Abstract
Cephalosporins are major antimicrobials used to treat serious infections. However, their effectiveness is being compromised by the emergence of extended-spectrum β-lactamases (ESBLs). A total of 138 enteric bacteria were isolated from 53 faecal samples of pigs collected from different districts of Mizoram, of which 102 (73.91 %) were Escherichia coli, 26 (18.84 %) were Salmonella spp. and 10 (7.25 %) were Klebsiella pneumoniae. Phenotypic confirmatory test (Double Discs Synergy Test) showed that 8 (5.80 %) E. coli isolates were ESBLs producer. PCR analysis confirmed that out of the eight isolate, 7 (5.07 %) harboured blaCTX-M-1 gene and/or blaTEM gene. Of the eight positive isolates, 7 (5.07 %) and 3 (2.17 %) were found to be positive for blaCTX-M-1 gene and blaTEM gene, respectively, of which 3 (2.17 %) isolates were positive for both the genes. Only 4 (2.90 %) E. coli isolates carried blaCTX-M-1 gene alone. Agarose gel electrophoresis showed that all the isolates were carrying plasmids ranging between 0.9 and ~30 kb. Out of the seven isolates positive for blaCTX-M-1 and/or blaTEM, 2 (1.84 %) isolates were confirmed for blaCTX-M-1 gene in their plasmid. Only one E. coli isolate was found to be positive for both the genes in its plasmid. The resistance plasmid could not be transferred to a recipient by in vitro horizontal gene transfer method.
Keywords: ESBLs, BlaCTX-M-1, BlaTEM, Pigs, North East India
Introduction
The rapid emergence of extended-spectrum β-lactamases (ESBLs) in the food producing animals has been recorded and published worldwide [1–5]. It is important to note its impact on the treatment and therapeutic strategy of serious infections [6–10]. Food animals, including pigs are one of the most important sources of development of multi-drug resistant (MDR) bacteria because of continuous use of antibiotics as feed additives and growth promoting factors in a sub-therapeutic level [11–13]. This practice may lead to selection of a resistant population in the native microbiota of the animal and the local environment due to shedding through faeces. The MDR bacteria may re-enter the human and animal populations through various routes including natural water, irrigation water, drinking water, vegetables and foods. The present study is carried out to record the prevalence of ESBLs producing Escherichia coli, Salmonella spp. and Klebsiella pneumoniae in pig population in North Eastern India.
Materials and Methods
Bacterial Isolates
A total of 53 faecal samples from pigs were collected from different districts of Mizoram between September 2011 and March 2012. Samples were collected from animal of either sex, irrespective of their age, sex or breed. Animals under the study were reared under different housing system and belonged to organized as well as unorganized farms. Samples were collected by dry rectal swabbing. However, for collection of samples from distant locations, a sterilized swab dipped in nutrient broth was used as transport medium. Each swab containing the collected sample was then inserted into separate sterilized test tube and carried to the laboratory under cold chain for further processing.
For isolation of E. coli and K. pneumoniae, the collected faecal samples were inoculated on MLA and single colonies were selected and confirmed by standard bacteriological technique. For isolation of Salmonellae, samples were first enriched in Selenite F broth, and streaked on SalmonellaShigella agar plate. Pure colonies were then selected and identified as per standard bacteriological technique [14].
Phenotypic Detection of ESBLs
All the isolates were subjected to in vitro antibiotic sensitivity test by disc diffusion method against commonly used antibiotics as per the recommendation of Clinical Laboratory Standard Institute [15]. The antibiotics used for the study were ceftriaxone, cephotaxime, cefixime, cefazolin, cephalexin, ampicillin, erythromycin, chlortetracycline, streptomycin, enrofloxacin, oxytetracycline and lincomycin. The isolates exhibiting resistance to the extended-spectrum cephalosporin group of antibiotics were selected for confirmation of ESBLs production by placing cefotaxime and cefotaxime/clavulanate discs on the inoculated Mueller–Hinton agar plate at a distance of 30 cm apart. It was incubated overnight and the increase in zone size of more than 5 mm was considered as positive for ESBLs production.
Characterization of ESBLs Producing Isolates
Bacterial lysate was prepared from all the isolates found to be positive for ESBLs production phenotypically, and were tested for the presence of blaCTX-M-1 and blaTEM genes by PCR assay using specific primers (Table 1). PCR was carried out in a 0.2 ml thin wall PCR tubes using the bacterial lysate as template DNA with a final volume of 25 μl containing 10× buffer, 1.5 mM MgCl2, 200 pM of each oligonucleotide primers, 200 μM of each dNTPS, 1 U of Taq polymerase and 4.0 μl DNA lysate. PCR was carried out in a thermal cycler and the cycling condition for blaCTX-M-1 was: initial denaturation at 94 °C for 7 min followed by 30 cycles of amplification with denaturation at 94 °C for 30 s, annealing at 57 °C for 30 s, and extension at 72 °C for 30 s, ending with a final extension at 72 °C for 5 min. For blaTEM gene, the annealing temperature was 53 °C.
Table 1.
Details of the oligonucleotide primers used in the present study
Multiplex PCR was carried out using the same composition of PCR reaction mixture mentioned above. However, the annealing temperature was set to 54 °C.
Extraction of Plasmid and Genomic DNA
Plasmid DNA was extracted as per the method described by Sambrook and Russel [16] and the chromosomal DNA was extracted as per the method of Nazik et al. [17] from the isolates harbouring the ESBLs genes. PCR was performed using the Plasmid and chromosomal DNA separately following the above mentioned settings to find out the location of the target genes.
Curing of Plasmid
All the isolates, carrying blaCTX-M-1 and/or blaTEM genes in their plasmid were subjected to curing using acridine orange as curing agent following the method described by Silhavy et al. [18] with suitable modifications. In brief, 0.2 ml of overnight culture was inoculated in 5 ml LB broth containing different concentrations (2.5, 1.25, 1.0, 0.7, 0.5, 0.25 and 0.1 mg/ml) of acridine orange. Positive control contained only cells without acridine orange, while negative control contained only acridine orange without cells. All the tubes were incubated (in dark) at 37 °C for overnight. Next day tubes containing the highest concentration of acridine orange showing growth were selected and loopful was streaked on Mac Conkey’s agar plates and incubated overnight.
Horizontal Gene Transfer
The ability of transfer of antibiotic resistance genes within Enterobacteriaceae group of bacteria was recorded by in vitro conjugation study. E. coli isolates harboring the ESBLs gene were used as donor and Salmonella enteritidis (ATCC 13076), which was made resistant to nalidixic acid was used as recipient strain. The recipient strain was sensitive to cefazolin, cephalexin, ceftriaxone and cefotaxime and was not carrying blaCTX-M-1 and/or blaTEM genes in its plasmid as confirmed by PCR analysis. In vitro mating experiments were performed by broth mating [19], filter paper mating [20] and plate mating [21]. Transconjugants were selected on Mac Conkey’s agar containing ceftriaxone (50 μg/ml) and nalidixic acid (100 μg/ml). Donor and recipient strains were grown separately in antibiotic free medium as well as antibiotic medium as control. Selected transconjugants were further characterized for their antimicrobial susceptibility, ESBLs phenotype and presence of blaCTX-M-1 and/or blaTEM genes by PCR.
Results
Bacterial Isolates
Out of the 53 faecal samples collected in this study, 27 (50.94 %) were collected from organized farms, while the remaining 26 (49.06 %) were from local backyard farms. A total of 138 bacteria were isolated, of which 102 (73.91 %) were E. coli, 26 (18.84 %) were Salmonella spp. and 10 (7.25 %) were K. pneumoniae as confirmed by standard bacteriological techniques.
Phenotypic Detection of ESBLs Production
Of the 138 isolates, 8 (5.8 %) E. coli isolates showed resistance to cephalosporin group of antibiotics, while no Salmonella spp. and K. pneumoniae isolates showed resistance against extended-spectrum cephalosporins. Of the eight E. coli isolates, 8 (100 %), 7 (87.5 %), 8 (100 %), 6 (75 %), 6 (75 %), 6 (75 %), 7 (87.5 %), 4 (50 %), 6 (75 %), 7 (87.5 %), 4 (50 %) and 5 (62.5 %) showed 100 % resistance to cefixime, cefazolin, cephalexin, ceftriaxone, cefotaxime, enrofloxacin, oxytetracycline, streptomycin, ampicillin, chlortetracycline, erythromycin and lincomycin, respectively (Table 2). All the 8 (5.8 %) E. coli isolates suspected for the ESBLs production by disc diffusion method were confirmed to be a ESBLs producer, based on the Double Discs Synergy Test.
Table 2.
Antimicrobial drug resistance pattern of selected bacterial isolates obtained from pigs of different districts of Mizoram
| Isolates | Zone of inhibition (in mm) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CZ | CTX | CTR | CFM | CN | EX | O | S | L | A | CT | E | |
| PE4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 13 |
| PE5 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 0 | 14 |
| PE6 | 0 | 15 | 17 | 0 | 0 | 12 | 0 | 14 | 12 | 13 | 0 | 13 |
| PE29 | 18 | 23 | 23 | 0 | 0 | 20 | 14 | 12 | 0 | 12 | 15 | 13 |
| PE76 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PE98 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| PE99 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 |
| PE100 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 |
P pig, EE. coli
Genotypic Characterization of β-Lactamase Genes
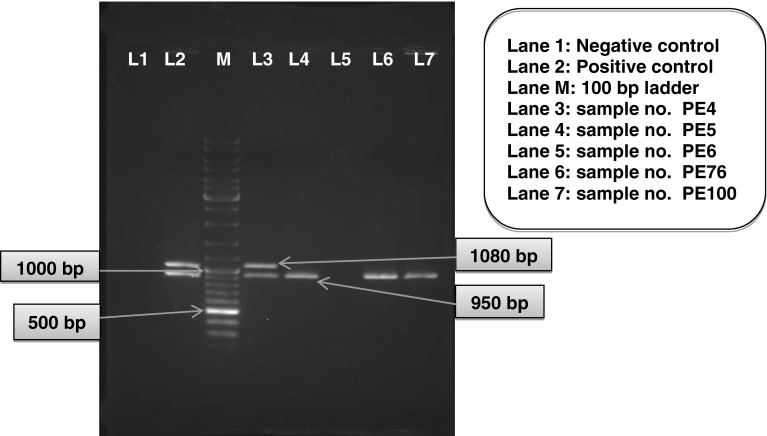
Out of the eight phenotypically positive isolates screened for the presence of bla genes by PCR using bacterial lysate as template DNA, 7 (5.07 %) were found to be positive for blaCTX-M-1 gene (950 bp) and/or blaTEM gene (1,080 bp). All the seven isolates were positive for blaCTX-M-1 gene and three of the isolates have an additional blaTEM gene (Table 3; Fig. 1).
Table 3.
PCR-based detection of blaCTX-M-1 and blaTEM genes in E. coli isolates obtained from pigs of different districts of Mizoram
| Isolates | blaCTX-M-1 (950 bp) | blaTEM (1080 bp) | ||||
|---|---|---|---|---|---|---|
| Lysate | Plasmid | Chromosomal | Lysate | Plasmid | Chromosomal | |
| PE4 | + | + | + | + | ||
| PE5 | + | + | ||||
| PE29 | + | + | ||||
| PE76 | + | + | + | + | ||
| PE98 | + | + | + | + | ||
| PE99 | + | + | ||||
| PE100 | + | + | ||||
P pig, EE. coli (None of the K. pneumoniae and Salmonella spp. isolates was found positive)
Fig. 1.
Multiplex PCR assay for detection of blaCTX-M-1 and blaTEM genes from phenotypically positive isolates
Plasmid Profiling and Curing
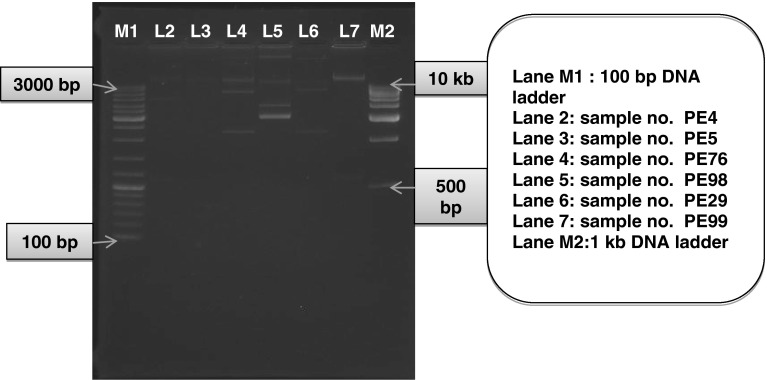
Agarose gel electrophoresis of the extracted plasmids showed that all the isolates were carrying plasmids ranging between 0.9 and ~30 kb (Fig. 2). Out of the seven isolates positive for blaCTX-M-1 and/or blaTEM genes, two isolates were confirmed to harbor the blaCTX-M-1 gene in their plasmid. Only one blaTEM gene was detected in plasmid.
Fig. 2.
Agarose gel electrophoresis for demonstration of plasmids extracted from the isolates positive for blaCTX-M-1 and/or blaTEM genes by PCR assay
Using acridine orange (1.25–1.5 mg/ml), the E. coli isolate (PE4) was successfully cured. Confirmation of curing was done by disc diffusion assay, where the organism showed 100 % sensitivity against all the antibiotics; plasmid extraction could not trace any plasmids and by PCR assay, no ESBLs genes could be detected.
In Vitro Horizontal Gene Transfer
The resistance trait from the donor isolates could not be transferred to the recipient isolate. There is no growth of transconjugants on the Mac Conkey’s agar plate. The recipient strain, after conjugation experiments remained susceptible to cefixime, cefazolin, cephalexin, ceftriaxone, cefotaxime, enrofloxacin, oxytetracycline, streptomycin, ampicillin, chlortetracycline, erythromycin and lincomycin and was also found negative for blaCTX-M-1 and blaTEM genes by PCR assay.
Discussion
The present study revealed that blaCTX-M-1 is the most abundant ESBLs type in this region, with E. coli being the major ESBLs producer, which is in accordance with the reports of other workers from different places of the world [1, 3, 23]. And also the presence of more than one bla genes is often reported worldwide [1, 23].
Prevalence of higher rate of CTX-M over TEM gene was recorded in this study, which is in agreement with the studies conducted by Ensor et al. [24] as well as Jones et al. [25]. CTX-M may be increased due to wide use of third generation cephalosporins, especially ceftriaxone and cefotaxime or may be associated with high mobilization of the encoding genes [26]. Barlow et al. [27] reported that the blaCTX-M genes have been mobilized to plasmid almost 10 times more frequently than other class A β-lactamases. The predominance of CTX-M type of ESBLs gene is may be an indication that this allele would now be common in North East Region of India. Muzaheed et al. [28] also reported high prevalence of CTX-M genes in K. pneumoniae and E. coli from Southern India.
Out of the seven ESBLs positive isolates, six were isolated from organized farm. Only one positive isolate (PE29) was observed from the sample collected from local backyard farm. It may be due to the frequent and routine use of third generation antibiotics in the organized farm. The prevalence of ESBLs in the farm animals is also reported by other workers from India and abroad [1–3].
Most of the ESBLs producing organisms under this study were also found to be co-resistant to fluoroquinolones, aminoglycosides as well as co-trimoxazole, which corroborate with the study done by Denholm et al. [29] and Jabeen et al. [30]. Perez et al. [9] also reported similar kind of information, where the ESBLs producing enteric bacteria are also resistant to other group of antibiotics including aminoglycosides, tetracycline, sulfonamides, trimethoprim and chloramphenicol. Development of co-resistance against other antibiotics along with β-lactam antibiotics by the ESBLs producing organisms generally appeared in the large plasmids, where most of the resistant genes may co-exist.
In the present study, the resistant plasmids could be successfully cured by acridine orange. Although curing provides only the preliminary evidence that genetic traits are of extra-chromosomal nature but loss of growth on antibiotic containing plates also shows that the MDR genes may be plasmid borne. The resistance determining traits are often transposable, which exist in both plasmid and chromosomal locations (flip–flop mechanism) [31]. It is however, important to note that not all antibiotic resistance genes are plasmid mediated [32] and copies of the plasmid lying closer to the membranes are readily eliminated by chemical agents, while those lying closer to the nucleus may escape the curing effect, thereby; one may observe partial curing [33].
During conjugation study neither of the plasmids carrying any one of the target gene could be transferred horizontally to the recipient isolate. Similarly, low transconjugation success was also reported by other workers in Switzerland and Germany [19, 34]. Franiczek et al. [35] also reported that none of the four E. coli isolates could transfer their resistance gene to other recipient strains. Yuan et al. [36] reported that plasmids are transferred under the influence of environmental condition (in vitro vs in vivo). In vitro experiment showed transfer of the plasmids ranging from 108 to 157 kb, while in vivo conjugation experiment showed a transfer of smaller sized plasmids. Failure of conjugation in the present study may be because of the small size plasmids carried by the donors. It is suggestive to study the involvements of the insertion sequence ISEcp1 [37, 38], as well as assessment of the incompatibility group [39, 40] of the plasmid may help to understand the failure.
In summary, we reported the presence of ESBLs in Enterobacteriaceae from pigs in NER India. The findings are worrisome as transmission to human via the food chain of bacteria resistant to practically all antimicrobial classes cannot be dismissed. More strict veterinary antibiotic policies are needed in order to prevent emergence and dissemination of these strains among animals and humans, limiting future problems of therapy failure.
Acknowledgments
The authors are highly thankful to the “Institutional Biotech Hub” CVSc and AH, CAU, Aizawl funded by DBT, GOI for providing the required instrumentation facilities; DBT sponsored Twinning Project on “Epidemiology of ESBLs in Enterobacterioceae group of bacteria isolated from swine in NER (Assam, Arunachal Pradesh, Manipur, Meghalaya, Mizoram, Nagaland, Sikkim and Tripura) and swine and poultry in West Bengal” for providing financial assistance; and the Dean, CVSc and AH, CAU, Aizawl, Mizoram for providing infrastructure facilities to conduct the research work.
Contributor Information
H. Lalzampuia, Email: ateahlawndo@gmail.com
T. K. Dutta, Email: tapandutta@rediffmail.com
References
- 1.Smet A, Martel A, Persoons D, Dewulf J, Heyndrickx M, Catry B, Herman L, Haesebrouck F, Butaye P. Diversity of extended-spectrum β-lactamases and class C β-lactamases among cloacal Escherichia coli isolates in Belgian broiler farms. Antimicrob Agents Chemother. 2008;52:1238–1243. doi: 10.1128/AAC.01285-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moodley A, Guardabassi L. Transmission of IncN plasmids carrying blaCTX-M-1 between commensal Escherichia coli in pigs and farm workers. Antimicrob Agents Chemother. 2009;53:1709–1711. doi: 10.1128/AAC.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Horton RA, Randall LP, Snary EL, Cockrem H, Lotz S, Wearing H, Duncan D, Rabie A, McLaren I, Watson E, La Ragione RM, Coldham NG. Fecal carriage and shedding density of CTX-M extended-spectrum-β-lactamase-producing Escherichia coli in cattle, chickens, and pigs: implications for environmental contamination and food production. Appl Environ Microbiol. 2011;77:3715–3719. doi: 10.1128/AEM.02831-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hiroi M, Harada T, Kawamori F, Takahashi N, Kanda T, Sugiyama K, Masuda T, Yoshikawa Y, Ohashi N. A survey of β-lactamase-producing Escherichia coli in farm animals and raw retail meat in Shizuoka Prefecture, Japan. Jpn J Infect Dis. 2011;64:153–155. [PubMed] [Google Scholar]
- 5.Yuan L, Liu J-H, Hu G-Z, Pan Y-S, Liu Z-M, Mo J, Wei Y-J. Molecular characterization of extended-spectrum β-lactamase-producing Escherichia coli isolates from chickens in Henan Province, China. J Med Microbiol. 2009;58:1449–1453. doi: 10.1099/jmm.0.012229-0. [DOI] [PubMed] [Google Scholar]
- 6.Bonnet R. Growing group of extended-spectrum beta-lactamases: the CTX-M enzymes. Antimicrob Agents Chemother. 2004;48:1–14. doi: 10.1128/AAC.48.1.1-14.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Canton R, Novais A, Valverde A. Prevalence and spread of extended spectrum β-lactamase-producing Enterobacteriaceae in Europe. Clin Microbiol Infect. 2008;14:144–153. doi: 10.1111/j.1469-0691.2007.01850.x. [DOI] [PubMed] [Google Scholar]
- 8.Coque TM, Baquero F, Canton R. Increasing prevalence of ESBL-producing Enterobacteriaceae in Europe. Eurosurveillance. 2008;13:19044. [PubMed] [Google Scholar]
- 9.Perez F, Endimiani A, Hujer KM, Bonomo RA. The continuing challenge of ESBLs. Curr Opin Pharmacol. 2007;7(5):459–469. doi: 10.1016/j.coph.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rodriguez-Bano J, Alcala JC, Cisneros JM. Community infections caused by extended-spectrum β-lactamase producing Escherichia coli. Arch Intern Med. 2008;168:1897–1902. doi: 10.1001/archinte.168.17.1897. [DOI] [PubMed] [Google Scholar]
- 11.Willis C. Antibiotics in the food chain: their impact on the consumer. Rev Med Microbiol. 2000;11:153–160. doi: 10.1097/00013542-200011030-00005. [DOI] [Google Scholar]
- 12.Aarestrup FM. Antimicrobial resistance in bacteria of animal origin. Washington, DC: ASM Press; 2006. [DOI] [PubMed] [Google Scholar]
- 13.Prescott JF, Baggot JD, Walker RD. Antimicrobial therapy in veterinary medicine. 3. Ames: Iowa State University Press; 2000. [Google Scholar]
- 14.Quinn PJ, Carter ME, Markey B, Carter GR. Clinical veterinary microbiology. Spain: Wolfe Publishing, Grafos, S. A. Arte Sobre Papel; 1994. [Google Scholar]
- 15.Performance standards for antimicrobial susceptibility testing, twentieth informational supplement, CLSI Document M100-S20. Wayne: Clinical and Laboratory Standards Institute; 2010. [Google Scholar]
- 16.Sambrook J, Russel DW. Plasmid and their usefulness in molecular cloning. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbour Laboratory Press; 2001. p. 1.32. [Google Scholar]
- 17.Nazik H, Öngen B, Kuvat N. Investigation of plasmid-mediated quinolone resistance among isolates obtained in a Turkish intensive care unit. Jpn J Infect Dis. 2008;61(4):310–312. [PubMed] [Google Scholar]
- 18.Silhavy TJ, Berman ML, Enquist LW. Experiments with gene fusions. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 1984. [Google Scholar]
- 19.Schmitt J, Jacobs E, Schmidt H. Molecular characterization of extended-spectrum β-lactamases in Enterobacteriaceae from patients of two hospitals in Saxony, Germany. J Med Microbiol. 2007;56:241–249. doi: 10.1099/jmm.0.46670-0. [DOI] [PubMed] [Google Scholar]
- 20.Sun Y, Zeng Z, Chen S, Ma J, He L, Liu Y, Deng Y, Lei T, Zhao J, Liu J-H. High prevalence of blaCTX-M extended spectrum β-lactamase genes in Escherichia coli isolates from pets and emergence of CTX-M-64 in China. Clin Microbiol Infect. 2010;16:1475–1481. doi: 10.1111/j.1469-0691.2010.03127.x. [DOI] [PubMed] [Google Scholar]
- 21.Gniadkowski M. Evolution and epidemiology of extended spectrum β-lactamases (ESBLs) and ESBL-producing microorganisms. Clin Microbiol Infect. 2001;7:597–608. doi: 10.1046/j.1198-743x.2001.00330.x. [DOI] [PubMed] [Google Scholar]
- 22.Weill FX, Demartin M, Laetitia Fabre L, Grimont Patrick AD. Extended-spectrum-β-lactamase (TEM-52)-producing strains of Salmonella enterica of various serotypes isolated in France. J Clin Microbiol. 2004;42:3359–3362. doi: 10.1128/JCM.42.7.3359-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meunier D, Jouy E, Lazizzera C, Kobisch M, Madec JY. CTX-M-1- and CTX-M-15-type β-lactamases in clinical Escherichia coli isolates recovered from food-producing animals in France. Int J Antimicrob Agents. 2006;28:402–407. doi: 10.1016/j.ijantimicag.2006.08.016. [DOI] [PubMed] [Google Scholar]
- 24.Ensor VM, Shahid M, Evans JT, Hawkey PM. Occurrence, prevalence and genetic environment of CTX-M beta lactamases in Enterobacteriaceae from Indian hospitals. J Antimicrob Chemother. 2006;58:1260–1263. doi: 10.1093/jac/dkl422. [DOI] [PubMed] [Google Scholar]
- 25.Jones CH, Tuckman M, Keeney D, Ruzin A, Bradford PA. Characterization and sequence analysis of extended-spectrum beta-lactamase encoding genes from Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis isolates collected during tigecycline phase 3 clinical trials. Antimicrob Agents Chemother. 2009;53(2):465–475. doi: 10.1128/AAC.00883-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barguigua A, El Otmani F, Talmi M, Bourjilat F, Haouzane F, Zerouali K, Timinouni M. Characterization of ESBL-producing Escherichia coli and Klebsiella pneumoniae isolates from community in Morocco. J Med Microbiol. 2011;60:1344–1352. doi: 10.1099/jmm.0.032482-0. [DOI] [PubMed] [Google Scholar]
- 27.Barlow M, Reik RA, Jacobs SD, Medina M, Meyer MP. High rate of mobilization for blaCTX-M. Emerg Infect Dis. 2008;14:423–428. doi: 10.3201/eid1403.070405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muzaheed, Yohei Doi JM, Adams-Haduch JM, Shivannavar CT, Paterson DL, Gaddad SM. Faecal carriage of CTX-M-15 producing Klebsiella pneumoniae in patients with acute gastroenteritis. Indian J Med Res. 2009;129:599–602. [PubMed] [Google Scholar]
- 29.Denholm JT, Huysmans M, Spelman D. Community acquisition of ESBL-producing Escherichia coli: a growing concern. Med J Aust. 2009;190(1):45–46. doi: 10.5694/j.1326-5377.2009.tb02266.x. [DOI] [PubMed] [Google Scholar]
- 30.Jabeen K, Zafar A, Hasan R. Frequency and sensitivity pattern of extended spectrum beta-lactamase producing isolates in a tertiary care hospital laboratory of Pakistan. J Pak Med Assoc. 2005;55:436–439. [PubMed] [Google Scholar]
- 31.DeFlaun MF, Levy SB. In: Gene transfer in the environment. Levy SD, Miller RV, editors. New York: McGraw-Hill Publishing Company; 1989. pp. 1–32. [Google Scholar]
- 32.Shoemaker NB, Wang GR, Salyers AA. Evidence for natural transfer of a tetracycline resistance gene between bacteria from the human colon and bacteria from the bovine lumen. Appl Environ Microbiol. 1992;58:1313–1320. doi: 10.1128/aem.58.4.1313-1320.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenks PJ, Hu YM, Danel F, Mehtar S, Livermore DM. Plasmid mediated production of class I (AmpC) β-lactamase by two Klebsiella pneumoniae. J Antimicrob Chemother. 1995;35:235–236. doi: 10.1093/jac/35.1.235. [DOI] [PubMed] [Google Scholar]
- 34.Nuesch-Inderbinen M, Hachler H, Kayser FH. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur J Clin Microbiol Infect Dis. 1996;15:398–402. doi: 10.1007/BF01690097. [DOI] [PubMed] [Google Scholar]
- 35.Franiczek, Roman, Dolna I, Krzyzanowska B, Szufnarowski K, Krochmal BK. Conjugative transfer of multiresistance plasmids from ESBL-positive Escherichia coli and Klebsiella spp. clinical isolates to Escherichia coli strain K12 C600. Adv Clin Exp Med. 2007;16(2):239–247. [Google Scholar]
- 36.Yuan M, Hall LMC, Savelkoul PHM, Vandenbroucke-Grauls CMJE, Livermore DM. SHV-13, a novel extended-spectrum β-lactamase, in Klebsiella pneumoniae isolates from patients in an intensive care unit in Amsterdam. Antimicrob Agents Chemother. 2000;44:1081–1084. doi: 10.1128/AAC.44.4.1081-1084.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cao V, Lambert T, Nhu DQ. Distribution of extended-spectrum beta-lactamases in clinical isolates of Enterobacteriaceae in Vietnam. Antimicrob Agents Chemother. 2002;46:3739–3743. doi: 10.1128/AAC.46.12.3739-3743.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotimi VO, Jamal W, Pal T, Sovenned A, Albert MJ. Emergence of CTX-M-15 type extended-spectrum β-lactamase-producing Salmonella spp. in Kuwait and the United Arab Emirates. J Med Microbiol. 2008;57:881–886. doi: 10.1099/jmm.0.47509-0. [DOI] [PubMed] [Google Scholar]
- 39.Cloeckaert A, Praud K, Lefevre M, Doublet B, Pardos M, Granier SA, Brisabois A, Weill F-X. IncI1 plasmid carrying extended-spectrum-β-lactamase gene blaCTX-M-1 in Salmonella enterica isolates from poultry and humans in France, 2003 to 2008. Antimicrob Agents Chemother. 2010;54:4484–4486. doi: 10.1128/AAC.00460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Girlich D, Poirel L, Carattoli A, Kempf I, Lartigue M-F, Bertini A, Nordmann P. Extended-spectrum β-lactamase CTX-M-1 in Escherichia coli isolates from healthy poultry in France. Appl Environ Microbiol. 2007;73:4681–4685. doi: 10.1128/AEM.02491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]