Abstract
DNA barcoding is a novel molecular identification method that aids in identifying traditional Chinese materia medica using traditional identification techniques. However, further study is needed to assess the stability and accuracy of DNA barcoding. Flos Lonicerae Japonicae, a typical medicinal flower, is widely used in China, Korea, and other Southeast Asian countries. However, Flos Lonicerae Japonicae and its closely related species have been misused and traded at varying for a wide range of prices. Therefore, Flos Lonicerae Japonicae must be accurately identified. In this study, the ITS2 and psbA-trnH regions were amplified by polymerase chain reaction (PCR). Sequence assembly was performed using CodonCode Aligner V 3.5.4. The intra- versus inter-specific variations were assessed using six metrics and “barcoding gaps.” Species identification was conducted using BLAST1 and neighbor-joining (NJ) trees. Results reveal that ITS2 and psbA-trnH exhibited an average intraspecific divergence of 0.001 and 0, respectively, as well as an average inter-specific divergence of 0.0331 and 0.0161. The identification efficiency of ITS2 and psbA-trnH evaluated using BLAST1 was 100%. Flos Lonicerae Japonicae was formed into one clade through the NJ trees. Therefore, Flos Lonicerae Japonicae can be stably and accurately identified through the ITS2 and psbA-trnH regions, respectively.
1. Introduction
Lonicera japonica Thunb. (Caprifoliaceae) is a commonly used herb that grows widely in China, Japan, Korea, and Southeast Asian countries [1]. The flower bud, called Flos Lonicerae Japonicae, is the most important part of the plant for medicinal use [2, 3], with more than 3000 years of medical history. Flos Lonicerae Japonicae was first reported in one of the earliest pharmacopoeias worldwide, Shen-Nong's Herbals Classic [4]. Flos Lonicerae Japonicae is considered a herbal medicine with heat-clearing, detoxifying, and anti-inflammatory effects [5, 6]. In addition, extracts of Flos Lonicerae Japonicae have shown efficacy against various diseases such as H1N1 influenza [4, 7–9], hand-foot-and-mouth disease (HFMD) [4, 10], and severe acute respiratory syndromes, among others [11–13]. In recent years, Flos Lonicerae Japonicae has also become the main material of a Chinese “cooling” beverage [14]. Flos Lonicerae, which comes from L. macranthoides Hand-Mazz, L. fulvotomentosa Hsu et S.C. Cheng, L. confusa (Sweet) DC., and L. hypoglauca Miq., is another medicinal material from Lonicera [6]. More than 30% of the current traditional Chinese medicine prescriptions have recently used Flos Lonicerae Japonicae [4]. Consequently, Flos Lonicerae and other species of Lonicera, such as L. japonica var. chinensis, L. similis, and L. acuminata, have been misused as Flos Lonicerae Japonicae because of high market demand for Flos Lonicerae Japonicae.
High-quality botanical materials are important for the safe and effective use of herbal products [15]. Several criteria and techniques have been used in the identification of Flos Lonicerae Japonicae to ensure its quality and therapeutic effects [16–19]. Li et al. introduced the morphologic and microstructural methods for identifying L. japonica and Lonicerae [20]. Despite its ability to preliminarily identify L. japonica, the method needs the integrity of the material structure. The geoherbalism of L. japonica has been identified by sequence divergence of the 5S-rRNA gene spacer region [21]. However, Flos Lonicerae Japonicae is difficult to identify according to morphology, microstructural methods, and certain molecular markers.
DNA barcoding can quickly and reliably identify species using a standard genetic sequence that can be amplified by universal primers from a small fragment of the genome [22–25]. Several DNA regions have been evaluated for potential application as DNA barcodes, such as matK, rbcL, psbA-trnH, and ITS2 [26–28]. In the recent years, the DNA barcoding technology has been increasingly used in several fields including species identification [29–33], biodiversity study [34, 35], misidentification detection [36], food safety testing [14], and tackling illegal trade of endangered species [37, 38]. Chen et al. suggested that ITS2 can be used as a novel DNA barcode for the identification of medicinal plants and their closely related species [27]. The potential identification ability of ITS2 in plants has been assessed by analyzing 50,790 plant ITS2 sequences [28]. The China Plant BOL Group has also shown that ITS2 can be used to identify seed plants [39]. ITS2 sequences are potential general phylogenetic markers and have been widely used for phylogenetic reconstructions at both the genus and the species levels [40, 41]. Previous studies used the psbA-trnH intergenic spacer region from plastid DNA to successfully resolve a number of species identification cases [42–45]. Chen et al. recommended psbA-trnH as a complementary barcode to ITS2 for a broad series of plant taxa [27].
In our previous study, the genomic DNA extracted from the leaves of L. japonica was used for genomic identification. Sun et al. tested several potential DNA regions and proposed that psbA-trnH can be used for the identification of L. japonica and its closely related species [46]. In the present study, the ITS2 and psbA-trnH barcoding regions of Flos Lonicerae Japonicae and closely related species were evaluated for the suitability for species discrimination.
2. Materials and Methods
2.1. Materials
Flower bud samples from 45 individuals belonging to seven species, which were used as Flos Lonicerae Japonicae and Flos Lonicerae in Chinese markets, were collected from various geographical areas in China, including Henan, Shandong, and Guizhou (Table 1). All samples were identified by Professor Yulin Lin from the Institute of Medicinal Plant Development (IMPLAD), Chinese Academy of Medical Sciences. Voucher specimens were deposited in the IMPLAD herbarium in Beijing, China. Subsequently, eight ITS2 sequences of L. japonica were downloaded from GenBank for further analysis.
Table 1.
Samples used in the present study.
| Latin name | Haplotype (for ITS2) | Voucher no. | GenBank no. ITS2, psbA-trnH |
Locality |
|---|---|---|---|---|
| L. japonica | A1 | YC0014MT15 | JX669516, JX885527 | Pingyi, Shandong |
| L. japonica | A1 | YC0014MT60 | JX682917, JX885540 | Pingyi, Shandong |
| L. japonica | A1 | YC0014MT91 | JX682918, JX885545 | Kaixian, Chongqing |
| L. japonica | A1 | YC0014MT104 | JX682919, JX885525 | Huanggang, Hubei |
| L. japonica | A2 | YC0014MT16 | JX669517, JX885528 | Pingyi, Shandong |
| L. japonica | A2 | YC0014MT29 | JX682898, JX885529 | Xixia, Henan |
| L. japonica | A2 | YC0014MT31 | JX682899, JX885530 | Xixia, Henan |
| L. japonica | A2 | YC0014MT32 | JX682900, JX885531 | Xixia, Henan |
| L. japonica | A2 | YC0014MT33 | JX682901, JX885532 | Xixia, Henan |
| L. japonica | A2 | YC0014MT41 | JX682902, JX885533 | Fengqiu, Henan |
| L. japonica | A2 | YC0014MT43 | JX682903, JX885534 | Fengqiu, Henan |
| L. japonica | A2 | YC0014MT54 | JX682904, JX885535 | Donghai, Jiangsu |
| L. japonica | A2 | YC0014MT55 | JX682905, JX885536 | Donghai, Jiangsu |
| L. japonica | A2 | YC0014MT57 | JX682906, JX885537 | Donghai, Jiangsu |
| L. japonica | A2 | YC0014MT58 | JX682907, JX885538 | Donghai, Jiangsu |
| L. japonica | A2 | YC0014MT59 | JX682908, JX885539 | Donghai, Jiangsu |
| L. japonica | A2 | YC0014MT62 | JX682909, JX885541 | Pingyi, Shandong |
| L. japonica | A2 | YC0014MT63 | JX682910, JX885542 | Pingyi, Shandong |
| L. japonica | A2 | YC0014MT64 | JX682911, JX885543 | Nanning, Guangxi |
| L. japonica | A2 | YC0014MT81 | JX682912, JX885544 | Liupanshui, Guizhou |
| L. japonica | A2 | YC0014MT92 | JX682913, JX885546 | Wanzhou, Chongqing |
| L. japonica | A2 | YC0014MT98 | JX682914, JX885547 | Zhoukou, Henan |
| L. japonica | A2 | YC0014MT101 | JX682915, JX885524 | Shucheng, Anhui |
| L. japonica | A2 | YC0014MT108 | JX682916, JX885526 | Julu, Hebei |
| L. japonica | A2 | — | JQ731712 | GenBank |
| L. japonica | A2 | — | JQ731715 | GenBank |
| L. japonica | A2 | — | JQ780993 | GenBank |
| L. japonica | A2 | — | JQ780991 | GenBank |
| L. japonica | A2 | — | JQ780986 | GenBank |
| L. japonica | A2 | — | JQ780987 | GenBank |
| L. japonica | A2 | — | JQ780988 | GenBank |
| L. japonica | A2 | — | JQ780989 | GenBank |
| L. macranthoides | B1 | YC0163MT10 | JX669518, JX885548 | Liuzhi, Guizhou |
| L. macranthoides | B1 | YC0163MT20 | JX682920, JX885549 | Banan, Chongqing |
| L. macranthoides | B1 | YC0163MT21 | JX682921, JX885550 | Longhui, Hunan |
| L. macranthoides | B1 | YC0163MT25 | JX682922, JX885551 | Nanjiang, Sichuan |
| L. macranthoides | B1 | YC0163MT26 | JX682923, JX885552 | Puer, Yunan |
| L. fulvotomentosa | B2 | YC0164MT01 | JX669519, JX885553 | Anlong, Guizhou |
| L. fulvotomentosa | B2 | YC0164MT07 | JX682924, JX885554 | Xingren, GuiZhou |
| L. fulvotomentosa | B2 | YC0164MT08 | JX682925, JX885555 | Zhenfeng, GuiZhou |
| L. fulvotomentosa | B2 | YC0164MT09 | JX682926, JX885556 | zerong, GuiZhou |
| L. fulvotomentosa | B2 | YC0164MT11 | JX682927, JX885557 | Xingyi, GuiZhou |
| L. hypoglauca | B3 | YC0165MT01 | JX669520, JX885558 | Nanning, Guangxi |
| L. hypoglauca | B3 | YC0165MT02 | JX682928, JX885559 | Hezhou, Guangxi |
| L. confusa | B4 | YC0166MT04 | JX669521, JX885560 | Shipan, Sichuan |
| L. confusa | B4 | YC0166MT07 | JX682929, JX885561 | Jianyang, Sichuan |
| L. similis | B5 | YC0169MT04 | JX669522, JX885562 | Chengkou, Chongqing |
| L. similis | B5 | YC0169MT07 | JX682930, JX885563 | Kaixian, Chongqing |
| L. similis | B5 | YC0169MT10 | JX682931, JX885564 | Chengkou, Chongqing |
| L. similis | B5 | YC0169MT11 | JX682932, JX885565 | Chengkou, Chongqing |
| L. similis | B5 | YC0169MT16 | JX682933, JX885566 | Kaixian, Chongqing |
| L. acuminata | B6 | YC0170MT01 | JX669523, JX885567 | Jianyang, Sichuan |
| L. acuminata | B6 | YC0170MT04 | JX682934, JX885568 | Guangyuan, Sichuan |
2.2. DNA Extraction, PCR Amplification, and Sequencing
Total genomic DNA was extracted from 25 mg of dry flower bud by the Plant Genomic DNA Kit (Tiangen Biotech Co., Beijing, China) after grinding for 1 min at 30 oscillations/s using a DNA Extraction Grinder (MM 400; Retsch, Haan, Germany). PCR amplifications of the ITS2 region were performed in a Peltier Thermal Cycler PTC0200 (BioRad Lab Inc., USA) using a pair of primer ITS2F (5′-ATGCGATACTTGGTGTGAAT-3′) and ITS3R (5′-GACGCTTCTCCAGACTACAAT-3′) [27]. PCR reactions were performed in a 25 μL volume containing approximately 20–50 ng of genomic DNA, 2× EasyTaq PCR SuperMix (TransGen Biotech Co., China), 1.0 μL of each primer (2.5 μM), and distilled deionized water. The reaction conditions were as follows: 95°C for 5 min, 40 cycles at 94°C for 30 s, 58°C for 30 s, 72°C for 45 s, and 72°C for 10 min. The psbA-trnH was conducted as described previously [27, 42]. The desired PCR products were separated by electrophoresis in 1.2% agarose gels. The purified PCR products were directly sequenced bidirectionally by the sequencing center at Molecular Biology and Chemical (Mbchem, Shanghai, China).
2.3. Data Analysis
The forward and reverse trace files were trimmed and assembled after sequencing using the CodonCode Aligner V 3.5.4 (CodonCode Co., USA). The ITS2 region was annotated with the HMMer software using ITS2 database [47]. The genetic distances were then calculated using MEGA 5.0 [48], according to the Kimura 2-Parameter (K2P) model. The average intra-specific distance, theta, and coalescent depth were computed to evaluate the intraspecific variation by the K2P model [49]. The average interspecific distance, minimum interspecific distance, and theta primer were used to represent interspecific divergences [24, 27]. The distribution of intra- versus inter-specific variability was assessed using the DNA barcoding gaps [50]. Species identification efficiency was determined based on BLAST1 method [51]. NJ trees were constructed to identify Flos Lonicerae Japonicae and its closely related species.
3. Results
3.1. Efficiency of PCR Amplification
DNA was isolated from 45 individuals. Some smear bands can only be found by DNA gel electrophoresis because of DNA degradation. The PCR amplification success rates for ITS2 and psbA-trnH were both 100% (Figure 1). The sequencing results showed that high-quality bidirectional trace files were obtained from the ITS2 regions.
Figure 1.
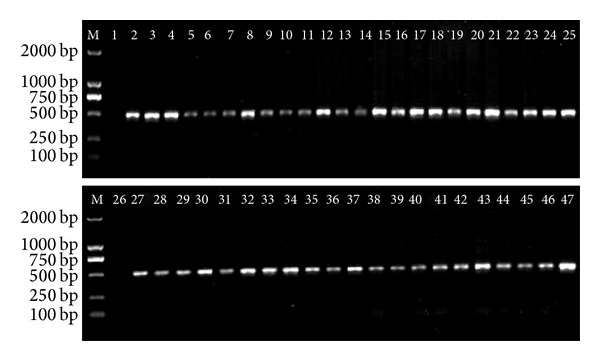
PCR amplification results of the ITS2 regions of L. japonica and its related species. M: marker; 1 and 26: negative control (CK); 2–25: L. japonica; 27–31: L. macranthoides; 32–36: L. fulvotomentosa; 37-38: L. hypoglauca; 39-40: L. confuse; 41–45: L. similis; 46-47: L. acuminate.
3.2. Analysis of DNA Divergence
The ITS2 sequence lengths of 24 Flos Lonicerae Japonicae samples were 228 bp, and the average GC content was 75.4%. Only one nucleotide variation site was found at the 27th base, which is the substitution of C/T. Two new haplotypes of ITS2 sequences generated in this study were submitted to GenBank, with the following accession numbers: JX669516 and JX669517. The ITS2 regions of haplotype A1 were the same as that of JX669516, whereas that of haplotype A2 was the same as that of JX669517. According to the K2P model, the intraspecific distance of Flos Lonicerae Japonicae ITS2 ranged from 0 to 0.0044. The ITS2 sequence length of Flos Lonicerae and other samples ranged from 228 bp to 229 bp; the average GC content was 73.9%. A total of 14 nucleotide variation sites and one insertion/deletion mutation were detected in 53 ITS2 sequences (Table 2). In this study, the ITS2 region of the other Lonicera species except L. japonica can be divided into six haplotypes (Table 1). These ITS2 regions were also submitted to GenBank, and GenBank accession numbers were obtained (Table 1).
Table 2.
Sequence characteristics of ITS2 and psbA-trnH of Flos Lonicerae Japonicae and its related species.
| Sequence characteristics | ITS2 | psbA-trnH |
|---|---|---|
| Length range in Flos Lonicerae Japonicae (bp) | 228 | 339 |
| Length in all taxa (bp) | 228-229 | 332–356 |
| Average of GC content in Flos Lonicerae Japonicae (%) | 75.4 | 29.5 |
| Average of GC content in all taxa (%) | 73.9 | 29.6 |
| No. of variable sites in Flos Lonicerae Japonicae | 1 | 0 |
| No. of variable sites in all taxa | 14 | 10 |
Using the same approach, also psbA-trnH sequences of the 45 Lonicera samples were analyzed (Table 2). The sequence length of the psbA-trnH of Flos Lonicerae Japonicae was 339 bp, and the average GC content was 29.5%. No variation sites were found in the psbA-trnH regions. Additional characteristics of the psbA-trnH sequences of 45 samples are shown in Table 2.
Ideal barcode sequences should provide a distinct inter-specific divergence and relatively small intra-specific variation. They must concurrently show significant differences between these two types of variation to distinguish individual intra-specific variation from variation between species. According to the K2P model, the genetic distance of Flos Lonicerae Japonicae from its closely related species was evaluated using the ITS2 regions. The inter-specific distance was 0.0331, which is larger than the intra-specific distance of 0.0010. The minimum inter-specific distance was 0.0142, which is larger than coalescent depth of 0.0019 (Table 3). Likewise, the minimum inter-specific distance for the psbA-trnH was 0.0060, which is larger than coalescent depth of 0.0004 (Table 3).
Table 3.
Inter- and intra-specific genetic divergence and identification efficiency in ITS2 and psbA-trnH sequences of L. japonica and its related species.
| Parameter | ITS2 | psbA-trnH |
|---|---|---|
| All inter-specific distance | 0.0331 ± 0.0119 | 0.0161 ± 0.0066 |
| Theta primer | 0.0253 ± 0.0093 | 0.0126 ± 0.0050 |
| The minimum inter-specific distance | 0.0142 ± 0.0105 | 0.0060 ± 0.0060 |
| All intra-specific distance | 0.0010 ± 0.0018 | 0.0000 ± 0.0003 |
| Theta | 0.0011 ± 0.0093 | 0.0002 ± 0.0004 |
| Coalescent depth | 0.0019 ± 0.0024 | 0.0004 ± 0.0011 |
| Identification efficiency (BLAST1) | 100% | 100% |
3.3. Assessment of Barcoding Gap
We investigated the distribution of genetic distance in classes of 0.010 distance units for ITS2 and psbA-trnH sequences to perform a preliminary examination of intra- versus inter-specific variation. The results showed apparent gaps between the intra- and inter-specific variability of ITS2. Obvious gaps were also found between the intra- and inter-specific variability of psbA-trnH using the same method (Figure 2).
Figure 2.
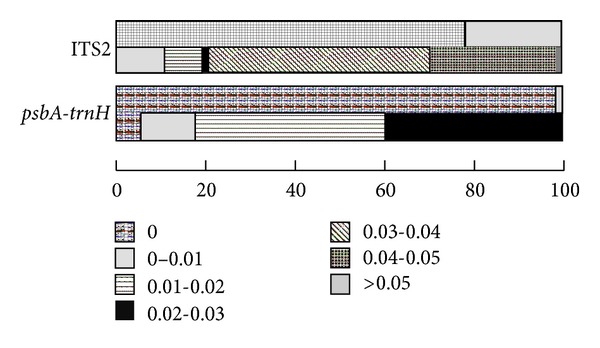
Relative distribution of inter-specific divergence between Flos Lonicerae Japonicae and intra-specific variation for ITS2 and psbA-trnH sequences. The two bars in the box represent intra-specific (above) and inter-specific (below) genetic distances.
3.4. Applicability for Species Discrimination
Two methods of BLAST1 and NJ tree techniques were used to evaluate the reliability of species identification using DNA barcoding. The results indicated that the ITS2 and psbA-trnH regions exhibited the highest identification efficiency (100%) by BLAST1 (Table 3). NJ tree effectively determines the power of a given locus combination to discriminate among species. In this study, a phylogenetic tree was constructed using the NJ method, with 1000 bootstrap replicates for the ITS2 and psbA-trnH sequences (Figure 3, Supplementary Figure 1 available online at: http://dx.doi.org/10.1155/2013/549037). The results showed that L. japonica formed into one clade, which can be distinguished successfully from its closely related species.
Figure 3.
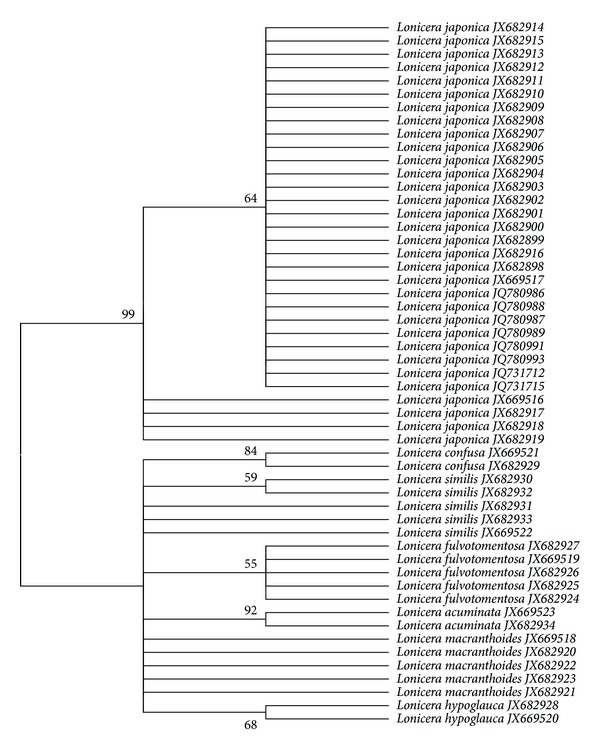
Phylogenetic tree of Flos Lonicerae Japonicae and its closely related species constructed with the ITS2 sequences using NJ method. The bootstrap scores (1 000 replicates) are shown (≥50%) for each branch.
4. Discussion
An ideal barcoding system should have sufficient variation among the sequences to discriminate species; however, it also needs to be sufficiently conserved to achieve less variability within species than between species [52]. In addition, the DNA barcode should be easily amplified using one pair of universal primers; identification should be rapid, accurate, cost effective, and easy to use even for a nonspecialist [53]. The applicability of the ITS2 regions for species identification has been tested by Chen et al. [27]. In recent years, the DNA barcoding technique has become the main method for species identification [54–56].
In the present study, the ITS2 region was used for the first time to identify Flos Lonicerae Japonicae and its closely related species. The samples were collected from the main producing areas of the species. All samples were obtained from the flower bud, which is the medicinal part of L. japonica. The following results were obtained from this study. First, the length of the ITS2 of Flos Lonicerae Japonicae was 228 bp, which is short and relatively easy to amplify using one pair of universal primers. Second, the ITS2 regions of Flos Lonicerae Japonicae obtained from different areas were similar; only one variable site was found. The minimum inter-specific distance was 0.0142, which is significantly larger than coalescent depth of 0.0019. Divergence at the inter-specific level and relatively low divergence at the intra-specific level (Table 3) were found. Third, the identification efficiency of the 53 samples was 100% by the BLAST1 method. The results obtained using the NJ tree can also be used to distinguish Flos Lonicerae Japonicae successfully. In summary, the ITS2 sequence can be used as a DNA barcode for identifying Flos Lonicerae Japonicae and its closely related species. To date, ITS2 regions have been widely used to distinguish related species. The study by Gao et al. [31] indicated that ITS2 regions can be used as an efficient and powerful marker for distinguishing various species in Fabaceae. Pang et al. accurately identified the Ephedrae herb and its closely related species by the ITS2 DNA barcoding technique [57]. The stability and accuracy of identifying Notopterygii Rhizoma et Radix has been tested using the ITS/ITS2 barcodes [55]. All results consistently showed that the ITS2 DNA barcode exhibits good stability and accuracy in identifying Chinese materia medica.
A previous study by Sun et al. used the leaves of L. japonica and proposed the psbA-trnH regions for the identification of L. japonica and its related species [46]. We obtained genomic DNA from the flowers of Flos Lonicerae Japonicae and assessed their PCR amplification and identification efficiency to test the psbA-trnH regions. The results revealed that psbA-trnH can also be used to identify Flos Lonicerae Japonicae and verified the results of Sun et al. [46]. This study simultaneously showed that ITS2 regions can be used to distinguish Flos Lonicerae Japonicae. However, in the study by Sun et al., the resulting PCR product of ITS2 was not amplified using the approach employed by Chen et al. [27]. Compared with the study by Sun et al., the present study obtained two findings. One is that genomic DNA, which can be improved for follow-up experiments, was extracted by extending the pyrolysis time. The other is that the ITS2 regions were amplified successfully by increasing the annealing temperature from 56°C to 58°C using a universal primer and a 2× EasyTaq PCR SuperMix (TransGen Biotech Co., China). Sequence analysis indicated that the average GC contents of Flos Lonicerae Japonicae was 75.4%, which may be that reason Sun et al. failed to obtain the ITS2 regions by the Chen et al. method [27].
Several studies have reported the existence of multiple copies of ITS2 in plants and animals [58]. Questions have been raised whether the sequence obtained by PCR would be stable and representative as a DNA marker. Alvarez and Wendel suggested that the presence of different copies of ITS2 can result in misleading phylogenetic inferences [58]. However, Coleman indicated that ITS2 can be effectively treated as a single locus [59]. The most recent study also revealed that using the major variants alone is sufficient for phylogeny construction and species determination in most cases although intragenomic multiple variants are frequently found within each genome. Inclusion of minor variants further improves the resolution of species identification [60]. In the present study, the PCR amplification success rates for ITS2 was 100% and high-quality sequence stability from the 45 samples was obtained. This result indicates that the major variants alone are sufficient for PCR amplification of the ITS2 region. Therefore, the ITS2 DNA barcode can be used to identify Flos Lonicerae Japonicae and its closely related species.
The results of this study can help identify Flos Lonicerae Japonicae and Flos Lonicerae in the market and ensure safe and stable supply of traditional medicines derived from L. japonica. These findings can be used as reference for the authentication of other medicinal herbs, especially the flower types. The results can also be valuable in resource protection, industrial production, customs, forensic examination, and plant quarantine.
5. Conclusions
ITS2 and psbA-trnH were examined for their stability and accuracy in identifying Chinese materia medica in the case study with Flos Lonicerae Japonicae. The results indicate that using the ITS2 and psbA-trnH regions as DNA barcodes can stably and accurately distinguish Chinese materia medica as well as provide a new technique to ensure clinical safety in using traditional Chinese medicines.
Supplementary Material
Flos Lonicerae Japonicae is a traditional Chinese materia medica which has an important function in the treatment of H1N1 influenza, hand-foot-and-mouth disease (HFMD), and severe acute respiratory syndromes (SARS). However, Flos Lonicerae Japonicae and its closely related species have been misused. Therefore, it is imperative to identify them accurately. DNA barcoding is a novel molecular identification method that provides a turning point in solving the difficulty in identifying traditional Chinese materia medica using traditional identification techniques. However, the stability and accuracy of DNA barcoding need further study. The present study aims to examine the stability and accuracy of the ITS2 and psbA-trnH regions in the identification of Flos Lonicerae Japonicae and its closely related species. The results showed that both regions can stably and accurately distinguish Flos Lonicerae Japonicae and its closely related species.
Conflict of Interests
The authors have declared that no competing interests exist in this paper.
Acknowledgments
The authors thank Y. L. Lin (Chinese Academy of Medical Sciences, Beijing, China) for morphological confirmation of medicinal materials. This work is supported by the National High-tech R&D Program of China (863 Program) (no. 2012AA021602) and supported by the Fundamental Research Funds for the Central Universities (no. 2012C02). This work is also supported by Grants from the National Natural Science Foundation of China (no. 81073001).
References
- 1.Park E, Kum S, Wang C, Park SY, Kim BS, Schuller-Levis G. Anti-inflammatory activity of herbal medicines: inhibition of nitric oxide production and tumor necrosis factor-α secretion in an activated macrophage-like cell line. American Journal of Chinese Medicine. 2005;33(3):415–424. doi: 10.1142/S0192415X05003028. [DOI] [PubMed] [Google Scholar]
- 2.Lu H, Jiang Y, Chen F. Application of preparative high-speed counter-current chromatography for separation of chlorogenic acid from Flos Lonicerae . Journal of Chromatography A. 2004;1026(1-2):185–190. doi: 10.1016/j.chroma.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 3.Choi C, Hyun AJ, Sam SK, Jae SC. Antioxidant constituents and a new triterpenoid glycoside from Flos Lonicerae . Archives of Pharmacal Research. 2007;30(1):1–7. doi: 10.1007/BF02977770. [DOI] [PubMed] [Google Scholar]
- 4.Yuan Y, Song LP, Li MH, et al. Genetic variation and metabolic pathway intricacy govern the active compound content and quality of the Chinese medicinal plant Lonicera japonica thumb. BMC Genomics. 2012;13, article 195 doi: 10.1186/1471-2164-13-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yoo H, Kang H, Yun SS, Park E, Lim C. Anti-angiogenic, antinociceptive and anti-inflammatory activities of Lonicera japonica extract. Journal of Pharmacy and Pharmacology. 2008;60(6):779–786. doi: 10.1211/jpp.60.6.0014. [DOI] [PubMed] [Google Scholar]
- 6.Chinese Pharmacopoeia Commission. The Pharmacopoeia of the People’s Republic of China, Volume 1. Beijing, China: China Medical Science Press; 2010. [Google Scholar]
- 7.Ko H, Wei B, Chiou W. The effect of medicinal plants used in Chinese folk medicine on RANTES secretion by virus-infected human epithelial cells. Journal of Ethnopharmacology. 2006;107(2):205–210. doi: 10.1016/j.jep.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Jin JH, Hyun L, Kwon SY, Son KH, Kim HP. Anti-inflammatory activity of the total flavonoid fraction from Broussonetia papyrifera in combination with Lonicera japonica . Biomolecules and Therapeutics. 2010;18(2):197–204. [Google Scholar]
- 9.Tian J, Che HL, Ha D, Wei YP, Zheng SY. Characterization and anti-allergic effect of a polysaccharide from the flower buds of Lonicera japonica . Carbohydrate Polymers. 2012;90:1642–1647. doi: 10.1016/j.carbpol.2012.07.044. [DOI] [PubMed] [Google Scholar]
- 10.Cao H, Liu Z, Steinmann P, Mu Y, Luo H, Liu J. Chinese herbal medicines for treatment of hand, foot and mouth disease: a systematic review of randomized clinical trials. European Journal of Integrative Medicine. 2012;4(1):e85–e111. [Google Scholar]
- 11.Lin L, Xu Y, He D, et al. A retrospective study on clinical features of and treatment methods for 77 severe cases of SARS. American Journal of Chinese Medicine. 2003;31(6):821–839. doi: 10.1142/S0192415X03001521. [DOI] [PubMed] [Google Scholar]
- 12.Li XH, Zhang K, Hu JH, et al. Clinical evaluation of integrative Chinese and western medicine in treating SARS. Chinese Journal of Integrative Medicine. 2003;9:181–184. [Google Scholar]
- 13.Shang X, Pan H, Li M, Miao X, Ding H. Lonicera japonica Thunb.: ethnopharmacology, phytochemistry and pharmacology of an important traditional Chinese medicine. Journal of Ethnopharmacology. 2011;138(1):1–21. doi: 10.1016/j.jep.2011.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li M, Wong K, Chan W, et al. Establishment of DNA barcodes for the identification of the botanical sources of the Chinese “cooling” beverage. Food Control. 2012;25(2):758–766. [Google Scholar]
- 15.Chung S, Cheng F, Lee M, Lin J, Lin M, Wang M. Ginkgo biloba leaf extract (EGb761) combined with neuroprotective agents reduces the infarct volumes of gerbil ischemic brain. American Journal of Chinese Medicine. 2006;34(5):803–817. doi: 10.1142/S0192415X06004302. [DOI] [PubMed] [Google Scholar]
- 16.Wang C, Li P, Ding J, Fishbein A, Yuan C. Discrimination of Lonicera japonica THUNB. from different geographical origins using restriction fragment length polymorphism analysis. Biological and Pharmaceutical Bulletin. 2007;30(4):779–782. doi: 10.1248/bpb.30.779. [DOI] [PubMed] [Google Scholar]
- 17.Peng X, Li W, Wang W, Bai G. Identification of Lonicera japonica by PCR-RFLP and allele-specific diagnostic PCR based on sequences of internal transcribed spacer regions. Planta Medica. 2010;76(5):497–499. doi: 10.1055/s-0029-1186235. [DOI] [PubMed] [Google Scholar]
- 18.Fu S, Zhang T, Lü J, Guo J, Yuan H, Xiao X. Comparison of microcalorimetric fingerprint profiles of Lonicerae Japonicae Flos and Lonicerae Flos. Yaoxue Xuebao. 2011;46(10):1251–1256. [PubMed] [Google Scholar]
- 19.Jiang C, Yuan Y, Liu GM, et al. EST-SSR identification of Lonicera japonica Thunb. Acta Pharmaceutica Sinica. 2012;47:803–810. [PubMed] [Google Scholar]
- 20.Li JJ, Li JF, Li JY, Zhou YQ, Chen XJ. Identification methods of Lonicera japonica and Lonicera confusa . Journal of Henan Agricultural Sciences. 2011;40:134–137. [Google Scholar]
- 21.Li P, Cai CH, Xing JP. Preliminary attempt to identify geoherbalism of Flos Lonicerae by sequence divergence of 5S-rRNA gene spacer region. Chinese Traditional and Herbal Drugs. 2001;32:834–837. [Google Scholar]
- 22.Hebert PDN, Ratnasingham S, DeWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proceedings of the Royal Society B. 2003;270(1):S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebert PDN, Cywinska A, Ball SL, DeWaard JR. Biological identifications through DNA barcodes. Proceedings of the Royal Society B. 2003;270(1512):313–321. doi: 10.1098/rspb.2002.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lahaye R, Van Der Bank M, Bogarin D, et al. DNA barcoding the floras of biodiversity hotspots. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(8):2923–2928. doi: 10.1073/pnas.0709936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Erickson DL, Spouge J, Resch A, Weigt LA, Kress WJ. DNA barcoding in land plants: developing standards to quantify and maximize success. Taxon. 2008;57(4):1304–1316. [PMC free article] [PubMed] [Google Scholar]
- 26.Hollingsworth PM, Forrest LL, Spouge JL, et al. A DNA barcode for land plants. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(31):12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Yao H, Han J, et al. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE. 2010;5(1) doi: 10.1371/journal.pone.0008613.e8613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao H, Song J, Liu C, et al. Use of ITS2 region as the universal DNA barcode for plants and animals. PLoS ONE. 2010;5(10) doi: 10.1371/journal.pone.0013102.e13102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luo K, Chen SL, Chen KL, et al. Assessment of candidate plant DNA barcodes using the Rutaceae family. Science China Life Sciences. 2010;53(6):701–708. doi: 10.1007/s11427-010-4009-1. [DOI] [PubMed] [Google Scholar]
- 30.Shi L, Zhang J, Han J, et al. Testing the potential of proposed DNA barcodes for species identification of Zingiberaceae. Journal of Systematics and Evolution. 2011;49(3):261–266. [Google Scholar]
- 31.Gao T, Yao H, Song J, Zhu Y, Liu C, Chen S. Evaluating the feasibility of using candidate DNA barcodes in discriminating species of the large Asteraceae family. BMC Evolutionary Biology. 2010;10(1, article 324) doi: 10.1186/1471-2148-10-324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Wang F, Yan H, Hao G, Hu C, Ge X. Testing DNA barcoding in closely related groups of Lysimachia L. (Myrsinaceae) Molecular Ecology Resources. 2012;12(1):98–108. doi: 10.1111/j.1755-0998.2011.03076.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu Y, Zhang L, Liu Z, Luo K, Chen S, Chen K. Species identification of Rhododendron (Ericaceae) using the chloroplast deoxyribonucleic acid PsbA-trnH genetic marker. Pharmacognosy Magazine. 2012;8(29):29–36. doi: 10.4103/0973-1296.93311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ebihara A, Nitta JH, Ito M. Molecular species identification with rich floristic sampling: DNA barcoding the pteridophyte flora of Japan. PLoS ONE. 2010;12 doi: 10.1371/journal.pone.0015136.e15136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kress WJ, Erickson DL, Swenson NG, Thompson J, Uriarte M, Zimmerman JK. Advances in the use of DNA barcodes to build a community phylogeny for tropical trees in a Puerto Rican forest dynamics plot. PloS ONE. 2010;5(11):p. e15409. doi: 10.1371/journal.pone.0015409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pryer KM, Schuettpelz E, Huiet L, et al. DNA barcoding exposes a case of mistaken identity in the fern horticultural trade. Molecular Ecology Resources. 2010;10(6):979–985. doi: 10.1111/j.1755-0998.2010.02858.x. [DOI] [PubMed] [Google Scholar]
- 37.Hebert PDN, Penton EH, Burns JM, Janzen DH, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(41):14812–14817. doi: 10.1073/pnas.0406166101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ragupathy S, Newmaster SG, Murugesan M, Balasubramaniam V. DNA barcoding discriminates a new cryptic grass species revealed in an ethnobotany study by the hill tribes of the Western Ghats in southern India. Molecular Ecology Resources. 2009;9(1):164–171. doi: 10.1111/j.1755-0998.2009.02641.x. [DOI] [PubMed] [Google Scholar]
- 39.China Plant BOL Group. Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(49):19641–19646. doi: 10.1073/pnas.1104551108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miao M, Warren A, Song W, Wang S, Shang H, Chen Z. Analysis of the internal transcribed spacer 2 (ITS2) region of Scuticociliates and related taxa (Ciliophora, Oligohymenophorea) to infer their evolution and phylogeny. Protist. 2008;159(4):519–533. doi: 10.1016/j.protis.2008.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Schultz J, Wolf M. ITS2 sequence-structure analysis in phylogenetics: a how-to manual for molecular systematics. Molecular Phylogenetics and Evolution. 2009;52(2):520–523. doi: 10.1016/j.ympev.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 42.Kress WJ, Wurdack KJ, Zimmer EA, Weigt LA, Janzen DH. Use of DNA barcodes to identify flowering plants. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(23):8369–8374. doi: 10.1073/pnas.0503123102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao H, Song J, Ma X, et al. Identification of Dendrobium species by a candidate DNA barcode sequence: the chloroplast PsbA-trnH intergenic region. Planta Medica. 2009;75(6):667–669. doi: 10.1055/s-0029-1185385. [DOI] [PubMed] [Google Scholar]
- 44.Song J, Yao H, Li Y, et al. Authentication of the family Polygonaceae in Chinese pharmacopoeia by DNA barcoding technique. Journal of Ethnopharmacology. 2009;124(3):434–439. doi: 10.1016/j.jep.2009.05.042. [DOI] [PubMed] [Google Scholar]
- 45.Ma X, Xie C, Liu C, et al. Species identification of medicinal pteridophytes by a DNA barcode marker, the chloroplast PsbA-trnH intergenic region. Biological and Pharmaceutical Bulletin. 2010;33(11):1919–1924. doi: 10.1248/bpb.33.1919. [DOI] [PubMed] [Google Scholar]
- 46.Sun Z, Gao T, Yao H, Shi L, Zhu Y, Chen S. Identification of Lonicera japonica and its related species using the DNA barcoding method. Planta Medica. 2011;77(3):301–306. doi: 10.1055/s-0030-1250324. [DOI] [PubMed] [Google Scholar]
- 47.Koetschan C, Förster F, Keller A, et al. The ITS2 Database III—sequences and structures for phylogeny. Nucleic Acids Research. 2010;38(1):D275–D279. doi: 10.1093/nar/gkp966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer CP, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biology. 2005;3:2229–2238. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gao T, Yao H, Song J, et al. Identification of medicinal plants in the family Fabaceae using a potential DNA barcode ITS2. Journal of Ethnopharmacology. 2011;130(1):116–121. doi: 10.1016/j.jep.2010.04.026. [DOI] [PubMed] [Google Scholar]
- 51.Ross HA, Murugan S, Li WLS. Testing the reliability of genetic methods of species identification via simulation. Systematic Biology. 2008;57(2):216–230. doi: 10.1080/10635150802032990. [DOI] [PubMed] [Google Scholar]
- 52.Taberlet P, Coissac E, Pompanon F, et al. Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Research. 2007;35(3, article e14) doi: 10.1093/nar/gkl938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stockinger H, Krüger M, Schüßler A. DNA barcoding of arbuscular mycorrhizal fungi. New Phytologist. 2010;187(2):461–474. doi: 10.1111/j.1469-8137.2010.03262.x. [DOI] [PubMed] [Google Scholar]
- 54.Pang X, Song J, Zhu Y, Xu H, Huang L, Chen S. Applying plant DNA barcodes for Rosaceae species identification. Cladistics. 2011;27(2):165–170. doi: 10.1111/j.1096-0031.2010.00328.x. [DOI] [PubMed] [Google Scholar]
- 55.Xin TY, Yao H, Luo K, et al. Stability and accuracy of the identification of Notopterygii Rhizoma et Radix using the ITS/ITS2 barcodes. Acta Pharmaceutica Sinica. 2012;47:1098–1105. [PubMed] [Google Scholar]
- 56.Hou DY, Song JY, Yao H, et al. Molecular identification of Corni Fructus and its adulterants by ITS/ITS2 sequences. Chinese Journal of Natural Medicines. 2013;11:121–127. doi: 10.1016/S1875-5364(13)60038-2. [DOI] [PubMed] [Google Scholar]
- 57.Pang XH, Song JY, Xu HB, Yao H. Using ITS2 barcode to identify ephedrae herba. China Journal of Chinese Materia Medica. 2012;37:1118–1121. [PubMed] [Google Scholar]
- 58.Álvarez I, Wendel JF. Ribosomal ITS sequences and plant phylogenetic inference. Molecular Phylogenetics and Evolution. 2003;29(3):417–434. doi: 10.1016/s1055-7903(03)00208-2. [DOI] [PubMed] [Google Scholar]
- 59.Coleman AW. Pan-eukaryote ITS2 homologies revealed by RNA secondary structure. Nucleic Acids Research. 2007;35(10):3322–3329. doi: 10.1093/nar/gkm233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song JY, Shi LC, Li DZ, et al. Extensive pyrosequencing reveals frequent intra-genomic variations of internal transcribed spacer regions of nuclear ribosomal DNA. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0043971.e43971 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flos Lonicerae Japonicae is a traditional Chinese materia medica which has an important function in the treatment of H1N1 influenza, hand-foot-and-mouth disease (HFMD), and severe acute respiratory syndromes (SARS). However, Flos Lonicerae Japonicae and its closely related species have been misused. Therefore, it is imperative to identify them accurately. DNA barcoding is a novel molecular identification method that provides a turning point in solving the difficulty in identifying traditional Chinese materia medica using traditional identification techniques. However, the stability and accuracy of DNA barcoding need further study. The present study aims to examine the stability and accuracy of the ITS2 and psbA-trnH regions in the identification of Flos Lonicerae Japonicae and its closely related species. The results showed that both regions can stably and accurately distinguish Flos Lonicerae Japonicae and its closely related species.


