Abstract
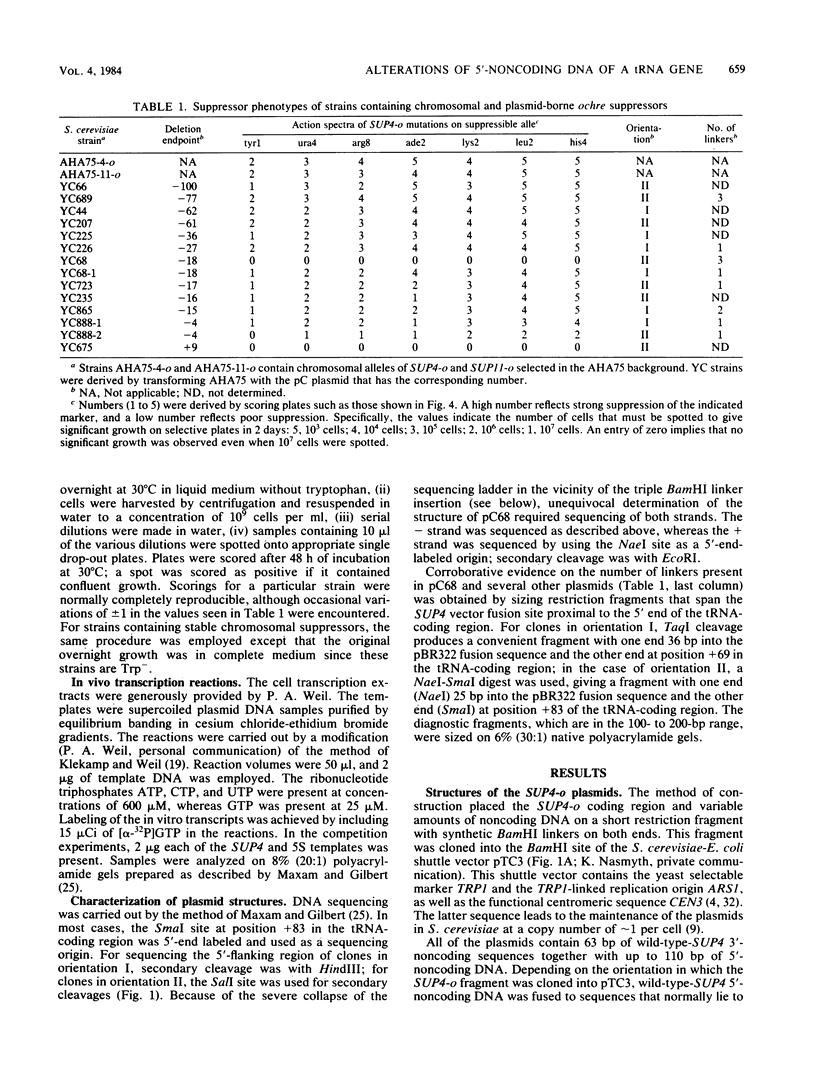
Deletion mutations ending in the 5'-flanking sequences of the Saccharomyces cerevisiae SUP4-o gene have been analyzed for their effects on gene expression. This ochre-suppressing tRNATyr gene was cloned into a S. cerevisiae centromeric plasmid, and its level of in vivo expression was monitored by observing the suppressor phenotype of the gene after transformation into S. cerevisiae. A deletion mutant that retains only four base pairs of the 5'-flanking sequence is profoundly deficient in expression; deletion mutants extending to positions -18, -17, -16, or -15 are moderately deficient; deletion mutants extending to positions -36 or -27 are slightly defective; and mutants retaining more than 60 base pairs of the original 5'-flanking DNA are expressed normally. In some cases, the cloning procedure led to the introduction of multiple BamHI linkers at the SUP4-o-vector fusion site, and in one instance, the resulting structure dramatically affects gene function: the presence of three linkers abutting a -18 deletion completely inhibits the in vivo expression of SUP4-o. In contrast, three linkers that abut a -77 deletion have no effect on in vivo expression. The template properties of these plasmids in a homologous in vitro transcription system parallel the levels of in vivo expression, suggesting that the mutations predominantly affect transcription. The data demonstrate that there are significant functional constraints on the 5'-flanking sequences of this RNA polymerase III-transcribed gene. The dramatic effects of the multiple linker insertion at position -18 suggest that there may be extensive melting of the DNA in this region during normal transcription initiation.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Betlach M. C., Boyer H. W. Construction and characterization of new cloning vehicles. I. Ampicillin-resistant derivatives of the plasmid pMB9. Gene. 1977;2(2):75–93. doi: 10.1016/0378-1119(77)90074-9. [DOI] [PubMed] [Google Scholar]
- Ciliberto G., Raugei G., Costanzo F., Dente L., Cortese R. Common and interchangeable elements in the promoters of genes transcribed by RNA polymerase iii. Cell. 1983 Mar;32(3):725–733. doi: 10.1016/0092-8674(83)90058-2. [DOI] [PubMed] [Google Scholar]
- Clarke L., Carbon J. Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature. 1980 Oct 9;287(5782):504–509. doi: 10.1038/287504a0. [DOI] [PubMed] [Google Scholar]
- DeFranco D., Schmidt O., Söll D. Two control regions for eukaryotic tRNA gene transcription. Proc Natl Acad Sci U S A. 1980 Jun;77(6):3365–3368. doi: 10.1073/pnas.77.6.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFranco D., Sharp S., Söll D. Identification of regulatory sequences contained in the 5'-flanking region of Drosophila lysine tRNA2 genes. J Biol Chem. 1981 Dec 10;256(23):12424–12429. [PubMed] [Google Scholar]
- Dinces N. B., Milman G. Viable polyoma virus with four BamHI decanucleotide linkers inserted at the 26-minute HincII site. J Mol Appl Genet. 1982;1(5):395–404. [PubMed] [Google Scholar]
- Engelke D. R., Ng S. Y., Shastry B. S., Roeder R. G. Specific interaction of a purified transcription factor with an internal control region of 5S RNA genes. Cell. 1980 Mar;19(3):717–728. doi: 10.1016/s0092-8674(80)80048-1. [DOI] [PubMed] [Google Scholar]
- Fitzgerald-Hayes M., Clarke L., Carbon J. Nucleotide sequence comparisons and functional analysis of yeast centromere DNAs. Cell. 1982 May;29(1):235–244. doi: 10.1016/0092-8674(82)90108-8. [DOI] [PubMed] [Google Scholar]
- Fowlkes D. M., Shenk T. Transcriptional control regions of the adenovirus VAI RNA gene. Cell. 1980 Nov;22(2 Pt 2):405–413. doi: 10.1016/0092-8674(80)90351-7. [DOI] [PubMed] [Google Scholar]
- Galli G., Hofstetter H., Birnstiel M. L. Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature. 1981 Dec 17;294(5842):626–631. doi: 10.1038/294626a0. [DOI] [PubMed] [Google Scholar]
- Gergen J. P., Stern R. H., Wensink P. C. Filter replicas and permanent collections of recombinant DNA plasmids. Nucleic Acids Res. 1979 Dec 20;7(8):2115–2136. doi: 10.1093/nar/7.8.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman H. M., Olson M. V., Hall B. D. Nucleotide sequence of a mutant eukaryotic gene: the yeast tyrosine-inserting ochre suppressor SUP4-o. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5453–5457. doi: 10.1073/pnas.74.12.5453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall B. D., Clarkson S. G., Tocchini-Valentini G. Transcription initiation of eucaryotic transfer RNA genes. Cell. 1982 May;29(1):3–5. doi: 10.1016/0092-8674(82)90083-6. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., Leupold U. Suppressors in yeast. Curr Top Microbiol Immunol. 1974;64(0):1–47. doi: 10.1007/978-3-642-65848-8_1. [DOI] [PubMed] [Google Scholar]
- Hawthorne D. C., Mortimer R. K. Genetic mapping of nonsense suppressors in yeast. Genetics. 1968 Dec;60(4):735–742. doi: 10.1093/genetics/60.4.735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopper A. K., Kurjan J. tRNA synthesis: identification of in vivo precursor tRNAs from parental and mutant yeast strains. Nucleic Acids Res. 1981 Feb 25;9(4):1019–1029. doi: 10.1093/nar/9.4.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klekamp M. S., Weil P. A. Specific transcription of homologous class III genes in yeast-soluble cell-free extracts. J Biol Chem. 1982 Jul 25;257(14):8432–8441. [PubMed] [Google Scholar]
- Klemenz R., Stillman D. J., Geiduschek E. P. Specific interactions of Saccharomyces cerevisiae proteins with a promoter region of eukaryotic tRNA genes. Proc Natl Acad Sci U S A. 1982 Oct;79(20):6191–6195. doi: 10.1073/pnas.79.20.6191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koski R. A., Allison D. S., Worthington M., Hall B. D. An in vitro RNA polymerase III system from S. cerevisiae: effects of deletions and point mutations upon SUP4 gene transcription. Nucleic Acids Res. 1982 Dec 20;10(24):8127–8143. doi: 10.1093/nar/10.24.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer R. A., Philippsen P., Davis R. W. Divergent transcription in the yeast ribosomal RNA coding region as shown by hybridization to separated strands and sequence analysis of cloned DNA. J Mol Biol. 1978 Aug 15;123(3):405–416. doi: 10.1016/0022-2836(78)90087-6. [DOI] [PubMed] [Google Scholar]
- Kurjan J., Hall B. D. Mutations at the Saccharomyces cerevisiae SUP4 tRNA(Tyr) locus: isolation, genetic fine-structure mapping, and correlation with physical structure. Mol Cell Biol. 1982 Dec;2(12):1501–1513. doi: 10.1128/mcb.2.12.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison J. T., Kung H. K. Large oligonucleotides isolated from yeast tyrosine transfer ribonucleic acid after partial digestion with ribonuclease T1. J Biol Chem. 1967 Mar 25;242(6):1324–1330. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- ROMAN H. Studies of gene mutation in Saccharomyces. Cold Spring Harb Symp Quant Biol. 1956;21:175–185. doi: 10.1101/sqb.1956.021.01.015. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J., Esposito R. E., Esposito M. S. The effect of ochre suppression on meiosis and ascospore formation in Saccharomyces. Genetics. 1977 Jan;85(1):35–54. doi: 10.1093/genetics/85.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakonju S., Bogenhagen D. F., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: I. The 5' border of the region. Cell. 1980 Jan;19(1):13–25. doi: 10.1016/0092-8674(80)90384-0. [DOI] [PubMed] [Google Scholar]
- Sakonju S., Brown D. D. Contact points between a positive transcription factor and the Xenopus 5S RNA gene. Cell. 1982 Dec;31(2 Pt 1):395–405. doi: 10.1016/0092-8674(82)90133-7. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Hagenbüchle O., Zuniga M. C. The nucleotide sequence of two silk gland alanine tRNAs: implications for fibroin synthesis and for initiator tRNA structure. Cell. 1977 Jul;11(3):561–570. doi: 10.1016/0092-8674(77)90074-5. [DOI] [PubMed] [Google Scholar]
- Sprague K. U., Larson D., Morton D. 5' flanking sequence signals are required for activity of silkworm alanine tRNA genes in homologous in vitro transcription systems. Cell. 1980 Nov;22(1 Pt 1):171–178. doi: 10.1016/0092-8674(80)90165-8. [DOI] [PubMed] [Google Scholar]
- Stinchcomb D. T., Struhl K., Davis R. W. Isolation and characterisation of a yeast chromosomal replicator. Nature. 1979 Nov 1;282(5734):39–43. doi: 10.1038/282039a0. [DOI] [PubMed] [Google Scholar]
- Struhl K., Stinchcomb D. T., Scherer S., Davis R. W. High-frequency transformation of yeast: autonomous replication of hybrid DNA molecules. Proc Natl Acad Sci U S A. 1979 Mar;76(3):1035–1039. doi: 10.1073/pnas.76.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. Complete nucleotide sequence of the Escherichia coli plasmid pBR322. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 1):77–90. doi: 10.1101/sqb.1979.043.01.013. [DOI] [PubMed] [Google Scholar]
- Thimmappaya B., Jones N., Shenk T. A mutation which alters initiation of transcription by RNA polymerase III on the Ad5 chromosome. Cell. 1979 Dec;18(4):947–954. doi: 10.1016/0092-8674(79)90207-1. [DOI] [PubMed] [Google Scholar]