Background: LAMTOR3 is an endosomal MAPK scaffold recently shown also to modulate the mTORC1 pathway.
Results: Association to LAMTOR2 and recruitment to the endosomal membrane stabilize LAMTOR3 and prevent the premature degradation of the protein by the proteasome.
Conclusion: The ubiquitin-proteasome pathway and LAMTOR complex assembly tightly control LAMTOR3 abundance.
Significance: Cells may modulate the dual ERK/mTORC1 activity of LAMTOR3 by tightly controlling its intracellular protein level.
Keywords: MAP Kinases (MAPKs), mTOR, Protein Degradation, Scaffold Proteins, Signal Transduction, LAMTOR1 (p18), LAMTOR2 (p14), LAMTOR3 (MP1), LAMTOR4 (c7orf59), and LAMTOR5 (HBXIP)
Abstract
LAMTOR3 (MP1) and LAMTOR2 (p14) form a heterodimer as part of the larger Ragulator complex that is required for MAPK and mTOR1 signaling from late endosomes/lysosomes. Here, we show that loss of LAMTOR2 (p14) results in an unstable cytosolic monomeric pool of LAMTOR3 (MP1). Monomeric cytoplasmic LAMTOR3 is rapidly degraded in a proteasome-dependent but lysosome-independent manner. Mutational analyses indicated that the turnover of the protein is dependent on ubiquitination of several lysine residues. Similarly, other Ragulator subunits, LAMTOR1 (p18), LAMTOR4 (c7orf59), and LAMTOR5 (HBXIP), are degraded as well upon the loss of LAMTOR2. Thus the assembly of the Ragulator complex is monitored by cellular quality control systems, most likely to prevent aberrant signaling at the convergence of mTOR and MAPK caused by a defective Ragulator complex.
Introduction
Scaffold proteins serve as docking platforms that recruit specific components of signaling cascades into defined subcellular locations. That in turn determines different signaling outputs by influencing the subset of effectors that are efficiently activated. Furthermore, scaffolds exert an isolating function protecting the module from irrelevant stimuli, and they may also modulate signaling kinetics by, for instance, recruiting phosphatases (1). In addition, increasing levels of the scaffold effect the signaling cascade in a bell-shaped curve manner, emphasizing the biphasic character of this regulation (2). Because scaffolds influence the biological fate of cells by modulating the kinetics of their respective signaling pathways, the function and the localization of scaffold proteins require tight control.
At present, several ERK scaffold proteins have been identified: KSR1, LAMTOR3, SEF, paxillin, β-arrestin, IQGAP1, and MORG1 (3, 4). The body of evidence on the function of these proteins has increased over the last years. The regulation of KSR1 and its localization are particularly well characterized. KSR1 degradation depends on Thr-274 phosphorylation that is mediated by RAS and Ser-392 phosphorylation that is induced by Cdc25C-associated kinase 1 (C-TAK1) or the nucleoside diphosphate kinase mitochondrial-23 (Nm23). Only when both residues on KSR1 are phosphorylated can the protein be degraded (1). KSR1 proteolysis seems to be regulated by Nm23, which favors KSR1 interaction with the heat shock protein HSP90 (5).
LAMTOR3 was initially identified in a two-hybrid screen as a MEK1 interacting protein. LAMTOR3 was shown to selectively interact with MEK1/ERK1 but not MEK2/ERK2 and to favor MAPK activation (6). We have shown previously that LAMTOR2, a small protein peripherally associated with the cytosolic membrane of late endosomes, is required and sufficient to localize LAMTOR3 to the surface of endosomes (7). The LAMTOR2-LAMTOR3 complex plays a crucial role in EGF and granulocyte colony-stimulating factor-dependent ERK1/2 activation and EGFR degradation. In addition, LAMTOR2 and LAMTOR3 are required in vivo for cellular proliferation and tissue homeostasis (8, 9). LAMTOR2 and LAMTOR3 also contribute to the biogenesis of late endosomes/lysosomes and lysosome-related organelles, like phagosomes (9, 10). As a consequence, LAMTOR2 deficiency compromises the activity of neutrophils, B cells, cytotoxic T cells, and melanocytes (9). Recently LAMTOR1, also known as p27RF-Rho, was identified as the membrane anchor of the LAMTOR2-LAMTOR3 complex. This protein is recruited to late endosomal lipid rafts by N-terminal myristoylation and palmitoylation (11). Finally, it was shown that the LAMTOR1-LAMTOR2-LAMTOR3 complex mediates via the Rag GTPases the activation of mTOR1 signaling on late endosomes/lysosomal membranes (12), and the complex was renamed Ragulator (12). Taken together, these data highlight the role of the endosomal scaffold complex LAMTOR1-LAMTOR2-LAMTOR3 as a convergence point of signaling pathways controlling proliferation and tissue homeostasis.
Interestingly, we found a close relation between intracellular levels of LAMTOR3 and those of its heterodimeric partner, LAMTOR2. First, the requirement for both LAMTOR3 and LAMTOR2 to be present at equivalent amounts was also evident in our previous crystallography work (13). Second, we have shown (16) an equimolar ratio of the endogenous LAMTOR3 and LAMTOR2 on endosomes by absolute quantification of the proteins by quantitative mass spectrometry. Third, deletion of LAMTOR2 causes reduced LAMTOR3 protein levels in all cellular and animal LAMTOR2 knock-out models generated (8). Furthermore, analysis of B cells obtained from patients with a 3′-UTR mutation on LAMTOR2 revealed a LAMTOR2 hypomorphic phenotype and consequently reduced LAMTOR3 protein levels (9). Here, we demonstrate that a loss of LAMTOR2 causes a severe decrease in all remaining LAMTOR components, namely LAMTOR1, LAMTOR3, and the recently identified LAMTOR4 and LAMTOR5 (14). In particular, we show that the absence of LAMTOR2 results in an unstable cytosolic monomeric pool of LAMTOR3. This monomeric cytoplasmic LAMTOR3 is rapidly degraded in a proteasome-dependent but lysosome-independent manner. Based on these results, we suggest that the assembly of the Ragulator complex is tightly controlled to prevent aberrant signaling from a dysfunctional late endosomal scaffold complex.
EXPERIMENTAL PROCEDURES
Reagents, Antibodies, and Constructs
Velcade was obtained from Millennium Pharmaceuticals, and MG132 was from Sigma. Tissue culture grade reagents and media were purchased from Aplichem or Sigma. Lipofectamine 2000 was obtained from Invitrogen. Triton X-100 was purchased from Pierce, and Mowiol medium was from Carl Roth.
Primary antibodies were always used according to the manufacturer's instructions. The phospho-ERK1/2, ERK1/2, MEK1/2, ubiquitin (P4D1), and p27 antibodies were purchased from Cell Signaling. The p21 antibody and the human and mouse CD107a (LAMP1) were obtained from Pharmingen. The GFP and the His antibodies were purchased from Clonthec, α-tubulin was from Sigma, and phospho-p38 was from New England Biolabs. Anti-Xpress antibody was obtained from Invitrogen, and the HA.11 was from Covance. For immunofluorescence experiments, the purified HA.11 clone 16B12 was used. The EEA1, MEK1, and caveolin I antibodies were purchased from Transduction Laboratories. Anti-LAMTOR1 and -LAMTOR4 were obtained from Atlas antibodies, the LAMTOR5 antibody was purchased from Santa Cruz, and anti-LC3B was from Novus Biologicals. The anti-LAMTOR2 and anti-LAMTOR3 antibodies were described previously (7). The transferrin receptor and Myc antibodies were generated in house: the transferrin receptor antibody was purified from a homemade hybridoma cell line, and the Myc antibody was from serum obtained by a rabbit immunization with the Myc peptide. The Alexa 488 and Alexa 568 secondary antibodies were from Molecular Probes. Rab7 and Rab5 antibodies were supplied by Dr. Angela Wandiger-Ness (University of New Mexico). XLAMTOR2, Myc6LAMTOR3, H6T7LAMTOR3, His6LAMTOR2LAMTOR3, and His6LAMTOR3LAMTOR2 were generated in our group and previously published (7, 8, 13, 15). The retroviral constructs MMP-IRES.GFP, MMP-LAMTOR2WT.GFP, and MMP-LAMTOR2Δb3.GFP were also previously published (9). The retroviral construct pEGFPLAMTOR2 was described elsewhere (16). The constructs expressing p21 and p27 under the T7 promoter were a gift from Dr. Ludger Hengst (Department of Medical Biochemistry, Biocenter, Innsbruck Medical University). A PCR product introducing BamHI and SalI sites on murine Myc6LAMTOR3 was amplified. Upon digestion with the same enzymes, the product was subcloned into the retroviral transfer vector pLIB-MCS2-iresPURO (17). Point mutations on lysine sites were introduced using the QuikChange multisite mutagenesis kit from Stratagene and Myc6LAMTOR3WT pLIB-MCS2-iresPURO as a template. PCR products inserting SalI and NotI+Stop into Myc6LAMTOR3wt-iresPURO, Myc6LAMTOR3NTERM-iresPURO, Myc6LAMTOR3MIDDLE-iresPURO, Myc6LAMTOR3CTERM-iresPURO, and Myc6LAMTOR3KO-iresPURO were amplified. Upon digestion with the same enzymes, the products were subcloned into SalI and NotI sites of pCMV-HA (Clontech). The remaining constructs were obtained by lysine to arginine mutations using the QuikChange multisite-directed mutagenesis kit on already existing clones. We used PCR to generate a LAMTOR3 Gateway entry clone. A recombination reaction using the Gateway® LR ClonaseTM Enzyme was performed to shuttle LAMTOR3 to the destination vector pcDNA5/FRT/TO/SH/GW (18). All of the generated constructs were sequence verified.
Cell Culture, Transfection, and Generation of Retrovirus
Cell lines were kept at 37 °C, in 5% CO2 and 98% humidity. HeLa cells and mouse embryonic fibroblasts (MEF)3 for LAMTOR2 (8) were grown in high glucose DMEM supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% FCS. 293gpg cells (19) were kept in culture in high glucose DMEM, supplemented with 50 IU/ml penicillin streptomycin and 10% FCS, 2 mm l-glutamine, 1 μg/ml tetracycline, 2 μg/ml puromycin, and 0.3 mg/ml G418. Phoenix cells were a kind gift from Dr. Stephan Geley (Division of Molecular Pathophysiology, Biocenter, Innsbruck Medical University). These cells were cultured in RPMI supplemented with 50 IU/ml penicillin/streptomycin and 10% FCS. HEK293 Flp-InTM T-RexTM cells containing a single FRT site and stably expressing the Tet repressor were obtained from Invitrogen and cultured in high glucose DMEM containing 10% (v/v) FBS, 50 IU/ml penicillin, 50 mg/ml streptomycin, 100 μg/ml Zeocin, and 15 μg/ml blasticidin. Transfected HEK293 Flp-InTM T-RexTM cells were selected and maintained in high glucose DMEM supplemented with 10% (v/v) FBS, 50 IU/ml penicillin, 50 mg/ml streptomycin, and 100 μg/ml hygromycin B. The Ts20 cell line derived from BALB/3T3 was kept at 32 °C, in 5% CO2 and 98% humidity. The cells were grown in DMEM supplemented with 50 IU/ml penicillin, 50 μg/ml streptomycin, and 10% FCS (20). The cells were transfected with the different constructs using Lipofectamine 2000. Amounts of DNA, Lipofectamine, and transfection medium were in accordance to the manufacturer's instructions. 48 h upon transfection, the cells were used for immunofluorescence or biochemical assays. HEK293 cells were co-transfected with SH-LAMTOR3 and pOG44 and selected in high glucose DMEM with 10% FBS, 50 μg/ml penicillin/streptomycin, and 100 μg/ml hygromycin. Following selection, the cells were tested for inducible bait expression in medium supplemented with different concentrations of tetracycline. The production of retroviral particles expressing IRES.GFP, LAMTOR2WT.GFP, and LAMTOR2Δb3.GFP was previously described (9). Retroviruses expressing murine LAMTOR3 and LAMTOR3 mutants were generated by co-transfection of Phoenix cells with the construct of interest and pVSVG. 48 h after changing the medium, the supernatant containing the virus was passed through a 0.22-μm filter and stored at −80 °C.
Infection of MEF and Generation of Stable Single Cell Clones
Polybrene was added to the virus (multiplicity of infection of 5) to a final concentration of 400 μg/ml immediately before infection. Infected plates were centrifuged at 2000 rpm for 20 min and then incubated at 37 °C, in 5% CO2 and 98% humidity. 48 h after infection, the cells were used for biochemical studies or antibiotic selection was started. The puromycin concentration for selection was titrated from 1 to 10 μg/ml. Selection was maintained for at least 6 weeks before freezing down the clones. For the generation of the LAMTOR2−/− LAMTOR2Δb3.GFP cell line, infected MEF were FACS-sorted to select for the presence of GFP. The cells were used on day 12 post-selection for size exclusion chromatography (96% GFP-positive cells as confirmed by FACS).
Preparation of Total Cell Lysates, Cytosol, and Total Membrane Fractions
Total cell lysates were prepared as described in Ref. 7. Cytosol and total membrane fractions were prepared by harvesting cells in PBS and homogenizing them in PBS supplemented with 250 mm sucrose, 3 mm imidazole, pH 7.4, 10 μg/ml aprotinin, 1 μg/ml pepstatin, 10 μg/ml leupeptin, and 1 mm Pefabloc, 1 mm Na3OV4, 10 mm β-glycerophosphate. Upon centrifugation, to discard nuclei and unbroken cells, the post nuclear supernatant was subjected to two runs of 100,000 × g for 1 h. The cytosolic fraction was transferred to a new tube, and the membrane pellet was resuspended in PBS, supplemented with 250 mm sucrose, 3 mm imidazole pH 7.4, 0.35% Triton X-100. Upon solubilization of the membranes, the samples were precipitated using trichloroacetic acid and deoxycholate. The precipitated samples were resuspended in total cell lysate lysis buffer (50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1% Triton X-100, 10% glycerol, 0.5 mm EDTA, 0.5 mm EGTA, 5 mm sodium phosphate, 10 μg/ml aprotinin, 1 μg/ml pepstatin, 10 μg/ml leupeptin, 1 mm Pefabloc, 2 mm Na3OV4, 50 mm NaF, and 10 mm β-glycerophosphate) supplemented with Laemmli sample buffer and boiled (21).
Size Exclusion Chromatography
Cytosol and total cell lysates were prepared as described above. Samples were run on an Äkta HIPrep1660-Sephacryl-S200HR column previously equilibrated in PBS supplemented with 250 mm sucrose, 3 mm imidazole, pH 7.4. The obtained 5-ml fractions were precipitated using trichloroacetic acid and deoxycholate. The precipitated samples were resuspended in Laemmli sample buffer and analyzed by immunoblotting. The molecular weight distribution over the column was determined by comparison with a calibration run of proteins of known size.
Immunoprecipitation of SH-LAMTOR3
HEK293 cells expressing SH-LAMTOR3 were lysed in radioimmunoprecipitation assay buffer (50 mm Tris-HCl, pH 8, 250 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm EDTA, 50 mm NaF, 1 mm Na3VO4, 0.5 mm PMSF, 0.5 mg/ml pepstatin, 5 mg/ml leupeptin, 1 mm Pefabloc SC, supplemented with 10 mm N-ethylmaleimide. After sonication on ice, the samples were centrifuged to remove insoluble material. Lysates (∼60 mg of total protein) were loaded on Biospin columns (Bio-Rad) containing Strep-Tactin beads (IBA protein TAGnologies) and gravity-drained. The beads were washed with radioimmunoprecipitation assay buffer, and the bound protein was eluted with freshly prepared 2.5 mm d-biotin in radioimmunoprecipitation assay buffer. TCA-precipitated protein samples were immunoblotted as described below.
Immunoblots, Statistical, and Semiquantitative Analysis of Images
Lysates or other protein samples were separated by SDS-PAGE, transferred on a semidry chamber into PVDF membranes, and probed with the respective antibodies. Western blot images were acquired using the FluorChem 5500 system (Alpha Innotech/Biozym). For the half-life estimation, we have calibrated the FluorChem 5500 system with increasing concentrations of recombinant His6LAMTOR2LAMTOR3 and confirmed that the signal intensity detected by the camera correlated linearly with LAMTOR3 amounts. For Western blot quantifications, the last exposure frame before signal saturation was used. The signal intensity detected by the camera was automatically converted into a numeric format. The values were inserted into an Excel worksheet. For each time point, background was subtracted, and each intensity value was normalized against tubulin levels. The value 1 was arbitrarily assigned to the normalized value obtained for each construct at 0 min. The remaining time point values were given using time point 0 as reference value. Zero arbitrary units correspond to the detection limit of the FluorChem 5500 system. The mean ± S.E. from three independent experiments was calculated using Excel functions. Two-tailed Student's t tests were used for statistical analyses. The difference was considered significant when p < 0.05.
Immunofluorescence and Image Processing
Immunofluorescence images were obtained as described in Ref. 7. The samples were observed using an epifluorescence microscope (Axio Imager.M1; Carl Zeiss Microimaging Inc.). The Axiovision Rel 4.8 image examiner software was used for image acquisition. Confocal image analysis was performed using the laser-scanning microscope LSM510 Meta Axiovert 200 (Carl Zeiss MicroImaging, Inc.) and a 63× plan-Apochromat NA 1.4 oil objective. Images were acquired using the LSM image examiner software (version 3.1.0.117). Independent of the acquisition strategy, all of the images were converted to Adobe Photoshop CS3, where brightness, contrast, or tonal values were adjusted.
Quantitative RT-PCR
Total RNA was isolated by using TRIzolTM reagent (Invitrogen). DNA was removed by treatment with DNase I (Fermentas) according to the manufacturer's instructions. RNA was reverse-transcribed using a Revert Aid first strand cDNA synthesis kit (Fermentas) following the manufacturer's instructions. Primers for RT-PCR for GenBankTM accession number NM019920 were 5′-ccctgagacctggcttcctatcc-3′ and 5′-gaggcccaggtgtgcatcagaa-3′. Normalization was against GAPDH with 5′-cggagtcaacggatttggtcgtat-3′ and 5′-agccttctccatggtggtgaagac-3′. SYBR-Green-detected PCR was performed using the DNA Engine Opticon Continuous Fluorescence Detector (MJ Research). Fluorescence was detected after each elongation step at an optimal temperature to obtain specific signal only. For the amplification the following program was used: denaturation for 2 min at 95 °C and amplification for 30 s at 95 °C, 30 s at 65 °C (55 °C), and 30 s at 72 °C. In all programs, 41 cycles were used. The amplification specificity was determined from the melting curve after each run. Correct amplicon size and identity were confirmed by gel analysis and sequencing. Fluorescence intensity was plotted against cycle number on a logarithmic scale. The samples were analyzed in triplicate, and three technical replicates were performed. Cycle number, when the fluorescent signal was first detected, was plotted against the copy number derived from standard curves. The average data for specific PCR products were normalized to values of GAPDH and also on the PCR efficiency using proprietary software from the company Biozym.
RESULTS
LAMTOR3 Protein Is Stabilized by Associating with LAMTOR2
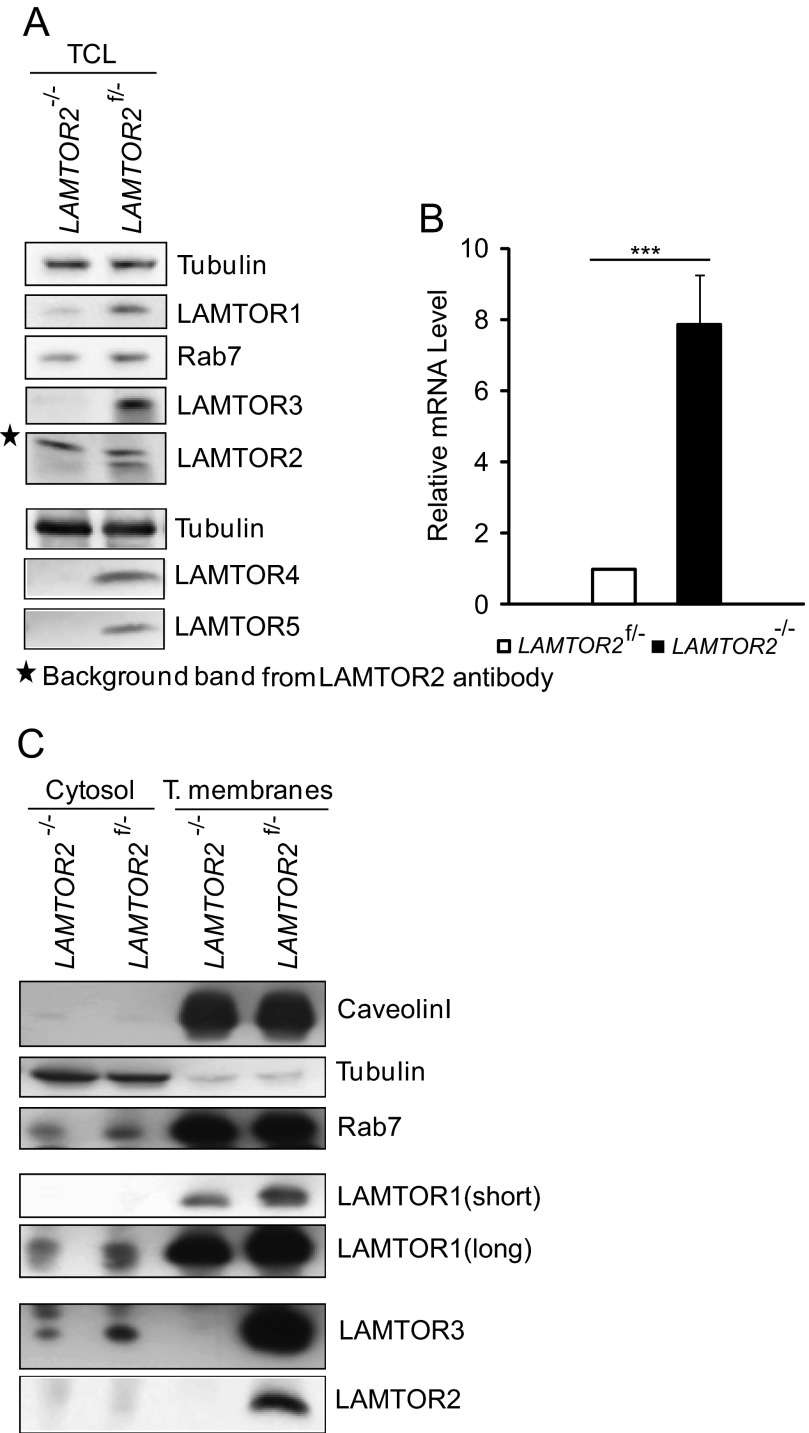
Immortalized LAMTOR2−/−MEF are devoid of LAMTOR2 mRNA and protein. In these cells, the endogenous protein levels of LAMTOR3 protein are severely reduced and sometimes even undetectable (Fig. 1A) (8). LAMTOR2 recruits LAMTOR3 to late endosomes, and we have previously reported that exogenously expressed LAMTOR3 mislocalizes to the cytoplasm in the absence of LAMTOR2 (8). Interestingly, despite the severe down-regulation of LAMTOR3 protein levels, the mRNA levels of LAMTOR3 were at least 5-fold increased in the LAMTOR2 deficient MEF (Fig. 1B).
FIGURE 1.
Endogenous LAMTOR3 protein is present in the cytosol of both LAMTOR2−/− and LAMTOR2f/−MEF, and its mRNA levels are up-regulated in LAMTOR2−/−MEF. A, total cell lysates (TCL) were prepared as described under “Experimental Procedures.” The samples were separated by SDS-PAGE and probed with the indicated antibodies. B, LAMTOR3 mRNA in LAMTOR2f/− and LAMTOR2−/−MEF. Random hexamer primers were used and normalized to GAPDH levels. The graph represents an average of three independent experiments (means ± S.E., n = 3). ***, p < 0.001. C, cytosol and total membrane fractions were prepared as described under “Experimental Procedures.” The samples were separated by SDS-PAGE and probed with the indicated antibodies.
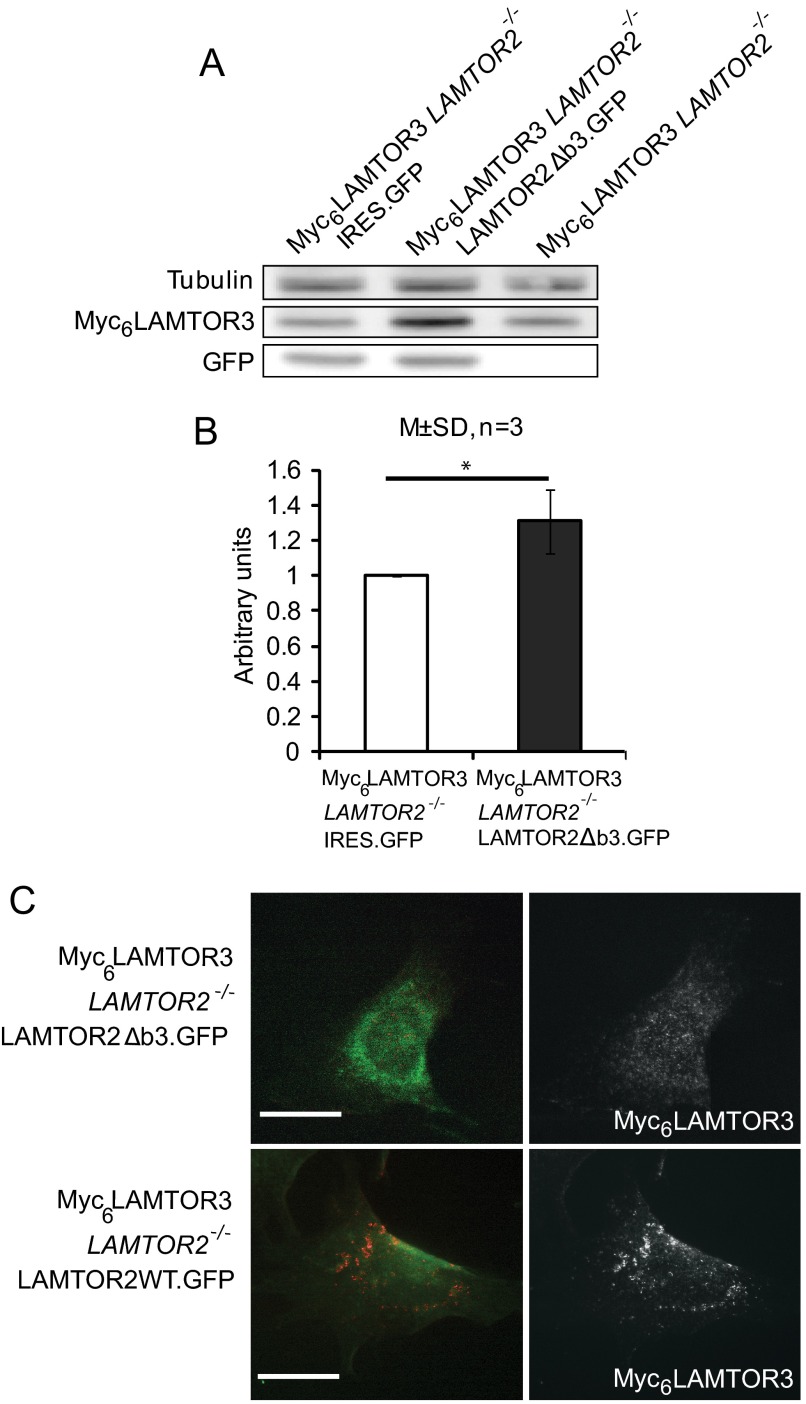
We hypothesized that the reduced levels of endogenous LAMTOR3 in LAMTOR2−/−MEF may arise from the lack of interaction with its partner LAMTOR2 and/or from the mislocalization of LAMTOR3 into the wrong subcellular compartment. Moreover, other Ragulator components, such as LAMTOR 1, 4, and 5, were also severely decreased in LAMTOR2−/−MEF (Fig. 1A). The structure of the LAMTOR2-LAMTOR3 heterodimer shows that both proteins contribute extensively to a larger interaction surface. Hence in absence of LAMTOR2, the interacting residues of LAMTOR3 may be exposed and destabilize LAMTOR3. To directly address whether the sole presence of LAMTOR2 without proper endosomal localization would be sufficient to stabilize LAMTOR3 protein, we have reconstituted LAMTOR2−/−MEF with a cytoplasmic mutant of LAMTOR2 (LAMTOR2Δb3) that no longer localizes to late endosomes but still interacts with LAMTOR3 (13). We have therefore used the retrovirus construct expressing bicistronically LAMTOR2Δb3 and GFP to infect a Myc6LAMTOR3LAMTOR2−/− stable clone (Fig. 2). The selected Myc6LAMTOR3 single cell clones on both LAMTOR2−/−MEF and controls were chosen because they showed very similar Myc6LAMTOR3 expression levels, and their overexpression levels were comparable to those of the endogenous LAMTOR3 protein (supplemental Fig. S1A). This mild expression of Myc6-tagged LAMTOR3 was not able to rescue the growth defect of the LAMTOR2−/−MEF (supplemental Fig. S1B). As a control for the infection experiment, we used a retroviral construct expressing GFP only. Interestingly, LAMTOR2Δb3 expression was sufficient to stabilize Myc6LAMTOR3, whereas GFP expression had no effect (Fig. 2, A and B). Furthermore, LAMTOR2WT.GFP restored Myc6LAMTOR3 localization to perinuclear, punctuated vesicles, whereas LAMTOR2Δb3.GFP did not (Fig. 2C). Taken together, we concluded that direct protein-protein interaction between LAMTOR2 and LAMTOR3 plays a major role in LAMTOR3 stability.
FIGURE 2.
LAMTOR3 protein is stabilized by associating with LAMTOR2. A, LAMTOR2Δb3.GFP and IRES.GFP retrovirus were used to infect the Myc6LAMTOR3LAMTOR2−/− stable clone. Infected cells were harvested 48 h afterward. The cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. B, graph shows the quantification of three independent experiments (means ± S.D. (M±SD); n = 3). *, p < 0.05). C, Myc6LAMTOR3LAMTOR2−/− cells were infected with LAMTOR2Δb3.GFP or LAMTOR2WT.GFP retrovirus. The cells were fixed in paraformaldehyde, and immunofluorescence analysis was performed as described under “Experimental Procedures” using anti-Myc antibodies. Bars, 20 μm.
In the Absence of LAMTOR2, LAMTOR3 Is Predominantly Found as a Cytosolic Monomeric Protein
We have purified cytosol and total membranes from LAMTOR2−/−MEF and LAMTOR2f/−MEF (Fig. 1C). The correct separation of the bulk of tubulin from the total membrane fraction containing caveolin-1 attested to the quality of our separation procedure. In addition, Rab7, a small GTPase and well established late endosomal marker, was enriched in the membrane fraction but was also present, although to a reduced extent, in the cytosol, consistent with the cytoplasmic recycling of Rab7/GDP (22). In accordance to previously published data, we found LAMTOR1 on the membrane fraction. Similarly to LAMTOR3, the protein levels of LAMTOR1, 4, and 5 were also reduced in the LAMTOR2−/−MEF (12, 14, 16). In our experiments, we found LAMTOR2 exclusively present in the total membrane fraction of LAMTOR2f/−MEF. Finally, we found endogenous LAMTOR3 predominantly associating with the membrane fraction in the presence of LAMTOR2. In LAMTOR2−/−MEF, we observed no recruitment of LAMTOR3 to total membranes despite the presence of LAMTOR1 in this fraction. In addition, for the first time we present here evidence of a small pool of LAMTOR3 in the cytoplasm of LAMTOR2 deficient cells, despite the presence of more LAMTOR3 mRNA.
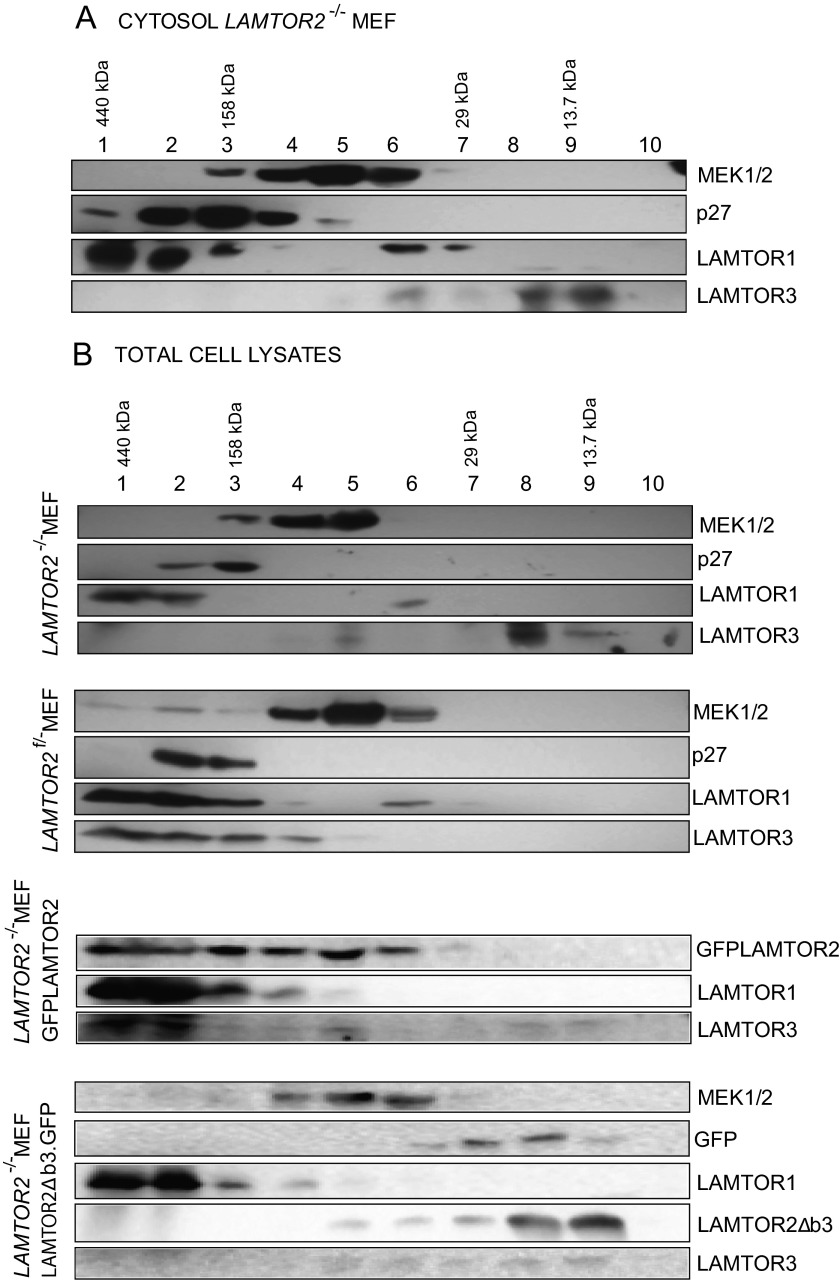
To test whether LAMTOR3 would associate with other proteins in the cytoplasm, we performed size exclusion chromatography (Fig. 3A). In the cytosol of LAMTOR2−/−MEF, LAMTOR3 elutes in a small molecular weight fraction consistent with the Mr of monomeric LAMTOR3. MEK1 and LAMTOR1, both known interactors of LAMTOR3, were found in high molecular weight complexes in the LAMTOR2−/− cytosol (6, 11). In accordance with the previously published shuttling of p27Kip1 between the nucleus and the cytoplasm, we detected p27Kip1 in purified cytosolic fractions (23). p27Kip1 was previously reported to interact with LAMTOR1, and we also found both proteins enriched in high molecular weight fractions (24) (Fig. 3A). Next we compared the size exclusion chromatography profiles obtained from LAMTOR2−/−MEF cytosol with those of total cell lysates of LAMTOR2−/−, LAMTOR2f/−, LAMTOR2−/−GFPLAMTOR2, and LAMTOR2−/−LAMTOR2Δb3.GFPMEF (Fig. 3B). The analysis of the total cell lysate of LAMTOR2−/−MEF showed LAMTOR3 present in low molecular weight fractions. This result was not surprising because LAMTOR3 in LAMTOR2-depleted MEF was exclusively cytosolic (Fig. 1B). In contrast, the analysis of LAMTOR2f/−MEF lysates revealed LAMTOR3 present in high molecular weight fractions. In addition, the exogenous expression of GFPLAMTOR2 in the LAMTOR2−/−MEF was sufficient to restore the presence of LAMTOR3 in high molecular weight complexes. Interestingly, in cells expressing the cytoplasmic LAMTOR2Δb3, LAMTOR3 was partially found in intermediate molecular weight complexes, the same fractions where LAMTOR2Δb3 could be detected. In these cells, LAMTOR3 can heterodimerize with LAMTOR2Δb3 in the cytoplasm (13) but does not form a functional Ragulator complex on late endosomes. Hence the LAMTOR2Δb3-LAMTOR3 heterodimer does not interact with other components such as LAMTOR1, which would allow the formation of a high molecular weight complex on late endosomes. Overall, we conclude that in absence of LAMTOR2, LAMTOR3 exists as an unstable monomer in the cytoplasm.
FIGURE 3.
In the absence of LAMTOR2, LAMTOR3 is predominantly found as a monomeric protein. A, size exclusion chromatography from LAMTOR2−/−MEF cytosol. Cytosolic fraction was prepared as described under “Experimental Procedures” and run on an Äkta HIPrep1660 Sephacryl S200HR column. The obtained 5-ml fractions were precipitated using trichloroacetic acid and deoxycholate and resuspended in Laemmli sample buffer. The samples were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. B, size exclusion chromatography from LAMTOR2−/−, LAMTOR2f/−, LAMTOR2−/−GFPLAMTOR2, and LAMTOR2−/−LAMTOR2Δb3.GFP MEF lysates. The same experimental procedure as described in A was performed with total cell lysates from the mentioned MEF cell lines. Images are representative examples from at least two independent experiments.
The Half-lives of Endogenous LAMTOR3 and Myc6LAMTOR3 Are Reduced in the Absence of LAMTOR2
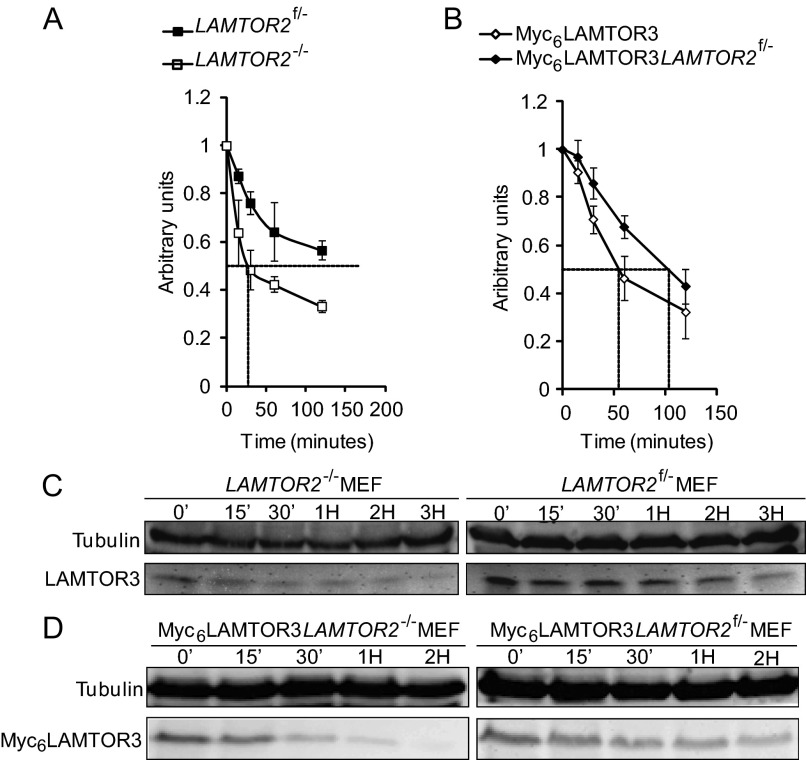
We have used cycloheximide, an inhibitor of eukaryotic protein synthesis, to estimate the rate of degradation of LAMTOR3, because LAMTOR3 is a poor labeling substrate for [35S]methionine/cysteine labeling (only one cysteine, Cys-73, and Met-1, the latter one is removed co-translationally; see supplemental Fig. S1C and our unpublished data).
We treated LAMTOR2−/−MEF, LAMTOR2f/−MEF, Myc6LAMTOR3LAMTOR2−/−MEF, and Myc6LAMTOR3LAMTOR2f/−MEF with cycloheximide for different times (Fig. 4 and supplemental Fig. S2). Tubulin, used as a control, had a half-life of ∼48 h (25). Endogenous LAMTOR3 in contrast, seemed to have a much faster turnover with a half-life slightly longer than 3 h in LAMTOR2f/−MEF but of only 30 min in LAMTOR2−/−MEF. Similarly, the half-life of overexpressed Myc6LAMTOR3 was significantly decreased in LAMTOR2−/−MEF when compared with control MEF. Interestingly, the half-life of overexpressed Myc6LAMTOR3 (approximately 100 min) in LAMTOR2f/− was also decreased when compared with endogenous LAMTOR3 levels.
FIGURE 4.
The half-lives of endogenous LAMTOR3 and Myc6LAMTOR3 are reduced in the absence of LAMTOR2. Cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. A and B, quantifications of the according Western blot are shown in A and B representing the averages of three independent experiments (means ± S.E., n = 3). C, 80% confluent LAMTOR2−/− and LAMTOR2f/−MEF were treated with 50 μg/ml cycloheximide for the corresponding time points. The cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. D, Myc6LAMTOR3LAMTOR2−/− and Myc6LAMTOR3LAMTOR2f/− stable clones were treated with 50 μg/ml cycloheximide for the corresponding time points.
Taken together, endogenous LAMTOR3 and Myc6LAMTOR3 are relatively short-lived proteins, and when not in a protein complex with LAMTOR2 they are rapidly degraded in the cytoplasm. Hence it seems that protein degradation is responsible for the reduced LAMTOR3 protein levels seen in the absence of LAMTOR2.
The Proteasome Mediates the Degradation of Both Endogenous LAMTOR3 and Myc6LAMTOR3
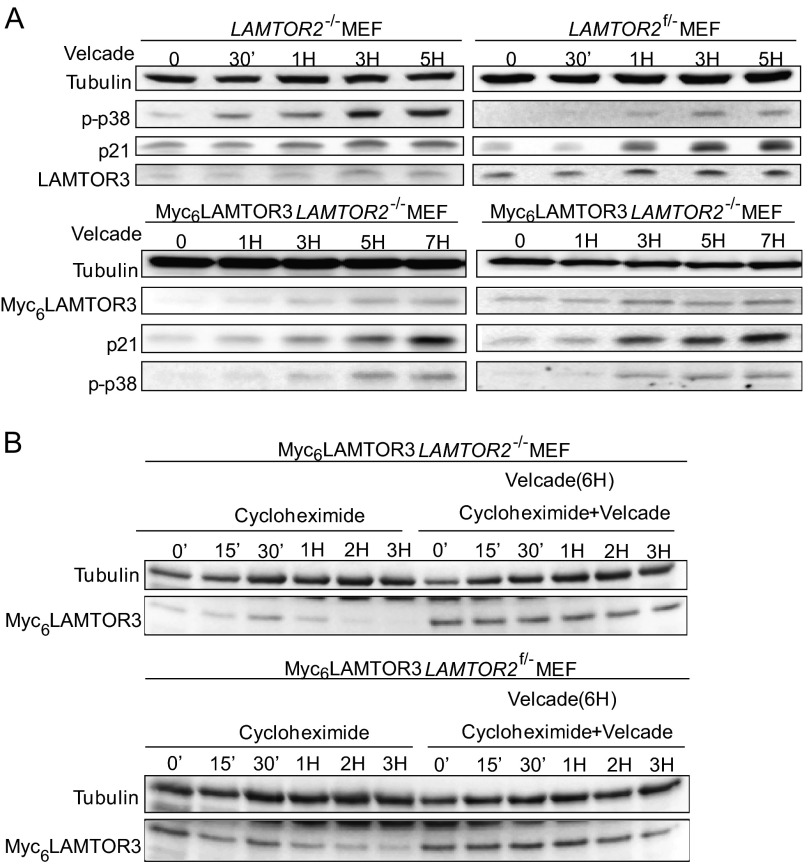
The cellular quality control system targets proteins destined for degradation either to the proteasome or to the lysosome. To test how LAMTOR3 is degraded, we treated LAMTOR2−/−MEF, LAMTOR2f/−MEF, Myc6LAMTOR3LAMTOR2−/−MEF, and Myc6LAMTOR3LAMTOR2f/−MEF with Velcade, an established proteasomal inhibitor (Fig. 5A). By blocking protein degradation, the accumulation observed at the protein level should reflect de novo protein synthesis. In Velcade-treated cells, the protein levels of endogenous LAMTOR3 and Myc6LAMTOR3 were stabilized in both LAMTOR2−/−MEF and controls (Fig. 5A). In the presence of cycloheximide alone, LAMTOR3 is quickly degraded (Figs. 4, C and D, and 5B). However, when LAMTOR2−/−MEF were first treated with Velcade, LAMTOR3 accumulated, and subsequent treatment with cycloheximide no longer resulted in LAMTOR3 degradation (Fig. 3B). These findings show that a substantial fraction of LAMTOR3 is degraded via the proteasome. Velcade treatment is known to induce apoptosis, so we included p38 activation as a positive control. In addition, we have also confirmed Velcade activity by probing for the stabilization of a known short-lived protein, p21 (26, 27). Finally, we have used tubulin as a loading control because the protein should remain stable for the length of the time course.
FIGURE 5.
The degradation of endogenous LAMTOR3 and Myc6LAMTOR3 is mediated by the proteasome. A, 80% confluent LAMTOR2−/−MEF, LAMTOR2f/−MEF, Myc6LAMTOR3LAMTOR2−/−, and Myc6LAMTOR3LAMTOR2f/− stable clones were treated with 50 nm Velcade for the corresponding time points. The cell lysates were separated by SDS-PAGE and probed with the indicated antibodies. B, Velcade and cycloheximide treatment of Myc6LAMTOR3LAMTOR2−/− and Myc6LAMTOR3LAMTOR2f/− stable clones. 80% confluent cells were pretreated with 50 nm Velcade for 6 h or left untreated. 50 μg/ml cycloheximide or 50 μg/ml cycloheximide plus 50 nm Velcade were then added to the cells for the corresponding time points. The cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies.
To further address the impact of Velcade treatment on LAMTOR3 protein, we have treated LAMTOR2−/−MEF and control cells with Velcade and performed size exclusion chromatography (supplemental Fig. S3). In brief, Velcade treatment of LAMTOR2−/−MEF recruited LAMTOR3 to higher molecular weight complexes. We speculate that under those conditions LAMTOR3 may be associating with chaperones and/or with the proteasome-mediated degradation machinery. In contrast in LAMTOR2f/−MEF LAMTOR3 remained in high molecular complexes independent of Velcade treatment (Fig. 5 and supplemental Fig. S3).
p21 and Rb are two examples of proteins that directly bind the proteasomal subunit C8 and are degraded in an ubiquitin-independent manner (28–31). Interestingly, LAMTOR3 has in its N terminus a stretch of residues that closely resembles the domain of p21 required for binding to C8 (29) (supplemental Fig. S4). To test whether LAMTOR3 could follow the same degradation route as p21, we have incubated purified recombinant LAMTOR3, or LAMTOR2-LAMTOR3 complexes, with pure 20S subunits (supplemental Fig. S4). As previously shown, in vitro transcribed and translated p21 could be degraded by 20S subunits, whereas p27 was resistant to 20S mediated degradation. LAMTOR3, in all combinations tested, was not degraded by purified 20S proteasomes (supplemental Fig. S4).
The Degradation of Endogenous LAMTOR3 Is Independent from Lysosome Function
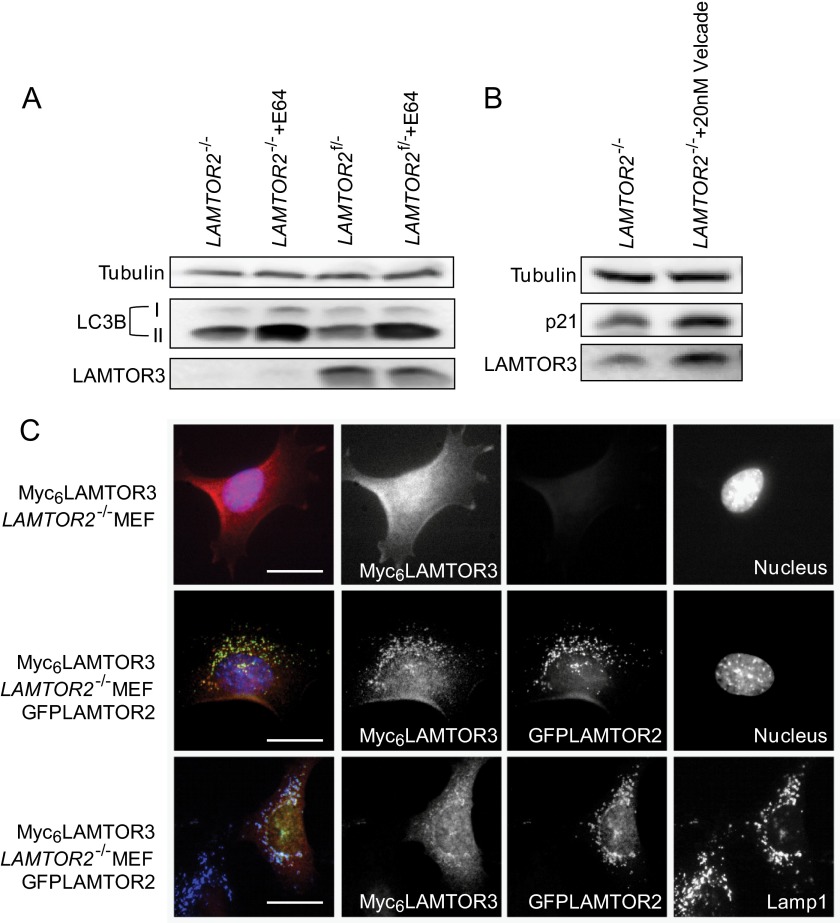
Next, we addressed whether lysosomal degradation could also partially contribute to LAMTOR3 turnover. We treated LAMTOR2−/−MEF and LAMTOR2f/−MEF with E64, a membrane-permeable inhibitor for lysosomal hydrolases for 7 h (Fig. 6A). LC3B-II, a key regulator for autophagy, is delivered together with other autophagosomal cargo into the lysosomes. Hence blocking lysosomal hydrolases stabilizes LC3B-II. As expected, E64 treatment resulted in the accumulation of the LC3B-II form, whereas no accumulation of the cytosolic variant (LC3B-I) was observed. However, under the conditions tested, E64 treatment showed no effect on endogenous LAMTOR3 levels. Furthermore, treatment of LAMTOR2f/−MEF with E64 and chloroquine for 3 h did not result in an increase in LAMTOR3 levels (supplemental Fig. S5A). The slight increase in LAMTOR3 levels upon treatment with leupeptin can be explained by the fact that leupeptin also blocks the trypsin-like proteolytic sites of the proteasome.
FIGURE 6.
The degradation of endogenous LAMTOR3 is lysosome-independent. A, LAMTOR2−/− and LAMTOR2f/−MEF were treated 100 μg/ml E64 for 7 h. Lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. B, LAMTOR2−/− and LAMTOR2f/−MEF were treated with 20 nm Velcade for 7 h. The lysates were separated by SDS-PAGE, analyzed by Western blotting and probed with the indicated antibodies. C, Myc6LAMTOR3LAMTOR2−/− and Myc6LAMTOR3LAMTOR2−/−GFPLAMTOR2 MEF were subjected to indirect immunofluorescence analysis using antibodies against Myc and LAMP1. Epifluorescence pictures are shown. Because of the reduced levels of the protein and the diffuse cytosolic localization, the Myc6 panel shown for the Myc6LAMTOR3LAMTOR2−/− cells was acquired with a higher exposure time than the remaining images. Bars, 20 μm.
In addition, we also performed chloroquine treatment in combination with EGF stimulation. Whereas chloroquine treatment for 7 h sustained MAPK activity consistent with a block in EGFR degradation, LAMTOR3 protein levels were not increased (supplemental Fig. S5B). For comparison, we treated LAMTOR2−/−MEF with Velcade for 7 h. In contrast to lysosomal inhibition, proteasomal inhibition resulted in accumulation of LAMTOR3 protein (Fig. 6B).
We have shown previously that Myc6LAMTOR3 mislocalizes to the cytoplasm in LAMTOR2-depleted MEF (8), distributing evenly throughout the cytoplasm (Fig. 6C). Myc6LAMTOR3 mislocalization could be rescued by expression of a GFPLAMTOR2 retroviral construct in LAMTOR2−/−MEF, indicating that Myc6LAMTOR3 was in principle functional. Overall, it seems very likely that cytoplasmic LAMTOR3 is degraded by the proteasome and not by the lysosome.
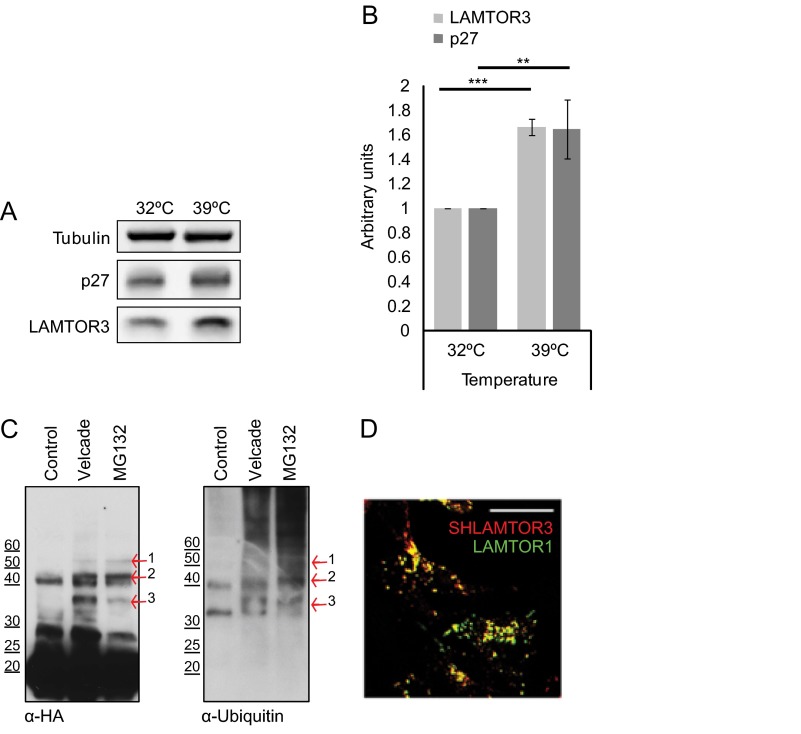
The Ubiquitin-mediated Proteasomal Degradation Pathway Controls LAMTOR3 Degradation
Most proteasomal substrates are first marked for degradation by covalent linkage of polyubiquitin chains to lysines. To substantiate a direct involvement of the ubiquitin proteasome pathway in LAMTOR3 degradation, we made use of the previously established BALB/3T3 mutant cell line carrying a temperature-sensitive ubiquitin-activating E1 enzyme (20). We compared the expression levels of LAMTOR3 in cells kept at the permissive temperature or shifted to the restrictive temperature for 16 h (Fig. 7, A and B). We have included p27 as a bona fide ubiquitin-proteasomal substrate. Both LAMTOR3 and p27 expression levels increased in cells kept at the restrictive temperature, consistent with the involvement of ubiquitination in LAMTOR3 degradation. Furthermore, we have affinity-purified SH-LAMTOR3 from HEK293 cells and could show that upon treatment with Velcade or MG132, SH-LAMTOR3 was found in high molecular weight bands, possibly representing ubiquitinated LAMTOR3. Furthermore, LAMTOR3-positive bands were also positive for ubiquitin after immunoblotting with anti-ubiquitin antibody (Fig. 7C). As a quality control, we confirmed by immunofluorescence analysis that SH-LAMTOR3 co-localized correctly with endogenous LAMTOR1 on punctuated perinuclear vesicles (Fig. 7D). In a proteome-wide quantitative survey, Lys-62 of LAMTOR3 was found to be ubiquitinated (32). In an attempt to identify whether this or other lysine residues mediate LAMTOR3 proteosomal degradation, we performed tandem affinity purification of Velcade-treated SH-LAMTOR3 (supplemental Fig. S6). As expected, ubiquitin-positive bands were enriched in the Velcade-treated sample. We selected five ubiquitin-positive bands and excised the corresponding regions of a silver-stained SDS-PAGE gel. We could identify by LC-MS/MS the presence of both LAMTOR3 and ubiquitin in several of the bands excised. Unfortunately, under the tested experimental conditions, we were unable to identify the exact ubiquitinated lysine residues in LAMTOR3.
FIGURE 7.
The ubiquitin-mediated proteasomal degradation pathway controls LAMTOR3 degradation. A, the temperature-sensitive Ts20 cells derived from BALB/3T3 cells were kept at the permissive temperature of 32 °C or incubated for 16 h at 39 °C. Lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. B, graphic shows the quantification of three independent experiments (means ± S.D., n = 3). **, p < 0.01; ***, p < 0.001. C, HEK293 SH-LAMTOR3 cells were induced with 400 ng/ml tetracycline for 24 h and supplemented with 200 nm Velcade or 10 μm MG132 for the final 7 h. The cells were lysed, and the obtained lysates were immobilized on a Strep-Tactin column. Samples were eluted with biotin, separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. The arrows highlight bands positive for both HA and ubiquitin. D, HEK293 SH-LAMTOR3 cells were induced with 1 μg/ml tetracycline for 24 h and subjected to indirect immunofluorescence analysis using antibodies against HA and LAMP1. Confocal images are shown. Bar, 10 μm.
The Proteasomal Degradation of LAMTOR3 Requires Ubiquitination
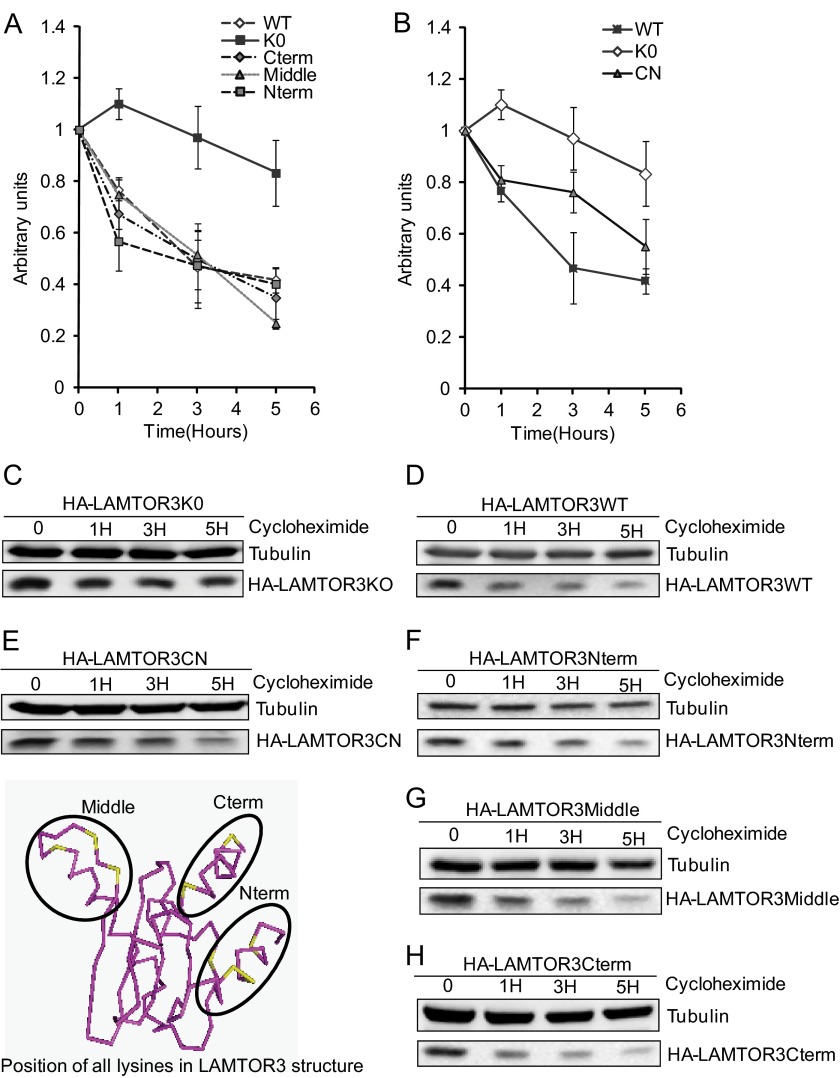
As a second strategy to identify the ubiquitinated lysine residues that contribute to the degradation of LAMTOR3, we have embarked on site-directed mutagenesis. The lysine residues on LAMTOR3 are clustered in three distinct regions: four lysines at the N terminus, two at the C terminus and interestingly, another three in the vicinity of the b3* loop (13) (Fig. 8). We have therefore generated mammalian expression constructs in which lysines, in each of these areas, had been mutated into arginine. They were designated HA-LAMTOR3Cterm, HA-LAMTOR3Middle, and HA-LAMTOR3Nterm. We also generated a lysine-less mutant of LAMTOR3 that we named HA-LAMTOR3K0. HA-LAMTOR3WT and the mutants were analyzed by transient transfection in HeLa cells and tested in a cycloheximide time course (Fig. 8).
FIGURE 8.
Redundancy in lysine contribution toward LAMTOR3 proteasomal degradation. Stability of the tested mutant proteins expressed in cycloheximide treated HeLa cells (80% confluent) is shown in A and B. 48 h after transfection, the cells were treated with 50 μg/ml cycloheximide for the corresponding times. The lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. The graphs represent average of three independent experiments (means ± S.E., n = 3). C, HA-LAMTOR3K0. D, HA-LAMTOR3WT. E, HA-LAMTOR3CN. F, HA-LAMTOR3Nterm. G, HA-LAMTOR3Middle. H, HA-LAMTOR3Cterm.
HA-LAMTOR3WT was degraded with a half-life of roughly 3 h. This was consistent with the observations made for endogenous LAMTOR3 in LAMTOR2f/−MEF. The cycloheximide treatment of HeLa cells transfected with HA-LAMTOR3Nterm, HA-LAMTOR3Cterm, and HA-LAMTOR3Middle showed kinetics of degradation similar to HA-LAMTOR3WT. Importantly the HA-LAMTOR3K0 mutant showed a half-life of more than 5 h.
Analysis of the crystal structure demonstrated that the C and N termini of LAMTOR3 define two putative binding pockets: one at the two-helix side of the complex and another at the four-helix side (13). Both of these hydrophobic pockets have lysine residues from the C and N termini in their vicinity. We therefore decided to create an additional mutant with all lysines in the C and N termini mutated. This mutant, called HA-LAMTOR3CN, still retained the lysines in the middle region. HeLa cells were transiently transfected with HA-LAMTOR3CN and tested in a cycloheximide time course. HA-LAMTOR3CN showed kinetics of degradation slower than HA-LAMTOR3WT and faster than HA-LAMTOR3K0. Taking into consideration the results obtained with HA-LAMTOR3Nterm and HA-LAMTOR3Cterm, in which there was no difference to HA-LAMTOR3WT, the effect observed with the HA-LAMTOR3CN mutant indicated that lysines within these regions might play a prevalent role as ubiquitin acceptors. The results also showed that the HA-LAMTOR3CN mutant was degraded faster than HA-LAMTOR3K0. We assumed that this was due to a contribution of the lysines in the middle region when all remaining lysines were mutated.
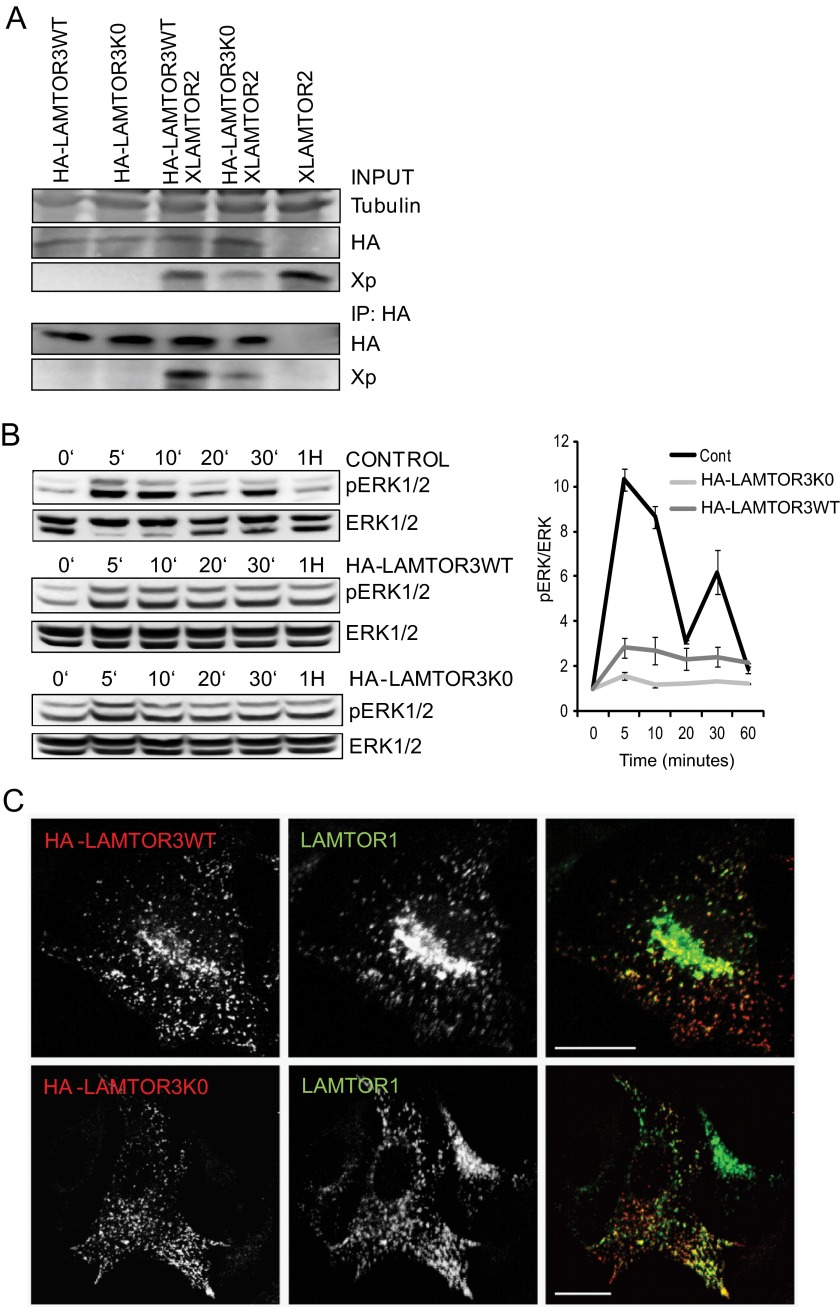
To rule out that the systematic mutation of all LAMTOR3 lysines to arginine had an overall effect on protein structure and consequently on LAMTOR3 function, we have further characterized the HA-LAMTOR3K0 mutant (Fig. 9). Correct interaction with LAMTOR2 was accessed by co-immunoprecipitation. We have overexpressed HA-LAMTOR3WT or HA-LAMTOR3K0 alone or in combination with XLAMTOR2 in HeLa cells (Fig. 9A). Immunoprecipitated HA-LAMTOR3K0 co-immunoprecipitated XLAMTOR2, thereby demonstrating that both proteins interacted with each other. As a control, the HA antibody was not able to immunoprecipitate XLAMTOR2 in cells where neither HA-LAMTOR3WT nor HA-LAMTOR3K0 had been expressed (Fig. 9A, last lane).
FIGURE 9.
Characterization of HA-LAMTOR3wt and HA-LAMTOR3K0. A, co-immunoprecipitation of XLAMTOR2 and HA-LAMTOR3wt or HA-LAMTOR3K0. HeLa cells were transfected with the corresponding constructs. 48 h after transfection, the cells were harvested, lysed, and subjected to immunoprecipitation (IP). The samples were loaded on SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. B, HeLa cells were mock transfected or transfected with HA-LAMTOR3wt or HA-LAMTOR3K0. The cells were serum-starved overnight and induced, 48 h after transfection, with 100 ng/ml EGF for the corresponding time points. Samples were then separated by SDS-PAGE and probed with the indicated antibodies. The graph represents averages of three independent experiments (S.D., n = 3). Cont, control. C, HeLa cells were transfected with HA-LAMTOR3wt or HA-LAMTOR3K0. The following day, the cells were split into coverslips and left to grow another 24 h. Transfected cells were fixed in paraformaldehyde, and immunofluorescence analysis was performed as described under “Experimental Procedures” using antibodies against HA and LAMTOR1. Bars, 20 μm.
Next, we addressed whether HA-LAMTOR3K0 was capable of promoting MAPK activation as demonstrated for wild type LAMTOR3. It should be emphasized that scaffold proteins influence signal propagation in a manner that is dependent on their intracellular concentration (2). Consistently, overexpression of HA-LAMTOR3WT dampened MAPK profile upon EGF stimulation (Fig. 9B). Interestingly, the HA-LAMTOR3K0 had a strong negative effect on MAPK activation. One possible explanation is that HA-LAMTOR3K0 is an effective MAPK scaffold and that therefore overexpression leads to dampening of MAPK activation. However, it could also be that the lysine to arginine mutations affect the recruitment of the kinases to LAMTOR3. Under this scenario, HA-LAMTOR3K0 would function as a dominant negative mutant version of LAMTOR3, thereby dampening ERK activation.
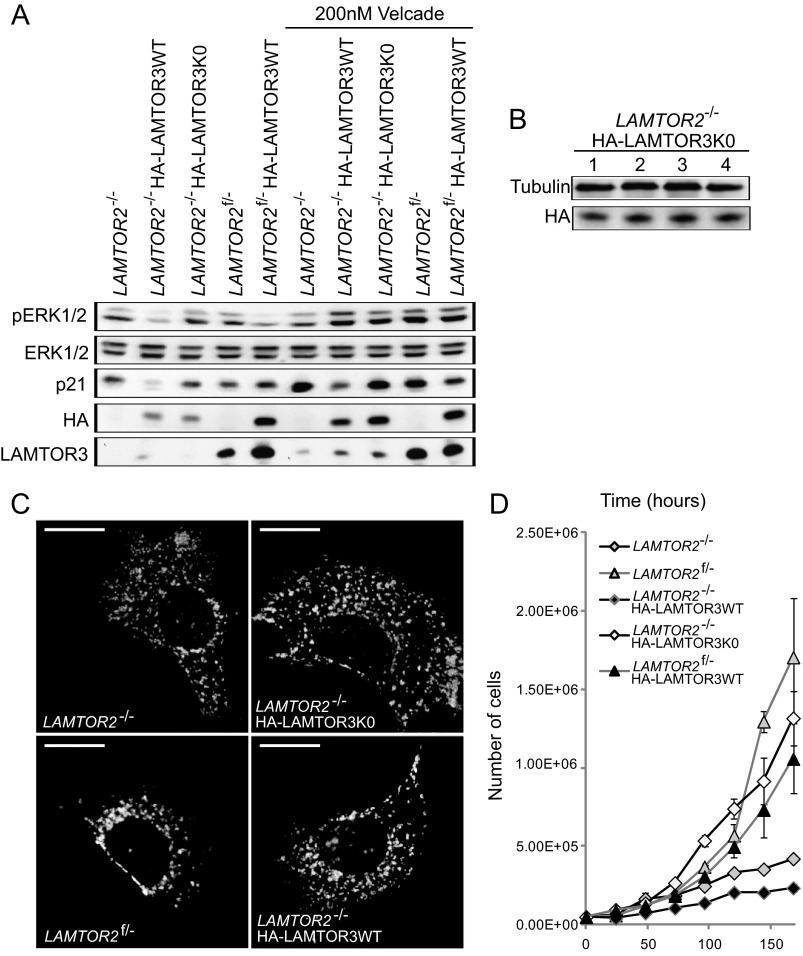
In addition, we also addressed the subcellular localization of HA-LAMTOR3K0. The lysine-less mutant correctly co-localized with LAMTOR1, the anchor protein of the complex, in organelle like vesicles (Fig. 9C). The same co-localization pattern was observed for HA-LAMTOR3WT. To further address the functional consequences of a degradation-deficient mutant of LAMTOR3, we have generated stable cell lines of HA-LAMTOR3WT in the background of control and LAMTOR2 knock-out MEF (Fig. 10A). In addition, we have generated a stable cell line expressing HA-LAMTOR3K0 in LAMTOR2−/−MEF. Analysis of total cell lysates from LAMTOR2−/−, LAMTOR2−/−HA-LAMTOR3WT, LAMTOR2−/−HA-LAMTOR3K0, LAMTOR2f/−, and LAMTOR2f/−HA-LAMTOR3WT revealed several interesting results. First of all, expression of HA-LAMTOR3 resulted in higher levels of the endogenous protein when compared with the parental cell lines. Second, the expression of HA-LAMTOR3WT in both LAMTOR2−/− and LAMTOR2f/− resulted in reduced MAPK activity, whereas expression of HA-LAMTOR3K0 in LAMTOR2−/−MEF had no effect on ERK phosphorylation. This surprising observation may indicate that HA-LAMTOR3K0 is not a functional MAPK scaffold. From endosomes, (overexpression in HeLa cells; Fig. 9), the lysine-less mutant could compete with the endogenous protein for binding to other LAMTOR components and therefore negatively affect the scaffolding function of the complex. In contrast, HA-LAMTOR3K0 expression in LAMTOR2−/− is restricted to the cytoplasm where LAMTOR3 is mostly found as a monomer. Because the mutant may not be able to recruit the kinases properly or in the right conformation, MAPK output remains stable. The expression of HA-LAMTOR3WT in LAMTOR2−/− is also restricted to the cytoplasm, but in contrast to the mutant, HA-LAMTOR3WT may sequester MEK from other functional MAPK scaffolds, thereby explaining the reduction in ERK activation. In addition, we treated the stable cell lines with 200 nm Velcade for 7 h, and interestingly we observed that both HA-LAMTOR3WT and HA-LAMTOR3K0 accumulated upon proteasomal inhibition. The lysine-less mutant may be degraded in a proteasome-dependent but ubiquitin-independent manner, or it may happen that under these nonphysiological conditions, unconventional ubiquitin acceptor sites are used. As reported in supplemental Fig. S4, purified 20S proteasome is not able to degrade recombinant LAMTOR3. However, we cannot formally exclude a contribution of the ubiquitin-independent proteasome pathway toward endogenous LAMTOR3 degradation, in particular because the lysine-less mutant reacts to proteasomal inhibition. Because of this unexpected result, we also confirmed that the expression levels of HA-LAMTOR3K0 in LAMTOR2−/− were stable for several days (Fig. 10B). In addition, we also addressed whether expression of HA-LAMTOR3WT or HA-LAMTOR3K0 was able to restore the defect in late endosomal localization observed in the LAMTOR2−/−MEF (Fig. 10C) (8). In contrast to the perinuclear enrichment observed in control MEF, all remaining cell lines showed LAMP1-positive late endosomes/lysosomes redistributed toward the periphery of the cell, indicating that neither HA-LAMTOR3WT nor HA-LAMTOR3K0 expression restored late endosomal positioning in LAMTOR2−/−MEF. Finally, we tested the growth kinetics of the different cell lines (Fig. 10D). Interestingly the growth defect of LAMTOR2−/−MEF was partially rescued by the expression of HA-LAMTOR3K0, whereas the endosomal mislocalization defect was not. These results indicate that the increased stability of the lysine-less mutant may allow cytosolic LAMTOR3 to support cellular proliferation through yet unclear mechanisms.
FIGURE 10.
Expression of HALAMTOR3K0 partially rescues LAMTOR2−/−MEF cell growth defect, but it does not restore late endosomal positioning. A, LAMTOR2−/−HA-LAMTOR3WT, LAMTOR2f/−HA-LAMTOR3WT, and LAMTOR2−/−HA-LAMTOR3K0 stable clones were generated as described under “Experimental Procedures.” LAMTOR2−/−, LAMTOR2f/−, LAMTOR2−/−HA-LAMTOR3WT, LAMTOR2f/−HA-LAMTOR3WT, and LAMTOR2−/−HA-LAMTOR3K0 MEF were treated with 200 nm Velcade for 7 h. Total cell lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. B, LAMTOR2−/−HA-LAMTOR3K0 cells were harvested in 4 consecutive days. The lysates were separated by SDS-PAGE, analyzed by Western blotting, and probed with the indicated antibodies. C, LAMTOR2−/−, LAMTOR2f/−, LAMTOR2−/−HA-LAMTOR3WT, and LAMTOR2−/−HA-LAMTOR3K0 MEF were fixed in paraformaldehyde and subjected to indirect immunofluorescence analysis as described under “Experimental Procedures” using LAMP1 antibody. Epifluorescence pictures are shown. Bars, 20 μm. D, growth curves of LAMTOR2−/−, LAMTOR2f/−, and stable clones from LAMTOR2−/−HA-LAMTOR3WT, LAMTOR2−/−HA-LAMTOR3K0, and LAMTOR2f/−HA-LAMTOR3WT MEF. 5 × 104 cells were plated into 10-cm dishes. The cells were counted using a Casy cell counter from Schärfe system GmbH at the indicated times (hours). The graph represents averages of three independent experiments (means ± S.D., n ≥ 3).
DISCUSSION
Here we describe that loss of LAMTOR2 results in the mislocalization of monomeric LAMTOR3 to the cytoplasm, which in turn triggers its rapid degradation through the proteasome. Interestingly, we observed a cytosolic pool of LAMTOR3 protein in LAMTOR2-depleted MEF, whereas endosomal LAMTOR3 was only observed in LAMTOR2f/−MEF. Cytosolic LAMTOR3 was almost exclusively found as a monomer. Because monomeric LAMTOR3 does not exert its functions from the cytoplasm, we assume that this pool of LAMTOR3 may be in transit and en route to endosomes or already committed to degradation.
Previous publications have shown that in vitro, LAMTOR3 promotes the formation of a RAF, MEK, and ERK signaling module to promote ERK activation and prevent negative feedback loops (33, 34). In cells, the situation may be similar but might be harder to detect because of the transient kiss and run interaction between LAMTOR3 and MEK/ERK. A recent report demonstrated that ERK substrate specificity depends largely on the membrane microdomain where RAS is activated and on the choice and localization of the scaffold protein mediating signal propagation (35). The LAMTOR1/2/3/4/5 (LAMTOR1-LAMTOR2-LAMTOR3-LAMTOR4-LAMTOR5) complex provides this spatial information required for MAPK activation on late endosomes. Hence, it seems reasonable to assume that cells monitor the localization of LAMTOR3 and aim to prevent its accumulation in the cytoplasm. LAMTOR2−/−MEF have a cell cycle defect with delayed entry into mitosis (8), endosomal biogenesis defects (8, 9), and compromised MAPK (7) and mTOR activity (12). Here we show that expression of a LAMTOR3 lysine-less mutant in LAMTOR2 ablated cells leads to a partial growth rescue of the LAMTOR2−/−MEF but does not restore the endosomal biogenesis defect. We therefore propose that through tight proteasome-mediated degradation and interaction with LAMTOR2, cells ensure that LAMTOR3 functions are only exerted from late endosomes, where the convergence of different inputs ensures a correct biological response.
We also demonstrated that endogenous LAMTOR3 protein has a half-life of ∼3 h. When LAMTOR3 is unable to associate with LAMTOR2 and hence remains in the cytoplasm, the half-life of LAMTOR3 drops below 50 min. Interestingly, the LAMTOR3 mRNA levels are higher in LAMTOR2-depleted MEF when compared with controls, pointing to possible additional regulatory mechanisms. Either way, the results presented further strengthen the model that LAMTOR3 protein instability is the main cause of reduced LAMTOR3 protein levels observed in LAMTOR2−/−MEF.
In the particular case of LAMTOR3, we speculate that the proteasomal turnover might be used as a biological mechanism to prevent the accumulation of a mislocalized monomeric cytoplasmic pool. Similarly the protein levels of LAMTOR1, -4, and -5 were also clearly decreased, emphasizing that the entire Ragulator complex might be destabilized when a proper complex cannot be form in vivo (e.g., under conditions where LAMTOR2, for instance, is missing).
A cytosolic mutant of LAMTOR2 confirmed that heterodimer formation was critical for LAMTOR3 stability, and the cycloheximide time course results indicated that LAMTOR3 turnover was faster in the absence of LAMTOR2. The crystal structure of LAMTOR2-LAMTOR3 suggests the formation of a very tight complex with a large interaction surface (13, 36). Thus far, there has been no clear demonstration of the three-dimensional folding of monomeric LAMTOR3 in solution. However, the folding of LAMTOR2 in solution is identical to the conformation found in the heterodimer complex (37). In addition, LAMTOR2 NMR revealed that the protein is not forming homodimers (37). Furthermore, computational analysis of structural dynamic fluctuations predicts that unbound LAMTOR3 differs from its heterodimer conformation only in the positioning of the b3 loop and on its C- and N-terminal tails (38). Based on this indirect evidence and on the high similarity LAMTOR2 and LAMTOR3 present at the structural level, we presume that cytosolic monomeric LAMTOR3 shows the same overall conformation as it does in the heterodimer. However, we could show that monomeric cytosolic LAMTOR3 is a potential client for protein quality control. Cytosolic quality control defines a series of triage decisions mediated by a dedicated pool of chaperones and E3 ligases that ultimately direct a polypeptide to either refolding or degradation (39). Free complex subunits, synthesized in the absence of cofactors or complementary subunits, may expose hydrophobic surfaces causing toxic protein aggregation events that would therefore favor rapid degradation (40). In the two-helix side, the absence of LAMTOR2 would expose Phe-49 and Phe-53 LAMTOR3 residues normally contributing to the electropositive channel found in the middle of the complex. More importantly, the b3 strand of LAMTOR3 has several hydrophobic residues that intertwine with residues on LAMTOR2 b3 strand stabilizing the heterodimer. In the absence of LAMTOR2, Leu-63, Leu-65, Ile-71, Ile-72, and Cys-73 would form a relatively big hydrophobic surface on LAMTOR3 facing the intracellular milieu. In addition, it is well established that quality control-mediated degradation is mainly performed by the ubiquitin-proteasome system (41–43). Taken together, we suggest that monomeric cytosolic LAMTOR3 might be subject to protein quality control.
To define the degradation pathway mediating LAMTOR3 decay, we have made use of a cell line carrying a temperature-sensitive E1 ubiquitin-activating enzyme, as well as inhibitors of both lysosomal and proteasomal degradation. Because of the intrinsic interdependence of both pathways, we kept the incubation times relatively short. We found that LAMTOR3 degradation was mainly mediated by the proteasome. In addition, we could show that upon proteasomal inhibition SH-LAMTOR3 accumulated at higher molecular weight bands, in which ubiquitin was also present. Although we failed to pinpoint one specific lysine residue that was essential for ubiquitination, mutation of all lysines to arginines resulted in a mutant protein with increased stability.
One attractive mechanism to regulate LAMTOR3 abundance may be the time/stimulus-dependent association of additional interacting partners and/or post-translational modifications on the protein that might favor its ejection from the LAMTOR2 complex. Furthermore, because LAMTOR3 is also regulated on mRNA level, an additional yet unknown regulatory mechanism appears to monitor the function of this scaffold complex. Nevertheless, we have shown here that once segregated to the cytoplasmic fraction, monomeric LAMTOR3 could be ubiquitinated by an unknown cytosolic quality control E3 ligase and recruited to the proteasome for degradation.
Supplementary Material
Acknowledgments
We are grateful to Dr. Angela Wandiger-Ness for sharing the Rab7 and Rab5 antibodies and Matthias Gstaiger for providing the pcDNA5/FRT/TO/SH/GW destination vector. We also thank Dr. Ludger Hengst for the p21 and p27 constructs and for many helpful discussions. The Phoenix cells were a kind gift from Dr. Stephan Geley.
This work was supported by FWF Austrian Science Fund Special Research Program “Cell Proliferation and Cell Death in Tumors” Grant SFB021 (to L. A. H.) and Grant P19486-B12 (to M. W. H.) and by funds from the Austrian Proteomics Platform APP Austrian Genome Program, GEN-AU (to L. A. H.).

This article contains supplemental text and Figs. S1–S6.
- MEF
- mouse embryonic fibroblast(s).
REFERENCES
- 1. Kolch W. (2005) Coordinating ERK/MAPK signalling through scaffolds and inhibitors. Nat. Rev. Mol. Cell Biol. 6, 827–837 [DOI] [PubMed] [Google Scholar]
- 2. Levchenko A., Bruck J., Sternberg P. W. (2000) Scaffold proteins may biphasically affect the levels of mitogen-activated protein kinase signaling and reduce its threshold properties. Proc. Natl. Acad. Sci. U.S.A. 97, 5818–5823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McKay M. M., Morrison D. K. (2007) Integrating signals from RTKs to ERK/MAPK. Oncogene 26, 3113–3121 [DOI] [PubMed] [Google Scholar]
- 4. Brown M. D., Sacks D. B. (2009) Protein scaffolds in MAP kinase signalling. Cell Signal. 21, 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Salerno M., Palmieri D., Bouadis A., Halverson D., Steeg P. S. (2005) Nm23-H1 metastasis suppressor expression level influences the binding properties, stability, and function of the kinase suppressor of Ras1 (KSR1) Erk scaffold in breast carcinoma cells. Mol. Cell. Biol. 25, 1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schaeffer H. J., Catling A. D., Eblen S. T., Collier L. S., Krauss A., Weber M. J. (1998) MP1. A MEK binding partner that enhances enzymatic activation of the MAP kinase cascade. Science 281, 1668–1671 [DOI] [PubMed] [Google Scholar]
- 7. Teis D., Wunderlich W., Huber L. A. (2002) Localization of the MP1-MAPK scaffold complex to endosomes is mediated by p14 and required for signal transduction. Dev. Cell 3, 803–814 [DOI] [PubMed] [Google Scholar]
- 8. Teis D., Taub N., Kurzbauer R., Hilber D., de Araujo M. E., Erlacher M., Offterdinger M., Villunger A., Geley S., Bohn G., Klein C., Hess M. W., Huber L. A. (2006) p14-MP1-MEK1 signaling regulates endosomal traffic and cellular proliferation during tissue homeostasis. J. Cell Biol. 175, 861–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bohn G., Allroth A., Brandes G., Thiel J., Glocker E., Schäffer A. A., Rathinam C., Taub N., Teis D., Zeidler C., Dewey R. A., Geffers R., Buer J., Huber L. A., Welte K., Grimbacher B., Klein C. (2007) A novel human primary immunodeficiency syndrome caused by deficiency of the endosomal adaptor protein p14. Nat. Med. 13, 38–45 [DOI] [PubMed] [Google Scholar]
- 10. Taub N., Nairz M., Hilber D., Hess M. W., Weiss G., Huber L. A. (2012) The late endosomal adaptor p14 is a macrophage host-defense factor against Salmonella infection. J. Cell Sci. 125, 2698–2708 [DOI] [PubMed] [Google Scholar]
- 11. Nada S., Hondo A., Kasai A., Koike M., Saito K., Uchiyama Y., Okada M. (2009) The novel lipid raft adaptor p18 controls endosome dynamics by anchoring the MEK-ERK pathway to late endosomes. EMBO J. 28, 477–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sancak Y., Bar-Peled L., Zoncu R., Markhard A. L., Nada S., Sabatini D. M. (2010) Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell 141, 290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kurzbauer R., Teis D., de Araujo M. E., Maurer-Stroh S., Eisenhaber F., Bourenkov G. P., Bartunik H. D., Hekman M., Rapp U. R., Huber L. A., Clausen T. (2004) Crystal structure of the p14/MP1 scaffolding complex. How a twin couple attaches mitogen-activated protein kinase signaling to late endosomes. Proc. Natl. Acad. Sci. U.S.A. 101, 10984–10989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bar-Peled L., Schweitzer L. D., Zoncu R., Sabatini D. M. (2012) Ragulator is a GEF for the Rag GTPases that signal amino acid levels to mTORC1. Cell 150, 1196–1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wunderlich W., Fialka I., Teis D., Alpi A., Pfeifer A., Parton R. G., Lottspeich F., Huber L. A. (2001) A novel 14-kilodalton protein interacts with the mitogen-activated protein kinase scaffold mp1 on a late endosomal/lysosomal compartment. J. Cell Biol. 152, 765–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stasyk T., Holzmann J., Stumberger S., Ebner H. L., Hess M. W., Bonn G. K., Mechtler K., Huber L. A. (2010) Proteomic analysis of endosomes from genetically modified p14/MP1 mouse embryonic fibroblasts. Proteomics 10, 4117–4127 [DOI] [PubMed] [Google Scholar]
- 17. Obexer P., Geiger K., Ambros P. F., Meister B., Ausserlechner M. J. (2007) FKHRL1-mediated expression of Noxa and Bim induces apoptosis via the mitochondria in neuroblastoma cells. Cell Death Differ. 14, 534–547 [DOI] [PubMed] [Google Scholar]
- 18. Glatter T., Wepf A., Aebersold R., Gstaiger M. (2009) An integrated workflow for charting the human interaction proteome. Insights into the PP2A system. Mol. Syst. Biol. 5, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ory D. S., Neugeboren B. A., Mulligan R. C. (1996) A stable human-derived packaging cell line for production of high titer retrovirus/vesicular stomatitis virus G pseudotypes. Proc. Natl. Acad. Sci. U.S.A. 93, 11400–11406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chowdary D. R., Dermody J. J., Jha K. K., Ozer H. L. (1994) Accumulation of p53 in a mutant cell line defective in the ubiquitin pathway. Mol. Cell. Biol. 14, 1997–2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 22. Gonzalez L., Jr., Scheller R. H. (1999) Regulation of membrane trafficking. Structural insights from a Rab/effector complex. Cell 96, 755–758 [DOI] [PubMed] [Google Scholar]
- 23. Connor M. K., Kotchetkov R., Cariou S., Resch A., Lupetti R., Beniston R. G., Melchior F., Hengst L., Slingerland J. M. (2003) CRM1/Ran-mediated nuclear export of p27Kip1 involves a nuclear export signal and links p27 export and proteolysis. Mol. Biol. Cell 14, 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoshino D., Koshikawa N., Seiki M. (2011) A p27(kip1)-binding protein, p27RF-Rho, promotes cancer metastasis via activation of RhoA and RhoC. J. Biol. Chem. 286, 3139–3148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson W. C., Deanin G. G., Gordon M. W. (1979) Intact microtubules are required for rapid turnover of carboxyl-terminal tyrosine of α-tubulin in cell cultures. Proc. Natl. Acad. Sci. U.S.A. 76, 1318–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bloom J., Amador V., Bartolini F., DeMartino G., Pagano M. (2003) Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell 115, 71–82 [DOI] [PubMed] [Google Scholar]
- 27. Coulombe P., Rodier G., Bonneil E., Thibault P., Meloche S. (2004) N-terminal ubiquitination of extracellular signal-regulated kinase 3 and p21 directs their degradation by the proteasome. Mol. Cell. Biol. 24, 6140–6150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jin Y., Lee H., Zeng S. X., Dai M. S., Lu H. (2003) MDM2 promotes p21waf1/cip1 proteasomal turnover independently of ubiquitylation. EMBO J. 22, 6365–6377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Touitou R., Richardson J., Bose S., Nakanishi M., Rivett J., Allday M. J. (2001) A degradation signal located in the C-terminus of p21WAF1/CIP1 is a binding site for the C8 alpha-subunit of the 20S proteasome. EMBO J. 20, 2367–2375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chen X., Chi Y., Bloecher A., Aebersold R., Clurman B. E., Roberts J. M. (2004) N-Acetylation and ubiquitin-independent proteasomal degradation of p21Cip1. Mol. Cell 16, 839–847 [DOI] [PubMed] [Google Scholar]
- 31. Sdek P., Ying H., Chang D. L., Qiu W., Zheng H., Touitou R., Allday M. J., Xiao Z. X. (2005) MDM2 promotes proteasome-dependent ubiquitin-independent degradation of retinoblastoma protein. Mol. Cell 20, 699–708 [DOI] [PubMed] [Google Scholar]
- 32. Wagner S. A., Beli P., Weinert B. T., Nielsen M. L., Cox J., Mann M., Choudhary C. (2011) Mol. Cell. Proteomics 10.1074/mcp.M111013284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Catling A. D., Schaeffer H. J., Reuter C. W., Reddy G. R., Weber M. J. (1995) A proline-rich sequence unique to MEK1 and MEK2 is required for raf binding and regulates MEK function. Mol. Cell. Biol. 15, 5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Brahma A., Dalby K. N. (2007) Regulation of protein phosphorylation within the MKK1-ERK2 complex by MP1 and the MP1*P14 heterodimer. Arch. Biochem. Biophys. 460, 85–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Casar B., Arozarena I., Sanz-Moreno V., Pinto A., Agudo-Ibáñez L., Marais R., Lewis R. E., Berciano M. T., Crespo P. (2009) Ras subcellular localization defines extracellular signal-regulated kinase 1 and 2 substrate specificity through distinct utilization of scaffold proteins. Mol. Cell. Biol. 29, 1338–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lunin V. V., Munger C., Wagner J., Ye Z., Cygler M., Sacher M. (2004) The structure of the MAPK scaffold, MP1, bound to its partner, p14. A complex with a critical role in endosomal MAP kinase signaling. J. Biol. Chem. 279, 23422–23430 [DOI] [PubMed] [Google Scholar]
- 37. Qian C., Zhang Q., Wang X., Zeng L., Farooq A., Zhou M. M. (2005) Structure of the adaptor protein p14 reveals a profilin-like fold with distinct function. J. Mol. Biol. 347, 309–321 [DOI] [PubMed] [Google Scholar]
- 38. Cui Q., Sulea T., Schrag J. D., Munger C., Hung M. N., Naïm M., Cygler M., Purisima E. O. (2008) Molecular dynamics-solvated interaction energy studies of protein-protein interactions. The MP1-p14 scaffolding complex. J. Mol. Biol. 379, 787–802 [DOI] [PubMed] [Google Scholar]
- 39. Cyr D. M., Höhfeld J., Patterson C. (2002) Protein quality control. U-box-containing E3 ubiquitin ligases join the fold. Trends Biochem. Sci. 27, 368–375 [DOI] [PubMed] [Google Scholar]
- 40. Goldberg A. L. (2003) Protein degradation and protection against misfolded or damaged proteins. Nature 426, 895–899 [DOI] [PubMed] [Google Scholar]
- 41. Hershko A., Ciechanover A. (1998) The ubiquitin system. Annu. Rev. Biochem. 67, 425–479 [DOI] [PubMed] [Google Scholar]
- 42. Wolf D. H., Hilt W. (2004) The proteasome. A proteolytic nanomachine of cell regulation and waste disposal. Biochim. Biophys. Acta 1695, 19–31 [DOI] [PubMed] [Google Scholar]
- 43. McClellan A. J., Tam S., Kaganovich D., Frydman J. (2005) Protein quality control. Chaperones culling corrupt conformations. Nat. Cell Biol. 7, 736–741 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.