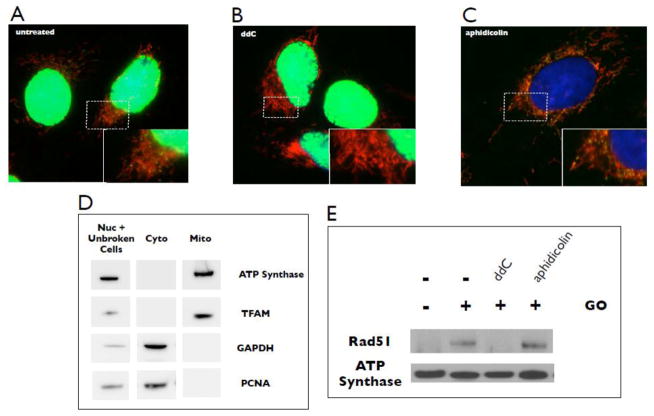
We previously demonstrated that mitochondria show an increase in levels of Rad51 protein following exposure to oxidative stress (Sage et al., 2010), suggesting that Rad51 may be limiting for a required recombination function under these conditions. As this type of stress induces an increase in mtDNA content (Lee and Wei, 2005; Lee et al., 2000; Lee et al., 2002; Wei et al., 2001), it is possible that Rad51 functions to support productive mtDNA replication following DNA damage. To explore this possibility, we employed a series of experiments to investigate the relationship between Rad51 and mtDNA replication. Using bromodeoxyuridine (BrdU) incorporation as a readout for DNA replication, we show that a 2 h pulse results in clear incorporation of the label into both nuclear and mtDNA (Fig. 1A). Similar to previous studies (Davis and Clayton, 1996), centers of mtDNA replication appear as yellow/green dots (BrdU) embedded in mitochondria, which have been stained using MitoTracker Deep Red (see Materials and methods). The nucleoside analogue dideoxycytidine (ddC) is a potent and specific inhibitor of mtDNA replication at low concentrations (Feng et al., 2001), and treatment of cells with 20 μM ddC for 2 h prior to the BrdU pulse prevents detectable incorporation into mtDNA while allowing nuclear DNA replication to proceed (Fig. 1B). In contrast, aphidicolin is a potent inhibitor of nuclear DNA polymerase alpha while having no effect on mitochondrial DNA polymerase gamma (Zimmermann et al., 1980). Consistent with this, we find that treatment of cells with 3 μM aphidicolin for 2 h prior to BrdU treatment prevents labeling of nuclear DNA while leaving mtDNA synthesis unaffected (Fig 1C). Therefore, using these tools we can specifically and selectively inhibit either mitochondrial or nuclear DNA replication.
Fig. 1.
Recruitment of Rad51 to the mitochondria in response to stress is suppressed by specific inhibition of mtDNA replication. (A) U20S cells were treated for 2 h with 10 μg/mL BrdU, then stained with MitoTracker Deep Red and anti-BrdU (green). Yellow/green spots embedded in mitochondria indicate sites of nascent mtDNA synthesis. (B) Inhibition of mtDNA labeling by treatment with 20 μM ddC 2 h prior to the BrdU pulse. (C) Inhibition of nuclear DNA labeling by treatment with 3 μM aphidicolin 2 h prior to the BrdU pulse. The nuclear compartment is stained with DAPI. Images are representative of 250 cells examined. (D) Western blot analysis indicates enrichment of mitochondrial protein fraction and the absence of contaminating cytosolic and nuclear proteins. Marker proteins: TFAM and ATP synthase (mitochondrial); PCNA (nuclear); GAPDH (cytosolic). (E) Western blot of Rad51 in purified mitochondrial fractions following treatment of cells with 10 milliunits/mL GO with or without a 2 h pre-treatment with either 20 μM ddC or 3 μM aphidicolin. ATP synthase serves as the loading control. Blots are representative of three independent experiments.
To evaluate the role of mtDNA replication in the recruitment of Rad51 to the mitochondria, we utilized our subcellular fractionation procedure optimized to isolate a mitochondrial protein fraction enriched for the mitochondrial markers TFAM and ATP synthase, and free of detectible levels of the nuclear marker PCNA and the cytosolic marker GAPDH (Sage et al., 2010) (Fig 1D). Our earlier immunofluorescence studies showed a significant and diffuse staining of Rad51 in the cytosol (Bennett and Knight, 2006). Therefore, to provide a more direct detection of mitochondrially-localized Rad51 and stress related changes in mitochondrial levels of Rad51 we developed this fractionation and blotting protocol (Sage et al. 2010). A similar concern regarding immunofluorescence detection of relatively low levels of mitochondrial p53 was also addressed by Marchenko et al. (2000). Increased oxidative stress was generated by adding the enzyme glucose oxidase (GO) directly to the culture medium (Sage et al., 2010). This allows a consistent production of hydrogen peroxide over an extended time course at concentrations shown to preferentially damage mtDNA (Salazar and Van Houten, 1997). Using this fractionation procedure we show that treatment of cells with oxidative stress induces an increase in mitochondrial Rad51 levels (Fig. 1E). Total cellular Rad51 levels do not change under these conditions (not shown), consistent with other studies showing that DNA damage does not affect Rad51 protein levels (Chen et al., 1997; Henson et al., 2006; Raderschall et al., 2002; Vispe et al., 1998). Strikingly, inhibiting mtDNA replication by treating cells with ddC for 2 h prior to oxidative stress completely abrogates this increase. In contrast, inhibiting nuclear DNA replication with aphidicolin does not affect the stress-induced increase in mitochondrial Rad51 (Fig. 1E). These results indicate that recruitment of Rad51 to mitochondria in response to oxidative stress requires ongoing mtDNA replication, and second, that oxidatively damaged DNA itself is not the signal for recruitment.
Previously, we used a protease protection assay as well as a mtDNA-immunoprecipitation assay to show that mitochondrial Rad51 is largely confined to the matrix and physically associates with mtDNA (Sage et al., 2010). However, the mechanism by which Rad51 is recruited to the mitochondrion following oxidative stress is currently unknown. As for p53, another protein recruited to mitochondria under stress conditions (Ferecatu et al., 2009), Rad51 bears no evident N-terminal mitochondrial targeting sequence when analyzed using prediction software such as MitoProt II and Predotar (Sage et al., 2010). It is possible that a stress-activated chaperone facilitates this process; possibly the known Rad51 binding partner, HSP90 (Ko et al., 2012). Future studies will include efforts to understand the mechanisms that underlie this adaptive stress response.
3.2 Post-DNA damage mtDNA replication inhibition relieves Rad51 requirement for mtDNA copy number stabilization
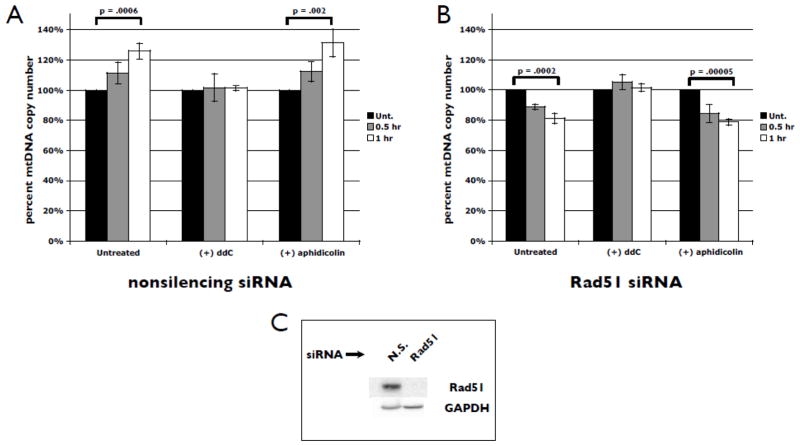
An important measure of efficient mtDNA replication is the maintenance of the number of copies of the genome per cell (mtDNA copy number). MtDNA copy number is highly regulated and cell type specific, and defects have been linked to a variety of human diseases (Graziewicz et al., 2006). We have shown that the maintenance of mtDNA levels, specifically under oxidative stress conditions, requires the presence of Rad51 (Sage et al., 2010). To evaluate the potential interplay between mtDNA replication and Rad51 in this maintenance process, we first monitored changes in mtDNA copy number by qPCR in the presence or absence of endogenous levels of Rad51. In cells expressing normal levels of Rad51 [Fig. 2A (−) pol inhibitors], exposure to GO for 0.5 or 1 h induces an increase in copy number of approximately 25–30%, consistent with studies from our group and others (Lee and Wei, 2005; Lee et al., 2000; Lee et al., 2002; Wei et al., 2001). However, when cells have been depleted of Rad51, copy number drops rapidly in response to stress [approximately 20% following 1 h of GO treatment, Fig. 2B (−) pol inhibitors]. Western blot analysis showed no detectable Rad51 in siRNA-treated total cell extracts relative to nonsilencing control siRNA treatment (Fig. 2C). Therefore, under these stress conditions the absence of Rad51 results in an overall decrease in mtDNA copy number by approximately 45–50%. We repeated this experiment using a pre-treatment step with ddC to block mtDNA replication. Interestingly, we find that this completely prevents both the increase in copy number seen when Rad51 is present as well as the drop observed when Rad51 has been depleted prior to stress [Fig. 2A, B (+) ddC]. In contrast, pre-treatment with aphidicolin fails to prevent either the increase in copy number when Rad51 is present or the drop in copy number when Rad51 is siRNA-depleted [Fig. 2A, B (+) aphidicolin]. Similar to our findings in the absence of replication inhibitors, we observe an overall stress-induced decrease in mtDNA copy number of approximately 45–50% following inhibition of nuclear DNA replication and depletion of Rad51. These results suggest that successful mtDNA replication under stress conditions requires Rad51.
Fig. 2.
Stress-induced changes in mtDNA copy number requires ongoing mtDNA replication. U20S cells were transfected with nonsilencing (A) or Rad51-specific (B) siRNAs (10 nm). 48 h following transfection, cells were treated with 5 milliunits/ml GO or treated with GO following a 2 h pre-treatment with 20 μM ddC or 3 μM aphidicolin. DNA was isolated at 0, 0.5, and 1 h after GO addition. MtDNA copy number was determined by qPCR, with changes calculated relative to unstressed controls and normalized to the 18S rRNA gene. Error bars indicate S.E.M. (n = 4). (C) Western blot analysis of Rad51 in total cell lysates following 48 h of nonsilencing (N.S.) or Rad51-specific siRNA treatment. GAPDH serves as the loading control. Blots are representative of 4 independent experiments.
3.3 Rad51 is recruited to mitochondria of cells recovering from mtDNA depletion
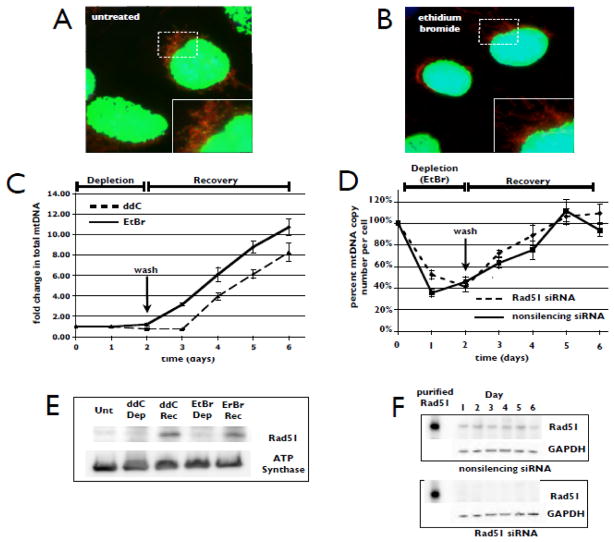
While the GO-induced oxidative stress used here introduces DNA lesions that are potential blocks to replication fork progression, these lesions are continuously present at lower levels under normal culture conditions. To investigate the role of Rad51 in mtDNA replication and copy number maintenance in the absence of exogenous DNA damage, we treated cells with ddC or ethidium bromide (EtBr) to inhibit mtDNA replication. As with ddC treatment (Fig. 1B), pre-treatment with EtBr (50 ng/ml, 2 h) prevents detectible incorporation of BrdU into mtDNA, while allowing nuclear DNA replication to proceed (Fig. 3A, B). Variations observed in intensity of nuclear BrdU staining on a cell-to-cell basis likely indicate differences in cell cycle phase in the asynchronous culture. Over two-days of EtBr treatment, the total amount of mtDNA in the entire cell population does not change, indicating both the inhibition of synthesis and the lack of degradation over the course of two cell doubling times (Fig. 3C days 0–2). However, because cell growth continues, the mtDNA copy number, which is calculated on a per cell basis relative to the 18S rRNA gene, decreases as a result of dilution through cell division (Fig. 3D days 0–2). We refer to this stage of treatment as “Depletion” (the distinction between total mtDNA in the cell population vs. mtDNA copy number per cell will become important in the next section when we describe experiments performed following RNAi-mediated knockdown of Rad51). EtBr is then washed away, allowing cells to resume mtDNA replication and regenerate mtDNA copy number (Fig. 3C, D days 2–6). We refer to this phase as “Recovery”. Thus, this experimental system generates two distinct phases of replication demand, “Depletion” where mtDNA replication is inhibited, and “Recovery” where the requirement for mtDNA replication is significantly higher than under normal culture conditions (hyper-replication). Western blot analysis of mitochondrial protein fractions reveals low levels of Rad51 during the “Depletion” phase (Fig. 3E, ddC/Dep and EtBr/Dep). Importantly however, when mitochondria enter a hyper-replication mode during “Recovery”, levels of Rad51 in the mitochondria rise dramatically (Fig. 3E, ddC/Rec and EtBr/Rec). Total cellular Rad51 levels do not change under these conditions (Fig. 3F). Therefore, the data suggest that recruitment of Rad51 to mitochondria is a component of the response to an increased demand for mtDNA replication in the absence of exogenous DNA damage.
Fig. 3.
Recovery of mtDNA copy number following mtDNA depletion recruits Rad51 to the mitochondria. (A) U20S cells were treated for 2 h with 10 μg/mL BrdU, then stained with MitoTracker Deep Red and anti-BrdU (green). Yellow/green spots embedded in mitochondria indicate sites of nascent mtDNA synthesis. (B) Inhibition of mtDNA labeling was achieved with a 2 h pre-treatment with 50 ng/mL EtBr prior to the BrdU pulse. (C) U20S cells were treated with either 20 μM ddC or 50 ng/mL EtBr for 2 days, then washed and allowed to recover for 4 days. Total DNA was isolated every 24 h for qPCR determination of copy number per cell with changes being calculated relative to unstressed controls and normalized to the 18S rRNA gene. Fold change in total mtDNA in each population was determined by multiplying copy number per cell by relative cell count for each day. Error bars indicate S.E. (n = 4). (D) U20S cells were transfected with either nonsilencing or Rad51-specific siRNAs. Both populations of cells were treated with 50 ng/mL EtBr for 2 days, then washed and allowed to recover for 4 days. Total DNA was isolated every 24 h for qPCR determination of mtDNA copy number per cell. Error bars indicate S.E. (n = 4). (E) Western blot of Rad51 in purified mitochondrial fractions isolated from cells not treated with polymerase inhibitors (untreated, Unt), cells treated with either inhibitor during the first 24 h of the depletion phase (Dep) and cells 24 h post drug removal (Rec, day 3 in panels C and D). ATP synthase serves as the loading control. (F) Western blot analysis of Rad51 in total cell lysates that were prepared every 24 h during the depletion and recovery phases. GAPDH serves as the loading control. All blots are representative of 4 independent experiments.
3.4 In the absence of exogenous DNA damage Rad51 depletion reduces total mtDNA corresponding to a coincident decrease in cell growth rate
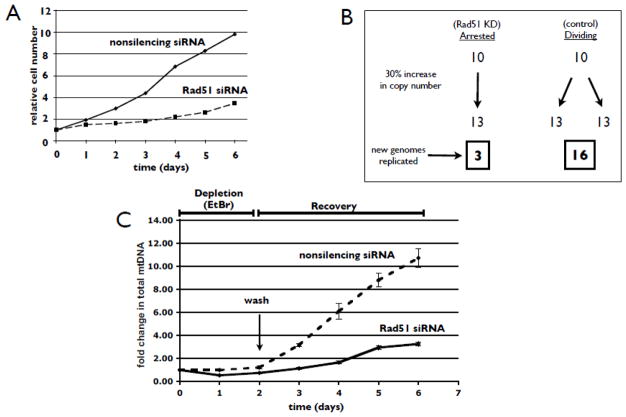
As shown above while inhibition of mtDNA synthesis with ddC or EtBr did not significantly change the total levels of mtDNA in the entire cell population (Fig. 3C days 0–2), mtDNA copy number per cell is reduced sharply as the now limited number of mitochondrial genomes are diluted into the expanding cell population (Fig. 3D days 0–2). The rate and level of mtDNA depletion, approximately 60% at 48 h, is similar to that observed by other groups using ddC and EtBr at these concentrations (Brown and Clayton, 2002; Stewart et al., 2011). This phenomenon highlights the fact that the major demand for synthesis of mtDNA is the result of ongoing cell division. Removal of polymerase gamma inhibitors adds a further demand to synthesize new genomes as the cells adjust mtDNA replication to recover the steady-state copy number as they continue to divide (Fig. 3C, D days 2–6). Therefore, the condition of greatest replication demand is generated in cells that are actively dividing and simultaneously recovering from mtDNA depletion. When we examine the rate of mtDNA copy number recovery on a per cell basis following siRNA-mediated knockdown of Rad51, we find no significant difference compared to the rate of copy number recovery when Rad51 is present (Fig. 3D days 2–6; Fig. 3F shows Rad51 knockdown control). However, it is important to note that in the absence of Rad51, cells display a dramatic reduction in the rate of cell division (Fig. 4A). Therefore, the total cell number in the Rad51 siRNA-treated population in Fig. 3D is significantly less than in the population treated with nonsilencing siRNA. This difference in cell division rate has a profound effect on mtDNA replication demand during recovery from mtDNA depletion. For example, an actively cycling cell displaying a 30% copy number increase over one doubling period, (see Fig. 3D days 2–3), must synthesize greater than 5 times the number of mitochondrial genomes than an arrested cell to show the same increase in mtDNA copy number per cell (Fig. 4B). When the difference in cell proliferation is taken into account, we find that cells depleted of Rad51 synthesize approximately 8 fold less mtDNA following 4 days of recovery (Fig. 4C). Therefore, the hyper-replication condition under which Rad51 is recruited to mitochondria (Fig. 3E) cannot be appropriately reproduced in cells depleted of Rad51 as these cells divide far more slowly than their control counterparts (Fig. 4A). While this highlights a technical challenge in studying the mitochondrial function of a protein with other critical activities in the cell, these data are consistent with our other results suggesting an important role for Rad51 in promoting mtDNA synthesis under conditions where there is increased replication demand.
Fig. 4.
RNAi-mediated knockdown of Rad51 significantly slows cell division and mtDNA replication demand during recovery from mtDNA depletion. (A) U20S cells were transfected with nonsilencing or Rad51-specific siRNAs (10 nm), and simultaneously treated with 50 ng/mL EtBr. Two days following treatment, cells were washed and allowed to recover for 4 days. Cells were trypsinized and counted every 24 h. Cell numbers are displayed relative to initial seeding. (B) Model illustrating the difference in DNA replication demand between cycling and non-cycling cells. During the course of one population doubling time, an arrested cell must only replicate a number of genomes equivalent to the observed copy number increase (left pathway). An actively dividing cell must duplicate its mtDNA compliment in addition to replication required to account for the increase in copy number observed (right pathway). (C) U20S cells were transfected with nonsilencing or Rad51-specific siRNAs and treated with EtBr as in (A). Total DNA was isolated every 24 h for qPCR determination of copy number per cell. Fold change in total mtDNA in each population was determined by multiplying copy number per cell by relative cell count for each day. Error bars indicate S.E. (n = 4).
Studies using a variety of cell types including bacteria (Heller and Marians, 2006; Kreuzer, 2005; McGlynn and Lloyd, 2002), yeast (Davis and Symington, 2004; Flores-Rozas and Kolodner, 2000), and human (Kim et al., 2012; Petermann and Helleday, 2010) all reveal that recombination is critical to the recovery of stalled or collapsed forks during genomic DNA replication. It was reported that as many as 50% of bacterial cells require fork restart during a single round of DNA replication (Cox et al., 2000). While the frequency of fork collapse during mammalian mtDNA replication is unknown, the constant exposure to oxidative damage likely makes replication restart a common event. There is also evidence that recombination events occur in mammalian mitochondria. For example, both intra- and inter-molecular recombination can be detected following restriction enzyme-mediated introduction of DSBs into mouse mtDNA (Bacman et al., 2009). Additionally, 4-way junction structures are prevalent in human heart mtDNA (Kajander et al., 2001), and it was proposed that these derive from recombination activity associated with replication initiation and processivity in a tissue with such high metabolic demand (Pohjoismaki and Goffart, 2011; Pohjoismaki et al., 2009).
Previously, we identified a critical need for Rad51 in the mitochondrial response to oxidative stress (Sage et al., 2010). While the majority of this damage can be repaired by base excision repair, all of these DNA lesions represent potent blocks to replication. The restart of stalled or collapsed replication forks is critical for the maintenance of genomic DNA integrity and this is a process in which Rad51 is intimately involved (Helleday, 2003; Petermann and Helleday, 2010). In the present study, we show that while oxidatively damaged DNA by itself is insufficient to recruit Rad51 to the mitochondria, it is ongoing mtDNA replication in the presence of this damage that is required for the recruitment of Rad51.
The stress-induced decrease in mtDNA copy number observed when Rad51 has been depleted can be completely prevented by halting mtDNA replication prior to the application of stress (Fig. 2B). This copy number decrease in the absence of Rad51 is rapid relative to the more gradual reduction in mtDNA content observed during inhibitor-mediated depletion (Fig. 3D). Recently, a very complementary report showed a critical role for Rad51 in mitigating post-DNA damage replication toxicity following IR treatment (Groth et al., 2012). In addition to primary DNA lesions induced by IR, they characterized secondary DSBs resulting from ongoing replication of damaged DNA. Our results mirror this role of Rad51 in mitochondria, and further establish the recombinase as a critical adaptive replication factor in the maintenance of genome integrity under conditions when replication demand is high or when DNA lesions are present.
Overall, our observations reveal a broader role for Rad51 than previously appreciated regarding its importance in maintaining the complete genomic content of human cells.
Highlights.
Recruitment of Rad51 to mitochondria under stress conditions requires ongoing mtDNA replication
Mitochondrial Rad51 levels increase during recovery from mtDNA depletion under non-stress conditions
Rad51 promotes mtDNA synthesis under conditions where there is increased replication demand
Rad51 plays a critical role in maintaining the complete genomic content of human cells
Acknowledgments
The authors thank Dr. Anthony Cura for valuable discussions. This work was supported by the National Institutes of Health [RO-1 GM44772 to KLK].
Abbreviations
- BrdU
5-bromo-2′-deoxyuridine
- ddC
2′-3′-dideoxycytidine
- DSB
double strand break
- EtBr
ethidium bromide
- GO
glucose oxidase
- HR
homologous recombination
- mtDNA
mitochondrial DNA
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J Mol Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- Bacman SR, Williams SL, Moraes CT. Intra- and inter-molecular recombination of mitochondrial DNA after in vivo induction of multiple double-strand breaks. Nucleic Acids Res. 2009;37:4218–4226. doi: 10.1093/nar/gkp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett BT, Knight KL. Cellular localization of human Rad51C and regulation of ubiquitin-mediated proteolysis of Rad51. J Cell Biochem. 2005;96:1095–1109. doi: 10.1002/jcb.20640. [DOI] [PubMed] [Google Scholar]
- Brown TA, Clayton DA. Release of replication termination controls mitochondrial DNA copy number after depletion with 2′,3′-dideoxycytidine. Nucleic Acids Res. 2002;30:2004–2010. doi: 10.1093/nar/30.9.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Nastasi A, Shen Z, Brenneman M, Crissman H, Chen DJ. Cell cycle-dependent protein expression of mammalian homologs of yeast DNA double-strand break repair genes Rad51 and Rad52. Mutat Res. 1997;384:205–211. doi: 10.1016/s0921-8777(97)00020-7. [DOI] [PubMed] [Google Scholar]
- Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- Davis AF, Clayton DA. In situ localization of mitochondrial DNA replication in intact mammalian cells. J Cell Biol. 1996;135:883–893. doi: 10.1083/jcb.135.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AP, Symington LS. RAD51-dependent break-induced replication in yeast. Mol Cell Biol. 2004;24:2344–2351. doi: 10.1128/MCB.24.6.2344-2351.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng JY, Johnson AA, Johnson KA, Anderson KS. Insights into the molecular mechanism of mitochondrial toxicity by AIDS drugs. J Biol Chem. 2001;276:23832–23837. doi: 10.1074/jbc.M101156200. [DOI] [PubMed] [Google Scholar]
- Ferecatu I, Bergeaud M, Rodriguez-Enfedaque A, Le Floch N, Oliver L, Rincheval V, Renaud F, Vallette FM, Mignotte B, Vayssiere JL. Mitochondrial localization of the low level p53 protein in proliferative cells. Biochem Biophys Res Commun. 2009;387:772–777. doi: 10.1016/j.bbrc.2009.07.111. [DOI] [PubMed] [Google Scholar]
- Flores-Rozas H, Kolodner RD. Links between replication, recombination and genome instability in eukaryotes. Trends Biochem Sci. 2000;25:196–200. doi: 10.1016/s0968-0004(00)01568-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziewicz MA, Longley MJ, Copeland WC. DNA polymerase gamma in mitochondrial DNA replication and repair. Chem Rev. 2006;106:383–405. doi: 10.1021/cr040463d. [DOI] [PubMed] [Google Scholar]
- Groth P, Orta ML, Elvers I, Majumder MM, Lagerqvist A, Helleday T. Homologous recombination repairs secondary replication induced DNA double-strand breaks after ionizing radiation. Nucleic Acids Res. 2012;40:6585–6594. doi: 10.1093/nar/gks315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartlerode A, Odate S, Shim I, Brown J, Scully R. Cell cycle-dependent induction of homologous recombination by a tightly regulated I-SceI fusion protein. PLoS ONE. 2011;6:e16501. doi: 10.1371/journal.pone.0016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T. Pathways for mitotic homologous recombination in mammalian cells. Mutat Res. 2003;532:103–115. doi: 10.1016/j.mrfmmm.2003.08.013. [DOI] [PubMed] [Google Scholar]
- Heller RC, Marians KJ. Replisome assembly and the direct restart of stalled replication forks. Nat Rev Mol Cell Biol. 2006;7:932–943. doi: 10.1038/nrm2058. [DOI] [PubMed] [Google Scholar]
- Henson SE, Tsai SC, Malone CS, Soghomonian SV, Ouyang Y, Wall R, Marahrens Y, Teitell MA. Pir51, a Rad51-interacting protein with high expression in aggressive lymphoma, controls mitomycin C sensitivity and prevents chromosomal breaks. Mutat Res. 2006;601:113–124. doi: 10.1016/j.mrfmmm.2006.06.016. [DOI] [PubMed] [Google Scholar]
- Jeng JY, Yeh TS, Lee JW, Lin SH, Fong TH, Hsieh RH. Maintenance of mitochondrial DNA copy number and expression are essential for preservation of mitochondrial function and cell growth. J Cell Biochem. 2008;103:347–357. doi: 10.1002/jcb.21625. [DOI] [PubMed] [Google Scholar]
- Kajander OA, Karhunen PJ, Holt IJ, Jacobs HT. Prominent mitochondrial DNA recombination intermediates in human heart muscle. EMBO Rep. 2001;2:1007–1012. doi: 10.1093/embo-reports/kve233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TM, Ko JH, Hu L, Kim SA, Bishop AJ, Vijg J, Montagna C, Hasty P. RAD51 mutants cause replication defects and chromosomal instability. Mol Cell Biol. 2012;32:3663–3680. doi: 10.1128/MCB.00406-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko JC, Chen HJ, Huang YC, Tseng SC, Weng SH, Wo TY, Huang YJ, Chiu HC, Tsai MS, Chiou RYY, Lin YW. HSP90 inhibition induces cytotoxicity via down-regulation of Rad51 expression and DNA repair capacity in non-small cell lung cancer cells. Regul Toxicol Pharmacol. 2012;64:415–424. doi: 10.1016/j.yrtph.2012.10.003. [DOI] [PubMed] [Google Scholar]
- Kreuzer KN. Interplay between DNA replication and recombination in prokaryotes. Annu Rev Microbiol. 2005;59:43–67. doi: 10.1146/annurev.micro.59.030804.121255. [DOI] [PubMed] [Google Scholar]
- Lee HC, Wei YH. Mitochondrial biogenesis and mitochondrial DNA maintenance of mammalian cells under oxidative stress. Int J Biochem Cell Biol. 2005;37:822–834. doi: 10.1016/j.biocel.2004.09.010. [DOI] [PubMed] [Google Scholar]
- Lee HC, Yin PH, Chi CW, Wei YH. Increase in mitochondrial mass in human fibroblasts under oxidative stress and during replicative cell senescence. J Biomed Sci. 2002;9:517–526. doi: 10.1007/BF02254978. [DOI] [PubMed] [Google Scholar]
- Lee HC, Yin PH, Lu CY, Chi CW, Wei YH. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem J. 2000;348(Pt 2):425–432. [PMC free article] [PubMed] [Google Scholar]
- Liu P, Demple B. DNA repair in mammalian mitochondria: Much more than we thought? Environ Mol Mutagen. 2010;51:417–426. doi: 10.1002/em.20576. [DOI] [PubMed] [Google Scholar]
- Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol Cell Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin C, Schultz N, Arnaudeau C, Mohindra A, Hansen LT, Helleday T. RAD51 is involved in repair of damage associated with DNA replication in mammalian cells. J Mol Biol. 2003;328:521–535. doi: 10.1016/s0022-2836(03)00313-9. [DOI] [PubMed] [Google Scholar]
- Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat Rev Mol Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- Mineri R, Pavelka N, Fernandez-Vizarra E, Ricciardi-Castagnoli P, Zeviani M, Tiranti V. How do human cells react to the absence of mitochondrial DNA? PLoS ONE. 2009;4:e5713. doi: 10.1371/journal.pone.0005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat Rev Mol Cell Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S. Of circles, forks and humanity: Topological organisation and replication of mammalian mitochondrial DNA. BioEssays. 2011;33:290–299. doi: 10.1002/bies.201000137. [DOI] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Taylor RW, Turnbull DM, Suomalainen A, Jacobs HT, Karhunen PJ. Developmental and pathological changes in the human cardiac muscle mitochondrial DNA organization, replication and copy number. PLoS ONE. 2010;5:e10426. doi: 10.1371/journal.pone.0010426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjoismaki JL, Goffart S, Tyynismaa H, Willcox S, Ide T, Kang D, Suomalainen A, Karhunen PJ, Griffith JD, Holt IJ, Jacobs HT. Human heart mitochondrial DNA is organized in complex catenated networks containing abundant four-way junctions and replication forks. J Biol Chem. 2009;284:21446–21457. doi: 10.1074/jbc.M109.016600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- Roseaulin L, Yamada Y, Tsutsui Y, Russell P, Iwasaki H, Arcangioli B. Mus81 is essential for sister chromatid recombination at broken replication forks. EMBO J. 2008;27:1378–1387. doi: 10.1038/emboj.2008.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K, Kruger I, Thompson LH, Lobrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage JM, Gildemeister OS, Knight KL. Discovery of a novel function for human Rad51: maintenance of the mitochondrial genome. J Biol Chem. 2010;285:18984–18990. doi: 10.1074/jbc.M109.099846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salazar JJ, Van Houten B. Preferential mitochondrial DNA injury caused by glucose oxidase as a steady generator of hydrogen peroxide in human fibroblasts. Mutat Res. 1997;385:139–149. doi: 10.1016/s0921-8777(97)00047-5. [DOI] [PubMed] [Google Scholar]
- Saleh-Gohari N, Bryant HE, Schultz N, Parker KM, Cassel TN, Helleday T. Spontaneous homologous recombination is induced by collapsed replication forks that are caused by endogenous DNA single-strand breaks. Mol Cell Biol. 2005;25:7158–7169. doi: 10.1128/MCB.25.16.7158-7169.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shokolenko I, Venediktova N, Bochkareva A, Wilson GL, Alexeyev MF. Oxidative stress induces degradation of mitochondrial DNA. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart JD, Schoeler S, Sitarz KS, Horvath R, Hallmann K, Pyle A, Yu-Wai-Man P, Taylor RW, Samuels DC, Kunz WS, Chinnery PF. POLG mutations cause decreased mitochondrial DNA repopulation rates following induced depletion in human fibroblasts. Biochim Biophys Acta. 2011;1812:321–325. doi: 10.1016/j.bbadis.2010.11.012. [DOI] [PubMed] [Google Scholar]
- Vispe S, Cazaux C, Lesca C, Defais M. Overexpression of Rad51 protein stimulates homologous recombination and increases resistance of mammalian cells to ionizing radiation. Nucleic Acids Res. 1998;26:2859–2864. doi: 10.1093/nar/26.12.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei YH, Lee CF, Lee HC, Ma YS, Wang CW, Lu CY, Pang CY. Increases of mitochondrial mass and mitochondrial genome in association with enhanced oxidative stress in human cells harboring 4,977 BP-deleted mitochondrial DNA. Ann N Y Acad Sci. 2001;928:97–112. doi: 10.1111/j.1749-6632.2001.tb05640.x. [DOI] [PubMed] [Google Scholar]
- Zimmermann W, Chen SM, Bolden A, Weissbach A. Mitochondrial DNA replication does not involve DNA polymerase alpha. J Biol Chem. 1980;255:11847–11852. [PubMed] [Google Scholar]