Abstract
Background
Protein-Protein Interactions (PPIs) are key for many cellular processes. The characterization of PPI interfaces and the prediction of putative ligand binding sites and hot spot residues are essential to design efficient small-molecule modulators of PPI. Terphenyl and its derivatives are small organic molecules known to mimic one face of protein-binding alpha-helical peptides. In this work we focus on several PPIs mediated by alpha-helical peptides.
Method
We performed computational sequence- and structure-based analyses in order to evaluate several key physicochemical and surface properties of proteins known to interact with alpha-helical peptides and/or terphenyl and its derivatives.
Results
Sequence-based analysis revealed low sequence identity between some of the analyzed proteins binding alpha-helical peptides. Structure-based analysis was performed to calculate the volume, the fractal dimension roughness and the hydrophobicity of the binding regions. Besides the overall hydrophobic character of the binding pockets, some specificities were detected. We showed that the hydrophobicity is not uniformly distributed in different alpha-helix binding pockets that can help to identify key hydrophobic hot spots.
Conclusions
The presence of hydrophobic cavities at the protein surface with a more complex shape than the entire protein surface seems to be an important property related to the ability of proteins to bind alpha-helical peptides and low molecular weight mimetics. Characterization of similarities and specificities of PPI binding sites can be helpful for further development of small molecules targeting alpha-helix binding proteins.
Background
Protein-Protein Interactions (PPIs) are key to many cellular processes. Abnormal PPIs contribute to many disease states and as such, PPIs represent today a new class of drug targets essentially unexploited for drug discovery. Indeed, the size of the human interactome has been estimated to be between 300,000 [1] and 650,000 interactions [2]. In the last decade many studies have been performed in order to target PPIs [3]. Several small-molecule inhibitors of PPIs have been demonstrated therapeutic potential [4-8]. However, efficient targeting of PPIs is still being considered as an important challenge [3,9,10]. In contrast to enzyme-substrate interactions, protein-protein recognition often occurs through flat surfaces or wide shallow grooves. Recent structural analyses of PPI interfaces and small molecules disrupting PPIs suggested that such ligands might mimic the structural characteristics of the protein partner [6,11]. To facilitate the discovery of new PPI small-molecule inhibitors, the characterization of PPI interfaces [12,13] and the prediction of putative ligand binding sites are essential. Physicochemical properties of both ligand and protein are key to mediate the binding [14], such as cavity sizes, shape complementarity, electrostatic potential and hydrophobicity [12,15].
The role of alpha-helical peptides in mediating many PPIs is well demonstrated and development of small organic molecules mimicking such peptides becomes important [16]. Recent studies have been carried out on the whole Protein Data Bank (PDB) in order to establish a druggability profile of alpha-helix mediated PPIs and to predict which of them could bind a small molecule [17]. More specifically, terphenyl and its derivates are small organic molecules [18-26] mimicking one face of an alpha-helical peptide, i.e. the side chains of three key residues occupying positions i, i+3 and i+7[25,26] or i, i+4 and i+7[20] of the bound helix. It has been suggested that terphenyl compounds can serve as pharmacological probes because they are membrane permeable [22]. Terphenyl 1 and 2, which mimic the calmodulin binding face of smooth muscle myosin light chain kinase (smMLCK), have been shown to inhibit the interactions of calmodulin (CaM) with the enzyme 3'-5'-cyclic nucleotide phosphodiesterase (PDE) and with the helical peptide C20W of the plasma membrane calcium pumps [18]. Following the similarity between the calmodulin and human centrin 2 (HsCen2) alpha-helix binding sites, we recently suggested that terphenyl 2 might also inhibit the interaction between HsCen2 and a 17 residues peptide of Xeroderma Pigmentosum Group C (XPC) protein [27]. Terphenyl derivates mimicking the alpha-helical structure of p53 N-terminal peptide inhibit the p53-MDM2 [22] and the p53-HDM2 interactions [21]. These molecules also mimic the alpha-helical region of Bak BH3 domain, which binds BCL-X2, thus disrupting the BCL-X2/Bak interaction [19,20,24].
In this work we performed a computational analysis in order to evaluate several key physicochemical and surface properties of proteins known to interact with alpha-helical peptides or to bind terphenyl and its derivatives. We calculated the binding pocket volumes and the fractal dimensions of the surface cavities for the entire protein and for the binding pockets. We identified several similarities and specificities characterizing such protein binding sites that can be helpful for future development of more efficient small-molecule inhibitors targeting alpha-helix binding proteins.
Methods
In this study we compared the sequence and surface properties of the investigated proteins. In order to analyze the sequence similarities we performed sequence alignment using the CLUSTALW software [28]. Interacting residues at the protein-protein interface in terms of contact distances were found using the ContPro online freely available tool [29]. We identified the protein residues interacting with the three key residues of the alpha-helical peptide (occupying positions i, i+3 and i+7 or i, i+4 and i+7) those relative positions are mimicked by terphenyl and its derivatives. The distance threshold was set to 5 Å for the side chain atoms.
In order to evaluate the protein surface properties, the bound peptide was removed for each complex. The surface characteristics of the entire protein and those of the peptide-binding cavity were analyzed. Using the approach of the fractal geometry we quantitatively described the surface roughness for the entire protein and for the binding cavity, expressed by global surface fractal dimension (DS) and local surface fractal dimension (DL), respectively. In order to calculate the surface fractal dimension we used the method proposed by Lewis and Rees [30] based on the scaling law between the surface area (SA) and the radius of the rolling probe molecule (R) on the surface, i.e. SA is proportional to the radius to the power 2-Ds:
| (1) |
The surface fractal dimension was determined from the slope of the double logarithmical plot of SA versus R. The surface area of the protein was computed using the on-line available software GETAREA [31]. Probe radii of 1, 1.2, 1.4, 1.6, 1.8 and 2 Å were used. For the proteins cavities, the same algorithm was employed using the CASTp software [32]. Hydrophobicity and local hydrophobic density for binding pockets were determined using Fpocket [33]. Pocket volumes were computed using CASTp [32].
Molecular docking of terphenyl 2 was performed into the alpha-helical binding sites of calmodulin (code entry 2O5G) and troponin C (code entry 1A2X) using AutoDock 4.2 [34]. The input files preparation and docking analysis were carried out using AutoDockTools. Grid maps were centered in the alpha-helix binding site for both structures. Grids sizes were 126 Å x 126 Å x 126 Å with a grid spacing of 0.33 Å for calmodulin and 126 Å x 126 Å x 126 Å with a grid spacing of 0.28 Å for troponin C. Ligand conformational searching was performed using Lamarckian genetic algorithm and all ligand torsion angles were flexible. The following docking parameters were used: 250 Lamarckian genetic algorithm runs, a population size of 250, a maximum of 2 500 000 energy evaluations and a maximum of 27000 generations.
Figures were prepared using PyMol [35] and CHIMERA software [36].
Results and discussions
Sequence-based analysis
We analyze several proteins interacting with alpha-helical peptides, some of them being known to bind also terphenyl and/or its derivatives. To characterize and compare their surface properties we examine the sequences and the three dimensional (3D) structures of the complexes formed by the protein and the bound peptide. The 3D structures are retrieved from the PDB [37], the entry codes being presented in Table 1. Most of the structures are crystallographic. Two NMR structures are also used: the C-terminal domain of human centrin 2 in complex with the repeat sequence of human Sfi 1 and the human BCL-XL in complex with the BAK peptide.
Table 1.
Protein – alpha-helical peptide complexes
| Protein complex | PDB code Resolution | SwissProt code | Interacting residues of the bound alpha-helix |
|---|---|---|---|
| Chicken calmodulin in complex with smooth muscle myosin light chain kinase (smMLCK) |
2O5G* |
P62149 |
TRP5, THR8, VAL12 |
| 1.08 Å | |||
| Human calmodulin in complex with a mutant peptide of human DRP-1 kinase |
1ZUZ |
P62158 |
TRP305, PHE309, VAL312 |
| 1.91 Å | |||
| Human calmodulin in complex with CAV1.1 IQ peptide |
2VAY* |
P62158 |
THR526, ILE529, PHE533 |
| 1.94 Å | |||
| Human calmodulin in complex with CAV2.2 IQ peptide |
3DVE |
P62158 |
MET854, VAL857, MET161 |
| 2.35 Å | |||
| E Coli calmodulin in complex with RS20 peptide of smMLCK |
1QTX |
- |
TRP5, THR8, VAL12 |
| 1.65 Å | |||
| Rat calmodulin in complex with NMDA receptor NR1C1peptide |
2HQW |
P62161 |
PHE880, THR884, LEU887 |
| 1.90 Å | |||
| Human centrin 2 in complex with the centrin binding region of XPC protein |
2GGM |
P41208 |
TRP848, LEU851, LEU855 |
| 2.35 Å | |||
| C-terminal domain of human centrin 2 in complex with a repeat sequence of human Sfi 1 |
2K2I |
P41208 |
LEU651, LEU655, TRP658 |
| NMR | |||
| Scherffelia dubia centrin in complex with smMLCK peptide |
3KF9 |
Q06827 |
TRP4, PHE8, VAL11 |
| 2.60 Å | |||
| Human BCL-XL in complex with BAK peptide |
1BXL* |
Q07817 |
VAL574, LEU578, ILE581 |
| NMR | |||
| Human E3 ubiquitin-protein ligase MDM2 in complex with p53 tumor transactivation domain (fragment 17-125) |
1YCR* |
Q00987 |
PHE19, TRP23, LEU26 |
| 2.60 Å | |||
| Rabbit cardiac troponin C in complex with a fragment (residues 1-47) of cardiac troponin I |
1A2X |
P02586 |
LEU17, MET21, ILE24 |
| 2.30 Å |
*known to be disrupted by terphenyl or its derivatives.
Multiple sequences alignment (Figure 1) shows low sequence identity for the most of the analyzed proteins (shown in Table 2) both for the entire sequences and for the binding areas. The binding areas included all residues of the protein interacting with the alpha-helical peptide. Chicken, human, E. coli and rat calmodulin have very similar sequences (rat, chicken and human calmodulin are 100% identical; E coli has 98% identity with the others). For BCL-XL and human ubiquitin carboxyl-terminal hydrolase MDM2 only those fragments of sequences that are present in the 3D structures are considered. There is a high similarity only between the calmodulin, centrin 2 and troponin C sequences.
Figure 1.
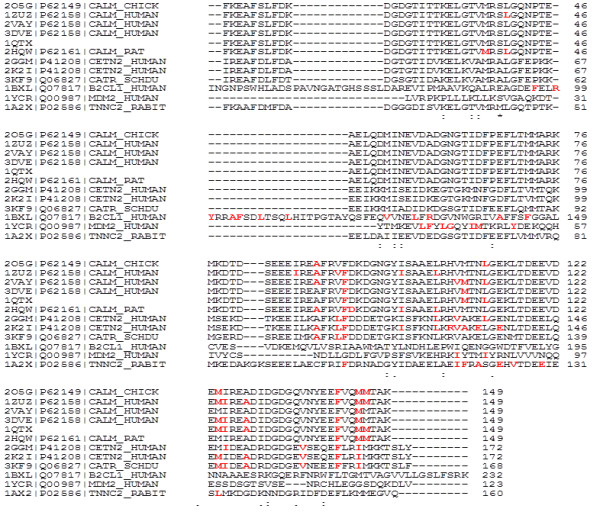
Sequence alignment of alpha-helix binding proteins. The amino acid residues interacting with alpha-helical peptides are presented in red.
Table 2.
Sequence identity (in %) between the considered proteins (the binding area/entire protein)
| Protein/ sequence identity | Human calmodulin | Human centrin 2 | Scherffelia dubia centrin | Human BCL-X2 | Human E3 ubiquitin-protein ligase MDM2 |
|---|---|---|---|---|---|
| Human centrin 2 |
54/50 |
|
|
|
|
| Scherffelia dubia centrin |
56/55 |
90/74 |
|
|
|
| Human BCL-X2 |
5/7 |
5/5 |
5/8 |
|
|
| Human E3 ubiquitin-protein ligase MDM2 |
5/4 |
5/10 |
7/6 |
9/5 |
|
| Rabbit cardiac troponin C | 57/51 | 57/34 | 37/32 | 5/9 | 5/19 |
The binding area was defined here as all residues of the protein interacting with the helical peptide.
Structure-based analysis
Figure 2 illustrates the complexes’ structures of six alpha-helix binding proteins. In all shown complexes, bulky hydrophobic residues of the bound peptide anchor into the protein binding pocket. Following the sequence similarities we superimposed the alpha-helix binding regions structures of calmodulin, human centrin 2, scherffelia dubia centrin and rabbit troponin C (Figure 3a). Strong structural homology for binding regions is seen following the sequence similarity of these proteins. Figure 3b and 3c illustrate the binding pockets of BCL-XL and human E3 ubiquitin-protein ligase MDM2, respectively.
Figure 2.
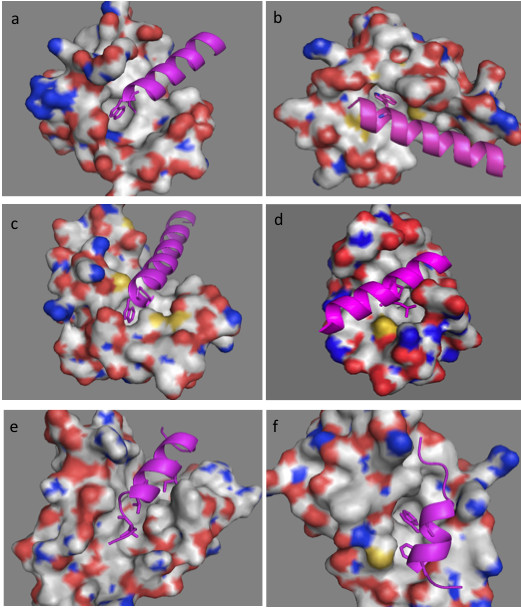
3D structures of the complexes formed by: (a) human centrin 2 and a 10 residue peptide of Xeroderma Pigmentosum group C protein, code entry 2GGM. (b) chicken calmodulin and smooth muscle myosin light chain kinase (smMLCK), code entry 2O5G. (c) scherffelia dubia centrin and smMLCK peptide, code entry 3KF9. (d) rabbit cardiac troponin C and a fragment of cardiac troponin I, code entry 1A2X. (e) human BCL-XL and BAK peptide, code entry 1BXL. (f) human E3 ubiquitin-protein ligase MDM2 and p53 tumor transactivation domain, code entry 1YCR. All proteins are shown as surface in atom color type (C and H-white, N – blue, O -red, S – yellow) and ligands are shown in magenta cartoon with hydrophobic interacting residues given as sticks.
Figure 3.
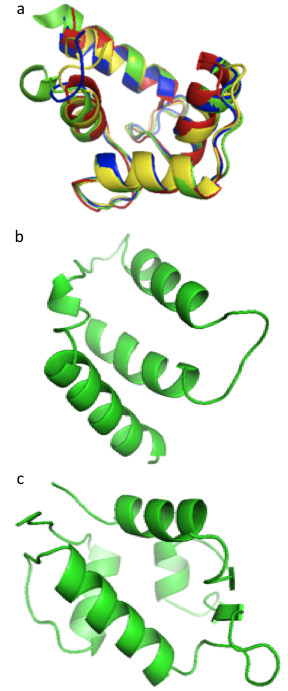
3D structures of alpha-helix binding domains. (a) Superposition of the alpha-helix binding regions of chicken calmodulin (red, code entry 2O5G), HsCen2 (blue, code entry 2GGM), scherffelia dubbia centrin (green, code entry 3KF9) and rabbit troponin C (yellow, code entry 1A2X). (b) Structure of human BCL-XL binding domain (code entry 1BXL). (c) Structure of human E3 ubiquitin-protein ligase MDM2 (code entry 1YCR) binding domain.
The interacting residues of the proteins and bound peptides, identified with ContPro [29], are shown in Figures 1 and 4 and Table 1. The results reveal that usually hydrophobic residues such as TRP, LEU, ILE, PHE, VAL, MET are involved in the interactions. The presence of hydrophobic residues suggests a favorable interaction with terphenyl-like molecules anchoring in the hydrophobic cavities. Most of the residues involved in the interactions between the proteins and alpha-helices are hydrophobic for both partners, as also observed in other studies [38]. We notice several key residues involved in the interaction of the same protein with different peptide partners. For example, in the case of calmodulin, PHE92, MET124, PHE141, MET144 and MET145 are involved in most of the peptides’ interactions. These residues can thus be considered as key for the interaction with terphenyl and its derivatives, or other alpha-helix mimetics. We noticed the presence of MET residues in most of the alpha-helix binding pockets analyzed here. In a recent study, MET residues have not been identified to be a part of hot spot amino acids, in particular in alpha-helix mediated protein interfaces [39]. However, our analysis clearly indicates their presence in positions that are key for the interaction with the alpha-helical partner. Furthermore, Ma and Nussinov [40] have also concluded that the amino acids TRP, MET, and PHE are important for protein-protein interactions. They showed that TRP/MET/PHE residues play roles in the dimerization of the transcriptase (p51/p66) and in cell-fusion processes, including the gp120-CD4 interaction and the gp41 six-helix bundle formation. They suggested that polarizability of MET allows it to assume roles of both hydrophobic and hydrophilic residues [40]. Further, its larger flexibility compared to other hydrophobic residues may facilitate the plasticity of hydrophobic binding pockets allowing to accommodate different ligands [27].
Figure 4.
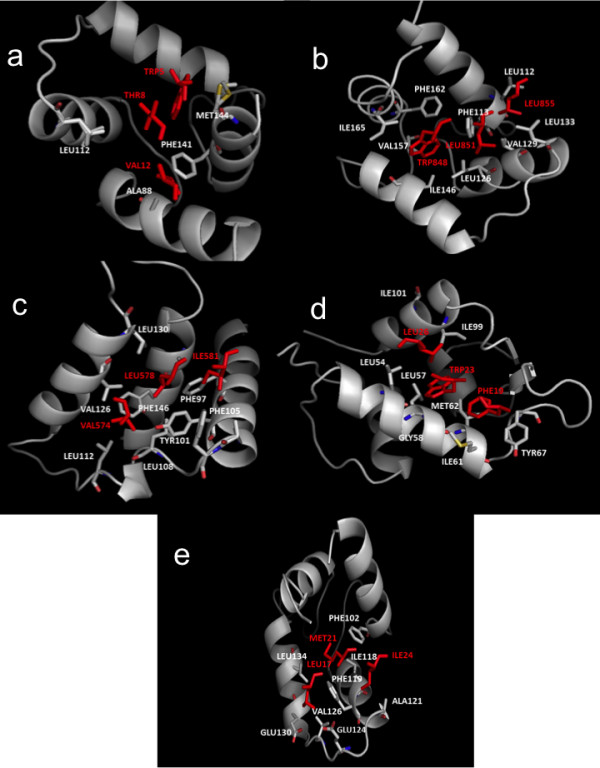
Illustration of the interacting residues (in sticks) of the protein (atom color type) and the bound peptide (red): (a) chicken calmodulin and smMLCK (code entry 2O5G), (b) human centrin 2 and the centrin binding region of XPC (code entry 2GGM), (c) human BCL-XL protein and BAK (code entry 1BXL), (d) human E3 ubiquitin- protein ligase MDM2 and p53 tumor transactivation domain (code entry 1YCR), (e) rabbit cardiac troponin C and cardiac troponin I (code entry 1A2X).
We used Fpocket [33] and CASTp [32] to calculate geometrical and physicochemical characteristics of the binding pockets taking into account the protein residues interacting with the alpha-helical peptides. The overall hydrophobic character of the binding pockets is again clearly identified. Yet, some specificity is also observed, several pockets show high hydrophobicity score but low local hydrophobic density, or vice versa, demonstrating that the hydrophobic patches are not always regularly distributed in the binding pockets. For example, 1YCR and 3KF9 have similar hydrophobicity scores but high and low calculated hydrophobic density, respectively. The differences of the hydrophobicity distribution are illustrated in Figure 5.
Figure 5.
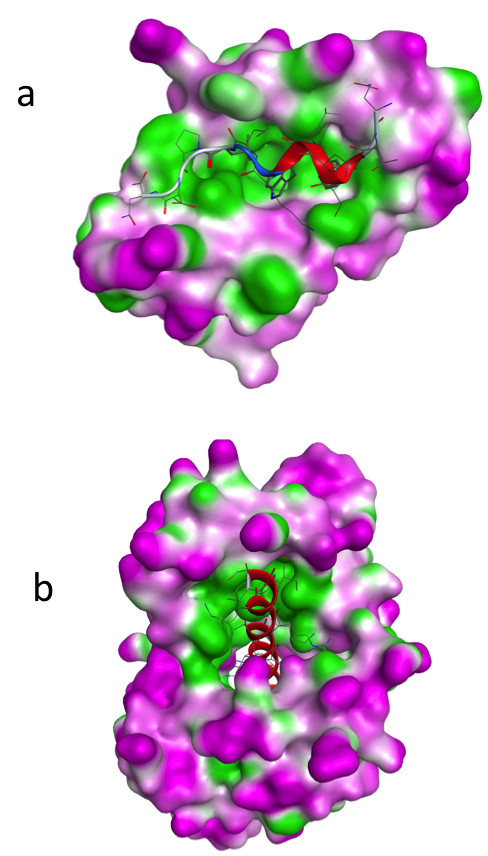
Surface lipophilicity (shown in green) of alpha-helix binding proteins computed using MOE. (a) Human E3 ubiquitin-protein ligase MDM2 (PDB code 1YCR), (b) Scherffelia dubia centrin (PDB code 3KF9).
The volumes of the detected pockets in the peptide-binding regions computed with CASTp are given in Table 3. The average volume of the sub-cavities present at the PPI interfaces found by Fuller et al [41] was ~60 Å3. Sonavane & Chakrabarti [42] found PPI pocket volumes to be up to ~330 Å3. We found similar volumes to those reported in Bourgeas et al. [43]. Taking into account the various algorithms and different concepts for binding pocket definition, such differences for the computed volumes can be expected. Several small cavities are present in the binding region (seen in Figure 2 and Figure 5), as it has been previously observed for other targeted PPI interfaces [39]. For the proteins studied here, the presence of several small hydrophobic cavities in the alpha-helix binding region seems to be a typical surface feature guiding the anchoring of hydrophobic residues from the peptide side. Such characteristics can also facilitate targeting PPI mediated by alpha-helices by small molecules containing hydrophobic anchors (as terphenyl or other mimetics).
Table 3.
Geometrical and physicochemical characteristics of the identified pockets
| Protein code PDB | Volume (Å3) | Hydrophobicity score | Local hydrophobic density |
|---|---|---|---|
| Chicken calmodulin |
312.0 |
68.86 |
43.00 |
| 2O5G | |||
| Human calmodulin |
203.0 |
68.86 |
42.00 |
| 1ZUZ | |||
| Human calmodulin |
219.8 |
59.62 |
40.00 |
| 2VAY | |||
| Human calmodulin |
226.4 |
61.00 |
39.32 |
| 3DVE | |||
| E.coli calmodulin |
317.9 |
56.63 |
40.15 |
| 1QTX | |||
| Rat calmodulin |
310.6 |
56.62 |
43.78 |
| 2HQW | |||
| Human centrin 2 |
147.9 |
41.47 |
32.00 |
| 2GGM | |||
| Human centrin 2 |
210.9 |
39.93 |
35.08 |
| 2K2I | |||
| Scherffelia dubia centrin |
221.5 |
58.19 |
31.00 |
| 3KF9 | |||
| Human BCL-XL |
321.5 |
36.91 |
42.04 |
| 1BXL | |||
| Human E3 ubiquitin-protein ligase MDM2 |
201.9 |
51.18 |
55.20 |
| 1YCR |
|
|
|
| Rabbit cardiac troponin C |
213.1 |
63.07 |
39.15 |
| 1A2X |
Further, we decided to explore the roughness of the alpha-helix binding sites. The methodology implemented to calculate the fractal surface dimensions, used for the roughness evaluation, is illustrated in Figure 6 for the global surface roughness of chicken calmodulin. The fractal global surface dimension and the fractal local surface dimension for the binding site of chicken calmodulin are calculated to be DS=2.238; ± 0.006 and DL= 2.616 ± 0.072, respectively. The global and local fractal dimensions for the other proteins are given in Table 4. Our results and other previously published data [44-47] suggest that the global fractal dimension of protein surface is about 2. The local surface fractal dimensions for the binding cavities are computed to be larger than the global surface fractal dimensions for all studied proteins. This reflects the higher roughness of the binding site and its more complex shape and that can be considered as important for ligand binding. The most important differences between DS and DL are obtained for human calmodulin (2VAY), centrin (3KF9, 2K2I), BCL-XL (1BXL), MDM2 (1YCR) and troponin C (1A2X). It has been experimentally demonstrated that human calmodulin [18], BCL-XL [19,20] and MDM2 [21,22] interact with terphenyl or its derivatives. Recently, we suggested a possible binding of terphenyl 2, which mimics the relative positions of the side chains of residues TRP848, LEU851, LEU855 of the XPC peptide, into human centrin 2 following our energetic and conformational flexibility analysis performed for the alpha-helical peptide-binding pocket of centrin 2 [27]. The DL value for the peptide-binding site of troponin C shows rougher surface than the entire protein, similarly to the above listed terphenyl-binding proteins.
Figure 6.
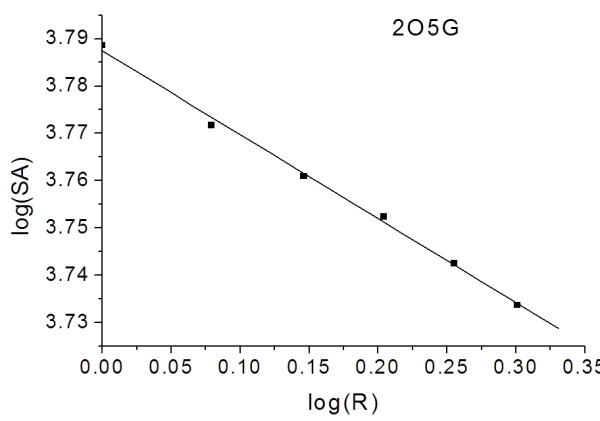
Double logarithmical plot of the surface area versus probe radii for chicken calmodulin (PDB code 2O5G).
Table 4.
Global (DS) and local (DL) surface fractal dimensions of investigated proteins
| Code PDB | DS | DL |
|---|---|---|
| 2O5G |
2.238 ± 0.006 |
2.616 ± 0.072 |
| 1ZUZ |
2.181 ± 0.007 |
2.487 ± 0.058 |
| 2VAY |
2.183 ± 0.006 |
2.757 ± 0.108 |
| 3DVE |
2.217 ± 0.003 |
2.418 ± 0.040 |
| 1QTX |
2.302 ± 0.002 |
2.494 ± 0.069 |
| 2HQW |
2.172 ± 0.002 |
2.454 ± 0.082 |
| 2GGM |
2.247 ± 0.004 |
2.373 ± 0.018 |
| 2K2I |
2.167 ± 0.008 |
2.892 ± 0.124 |
| 3KF9 |
2.179 ± 0.006 |
2.892 ± 0.153 |
| 1BXL |
2.230 ± 0.007 |
2.696 ± 0.225 |
| 1YCR |
2.173 ± 0.014 |
2.708 ± 0.055 |
| 1A2X | 2.177 ± 0.005 | 2.624 ± 0.032 |
Taking into consideration the sequence and structural homology of troponin C and calmodulin and other physicochemical similarities of the binding sites as discussed above, we decided to probe putative terphenyl binding into troponin C. We performed docking of terphenyl 2 into the peptide-binding sites of calmodulin and troponin C using AutoDock. The best scored docking poses are shown in Figure 7. The terphenyl orientations in the best scored poses correspond to the position of the bound alpha-helical peptides shown in Figure 2. The predicted interaction energies of -7.98 and -8.18 kcal/mol for terphenyl binding in calmodulin and troponin C, respectively, suggest favorable interactions with the two proteins.
Figure 7.
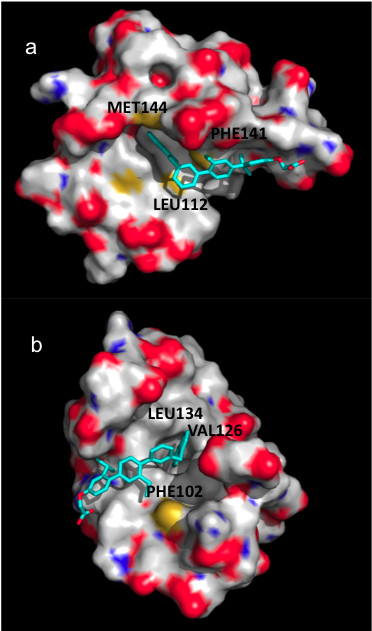
Best scored docking poses of terphenyl. The poses after docking-scoring with AutoDock are shown in cyan. (a) chicken calmodulin, code entry 2O5G, (b) rabbit cardiac troponin C, code entry 1A2X.
In the light of the results obtained here, it is now interesting to discuss the physicochemical properties of known PPI modulators, such as terphenyl. In a previous work [10] we gathered a set of 66 PPI inhibitors among which some terphenyl derivatives and other inhibitors of alpha-helix mediated PPI were present. In that work we demonstrated the more hydrophobic character of these compounds but also their bigger size. Interestingly, we also showed the importance of a critical number of aromatic bonds and some specific molecular shapes (T-shaped, star-shaped, or L-shaped compounds), among which some correspond to terphenyl derivatives. The present work therefore confirms that such genuine properties on the ligand side seem to be cavity-driven, and that these small molecules must possess certain properties in order to efficiently modulate an alpha-helix mediated PPI and to mimic the native partner and its properties.
Conclusions
Modulating protein-protein interactions using small molecules based on surface recognition has been a field of increasing interest during the last decade. PPI interfaces are very complex and need to be analyzed in order to be efficiently targeted for drug discovery purposes. Designed compounds must bind with high affinity and selectivity to the target protein. The low sequence identity found between some of the analyzed proteins suggests that there are no sequence requirements for the ability of proteins to bind alpha-helical peptides and consequently small-molecule mimetics.
From the structural point of view, all investigated proteins show larger surface fractal dimensions for the peptide-binding pockets than the entire protein surface reflecting the higher complexity of the shape of the binding sites. Also, the presence of several hydrophobic patches at the protein surface seems to be an important property related to the ability of the protein to bind alpha-helical peptides and mimetics. Furthermore, we showed that hydrophobicity is not uniformly distributed across different alpha-helix binding pockets and that its distribution can be used to identify hydrophobic hot spots.
Many similarities between the binding sites studied here are observed and terphenyl or its derivatives binding to various alpha-helix binding proteins can be suggested. However, targeting various PPI complexes by similar small molecules can rise selectivity problems in the context of drug discovery or chemical biology projects. Thus, the specificities found here for different binding sites, e.g. key residues, roughness and local hydrophobic density, can be further exploited to optimize terphenyl-like ligands in order to improve their selectivity.
Abbreviations
PPI: Protein-Protein interactions; smMLCK: smooth muscle myosin light chain kinase; CaM: Calmodulin; HsCen2: Human centrin 2; PDE: 3'-5'-cyclic nucleotide phosphodiesterase; XPC: Xeroderma pigmentosum group C.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
AI carried out the sequence alignment and binding pockets analysis. DC carried out the fractal calculations. AI and DC drafted the manuscript. VM carried out the volume calculations and docking analysis. OS participated in the protein-protein interface analysis and discussion writing. MAM designed and coordinated the study. All authors participated in manuscript writing and approved the final manuscript.
Pre-publication history
The pre-publication history for this paper can be accessed here:
Contributor Information
Adriana Isvoran, Email: adriana.isvoran@cbg.uvt.ro.
Dana Craciun, Email: craciundana@gmail.com.
Virginie Martiny, Email: virginie.martiny@univ-paris-diderot.fr.
Olivier Sperandio, Email: olivier.sperandio@inserm.fr.
Maria A Miteva, Email: maria.miteva@univ-paris-diderot.fr.
Acknowledgments
The financial support from the West University of Timisoara, the Inserm institute and the University Paris Diderot is greatly appreciated.
References
- Hunter T, Maniatis T, Califano A, Honig B, Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556–660. doi: 10.1038/nature11503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf MP, Thorne T, de Silva E, Stewart R, An HJ, Lappe M, Wiuf C. Estimating the size of the human interactome. Proc Natl Acad Sci USA. 2008;105:6959–6964. doi: 10.1073/pnas.0708078105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockwell BR. Exploring biology with small organic molecules. Nature. 2004;432:846–854. doi: 10.1038/nature03196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson CG, Arkin MR. Small-molecule inhibitors of IL-2/IL-2R: Lessons learned and applied. Curr Top Microbiol Immunol. 2011;348:25–29. doi: 10.1007/82_2010_93. [DOI] [PubMed] [Google Scholar]
- Villoutreix BO, Bastard K, Sperandio O, Fahraeus R, Poyet JL, Calvo F, Deprez B, Miteva MA. In silico-in vitro screening of protein-protein interactions: towards the next generation of therapeutics. Curr Pharm Biotechnol. 2008;9:103–122. doi: 10.2174/138920108783955218. [DOI] [PubMed] [Google Scholar]
- Fry DC. Protein-protein interactions as targets for small molecule drug discovery. Biopolymers. 2006;84:535–552. doi: 10.1002/bip.20608. [DOI] [PubMed] [Google Scholar]
- Gautier B, Miteva MA, Goncalves V, Huguenot F, Coric P, Bouaziz S, Seijo B, Gaucher JF, Broutin I, Garbay C, Lesnard A, Rault S, Inguimbert N, Villoutreix BO, Vidal M. Targeting the proangiogenic VEGF-VEGFR protein-protein interface with drug-like compounds by in silico and in vitro screening. Chem Biol. 2011;18(12):1631–1639. doi: 10.1016/j.chembiol.2011.10.016. [DOI] [PubMed] [Google Scholar]
- Villoutreix BO, Laconde G, Lagorce D, Martineau P, Miteva MA, Dariavach P. Tyrosine kinase syk non-enzymatic inhibitors and potential anti-allergic drug-like compounds discovered by virtual and in vitro screening. PLoS One. 2011;6(6):e21117. doi: 10.1371/journal.pone.0021117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein-protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- Sperandio O, Reynes CH, Camproux AC, Villoutreix BO. Rationalizing the chemical space of protein-protein interaction inhibitors. Drug Discov Today. 2010;15:220–229. doi: 10.1016/j.drudis.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Morelli X, Bourgeas R, Roche P. Chemical and structural lessons from recent successes in protein-protein interaction inhibition (2P2I) Curr Opin Chem Biol. 2011;15:475–481. doi: 10.1016/j.cbpa.2011.05.024. [DOI] [PubMed] [Google Scholar]
- Tripathi A, Kellogg GE. A Novel and Efficient Tool for Locating and Characterizing Protein Cavities and Binding Sites. Proteins. 2010;78(4):825–842. doi: 10.1002/prot.22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosdidier S, Fernández-Recio J. Protein-protein Docking and Hot-spot Prediction for Drug Discovery. Curr Pharm Des. 2012;18(30):4607–4618. doi: 10.2174/138161212802651599. [DOI] [PubMed] [Google Scholar]
- Andersson CD, Chen BY, Linusson A. Mapping of ligand-binding cavities in proteins. Proteins. 2010;78(6):1408–1422. doi: 10.1002/prot.22655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koes D, Khoury K, Huang Y, Wang W, Bista M, Popowicz GM, Wolf S, Holak TA, Domling A, Camacho CJ. Enabling Large-Scale Design, Synthesis and Validation of Small Molecule Protein-Protein Antagonists. PLoS One. 2012;7(3):e32839. doi: 10.1371/journal.pone.0032839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petsko GA, Ringe D. Protein structure and function. London: New Science Press Ltd; 2004. [Google Scholar]
- Bullock BN, Jochim AL, Arora PS. Assessing helical protein interfaces for inhibitor Design. J Am Chem Soc. 2011;133(36):14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orner BP, Ernst JT, Hamilton AD. Towards Proteomimetics: Terphenyl Derivatives as Structural and Functional Mimics of Extended Regions of an a-Helix. J Am Chem Soc. 2001;123(22):5382–5383. doi: 10.1021/ja0025548. [DOI] [PubMed] [Google Scholar]
- Kutzki O, Park HS, Ernst JT, Orner BP, Yin H, Hamilton AD. Development of a Potent Bcl-xL Antagonist Based on r-Helix Mimicry. J Am Chem Soc. 2002;124(40):11838–11839. doi: 10.1021/ja026861k. [DOI] [PubMed] [Google Scholar]
- Yin H, Lee GI, Sedey KA, Kutzki O, Park HS, Orner BP, Ernst JT, Wang HG, Sebti SM, Hamilton AD. Terphenyl-Based Bak BH3 alpha-helical proteomimetics as low-molecular-weight antagonists of Bcl-xL. J Am Chem Soc. 2005;127(29):10191–10196. doi: 10.1021/ja050122x. [DOI] [PubMed] [Google Scholar]
- Yin H, Lee G, Park HS, Payne GA, Rodriguez JM, Sebti SM, Hamilton AD. Terphenyl-Based Helical Mimetics That Disrupt the p53/HDM2 Interaction. Angew Chem. 2005;117(18):2764–2767. doi: 10.1002/ange.200462316. [DOI] [PubMed] [Google Scholar]
- Chen L, Yin H, Farooqi B, Sebti S, Hamilton AD, Chen J. p53 alpha-Helix mimetics antagonize p53/MDM2 interaction and activate p53. Mol Canc Ther. 2005;4(6):1019–1025. doi: 10.1158/1535-7163.MCT-04-0342. [DOI] [PubMed] [Google Scholar]
- Che Y, Brooks BR, Marshall GR. Protein recognition motifs: design of peptidomimetics of helix surfaces. Biopolymers. 2007;86(4):288–297. doi: 10.1002/bip.20744. [DOI] [PubMed] [Google Scholar]
- Becerril J, Rodriguez JM, Wyrembak PN, Hamilton AD. In: Protein Surface Recognition: Approaches for Drug Discovery. Giralt E, Peczuh M, Salvatella X, editor. London: John Willey&Sons; 2011. Inhibition of protein-protein interaction by peptide mimics. [Google Scholar]
- Fairlie DP, West ML, Wong AK. Towards protein surface mimetics. Curr Med Chem. 1998;5(1):29–62. [PubMed] [Google Scholar]
- Maity P, König B. Synthesis and structure of 1,4-dipiperazino benzenes: chiral terphenyl-type peptide helix mimetics. Org Lett. 2008;10(7):1473–1476. doi: 10.1021/ol8002749. [DOI] [PubMed] [Google Scholar]
- Isvoran A, Badel A, Craescu CT, Miron S, Miteva MA. Exploring NMR ensembles of calcium binding proteins: Perspectives to design inhibitors of protein-protein interactions. BMC Struct Biol. 2011;11:24. doi: 10.1186/1472-6807-11-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: lmproving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firoz A, Malik A, Afzal O, Jha V. ContPro: A web tool for calculating amino acid contact distances in protein from 3D –structure at different distance threshold. Bioinformation. 2010;5(2):55–57. doi: 10.6026/97320630005055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M, Rees DC. Fractal surfaces of proteins. Science. 1985;230:1163–1165. doi: 10.1126/science.4071040. [DOI] [PubMed] [Google Scholar]
- Fraczkiewicz R, Braun W. Exact and efficient analytical calculation of accesible surface areas and their gradients for macromolecules. J Compl Chem. 1998;19:319–333. doi: 10.1002/(SICI)1096-987X(199802)19:3<319::AID-JCC6>3.0.CO;2-W. [DOI] [Google Scholar]
- Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated resiudes. Nucleic Acid Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guilleoux VL, Schmidtke P, Tuffery P. Fpocket; An open source platform for ligand binding pocket detection. BMC Bioinforma. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J Comput Chem. 2009;30(16):2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. The PyMol molecular graphics system. San Carlos: DeLano Scientific; 2002. [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira IS, Fernandes PA, Ramos MJ. Hot spots—A review of the protein–protein interface determinant amino-acid residues. Proteins Struct Funct Bioinform. 2007;68(4):803–812. doi: 10.1002/prot.21396. [DOI] [PubMed] [Google Scholar]
- Brooke N, Bullock A, Paramjit SA. Assessing Helical Protein Interfaces for Inhibitor Design. J Am Chem Soc. 2011;133(36):14220–14223. doi: 10.1021/ja206074j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma B, Nussinov R. Trp/Met/Phe Hot Spots in Protein-Protein Interactions: Potential Targets in Drug Design. Curr Top Med Chem. 2007;7:999–1005. doi: 10.2174/156802607780906717. [DOI] [PubMed] [Google Scholar]
- Fuller JC, Burgoyne NJ, Jackson RM. Predicting druggable binding sites at the protein–protein interface. Drug Discov Today. 2009;14(3–4):155–161. doi: 10.1016/j.drudis.2008.10.009. [DOI] [PubMed] [Google Scholar]
- Sonavane S, Chakrabarti P. Cavities and Atomic Packing in Protein Structures and Interfaces. PLoS Comput Biol. 2008;4:e100001188. doi: 10.1371/journal.pcbi.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourgeas R, Basse MJ, Morelli X, Roche P. Atomic Analysis of Protein-Protein Interfaces with Known Inhibitors: The 2P2I Database. PLoS One. 2010;3:e9598. doi: 10.1371/journal.pone.0009598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetze T, Brickmann J. Self similarity of protein surfaces. Biophys J. 1992;61:109–118. doi: 10.1016/S0006-3495(92)81820-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettit FK, Bowie JU. Protein surface roughness and small molecular binding sites. J Mol Biol. 1999;285(4):1377–1382. doi: 10.1006/jmbi.1998.2411. [DOI] [PubMed] [Google Scholar]
- Stawiski EW, Baucom AE, Lohr SC, Gregoret LM. Predicting protein function from structure: Unique structural features of proteases. Proc Natl Acad Sci. 2000;97:3954–3958. doi: 10.1073/pnas.070548997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stawiski EW, Mandel-Goutfreund Y, Lowenthal AC, Gregoret LM. Progress in predicting protein structure from sequence: unique features of O-glycosidases. Pacific Symp Biocomput. 2002;7:637–648. doi: 10.1142/9789812799623_0059. [DOI] [PubMed] [Google Scholar]


