Background: Cyanobacterial uptake hydrogenases perform hydrogen oxidation in nitrogen-fixing cyanobacteria, but their biophysical properties are unknown.
Results: The small subunit, HupS, from the Nostoc punctiforme uptake hydrogenase was heterologously expressed and spectroscopically characterized in different redox conditions.
Conclusion: Recombinant HupS incorporates three iron-sulfur clusters with unusual iron coordination.
Significance: We provide the foundation for engineering of cyanobacterial uptake hydrogenases.
Keywords: Cyanobacteria, Electron Paramagnetic Resonance (EPR), Hydrogenase, Iron-Sulfur Protein, Redox
Abstract
In nitrogen-fixing cyanobacteria, hydrogen evolution is associated with hydrogenases and nitrogenase, making these enzymes interesting targets for genetic engineering aimed at increased hydrogen production. Nostoc punctiforme ATCC 29133 is a filamentous cyanobacterium that expresses the uptake hydrogenase HupSL in heterocysts under nitrogen-fixing conditions. Little is known about the structural and biophysical properties of HupSL. The small subunit, HupS, has been postulated to contain three iron-sulfur clusters, but the details regarding their nature have been unclear due to unusual cluster binding motifs in the amino acid sequence. We now report the cloning and heterologous expression of Nostoc punctiforme HupS as a fusion protein, f-HupS. We have characterized the anaerobically purified protein by UV-visible and EPR spectroscopies. Our results show that f-HupS contains three iron-sulfur clusters. UV-visible absorption of f-HupS has bands ∼340 and 420 nm, typical for iron-sulfur clusters. The EPR spectrum of the oxidized f-HupS shows a narrow g = 2.023 resonance, characteristic of a low-spin (S = ½) [3Fe-4S] cluster. The reduced f-HupS presents complex EPR spectra with overlapping resonances centered on g = 1.94, g = 1.91, and g = 1.88, typical of low-spin (S = ½) [4Fe-4S] clusters. Analysis of the spectroscopic data allowed us to distinguish between two species attributable to two distinct [4Fe-4S] clusters, in addition to the [3Fe-4S] cluster. This indicates that f-HupS binds [4Fe-4S] clusters despite the presence of unusual coordinating amino acids. Furthermore, our expression and purification of what seems to be an intact HupS protein allows future studies on the significance of ligand nature on redox properties of the iron-sulfur clusters of HupS.
Introduction
Hydrogen (H2) as a fuel and general energy carrier has boosted the interest in biological H2 production as a renewable energy source (1, 2). Hydrogenases are metalloenzymes that occur in a wide variety of microorganisms, which catalyze the reversible oxidation of H2: H2 ⇋ 2H+ + 2e−. Engineering hydrogenases for applications in biotechnological H2 production, is one strategy for increasing H2 output that has attracted increasing research ventures in later years (2–4).
Most hydrogenases fall into two main classes: the nickel-iron (NiFe)3 hydrogenases, containing a NiFe complex in the catalytic site, and the Fe2 hydrogenases, containing a binuclear iron complex. A third class contains a mononuclear iron center. To date, several crystal structures have been determined for different prokaryotic NiFe hydrogenases, e.g. from the Desulfovibrio genus, from Ralstonia eutropha and Allochromatium vinosum, revealing several shared features (5–11). Common for all NiFe hydrogenases are two protein subunits, referred to as the large and small subunit, respectively. The large subunit contains the active site where H2 oxidation or production is catalyzed by an inorganic NiFe complex.
The small subunit harbors, in most cases, three iron-sulfur (FeS) clusters: a proximal (closest to the active site on the large subunit) [4Fe-4S], a medial [3Fe-4S], and a distal [4Fe-4S] complex. The three FeS clusters are aligned so as to form an electron conduit between the protein surface and the active NiFe site across a total distance of ∼30 Å (Fig. 1A). Depending on the nature of the hydrogenase, the electrons can be transported to or away from the active site. In enzymes where H2 is oxidized, so-called uptake hydrogenases, the electrons are directed via the FeS clusters to the surface. In so-called bidirectional hydrogenases, the FeS clusters can drive electron flow from either toward or away from the active site, depending on the environmental and metabolic conditions of their host. The medial location of the [3Fe-4S] cluster in the small subunit is puzzling as this type of cluster usually presents a higher reduction potential than [4Fe-4S] clusters. It may thus act as an electron trap in the electron transfer chain (12, 13).
FIGURE 1.
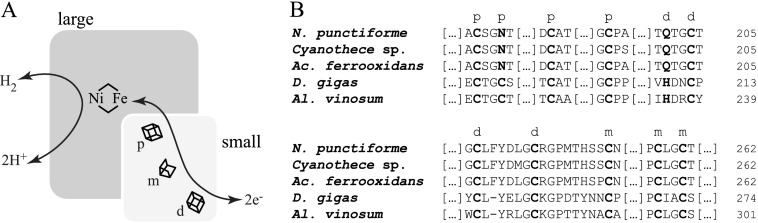
A, schematic representation of the spatial distribution of metal centers in a NiFe hydrogenase. The large subunit contains the NiFe active site where H2 is either formed or oxidized. The small subunit contains typically three FeS clusters: a proximal [4Fe-4S] (p) nearer the active site on the large subunit, a medial [3Fe-4S] (m), and a distal [4Fe-4S] (d), which participate in electron transfer. B, comparison between FeS cluster binding motifs of HupS from N. punctiforme ATCC 29133 and: Cyanothece sp. ATCC 51142 HupS; Acidithiobacillus (Ac.) ferrooxidans ATCC 23270 HoxK; D. gigas HydA; Allochromatium (Al.) vinosum DSM 180 HydA. Letters in boldface type indicate amino acids involved in coordination of the FeS clusters, marked as a proximal (p), medial (m), or distal (d) cluster, respectively. The proximal cluster in N. punctiforme HupS, and in other cyanobacterial-like small subunits, contains an asparagine instead of one of the cysteines, and the distal cluster presents a glutamine replacing histidine.
Cyanobacteria are phototrophic microorganisms that can produce H2 from solar energy and water. They are therefore attractive targets for efforts to improve their productivity via genetic engineering. Most cyanobacteria possess at least one copy each of the uptake and bidirectional hydrogenases (14). To this date, all known cyanobacterial hydrogenases are predicted to be NiFe hydrogenases based on sequence homology (14, 15). Only one cyanobacterial hydrogenase, the bidirectional hydrogenase from Synechocystis strain PCC 6803, has so far been isolated and characterized (16).
Only the uptake hydrogenase, HupSL, is found in the heterocyst-forming, nitrogen-fixing cyanobacterium Nostoc punctiforme ATCC 29133 (identical with strain PCC 73102, and henceforth in this work referred to as N. punctiforme). The uptake hydrogenase in filamentous strains has been found by immunogold labeling and immunolocalization in both heterocysts, which provide a microaerobic environment, and vegetative cells under N2-fixing conditions (17, 18), but was suggested to be in an inactive form in the vegetative cells (17). Most uptake hydrogenases investigated so far are rapidly inhibited by molecular oxygen (13), and a cyanobacterial uptake hydrogenase localized in the vegetative cells would undoubtedly be inactivated by photosynthetic oxygen evolution (14). Recently, it was demonstrated that the active enzyme is produced solely in heterocysts under N2-fixing conditions (19). The maturation system and genomic context of HupSL in N. punctiforme have been investigated extensively. The hupSL promoter region and binding sites for the transcriptional regulator NtcA have been identified (20); the extended hyp operon region, comprising the assembly and maturation system of HupSL, has been shown to be regulated by the transcriptional regulator CalA (19); and HupW, a protease needed for the cleavage of a C-terminal peptide from the large subunit HupL, has been shown to be transcribed in N2-fixing cultures (20). In a related organism, Nostoc (Anabaena) sp. PCC 7120, HupW has been shown to specifically cleave HupL (21).
Because the uptake hydrogenase of N. punctiforme is an H2-oxidizing enzyme, the electron transfer in the small subunit, HupS, is expected to be directed away from the active site. Following oxidation of H2, electrons are presumed to first move from the active site to the proximal cluster, then to the medial and distal clusters, before they reach the native redox partner protein. The relative reduction potentials of the three FeS clusters have been suggested to play an important role in steering this directionality (9, 22, 23). In contrast with the structurally more well studied hydrogenases from, e.g. sulfate-reducing bacteria, the FeS clusters of cyanobacterial uptake hydrogenases share unusual FeS cluster binding motifs involving non-cysteine residues: an asparagine instead of a cysteine in the proximal cluster and a glutamine instead of a histidine in the distal one (Fig. 1B). Most studies on NiFe hydrogenase metallic centers have been either focused on characterizing the diverse states of the catalytic NiFe site in the large subunit or on cluster conversion between [3Fe-4S] and [4Fe-4S]. However, little is known about how non-cysteinyl coordination of the FeS clusters affects the activity of the enzyme. These differences, together with the fact that HupSL is the only hydrogenase present in N. punctiforme, make this organism an attractive model system for studies of how modulation of the FeS cluster environment affects the rate of hydrogen uptake.
In this work, we report the cloning and heterologous expression of HupS from N. punctiforme ATCC 29133, presenting UV-visible absorption and EPR spectroscopy data. HupS was expressed as a soluble fusion protein, f-HupS, in Escherichia coli with the purpose of investigating the nature of FeS clusters in cyanobacterial uptake hydrogenases. To our knowledge, this constitutes the first report on spectroscopic data from a FeS cluster-containing subunit from a hydrogenase without the presence of the nickel-iron-containing large subunit, and therefore without overlapping interfering features from the nickel-iron active site.
EXPERIMENTAL PROCEDURES
Cloning
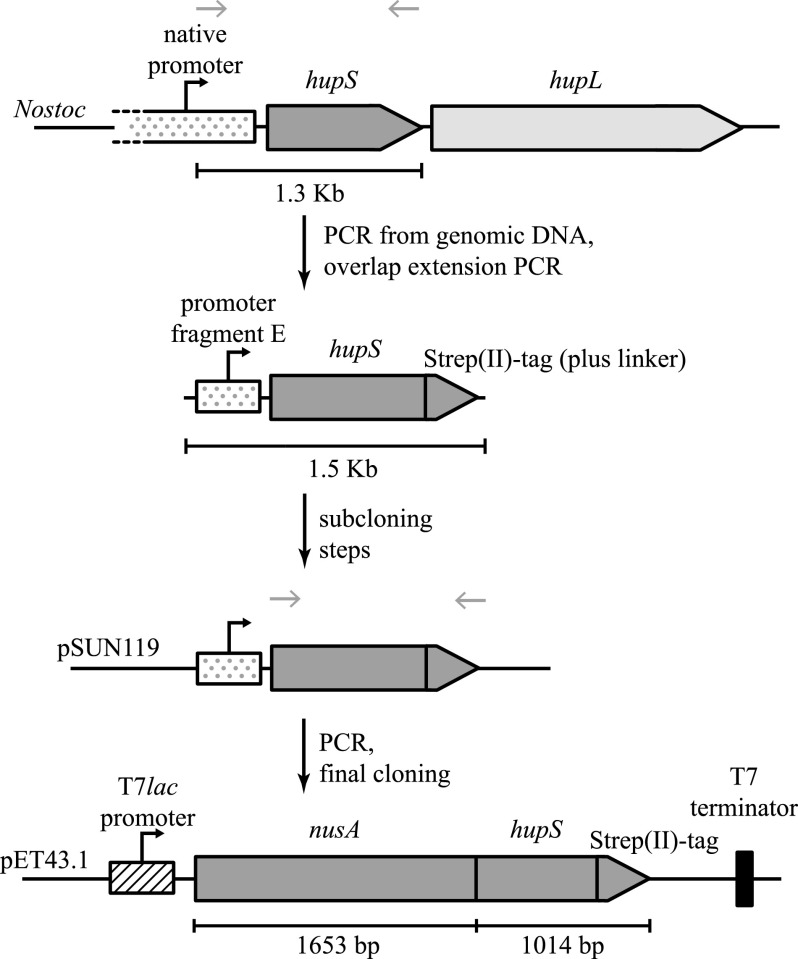
A 1.3-kb fragment containing hupS and the upstream region including promoter fragment E (20) was amplified from N. punctiforme ATCC 29133 genomic DNA using PCR. After gel purification, the fragment was used in overlap extension PCR to add the sequence for a (Gly3Ser)2Gly linker and a Strep(II)-tag on the 3′ end of hupS (using primers 5′-CGC CTG CAG TTC ACC TTT AAA ATC-3′ and 5′-GTA CCT ATT TTT TCT AAA TTG CGG GGA CTC CAG CCA GAA CCT CCT CCA GAA C-3′). The 1.5-kb fused product, including an upstream PstI and a downstream SacI site, was further amplified by PCR, gel-purified, and then blunt-end ligated into pJET1.2 (Fermentas). The ligation product was transformed into TOP10 cells (Invitrogen), and these were plated on LB-agar plates containing 50 μg/ml ampicillin. Selected positive clones were confirmed by sequencing (Macrogen) and digested with PstI and SacI (Fermentas) to yield a 1.4-kb fragment that was gel-purified and ligated into pSUN119 (24) using T4 ligase (Fermentas).
The resulting vector, pSUN119HupSStrepII, was used as template for PCR using DreamTaq polymerase (Fermentas) to amplify a fragment containing only the sequences for HupS, linker, and Strep(II)-tag (but not promoter fragment E), flanked by a 5′ SacI restriction site just upstream of the hupS start codon and a 3′ HindIII site after the stop codon (using primers 5′-AAC AGA GCT CCC ATG ACT AAC G-3′ and 5′-CTA GCG AAG CTT TTA TTT TTC AAA TTG-3′). After gel purification, both the resulting 1-kb fragment and pET43.1a(+) (Novagen) were digested with SacI and HindIII (FastDigest, Fermentas), gel-purified, and used for ligation using the Quick Ligation kit (New England Biolabs). The construct was made so as to express the polypeptide HupS-linker-Strep(II)-tag fused to the C terminus of the solubilization protein NusA (Nus·TagTM) present in the commercial vector; the resulting fusion protein, f-HupS, contains protease recognition sites for both thrombin and enterokinase between NusA and HupS. The ligation product was transformed into DH5α cells, which were then plated on LB-agar plates containing 50 μg/ml ampicillin. Colonies were screened for positive clones by colony PCR, which were then sequenced. A positive clone, pET431HupS, was subsequently used for protein expression of f-HupS. An overview of the cloning process is found in Fig. 2. Sequence alignment was performed using ClustalW (25).
FIGURE 2.
Overview of cloning process of hupS in the pET43.1 plasmid. N. punctiforme hupS was amplified from N. punctiforme. A Strep(II)-tag was subsequently added, and after a cloning step in pSUN119, hupS was cloned downstream of the Nus·Tag coding region as a fusion construct. The construct is under the control of a T7lac promoter, and uses a T7 transcription terminator. The fusion construct, including the Strep(II)-tag and stop codon, is 2667 bp long, giving rise to a 888-aa-long polypeptide, f-HupS. Gray arrows show primer annealing location.
Protein Expression and Purification
All solutions used in anaerobic work were purged for at least 30 min with N2 prior to use. Protein purification was carried out in a glove box (MBraun) under an argon atmosphere. All manipulations were done at 4 °C except where otherwise stated.
50-ml pre-cultures of transformed E. coli BL21(DE3) (Novagen) were grown overnight at 37 °C, 200 rpm shaking, in LB media containing 50 μg/ml ampicillin, and used to inoculate 9 liters of autoinduction medium ZYP-5052 (24), 1.5 liters per 3-liter Erlenmeyer flask, also supplemented with 50 μg/ml ampicillin. Cultures were grown aerobically at 37 °C for 2 h and then at 20 °C for another 18–22 h. Cells were then collected by centrifugation at 6000 × g, pellets were resuspended in cold buffer W (100 mm Tris-HCl, pH 7.5 + 150 mm NaCl), centrifuged again at 4500 × g, and frozen at −20 °C.
The day before EPR experiments were performed, cells were quickly thawed and resuspended in buffer W containing 1 mm MgCl2 and protease inhibitor according to the manufacturer's instructions (cOmplete EDTA-free mixture tablets, Roche Applied Science). The buffer also contained 10 mm glucose, 0.5 units/ml glucose oxidase (Sigma-Aldrich) and 0.5 units/ml catalase (Sigma-Aldrich) to maintain anaerobic conditions (26). Cells were broken by sonication in a Sonics Vibra-Cell VCX750 at 750 watts for 10 min, using 10-s pulses at 70% amplitude. DNase I (20 μg/ml; Sigma-Aldrich), RNase A (40 μg/ml; Sigma-Aldrich), and avidin (0.5 mg/liter culture; ProSpec) were then added, and the resulting crude extract was centrifuged at 184,000 × g for 2 h. The supernatant (soluble fraction) was immediately transferred to a vial on ice that was capped and kept anaerobic under a constant flow of N2 for 30 min. The soluble fraction was then introduced in the glove box and applied onto a pre-equilibrated Strep-Tactin column (IBA) (in buffer W containing 0.5 mg/ml avidin) at room temperature. After application of the soluble fraction, the column was washed with five bed volumes of buffer W plus avidin, then two bed volumes of buffer W alone. The protein was eluted with three bed volumes of buffer W containing 5 mm desthiobiotin (IBA). The fraction containing the highest amount of protein was collected and transferred to EPR tubes (150 μl/tube). Remaining fractions were stored anaerobically at −80 °C.
SDS-PAGE and Western Blotting
The protein contents of different purification fractions were analyzed by 10% SDS-PAGE minigels using Laemmli's buffer system (27) in either a SE250 Mighty Small II unit (Hoefer) or a Mini-Protean Tetra Electrophoresis system (Bio-Rad). Total protein was detected directly on gels using PageBlue protein staining (Thermo Scientific). For identification of f-HupS in the same fractions, minigels were blotted onto nitrocellulose membranes using either a TE22 Mini Tank Transfer Unit (GE Healthcare) or a Trans-Blot Turbo Transfer System (Bio-Rad). Membranes were treated with Strep-Tactin-HRP conjugate (IBA) for chemiluminiscence detection according to the manufacturer's instructions. Chemiluminiscence was detected after treatment with Immun-Star-HRP Chemiluminiscent kit (Bio-Rad) in a ChemiDoc XRS system (Bio-Rad), using exposure times between 10 s and 3 min.
Protein Quantification
Protein quantification was performed using the Coomassie (Bradford) protein assay kit (Thermo Scientific), using bovine serum albumin as a standard according to the manufacturer's instructions.
UV-visible Spectrophotometry
Samples were monitored in anaerobic 1-ml quartz cuvettes on a Varian Cary 50 Bio UV-visible spectrophotometer. Empty sealed cuvettes were purged with nitrogen gas for at least 10 min. f-HupS (1.9 mg protein/ml) was then added through a rubber septum, and spectra were taken immediately.
EPR Spectroscopy
Samples were investigated by continuous wave X-band EPR either directly as purified or after reduction or oxidation by addition of either sodium dithionite (Sigma-Aldrich) or potassium hexacyanoferrate(III) (ferricyanide; Merck), respectively, directly into the EPR tube. EPR tubes were capped, brought out of the glove box, and immediately frozen in liquid nitrogen; dithionite-reduced samples were left to incubate on ice up to 30 min before freezing. The protein concentration was 1 mg/ml. Final concentrations of added reagents varied between equimolar to 20-fold of protein concentration and were added so as to not change the final sample volume >5%. Measurements were performed on a Bruker ELEXYS E500 spectrometer using an ER049X SuperX microwave bridge in a Bruker SHQ0601 cavity equipped with an Oxford Instruments continuous flow cryostat and using an ITC 503 temperature controller (Oxford Instruments). Measurement temperatures ranged from 4.5 to 25 K, using liquid helium as coolant. The spectrometer was controlled by the Xepr software package (Bruker). EPR spectral simulations were run on WINEPR SimFonia (version 1.26, Bruker).
RESULTS
Heterologous Expression of HupS
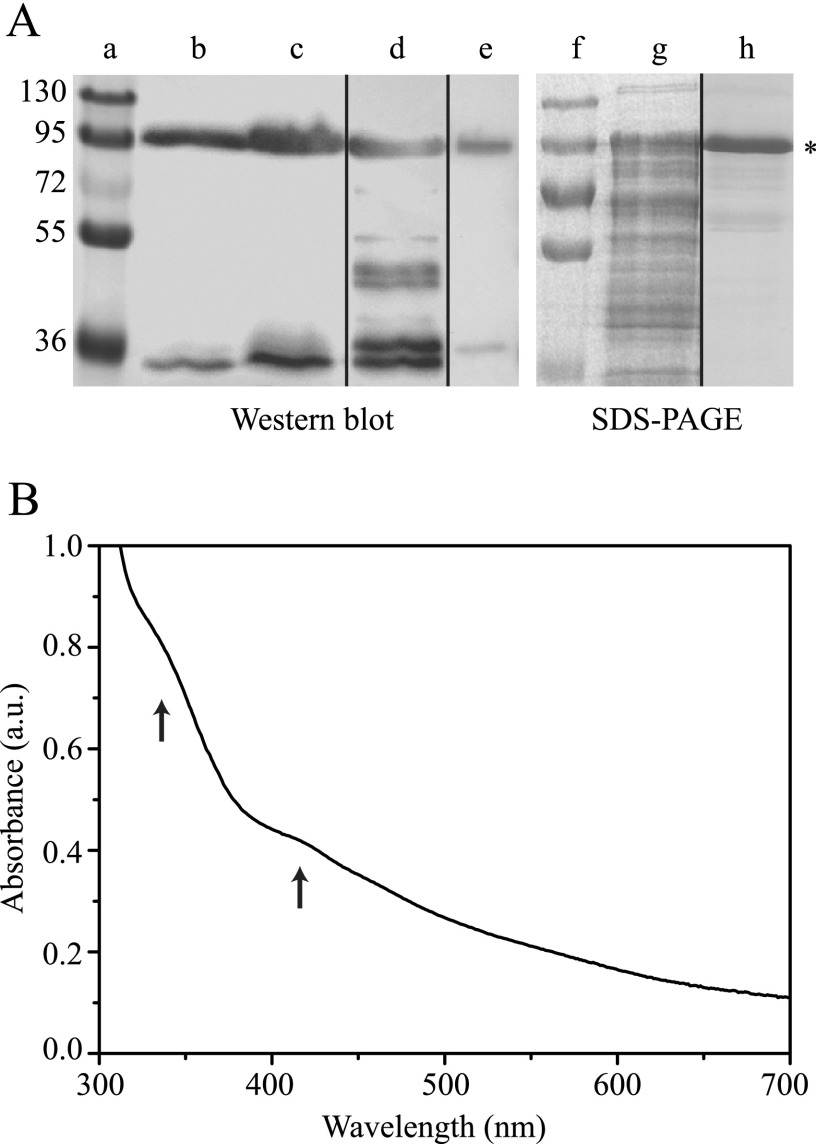
Previous attempts of expression of HupS under the T7lac promoter were not successful because the purified protein was insoluble.4 We therefore made a construct, f-HupS, where HupS was fused to the C terminus of NusA (Nus·Tag), a protein tag specifically developed for its ability to solubilize difficult target proteins that are otherwise prone to aggregate (28). The construct was made by cloning the N. punctiforme ATCC 29133 hupS open reading frame into pET43.1a(+) downstream of the vector-encoded Nus·Tag and including a C-terminal Strep(II) tag to allow purification via affinity chromatography (Fig. 2). Expression of f-HupS was confirmed by SDS-PAGE analysis of whole cell contents, as judged by the appearance of a ∼97-kDa polypeptide after 20 h of growth (data not shown).
The involvement of the Nus·Tag was quite successful, and f-HupS was partially soluble (∼40–50%, as judged by SDS-PAGE, data not shown). It could therefore be produced in amounts allowing spectroscopic analysis. When the protein was purified anaerobically under an argon atmosphere, typically 0.6–1.2 mg of pure, soluble f-HupS were obtained per liter of culture (Fig. 3A, lanes e and h). The purification of f-HupS could be achieved to near homogeneity, as judged by 10% SDS-PAGE (Fig. 3A, lanes g and h). The degree of purification is determined to be ∼30-fold, compared with the crude extract. Attempts to purify f-HupS aerobically resulted in an unstable protein that suffered proteolysis, as judged by the presence of multiple bands in Western blots when using a detection system for the Strep(II)-tag (Fig. 3A, lane d).
FIGURE 3.
Analysis of the purified f-HupS. A, lanes a–e: Western blot of purification fractions, using chemiluminiscent detection of the Strep(II)-tag. Lane a, protein markers (size shown on left side in kDa). Lane b, crude cell extract, after cell disruption (30 μg total protein). Lane c, supernatant after centrifugation (30 μg of total protein). Lane d, aerobically purified f-HupS (3 μg). Lane e, anaerobically purified f-HupS (2 μg). Lanes f–h, SDS-PAGE of crude extract and anaerobically purified f-HupS. Lane f, protein markers. Lane g, crude cell extract (30 μg total protein). Lane h, anaerobically purified f-HupS (5 μg). Asterisk denotes polypeptide corresponding to full-length f-HupS. Vertical lines denote non-contiguous lanes. Aerobically purified f-HupS presented multiple bands due to protein degradation (lane d), which were absent from anaerobically purified f-HupS (lanes e and h). B, UV-visible absorption spectrum of the anaerobically purified f-HupS (1.9 mg protein/ml). The spectrum was taken in a sealed cuvette, previously purged with nitrogen gas. The arrows mark absorption bands with maxima at ∼340 and 420 nm, which are typically seen in FeS proteins. a.u.: arbitrary units.
UV-visible Spectrophotometry
NusA lacks any cofactors that could interfere with visible absorption or EPR spectroscopy of the FeS clusters in HupS. The f-HupS fusion protein had a light brown color and was analyzed by UV-visible spectrophotometry as anaerobically purified (Fig. 3B). The protein presents two discernible absorption bands in the 300–500-nm region with maxima at ∼340 and 420 nm, respectively, as well as a broad shoulder in the 500–600-nm region. The shape and location of these bands are typical of broad ligand-to-metal charge transfer in iron-sulfur proteins such as ferredoxins (29, 30) and indicated that iron-sulfur clusters had been successfully incorporated into f-HupS. This is further substantiated by the 420/315 nm absorbance ratio of 0.43, which is comparable with the corresponding ratio 0.68 found in ferredoxin II from Desulfovibrio gigas (29). In contrast, the often used 420/280 nm ratio is not a useful measure of purity due to the presence of NusA, which has a large contribution at 280 nm and necessarily leads to a lower 420/280 nm absorbance ratio than in ferredoxins.
EPR Spectroscopy
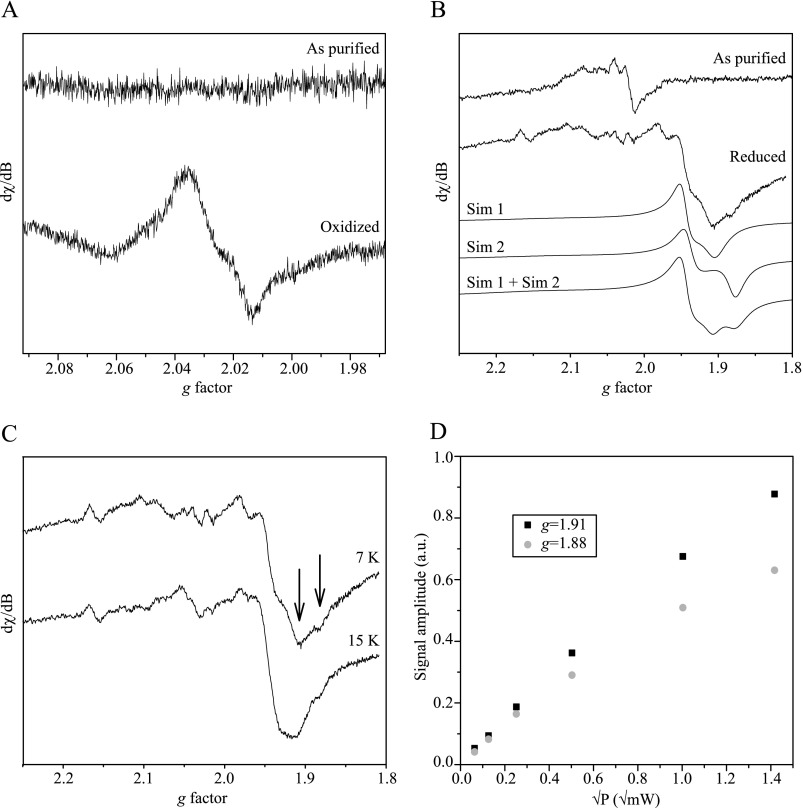
Fig. 4 shows EPR spectra of anaerobically purified f-HupS. f-HupS presented no distinguishable EPR features at low (8 μW) microwave power when taken immediately after purification under anaerobic conditions (Fig. 4A, top). Upon oxidation with equimolar amounts of ferricyanide, a narrow g = 2.023 resonance appeared in the low microwave power spectrum (Fig. 4A, bottom). This resonance was easily saturated at low microwave power between 4 and 25 K, and the signal intensity quickly decreased at above 15 K (not shown). We attribute the signal at g = 2.023 to a [3Fe-4S]+ cluster in a low spin (S = 1/2) state, in analogy to very similar spectra that have been observed for other [3Fe-4S] proteins such as ferredoxin II from D. gigas (31) and the oxidized form of Desulfovibrio africanus ferredoxin III (32).
FIGURE 4.
X-band EPR spectra of f-HupS. A, top spectrum, f-HupS as purified; bottom spectrum, after oxidation of the sample with ferricyanide. EPR conditions were as follows: 8 microwatts of applied microwave power; temperature = 7 K. B, top spectrum, f-HupS as purified; second spectrum, after reduction with dithionite. EPR conditions: 2 milliwatts of applied microwave power; temperature = 7 K. Third and fourth spectrum (Sim 1, Sim 2), simulated spectra of the two components of the reduced sample spectrum (see main text for details). Bottom spectrum (Sim1+sim2), mathematical addition of sim1 and sim2 simulations, weighed 50% each. C, dithionite-reduced f-HupS measured at different temperatures, at the same microwave power (2 milliwatts). Top spectrum, 15 K. Bottom spectrum, 7 K. The arrows point at spectral features belonging to the two spectral components that change differently with temperature. D, variation in EPR signal amplitudes with different applied microwave power (P) in dithionite-reduced f-HupS, measured at temperature = 15 K. The amplitudes were measured from the resonances indicated with arrows in C. Gray circles, the g = 1.91 resonance; Black squares, the g = 1.88 resonance. The modulation amplitude in all measurements was 10 G. Protein concentration was 9 mg/ml.
Using higher microwave power (2 milliwatts), some small features became visible in the anaerobically purified sample: a signal around g = 2.02, and smaller signals at g ≥ 2.04 (Fig. 4B, top). The minor signals with g ≥ 2.04 are attributable to a small contamination of adventitiously bound manganese(II).
Upon reduction with dithionite, the g = 2.023 resonance disappeared, and instead, a wider and more complex spectrum was observed (Fig. 4B, second trace). In particular, we observed a large new resonance centered on g = 1.94, with two additional signals at g = 1.91 and g = 1.88. By investigation of the temperature variation and microwave power saturation of this spectrum, we could distinguish between two separate spectroscopic species (Fig. 4, C and D). When the temperature was raised from 7 to 15 K, the g = 1.91 resonance increased in intensity relative to the base line, whereas the g = 1.88 resonance was almost unchanged (Fig. 4C). By comparing the g = 1.91 and the g = 1.88 intensities at different microwave powers at a fixed temperature (15 K), we could observe that the two resonances behave differently (Fig. 4D). These different behaviors in temperature and microwave power dependence demonstrate the presence of two magnetically distinct species. This is important since it shows that our preparation of f-HupS contains two different low-potential [4Fe-4S] clusters.
Electron transfer from dithionite to FeS clusters is known to be sluggish in the absence of redox mediators (27). Therefore, the sample was reduced by treatment with dithionite for varying times, from below 1 min up to 30 min. We could observe the signal of the reduced protein already after a 10-min incubation (data not shown), and its intensity increased further at the longer treatment time. At least 90% of the sample was reduced after 10 min, and a treatment of less than one minute was insufficient to reduce all FeS clusters (data not shown). We have therefore chosen to treat reduced samples with dithionite for 30 min to ensure that we could obtain the maximum possible intensity from the observed resonances.
The spectrum from the reduced sample could be simulated as a superposition of two S = ½ species. This was achieved by mathematical addition of two separate simulations, both for S = ½ species, with a 50% contribution of each simulation to the final result (Fig. 4B, three last traces). The first simulation includes gx = 1.905 (40 G width) and gy = 1.946 (25 G width) components, whereas the second includes gx = 1.877 (30 G width) and gy = 1.940 (30 G width) components (the gz components in the acquired spectra were hidden under manganese contamination and were therefore broadened to 200 G at g = 2.14). This result indicates that the experimental spectrum arises from the overlap of two different S = ½ species. Thus, the simulations corroborate our conclusion that two distinct [4Fe-4S] clusters are observed in the reduced sample spectrum.
When we analyzed aerobically purified f-HupS, we observed a spectral feature at g = 2.023 similar to the one from the [3Fe-4S] cluster observed in the oxidized anaerobic preparations, as well as a sharp feature at g = 4.3 typical of unspecifically bound mononuclear iron (data not shown). In contrast to the anaerobically purified protein, no resonances attributable to [4Fe-4S] clusters were detected in the aerobically purified sample after reduction by dithionite (data not shown). Part of the observed [3Fe-4S] signal in this case may therefore arise from [4Fe-4S] clusters that have undergone oxidation and degradation. This phenomenon is common in [4Fe-4S]-containing proteins when they are exposed to dioxygen (e.g. Refs. 33–36). We also observed proteolysis as evaluated by SDS-PAGE and Western blotting of those samples, where several bands reacted with the antibody (Fig. 3A, lane d). These results show that HupS is an oxygen-sensitive protein, at least in the absence of HupL. However, aerobic conditions during the expression and initial purification phases were feasible due to the reducing intracellular environment of E. coli (37). This was reinforced during cell lysis by the use of a glucose oxidase/catalase system (26) as an extra precaution against the deleterious effects of dioxygen.
DISCUSSION
Previous studies of NiFe hydrogenases have been mainly focusing on the structure and reactivity of the catalytic NiFe site on the large subunit. More recently, some interest has also been directed to how the FeS clusters in the small subunit modulate the overall enzyme activity, mainly through mutagenesis studies targeting cluster-coordinating amino acids (9, 38, 39). However, spectroscopic characterization of the FeS clusters in the holoenzyme is difficult due to interference from the NiFe site, both due to overlapping signals in EPR spectroscopy, and magnetic coupling between the NiFe site and the FeS clusters. In addition, the enzyme we targeted for our studies, Nostoc punctiforme HupSL, is expressed only in heterocysts, under nitrogen-fixing conditions (20, 40). This precludes its purification in adequate yields for biophysical characterization. We therefore chose to express the small subunit HupS in E. coli and characterize it separately from the large subunit.
By using the Nus·Tag, we found that heterologous expression of soluble HupS is possible when using a solubilization fusion protein. This is different from heterologous expression of the HupS alone, which rendered a non-soluble product. The fusion protein, f-HupS, yielded a partially soluble product, but part of the protein was found in insoluble fractions of the cells. Interestingly, it has been shown that heterologous expression of HupL from Lyngbya majuscula CCAP 1446/4 in E. coli also results in an insoluble product (41). Thus, it seems that both the large (HupL) and the small (HupS) subunit of HupSL are unstable on their own and difficult to obtain in soluble form when purified alone. This probably reflects that the contact interface between small and large subunits of known NiFe hydrogenases is rather large (12). In part, this was possible to overcome by use of the Nus·Tag to create the fusion protein f-HupS. The large extra protein obviously assists in maintaining HupS in a soluble form, accessible for spectroscopy. This is a useful result, but due to the still rather poor solubility and stability of the fusion protein, we chose to not cleave HupS from its solubilization fusion partner NusA.
The preparation yielded soluble protein with a brown color reminiscent of other iron-sulfur proteins, notably ferredoxins and was studied by UV-visible and EPR spectroscopies. Both confirmed that f-HupS incorporated FeS centers. In addition, the EPR spectra of anaerobically purified f-HupS allowed us to distinguish two different types of clusters. We assigned the two species to [3Fe-4S] and [4Fe-4S] clusters, which is in accordance with structurally known hydrogenases.
The EPR signature from a [3Fe-4S]+ cluster in f-HupS appeared upon oxidation with ferricyanide. After incubation with dithionite, two different EPR signals from two distinct [4Fe-4S]+ clusters were observed. In hydrogenases with known structure, the medial [3Fe-4S]+ cluster has been reported to have a relatively high redox potential and is EPR active in its oxidized form (9, 42). The distal and proximal clusters on the other hand, have more negative potentials and would be observed by EPR only after reduction. Therefore, our results show that our purified f-HupS containts three FeS centers with similar properties as the small subunits in intact NiFe hydrogenases.
No cyanobacterial uptake hydrogenase has so far been spectroscopically or structurally characterized. Two characteristic deviances in the small subunit amino acid sequence distinguish the cyanobacterial uptake hydrogenases from their more well studied cousins in e.g. the Desulfovibrio family. The first is the absence of a cysteine in the proximal cluster motif. The corresponding position of this residue would be number 15 in the N. punctiforme sequence. This residue is found to be an asparagine in all the cyanobacterial HupS proteins. The second difference is a glutamine instead of a histidine at position 100 (Fig. 1B, also cf. Ref. 44) in the distal cluster binding motif in N. punctiforme. From the amino acid sequence, it could be argued that the lack of conventional ligands for both the proximal and distal clusters, would lead to a different cluster composition than in previously known hydrogenases. However, we observe two distinct [4Fe-4S] cluster EPR signals from the cyanobacterial HupS, which strongly supports the existence of both a proximal and a distal cluster despite the unusual ligand configuration.
In the absence of the normal cysteine ligand in position 15, the proximal cluster could be coordinated by a cysteine residue located 13 amino acids downstream of the usual motif (Cys-27). However, we find this unlikely because a comparison with known crystal structures reveals that this downstream cysteine will be located in a loop ∼20 Å away from the proximal cluster and therefore not in a coordinating distance. Instead, we suggest that Asn-15 is a coordinating residue for the proximal cluster. In addition, we suggest that Gln-100 coordinates the distal cluster in the absence of a histidine in that position.
The cyanobacterial-like uptake hydrogenase HoxKG from Acidithiobacillus ferrooxidans, a chemolithotrophic, aerobic bacterium, has been purified (43) and studied by EPR (44). The small subunit HoxK and the cyanobacterial HupS share the amino acid signatures described above in their FeS cluster binding motifs. From EPR spectroscopy in the Acidithiobacillus enzyme, the magnetic interaction between the proximal FeS cluster and the NiFe active site was found to be different from the “standard” hydrogenases from A. vinosum and D. gigas. It was suggested that this is due to an unusual ligand sphere in the A. ferrooxidans HoxKG proximal cluster, although the authors did not go so far as to suggest coordination by asparagine (44). Furthermore, the low iron:protein ratio made the existence of the distal cluster unclear, and the authors suggested that the distal cluster was lacking due to the absence of adequate protein ligands. Whether or not A. ferrooxidans HoxKG may lack one of the clusters, the situation is different in our purified f-HupS protein which clearly contains three FeS clusters.
In summary, we have for the first time isolated, through heterologous expression, the small subunit of a cyanobacterial uptake hydrogenase. This enzyme is only found in the heterocysts in N. punctiforme, which limits the availability for spectroscopic characterization of this enzyme. Furthermore, heterologous expression of HupS provides a foundation for engineering of the electron transfer chain for introduction in heterocysts. The protein was found to contain three FeS clusters, in accordance with previously isolated enzymes from other bacterial species. Although the FeS binding motifs show differences from earlier studied enzymes, their EPR signatures show that the protein nevertheless contains two [4Fe-4S] and one [3Fe-4S] cluster. To clarify the role of the coordinating ligands of N. punctiforme HupS, we are currently performing mutagenesis studies. We also further investigate how different FeS cluster binding motifs affect the redox potentials of the clusters.
Acknowledgment
We thank Dr. Ping Huang for assistance with the EPR spectroscopy analysis.
This work was supported by the Knut and Alice Wallenberg Foundation and the Swedish Energy Agency.
D. Camsund and P. Lindblad, personal communication.
- NiFe
- nickel-iron
- FeS
- iron-sulfur
- f-HupS
- recombinant HupS-NusA fusion protein.
REFERENCES
- 1. Styring S. (2012) Artificial photosynthesis for solar fuels. Faraday Discuss. 155, 357–376 [DOI] [PubMed] [Google Scholar]
- 2. Dasgupta C. N., Jose Gilbert J., Lindblad P., Heidorn T., Borgvang S. A., Skjanes K., Das D. (2010) Recent trends on the development of photobiological processes and photobioreactors for the improvement of hydrogen production. Int. J. Hydrogen Energy 35, 10218–10238 [Google Scholar]
- 3. Lee H. S., Vermaas W. F., Rittmann B. E. (2010) Biological hydrogen production: prospects and challenges. Trends Biotechnol. 28, 262–271 [DOI] [PubMed] [Google Scholar]
- 4. Armstrong F. A., Belsey N. A., Cracknell J. A., Goldet G., Parkin A., Reisner E., Vincent K. A., Wait A. F. (2009) Dynamic electrochemical investigations of hydrogen oxidation and production by enzymes and implications for future technology. Chem. Soc. Rev. 38, 36–51 [DOI] [PubMed] [Google Scholar]
- 5. Fritsch J., Scheerer P., Frielingsdorf S., Kroschinsky S., Friedrich B., Lenz O., Spahn C. M. (2011) The crystal structure of an oxygen-tolerant hydrogenase uncovers a novel iron-sulphur centre. Nature 479, 249–252 [DOI] [PubMed] [Google Scholar]
- 6. Matias P. M., Soares C. M., Saraiva L. M., Coelho R., Morais J., Le Gall J., Carrondo M. A. (2001) [NiFe] hydrogenase from Desulfovibrio desulfuricans ATCC 27774: gene sequencing, three-dimensional structure determination and refinement at 1.8 Å and modelling studies of its interaction with the tetrahaem cytochrome c3. J. Biol. Inorg. Chem. 6, 63–81 [DOI] [PubMed] [Google Scholar]
- 7. Ogata H., Kellers P., Lubitz W. (2010) The Crystal Structure of the [NiFe] Hydrogenase from the Photosynthetic Bacterium Allochromatium vinosum: Characterization of the Oxidized Enzyme (Ni-A State). J. Mol. Biol. 402, 428–444 [DOI] [PubMed] [Google Scholar]
- 8. Ogata H., Mizoguchi Y., Mizuno N., Miki K., Adachi S., Yasuoka N., Yagi T., Yamauchi O., Hirota S., Higuchi Y. (2002) Structural Studies of the Carbon Monoxide Complex of [NiFe] hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen. J. Am. Chem. Soc. 124, 11628–11635 [DOI] [PubMed] [Google Scholar]
- 9. Rousset M., Montet Y., Guigliarelli B., Forget N., Asso M., Bertrand P., Fontecilla-Camps J. C., Hatchikian E. C. (1998) [3Fe-4S] to [4Fe-4S] cluster conversion in Desulfovibrio fructosovorans [NiFe] hydrogenase by site-directed mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 95, 11625–11630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Volbeda A., Charon M. H., Piras C., Hatchikian E. C., Frey M., Fontecilla-Camps J. C. (1995) Crystal structure of the nickel-iron hydrogenase from Desulfovibrio gigas. Nature 373, 580–587 [DOI] [PubMed] [Google Scholar]
- 11. Volbeda A., Martin L., Cavazza C., Matho M., Faber B. W., Roseboom W., Albracht S. P., Garcin E., Rousset M., Fontecilla-Camps J. C. (2005) Structural differences between the ready and unready oxidized states of [NiFe] hydrogenases. J. Biol. Inorg. Chem. 10, 239–249 [DOI] [PubMed] [Google Scholar]
- 12. Fontecilla-Camps J. C., Volbeda A., Cavazza C., Nicolet Y. (2007) Structure/function relationships of [NiFe]- and [FeFe]-hydrogenases. Chem. Rev. 107, 4273–4303 [DOI] [PubMed] [Google Scholar]
- 13. Vincent K. A., Parkin A., Armstrong F. A. (2007) Investigating and exploiting the electrocatalytic properties of hydrogenases. Chem. Rev. 107, 4366–4413 [DOI] [PubMed] [Google Scholar]
- 14. Tamagnini P., Leitão E., Oliveira P., Ferreira D., Pinto F., Harris D. J., Heidorn T., Lindblad P. (2007) Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol. Rev. 31, 692–720 [DOI] [PubMed] [Google Scholar]
- 15. Vignais P. M., Billoud B. (2007) Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272 [DOI] [PubMed] [Google Scholar]
- 16. Germer F., Zebger I., Saggu M., Lendzian F., Schulz R., Appel J. (2009) Overexpression, Isolation, and Spectroscopic Characterization of the Bidirectional [NiFe] Hydrogenase from Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 36462–36472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Seabra R., Santos A., Pereira S., Moradas-Ferreira P., Tamagnini P. (2009) Immunolocalization of the uptake hydrogenase in the marine cyanobacterium Lyngbya majuscula CCAP 1446/4 and two Nostoc strains. FEMS Microbiol. Lett. 292, 57–62 [DOI] [PubMed] [Google Scholar]
- 18. Lindblad P., Sellstedt A. (1990) Occurrence and localization of an uptake hydrogenase in the filamentous heterocystous cyanobacterium Nostoc PCC 73102. Protoplasma 159, 9–15 [Google Scholar]
- 19. Camsund D., Devine E., Holmqvist M., Yohanoun P., Lindblad P., Stensjö K. (2011) A HupS-GFP fusion protein demonstrates a heterocyst-specific localization of the uptake hydrogenase in Nostoc punctiforme. FEMS Microbiol. Lett. 316, 152–159 [DOI] [PubMed] [Google Scholar]
- 20. Devine E., Holmqvist M., Stensjö K., Lindblad P. (2009) Diversity and transcription of proteases involved in the maturation of hydrogenases in Nostoc punctiforme ATCC 29133 and Nostoc sp. strain PCC 7120. BMC Microbiol. 9, 53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindberg P., Devine E., Stensjö K., Lindblad P. (2012) HupW protease specifically required for processing of the catalytic subunit of the uptake hydrogenase in the cyanobacterium Nostoc sp. strain PCC 7120. Appl. Environ. Microbiol. 78, 273–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Bingemann R., Klein A. (2000) Conversion of the central [4Fe-4S] cluster into a [3Fe-4S] cluster leads to reduced hydrogen-uptake activity of the F-420-reducing hydrogenase of Methanococcus voltae. Eur. J. Biochem. 267, 6612–6618 [DOI] [PubMed] [Google Scholar]
- 23. Page C. C., Moser C. C., Chen X., Dutton P. L. (1999) Natural engineering principles of electron tunnelling in biological oxidation-reduction. Nature 402, 47–52 [DOI] [PubMed] [Google Scholar]
- 24. Summers M. L., Wallis J. G., Campbell E. L., Meeks J. C. (1995) Genetic evidence of a major role for glucose-6-phosphate dehydrogenase in nitrogen fixation and dark growth of the cyanobacterium Nostoc sp. strain ATCC 29133. J. Bacteriol. 177, 6184–6194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Clustal-W - Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Uppoor R., Niebergall P. J. (1996) β-D(+) Glucose-Glucose Oxidase-Catalase for Use as an Antioxidant System. Pharm. Dev. Technol. 1, 127–134 [DOI] [PubMed] [Google Scholar]
- 27. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 28. Davis G. D., Elisee C., Newham D. M., Harrison R. G. (1999) New fusion protein systems designed to give soluble expression in Escherichia coli. Biotechnol. Bioeng. 65, 382–388 [PubMed] [Google Scholar]
- 29. Moura J. J. G., Macedo A. L., Goodfellow B. J., Moura I. (2001) Ferredoxins containing one [3Fe-4S] cluster. Desulfovibrio gigas ferredoxin II - solution structure. in Handbook of Metalloproteins (Messerschmidt A. H., Poulos T., Wieghardt K., eds) pp. 553–559, John Wiley & Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 30. Stout C. D. (2001) Ferredoxins containing two different Fe/S centers of the forms [4Fe-4S] and [3Fe-4S]. in Handbook of Metalloproteins (Messerschmidt A. H., R., Poulos T., Wieghardt K., eds), pp. 560–573, John Wiley & Sons, Ltd., Chichester, United Kingdom [Google Scholar]
- 31. Cammack R., Rao K. K., Hall D. O., Moura J. J., Xavier A. V., Bruschi M., Le Gall J., Deville A., Gayda J. P. (1977) Spectroscopic studies of the oxidation-reduction properties of three forms of ferredoxin from Desulphovibrio gigas. Biochim. Biophys. Acta 490, 311–321 [DOI] [PubMed] [Google Scholar]
- 32. Busch J. L., Breton J. L., Bartlett B. M., Armstrong F. A., James R., Thomson A. J. (1997) [3Fe-4S] <–> [4Fe-4S] cluster interconversion in Desulfovibrio africanus ferredoxin III: properties of an Asp14 → Cys mutant. Biochem. J. 323, 95–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Clements A. P., Kilpatrick L., Lu W. P., Ragsdale S. W., Ferry J. G. (1994) Characterization of the iron-sulfur clusters in ferredoxin from acetate-grown Methanosarcina thermophila. J. Bacteriol. 176, 2689–2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hilberg M., Pierik A. J., Bill E., Friedrich T., Lippert M. L., Heider J. (2012) Identification of FeS clusters in the glycyl-radical enzyme benzylsuccinate synthase via EPR and Mössbauer spectroscopy. J. Biol. Inorg. Chem. 17, 49–56 [DOI] [PubMed] [Google Scholar]
- 35. Green J., Bennett B., Jordan P., Ralph E. T., Thomson A. J., Guest J. R. (1996) Reconstitution of the [4Fe-4S] cluster in FNR and demonstration of the aerobic-anaerobic transcription switch in vitro. Biochem. J. 316, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rather L. J., Bill E., Ismail W., Fuchs G. (2011) The reducing component BoxA of benzoyl-coenzyme A epoxidase from Azoarcus evansii is a [4Fe–4S] protein. Biochim. Biophys. Acta 1814, 1609–1615 [DOI] [PubMed] [Google Scholar]
- 37. Derman A. I., Prinz W. A., Belin D., Beckwith J. (1993) Mutations that allow disulfide bond formation in the cytoplasm of Escherichia coli. Science 262, 1744–1747 [DOI] [PubMed] [Google Scholar]
- 38. Dementin S., Belle V., Bertrand P., Guigliarelli B., Adryanczyk-Perrier G., De Lacey A. L., Fernandez V. M., Rousset M., Léger C. (2006) Changing the ligation of the distal [4Fe4S] cluster in NiFe hydrogenase impairs inter- and intramolecular electron transfers. J. Am. Chem. Soc. 128, 5209–5218 [DOI] [PubMed] [Google Scholar]
- 39. Dementin S., Belle V., Champ S., Bertrand P., Guigliarelli B., De Lacey A. L., Fernandez V. M., Léger C., Rousset M. (2008) Molecular modulation of NiFe hydrogenase activity. Int. J. Hydrogen Energy 33, 1503–1508 [Google Scholar]
- 40. Hansel A., Axelsson R., Lindberg P., Troshina O. Y., Wünschiers R., Lindblad P. (2001) Cloning and characterisation of a hyp gene cluster in the filamentous cyanobacterium Nostoc sp. strain PCC 73102. FEMS Microbiol. Lett. 201, 59–64 [DOI] [PubMed] [Google Scholar]
- 41. Leitão E., Oxelfelt F., Oliveira P., Moradas-Ferreira P., Tamagnini P. (2005) Analysis of the hupSL operon of the nonheterocystous cyanobacterium Lyngbya majuscula CCAP 1446/4: Regulation of transcription and expression under a light-dark regimen. Appl. Environ. Microbiol. 71, 4567–4576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Teixeira M., Moura I., Xavier A. V., Moura J. J., LeGall J., DerVartanian D. V., Peck H. D., Jr., Huynh B. H. (1989) Redox intermediates of Desulfovibrio gigas [NiFe] hydrogenase generated under hydrogen. Mössbauer and EPR characterization of the metal centers. J. Biol. Chem. 264, 16435–16450 [PubMed] [Google Scholar]
- 43. Fischer J., Quentmeier A., Kostka S., Kraft R., Friedrich C. G. (1996) Purification and characterization of the hydrogenase from Thiobacillus ferrooxidans. Arch. Microbiol. 165, 289–296 [DOI] [PubMed] [Google Scholar]
- 44. Schröder O., Bleijlevens B., de Jongh T. E., Chen Z., Li T., Fischer J., Förster J., Friedrich C. G., Bagley K. A., Albracht S. P., Lubitz W. (2007) Characterization of a cyanobacterial-like uptake [NiFe] hydrogenase: EPR and FTIR spectroscopic studies of the enzyme from Acidithiobacillus ferrooxidans. J. Biol. Inorg. Chem. 12, 212–233 [DOI] [PubMed] [Google Scholar]